Abstract
Mg2+-dependent oligomerization of nucleosomal arrays is correlated with higher order folding transitions that stabilize chromosome structure beyond the 30-nm diameter fiber. In the present studies, we have employed a novel mutagenesis-based approach to identify the macromolecular determinants that control H4 N-terminal domain (NTD) function during oligomerization. Core histones were engineered in which 1) the H2A, H2B, and H3 NTDs were swapped onto the H4 histone fold; 2) the length of the H4 NTD and the H2A NTD on the H4 histone fold, were increased; 3) the charge density of the NTDs on the H4 histone fold was increased or decreased; and 4) the H4 NTD was placed on the H2B histone fold. Model nucleosomal arrays were assembled from wild type and mutant core histone octamers, and Mg2+-dependent oligomerization was characterized. The results demonstrated that the H2B and H3 NTDs could replace the H4 NTD, as could the H2A NTD if it was duplicated to the length of the native H4 NTD. Arrays oligomerized at lower salt concentrations as the length of the NTD on the H4 histone fold was increased. Mutations that decreased the NTD charge density required more Mg2+ to oligomerize, whereas mutants that increased the charge density required less salt. Finally, the H4 NTD functioned differently when attached to the H2B histone fold than the H4 histone fold. These studies have revealed new insights into the biochemical basis for H4 NTD effects on genome architecture as well as the protein chemistry that underlies the function of the intrinsically disordered H4 NTD.
Chromatin fibers consist of linear polymers of nucleosomes, termed nucleosomal arrays, together with a specific complement of associated proteins, e.g. linker histones, chromatin architectural proteins, and transcription factors. Each nucleosome in the array contains 147 bp of DNA wrapped 1.75 times around a core histone octamer and is separated from its neighbor by 10–80 bp of linker DNA (1). The histone octamer is composed of two each of the four core histones, H2A, H2B, H3, and H4. The α-helical histone fold portions of the core histones constrain the superhelical turns of the nucleosomal DNA, largely through hydrogen bonding between the proteins and DNA (2). Unlike the structured histone fold domains, the highly basic core histone N-terminal “tail” domains (NTDs)2 are found on the outside of the nucleosome (3, 4) and are intrinsically disordered (5, 6). Chromatin fibers are hierarchically organized within interphase chromosomes. Extended 10-nm diameter beads-on-a-string arrays are locally compacted into 30-nm diameter fibers, which are subsequently coiled further to form extensively condensed, large scale “chromonema” fibers that are ∼130 nm in diameter (7, 8). Chromonema fibers are condensed even further to form 200–400-nm diameter interphase chromatids (7).
Compositionally defined and length-defined model nucleosomal arrays have been extensively used to probe chromatin condensation transitions in vitro and to relate them to the structure and stability of interphase chromosomes (9, 10). It is well established that condensation of 12-mer nucleosomal arrays and linker histone-bound chromatin fibers involves two distinct structural transitions: folding and oligomerization (10–15). Folding results from local nucleosome-nucleosome interactions in cis and ultimately leads to formation of shortened fibers that are ∼30 nm in diameter (10, 16, 17). Folding is induced by low levels of monovalent or divalent cations, which screen linker DNA charge (18) and allow close approach of neighboring nucleosomes. The oligomerization transition is cooperative (10, 19, 20) and involves both intra-array nucleosome-nucleosome interactions in cis and inter-array interactions in trans (21, 22). Oligomerization occurs at slightly higher concentrations of divalent cations than those that induce folding (10–15) and also is selectively induced by polyamines (23). Oligomerization produces supramolecular assemblies with sedimentation coefficients >100 S that dissociate into beads-on-a-string nucleosomal arrays when the divalent cations are removed from the buffer, indicating that both the oligomerization and folding transitions are freely reversible (19).
Considerable evidence indicates that divalent cation-dependent oligomerization of short nucleosomal array fragments is an in vitro manifestation of the long range fiber-fiber interactions that help stabilize chromosomal structures beyond the 30-nm fiber. Early studies examined the effects of salts on chromosomal morphology in intact isolated rat liver nuclei (24). Dense, granular chromatin (heterochromatin) was present when the nuclei were isolated in buffers containing >2 mm Mg2+. However, washing the nuclei in buffer lacking divalent cations led to rapid nuclear swelling and spectral changes indicative of chromosome decondensation. Returning the nuclei to buffer containing 2 mm MgCl2 led to reformation of the dense granules, indicating that the Mg2+-dependent chromosomal transition(s) were reversible. Similar results subsequently were obtained by other investigators (25–27). When interphase chromosomes were isolated in buffers containing polyamines and divalent cations, highly condensed 130-nm diameter chromonema fibers were observed by electron microscopy. However, when isolated in buffers lacking polyamines and divalent cations, no condensed structures were observed beyond the 30-nm fiber (7). Taken together, these early seminal studies established that divalent cations or polyamines are required to maintain extensively condensed chromosomal structures and strongly argue that there is a close relationship between model system oligomerization in vitro and higher order chromatin fiber folding beyond the 30-nm fiber.
The core histone NTDs provide major macromolecular conduits through which the structure and function of the chromatin fiber is regulated (10, 28–31). Such intrinsically disordered domains lack a native secondary structure, yet are able to participate in specific macromolecular interactions, often with multiple partners (6, 32–37). Unlike the structured histone fold domains, very little is known about the protein chemistry that dictates how the NTDs function. The core histone NTDs are essential mediators of chromatin condensation (10). Without the NTDs, nucleosomal arrays (11, 38, 39) and linker histone-bound chromatin fibers (40) do not condense beyond the 10-nm diameter beads-on-a-string structure in the presence of ionic conditions that induce folding and oligomerization of native nucleosomal arrays. The histone H4 NTD contributes the primary role in the folding transition (11). In contrast, oligomerization is mediated through the independent action of all four core histone NTDs (41).
The biochemical mechanisms that dictate NTD function during nucleosomal array oligomerization are poorly understood. Because of their high positive charge density, the NTDs have been proposed to bind to DNA and subsequently neutralize backbone charge (42, 43). Cross-linking of the H3 and H4 NTDs to DNA has been observed recently, concomitant with Mg2+-dependent oligomerization (21, 22, 44). However, other evidence suggests that a DNA charge neutralization-based mechanism is too simple. For example, the H3 and H4 NTDs alone can mediate oligomerization, but the H2A and H2B NTDs cannot (41), even though all four NTDs have essentially the same charge density. Acetylation of Lys-16 on the H4 NTD and neutralization of a single positive charge have the same effect on oligomerization as removal of the entire H4 NTD with nine positive charges (45). Consequently, a direct study of the determinants of NTD function during oligomerization was warranted. When selectively removed from nucleosomal arrays, the H4 NTD has the largest effect on oligomerization (11, 41). Arrays lacking the H4 NTD still oligomerize (due to the action of the other NTDs) but require higher amounts of Mg2+ than the wild type to achieve half-maximal oligomerization. In the present work, we have performed a novel mutagenesis-based analysis of H4 NTD function. Four possible determinants were examined: primary sequence, NTD length, charge density, and the histone fold attachment site. Novel H4 proteins were constructed in which the NTDs of the other three core histones were swapped into the H4 histone fold; the H4 and H2A NTD were tandemly repeated to increase the length of the NTD; the positive charge density was altered via site-directed mutagenesis; and the H4 NTD was attached to the H2B histone fold. Our results indicate that although the primary sequence appears inconsequential, the H4 NTD length, charge density, and histone fold attachment site all are all important determinants of the function of the H4 NTD during Mg2+-dependent oligomerization. Amino acid composition also appears to contribute to NTD function. These results significantly advance our understanding of both the biochemical mechanism of stabilization of chromosome structure beyond the 30-nm fiber and the unique protein chemistry that enables H4 NTD function during chromatin condensation.
EXPERIMENTAL PROCEDURES
Cloning of Mutant Core Histones
Polymerase chain reaction (PCR) was performed using the cDNA encoding the Xenopus laevis core histones (cloned into pET3A (Novagen)) as templates (46–48). PCR primers were designed to amplify fragments from these templates and included specific restriction sites. For tail swap mutants, amplified NTD fragments were flanked by Nde-1 at the 5′ end and one of the following sites at the 3′ end: Nae-1, Nhe-1, and Stu-1. Amplified histone fold fragments were flanked by 5′ restriction sites complementary to those at the 3′ end of the prospective NTD fragment and with BamH1 sites at the 3′ ends. The restriction enzymes used at the union of the NTD and histone-fold fragments were chosen such that the composition of the amino acids at the union was maintained closely to the parent histone amino acids. For example, the Nae-1 site (5′-GCCGGC-3′) codes for alanine and glycine; both are among the amino acids commonly found in the H2A and H4 NTDs (see Table 1). Tandemly repeated (doubled and tripled) H2A NTDs cloned onto the H4 histone-fold domain were generated in steps by repeatedly cloning an additional H2A NTD upstream of the T2AHF4 clone3 using PCR methods similar to those described above. The sequences of the wild type (WT) and various mutant NTDs are shown in Fig. 1.
TABLE 1.
Core histone properties
| Lengtha | Charge densityb | Amino acid composition |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lys | Arg | Gly | Ala | Thr | Pro | Glu | Asp | Asn | Gln | Ser | Cys | Met | His | Val | Ile | Leu | Phe | Trp | Tyr | |||
| % of total composition | ||||||||||||||||||||||
| H2A | 13 | 0.46 | 23 | 15 | 31 | 8 | 8 | 0 | 0 | 0 | 0 | 8 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H4 | 27 | 0.33 | 18 | 15 | 30 | 4 | 0 | 0 | 0 | 4 | 4 | 4 | 4 | 0 | 0 | 4 | 4 | 4 | 7 | 0 | 0 | 0 |
| H2B | 25 | 0.36 | 33 | 4 | 8 | 16 | 8 | 8 | 0 | 4 | 0 | 8 | 8 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| H3 | 37 | 0.35 | 22 | 11 | 11 | 22 | 14 | 5 | 0 | 0 | 0 | 5 | 5 | 0 | 0 | 0 | 3 | 0 | 3 | 0 | 0 | 0 |
a Number of amino acid residues, not including the N-terminal methionine.
b Charge density is calculated as the total net NTD charge divided by the length. The N-terminal backbone amino group is included in the calculation.
FIGURE 1.
Amino acid sequences of mutant NTDs. Initiating methionines were removed by bacterial processing. The dots denote links between tandem-repeat NTDs. Amino acids in boldface differ from the native amino acids as a result of the cloning process (see “Experimental Procedures”). The amino acid substitutions made in the charge density mutants are underlined.
The resulting PCR fragments were purified using the QIAquick PCR purification kit (Qiagen), digested with the appropriate restriction enzymes, gel-purified on 1.5% agarose, and extracted using the QIAquick gel extraction kit (Qiagen). The fragments were ligated into appropriately digested pET3a (or previously cloned histone chimeras; see above) and transformed into Escherichia coli DH5α. Clones were screened by the appearance of a band of the appropriate length following digestion with Nde-1 and BamH-1 and agarose electrophoresis. The fidelity of the clones was confirmed by sequencing (Macromolecular Resource Facility, Colorado State University).
Histone Octamer Assembly and Purification
BL21(DE3)pLysS cells were transformed with the pET plasmids, and the core histones were expressed and purified as described previously (41, 47, 48). Lyophilized core histones were mixed at equimolar ratio following dissolution in 6 m guanidine-HCl (13, 41, 48). Following dialysis, the sample was subjected to gel filtration chromatography using a Superdex S-200 prep grade 16/60 column (GE Healthcare) as described (41). The peak corresponding to the intact histone octamer was pooled and checked for purity and composition by SDS-PAGE and Coomassie staining (41). Octamers were stored at 4 °C until use.
Reconstitution of Nucleosomal Arrays
208-12 DNA was prepared as described (41, 49). Nucleosomal arrays were reconstituted from 208-12 DNA and purified the mutant wild type and mutant core histone octamers using salt dialysis as described (41, 50).
Sedimentation Velocity
Sedimentation velocity experiments were performed as described (13, 41, 51). Data were analyzed using the method of Demeler and van Holde (52) using Ultrascan software. The extent of nucleosome saturation of the 208-12 DNA template was determined from the sedimentation coefficient at boundary fraction = 0.5 as described (41, 50).
Oligomerization Assay
The differential centrifugation oligomerization assay was performed as a function of MgCl2 concentration as described (13, 19, 41). Each data point reflects the mean of 3–12 independent assays utilizing at least two independent preparations of nucleosomal arrays. The Mg50, defined as the MgCl2 concentration at which 50% of the nucleosomal array was in the supernatant, was determined from the midpoint of the oligomerization curves.
RESULTS
Many studies have been directed toward defining the roles of the core histone NTDs in chromatin condensation (10, 11, 15, 38, 39, 42, 43, 45, 53–58). Although the involvement of NTDs is well established, the molecular basis of how they function is much less understood. Consequently, in the present work, we have investigated the macromolecular determinants through which the H4 NTD mediates the nucleosome-nucleosome interactions involved in salt-dependent nucleosomal array oligomerization. To accomplish this, we engineered recombinant X. laevis core histone H4 proteins with novel NTDs (Fig. 1). These mutant histones were then expressed and purified from E. coli and assembled onto 12-mer 5 S ribosomal DNA templates to yield the model nucleosomal arrays used in the oligomerization studies. Importantly, because each core histone NTD functions independently during oligomerization (41), our experiments only assay the effects of the mutations on the action of the H4 NTD.
Assembly of Equally Saturated Chimeric Nucleosomal Arrays
The number of octamers assembled on a 12-mer 5 S ribosomal DNA template will influence the MgCl2-dependent oligomerization curve and the Mg50. Specifically, the magnitude of the Mg50 increases as the number of nucleosomes on the template decreases (19). Thus, as controls to test for nucleosome saturation level after reconstitution, all nucleosomal arrays used in these studies were first analyzed in low salt by sedimentation velocity (41, 50). Data were analyzed using the modified method of Demeler and van Holde (52) to yield the integral distribution of sedimentation coefficients, G(s). Average sedimentation coefficients were obtained from boundary fraction = 50% (save). The wild type arrays had an save of 24.6 S (supplemental Fig. 1), indicating that the sample contained an average of 10 nucleosomes/template (50). The nucleosomal arrays lacking the H4 NTD sedimented at 24.1 S at the boundary midpoint (supplemental Fig. 1), consistent with previous studies showing that the loss of a single NTD leads to an ∼1 S decrease in the array sedimentation coefficient (41). All of the mutant nucleosomal arrays with swapped and lengthened NTDs had an save of 23–26 S (supplemental Fig. 1). The sedimentation data provide an important control and indicate that wild type and mutant arrays utilized in these studies all contained an average of 9–10 nucleosomes/208-12 DNA template. Consequently, any significant differences observed in the oligomerization properties will be due to the mutated histones and not to differences in template saturation.
The H2A, H2B, and H3 NTDs Can Replace the H4 NTD When Fused to the H4 Histone Fold Domain
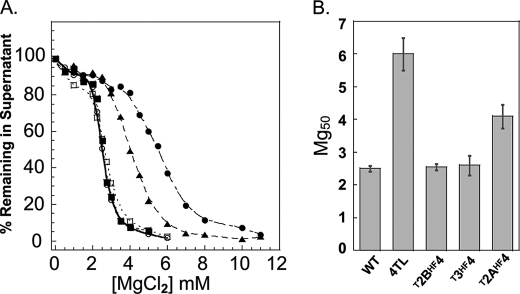
Nucleosomal arrays were assayed for MgCl2-dependent oligomerization using a well established differential centrifugation assay (13, 19, 41, 59). This assay yields a plot of the fraction of arrays remaining in the supernatant versus the concentration of MgCl2. Fig. 2A shows such plots for control nucleosomal arrays assembled with WT H4 tail-less (TL) octamers and for mutant arrays in which the H4 NTD has been replaced by the NTDs of the other three core histones3 (T2AHF4, T2BHF4, and T3HF4). The WT nucleosomal arrays showed cooperative oligomerization behavior, as observed previously (13, 19, 41, 59). Under these conditions, the Mg50 was 2.5 mm (Fig. 2B). Consistent with previous results, the Mg50 of the H4 TL nucleosomal arrays was much higher (6.5 mm). Unexpectedly, when either the H3 or H2B NTDs were attached to the H4 histone fold, the Mg50 of the resulting T2BHF4 and T3HF4 chimeric arrays (2.6 mm) was essentially identical to that of the WT array (Fig. 2B). The oligomerization of the T2AHF4 array remained cooperative but yielded an Mg50 (4 mm) intermediate between the WT and H4 TL arrays (Fig. 2B). Thus, the H3 and H2B NTDs provided all of the requirements needed to replace the native H4 NTD during oligomerization, whereas the H2A NTD partially satisfied these requirements.
FIGURE 2.
Oligomerization of nucleosomal arrays with H2A, H2B, or H3 NTDs on the H4 histone fold domain. A, oligomerization as a function of MgCl2 concentration as determined by differential centrifugation (see “Experimental Procedures”) for nucleosomal arrays in which the NTD on histone H4 was replaced by the NTD of H2A (▲), H2B (■), and H3 (○). The oligomerization plots for the WT (□) and H4 TL (•) are also shown. B, Mg50 values derived from the data in A. Error bars represent the S.D. of 3–12 independent oligomerization assays.
Changes in H4 NTD Length Influence Mg50
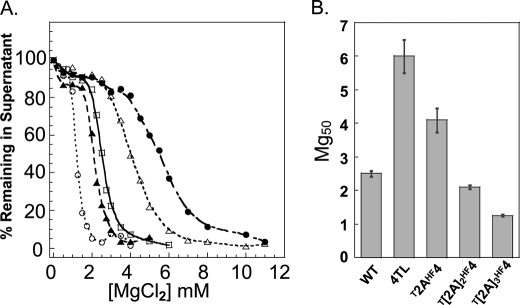
We were curious why the H2B and H3 NTDs could fully substitute for the H4 NTD, but the effect of the H2A NTD was partial. We noted that the H2A NTD is by far the shortest of the four core histone NTDs, being only 13 amino acids in length.4 In contrast, the H2B, H3, and H4 NTDs contain 25, 37, and 27 amino acids, respectively. Thus, we next sought to determine whether the length of the H2A NTD on the H4 histone fold domain influenced nucleosomal array oligomerization. To do so, we created mutants in which the H2A NTD sequence was repeated two and three times and cloned on the histone fold of H4, such that the resulting NTDs were 26 (T[2A]2HF4) and 39 (T[2A]3HF4) amino acids in length. Importantly, the amino acid composition and charge density of the H2A NTD are similar to that of the H4 NTD (Table 1); thus, NTD length was the primary variable in this experiment. Fig. 3A shows the oligomerization profiles of the T[2A]2HF4 and T[2A]3HF4 arrays, along with the T2AHF4, H4 TL, and WT control arrays. The Mg50 values derived from these curves are shown in Fig. 3B. A clear trend was observed in this experiment; as the length of the H2A NTD fused to the H4 was increased, the Mg50 decreased. Interestingly, the T[2A]2HF4 construct, which has an “H4 tail” that is nearly the same length and has a very similar amino acid composition and charge density (Table 1) as the native H4 NTD, behaved similarly (Mg50 = 2.1 mm) to the WT array. Consistent with this trend, the longer T[2A]3HF4 array had an even lower Mg50 (1.25 mm).
FIGURE 3.
Oligomerization of nucleosomal arrays with lengthened H2A NTDs on the H4 histone fold domain. A, oligomerization as a function of MgCl2 concentration for nucleosomal arrays in which the NTD on histone H4 was replaced by a double H2A NTD (▲, T[4]2HF4) and a triple H2A NTD (○, T[4]3HF4). The oligomerization plots for the WT (□) and H4 TL (•) and T2AHF4 (△) arrays were taken from Fig. 2 and shown as controls. B, Mg50 values derived from the data in A. Error bars represent the S.D. of 3–12 independent oligomerization assays.
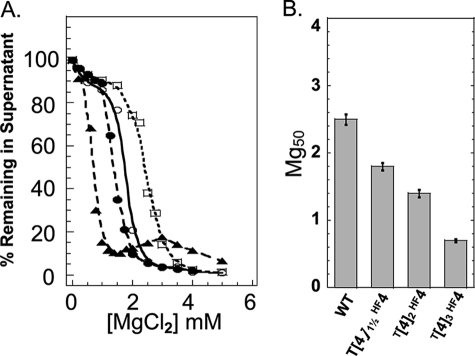
To further investigate the effect of domain length on H4 NTD function, we created tandemly repeated H4 NTD mutants in which the NTDs were 42 (T[4]1½HF4), 54 (T[4]2HF4), and 81 (T[4]3HF4) amino acids in length. The T[4]1½HF4 mutant was constructed so that native H4 NTD residues 1–28 amino acids abut the nucleosome, whereas the added residues (i.e. 15–28) are distal to the first H4 NTD. The results of oligomerization assays with these and WT arrays are shown in Fig. 4, A and B. As we observed with the H2A-based NTDs, the Mg50 decreased continuously as the H4 NTD length increased. The extra 14 amino acids present in T[4]1½HF4 arrays decreased the Mg50 from 2.5 mm (WT) to 1.75 mm, whereas addition of 12 more amino acids (T[4]2HF4) decreased the Mg50 to 1.4 mm MgCl2. The 81-amino acid T[4]3HF4 array had the lowest Mg50 measured in our studies (0.75 mm).
FIGURE 4.
Oligomerization of nucleosomal arrays with lengthened H4 NTDs on the H4 histone fold domain. A, oligomerization as a function of MgCl2 concentration for nucleosomal arrays in which the NTD on histone H4 was extended with the first 14 amino acids of the H4 NTD (○, T[4]½HF4) and tandemly repeated twice (•, T[4]2 HF4) or tandemly repeated three times (▲, T[4]3 HF4). The oligomerization plot for the WT (□) array was taken from Fig. 2 and is shown as a control. B, Mg50 values derived from the data in A. Error bars represent the S.D. of 3–12 independent oligomerization assays.
Changes in Charge Density of H4 NTD Influence Mg50
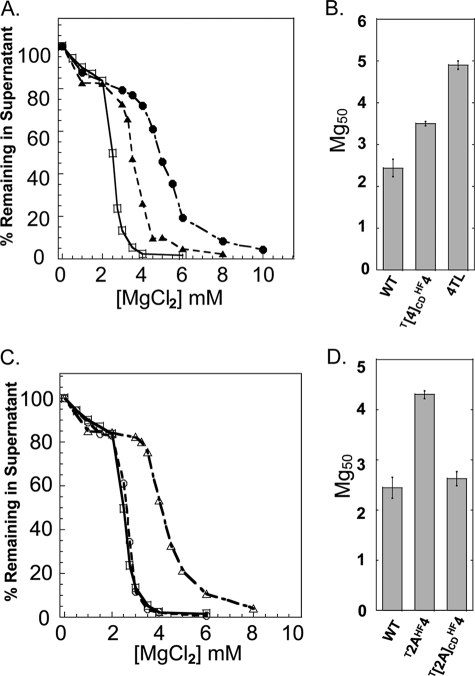
The positive charge density of the four core histone NTDs is high, ranging from 3.2–4.6 net positive charges/10 residues (Table 1). Thus, although the absolute charge of the increased length NTDs changed, all of the mutant nucleosomal arrays studied thus far had H4 NTDs with a similar charge density. To test directly how charge density influences H4 NTD function, we constructed two mutant H4 proteins. T[4]CDHF4 had a native length H4 NTD with only 1.9 positive charges/10 residues, whereas T[2A]CDHF4 had a native length H2A NTD with 6.9 positive charges/10 residues. The mutations were made in a way that kept the amino acid composition of the NTD as close as possible to the wild type (Table 1 and Fig. 1). Fig. 5 shows the results of the oligomerization assays. The T[4]CDHF4 mutant arrays with a reduced H4 NTD charge density displayed an intermediate Mg50 compared with WT and H4 tail-less control arrays (Fig. 5, A and B). The T[2A]CDHF4 mutant with higher charge density had an Mg50 that was significantly lower than the T[2A]HF4 control (Fig. 5, C and D). Together with the data in Figs. 3 and 4, these experiments indicate that both charge density and NTD length are major determinants of H4 NTD function.
FIGURE 5.
Oligomerization of nucleosomal arrays with altered NTD charge density. A, oligomerization as a function of MgCl2 concentration for nucleosomal arrays in which the charge density of the native H4 NTD was reduced (▲, T[4]CDHF4). The oligomerization plots for the WT (□) and H4 TL (•) arrays were taken from Fig. 2 and shown as controls. B, Mg50 values derived from the data in A. Error bars represent the S.D. of 3–12 independent oligomerization assays. C, oligomerization as a function of MgCl2 concentration for nucleosomal arrays in which the charge density of the H2A NTD on the H4 histone fold was increased (○, T[2A]CDHF4). The oligomerization plots for the WT (□) and T2AHF4 (△)arrays were taken from Fig. 2 and shown as controls. D, Mg50 values derived from the data in C. Error bars represent the S.D. of 3–12 independent oligomerization assays.
H4 NTD Placed on H2B Histone Fold Does Not Lower Mg50 Relative to WT
Removal of the H4 NTDs from nucleosomal arrays leads to a much larger increase in Mg50 than removal of the H2B NTDs (41). We were therefore curious whether placement of the H4 NTD on the H2B histone fold would lead to arrays that oligomerized at a lower Mg50 than the WT. To test this question, we constructed a recombinant H2B histone in which the 24-residue NTD of H2B was replaced with the 27-residue H4 NTD (T4HFB). The results of oligomerization assays performed with these arrays are shown in Fig. 6 and demonstrated that the T4HF2B arrays behaved identically (Mg50 = 2.6 mm) to WT arrays. This indicated that the H4 NTD substituted equally well for the H2B NTD when fused to the H2B histone fold domain but did not decrease the Mg50 below that of the WT. Thus, the effect of the H4 NTD on the Mg50 was influenced by the histone fold to which it was attached.
FIGURE 6.
Oligomerization of nucleosomal arrays with H4 NTDs on the H2B histone fold domain. A, oligomerization as a function of MgCl2 concentration for nucleosomal arrays in which the NTD on histone H2B was replaced by the NTD of H4 (▲, T4HFB). The oligomerization plot for the WT (□) array was taken from Fig. 2 and shown as a control. B, Mg50 values derived from the data in A. Error bars represent the S.D. of 3–12 independent oligomerization assays.
DISCUSSION
This work has yielded new observations that are relevant to both the biochemical basis of chromosome structure and stability and the unusual protein chemistry that underlies H4 NTD function. The histone NTDs are among the most conserved amino acid sequences found in eukaryotic proteins. The H4 NTD, in particular, is >95% identical across all known species (60). Nucleosomal array oligomerization is mediated by NTD-dependent nucleosome-nucleosome interactions within the same fiber in cis and between different fibers in trans (21, 22, 44). Thus, our working assumption was that the H4 NTD had evolved to be of optimal length needed to maximize NTD interactions with their chromatin targets, and we speculated that lengthening the NTD might interfere with WT oligomerization through a steric clash. However, the Mg50 decreased as the length of the NTD fused to the H4 histone fold was increased (Figs. 3 and 4). This indicates that at any given salt concentration, the arrays with longer NTDs fused to the H4 histone fold were more extensively oligomerized than WT arrays. The same relationship was observed with the NTD charge density; as the positive charge density increased, the Mg50 decreased (Fig. 5). From a biological perspective, the length and charge density dependences identified here have profound implications for understanding genome structure and stability. The correlations between in vitro nucleosomal array oligomerization and the stability of chromosome fiber structures beyond the 30-nm fiber are extensive and well documented (9, 10, 19, 24–27) (see Introduction). By lowering the Mg50, the effect of increasing either the H4 NTD length or charge density at physiological salt concentrations is to shift the equilibrium toward stabilizing more extensively condensed chromosomal fibers (Figs. 3–5). Thus, the H4 NTD has presumably evolved its native length and charge density to maintain a fine tuned balance between the chromosomal fiber condensation and the decondensation that is needed for proper biological functioning of the genome. If the NTD was longer and/or had a higher charge density, condensed chromosomal fibers would be too stable, and genomic DNA would be less accessible. In contrast, if the H4 NTD length was shorter, or if its charge density was lower, chromosomal fibers would be too unstable and incapable of properly condensing the genome. Given that the charge density of the core histone NTDs is nearly a constant physical property, it will be important to determine whether the relationships established for the H4 NTD are general and whether they also apply to the other core histone NTDs.
Because of its high degree of sequence conservation (60), we presumed that the specific amino acid sequence of the H4 NTD would be crucial for function and expected that the domain swap mutants would behave as H4 TL arrays. Instead, we found that the H2B and H3 NTDs fused to the H4 histone fold domain functioned nearly identically as the H4 NTD (Fig. 2), as did the H2A NTD, if it was duplicated to be nearly the same length as the others (Fig. 3). These results demonstrate that the primary sequence does not determine H4 NTD function during oligomerization. Instead, our explanation for the observed NTD interchangeability centers on the unique amino acid composition of the core histone NTDs. Comparison of the H2A and H4 NTDs provides the most direct test of this hypothesis, as their amino acid composition is specific and nearly the same (Table 1). When fused to the H4 histone fold domain, the double length H2A and H4 NTDs yielded almost identical oligomerization curves and Mg50 values (Figs. 2 and 3). Furthermore, increasing the length of the H2A NTD on the H4 histone fold domain lowered the Mg50 similar to lengthening the native H4 NTD (Figs. 3 and 4). Thus, amino acid composition and function are well correlated for the H2A and H4 NTDs. Except for a switch in the occurrence of Ala/Thr for Gly, the composition of the H2B and H3 NTDs also is very similar to that of H4 and H2A. Each of the NTDs is also enriched in positively charged residues and highly deficient in nonpolar amino acids (Table 1), properties that are characteristic of intrinsically disordered protein regions (6, 33, 61). The specific amino acid composition of the core histone NTDs not only makes them disordered but also flexible. Of note, amino acid composition has recently been shown to be important for the function of other intrinsically disordered protein regions including yeast prion domains (62) and the linker histone CTD (63). Why, then, are the core histone NTD primary sequences so highly conserved throughout evolution? We hypothesize that this relates to the need to preserve specific functional post-translational modification patterns (28–31, 64–67), which presumably evolved after the functions of the unmodified NTDs and their role in genome architecture.
All four core histone NTDs contribute independent functions during oligomerization. However, deletion of the individual NTDs leads to a differential increase in the Mg50, with loss of H4 > H3 > H2A ≈ H2B (41). One interpretation of this result is that the H4 NTD is the most “potent” NTD. However, when fused to the H4 histone fold domain, the H3 and H2B and double length H2A NTDs were as equally effective as the H4 NTD at inducing oligomerization (Fig. 2). Similarly, the H4 NTD fused to the H2B histone fold domain yielded arrays with the same Mg50 as the WT (Fig. 6) and not the lower Mg50 that might be predicted from the deletion result. Thus, the NTD deletion data (41), together with the results shown in Figs. 2 and 6, collectively indicate that the effect of the core NTDs on the Mg50 also is influenced by the histone fold domain to which they are attached. The histone fold attachment site will influence the spatial orientations of the NTD within both a monomeric and oligomeric nucleosomal array, suggesting in turn that each histone fold directs its NTD toward one or more chromatin binding sites.
What mechanisms govern H4 NTD action during oligomerization? Given the importance of the high NTD positive charge density, an obvious possibility is DNA charge neutralization. The N-terminal end of the H4 NTD has been cross-linked to DNA during the oligomerization transition (44). Also, the ability of all of the NTDs to substitute for each other when fused to the H4 histone fold is consistent with a role for nonspecific DNA binding. However, we emphasize that other observations suggest more complex mechanisms of NTD function. For instance, specific acetylation of Lys-16 is sufficient to disrupt H4 NTD-mediated oligomerization to the same extent as removing the entire NTD (17, 45, 68, 69), even though only 1 of 10 NTD positive charges are neutralized. The H3 and H4 NTDs can singly mediate oligomerization, whereas the H2A and H2B NTDs cannot (41), even though all four NTDs have a similar charge density. If all the NTDs are removed, very high MgCl2 concentrations (e.g. 40 mm) will not induce oligomerization. Thus, determinants other than DNA charge neutralization must contribute to the mechanism of H4 NTD function. In this regard, H4 NTD-dependent protein-protein interactions participate in local folding of nucleosomal arrays (11), and it is possible analogous interactions occur during oligomerization. In terms of NTD length, the increased potency of the longer NTDs could be due to an increased number of binding contacts with chromatin sites or might reflect an enhanced ability to find a single site via a “fly-casting” mechanism (6, 70). Ultimately, the involvement of the H4 NTD in promoting oligomerization seems to have both electrostatic and nonelectrostatic components and extends beyond simple mechanisms such as DNA charge neutralization.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM45916 (to J. C. H.).
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
3 The nomenclature used for the mutant chimeric histones and nucleosomal arrays is as follows. The specific N-terminal tail domain will be preceded by the superscript “T,” and the “histone fold” portion will be preceded by the superscript “HF.” Thus, the NTD of H2A cloned on the histone fold of H4 is written as T2AHF4, whereas two H2A NTDs fused to the H4 histone fold are written as T[2A]2HF4.
4 The lengths of the NTDs expressed herein reflect the loss of the initiating methionine following translation, a result confirmed by mass spectrometry of each of the purified recombinant histones (data not shown).
- NTD
- N-terminal domain
- WT
- wild type
- TL
- tail-less
- save
- average sedimentation coefficient
- CD
- charge density.
REFERENCES
- 1.Felsenfeld G., Groudine M. ( 2003) Nature 421, 448– 453 [DOI] [PubMed] [Google Scholar]
- 2.Luger K., Richmond T. J. ( 1998) Curr. Opin. Struct. Biol. 8, 33– 40 [DOI] [PubMed] [Google Scholar]
- 3.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. ( 1997) Nature 389, 251– 260 [DOI] [PubMed] [Google Scholar]
- 4.Luger K., Richmond T. J. ( 1998) Curr. Opin. Genet. Dev. 8, 140– 146 [DOI] [PubMed] [Google Scholar]
- 5.Hansen J. C., Tse C., Wolffe A. P. ( 1998) Biochemistry 37, 17637– 17641 [DOI] [PubMed] [Google Scholar]
- 6.Hansen J. C., Lu X., Ross E. D., Woody R. W. ( 2006) J. Biol. Chem. 281, 1853– 1856 [DOI] [PubMed] [Google Scholar]
- 7.Belmont A. S., Braunfeld M. B., Sedat J. W., Agard D. A. ( 1989) Chromosoma 98, 129– 143 [DOI] [PubMed] [Google Scholar]
- 8.Belmont A. S., Bruce K. ( 1994) J. Cell Biol. 127, 287– 302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fletcher T. M., Hansen J. C. ( 1996) Crit. Rev. Eukaryot. Gene Expr. 6, 149– 188 [DOI] [PubMed] [Google Scholar]
- 10.Hansen J. C. ( 2002) Annu. Rev. Biophys. Biomol. Struct. 31, 361– 392 [DOI] [PubMed] [Google Scholar]
- 11.Dorigo B., Schalch T., Bystricky K., Richmond T. J. ( 2003) J. Mol. Biol. 327, 85– 96 [DOI] [PubMed] [Google Scholar]
- 12.Fan J. Y., Gordon F., Luger K., Hansen J. C., Tremethick D. J. ( 2002) Nat. Struct. Biol. 9, 172– 176 [DOI] [PubMed] [Google Scholar]
- 13.Chodaparambil J. V., Barbera A. J., Lu X., Kaye K. M., Hansen J. C., Luger K. ( 2007) Nat. Struct. Mol. Biol. 14, 1105– 1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou J., Fan J. Y., Rangasamy D., Tremethick D. J. ( 2007) Nat. Struct. Mol. Biol. 14, 1070– 1076 [DOI] [PubMed] [Google Scholar]
- 15.Lu X., Simon M. D., Chodaparambil J. V., Hansen J. C., Shokat K. M., Luger K. ( 2008) Nat. Struct. Mol. Biol. 15, 1122– 1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Widom J. ( 1998) Annu. Rev. Biophys. Biomol. Struct. 27, 285– 327 [DOI] [PubMed] [Google Scholar]
- 17.Tremethick D. J. ( 2007) Cell 128, 651– 654 [DOI] [PubMed] [Google Scholar]
- 18.Clark D. J., Kimura T. ( 1990) J. Mol. Biol. 211, 883– 896 [DOI] [PubMed] [Google Scholar]
- 19.Schwarz P. M., Felthauser A., Fletcher T. M., Hansen J. C. ( 1996) Biochemistry 35, 4009– 4015 [DOI] [PubMed] [Google Scholar]
- 20.Lu X., Klonoski J. M., Resch M. G., Hansen J. C. ( 2006) Biochem. Cell Biol. 84, 411– 417 [DOI] [PubMed] [Google Scholar]
- 21.Kan P. Y., Lu X., Hansen J. C., Hayes J. J. ( 2007) Mol. Cell. Biol. 27, 2084– 2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X., Hayes J. J. ( 2008) Mol. Cell. Biol. 28, 227– 236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pollard K. J., Samuels M. L., Crowley K. A., Hansen J. C., Peterson C. L. ( 1999) EMBO J. 18, 5622– 5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olins D. E., Olins A. L. ( 1972) J. Cell Biol. 53, 715– 736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aaronson R. P., Woo E. ( 1981) J. Cell Biol. 90, 181– 186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weith A. ( 1983) Exp. Cell Res. 146, 199– 203 [DOI] [PubMed] [Google Scholar]
- 27.Dixon D. K., Burkholder G. D. ( 1985) Eur. J. Cell Biol. 36, 315– 322 [PubMed] [Google Scholar]
- 28.Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., Patel D. J. ( 2007) Nat. Struct. Mol. Biol. 14, 1025– 1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner B. M. ( 2007) Nat. Cell Biol. 9, 2– 6 [DOI] [PubMed] [Google Scholar]
- 30.Shilatifard A. ( 2008) Curr. Opin. Cell Biol. 20, 341– 348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bártová E., Krejcí J., Harnicarová A., Galiová G., Kozubek S. ( 2008) J. Histochem. Cytochem. 56, 711– 721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunker A. K., Brown C. J., Lawson J. D., Iakoucheva L. M., Obradoviæ Z. ( 2002) Biochemistry 41, 6573– 6582 [DOI] [PubMed] [Google Scholar]
- 33.Tompa P. ( 2002) Trends Biochem. Sci. 27, 527– 533 [DOI] [PubMed] [Google Scholar]
- 34.Dyson H. J., Wright P. E. ( 2005) Nat. Rev. Mol. Cell Biol. 6, 197– 208 [DOI] [PubMed] [Google Scholar]
- 35.Mészáros B., Tompa P., Simon I., Dosztányi Z. ( 2007) J. Mol. Biol. 372, 549– 561 [DOI] [PubMed] [Google Scholar]
- 36.Tompa P., Fuxreiter M. ( 2008) Trends Biochem. Sci. 33, 2– 8 [DOI] [PubMed] [Google Scholar]
- 37.Fuxreiter M., Tompa P., Simon I., Uversky V. N., Hansen J. C., Asturias F. J. ( 2008) Nat. Chem. Biol. 4, 728– 737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia-Ramirez M., Dong F., Ausio J. ( 1992) J. Biol. Chem. 267, 19587– 19595 [PubMed] [Google Scholar]
- 39.Tse C., Hansen J. C. ( 1997) Biochemistry 36, 11381– 11388 [DOI] [PubMed] [Google Scholar]
- 40.Carruthers L. M., Hansen J. C. ( 2000) J. Biol. Chem. 275, 37285– 37290 [DOI] [PubMed] [Google Scholar]
- 41.Gordon F., Luger K., Hansen J. C. ( 2005) J. Biol. Chem. 280, 33701– 33706 [DOI] [PubMed] [Google Scholar]
- 42.Arya G., Zhang Q., Schlick T. ( 2006) Biophys. J. 91, 133– 150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma S., Ding F., Dokholyan N. V. ( 2007) Biophys. J. 92, 1457– 1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kan P. Y., Caterino T. L., Hayes J. J. ( 2009) Mol. Cell. Biol. 29, 538– 546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shogren-Knaak M., Ishii H., Sun J. M., Pazin M. J., Davie J. R., Peterson C. L. ( 2006) Science 311, 844– 847 [DOI] [PubMed] [Google Scholar]
- 46.Luger K., Rechsteiner T. J., Flaus A. J., Waye M. M., Richmond T. J. ( 1997) J. Mol. Biol. 272, 301– 311 [DOI] [PubMed] [Google Scholar]
- 47.Luger K., Rechsteiner T. J., Richmond T. J. ( 1999) Methods Mol. Biol. 119, 1– 16 [DOI] [PubMed] [Google Scholar]
- 48.Luger K., Rechsteiner T. J., Richmond T. J. ( 1999) Methods Enzymol. 304, 3– 19 [DOI] [PubMed] [Google Scholar]
- 49.Hansen J. C., Ausio J., Stanik V. H., van Holde K. E. ( 1989) Biochemistry 28, 9129– 9136 [DOI] [PubMed] [Google Scholar]
- 50.Hansen J. C., Lohr D. ( 1993) J. Biol. Chem. 268, 5840– 5848 [PubMed] [Google Scholar]
- 51.Carruthers L. M., Schirf V. R., Demeler B., Hansen J. C. ( 2000) Methods Enzymol. 321, 66– 80 [DOI] [PubMed] [Google Scholar]
- 52.Demeler B., van Holde K. E. ( 2004) Anal. Biochem. 335, 279– 288 [DOI] [PubMed] [Google Scholar]
- 53.Garcia-Ramirez M., Rocchini C., Ausio J. ( 1995) J. Biol. Chem. 270, 17923– 17928 [DOI] [PubMed] [Google Scholar]
- 54.Fletcher T. M., Hansen J. C. ( 1995) J. Biol. Chem. 270, 25359– 25362 [DOI] [PubMed] [Google Scholar]
- 55.Moore S. C., Ausió J. ( 1997) Biochem. Biophys. Res. Commun. 230, 136– 139 [DOI] [PubMed] [Google Scholar]
- 56.Tse C., Sera T., Wolffe A. P., Hansen J. C. ( 1998) Mol. Cell. Biol. 18, 4629– 4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng C., Hayes J. J. ( 2003) J. Biol. Chem. 278, 24217– 24224 [DOI] [PubMed] [Google Scholar]
- 58.Robinson P. J., An W., Routh A., Martino F., Chapman L., Roeder R. G., Rhodes D. ( 2008) J. Mol. Biol. 381, 816– 825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwarz P. M., Hansen J. C. ( 1994) J. Biol. Chem. 269, 16284– 16289 [PubMed] [Google Scholar]
- 60.Baxevanis A. D., Landsman D. ( 1996) Nucleic Acids Res. 24, 245– 247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dunker A. K., Lawson J. D., Brown C. J., Williams R. M., Romero P., Oh J. S., Oldfield C. J., Campen A. M., Ratliff C. M., Hipps K. W., Ausio J., Nissen M. S., Reeves R., Kang C., Kissinger C. R., Bailey R. W., Griswold M. D., Chiu W., Garner E. C., Obradovic Z. ( 2001) J. Mol. Graph. Model. 19, 26– 59 [DOI] [PubMed] [Google Scholar]
- 62.Ross E. D., Baxa U., Wickner R. B. ( 2004) Mol. Cell. Biol. 24, 7206– 7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu X., Hamkalo B., Parseghian M. H., Hansen J. C. ( 2009) Biochemistry 48, 164– 172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peterson C. L., Laniel M. A. ( 2004) Curr. Biol. 14, R546– 551 [DOI] [PubMed] [Google Scholar]
- 65.Villar-Garea A., Imhof A. ( 2006) Biochim. Biophys. Acta 1764, 1932– 1939 [DOI] [PubMed] [Google Scholar]
- 66.Krebs J. E. ( 2007) Mol. Biosyst. 3, 590– 597 [DOI] [PubMed] [Google Scholar]
- 67.Cosgrove M. S., Wolberger C. ( 2005) Biochem. Cell Biol. 83, 468– 476 [DOI] [PubMed] [Google Scholar]
- 68.Shogren-Knaak M., Peterson C. L. ( 2006) Cell Cycle 5, 1361– 1365 [DOI] [PubMed] [Google Scholar]
- 69.Hansen J. C. ( 2006) ACS Chem. Biol. 1, 69– 72 [DOI] [PubMed] [Google Scholar]
- 70.Shoemaker B. A., Portman J. J., Wolynes P. G. ( 2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8868– 8873 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.