Abstract
We isolated an activation-tagged Arabidopsis (Arabidopsis thaliana) line, constitutive disease susceptibility2-1D (cds2-1D), that showed enhanced bacterial growth when challenged with various Pseudomonas syringae strains. Systemic acquired resistance and systemic PATHOGENESIS-RELATED GENE1 induction were also compromised in cds2-1D. The T-DNA insertion adjacent to NINE-CIS-EPOXYCAROTENOID DIOXYGENASE5 (NCED5), one of six genes encoding the abscisic acid (ABA) biosynthetic enzyme NCED, caused a massive increase in transcript level and enhanced ABA levels >2-fold. Overexpression of NCED genes recreated the enhanced disease susceptibility phenotype. NCED2, NCED3, and NCED5 were induced, and ABA accumulated strongly following compatible P. syringae infection. The ABA biosynthetic mutant aba3-1 showed reduced susceptibility to virulent P. syringae, and ABA, whether through exogenous application or endogenous accumulation in response to mild water stress, resulted in increased bacterial growth following challenge with virulent P. syringae, indicating that ABA suppresses resistance to P. syringae. Likewise ABA accumulation also compromised resistance to the biotrophic oomycete Hyaloperonospora arabidopsis, whereas resistance to the fungus Alternaria brassicicola was enhanced in cds2-1D plants and compromised in aba3-1 plants, indicating that ABA promotes resistance to this necrotroph. Comparison of the accumulation of salicylic acid and jasmonic acid in the wild type, cds2-1D, and aba3-1 plants challenged with P. syringae showed that ABA promotes jasmonic acid accumulation and exhibits a complex antagonistic relationship with salicylic acid. Our findings provide genetic evidence that the abiotic stress signal ABA also has profound roles in modulating diverse plant-pathogen interactions mediated at least in part by cross talk with the jasmonic acid and salicylic acid biotic stress signal pathways.
Plants rely on efficient resistance mechanisms that involve multiple layers of constitutive and induced defenses to protect themselves from pathogen attacks. Constitutive physical and chemical barriers on the plant surface prevent the establishment of pathogen infection structures, whereas the effect of induced defense is based on the ability to perceive and respond to pathogen-derived factors. The basal perception systems at the plant cell surface recognize general microbial invaders by detecting conserved microbe-associated molecular patterns (MAMPs), such as flagellin, a structural component of the bacterial flagellum (Gómez-Gómez and Boller, 2002). A multitude of plant major disease resistance (R) proteins specify recognition of pathogens carrying the corresponding avirulence (avr) genes. Recognition of the MAMPs- and avr-dependent signals may lead to activation of some overlapping inducible defenses, including synthesis or mobilization of antibiotic compounds and deposition of callose to reinforce the cell wall (Nurnberger et al., 2004; Clay et al., 2009). The MAMP-triggered basal defenses, however, do not always result in the development of localized programmed cell death (referred to as the hypersensitive response [HR]), a characteristic feature generally associated with R-gene-dependent defenses. HR also contributes to the establishment of the long-lasting systemic acquired resistance against subsequent attack by a broad range of normally virulent pathogens (Durrant and Dong, 2004).
Complex signaling networks orchestrate different types of plant-inducible defenses to prevent microbial growth. Pathogen recognition triggers a number of rapid cellular responses, including ionic changes, and phosphorylation cascades, which precede the accumulation of reactive oxygen species, nitric oxide, and salicylic acid (SA) and the transcriptional activation of defense-related genes. Interplay between reactive oxygen species, nitric oxide, and SA contributes to the establishment of HR. SA also has a key role in establishing local and systemic resistance to many virulent biotrophic pathogens, whereas jasmonic acid (JA) and ethylene (ET) are more often associated with resistance to necrotrophic pathogens. Considerable interactions occur within and between these hormone signaling networks, resulting in an overall mutual antagonism between SA and JA/ET signaling (Kunkel and Brooks, 2002; Beckers and Spoel, 2006). The similarities between the mechanisms of signaling and defense that underlie multilayer of disease resistance and the close interactions between different signaling pathways imply a fine-tuned deployment of conserved defense signals and effectors in different plant-pathogen interactions.
Genetic dissection of disease resistance in the model plant Arabidopsis (Arabidopsis thaliana) through loss-of-function mutagenesis has identified some important components of basal and R-gene-dependent defenses. For example, EDS1, PAD4, and MOS3 are essential for the resistance specified by the subclass of nucleotide-binding site Leu-rich repeat R proteins that contain an N-terminal Toll Interleukin1 receptor domain, and they are also required for basal resistance to virulent pathogens (Dangl and Jones, 2001; Glazebrook, 2001; Zhang and Li, 2005), while NPR1 was shown to mediate SA-dependent defense responses (Dong, 2001). However, many genes are hard to identify by this approach due to lethality or functional redundancy, especially in complex and reiterative signal networks (Dangl and Jones, 2001).
T-DNA activation tagging generates dominant, gain-of-function mutations that lead to enhanced expression of the tagged genes (Weigel et al., 2000). This approach has been used to investigate resistance mechanisms with the expectation that elevated expression of defense signaling molecules may lead to a quantitative impact on resistance even where there is significant cross talk and functional redundancy. Thus, several new components have been shown to regulate plant-pathogen interactions (Grant et al., 2003; Xia et al., 2004; Zhang et al., 2007). Aiming to uncover genes involved in SA-mediated signaling mechanisms, we carried out activation tagging in SA-deficient Arabidopsis plants that constitutively express the bacterial salicylate hydroxylase (NahG; Gaffney et al., 1993). No mutants were recovered that had enhanced resistance to infection by virulent bacteria; instead, we identified a mutant with enhanced disease symptoms that supported increased bacterial growth compared to NahG plants. Molecular characterization of the mutant revealed that massive activation of NINE-CIS-EPOXYCAROTENOID DIOXYGENASE5 (NCED5) led to constitutive accumulation of high levels of endogenous abscisic acid (ABA), which impacted multiple layers of defense against diverse plant pathogens. Further investigation showed that endogenous ABA synergizes with JA and exhibits a complex antagonistic relationship with SA during disease development. Our findings provide genetic evidence that physiological levels of ABA play an important role in modulating diverse plant-pathogen interactions and elaborate a link between abiotic stress and level of disease susceptibility.
RESULTS
cds2-1D Is a Dominant Mutation Conferring Enhanced Disease Susceptibility to Pseudomonas syringae
A collection of 8,000 activation tagged T1 plants in the NahG background were screened by hand-infiltration with virulent P. syringae pv maculicola ES4326 (Psm) chromosomally tagged with luxCDABE (Fan et al., 2008). While no mutant was found that enhanced basal resistance in the absence of SA, a mutant with increased disease severity was identified. In the F2 generation following back crossing to Columbia-0 (Col-0), three-quarters of the 40 plants in which NahG had been segregated out showed severe tissue maceration when challenged with Psm, while one-quarter developed only mild chlorosis akin to that in wild-type plants (Fig. 1A). Basta resistance, which is conferred by the selectable marker of the activation tagging vector, cosegregated with enhanced symptom development. Thus, the disease phenotype reflects a dominant mutation associated with the single T-DNA insertion and independent of the NahG transgene. We designated this mutant constitutive disease susceptibility2-1D (cds2-1D).
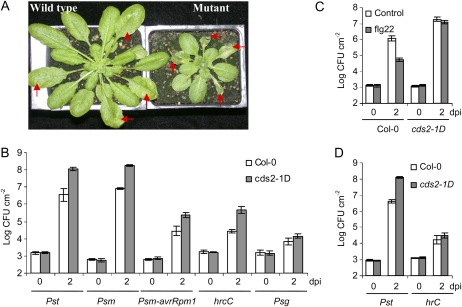
Figure 1.
Compromised disease resistance to bacterial infection in cds2-1D mutant. A, Disease symptom of wild-type Col-0 (left) and mutant cds2-1D (right) plants 3 d after hand-infiltration of Pst. The arrows indicate the inoculated leaves. B, Bacterial growth of various P. syringae strains in leaves from cds2-1D mutant (gray) and wild-type Col-0 (white) plants. C, Flagellin-induced restriction of Pst growth is attenuated in cds2-1D mutant. Leaves of Col-0 and cds2-1D plants were pretreated for 24 h by infiltration with 100 nm flg22 peptide (gray) or water as control (white) before bacterial challenge. D, Bacterial growth of wild-type Pst and the TTSS-deficient hrcC strain in detached leaves from Col-0 (white) and cds2-1D (gray) plants. The hand-infiltrated leaves were excised from plants and inserted in 0.7% water-agarose plate after the excessive water had dissipated. The plates were sealed with 3M Micropore surgical tape and incubated under the same condition as the in planta bacterial growth assay. In all the above experiments, leaves were infiltrated with 105 cfu mL−1 of bacteria and were collected 2 d after inoculation for growth assay. Data shown are means ± sd, and similar results were observed in three replicate experiments. dpi, Days post inoculation.
Disease Phenotypes of cds2-1D
Quantitative analysis of bacterial growth showed >30-fold greater growth of both virulent Psm and virulent P. syringae pv tomato DC3000 (Pst) in hand-infiltrated leaves of cds2-1D compared to wild-type plants, accompanied by severe tissue maceration. The growth of Psm carrying the avrRpm1 avirulence gene (Psm-avrRpm1), which triggers HR on the Col-0 genotype due to the cognate Rpm1 disease resistance gene, was 6-fold greater in cds2-1D than in wild-type plants, indicating a weakened resistance to a normally avirulent bacterial strain. The type three secretion system (TTSS)-deficient hrcC mutant of Pst DC3000 (hrcC) grew to 20-fold higher levels on cds2-1D plants compared to Col-0 plants, but the growth of this nonpathogenic mutant was weak compared to wild-type Pst and did not result in tissue maceration or development of visible lesions. Likewise, growth of P. syringae pv glycinea (Psg), which is nonpathogenic on Arabidopsis, was only enhanced 3-fold in cds2-1D compared to very low levels of growth in wild-type plants (Fig. 1B).
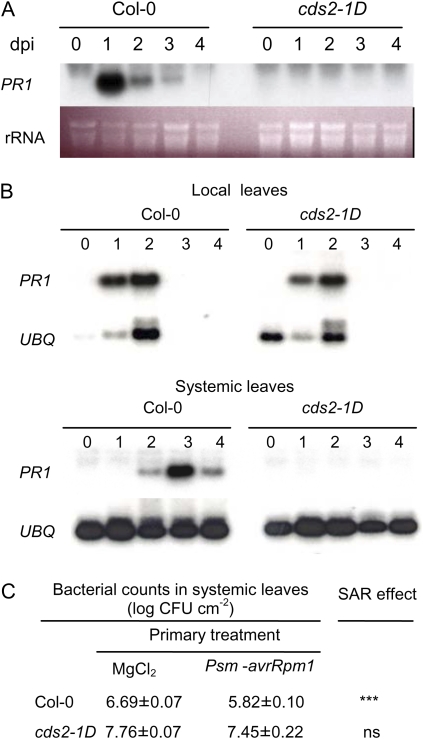
Accumulation of PATHOGENESIS-RELATED GENE1 (PR1) transcripts is a marker for SA signaling and establishment of systemic acquired resistance. Induction of PR1 by spraying plants with 1 mm SA was strongly suppressed in cds2-1D plants compared to wild-type Col-0 (Fig. 2A). Furthermore, while the local induction of PR1 at the site of inoculation with Psm-avrRpm1 was not affected in cds2-1D, the mutant plants showed no systemic accumulation of PR1 transcripts (Fig. 2B). Likewise, following local induction with avirulent Psm-avrRpm1, systemic acquired resistance of distant leaves subsequently challenged with virulent Pst was substantially reduced in cds2-1D plants compared to wild-type plants (Fig. 2C).
Figure 2.
SA induced PR1 expression and systemic acquired resistance (SAR) is suppressed in the cds2-1D mutant. Leaves were collected at 0, 1, 2, 3, and 4 d after SA treatment or bacterial challenge. A, RNA gel blot analysis of PR1 gene transcript levels in wild-type Col-0 and cds2-1D mutant plants sprayed with 1 mm SA. dpi, Days post inoculation. B, Systemic induction of PR1 expression is suppressed in the cds2-1D mutant plant. Two leaves of each plant were inoculated with 5 × 106 cfu mL−1 of Psm-avrRpm1. The inoculated local leaves and noninoculated systemic leaves were collected for RNA gel blot assay of PR1 transcript levels. C, Attenuated systemic acquired resistance in cds2-1D mutant. Treatments consist of inoculation of two leaves with 5 × 106 cfu mL−1 of avirulent Psm-avrRpm1, followed 2 d later with the second inoculation of systemic leaves with 105 cfu mL−1 of virulent Pst. Magnesium chloride (10 mm) was infiltrated as control to avirulent bacterial treatment. Growth of virulent Pst in systemic leaves was monitored 2 d after the second inoculation. Significant reduction of Pst growth in systemic leaves was observed in Col-0 plants between MgCl2 and avirulent Psm-avrRpm1 treatments (indicated by ***, t test, n = 6, P < 0.001), whereas the difference in cds2-1D plants was not statistically significant (indicated by “ns”). This experiment was repeated twice with similar results. [See online article for color version of this figure.]
The observation of enhanced growth of both virulent and hrcC strains on cds2-1D plants prompted us to check whether the mutation also affects MAMP-induced basal resistance, which requires SA signaling to suppress hrcC growth but is largely independent of SA pathways in suppressing growth of virulent bacteria (Zipfel et al., 2004; Tsuda et al., 2008). Infiltration of leaves with 100 nm flg22 peptide 1 d before bacterial challenge reduced the growth of virulent Pst >10-fold in wild-type Col-0 plants, while the same treatment had little or no effect on cds2-1D mutant plants (Fig. 1C), indicating compromised MAMP-triggered immunity.
Other cds2-1D Phenotypes
Apart from being more susceptible to Pst and Psm, cds2-1D plants showed delayed development and were smaller than the wild type. Moreover, when cds2-1D plants were well watered and kept in darkness, leaves became water soaked. To examine if this phenotype was associated with enhanced disease susceptibility, we tested bacterial growth in excised leaves, which do not develop water-soaked leaves. Wild-type Pst grew to 30-fold higher levels on detached cds2-1D leaves than equivalent wild-type leaves, but there was no significant difference between the weak growth of the hrcC mutant on detached leaves from cds2-1D and wild-type plants (Fig. 1D). Thus, most of the enhanced growth of Pst on cds2-1D was independent of the water-soaking phenotype but required a functional TTSS to be realized, with a smaller TTSS-independent component associated with the water-soaking phenotype (Fig. 1, B and D).
Molecular Characterization of the cds2-1D Mutation
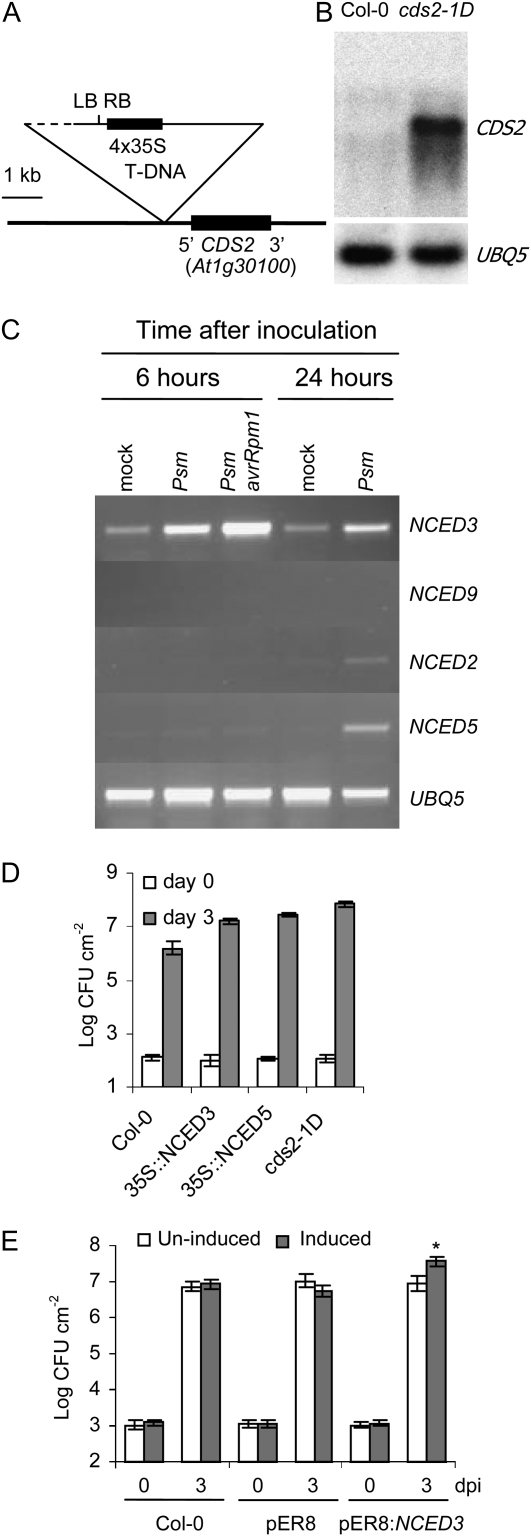
The cds2-1D phenotype and Basta resistance cosegregated as simple dominant Mendelian traits, suggesting that the cds2-1D locus was T-DNA tagged. Thermal asymmetric interlaced PCR (Liu et al., 1995) and inverse PCR (IPCR) failed to amplify Arabidopsis DNA sequences but revealed the presence of left and right T-DNA borders, indicating rearrangement at the insertion site. DNA gel blots hybridized with Cauliflower mosaic virus (CaMV) 35S enhancer sequences positioned the T-DNA region adjacent to the putative rearranged insertion site, and using IPCR with primers anchored in the identified T-DNA region, we were able to isolate Arabidopsis sequences flanking the insertion site. As illustrated in Figure 3A, the T-DNA had inserted approximately 400 bp upstream of the gene corresponding to locus At1g30100 such that the CaMV 35S enhancers were approximately 3.5 kb from the tagged gene. No other gene within 9 kb either side of this locus was annotated in The Arabidopsis Information Resource database. At1g30100 sequences hybridized to an approximately 2-kb transcript in RNA blots of total RNA from healthy leaves of cds2-1D plants, whereas little or no hybridization was observed with an equivalent RNA sample from wild-type plants (Fig. 3B).
Figure 3.
Functional analysis of cds2-1D allele and the related gene family members involved in Arabidopsis-P. syringae interaction. A, Structure of T-DNA insertion in cds2-1D mutant. The detected head-to-head T-DNA left and right borders are marked as LB and RB, respectively. The uncharacterized T-DNA region is indicated by dashed line. B, CDS2 expression is massively enhanced in the cds2-1D mutant. RNA gel blot assay of CDS2 gene transcript in healthy leaves from Col-0 and cds2-1D mutant plants. The bottom panel shows ubiquitin transcript levels of the same samples. C, Arabidopsis NCED genes are induced by bacterial infection. RT-PCR analysis of NCED transcripts levels in leaves from Col-0 plants that were infiltrated with 5 × 107 cfu mL−1 of virulent Psm or avirulent Psm-avrRpm1. Water was infiltrated as mock treatment. No PCR product could be observed when nonreverse-transcribed total RNA was used as negative controls to amplify NCED genes (data not shown). D, Constitutive overexpression of NCED genes enhanced susceptibility to bacterial infection. One of each representative transgenic lines constitutively overexpressing NCED3 or NCED5 genes, together with wild-type Col-0 and cds2-1D mutant plants, was hand-infiltrated with 104 cfu mL−1 of virulent Psm, and in planta bacterial growth were determined at 3 d after inoculation. E, Conditional overexpression of NCED3 enhances susceptibility to bacterial infection. Wild-type Col-0 plants and plants transformed with empty vector pER8 or pER8-NCED3 were sprayed with 10 μm β-estrodial 1 d before hand-infiltration of virulent Pst at 105 cfu mL−1. Water was sprayed as uninduced control. The in planta bacterial growth was determined 3 d after inoculation. Significant differences in bacterial growth were only detected between control and induced pER8:NCED3 plants (indicated by *, t test, P < 0.05). dpi, Days post inoculation. Data shown in the bacterial growth assays are means ± sd, and each experiment was repeated at least twice with similar results.
At1g30100 Is a Pathogen-Induced Member of the NCED Family
The At1g30100 locus is NCED5, one of six Arabidopsis genes encoding NCED. NCED catalyzes oxidative cleavage of 9-cis-xanthophylls to form xanthoxin, a key regulatory step of ABA biosynthesis. Among these six homologs, NCED3 is a major drought-induced NCED in Arabidopsis leaves, and NCED genes are developmentally regulated and associated with ABA synthesis in roots, flowers, and developing seeds (Iuchi et al., 2001; Tan et al., 2003). Based on sequence similarities NCED5 and NCED3 are members of a distinct subfamily of four of the NCED genes, and the transcription profiles of this subfamily in response to bacterial challenge were investigated by reverse transcription (RT)-PCR. As shown in Figure 3C, NCED5 and NCED2 transcripts accumulated in the later stages of the compatible interaction with Psm but were not induced by avirulent Psm-avrRpm1. In contrast, NCED3 showed strong early expression in both compatible and incompatible interactions, while NCED9 transcripts did not accumulate in response to bacterial inoculation.
Overexpression of NCED5 and NCED3 Enhances Susceptibility to P. syringae
To confirm that NCED overexpression indeed enhances disease susceptibility, the coding sequences of NCED5, the T-DNA-tagged gene, and NCED3, which is strongly induced by P. sryingae, were expressed in transgenic plants under the control of the CaMV 35S promoter. Multiple independent transgenic lines showing cds2-1D-like phenotypes were identified for both constructs, and growth of Psm was enhanced >10-fold in plants, showing strong constitutive expression of either NCED3 or NCED5 compared to growth on equivalent wild-type control plants (Fig. 3D).
To investigate whether enhanced disease susceptibility could be separated from the developmental phenotypes also associated with strong constitutive NCED overexpression, we made transgenic plants with NCED3 under the control of the estradiol-inducible XVE system (Zuo et al., 2000). No difference was observed in the development or morphology of pER8-NCED3 transgenic plants compared to the wild type. Spraying pER8-NCED3 lines with 10 μm of β-estradiol induced strong accumulation of NCED3 transcripts (data not shown), and challenge with Pst 24 h after estradiol treatment resulted in 4-fold greater bacterial growth than in equivalent uninduced plants. Estradiol treatment of wild-type or empty vector control transgenic plants had no effect on susceptibility to Pst (Fig. 3E).
Physiological Role of ABA in Bacterial Susceptibility
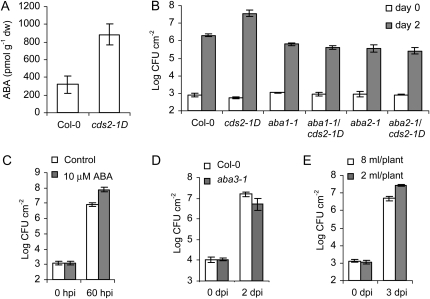
Overexpression of NCED enhances ABA levels (Thompson et al., 2000; Iuchi et al., 2001); likewise, leaves of healthy cds2-1D plants showed 3-fold elevation in the level of ABA (Fig. 4A). To further investigate if the enhanced susceptibility to bacterial infection in cds2-1D plants was tightly associated with the elevated level of endogenous ABA, we crossed cds2-1D with aba1-1 and aba2-1, which carry genetic lesions in the ABA biosynthetic pathway (Léon-Kloosterziel et al., 1996; Marin et al., 1996). All of the F1 plants derived from the crosses were Basta resistant and showed cds2-1D morphology, while in F2 progenies, aba genotyping analysis of the Basta-resistant plants showed that homozygosity of either aba locus resulted in the morphological phenotypes characteristic of the aba mutant. Similarly, both aba1-1 and aba2-1 mutations eliminated enhanced bacterial disease susceptibility of cds2-1D (Fig. 4B), indicating that the observed cds2-1D phenotypes are dependent on ABA biosynthesis. These findings, together with the observation that some NCED genes are pathogen induced, prompted further investigation of the role of ABA in bacterial pathogenesis.
Figure 4.
Physiological role of ABA in bacterial susceptibility. A, The cds2-1D mutant plants have increased basal level of free ABA in comparison to wild-type Col-0 plants. B, The enhanced bacterial growth in cds2-1D mutant is dependent on ABA biosynthesis pathway. Leaves of wild-type and homozygous mutant plants were hand-infiltrated with 105 cfu mL−1 of virulent Pst, and bacterial growth was determined 2 d after inoculation. C, Exogenous ABA treatment enhances bacterial susceptibility. Leaves of wild-type Col-0 plants were hand-infiltrated with 105 cfu mL−1 of virulent Pst before they were excised from the plants with a razor blade and fed through the petiole with sterilized water (white) or 10 μm ABA (gray). Bacterial number in the inoculated leaves was determined 60 h after inoculation. D, Bacterial susceptibility is attenuated in ABA-deficient mutant plants. Leaves from wild-type Col-0 (white) and aba3-1 mutant (gray) plants were hand-infiltrated with 106 cfu mL−1 of virulent Pst, and the leaf bacterial number was determined 2 d after inoculation. E, Water restriction enhanced bacterial susceptibility in Arabidopsis. Five-week-old Col-0 plants were supplied daily with sufficient level (8 mL/plant, white) or restricted level (2 mL/plant, gray) of water for 1 week and inoculated with 105 cfu mL−1 of virulent Pst. Bacterial number in the inoculated leaves was determined 3 d after inoculation. Data shown in the above assays are means ± sd; g−1dw is per gram dry weight. All the experiments were repeated at least twice with similar results. dpi, Days post inoculation; hpi, hours post inoculation.
First, we checked whether exogenous ABA promoted bacterial growth. To introduce ABA, leaves of wild-type Col-0 plants were detached after hand-infiltration with virulent Pst and petioles immersed in 10 μm ABA or water. After 2 d, bacterial growth was 10-fold greater in ABA-treated leaves compared to water-treated leaves (Fig. 4C). Similar effects could be observed in ABA-treated leaves on growth of the hrcC strain (Supplemental Fig. S1A).
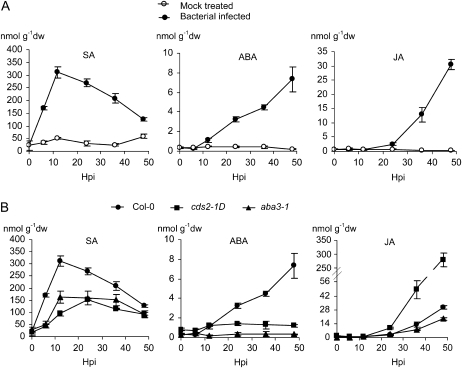
Next, we used liquid chromatography-mass spectrometry (LC-MS) to determine whether ABA accumulated during disease development. Marked accumulation of ABA was observed starting 12 h after inoculation with virulent Pst with continuing accumulation as chlorotic lesions developed. ABA accumulation preceded the onset of JA accumulation during lesion development but was slower than the transient accumulation of SA in the early stages of the compatible interaction. No significant changes in ABA, SA, or JA levels were observed in equivalent mock-inoculated plants (Fig. 5A).
Figure 5.
Accumulation of free SA, ABA, and JA in Arabidopsis leaves infected with P. syringae. Leaves of 5-week-old plants were hand-infiltrated with water or 5 × 106 cfu mL−1 of virulent Pst. The inoculated leaves were collected at 0, 6, 12, 24, 36, and 48 h post inoculation (Hpi) and the extracts of leaf tissue used for LC-MS quantification of free SA, ABA, and JA. A, SA, ABA, and JA induction by bacterial infection (solid circle) of wild-type Col-0 plants. Water was infiltrated as mock treatment (open circle). B, Comparison of SA, ABA, and JA induction in wild-type Col-0, aba3-1 (solid triangle), and cds2-1D (solid square) mutant plants in response to bacterial infection. For each data point, three biological replicates were assayed, and data shown are means ± sd; g−1dw is per gram dry weight. The experiments were repeated twice with similar outcomes.
Third, we investigated whether susceptibility to P. syringae was affected when ABA synthesis was blocked. The Arabidopsis mutant aba3-1 is blocked in the last step of ABA biosynthesis and fails to accumulate ABA in response to dehydration (Xiong et al., 2001). Three days after challenge with Pst, leaves of aba3-1 plants exhibited only weak symptom development compared to the chlorosis observed in equivalent leaves of wild-type controls (Supplemental Fig. S1B). Reduced symptom development was accompanied by 3-fold less bacterial growth in aba3-1 plants compared to the wild type (Fig. 4D).
Finally, we investigated whether mild water stress, a physiological condition that promotes accumulation of ABA, affected disease susceptibility. Wild-type plants were stressed by a reduced watering regimen for 7 d prior to challenge with Pst. Three days after inoculation bacterial growth was 5-fold greater than in unstressed plants (Fig. 4E).
Effect of ABA in Other Pathosystems
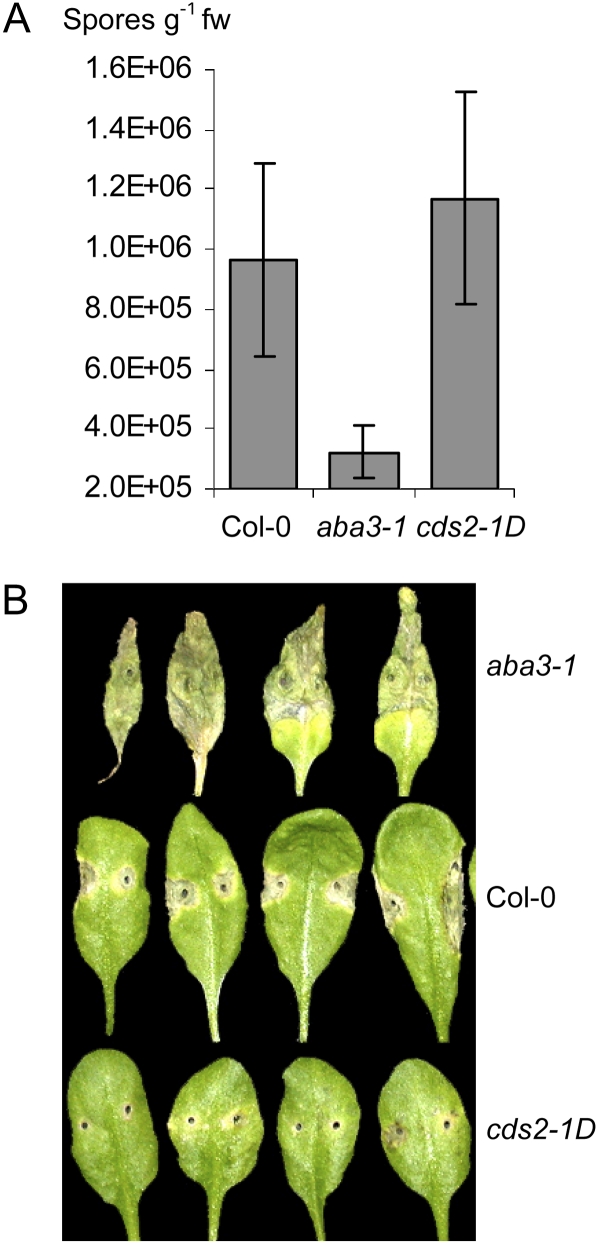
Taken in aggregate, these findings indicated that ABA enhanced susceptibility to the hemibiotrophic bacterial pathogen P. syringae. To extend the analysis, we investigated whether ABA also affected susceptibility in other pathosystems. Pathogenesis of the oomycete biotroph Hyaloperonospora arabidopsis was suppressed in leaves of aba3-1 plants with 3-fold less sporulation than in wild-type plants (Fig. 6A). While H. arabidopsis sporulation was not significantly enhanced in leaves of cds2-1D plants, hyphal growth was substantially more prolific in leaves of cds2-1D compared to wild-type plants (Fig. 6A; Supplemental Fig. S1C). In contrast, symptom development following inoculation with the fungal necrotroph Alternaria brassicicola was enhanced in aba3-1 plants and suppressed in cds2-1D plants compared to wild-type controls (Fig. 6B).
Figure 6.
ABA is involved in other Arabidopsis-microbial interactions. A, ABA biosynthesis is required for H. arabidopsis to attain full virulence on Arabidopsis. Three-week-old wild-type Col-0, aba3-1, and cds2-1D seedlings were sprayed with 4 × 104 spores mL−1 of H. arabidopsis, and the leaves were collected at 7 d after inoculation to determine levels of pathogen sporulation. Data shown are means ± sd; g−1fw is per gram fresh weight. B, ABA is required for resistance to A. brassicicola in Arabidopsis. Leaves of 6-week-old plants were inoculated with 5 μL of 105 spores mL−1 of A. brassicicola, and 7 d after the inoculated leaves from one representative plant of aba3-1 (top), Col-0 (middle), and cds2-1D (bottom) genotypes were excised for photography. These experiments were repeated twice with similar outcomes.
Endogenous ABA Synergizes with JA and Exhibits a Complex Antagonistic Relationship with SA during Disease Development
The observation that ABA reduced susceptibility to the necrotrophic pathogen A. brassicicola while promoting susceptibility to the biotrophic pathogens P. syringae and H. arabidopsis is reminiscent of some features of JA function in plant-pathogen interactions and the often antagonistic effects between JA and SA. We therefore examined the interplay between ABA and these two biotic stress signal molecules. In Pst-inoculated wild-type Col-0 plants following transient SA induction, ABA accumulation preceded the onset of JA induction (Fig. 5A), suggesting that ABA may promote JA accumulation and enhance JA action. To test this hypothesis, we examined JA accumulation in genotypes with differing ABA levels. Pst induction of JA accumulation was reduced in ABA-deficient aba3-1 plants compared to the wild type, whereas in cds2-1D plants, Pst induction of JA accumulation was accelerated and markedly potentiated (Fig. 5B). As expected, there was little accumulation of ABA in aba3-1 plants challenged with Pst, but interestingly, infection of cds2-1D plants did not enhance ABA levels beyond the elevated basal level in healthy leaves, suggesting that the levels of ABA in the early stages of infection are critical to the outcome and possible operation of a feedback loop (Fig. 5B). Building on this, and in contrast to the simple relationship between ABA and JA, there appeared to be a complex relationship between SA and ABA. Thus, while early SA accumulation was weaker in cds2-1D plants than the wild type, ABA-deficient plants also showed reduced SA accumulation in response to Pst challenge (Fig. 5B).
DISCUSSION
ABA is an important phytohormone that regulates many aspects of plant growth and development, especially responses to various abiotic stresses. Recently, several studies have demonstrated that ABA may also be widely involved in plant responses to biotic stresses caused by a broad range of plant pathogens (Mauch-Mani and Mauch, 2005; Asselbergh et al., 2008a) or insect herbivores (Thaler and Bostock, 2004; Bodenhausen and Reymond, 2007). Many early studies relied on the application of exogenous ABA to increase plant ABA levels (Asselbergh et al., 2008a), although exogenous ABA may not exert the same physiological effects as endogenous ABA (Christmann et al., 2005), and this may potentially complicate the interpretations of the roles of ABA in plant-pathogen interactions (Wasilewska et al., 2008). Our work on the cds2-1D mutant, which was identified from a forward genetic screen, showed that activation of NCED5, a key enzyme regulating de novo ABA biosynthesis, led to elevated endogenous ABA levels and to enhanced susceptibility to infection by P. syringae. The observations that overexpression of NCEDs in transgenic Arabidopsis recapitulated cds2-1D disease phenotype and that genetic lesions in the ABA biosynthesis pathway abolished the enhanced bacterial growth in the cds2-1D mutant pinpoint the role of ABA in promoting susceptibility to this hemibiotrophic bacterial pathogen. Susceptibility was increased by application of exogenous ABA at physiological levels or by endogenous accumulation in response to mild water stress, thereby demonstrating the physiological relevance of ABA-mediated suppression of resistance to this hemibiotrophic bacterial pathogen. Thus, our data help establish a potentially important link between abiotic stress and the level of host susceptibility to disease.
Compared with abiotic stresses, however, the role of ABA in biotic stresses may vary among different pathosystems. In mutants deficient in ABA biosynthesis, the resistance to the biotrophic pathogens Hyaloperonospora parasitica and Blumeria graminis is enhanced (Mohr and Cahill, 2003; Jensen et al., 2008), while resistance to the necrotrophic pathogens A. brassicicola and Pythium irregulare is compromised (Ton and Mauch-Mani, 2004; Adie, et al., 2007). Our data also demonstrate that ABA modulates pathogenesis in diverse plant-pathogen interactions with outcomes apparently influenced by the infection biology. Thus, the ABA biosynthesis pathway is also required for the full virulence of the obligate oomycete pathogen H. arabidopsis in terms of asexual spore production and proliferating hyphal growth, while ABA positively regulates resistance against necrotrophic A. brassicicola infection, indicating strikingly different roles of ABA in the interactions with bio/hemibiotrophs and necrotrophs. However, contradictory roles of ABA in other pathosystems have also been documented; for example, ABA suppresses resistance to necrotrophic pathogens, including Botrytis cinerea, Fusarium oxysporum, Plectosphaerella cucumerina, and Erwinia chrysanthemi (Audenaert et al., 2002; Anderson et al., 2004; Hernandez-Blanco et al., 2007; Asselbergh et al., 2008b), and ABA signaling may positively regulate the RLM1Col determined resistance to the hemibiotrophic fungal pathogen Leptosphaeria maculans and the edr1-mediated resistance to the obligate biotrophic powdery mildew pathogen Golovinomyces cichoracearum in Arabidopsis (Kaliff, et al., 2007; Wawrzynska et al., 2008). Hence, specific manifestations of the emerging new role for the abiotic stress signal ABA in modulating disease resistance may depend not only on pathogen lifestyle and overall infection biology but also on specialized features of each interaction, indicating that complex nuanced mechanisms underlie ABA modulation of plant biotic stresses.
ABA Modulation of Plant Disease Resistance Mechanisms
Plant receptors recognize extracellular MAMPs or intracellular virulence proteins derived from bacteria and activate multiple layers of defense to limit infection, whereas pathogenic bacteria are armed with a collection of effector molecules, including chemical and proteinacious factors to suppress plant defense responses and promote disease (Abramovitch et al., 2006). Recent work revealed that stomatal closure upon perception of MAMPs/bacteria is an important preinvasive innate immune response involving ABA signaling components to restrict bacterial entry (Melotto et al., 2006). However, at postinvasive stages, ABA biosynthesis and signaling pathways may be targeted by P. syringae TTSS effectors to suppress the plant defense response (de Torres-Zabala et al., 2007). Our study on the cds2-1D mutant showed that activation of ABA biosynthesis weakened several plant defense systems against bacterial infection. The significantly enhanced growth of both virulent (Pst and Psm) and nonpathogenic (hrcC) P. syringae strains on cds2-1D in comparison to wild-type Col-0 indicates an ABA effect on suppression of the nonspecific basal resistance against bacterial infection, which is consistent with the observation that suppression of Pst growth by treatment with bacterial MAMP (flg22 peptide) was attenuated in the cds2-D mutant. Previous studies showed that treatment of Arabidopsis plants with flg22 peptide or the nonpathogenic hrcC strain of Pst may lead to callose-associated cell wall modification (Gómez-Gómez et al., 1999; Hauck et al., 2003), and this extracellular defense response is suppressed by wild-type pathogenic bacteria or overexpression in planta of bacterial TTSS effectors (Hauck et al., 2003; Kim et al., 2005). Recent data showed that wild-type Pst enhances callose deposition in Arabidopsis mutants impaired in ABA biosynthesis or signaling, and exogenous ABA suppresses flg22 peptide induced callose deposition in wild-type Arabidopsis seedlings (de Torres-Zabala et al., 2007, 2009; Clay et al., 2009), indicating a negative role of ABA in activation of callose deposition. However, early studies also showed that an ABA-dependent defense pathway mediates priming of callose deposition in β-amino-butyric acid (BABA)-induced resistance against necrotrophic pathogens (Ton and Mauch-Mani, 2004). Work is ongoing to understand whether callose deposition has a role in the observed disease phenotypes in the cds2-1D mutant. The enhanced growth of P. syringae strains carrying avirulence genes, including avrRpm1, indicated suppression of R-gene-dependent resistance expression in cds2-1D mutant plants. The systemic induction of PR1 gene expression and acquired resistance to challenge by a normally virulent pathogen by local infection of avirulent Pst was significantly suppressed in cds2-1D mutant plants, indicating that ABA down-regulates systemic acquired resistance. Likewise, down-regulation of RPS2/avrRpt2 determined resistance and chemically induced systemic acquired resistance were observed in ABA-treated or environmentally stressed plants (Mohr and Cahill, 2003; Yasuda et al., 2008). These findings highlight the importance of further investigation of the molecular events underlying ABA-modulated defense mechanisms.
ABA has a pivotal role in protection against water loss in plants under desiccation. In well-watered conditions, cds2-1D plants developed a phenotype of abnormal water soaking leaves in the dark, suggesting that ABA may potentially modulate physiological conditions in the leaves to facilitate bacterial growth. However, several lines of evidence argue against this hypothesis: first, the enhanced bacterial growth in cds2-1D plants differed considerably between wild-type Pst, Psg, and the mutant hrcC strains, indicating that the ABA effect on bacterial disease susceptibility is due to specific factor(s); second, the enhanced Pst growth was uncoupled from the abnormal water-soaking cds2-1D phenotype in detached leaves; third, in drought-stressed wild-type Col-0 plants, where ABA is reported to accumulate due to water deficiency, bacterial growth was increased compared to well-watered plants. Hence, ABA may influence plant resistance to bacterial infection by interacting with defense signaling networks or modulating effectors of defenses, rather than through a more general effect on physiological status. Interestingly, the enhanced growth of the TTSS− strain hrcC in cds2-1D was largely abolished when inoculated leaves were detached, thereby revealing an apparent association with the water-soaking phenotype. Previous studies showed that constitutive overexpression of an NCED gene in tomato (Solanum lycopersicum) plants leads to increased root exudation and flooding of leaf intercellular spaces with fluid containing much higher levels of ABA (Thompson et al., 2007). We speculate that a similar mechanism produced the water-soaking phenotype in cds2-1D plant and that the higher levels of apoplastic ABA led to the enhanced growth of hrcC bacteria, whereas detaching the leaf prevented the high levels of apoplastic ABA sustained by root exudation and abolished the enhanced growth of hrcC bacteria. This hypothesis is supported by the observation that feeding detached Col-0 leaves with ABA significantly enhanced growth of hrcC (Supplemental Fig. S1A) without the water-soaking symptom. Hence, in detached cds2-1D leaves, the enhanced growth differed between wild-type Pst and the hrcC mutant, implying that distinct mechanisms mediate ABA modulation of plant basal defenses against bacterial infection.
Possible Mechanisms of ABA Effect on Disease Resistance Signaling
SA and JA are well-established signal molecules mediating plant disease resistance, and our study showed a striking sequential induction of SA, ABA, and JA in Pst-inoculated Col-0 plants. Further investigation on the impact of endogenous ABA on JA and SA accumulation demonstrated a clear role of ABA in JA accumulation. Thus, compared to wild-type Col-0, JA accumulation is attenuated in ABA-deficient aba3-1 mutant plants, whereas in cds2-1D, JA accumulation was substantially enhanced in response to bacterial infection. ABA is required for wound-induced JA accumulation in potato (Solanum tuberosum) and tomato plants (Penacortes et al., 1995), but the underlying mechanism is not well understood. Consistent with our findings, the attenuated JA accumulation was also observed in Arabidopsis aba2 mutant plants in response to infection by necrotrophic pathogen Pythium irregulare (Adie et al., 2007), implying a general role of ABA in modulating biotic stress-induced JA accumulation. Our data also showed that the potentiated JA induction in cds2-1D mutant plants was pathogenesis associated, and it is not yet clear whether this synergistic JA induction contributed to enhanced bacterial growth in cds2-1D, but it may help to explain the A. brassicicola disease phenotype of aba3-1 and cds2-1D mutant plants, where plant resistance had been shown to be dependent on the JA signaling pathway (Thomma et al., 1998). Earlier reports indicated that ABA antagonizes JA/ET signaling to suppress defense against F. oxysporum in Arabidopsis (Anderson et al., 2004), whereas recent investigations showed that this ABA repression only affects a subset of JA/ET-regulated genes and that ABA predominately activates many ABA-specific and ABA/JA-related genes to promote plant defenses (Adie, et al., 2007). Nevertheless, ABA-dependent BABA-induced resistance to A. brassicicola infection does not require COI1, a key component of JA signaling, suggesting that additional signaling pathways mediate BABA-induced resistance to this pathogen (Ton and Mauch-Mani, 2004). Hence, further work is necessary to dissect the enhanced resistance to A. brassicicola in cds2-1D plants.
In contrast to its clear synergistic effect on JA induction, ABA showed complex antagonistic effects on SA induction, as both aba3-1 and cds2-1D plants accumulated less SA in response to bacterial challenge. However, PR1 expression is suppressed in SA-treated leaves or in systemic leaves of cds2-1D plants inoculated with avirulent bacteria, suggesting a damping effect on SA signaling. ABA has been shown to negatively regulate resistance and SA-dependent defense pathways in tomato-Botrytis interaction (Audenaert et al., 2002). Similarly, ABA treatment reduces the levels of conjugated SA induced by avirulent Pseudomonas and suppresses chemical-induced systemic acquired resistance in Arabidopsis plants (Mohr and Cahill, 2007; Yasuda et al., 2008), whereas in compatible interactions, bacterial effectors promote virulence by targeting ABA biosynthesis and signaling that rapidly antagonize SA-mediated defenses (de Torres-Zabala et al., 2007, 2009). Our data also showed that ABA levels in the cds2-1D mutant after bacterial challenge were rather constant and moderate in comparison with that of the wild-type Col-0 plant, implying that the timing of ABA accumulation is probably crucial to its impact on modulation of SA signaling, JA induction, and the final disease outcome.
The fact that cds2-1D with the phenotype of enhanced bacterial growth was isolated from the NahG background indicates that ABA may be involved in suppression of SA-independent mechanisms that regulate plant resistance to bacterial infection. This accords with the observation that flg22 peptide induced basal resistance, which is largely independent of SA, JA, or ET signaling pathways in suppressing growth of virulent bacteria (Zipfel et al., 2004; Tsuda et al., 2008), was also suppressed in the cds2-1D mutant. Likewise, the enhanced resistance to the soil-borne bacterium Ralstonia solanacearum in Arabidopsis mutants compromised in secondary cell wall formation is not dependent on SA, JA, or ET but closely associated with ABA biosynthesis and signaling pathways (Hernandez-Blanco et al., 2007). Recent findings that ABA acts as a proinflammatory cytokine in granulocytes (Bruzzone et al., 2007), which requires cADP-Rib as second messenger, is a reminiscent of some aspects of ABA signaling in the plant response to abiotic stresses (Wu et al., 1997; Sanchez et al., 2004). This highlights the possibility that ABA may play a conserved role in modulating immunity across the plant and animal kingdoms. Future studies on the molecular mechanisms of ABA modulation of pathogenesis may shed light on how plants integrate and fine-tune the complex responses to diverse biotic and abiotic stresses.
MATERIALS AND METHODS
Activation Tagging Screening
The activation tagging vector pJFAT260 was constructed on the backbone of a streamlined mini binary vector, pCB302 (Xiang et al., 1999), with a signature sequence of approximately 180 bp engineered between the tetramerized CaMV 35S enhancer and the T-DNA right border to facilitate subsequent identification of flanking DNA by thermal asymmetric interlaced PCR or IPCR. The Agrobacterium tumefaciens strain GV3101 (pMP90) was transformed with pJFAT260, and the resulting strain was used for floral dip transformation (Clough and Bent, 1998) of Arabidopsis (Arabidopsis thaliana) NahG plants (Gaffney et al., 1993). The activation-tagged transformants were selected by spraying T1 seedlings with 0.1% (v/v) of herbicide CHALLENGE 60 (containing 60 g/L of glufosinate ammonium). Three weeks later, the herbicide-resistant plants were hand-infiltrated with the luxCDABE-tagged Psm, and in planta bacterial growth was assayed as described (Fan et al., 2008). The T2 seeds of plants showing aberrant bacterial growth were kept for further characterization.
Plant Material and Pathogen Inoculations
All plants used for disease tests were grown at 23°C under short-day conditions (9 h of light and 15 h of dark). Bacterial strains used in this study were Pseudomonas syringae pv maculicola ES4326 (Psm), Psm-avrRpm1, P. syringae pv tomato DC3000 (Pst), P. syringae pv. tomato DC3000 HrcC− (hrcC), and P. sryingae pv glycinea (Psg). All strains were grown at 28°C on King's B plates containing appropriate antibiotics for selection. The resulting bacteria were collected and washed twice with water before inoculation of plants using a 1-mL syringe. The inoculated plants were kept in a growth room at 23°C, and bacterial growth was determined as described by Whalen et al. (1991).
The Hyaloperonospora arabidopsis Noco2 was maintained on Arabidopsis accession Col-0. Freshly harvested conidiospores were suspended in water (4 × 104 spores mL−1) and sprayed onto 3-week-old plants. The inoculated plants were kept in a tray with a sealed lid to maintain high humidity in a short-day growth chamber at 19°C. Seven days after inoculation, leaves from four plants were pooled as one replicate and weighed in 15-mL tubes. Conidiospores were rinsed with 5 mL of distilled water by vortexing, and the spore numbers were determined using a hemocytometer. At least five replicates were analyzed in each assay.
The Alternaria brassicicola strain MUCL20297 was grown on potato dextrose agar for 10 d before harvest of conidiospores by washing the plate with distilled water. Fully expanded leaves of 5-week-old Arabidopsis plant were pierced with a pipet tip, and 5 μL of spore droplets (105 spores mL−1) were applied on the wounds. The inoculated plants were kept at 20°C and 100% relative humidity for 7 d before photography.
Histochemical Staining
The H. arabidopsis inoculated leaves were stained with lactophenol trypan blue and destained with saturated chloral hydrate. The material was subsequently mounted on a slide in 60% glycerol and examined using a light microscope to monitor mycelium development (Koch and Slusarenko, 1990).
Gene Expression Studies
Total RNA was extracted from Arabidopsis leaf samples with TRIzol reagent (Invitrogen). Transcripts levels of PR1 and CDS2-1D genes were determined by RNA gel blot analysis. Two micrograms of total RNA samples were resolved on formaldehyde-agarose gel and blotted onto nylon membrane and hybridized with 32P-labeled probes prepared with the Random Primer Kit (Amersham). A template for the PR1 probe was derived from an Arabidopsis cDNA clone (provided by K. Lawton, Syngenta), and CDS2 template was amplified from Arabidopsis genomic DNA with oligos used for RT-PCR assay (see below). RNA loading was monitored by ethidium bromide staining or hybridization with 32P-labeled ubiquitin gene probe. Expression of NCED genes was analyzed by RT-PCR with the following oligos: 5′-TCTTCTTACAATGCCGATGAGT-3′ and 5′-ACTCCGACGCCGTTTTGGTTG-3′ for NCED2 (At4g18350) gene; 5′-CTCCAACGAAGATCAACAAGTCA-3′ and 5′-CACACGACCTGCTTCGCCAAA-3′ for NCED3 (At3g14440) gene; 5′-GGAAATCCACACGCAGAACTA-3′ and 5′-TTGGTTTAAGCCTGGTTTAACAT-3′ for NCED5 (At1g30100) gene; and 5′-CCTTCTGTCCCAAGATGCTCA-3′ and 5′-AGGTTATGCACGACAGGTTTC-3′ for NCED9 (At1g78390) gene. Ten micrograms of total RNA was treated with the TURBO DNA-free kit (Ambion) to remove any contaminating genomic DNA, and 1 μg of treated RNA was reverse transcribed with SuperScript II reverse transcriptase (Invitrogen) in 20-μL reaction volume. Core mixtures that contain 1 μL of the resultant cDNA product were used to amplify individual NCED genes with PCR of 30 cycles. The ubiquitin gene was amplified with 25 cycles as reference to monitor cDNA input in each sample.
Construction of NCED Overexpression Transgenic Plants
The coding regions of Arabidopsis NCED3 and NCED5 gene were amplified with primers 5′-ATGGCTTCTTTCACGGCAACGGCT-3′/5′-ACTAGCAAACCGCACCCCAAAAG-3′ and 5′-ATACTCAAAATCTCTCGAGCTTC-3′/5′-ATTATGTGTCAACGTTTACTAGTT-3′, respectively, and cloned into pGEM T-easy vector for sequencing. The clones with correct sequence were used for subsequent cloning of the NCED genes into binary vector pCHF3 (Hajdukiewicz et al., 1994) for constitutive expression and pER8 (Zuo et al., 2000) for chemical-induced expression in Arabidopsis. The resultant constructs were used for floral dip transformation of Arabidopsis, and independent transgenic lines were selected by growing the T1 seeds on Murashige and Skoog plates containing appropriate antibiotics. Homozygous T2 lines with single T-DNA insertion indicated by segregation analysis were selected for further testing.
Extraction of Free SA, JA, and ABA
Arabidopsis leaves were snap-frozen with liquid nitrogen and lyophilized. About 10 mg of dried sample was ground to powder with liquid nitrogen and extracted at 4°C overnight with 4 mL of MeOH containing 40 ng of each d4-SA (catalog number D6322; CDN Isotopes) and d4-ABA (Plant Biotechnology Institute) and 100 ng dihydro JA (Tokyo Chemical Industry) as internal standards. The total extract was centrifuged (3,000g, 10 min), and the pellet was reextracted once with 2 mL of MeOH. The combined supernatant was dried with N2 gas, and resultant residue was acidified with 2.5 mL of 5% (w/v) trichloroacetic acid and extracted twice with 5 mL of ethylacetate and cyclopentane mixture (1:1, v/v). The organic phase in each extraction was separated with aqueous phase by centrifugation (3,000g, 10 min) and combined to be dried with N2 gas. The resulting residue was taken up in 500 μL of 20% MeOH for LC-MS analysis.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. ABA enhances disease susceptibility of Arabidopsis against P. syringae and H. arabidopsis infection.
Supplementary Material
Acknowledgments
We thank Drs. D.J. Oliver, N.H. Chua, and A. Robert-Seilaniantz for providing vector pCB302, pER8, and flg22 peptide, respectively, and Dr. G. Creissen for critical reading of the manuscript.
This work was supported by the Biotechnology and Biological Sciences Research Council.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Chris Lamb (chris.lamb@bbsrc.ac.uk).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Abramovitch RB, Anderson JC, Martin GB (2006) Bacterial elicitation and evasion of plant innate immunity. Nat Rev Mol Cell Biol 7 601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adie BAT, Perez-Perez J, Perez-Perez MM, Godoy M, Sanchez-Serrano JJ, Schmelz EA, Solano R (2007) ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19 1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K (2004) Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16 3460–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselbergh B, Achuo AE, Hofte M, Van Gijsegem F (2008. b) Abscisic acid deficiency leads to rapid activation of tomato defence responses upon infection with Erwinia chrysanthemi. Mol Plant Pathol 9 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselbergh B, De Vleesschauwer D, Hofte M (2008. a) Global switches and fine-tuning-ABA modulates plant pathogen defense. Mol Plant Microbe Interact 21 709–719 [DOI] [PubMed] [Google Scholar]
- Audenaert K, De Meyer GB, Hofte MM (2002) Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol 128 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers GJM, Spoel SH (2006) Fine-tuning plant defence signalling: salicylate versus jasmonate. Plant Biol 8 1–10 [DOI] [PubMed] [Google Scholar]
- Bodenhausen N, Reymond P (2007) Signaling pathways controlling induced resistance to insect herbivores in Arabidopsis. Mol Plant Microbe Interact 20 1406–1420 [DOI] [PubMed] [Google Scholar]
- Bruzzone S, Moreschi I, Usai C, Guida L, Damonte G, Salis A, Scarfi S, Millo E, De Flora A, Zocchi E (2007) Abscisic acid is an endogenous cytokine in human granulocytes with cyclic ADP-ribose as second messenger. Proc Natl Acad Sci USA 104 5759–5764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmann A, Hoffmann T, Teplova I, Grill E, Muller A (2005) Generation of active pools of abscisic acid revealed by in vivo Imaging of water-stressed Arabidopsis. Plant Physiol 137 209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG (2001) Plant pathogens and integrated defence responses to infection. Nature 411 826–833 [DOI] [PubMed] [Google Scholar]
- de Torres-Zabala M, Bennett MH, Truman WH, Grant MR (April 24, 2009) Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. Plant J http://dx.doi.org/10.1111/j.1365-313X.2009.03875.x [DOI] [PubMed]
- de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Egea PR, Bogre L, Grant M (2007) Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J 26 1434–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong XN (2001) Genetic dissection of systemic acquired resistance. Curr Opin Plant Biol 4 309–314 [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42 185–209 [DOI] [PubMed] [Google Scholar]
- Fan J, Crooks C, Lamb C (2008) High-throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE. Plant J 53 393–399 [DOI] [PubMed] [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261 754–756 [DOI] [PubMed] [Google Scholar]
- Glazebrook J (2001) Genes controlling expression of defense responses in Arabidopsis -- 2001 status. Curr Opin Plant Biol 4 301–308 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T (2002) Flagellin perception: a paradigm for innate immunity. Trends Plant Sci 7 251–256 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Felix G, Boller T (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18 277–284 [DOI] [PubMed] [Google Scholar]
- Grant JJ, Chini A, Basu D, Loake GJ (2003) Targeted activation tagging of the Arabidopsis NBS-LRR gene, ADR1, conveys resistance to virulent pathogens. Mol Plant Microbe Interact 16 669–680 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile Ppzp family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25 989–994 [DOI] [PubMed] [Google Scholar]
- Hauck P, Thilmony R, He SY (2003) A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc Natl Acad Sci USA 100 8577–8582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Blanco C, Feng DX, Hu J, Sanchez-Vallet A, Deslandes L, Llorente F, Berrocal-Lobo M, Keller H, Barlet X, Sanchez-Rodriguez C, et al (2007) Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19 890–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27 325–333 [DOI] [PubMed] [Google Scholar]
- Jensen MK, Hagedorn PH, de Torres-Zabala M, Grant MR, Rung JH, Collinge DB, Lyngkjaer MF (2008) Transcriptional regulation by an NAC (NAM-ATAF1,2-CUC2) transcription factor attenuates ABA signalling for efficient basal defence towards Blumeria graminis f. sp hordei in Arabidopsis. Plant J 56 867–880 [DOI] [PubMed] [Google Scholar]
- Kaliff M, Staal J, Myrenas M, Dixelius C (2007) ABA is required for Leptosphaeria maculans resistance via ABI1- and ABI4-dependent signaling. Mol Plant Microbe Interact 20 335–345 [DOI] [PubMed] [Google Scholar]
- Kim MG, da Cunha L, McFall AJ, Belkhadir Y, DebRoy S, Dangl JL, Mackey D (2005) Two Pseudomonas syringae type III effectors inhibit RIM-regulated basal defense in Arabidopsis. Cell 121 749–759 [DOI] [PubMed] [Google Scholar]
- Koch E, Slusarenko A (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5 325–331 [DOI] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JA, Koornneef M (1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10 655–661 [DOI] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8 457–463 [DOI] [PubMed] [Google Scholar]
- Marin E, Nussaume L, Quesada A, Gonneau M, Sotta B, Hugueney P, Frey A, Marion-Poll A (1996) Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J 15 2331–2342 [PMC free article] [PubMed] [Google Scholar]
- Mauch-Mani B, Mauch F (2005) The role of abscisic acid in plant-pathogen interactions. Curr Opin Plant Biol 8 409–414 [DOI] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126 969–980 [DOI] [PubMed] [Google Scholar]
- Mohr P, Cahill D (2007) Suppression by ABA of salicylic acid and lignin accumulation and the expression of multiple genes, in Arabidopsis infected with Pseudomonas syringae pv. tomato. Funct Integr Genomics 7 181–191 [DOI] [PubMed] [Google Scholar]
- Mohr PG, Cahill DM (2003) Abscisic acid influences the susceptibility of Arabidopsis thaliana to Pseudomonas syringae pv. tomato and Peronospora parasitica. Funct Plant Biol 30 461–469 [DOI] [PubMed] [Google Scholar]
- Nurnberger T, Brunner F, Kemmerling B, Piater L (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol Rev 198 249–266 [DOI] [PubMed] [Google Scholar]
- Penacortes H, Fisahn J, Willmitzer L (1995) Signals involved in wound-induced proteinase-inhibitor-II gene-expression in tomato and potato plants. Proc Natl Acad Sci USA 92 4106–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez JP, Duque P, Chua NH (2004) ABA activates ADPR cyclase and cADPR induces a subset of ABA-responsive genes in Arabidopsis. Plant J 38 381–395 [DOI] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu LJ, Li QB, Cline K, McCarty DR (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35 44–56 [DOI] [PubMed] [Google Scholar]
- Thaler JS, Bostock RM (2004) Interactions between abscisic-acid-mediated responses and plant resistance to pathogens and insects. Ecology 85 48–58 [Google Scholar]
- Thomma BPHJ, Eggermont K, Penninckx IAMA, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95 15107–15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Andrews J, Mulholland BJ, McKee JMT, Hilton HW, Black CR, Taylor IB (2007) Overproduction of abscisic acid in tomato increases transpiration efficiency and root hydraulic conductivity and influences leaf expansion. Plant Physiol 143 1905–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Jackson AC, Symonds RC, Mulholland BJ, Dadswell AR, Blake PS, Burbidge A, Taylor IB (2000) Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of abscisic acid. Plant J 23 363–374 [DOI] [PubMed] [Google Scholar]
- Ton J, Mauch-Mani B (2004) Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J 38 119–130 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Glazebrook J, Cohen JD, Katagiri F (2008) Interplay between MAMP-triggered and SA-mediated defense responses. Plant J 53 763–775 [DOI] [PubMed] [Google Scholar]
- Wasilewska A, Vlad F, Sirichandra C, Redko Y, Jammes F, Valon C, Frey NFD, Leung J (2008) An update on abscisic acid signaling in plants and more…. Mol Plant 1 198–217 [DOI] [PubMed] [Google Scholar]
- Wawrzynska A, Christiansen KM, Lan Y, Rodibaugh NL, Innes RW (2008) Powdery mildew resistance conferred by loss of the ENHANCED DISEASE RESISTANCE1 protein kinase is suppressed by a missense mutation in KEEP ON GOING, a regulator of abscisic acid signaling. Plant Physiol 148 1510–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al (2000) Activation tagging in Arabidopsis. Plant Physiol 122 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen MC, Innes RW, Bent AF, Staskawicz BJ (1991) Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Kuzma J, Marechal E, Graeff R, Lee HC, Foster R, Chua NH (1997) Abscisic acid signaling through cyclic ADP-Ribose in plants. Science 278 2126–2130 [DOI] [PubMed] [Google Scholar]
- Xia YJ, Suzuki H, Borevitz J, Blount J, Guo ZJ, Patel K, Dixon RA, Lamb C (2004) An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J 23 980–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang CB, Han P, Lutziger I, Wang K, Oliver DJ (1999) A mini binary vector series for plant transformation. Plant Mol Biol 40 711–717 [DOI] [PubMed] [Google Scholar]
- Xiong LM, Ishitani M, Lee H, Zhu JK (2001) The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13 2063–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda M, Ishikawa A, Jikumaru Y, Seki M, Umezawa T, Asami T, Maruyama-Nakashita A, Kudo T, Shinozaki K, Yoshida S, et al (2008) Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell 20 1678–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li X (2005) A putative nucleoporin 96 is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1,constitutive 1. Plant Cell 17 1306–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li Q, Li Z, Staswick PE, Wang M, Zhu Y, He Z (2007) Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol 145 450–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JDG, Felix G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428 764–767 [DOI] [PubMed] [Google Scholar]
- Zuo JR, Niu QW, Chua NH (2000) An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.