Abstract
Gene expression in nongreen plastids is largely uncharacterized. To compare gene expression in potato (Solanum tuberosum) tuber amyloplasts and leaf chloroplasts, amounts of transcripts of all plastid genes were determined by hybridization to plastome arrays. Except for a few genes, transcript accumulation was much lower in tubers compared with leaves. Transcripts of photosynthesis-related genes showed a greater reduction in tubers compared with leaves than transcripts of genes for the genetic system. Plastid genome copy number in tubers was 2- to 3-fold lower than in leaves and thus cannot account for the observed reduction of transcript accumulation in amyloplasts. Both the plastid-encoded and the nucleus-encoded RNA polymerases were active in potato amyloplasts. Transcription initiation sites were identical in chloroplasts and amyloplasts, although some differences in promoter utilization between the two organelles were evident. For some intron-containing genes, RNA splicing was less efficient in tubers than in leaves. Furthermore, tissue-specific differences in editing of ndh transcripts were detected. Hybridization of the plastome arrays with RNA extracted from polysomes indicated that, in tubers, ribosome association of transcripts was generally low. Nevertheless, some mRNAs, such as the transcript of the fatty acid biosynthesis gene accD, displayed relatively high ribosome association. Selected nuclear genes involved in plastid gene expression were generally significantly less expressed in tubers than in leaves. Hence, compared with leaf chloroplasts, gene expression in tuber amyloplasts is much lower, with control occurring at the transcriptional, posttranscriptional, and translational levels. Candidate regulatory sequences that potentially can improve plastid (trans)gene expression in amyloplasts have been identified.
Various plastid types with distinct functions are present in different organs and tissues of higher plants. Their biogenesis and differentiation are under the control of the nuclear genome and the environment. Plastid development is accompanied by changes in plastid gene expression as well as in plastid morphology and structure (Waters and Pyke, 2005; Pyke, 2007). Plastids of each type harbor multiple copies of the plastid genome (plastome), which contains approximately 120 genes. Plastid genes, which are mainly organized in operons, encode polypeptides or RNAs for the transcription/translation system, the photosynthetic apparatus, the biosynthesis of lipids, and perhaps other unknown functions (Wakasugi et al., 2001; Bock, 2007). Eighteen genes in the plastome contain one or more introns. The chloroplast genome of higher plants has a highly conserved organization, although sometimes structural changes (e.g. insertions and deletions) can be found even in closely related species (Daniell et al., 2006). Their role in gene expression, however, is largely unknown. Despite their prokaryotic origin, plant plastids show complex mechanisms controlling gene expression by adjusting the plastome copy number per cell or through changes at the transcriptional, posttranscriptional, and posttranslational levels. These complex regulatory patterns are often the result of the tightly coordinated expression of the nuclear and plastid genomes (Barkan and Goldschmidt-Clermont, 2000; Liere and Börner, 2007; Woodson and Chory, 2008).
In higher plants, plastid genes are transcribed by two distinct RNA polymerases: the PEP (for Plastid-Encoded Polymerase) is a multisubunit eubacteria-like RNA polymerase whose core subunits are encoded by the plastid rpoA, rpoB, rpoC1, and rpoC2 genes, while the NEP (for Nucleus-Encoded Polymerase) is a single-subunit phage-type RNA polymerase with different isoforms for the mitochondria and the plastids (RpoTm, RpoTp, and RpoTmp; Maliga, 1998; Liere and Börner, 2007). Plastid genes and operons often contain multiple promoters and can be grouped in three classes, based on the presence of either NEP or PEP promoters or both. PEP promoters are chiefly responsible for the transcription of photosynthetic genes in mature chloroplasts. On the other hand, genes encoding proteins required for housekeeping functions (in particular transcription and translation) are often transcribed by both PEP and NEP, while a few housekeeping genes appear to possess exclusively NEP promoters (Hajdukiewicz et al., 1997). Besides differential promoter usage in different plant species or tissues, it has been demonstrated that transcription of plastid genes responds to environmental and developmental cues by differential use of nucleus-encoded sigma factors conferring promoter specificity onto PEP as well as different forms of NEP (Hajdukiewicz et al., 1997; Allison, 2000; Lerbs-Mache, 2000; Toyoshima et al., 2005; Emanuel et al., 2006; Liere and Börner, 2007).
Recent results from analyses of gene expression and activity of plastid RNA polymerases in PEP-deficient tobacco (Nicotiana tabacum) plastids and in maize (Zea mays) leaves have indicated that, besides selective promoter utilization in different tissues and plastid types, mRNA turnover may play a significant role in determining transcript abundance in plastids (Krause et al., 2000; Legen et al., 2002; Cahoon et al., 2004).
Plastid gene expression is further regulated at the posttranscriptional level through the splicing and editing of various RNAs (Bock, 2000; Schmitz-Linneweber and Barkan, 2007). Both processes can be developmentally regulated and represent potential regulatory steps in plastid gene expression, which, at least in some cases, affect the production of functional polypeptides in different tissues and plastid types (Deng and Gruissem, 1988; Barkan, 1989; Karcher and Bock, 2002; Schmitz-Linneweber and Barkan, 2007; Kahlau and Bock, 2008). Differences between tobacco leaves and cultured cells in the editing efficiency of the rpoA mRNA encoding the α-subunit of PEP were observed, raising the possibility of RNA editing influencing RNA polymerase activity and thus transcriptional control (Hirose et al., 1999). Similarly, a link between the relative activities of PEP and NEP in different tissues and splicing efficiency of plastid genes has been hypothesized based on the different transcription elongation rates of the two polymerases (Schmitz-Linneweber and Barkan, 2007).
Very little is known about gene expression in potato (Solanum tuberosum) tuber amyloplasts. A lower level of gene expression in these organelles has been recently reported by Brosch and colleagues (2007), who indirectly inferred levels of gene transcription in chloroplasts and amyloplasts from hybridizing potato leaf- and tuber-derived RNAs to tobacco plastid DNA (ptDNA) fragments. In the same study, the presence of polysomes in amyloplasts was also shown, but specific mRNAs associated with such polysomes could not be detected. In previous studies with single genes, the gene for plastid ribosomal protein S16 (rps16) showed strong expression in chloroplasts and detectable transcripts in amyloplasts (Kang et al., 1995; Bae et al., 1998), while the accD gene involved in fatty acid biosynthesis displayed similar transcript levels in different tissues, environmental conditions, and developmental stages (Lee et al., 2004). Following the recent development of a potato plastid transformation procedure (Sidorov et al., 1999; Nguyen et al., 2005), the expression of heterologous transgenes appears to be much lower in tubers than in leaves. However, no specific investigations were carried out to establish at what level(s) gene expression is regulated in nongreen tuber plastids and whether any plastid regulatory sequences can be found that may improve (trans)gene expression in the potato tuber.
The achievement of high-level expression of transgenes in nongreen plastids requires a detailed understanding of the mechanisms involved in transcription, RNA processing, and translation. Microarrays permit the simultaneous analysis of the expression of all plastid genes (Nakamura et al., 2003). Oligonucleotide arrays have the additional advantage of being highly specific and sensitive, enabling the detection of mRNAs present only in a few copies per cell. This approach helps to overcome the potential problems originating from background amplification in reverse transcription (RT)-PCR and allows the selection of probes with a high specificity, thereby minimizing potential cross-hybridization artifacts (Bozdech et al., 2003).
The full sequence of the cultivated potato chloroplast genome has been recently determined in our and other laboratories (Gargano et al., 2005; Chung et al., 2006). In this study, this sequence information was used to design highly specific oligonucleotides that were spotted onto arrays for the analysis of the expression of all plastid genes. To our knowledge, this is the first comprehensive report on the expression of all plastid genes in potato. Detected differences in transcript levels between amyloplasts and chloroplasts were confirmed by northern-blot analyses. We show that variation in plastome copy number is unlikely to be the main cause of the observed differences in amounts of transcripts between chloroplasts and amyloplasts. Differences between the two organelle types were also found in promoter utilization, transcript splicing, mRNA editing, and polysome loading. A sample of nuclear genes involved in chloroplast biogenesis and plastid gene expression also showed significantly lower expression in tubers than in leaves. A small number of genes displaying relatively high amounts of ribosome-associated transcripts in tuber amyloplasts were identified, and their expression elements may serve as useful tools to optimize plastid transgene expression in potato tubers.
RESULTS
Transcript Accumulation
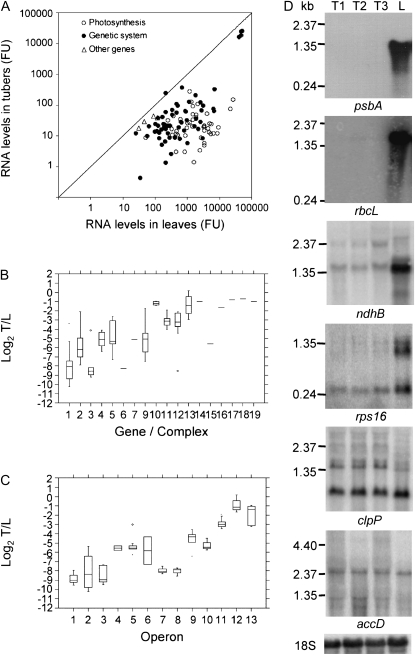
Arrays containing 70-mer oligonucleotides for all plastid genes (Kahlau and Bock, 2008) were used in hybridization experiments with total RNA from potato tubers and leaves (Supplemental Table S1). The results revealed strong differences in the amounts of transcripts of individual plastid genes in both tubers and leaves (Fig. 1A; Supplemental Table S1). After normalization of signals to internal standards, the signals from tubers ranged from 0.4 to 25,282.8 fluorescence units (mean = 811.5, median = 21.0). In comparison, the signals from leaves ranged from 25.9 to 52,077.1 fluorescence units (mean = 3,684.9, median = 857.9). These large differences suggest that transcription rates and/or transcript stability are generally much lower in amyloplasts than in chloroplasts. Genes for the rRNA components (rrn16, rrn23, rrn4.5, and rrn5) showed by far the highest transcript levels in tubers, with values being 2 to 3 orders of magnitude higher than those of other genes.
Figure 1.
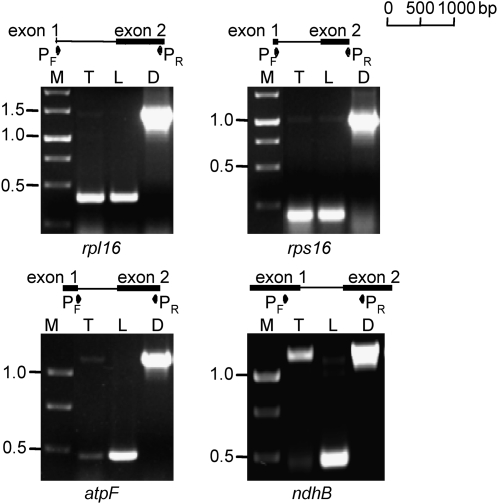
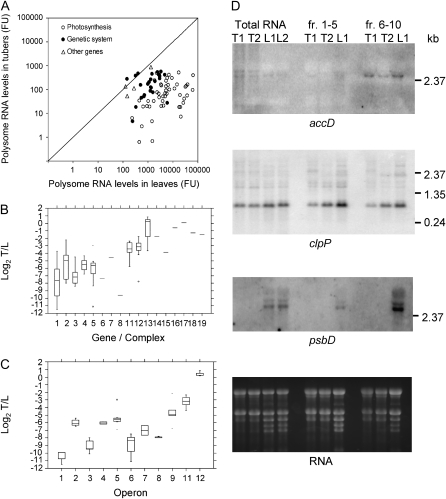
Transcript analysis of the plastid genome in potato tubers and leaves. A, Comparison of transcripts in leaves and tubers. Genes are grouped in three classes, as indicated in Supplemental Table S1. FU, Normalized fluorescence units. B, Box plots of log2 tuber/leaf signals from oligonucleotide arrays for different gene groups/protein complexes, as follows: 1 = psb, 2 = psa, 3 = pet, 4 = atp, 5 = ndh, 6 = rbcL, 7 = cemA, 9 = trn, 10 = rrn, 11 = rpl, 12 = rps, 13 = rpo, 14 = clpP, 15 = matK, 16 = accD, 17 = ycf1, 18 = ycf2, 19 = ycf15. C, Box plots of log2 tuber/leaf signals from oligonucleotide arrays for different operons (according to the operon structure of the tobacco plastome; Wakasugi et al., 1998), as follows: 1 = psbD-C-Z, 2 = psbE-F-L-J, 3 = psbB-T-H-petB-D, 4 = atpB-E, 5 = ndhH-A-I-G-E-psaC-ndhD, 6 = ndhC-K-J, 7 = psbK-I-trnGUCC, 8 = psaA-B-rps14, 9 = rps2-atpI-H-F-A, 10 = trnEUUC-YGUA-DGUC, 11 = rpl23-2-rps19-rpl22-rps3-rpl16-14-rps8-(ψinfA)-rps11-rpoA, 12 = rpoB-C1-C2, 13 = rrn16-trnIGAU-AUGC-rrn23-4.5-5. D, Northern-blot analyses for a subset of genes using 5 μg of total RNA from tubers at different stages of development (T1–T3, from small to big) and leaves (L). Details of array data for each gene are reported in Supplemental Table S1.
Overall, transcript accumulation in amyloplasts was much lower than in chloroplasts (Supplemental Fig. S1a). The mean and the median of the log2 of the ratio of the fluorescence signals from tubers versus leaves were −4.9, with the differences between the two tissues being statistically significant for all but seven genes (Supplemental Table S1). However, the expression patterns differed among the different gene classes (Fig. 1B). For photosynthetic genes, the ratio ranged from −10.2 to −2.1 (mean = −6.5), whereas for genes encoding components of the genetic system, it was between −8.6 and 0.2 (mean = −4.0). Although differences among individual genes were also evident, genes encoding subunits of PSII, the cytochrome b6f complex, and the large subunit of Rubisco (rbcL) showed the lowest transcript ratios (Fig. 1B; Supplemental Table S1). Among the genetic system genes, the genes for tRNAs, ribosomal proteins, and the MatK maturase showed the lowest transcript ratios, whereas those encoding rRNAs, PEP subunits, and the ClpP protease subunit were least affected. Transcripts of the accD gene, encoding the β-subunit of the fatty acid biosynthesis enzyme acetyl-CoA carboxylase, and the three conserved open reading frames ycf1, ycf2, and ycf15, encoding products with unknown function (Drescher et al., 2000), differed by less than a factor of 2 between leaves and tubers.
Considering the genes encoding subunits of PEP, significant differences were observed between the rpoB, rpoC2, and rpoC1 genes, which are cotranscribed and showed log2 ratios varying from 0.2 to −1.7, and rpoA, which is part of a distinct operon (comprising mainly ribosomal proteins genes) and showed a log2 value of −2.9. The latter value was in line with the values of the cotranscribed genes in the rpoA operon (Fig. 1C; Supplemental Table S1). In other cases, genes of the same operon showed similar regulatory patterns, suggesting substantial transcriptional control of gene expression.
Microarray experiments with tubers of different sizes did not show significant differences in transcript accumulation. Furthermore, results obtained with oligonucleotide arrays were in good agreement with those obtained with microarrays and macroarrays containing tobacco and potato PCR-amplified gene fragments, respectively (data not shown).
To obtain independent confirmation of the array data, RNA samples extracted from young leaves and from tubers at three different stages of development were blotted and hybridized to gene-specific probes selected on the basis of the array results (Fig. 1D; data not shown). Multiple transcripts, likely due to the use of alternative promoters and/or complex RNA processing patterns, were detected for some genes (e.g. rps16 and clpP) as already reported (Hajdukiewicz et al., 1997; Kuroda and Maliga, 2002; Kahlau and Bock, 2008). The results of northern-blot analyses confirmed the lower amounts of transcripts of the photosynthesis genes in potato tubers, with transcripts of rbcL, psbA, and many other genes encoding subunits of PSII, PSI, and the ATP synthase complex being barely detectable in tubers but very abundant in leaves. Transcripts of ndh genes for subunits of the NAD(P)H dehydrogenase complex, as well as those encoding ribosomal proteins, were also lower in tubers, but not to such a great extent. Interestingly, the clpP and accD genes displayed hybridization signals of comparable intensities in the two tissues. As observed before in our microarray experiments, no significant differences in transcript levels between the different stages of tuber development were found.
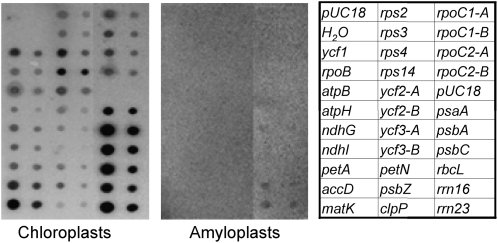
Transcription of 26 representative genes was also analyzed by run-on experiments (Fig. 2). These genes were selected on the basis of their transcript accumulation and function. Genes from different groups (e.g. psa, psb, ndh, atp, and rps), including those with very low (rpo genes) or high (rrn operon) transcript accumulation, were amplified by PCR and spotted onto nylon filters. Two dilutions of each PCR product were dotted in order to facilitate the comparison of hybridization intensities. The nylon filters were hybridized with [α-32P]UTP-labeled run-on transcripts derived from chloroplasts extracted from young leaves or intact amyloplasts purified from tubers. All analyzed genes showed clear signals in chloroplasts, although with different intensities. Transcription of genes of the photosynthetic system and of the rrn operon was higher than that of other genes. In tubers, only rrn16, rrn23, and psbA genes showed a weak signal, indicating very low transcription rates in amyloplasts.
Figure 2.
Run-on transcription activity for a sample of genes in leaf chloroplasts and tuber amyloplasts. Representative genes were selected on the basis of their transcript accumulation and function. Filters were spotted with PCR-amplified gene products from potato plastid DNA at two different concentrations (500 and 100 fmol), as indicated in the scheme at right, and hybridized with 32P-labeled run-on transcripts obtained with the two plastid types. Transcription in 2 × 107 lysed chloroplasts or amyloplasts was allowed to proceed for 5 min in 200-μL reactions supplemented with 200 μCi of [α-32P]UTP. Labeled run-on transcripts were hybridized to the nylon filters as described in “Materials and Methods.” Fragments amplified from distinct regions of the same gene are indicated by different letters.
Plastome Copy Number
To investigate plastid DNA content in potato tubers and leaves and to ascertain whether the observed differences in transcript accumulation were the result of different plastid genome copy numbers, Southern-blot and quantitative real-time (qRT)-PCR analyses were carried out.
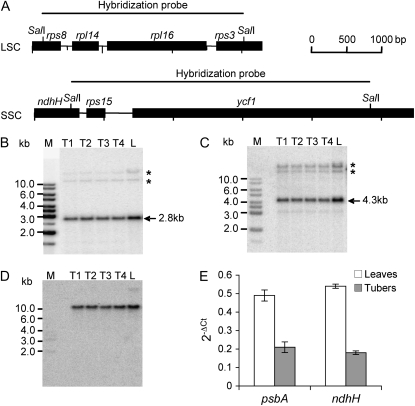
Membranes carrying SalI-digested total DNA were probed with two different probes: one for the large single copy (LSC) and one for the small single copy (SSC) region of the plastid genome. The former comprised rpl16, rpl14, and part of the rps8 and rps3 genes, while the latter included rps15 and part of the ndhH and ycf1 genes (Fig. 3A). Hybridization with LSC and SSC probes revealed the expected signals of 2.8 and 4.3 kb, respectively (Fig. 3, B and C). In both cases, additional signals were detected after extended exposure of the blot, most probably derived from promiscuous DNA (i.e. DNA of plastid origin integrated into the nuclear genome; Li et al., 2006). As a methylation-sensitive enzyme was used for digestion of the total DNA samples, the additional signals were visible in the high-Mr range representing largely undigested nuclear DNA (Fig. 3, B and C). These signals were used as an internal standard for checking the loading of the gel. In addition, membranes were hybridized with a nuclear actin gene probe (Fig. 3D). Hybridizations with both LSC and SSC probes showed comparable signal intensity in all samples. When the signals were normalized to the signals derived from high-Mr DNA or to those of the actin gene, a variation between samples of about 20% was observed, suggesting that plastid genome copy numbers were similar in leaves and tubers and did not change substantially with tuber development.
Figure 3.
Estimation of relative plastid genome copy numbers in tubers of different developmental stages (T1–T4, from small to big) and leaves (L). A, Physical maps of the LSC and SSC in the plastid genome of potato detected by Southern-blot analyses. Restriction sites used for generation of hybridization probes are shown. B and C, The labeled probes for LSC and SSC detect 2.8-kb (B) and 4.3-kb (C) fragments, respectively, in all samples. Asterisks indicate cross-hybridizing promiscuous DNA (Li et al., 2006). M, DNA size marker. D, Hybridization with the nuclear actin gene to control for equal loading of the lanes. E, Analysis of relative ptDNA contents in leaves and tubers by qRT-PCR amplification of the potato psbA and ndhH genes, located in the LSC and SSC, respectively. Data were normalized to the amount of the nuclear 18S rRNA gene and expressed as 2−ΔCt. Error bars indicate sd.
More quantitative analyses were carried out by qRT-PCR using primer pairs for the psbA and ndhH genes, located in the LSC and SSC, respectively. The data obtained indicate that plastome copy number in tubers decreased 2- to 3-fold compared with that in leaves (Fig. 3E).
Promoter Utilization
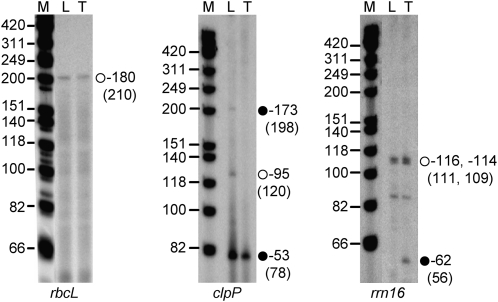
To characterize promoter utilization in potato tubers and detect possible differences in promoter usage in chloroplasts and amyloplasts, the 5′ ends of transcripts from selected genes were mapped by primer extension analysis. We selected the rrn16, clpP, and rbcL genes, as these were among the genes with the highest transcript levels in tubers (rrn16 and clpP) and/or leaves (rrn16 and rbcL; Fig. 4). The rrn16 and clpP genes are of particular interest because they contain promoters for both the NEP and the PEP, while the rbcL gene has a single PEP promoter (Hajdukiewicz et al., 1997).
Figure 4.
Primer extension analysis to map the rrn16, clpP, and rbcL transcription initiation sites in potato tubers (T) and leaves (L). Numbers on the right indicate the distance between the transcript's 5′ end and the first nucleotide of either the coding region (rbcL and clpP) or the mature transcript (rrn16). Numbers in parentheses refer to the size of the extension products. Black circles, NEP promoter; white circles, PEP promoter. M, DNA size marker.
For rbcL, we mapped one 5′ end 180 nucleotides upstream of the translation initiation codon (Fig. 4). This 5′ end was detected in both leaf chloroplasts and tuber amyloplasts. The same transcript was mapped in tobacco chloroplasts and shown to be derived from the PEP promoter of rbcL (Allison et al., 1996; Berg et al., 2004).
For the clpP gene, multiple transcripts derived from different promoters were mapped (Fig. 4), similar to previous findings in tobacco (Hajdukiewicz et al., 1997). In tubers, all clpP transcripts initiated from the strong −53 NEP promoter (Fig. 4, black circle). The same 5′ end was also mapped in leaves, confirming that the −53 transcriptional start site is active in both tissues. Transcripts from the second NEP promoter (position −173) and the PEP promoter of clpP (position −95) were detectable only in potato leaves.
For the highly transcribed rRNA operon, a 5′ end 114 nucleotides upstream of the mature 16S rRNA was mapped in leaves (Fig. 4). This −114 end originates from initiation at the well-characterized strong PEP (P1) promoter (Vera and Sugiura, 1995; Allison et al., 1996). The same signal was obtained using RNA purified from tobacco leaves (data not shown). This result was expected, as the rrn16 promoter sequences possess very high sequence similarity in tobacco and potato. In fact, only a single G upstream of the −10 TATAT sequence is replaced by an A in potato (Gargano et al., 2005; Chung et al., 2006). In potato tubers, we mapped, in addition to transcripts derived from the PEP promoter, a NEP-derived −62 end. This NEP (P2) promoter for transcription of the rRNA operon is normally silent in chloroplasts but was previously identified in tobacco BY2 cells and ΔrpoB plants (Vera and Sugiura, 1995; Allison et al., 1996).
Attempts to map the 5′ ends of some other NEP transcribed genes, such as atpB, atpI, and rps16, produced inconclusive results, probably due to the very low RNA accumulation levels in amyloplasts.
Transcript Processing
Eighteen plastid genes (six of them encoding tRNAs) contain one or more introns. We used RT-PCR analysis to investigate splicing as a key posttranscriptional RNA maturation step in plastids. In all experiments, RNA purified from tubers at three different stages of development and from two independent leaf samples was analyzed. Since no significant differences were obtained between samples of the same tissue, only representative results for the two tissues are displayed in Figure 5.
Figure 5.
RT-PCR analysis of splicing in the rpl16, rps16, atpF, and ndhB transcripts in tubers (T) and leaves (L). D, DNA control; M, DNA size marker. Approximate locations and orientations of the primers (P) used are indicated.
No differences in splicing between tubers and leaves were seen for the rpl16 and rps16 transcripts. In both cases, most transcript molecules were present in their spliced form (Fig. 5). Similar results were obtained for the rpl2, clpP, and ycf3 transcripts (data not shown). However, differences between leaves and tubers were evident for atpF and ndhB. In the former, virtually all transcripts were spliced in leaves, whereas approximately 50% of the transcripts remained unspliced in tubers. An even more dramatic difference was seen for ndhB, which was almost completely spliced in leaves but nearly fully unspliced in tubers (Fig. 5).
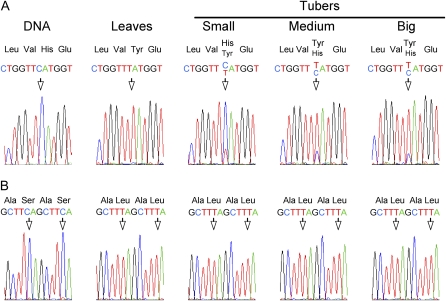
Potential differences in editing between leaves and tubers were investigated for the ndhB and ndhD genes, which are known to contain nine and seven editing sites, respectively, in tobacco and other Solanaceae species (Kahlau et al., 2006). In leaves, all sites except ndhD-1 and ndhD-3 were completely edited. Seven ndhB sites were fully edited in tubers of all sizes, but developmental variation in editing was observed for sites 3 and 9, corresponding to codons 196 and 494, respectively (Table I; Fig. 6). While nearly all transcripts were unedited in small tubers, the editing at ndhB site 3 increased in bigger tubers. No editing could be detected in tubers for ndhD site 1, corresponding to the translation initiation codon, while some reduction in editing efficiency was observed for ndhD sites 3, 6, and 7 (Table I).
Table I.
Comparison of RNA editing efficiency in ndhB and ndhD genes in leaf chloroplasts and tuber amyloplasts
| Site | Codon | Amino Acid Change | Leaves | Tubers
|
||
|---|---|---|---|---|---|---|
| Small | Medium | Big | ||||
| ndhB-1 | 50 | Ser to Leu | ++++a | ++++ | ++++ | ++++ |
| ndhB-2 | 156 | Pro to Leu | ++++ | ++++ | ++++ | ++++ |
| ndhB-3 | 196 | His to Tyr | ++++ | + | +++ | +++ |
| ndhB-4 | 204 | Ser to Leu | ++++ | ++++ | ++++ | ++++ |
| ndhB-5 | 246 | Pro to Leu | ++++ | ++++ | ++++ | ++++ |
| ndhB-6 | 249 | Ser to Phe | ++++ | ++++ | ++++ | ++++ |
| ndhB-7 | 277 | Ser to Leu | ++++ | ++++ | ++++ | ++++ |
| ndhB-8 | 279 | Ser to Leu | ++++ | ++++ | ++++ | ++++ |
| ndhB-9 | 494 | Pro to Leu | ++++ | +++ | +++ | +++ |
| ndhD-1 | 1 | Thr to Met | + | − | − | − |
| ndhD-2 | 128 | Ser to Leu | ++++ | ++++ | ++++ | ++++ |
| ndhD-3 | 200 | Ser to Leu | +++ | ++ | ++ | ++ |
| ndhD-4 | 225 | Ser to Leu | ++++ | ++++ | ++++ | ++++ |
| ndhD-5 | 293 | Ser to Leu | ++++ | ++++ | ++++ | ++++ |
| ndhD-6 | 433 | Ser to Leu | ++++ | +++ | +++ | +++ |
| ndhD-7 | 437 | Pro to Leu | ++++ | +++ | +++ | +++ |
+ and − indicate the degree of editing from completely edited (++++) to completely unedited (−).
Figure 6.
Analysis of ndhB mRNA editing in tubers and leaves. A, While all sites were completely edited in leaves, differences in the extent of editing were observed for codon 196 in tubers of different sizes. B, Two downstream editing sites (corresponding to codons 277 and 279) covered with the same sequencing reaction did not show any difference between leaves and tubers.
Translation
In order to obtain information about translation of all plastid genes, the ribosome association of all transcripts in chloroplasts and amyloplasts was compared by polysome analysis. Polysomes were extracted from potato tuber and leaf lysates fractionated in Suc gradients. Only RNA purified from the bottom fractions (corresponding to ribosomes loaded with mRNA) were labeled and used for hybridizations with oligonucleotide microarrays.
These experiments revealed that almost all plastid transcripts were highly ribosome associated in potato leaves (Fig. 7A; Supplemental Table S2). The highest values were obtained for rbcL, psbA, and several other genes involved in photosynthesis. Overall, transcripts for ribosomal proteins showed about 10-fold lower levels of ribosome association. Low levels of ribosome association were detected for transcripts of the rpoB-rpoC1-rpoC2 operon and some ycfs (ycf1, ycf2, and ycf15).
Figure 7.
Polysome-associated plastid transcripts in potato tubers and leaves. A, Comparison of polysome association in leaves and tubers. Genes are grouped in three classes, as indicated in Supplemental Table S2. FU, Normalized fluorescence units. B, Box plots of log2 tuber/leaf (T/L) signals from oligonucleotide arrays for different gene groups/protein complexes, as follows: 1 = psb, 2 = psa, 3 = pet, 4 = atp, 5 = ndh, 6 = rbcL, 7 = cemA, 8 = ccsA, 11 = rpl, 12 = rps, 13 = rpo, 14 = clpP, 15 = matK, 16 = accD, 17 = ycf1, 18 = ycf2, 19 = ycf15. C, Box plots of log2 tuber/leaf signals from oligonucleotide arrays for different operons (according to the operon structure in the tobacco plastome; Wakasugi et al., 1998), as follows: 1 = psbD-C-Z, 2 = psbE-F-L-J, 3 = psbB-T-H-petB-D, 4 = atpB-E, 5 = ndhH-A-I-G-E-psaC-ndhD, 6 = ndhC-K-J, 7 = psbK-I-(trnGUCC), 8 = psaA-B-rps14, 9 = rps2-atpI-H-F-A, 11 = rpl23-2-rps19-rpl22-rps3-rpl16-14-rps8-(ψinfA)-rps11-rpoA, 12 = rpoB-C1-C2. D, Northern-blot analysis of total RNA and RNA pools from different fractions after polysome separation on Suc gradients (fractions 1–5 and 6–10; see “Materials and Methods”). RNA samples from leaves (L1 and L2) or tubers (T1 and T2) were separated by electrophoresis and hybridized with 32P-labeled accD, clpP, and psbD gene-specific probes. Details of array data for each gene are reported in Supplemental Table S2.
In amyloplasts, the amounts of most plastid transcripts associated with polysomes were very low. Mean and median log2 ratios were −5.1 when comparing tubers with leaves (Supplemental Table S2). However, again, striking differences among gene categories and individual genes were obtained (Fig. 7B; Supplemental Fig. S1b). Ratios for photosynthesis genes ranged from −11.5 to −2.2 (mean = −6.7), whereas it was between −7.8 and 0.8 (mean = −3.0) for genetic system genes. Among the latter, the gene encoding ribosomal protein S14 (rps14) showed a particularly low ratio. Differences were evident between the cotranscribed rpoB-rpoC1-rpoC2 and rpoA, which is cotranscribed with several ribosomal protein genes. Besides the rpoB/C1/C2 operon, transcripts of other genes showing little difference in ribosome association in tubers compared with leaves were clpP, accD, ycf1, ycf2, and ycf15 (Fig. 7, B and C; Supplemental Table S2).
Microarray data obtained with Cy3-labeled RNA from polysomal fractions were further confirmed by northern-blot analyses (Fig. 7D). Hybridization with an accD probe demonstrated that, in both tubers and leaves, most transcripts were associated with ribosomes (fractions 6–10). Similar levels of ribosome-associated RNA in tubers and leaves were also observed for the clpP gene. In contrast to accD, however, nearly identical amounts of ribosome-associated (fractions 6–10) and ribosome-free (fractions 1–5) clpP transcripts were detected. Hybridizations with psbD, a photosynthesis gene that had shown much lower amounts of transcripts in tubers compared with leaves in microarray experiments (log2 = −9.9), revealed strong signals in the polysome fractions from leaves but an absence of both ribosome-bound and free psbD transcripts from tuber samples.
Nuclear Gene Expression
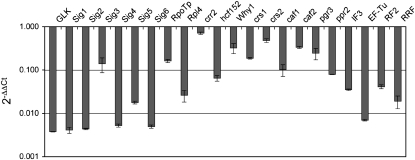
Representative nuclear genes involved in chloroplast biogenesis and plastid gene expression (Hashimoto et al., 2003; Williams and Barkan, 2003; Yamazaki et al., 2004; Liere and Börner, 2007; Marín-Navarro et al., 2007; Schmitz-Linneweber and Barkan, 2007; Prikryl et al., 2008; Waters et al., 2008) were analyzed by means of qRT-PCR (Fig. 8). Transcripts of all genes tested were lower in tubers compared with leaves. Some variation among genes was observed, however. Transcripts of the genes encoding a GLK transcription factor, PEP σ-factors, and other components of transcription/translation machineries (Rpl4, IF3, Ef-Tu, RF2, and RRF) were lowest in tubers compared with leaves. Slightly higher relative transcript levels were detected for Sig3, RpoTp (encoding the plastid-targeted phage-type RNA polymerase), and genes encoding factors involved in splicing (WHY1, CRS1, CRS2, CAF1, and CAF2), RNA processing (CRR2), and stability (PGR3) of plastid transcripts. hcf152 (involved in the processing of the psbB operon transcript) and ppr2 (functioning in the synthesis and/or assembly of the plastid translational machinery) displayed an intermediate reduction in gene expression.
Figure 8.
Expression of representative nuclear genes involved in chloroplast biogenesis and plastid gene expression in tubers. Transcript abundance was determined by qRT-PCR and is shown, after normalization to the elongation factor1-α gene and relative to leaves, as 2−ΔΔCt. Data are represented on a logarithmic scale. Error bars indicate sd.
DISCUSSION
In the course of this study, we have investigated global gene expression differences between potato leaf chloroplasts and tuber amyloplasts by analyzing the transcripts of all plastid genes on gene-specific oligonucleotide microarrays. So far, only a few studies have used array technologies to analyze the expression of higher plant plastid genomes (Legen et al., 2002; Nakamura et al., 2003; Magee et al., 2007; Kahlau and Bock, 2008; MacLean et al., 2008). The same oligonucleotide-based arrays employed in this study have been recently used to analyze changes in gene expression in tomato (Solanum lycopersicum) fruits (Kahlau and Bock, 2008). The availability of the potato plastome sequence (GenBank accession no. NC_008096; Gargano et al., 2005) allowed the design of reliable highly sensitive species-specific oligonucleotide arrays. Since most of the unidentified tobacco open reading frames are not conserved in other Solanaceae species and show frameshift mutations and/or larger deletions in potato (Gargano et al., 2005; Chung et al., 2006), it seems unlikely that they encode functional gene products; therefore, they were not included in our microarray analysis. In this study, microarray data sets were integrated with data on variation of plastome copy number, promoter usage, transcript splicing, and mRNA editing in potato chloroplasts and amyloplasts. Gene expression was generally much lower in potato tuber amyloplasts than in leaf chloroplasts. However, we detected striking differences among individual genes and operons and also in the relative contributions of transcriptional, posttranscriptional, and translational control mechanisms to the regulation of plastid gene expression. The results obtained could be directly compared with those recently produced in tomato (Kahlau and Bock, 2008), highlighting common mechanisms in tuber amyloplasts and fruit chromoplasts. This is interesting, because while tomato chromoplasts develop directly from chloroplasts, potato amyloplasts do not have a green past.
Global differences in transcript accumulation between green and nongreen plastids could depend on reduced transcription or increased RNA turnover. Based on run-on transcription experiments, we conclude that transcription in tuber amyloplasts is extremely low. Some low-level transcriptional activity was detectable only for the psbA and rrn genes. Similar results were reported for genes in sycamore (Acer pseudoplatanus), tobacco, and potato amyloplasts (Ngernprasirtsiri et al., 1990; Sakai et al., 1992; Brosch et al., 2007). Transcriptional control of gene expression in sycamore amyloplasts was suggested to be dependent on methylation of specific genome regions and not on rearrangements of the plastid genome (Ngernprasirtsiri et al., 1990). When we digested total leaf and tuber DNAs with the methylation-sensitive enzyme SalI, no significant differences were observed in the restriction patterns obtained, suggesting an overall similar cytosine methylation status of the two plastid genomes. More experiments based on parallel digestion with pairs of methylation-sensitive and insensitive isoschizomeres are needed to assess the possible role of methylation in transcriptional regulation of gene expression in tuber amyloplasts. Recent results, however, seem to exclude DNA methylation in the control of gene expression in plastids (Ahlert et al., 2009). Our results and those from earlier renaturation kinetics experiments (Steele Scott et al., 1984) indicate that the relative plastid DNA content does not change substantially in leaf and tuber cells. Thus, variation in plastome copy number is unlikely to be responsible for the much lower levels of transcripts observed in amyloplasts. Also in Arabidopsis (Arabidopsis thaliana), transcription was not regulated by plastome copy number in cotyledons or leaves of different ages (Zoschke et al., 2007). On the other hand, a contribution of plastid DNA copy number to plastid gene expression was suggested for root and cell suspension amyloplasts of spinach (Spinacia oleracea) and Arabidopsis (Aguettaz et al., 1987; Deng and Gruissem, 1988; Isono et al., 1997) as well as for developing barley (Hordeum vulgare) chloroplasts (Baumgartner et al., 1989).
In potato tubers, despite the much lower transcriptional activity, some transcripts could still be detected, suggesting higher transcript stability in amyloplasts than in chloroplasts, as proposed also by Brosch et al. (2007). The cis- and trans-acting factors (Barkan and Goldschmidt-Clermont, 2000; Monde et al., 2000) involved in the regulation of RNA stability are not known. A similar compensatory mechanism, however, was shown in transplastomic tobacco plants, in which lower rates of transcription in the dark were compensated by increased mRNA stability, provided that a segment encoding a stem-loop structure in the rbcL 5′ untranslated region was included in the construct (Shiina et al., 1998).
Differences in transcript levels detected between genes or operons are likely to depend on differences in the stability of the corresponding RNAs. On average, transcripts of photosynthesis genes and tRNA genes were much lower than those of genes encoding other complexes in amyloplasts. Lower accumulation of tRNAs probably reflects the generally reduced need for translation in amyloplasts. Similar results were obtained in chromoplasts of developing tomato fruits (Kahlau and Bock, 2008). In several cases, the genes cotranscribed from the same operon showed similar levels of transcripts, even if the gene products have different functions. For instance, the regulation of the transcripts of the rps14 and rpoA genes was more similar to that of other genes of the psaA and rpoA/rpl23 operons, respectively, than to genes encoding other Rps or Rpo subunits. In the case of the large rpoA/rpl23 operon, which comprises 11 genes and a pseudogene (ψinfA), it is remarkable that genes encoding proteins for three different complexes (Rpl, Rps, and Rpo) showed very similar regulation patterns. Variation in RNA stability was shown to be an important determinant of transcript levels during leaf and plastid development in higher plants, such as spinach, barley, and Arabidopsis (Mullet and Klein, 1987; Deng and Gruissem, 1988; Klaff and Gruissem, 1991; Kim et al., 1993; Zoschke et al., 2007) as well as in the unicellular green alga Chlamydomonas reinhardtii (Eberhard et al., 2002). Genes showing relatively higher levels of transcripts in tubers (e.g. rrn, clpP, accD, ycf1, and ycf2) contain either NEP or multiple PEP and NEP promoters (Hajdukiewicz et al., 1997; Legen et al., 2002). From the analysis of the 5′ end of transcripts, both polymerases were shown to be active in amyloplasts. Hence, based on the available data, it is not possible to attribute variation in transcript accumulation to differential activity of either PEP or NEP. Recent data, however, suggest that the two polymerases do not simply mediate gene class-specific transcription in different cells or plastid types but are also involved in differential processing, stability, and accumulation of the resulting transcripts and polypeptides (Legen et al., 2002; Cahoon et al., 2004; Zoschke et al., 2007).
The results available suggest that none of the mechanisms discussed is solely responsible for the lower plastid transcript levels in nongreen plastids. In tomato, although no attempt was made to distinguish between the effects of altered transcription and RNA stability, a significant reduction in transcript accumulation was observed in green fruits compared with leaves, suggesting that the down-regulation was triggered by the developmental program rather than the chloroplast-to-chromoplast conversion (Kahlau and Bock, 2008). As suggested previously (Deng and Gruissem, 1988; Brosch et al., 2007), the residual accumulation of transcripts in nongreen amyloplasts, particularly of those encoding components of the genetic machinery, is likely to be required to maintain some housekeeping functions and, perhaps, also to allow for the conversion of amyloplasts into chloroplasts in greening tubers and in green shoots differentiating from tuber cells cultivated in vitro (Jarret et al., 1980; Zhu et al., 1984).
The presence of polysomes had previously been observed in potato tuber amyloplasts, but specific mRNAs associated with polysomes were not identified (Brosch et al., 2007). In this study, much lower levels of polysome association in tubers were observed for transcripts of most genes and operons. Notable exceptions were the psbE/F/L/J and ndhC/K/J gene clusters, which showed decreased and increased levels of ribosome association, respectively, when polysome were analyzed. The Spearman rank correlation coefficient (Wilkinson et al., 1992) was 0.79 when 76 genes in leaf chloroplasts were ranked on the basis of their relative transcript abundances in total versus polysomal RNA. For tuber amyloplasts, the correlation coefficient was 0.52, suggesting that while, in leaf chloroplasts, the amount of RNA loaded onto polysomes is somehow related to the total amount of RNA present, this relationship is less strict in tuber amyloplasts. This indicates that, in amyloplasts, the association of mRNAs with polysomes may represent an additional regulatory step in gene expression. It is important to note that, in addition to polysome loading, other translational and posttranslational regulatory steps can also influence protein accumulation (Adam, 2007; Peled-Zehavi and Danon, 2007) and adjust subunit stoichiometries in multiprotein complexes in plastids.
Besides RNA accumulation and translation, the extent of some posttranscriptional processes, such as splicing and editing of (at least some) transcripts, was lower in tuber amyloplasts compared with leaf chloroplasts. Much lower levels of total and polysome-associated transcripts were seen for the matK gene, located in the trnKUUU intron and encoding a maturase-like protein presumably involved in intron excision from several plastid transcripts, including atpF (Barthet and Hilu, 2007). Interestingly, atpF was one of the two intron-containing transcripts that showed less splicing in potato tuber amyloplasts. matK expression was also found to be strongly down-regulated in tomato fruits (Kahlau and Bock, 2008) and, in tobacco and rice (Oryza sativa), appears to be regulated by light and developmental factors (Vogel et al., 1997; Nakamura et al., 2003; Barthet and Hilu, 2007).
Editing of ndh transcripts is known to be developmentally regulated in higher plant plastids (Karcher and Bock, 2002; Chateigner-Boutin and Hanson, 2003). In this study, a lower level of RNA editing in codons 196 and 494 of the ndhB gene was observed in developing tubers, with the unedited form being predominantly present in young tubers. As the editing events at both sites change the coding properties of the mRNA, the populations of NdhB proteins in leaves and young tubers should differ. Other changes relevant at the protein level occurred in the mRNA for the NdhD subunit. The apparent lack of editing of the first site in the ndhD mRNA results in the absence of the canonical AUG translational start codon. However, the physiological significance of the differential editing at this and other sites on the translation of ndh transcripts and/or the assembly and functionality of the NDH protein complex in tubers remains to be established. Complete editing of the other sites investigated suggests that, while all general components of the editing machinery are present in tuber amyloplasts, some site-specific editing factors may be missing or become rate limiting. Although some differences in tissue-specific editing patterns between potato tubers and tomato fruits were evident (e.g. in codon 277 of ndhB and codons 200 and 433 of ndhD), in general, similar site-specific changes in editing efficiency seem to occur during the development of the two organs (Kahlau and Bock, 2008), suggesting the presence of similar mechanisms regulating RNA editing activities in nongreen plastids of fruits and tubers.
Nuclear genes involved in various aspects of plastid gene expression and transcript maturation generally showed a concomitant decrease in expression, presumably as a result of cross talk between the organelles and the nucleus. At present, it is not clear whether this is a cause or a consequence of the reduced gene expression in amyloplasts. Based on our results on transcript accumulation in leaves and tubers, genes encoding factors involved in transcription and translation appear to be particularly strongly down-regulated in potato tubers. The reduction in the expression of plastid genes with PEP promoters (mainly photosynthesis-related genes) is likely linked to the strong reduction in transcript accumulation of the σ-factor (Sig) genes. A developmental stage-dependent regulation was reported for several of these genes. Sig3, for example, plays a distinctive role in the regulation of gene expression in some nongreen plastid types (Liere and Börner, 2007). Interestingly, in this study, Sig3 transcripts showed less reduction in tubers compared with other plastid σ-factors. Among the other nuclear genes analyzed here, it is interesting that, concomitant with reduced atpF splicing in amyloplasts, crs1 and Why1, two genes reported to be involved in atpF intron splicing (Schmitz-Linneweber and Barkan, 2007; Prikryl et al., 2008), also showed reduced expression in tubers. However, the direct causal relationship between plastid gene expression and the expression of nuclear genes in leaves versus tubers remains to be determined.
Despite recent progress, potato plastid transformation is still limited by low transformation frequencies and by low transgene expression levels in tubers of transplastomic plants (Sidorov et al., 1999; Nguyen et al., 2005). In this study, we have identified a number of plastid genes that were relatively highly expressed in potato tubers. These genes may provide a valuable source of 5′ and/or 3′ regulatory sequences that are potentially suitable to enhance (trans)gene expression in amyloplasts, although in some cases heterologous sequences were used to drive the expression of transgenes in nongreen plastids (Ruf et al., 2001; Kumar et al., 2004). accD, the only plastid gene involved in fatty acid biosynthesis, showed relatively high levels of total and ribosome-associated transcripts in amyloplasts, suggesting high transcript stability and efficient translation. Interestingly, enhanced stability of the accD transcript was previously found in PEP-deficient tobacco mutants, which showed decreased transcription rates but increased RNA levels (Hajdukiewicz et al., 1997; Krause et al., 2000; Legen et al., 2002). Reflecting the general need for lipid biosynthesis, the accD transcript constitutively accumulated in potato chloroplasts and amyloplasts (Lee et al., 2004) and sustained expression was also found in chromoplasts of developing tomato fruits (Kahlau and Bock, 2008). In tobacco plants, accD could not be deleted from the plastid genome, confirming that it is an essential gene for cell survival (Kode et al., 2005). Interestingly, several other essential genes (clpP, ycf1, and ycf2; Drescher et al., 2000; Kuroda and Maliga, 2003) were relatively highly expressed in potato tubers. In transient assays, the length of the accD 5′ untranslated region appeared to determine the expression level of a reporter gene in both nonphotosynthetic and photosynthetically competent tobacco plastids (Hirata et al., 2004). Stable transformation experiments are needed to confirm these results for potato amyloplasts and to evaluate the potential of the 5′ and 3′ regions from accD and the other genes found to be actively expressed in tubers to enhance (trans)gene expression in nongreen plastids. However, to generally increase the expression of transgenes in nongreen plastids, it may additionally be necessary to overexpress at least some of the nuclear genes involved in the plastid gene machinery. Future systematic investigation should be directed toward understanding the role of master genes controlling the expression of nuclear genes for plastid functions (López-Juez, 2007) as well as of mechanisms involved in nuclear-plastid cross talk in different organs (Woodson and Chory, 2008).
MATERIALS AND METHODS
Plant Material
Potato plants (Solanum tuberosum ‘Désirée’) were grown in vitro on Murashige and Skoog medium with B5 vitamins (Duchefa) and 30 g L−1 Suc under controlled conditions (16 h of light [40 μE m−2 s−1] and 8 h of dark at 24°C). After 3 to 4 weeks of culture in vitro, plants were transferred to soil and maintained in a growth chamber (14 h of light [200 μE m−2 s−1] at 25°C and 10 h of dark at 20°C) for tuberization. Fully expanded leaves were collected from 4-week-old plants. Tubers of different sizes were collected from 3-month-old plants. Three to 4 weeks before tuber harvest, pots were covered with black tissue to prevent light influence on gene expression in tubers developing near the soil surface. Collected tubers were initially sorted into very small (about 1.0 cm in diameter), small (1.5–2.0 cm), medium (2.5–3.0 cm), and big (about 4.0 cm) tubers. RNA extraction, amyloplast isolation, and run-on transcription analyses were done on the day of harvest. Remaining plant material was kept at −80°C.
RNA Isolation, Northern Blot, and Oligonucleotide Array Hybridizations
Total potato RNA was extracted from 2 g of tuber or leaf tissue according to the protocol described on The Institute for Genomic Research (TIGR) Web site (http://www.tigr.org/tdb/potato/images/SGED_SOP_3.2.1.pdf) and precipitated with LiCl. Purified RNA was treated with RNase-free DNase I (Promega) and subsequently precipitated with ethanol. For oligonucleotide array hybridizations, RNA was additionally purified through NucleoSpin RNA Clean-up columns (Macherey-Nagel).
For northern-blot analyses, 5 μg of total RNA was separated by electrophoresis on 1.2% agarose/formaldehyde gels and blotted onto Hybond N+ membranes (GE Healthcare). Gene-specific PCR products were used as probes. [α-32P]dCTP-labeled probes were generated by random priming (Prime-a-gene labeling system; Promega). Hybridizations were carried out at 42°C following the membrane manufacturer's instructions (GE Healthcare). Hybridization signals were quantified with a Typhoon 9200 Imager (GE Healthcare) and normalized to the 18S rRNA signal. Primers used to amplify the probes are listed in Supplemental Table S3.
Oligonucleotide microarrays containing 70-mer oligonucleotides for all genes in the tobacco (Nicotiana tabacum) plastome (accession no. NC_001879) were designed and printed as described by Kahlau and Bock (2008). For tomato (Solanum lycopersicum) and potato genes showing insufficient sequence identity with tobacco, species-specific oligonucleotides designed using the tomato (accession no. NC_007898) and potato (accession no. NC_008096) ptDNA sequences were also included. Twenty micrograms of total RNA and 10 μg of polysomal RNA, extracted from both tubers and leaves, were labeled with Cy3 fluorochrome using the SuperScript Indirect cDNA Labeling System (Invitrogen) and hybridized to the oligonucleotide array slides in a HybArray 12 station (Perkin-Elmer). Arrays were hybridized overnight at 41°C. Details about oligonucleotide sequences, production and hybridization of arrays, and data quantitation are as described by Kahlau and Bock (2008). All data were normalized to internal spike controls printed on the slides, and no dye swap was applied. Total transcript abundance of all genes was calculated as a mean of three independent biological replicates (RNA isolations) for leaves and four replicates for tubers. Polysome transcript abundance was calculated as a mean of two independent biological replicates (polysome extractions) for both leaves and tubers. Furthermore, data from each biological replicate are based on two arrays (additional technical replicates). Relative transcription and expression levels are presented as log2 ratios of tuber versus leaf data.
To analyze the statistical significance of transcript differences between tubers and leaves, the normalized microarray data were subjected to log(1+x) transformation, in order to reduce heterogeneity of error variances, and t tests were carried out for each gene. Raw P values from multiple t tests were controlled according to the false discovery rate procedure (Benjamini and Hochberg, 1995; Reiner et al., 2003), with q = 0.05.
Plastid Purification
Amyloplasts were purified from 30 g of potato tubers as described by Wischmann et al. (1999) and Brosch et al. (2007). Intactness of approximately 65% of the purified amyloplasts was estimated by microscopic observation and by determining the activity of ADPglucose pyrophosphorylase as a marker enzyme. Chloroplasts were purified from 5 g of leaf material according to Kemble (1987) using Suc gradient and high-speed centrifugation.
Run-On Transcription Analysis
Transcription in 2 × 107 lysed plastids was performed in 200-μL reactions supplemented with 200 μCi of [α-32P]UTP (specific activity, 800 Ci mmol−1) as described by Mullet and Klein (1987) and Berg et al. (2003). Transcription was carried out for 5 min, followed by RNA extraction with phenol/chloroform. Unincorporated [α-32P]UTP was removed by overnight precipitation at −20°C with 0.1 volume of sodium acetate and 2.5 volumes of ethanol. Labeled transcripts were hybridized to Hybond N+ filters carrying 30 PCR-amplified fragments from 26 plastid genes. All fragments were amplified from potato total DNA and spotted at two concentrations (500 and 100 fmol) onto Hybond N+ nylon filters according to the manufacturer's instructions. Hybridization and signal detection were done as described for northern-blot analyses.
Southern-Blot Analysis
Total genomic DNA was prepared from young potato leaves and tubers at four different stages of development. DNA was purified by a rapid miniprep procedure using sodium lauroyl sarcosine-based cell lysis (Pallotta et al., 2000). Five micrograms of RNase-treated DNA was digested with SalI, separated by electrophoresis on 0.8% (w/v) agarose gels, and blotted onto Hybond N+ membranes by capillary transfer under alkaline conditions according to the manufacturer's instructions. Two different probes, one for the LSC region (nucleotide positions 81,886–84,759) and one for the SSC region (nucleotide positions 124,270–128,580) of the potato plastid genome (GenBank accession no. NC_008096) were used for hybridization. Probes were generated by SalI digestion of recombinant pBR322 plasmids containing cloned SalI fragments of potato plastid DNA. The probe for the actin gene was PCR amplified from potato genomic DNA (primer sequences are listed in Supplemental Table S3). After agarose gel electrophoresis, fragments of interest were excised and purified with the GFX-PCR (DNA and Gel Band Purification) kit (GE Healthcare). Probes were labeled with [α-32P]dCTP as described above. Hybridizations were performed overnight at 65°C in Church buffer (Church and Gilbert, 1984). After hybridization, the blot was washed first with 2× SSC for 5 min followed by one wash with 2× SSC, 0.1% (w/v) SDS for 30 min and two washing steps with 0.1× SSC, 0.1% (w/v) SDS for 15 min each. The radioactivity was detected using a Typhoon 9200 Imager (GE Healthcare).
Primer Extension Analysis
Primer extension reactions were carried out with total leaf or tuber RNA samples using the Primer Extension System (Promega), as described by Allison and Maliga (1995) and Lee et al. (2004). Primers were labeled with [γ-32P]ATP using T4 polynucleotide kinase. Based on the approximate relative transcript abundances determined by northern-blot analyses, 1 μg (rrn16 and rbcL) or 30 μg (clpP) of RNA from leaves and 10 μg (rrn16) or 30 μg (clpP and rbcL) of RNA from tubers were used. Sequence ladders were generated with the same primers using the Sequenase II kit (USB). The sizes of the extension products were further determined by comparison with the radiolabeled ΦX174 HinfI DNA marker loaded on the same gel. The sequences of the primers used for mapping of the 5′ ends of the rrn16, clpP, and rbcL transcripts are listed in Supplemental Table S3.
cDNA Synthesis, PCR, and Sequencing
For semiquantitative analysis of plastid transcripts, 1 μg of DNase I-treated RNA from tubers and leaves was denatured in the presence of random primers for 10 min at 70°C. cDNA was prepared with the RevertAid H-minus first-strand cDNA synthesis kit (Fermentas) following the manufacturer's protocol. DNA and cDNA samples were amplified by standard PCR protocols (30 s at 94°C, 30 s at 58°C, and 1.5 min at 72°C for 28 or 30 cycles). PCR products were separated by electrophoresis on 1% agarose gels and quantified with the Typhoon 9200 Imager (GE Healthcare). Oligonucleotides used for RT-PCR and/or for sequencing the ndhB and ndhD regions containing the editing sites are listed in Supplemental Table S3.
Polysome Purification
Polysomes were purified as described by Kahlau and Bock (2008). Eight hundred milligrams of ground leaf or tuber material was treated with 4 mL of extraction buffer in conditions maintaining the integrity of the polysomes. After removal of the insoluble material, polysomes were fractionated on a continuous Suc gradient (15%–56%) by centrifugation at 200,000g for 80 min. Ten fractions, of 410 μL each, were collected, and the RNA was extracted with phenol/chloroform from each fraction. After ethanol precipitation, the RNA pellet was dissolved in 20 μL of 10 mm Tris-HCl, pH 7.6, and aliquots of 2 μL were analyzed by electrophoresis on 1.2% agarose/formaldehyde gels. Puromycin-treated lysates were used as a control (Kahlau and Bock, 2008). These control gradients were also fractionated, and the RNA was purified as described above.
For oligonucleotide array hybridizations, fractions 6 to 10 (containing the ribosome-loaded mRNAs) were pooled, precipitated twice, and dissolved in a minimal volume of 10 mm Tris-HCl, pH 7.6.
qRT-PCR
qRT-PCR was performed, in tubers and leaves, to analyze the relative changes of plastome copy number and the transcript levels of representative nuclear genes relevant to chloroplast biogenesis and plastid gene expression.
For analysis of the relative plastome DNA content, total DNA was extracted from potato tissues (young leaves and tubers of about 2 cm) using the DNeasy Plant Mini Kit and treated with RNaseA (Qiagen). Primers for the plastid psbA and ndhH genes were designed according to the potato plastome sequence (GenBank accession no. NC_008096; Supplemental Table S3) with Primer Express 2.0.0 software (Applied Biosystems), with optimal melting temperature at 60°C, GC content between 40% and 70%, and maximum length of amplified products of 140 bp, and validated using serial dilutions of plant total DNA. Real-time PCR efficiency was calculated for each gene and tissue with the slope of linear regression model according to the equation E = 10(1 − slope) (Nicot et al., 2005). Only oligonucleotides with amplification efficiency of 95% ± 5% were selected for further analyses. Dissociation curves were performed after each PCR to confirm single-product amplification. The size of the amplified products was further verified by electrophoresis on a 2% agarose gel. PCR was carried out for 10 min at 95°C followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. All reactions were performed on a 7900HT Fast Real-Time PCR system (Applied Biosystems). Plastome DNA content was determined as the number of cycles needed for the amplification to reach a threshold in the exponential phase of PCR (Ct; Livak and Schmittgen, 2001). The data obtained were analyzed using 7900HT System SDS software version 2.3. Two independent DNA preparations per tissue were analyzed in triplicate for each gene. For estimation of relative plastid DNA content, all quantifications were normalized against the amount of 18S rDNA as an internal standard, and data were expressed as 2−ΔCt (Livak and Schmittgen, 2001).
Total RNA was purified from leaf and tuber tissues with the NucleoSpin RNA Plant isolation kit and subsequently treated with RNase-free DNase (Macherey-Nagel). Two micrograms of RNA were reverse transcribed with the AccuScript High Fidelity First Strand cDNA Synthesis Kit (Stratagene) using oligo(dT)18 primers. The potato Why1 sequence was extracted from the GenBank database. For the other genes, the potato nucleotide sequences were obtained after BLAST of the corresponding genes in GenBank against the Solanum tuberosum database in TIGR (Supplemental Table S4). Gene-specific primers (Supplemental Table S3) were designed and validated using serial dilutions of plant cDNA as reported above. The PCR mixture contained 1 μL of 30-fold-diluted cDNA template and 0.3 μm primers. PCR was performed as for genomic DNA. Two independent RNA preparations per tissue were analyzed in triplicate for each gene. Expression levels were determined as described above for ptDNA determinations. Expression of the target genes in tubers and leaves was normalized to the amount of the transcript of the elongation factor1-α gene, which was shown to be the most stable among the housekeeping genes of potato (Nicot et al., 2005). Data for tubers relative to those for leaves were presented as 2−ΔΔCt (Livak and Schmittgen, 2001).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Graphical presentation of results from array experiments using MapMan software.
Supplemental Table S1. Data from microarray analysis with total RNA.
Supplemental Table S2. Data from microarray analysis with RNA from potato polysomes.
Supplemental Table S3. List of primers used for different analyses.
Supplemental Table S4. GenBank accession numbers of selected nuclear genes and corresponding potato TIGR Transcript Assemblies sequences used for qRT-PCR.
Supplementary Material
This work was supported by the European Union (FP6 Plastomics Project grant no. LSHG–CT–2003–503238). IGV Publication Number 338.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Teodoro Cardi (cardi@unina.it).
The online version of this article contains Web-only data.
References
- Adam Z (2007) Protein stability and degradation in plastids. Top Curr Genet 19 315–338 [Google Scholar]
- Aguettaz P, Seyer P, Pesey H, Lescure AM (1987) Relations between the plastid gene dosage and the levels of 16S rRNA and rbcL gene transcripts during amyloplast to chloroplast change in mixotrophic spinach cell suspensions. Plant Mol Biol 8 169–177 [DOI] [PubMed] [Google Scholar]
- Ahlert D, Stegemann S, Kahlau S, Ruf S, Bock R (2009) Insensitivity of chloroplast gene expression to DNA methylation. Mol Genet Genomics (in press) [DOI] [PMC free article] [PubMed]
- Allison LA (2000) The role of sigma factors in plastid transcription. Biochimie 82 537–548 [DOI] [PubMed] [Google Scholar]
- Allison LA, Maliga P (1995) Light-responsive and transcription-enhancing elements regulate the plastid psbD core promoter. EMBO J 14 3721–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison LA, Simon LD, Maliga P (1996) Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J 15 2802–2809 [PMC free article] [PubMed] [Google Scholar]
- Bae JM, Ahn MY, Harn CH, Jeong WJ, Jung M, Lim YP, Liu JR (1998) Cloning and characterization of plastid ribosomal protein S16 gene from potato (Solanum tuberosum L. cv Désirée). Mol Cells 8 466–470 [PubMed] [Google Scholar]
- Barkan A (1989) Tissue-dependent plastid RNA splicing in maize: transcripts from four plastid genes are predominantly unspliced in leaf meristems and roots. Plant Cell 1 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Goldschmidt-Clermont M (2000) Participation of nuclear genes in chloroplast gene expression. Biochimie 82 559–572 [DOI] [PubMed] [Google Scholar]
- Barthet MM, Hilu KW (2007) Expression of matK: functional and evolutionary implications. Am J Bot 94 1402–1412 [DOI] [PubMed] [Google Scholar]
- Baumgartner BJ, Rapp JC, Mullet JE (1989) Plastid transcription activity and DNA copy number increase early in barley chloroplast development. Plant Physiol 89 1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B Methodological 57 289–300 [Google Scholar]
- Berg S, Krause K, Krupinska K (2004) The rbcL genes of two Cuscuta species, C. gronovii and C. subinclusa, are transcribed by the nuclear-encoded plastid RNA polymerase (NEP). Planta 219 541–546 [DOI] [PubMed] [Google Scholar]
- Berg S, Krupinska K, Krause K (2003) Plastids of three Cuscuta species differing in plastid coding capacity have a common parasite-specific RNA composition. Planta 218 135–142 [DOI] [PubMed] [Google Scholar]
- Bock R (2000) Sense from nonsense: how the genetic information of chloroplasts is altered by RNA editing. Biochimie 82 549–557 [DOI] [PubMed] [Google Scholar]
- Bock R (2007) Structure, function, and inheritance of plastid genomes. Top Curr Genet 19 29–63 [Google Scholar]
- Bozdech Z, Zhu J, Joachimiak M, Cohen F, Pulliam B, DeRisi J (2003) Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol 4 R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch M, Krause K, Falk J, Krupinska K (2007) Analysis of gene expression in amyloplasts of potato tubers. Planta 227 91–99 [DOI] [PubMed] [Google Scholar]
- Cahoon AB, Harris FM, Stern DB (2004) Analysis of developing maize plastids reveals two mRNA stability classes correlating with RNA polymerase type. EMBO Rep 5 801–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Hanson MR (2003) Developmental co-variation of RNA editing extent of plastid editing sites exhibiting similar cis-elements. Nucleic Acids Res 31 2586–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HJ, Jung JD, Park HW, Kim JH, Cha HW, Min SR, Jeong WJ, Liu JR (2006) The complete chloroplast genome sequences of Solanum tuberosum and comparative analysis with Solanaceae species identified the presence of a 241-bp deletion in cultivated potato chloroplast DNA sequence. Plant Cell Rep 25 1369–1379 [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Lee SB, Grevich J, Saski C, Quesada-Vargas T, Guda C, Tomkins J, Jansen RK (2006) Complete chloroplast genome sequences of Solanum bulbocastanum, Solanum lycopersicum and comparative analyses with other Solanaceae genomes. Theor Appl Genet 112 1503–1518 [DOI] [PubMed] [Google Scholar]
- Deng XW, Gruissem W (1988) Constitutive transcription and regulation of gene expression in non-photosynthetic plastids of higher plants. EMBO J 7 3301–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drescher A, Ruf S, Calsa TJ, Carrer H, Bock R (2000) The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J 22 97–104 [DOI] [PubMed] [Google Scholar]
- Eberhard S, Drapier D, Wollman FA (2002) Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J 31 149–160 [DOI] [PubMed] [Google Scholar]
- Emanuel C, von Groll U, Müller M, Börner T, Weihe A (2006) Development- and tissue-specific expression of the RpoT gene family of Arabidopsis encoding mitochondrial and plastid RNA polymerases. Planta 223 998–1009 [DOI] [PubMed] [Google Scholar]
- Gargano D, Vezzi A, Scotti N, Gray JC, Valle G, Grillo S, Cardi T (2005) The complete nucleotide sequence genome of potato (Solanum tuberosum cv Désirée) chloroplast DNA. In Book of Abstracts of the 2nd Solanaceae Genome Workshop 2005, Ischia, Italy. 2nd Solanaceae Genome Workshop 2005 Organizing Committee, Ischia, Italy, p 107
- Hajdukiewicz PTJ, Allison LA, Maliga P (1997) The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J 16 4041–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Endo T, Peltier G, Tasaka M, Shikanai T (2003) A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J 36 541–549 [DOI] [PubMed] [Google Scholar]
- Hirata N, Yonekura D, Yanagisawa S, Iba K (2004) Possible involvement of the 5′-flanking region and the 5′UTR of plastid accD gene in NEP-dependent transcription. Plant Cell Physiol 45 176–186 [DOI] [PubMed] [Google Scholar]
- Hirose T, Kusumegi T, Tsudzuki T, Sugiura M (1999) RNA editing sites in tobacco chloroplast transcripts: editing as a possible regulator of chloroplast RNA polymerase activity. Mol Gen Genet 262 462–467 [DOI] [PubMed] [Google Scholar]
- Isono K, Niwa Y, Satoh K, Kobayashi H (1997) Evidence for transcriptional regulation of plastid photosynthesis genes in Arabidopsis roots. Plant Physiol 114 623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarret RL, Hasegawa PM, Erickson HT (1980) Factors affecting shoot initiation from tuber discs of potato (Solanum tuberosum). Physiol Plant 49 177–184 [Google Scholar]
- Kahlau S, Aspinall S, Gray JC, Bock R (2006) Sequence of the tomato chloroplast DNA and evolutionary comparison of solanaceous plastid genomes. J Mol Evol 63 194–207 [DOI] [PubMed] [Google Scholar]
- Kahlau S, Bock R (2008) Plastid transcriptomics and translatomics of tomato fruit development and chloroplast-to-chromoplast differentiation: chromoplast gene expression largely serves the production of a single protein. Plant Cell 20 856–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SG, Polking GF, Hannapel DJ (1995) Expression of a cDNA for chloroplast ribosomal protein S16 of potato (Solanum tuberosum L.). Plant Cell Rep 14 359–365 [DOI] [PubMed] [Google Scholar]
- Karcher D, Bock R (2002) The amino acid sequence of a plastid protein is developmentally regulated by RNA editing. J Biol Chem 277 5570–5574 [DOI] [PubMed] [Google Scholar]
- Kemble RJ (1987) A rapid, single leaf, nucleic acid assay for determining the cytoplasmic organelle complement of rapeseed and related Brassica species. Theor Appl Genet 73 364–370 [DOI] [PubMed] [Google Scholar]
- Kim M, Christopher DA, Mullet JE (1993) Direct evidence for selective modulation of psbA, rpoA, rbcL and 16S RNA stability during barley chloroplast development. Plant Mol Biol 22 447–463 [DOI] [PubMed] [Google Scholar]
- Klaff P, Gruissem W (1991) Changes in chloroplast mRNA stability during leaf development. Plant Cell 3 517–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kode V, Mudd EA, Iamtham S, Day A (2005) The tobacco plastid accD gene is essential and is required for leaf development. Plant J 44 237–244 [DOI] [PubMed] [Google Scholar]
- Krause K, Maier RM, Kofer W, Krupinska K, Herrmann RG (2000) Disruption of plastid-encoded RNA polymerase genes in tobacco: expression of only a distinct set of genes is not based on selective transcription of the plastid chromosome. Mol Gen Genet 263 1022–1030 [DOI] [PubMed] [Google Scholar]
- Kumar S, Dhingra A, Daniell H (2004) Plastid-expressed betaine aldehyde dehydrogenase gene in carrot cultured cells, roots, and leaves confers enhanced salt tolerance. Plant Physiol 136 2843–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Maliga P (2002) Overexpression of the clpP 5′-untranslated region in a chimeric context causes a mutant phenotype, suggesting competition for a clpP-specific RNA maturation factor in tobacco chloroplasts. Plant Physiol 129 1600–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Maliga P (2003) The plastid clpP1 protease gene is essential for plant development. Nature 425 86–89 [DOI] [PubMed] [Google Scholar]
- Lee SS, Jeong WJ, Bae JM, Bang JW, Liu JR, Harn CH (2004) Characterization of the plastid-encoded carboxyltransferase subunit (accD) gene of potato. Mol Cells 17 422–429 [PubMed] [Google Scholar]
- Legen J, Kemp S, Krause K, Profanter B, Herrmann RG, Maier RM (2002) Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J 31 171–188 [DOI] [PubMed] [Google Scholar]
- Lerbs-Mache S (2000) Regulation of rDNA transcription in plastids of higher plants. Biochimie 82 525–535 [DOI] [PubMed] [Google Scholar]
- Li W, Ruf S, Bock R (2006) Constancy of organellar genome copy numbers during leaf development and senescence in higher plants. Mol Genet Genomics 275 185–192 [DOI] [PubMed] [Google Scholar]
- Liere K, Börner T (2007) Transcription and transcriptional regulation in plastids. Top Curr Genet 19 121–174 [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- López-Juez E (2007) Plastid biogenesis, between light and shadows. J Exp Bot 58 11–26 [DOI] [PubMed] [Google Scholar]
- MacLean D, Jerome CA, Brown APC, Gray JC (2008) Co-regulation of nuclear genes encoding plastid ribosomal proteins by light and plastid signals during seedling development in tobacco and Arabidopsis. Plant Mol Biol 66 475–490 [DOI] [PubMed] [Google Scholar]
- Magee AM, MacLean D, Gray JC, Kavanagh TA (2007) Disruption of essential plastid gene expression caused by T7 RNA polymerase-mediated transcription of plastid transgenes during early seedling development. Transp Res 16 415–428 [DOI] [PubMed] [Google Scholar]
- Maliga P (1998) Two plastid RNA polymerases of higher plants: an evolving story. Trends Plant Sci 3 4–6 [Google Scholar]
- Marín-Navarro J, Manuell AL, Wu J, Mayfield SP (2007) Chloroplast translation regulation. Photosynth Res 94 359–374 [DOI] [PubMed] [Google Scholar]
- Monde RA, Greene JC, Stern DB (2000) The sequence and secondary structure of the 3′-UTR affect 3′-end maturation, RNA accumulation, and translation in tobacco chloroplasts. Plant Mol Biol 44 529–542 [DOI] [PubMed] [Google Scholar]
- Mullet JE, Klein RR (1987) Transcription and RNA stability are important determinants of higher plant chloroplast RNA levels. EMBO J 6 1571–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Furuhashi Y, Hasegawa K, Hashimoto H, Watanabe K, Obokata J, Sugita M, Sugiura M (2003) Array-based analysis on tobacco plastid transcripts: preparation of a genomic microarray containing all genes and all intergenic regions. Plant Cell Physiol 44 861–867 [DOI] [PubMed] [Google Scholar]
- Ngernprasirtsiri J, Kobayashi H, Akazawa T (1990) DNA methylation is a determinative element of photosynthesis gene expression in amyloplasts from liquid-cultured cells of sycamore (Acer pseudoplatanus L.). Cell Struct Funct 15 285–293 [DOI] [PubMed] [Google Scholar]
- Nguyen TT, Nugent G, Cardi T, Dix PJ (2005) Generation of homoplasmic plastid transformants of a commercial cultivar of potato (Solanum tuberosum L.). Plant Sci 168 1495–1500 [Google Scholar]
- Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56 2907–2914 [DOI] [PubMed] [Google Scholar]
- Pallotta MA, Graham RD, Langridge P, Sparrow DHB, Barker SJ (2000) RFLP mapping of manganese efficiency in barley. Theor Appl Genet 101 1100–1108 [Google Scholar]
- Peled-Zehavi H, Danon A (2007) Translation and translational regulation in chloroplasts. Top Curr Genet 19 249–281 [Google Scholar]
- Prikryl J, Watkins KP, Friso G, van Wijk KJ, Barkan A (2008) A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res 36 5152–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K (2007) Plastid biogenesis and differentiation. Top Curr Genet 19 1–28 [Google Scholar]
- Reiner A, Yekutieli D, Benjamini Y (2003) Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19 368–375 [DOI] [PubMed] [Google Scholar]
- Ruf S, Hermann M, Berger IJ, Carrer H, Bock R (2001) Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nat Biotechnol 19 870–875 [DOI] [PubMed] [Google Scholar]
- Sakai A, Kawano S, Kuroiwa T (1992) Conversion of proplastids to amyloplasts in tobacco cultured cells is accompanied by changes in the transcriptional activities of plastid genes. Plant Physiol 100 1062–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Barkan A (2007) RNA splicing and RNA editing in chloroplasts. Top Curr Genet 19 213–248 [Google Scholar]
- Shiina T, Allison L, Maliga P (1998) rbcL transcript levels in tobacco plastids are independent of light: reduced dark transcription rate is compensated by increased mRNA stability. Plant Cell 10 1713–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorov VA, Kasten D, Pang SZ, Hajdukiewicz PTJ, Staub JM, Nehra NS (1999) Stable chloroplast transformation in potato: use of green fluorescent protein as a plastid marker. Plant J 19 209–216 [DOI] [PubMed] [Google Scholar]
- Steele Scott N, Tymms MJ, Possingham JV (1984) Plastid-DNA levels in the different tissues of potato. Planta 161 12–19 [DOI] [PubMed] [Google Scholar]
- Toyoshima Y, Onda Y, Shiina T, Nakahira Y (2005) Plastid transcription in higher plants. Crit Rev Plant Sci 24 59–81 [Google Scholar]
- Vera A, Sugiura M (1995) Chloroplast rRNA transcription from structurally different tandem promoters: an additional novel-type promoter. Curr Genet 27 280–284 [DOI] [PubMed] [Google Scholar]
- Vogel J, Hübschmann T, Börner T, Hess WR (1997) Splicing and intron-internal RNA editing of trnK-matK transcripts in barley plastids: support for MatK as an essential splice factor. J Mol Biol 270 179–187 [DOI] [PubMed] [Google Scholar]
- Wakasugi T, Sugita M, Tsudzuki T, Sugiura M (1998) Update gene map of tobacco chloroplast DNA. Plant Mol Biol Rep 16 231–241 [Google Scholar]
- Wakasugi T, Tsudzuki T, Sugiura M (2001) The genomics of land plant chloroplasts: gene content and alteration of genomic information by RNA editing. Photosynth Res 70 107–118 [DOI] [PubMed] [Google Scholar]
- Waters M, Pyke K (2005) Plastid development and differentiation. In SG Møller, ed, Plastids. Blackwell Publishing, Oxford, pp 30–59
- Waters MT, Moylan EC, Langdale JA (2008) GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J 56 432–444 [DOI] [PubMed] [Google Scholar]
- Wilkinson L, Hill MA, Vang E (1992) SYSTAT: Statistics, Version 5.2. SYSTAT Inc., Evanston, IL
- Williams PM, Barkan A (2003) A chloroplast-localized PPR protein required for plastid ribosome accumulation. Plant J 36 675–686 [DOI] [PubMed] [Google Scholar]
- Wischmann B, Hamborg Nielsen T, Lindberg Møller B (1999) In vitro biosynthesis of phosphorylated starch in intact potato amyloplasts. Plant Physiol 119 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Chory J (2008) Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet 9 383–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Tasaka M, Shikanai T (2004) PPR motifs of the nucleus-encoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. Plant J 38 152–163 [DOI] [PubMed] [Google Scholar]
- Zhu YS, Merkle-Lehman DL, Kung SD (1984) Light-induced transformation of amyloplasts into chloroplasts in potato tubers. Plant Physiol 75 142–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R, Liere K, Börner T (2007) From seedling to mature plant: Arabidopsis plastidial genome copy number, RNA accumulation and transcription are differentially regulated during leaf development. Plant J 50 710–722 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.