Abstract
The nuclear receptor peroxisome proliferator-activated receptor γ (PPARγ) is a ligand-dependent transcription factor that acts as a primary regulator of adipogenesis and controls adipocyte metabolism and insulin action. Increased expression of tumor necrosis factor (TNFα) in adipose tissue of obese subjects potently suppresses the expression of PPARγ and attenuates adipocyte functions. Here we show that PPARγ is a substrate of caspase-3 and caspase-6 during TNFα receptor signaling in adipocytes, and the consequent PPARγ cleavage disrupts its nuclear localization. TNFα treatment of 3T3-L1 adipocytes decreases full-length PPARγ while increasing the level of a 45-kDa immunoreactive PPARγ fragment. Specific inhibitors of caspase-3 and caspase-6 attenuate the cleavage of PPARγ protein in response to TNFα in cultured adipocytes. Incubation of nuclear fractions with recombinant caspase-3 and caspase-6 also generates a 45-kDa PPARγ cleavage product. Dispersion of nuclear PPARγ to the cytoplasm in response to TNFα treatment occurs in parallel with detection of activated caspase-3. We suggest that activation of the caspase cascade by TNFα down-regulates PPARγ protein and PPARγ-mediated metabolic processes in adipose cells.
The development of insulin resistance in skeletal muscle of obese humans precedes and contributes to the onset of type 2 diabetes (1–3). This impaired responsiveness of muscle to insulin may result from high levels of circulating free fatty acids (FFAs)2 that disrupt insulin signaling pathways in muscle and other tissues (1, 4–9). Thus, the sequestration and storage of FFAs as triglycerides within adipose cells protects against the deleterious effect of circulating FFAs and, therefore, reduces insulin resistance in skeletal muscle. Adipose tissue also promotes insulin sensitivity in muscle by secreting adipokines including leptin and adiponectin, which promote fatty acid oxidation and decrease intracellular fatty acids (10, 11).
A large body of work has identified transcriptional regulators that participate in the control of adipocyte differentiation as well its metabolic and secretory functions (12, 13). The nuclear hormone receptor peroxisome proliferator-activated receptor γ (PPARγ) is a master regulator of adipocyte differentiation and plays an important role in glucose and lipid metabolism as well as insulin sensitivity in mature adipocytes (12–14). The essential role of PPARγ in adipocyte gene expression and differentiation has been firmly established by a number of observations, including the high level of PPARγ expression in adipose tissue and the onset of its expression coincident with early stages of adipogenesis in culture (15). Coordinated expression and actions of PPARγ with other factors, such as C/EBPα and C/EBPβ, during fat cell differentiation has been extensively documented (16–18). Additionally, it has been shown that activation of many adipocyte-specific genes occurs through binding of PPARγ to cis-acting promoter elements (19–21). Ectopic expression of PPARγ in fibroblasts induces adipogenesis (22, 23), whereas genetic ablation of the Pparγ gene disrupts adipogenesis, resulting in lipodystrophy, high levels of blood triglycerides and ectopic lipid deposition (24).
Overall, these observations are consistent with the notion that PPARγ is an essential transcriptional regulator of adipogenesis and is required for maintenance of adipocyte functions including FFA storage and lipid metabolism. Furthermore, the discovery that the insulin-sensitizing thiazolidinediones are high affinity ligands for PPARγ (25) suggested that the ability of thiazolidinediones to affect whole body glucose homeostasis is mediated at least in part through changes in PPARγ-dependent adipose gene expression. In fact, amelioration of insulin resistance in obese mice treated with rosiglitazone is coincident with changes in adipocyte gene expression including up-regulation of many genes important for lipid metabolism and storage of triglycerides, as well as glucose disposal (26). Most of these genes are similarly affected by thiazolidinedione treatment of adipocytes in culture (26, 27), suggesting that they reflect direct effects of PPARγ-dependent gene regulation in adipocytes. Thus, modulators of PPARγ functions in adipocytes are highly likely to play roles in regulation of adipocyte metabolism and in determining responses to insulin sensitizers, such as thiazolidinediones. Regulation of PPARγ expression and function in fat cells also needs to be identified and characterized to fully understand adipocyte biology and its role in whole body glucose and lipid homeostasis.
Development of obesity coincides with substantial infiltration of macrophages into adipose tissue, which is associated with increased expression of inflammatory cytokines including tumor necrosis factor α (TNFα). Large amounts of TNFα are secreted by adipocytes and macrophages within adipose tissue of obese subjects (28). These agents, in turn, up-regulate adipocyte lipolysis and down-regulate triglyceride storage and insulin signaling, contributing to the metabolic complications of obesity such as impaired glucose tolerance and peripheral insulin resistance. Interestingly, TNFα is known to be a potent negative regulator of adipogenesis and PPARγ function (29, 30). A marked down-regulation of PPARγ transcription in fat cells treated with TNFα has been demonstrated (29, 30) and NFκB signaling was proposed to mediate the TNFα inhibition of PPARγ expression in cultured cell systems (31).
Other signaling pathways triggered by TNFα may also influence PPARγ levels at the post-transcriptional level, as TNFα-activated protein kinases may also directly target PPARγ protein, influencing PPARγ activity and/or stability by phosphorylation. For instance, it has been proposed that phosphorylation of PPARγ promotes its degradation via a ubiquitin/proteasome-dependent pathway (32). Moreover, PPARγ phosphorylation has been claimed to decrease ligand-binding affinity, controlling interaction with its co-regulators (33). Together, these observations suggest that regulation of PPARγ activity and stability also appears to be negatively regulated by kinase-mediated phosphorylation (34) and ubiquitination (32) followed by PPARγ protein degradation via a ubiquitin/proteasome-dependent pathway (32). Combined, these pathways might be used by TNFα receptor signaling to decrease PPARγ protein levels in adipocytes from chronically inflamed adipose tissue in obese subjects (1). Additional pathways elicited by TNFα signaling could also be operating to down-regulate PPARγ function in fat cells (reviewed in Ref. 1). For instance, it has been demonstrated that TNFα-induced cleavage of insulin-signaling molecules, such as AKT, is mediated by a caspase-dependent pathway in adipocytes and seems to contribute to insulin resistance trigged by TNFα (35).
More recently, He and colleagues (36) demonstrated that caspase-1 mediates degradation of PPARγ proteins in adipocytes. The authors suggested that PPARγ levels could be modulated by TNFα-stimulated caspase-1 activity in adipose cells (36). In this study, we sought to extend these observations and to determine whether activation of the caspase cascade by TNFα plays a role in the reductions of PPARγ protein levels induced by this cytokine, similar to its role in the degradation of other signaling molecules in 3T3-L1 adipocytes. Here, we present evidence that TNFα triggers a caspase-dependent cleavage of PPARγ proteins in cultured adipocytes, which disrupts its nuclear localization. Caspase-3, -6, and -8, but not caspase-1, -2, -5, -7, or -9, are required for TNFα-elicited PPARγ cleavage in these cells. Based on these results, we postulate that a caspase-3/caspase-6-dependent pathway contributes to the mechanisms whereby TNFα suppresses PPARγ protein levels and its function in adipose cells.
EXPERIMENTAL PROCEDURES
Materials
Mouse anti-PPARγ monoclonal antibody (clone E-8, catalog number sc-7273) was purchased from Santa Cruz Biotechnology. Rabbit anti-caspase-8 (catalog number 4927), anti-caspase-3 (catalog number 9662), and anti-cleaved-(Asp175)-caspase-3 (catalog number 9661) polyclonal antibodies were from Cell Signaling Technologies. Mouse anti-β-actin (catalog number A2228) monoclonal antibody was from Sigma. Mouse anti-TATA-binding protein (catalog number ab818-100) monoclonal antibody was from Abcam. Hoechst 33342 trihydrochloride trihydrate (catalog number H-3570) was from Molecular Probes. Mouse recombinant TNFα (catalog number 654245), the proteasome inhibitors MG132 (catalog number 474791) and lactacystin (catalog number 426100), the recombinant caspases-1, -2, -3, -6, -7, -8, and -9, the general caspases inhibitor Boc-d-FMK (number 218745), and the specific caspase inhibitors (caspase inhibitor set II, number 218772) were from Calbiochem.
Cell Culture and Treatments
3T3-L1 fibroblasts were grown and differentiated into adipocytes as described (37). The 3T3-L1 adipocytes were cultured in complete Dubecco's modified Eagle's medium (10% fetal bovine serum, 50 units/ml penicillin, and 50 μg/ml streptomycin). For TNFα stimulation, 7 days post-differentiated 3T3-L1 adipocytes were treated with the indicated concentration of TNFα for the appropriate time intervals. Cells were then washed with ice-cold phosphate-buffered saline lysed in 1% SDS-containing lysing buffer on ice.
Preparation of Nuclear and Cytosolic Subcellular Fractions
Nuclear and cytosolic cell extracts were prepared from 3T3-L1 adipocytes by a modification (38) of the procedure of Dignam et al. (39) and described in Ref. 40. Briefly, cell monolayers were rinsed twice with ice-cold phosphate-buffered saline and once in hypotonic lysis buffer containing 20 mm Tris-HCl, pH 7.5, 10 mm NaCl, 3 mm MgCl2, 1 mm dithiothreitol, 0.1 mm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, 5 μg/ml leupeptin, and 5 μg/ml aprotinin. Cells were then harvested in hypotonic lysis buffer, incubated on ice for 5 min and homogenized with 16 strokes in a Dounce homogenizer by adding 0.1% Nonidet P-40 detergent. The resulting homogenate was centrifuged at 3500 × g at 4 °C for 5 min, and the supernatant was saved as cytosolic extract. The nuclear pellet was once resuspended in 0.5 volume of a hypotonic lysis buffer and centrifuged as before. The nuclear pellet was then resuspended in an extraction buffer containing 17.5 mm Hepes, pH 7.6, 330 mm NaCl, 1.1 m urea, 1.1% Nonidet P-40, 1 mm dithiothreitol, 1 mm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, 5 μg/ml leupeptin, and 5 μg/ml aprotinin. Nuclei were extracted for 30 min on ice. Finally, the sample was centrifuged at 13,000 × g for 10 min at 4 °C. The resulting nuclear extract and the previously obtained cytosolic extract were analyzed for protein content by BCA analysis (Pierce) according to the manufacturer's instructions and stored at 80 °C.
Immunoblotting
3T3-L1 adipocytes were incubated without or with the indicated TNFα or inhibitor concentrations for the indicated time, and then harvested with lysis buffer containing 1% SDS. Equal amounts of protein from total cell lysates were resolved by SDS-PAGE and electrotransferred to nitrocellulose membranes, which were incubated with the indicated antibodies overnight at 4 °C and then with horseradish peroxidase-linked secondary antibodies for 45 min at room temperature. Proteins were then detected with an enhanced chemiluminescence kit.
In Vitro Digestion with Recombinant Caspases
Aliquots of 40 μg of nuclear fractions isolated from 3T3-L1 adipocytes were incubated for 12 h at 37 °C in 50 μl of phosphate-buffered saline in the presence of 10 units of the indicated recombinant caspase. The reaction was stopped by the addition of electrophoresis sample buffer and samples were subsequently blotted for PPARγ protein.
Immunofluorescence
3T3-L1 adipocytes at day 7 were treated or not with TNFα, washed three times with ice-cold phosphate-buffered saline, and immediately fixed in 4% formaldehyde for 12 min at room temperature. Fixed adipocytes were permeabilized with 0.05% Triton X-100 and 0.05% Tween 20 and then stained with mouse monoclonal anti-PPARγ (clone E-8) and rabbit polyclonal anti-cleaved (Asp175)-caspase-3 antibodies for 12 h, followed by labeling with Hoechst 33342 for nuclear staining, Alexa 594-conjugated goat anti-mouse and Alexa 488-conjugated goat anti-rabbit secondary antibodies (Molecular Probes) for 30 min. The cells were preserved in Vectashield mounting medium for fluorescence (Vector Laboratories Inc.). The Images were obtained using a Nikon TE2000-E2 microscope with a Yokogawa CSU10b spinning disk confocal scan head and custom laser launch, NEOS AOTF and relay optics (Solamere Technology Group, Salt Lake City, UT). Multi-wavelength confocal z-series were acquired with a Nikon 40X Plan Apo oil objective (NA = 1.0) and a QImaging Rolera MGi camera using the standard digitizer. The final pixel size was 0.27 μm/pixel, the z step was 0.3 μm, and the exposure time was 1-s per image. Metamorph Software controlled the microscope hardware and image acquisition.
RESULTS
TNFα Decreases PPARγ Protein Levels, while Increasing the Formation of a 45-kDa Immunoreactive PPARγ Fragment
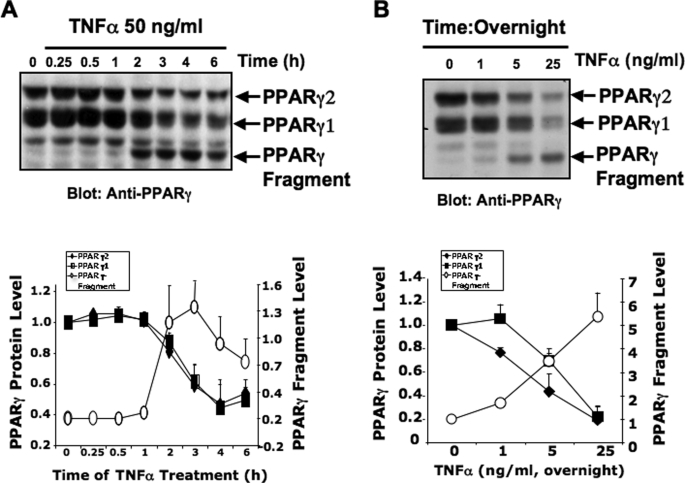
It has been previously shown that TNFα treatment of cultured adipocytes reduced PPARγ protein levels, presumably through suppression of Pparγ gene transcription via the NFκB signaling pathway (30, 31, 41). However, it is not fully understood whether the decrease in PPARγ protein levels induced by TNFα is also mediated by other TNFα-stimulated pathways in fat cells. Therefore, we conducted experiments to determine whether activation of TNFα receptors in 3T3-L1 adipocytes could lead to PPARγ protein degradation. Treatment of fully differentiated 3T3-L1 adipocytes (days 7–9) with TNFα for 16 h induced a dose-dependent decrease in PPARγ protein, with half-maximal and maximal effects occurring at 5 and ≈25–50 ng/ml, respectively (Fig. 1B). These concentrations of TNFα correspond to previously reported concentrations of TNFα that elicited maximal impairment of insulin signaling and inhibit PPARγ expression in 3T3-L1 adipocytes (42). Additionally, as depicted in Fig. 1A, treatment of cultured adipocytes with 50 ng/ml TNFα for 6 h caused a time-dependent 60% decrease in PPARγ protein. Immunoblotting with a monoclonal anti-PPARγ antibody raised against an epitope located at the C-terminal region of PPARγ protein also revealed a lower molecular mass band that migrated at ∼45 kDa in SDS-PAGE (Fig. 1, A and B). Incubation with TNFα increased the level of this 45-kDa PPARγ fragment in a dose- and time-dependent manner (Fig. 1, A and B). Thus, whereas TNFα treatment of cultured adipocytes reduces total PPARγ protein levels, it simultaneously induces the formation of a lower molecular weight fragment of PPARγ protein.
FIGURE 1.
Treatment of 3T3-L1 adipocytes with TNFα decreases PPARγ protein levels and induces the formation of a 45-kDa immunoreactive PPARγ fragment. A, upper panel, a representative experiment demonstrating the time course of the decrease and increase in PPARγ protein levels and a 45-kDa PPARγ fragment, respectively, upon treatment with TNFα. 3T3-L1 adipocytes were treated or not with 50 ng/ml TNFα for the indicated periods of time, lysed, and the PPARγ protein analyzed by Western blot, using a carboxyl-terminal anti-PPARγ monoclonal antibody (clone E-8). Densitometry analysis of four immunoblot experiments of 3T3-L1 adipocytes treated with TNFα for the indicated times is depicted in the lower panel. B, upper panel, dose-dependent relationship of TNFα treatment decrease in PPARγ protein and increase in the 45-kDa PPARγ-immunoreactive fragment. Lower panel shows the densitometry analysis of four immunoblot experiments, similar to the one depicted in the upper panel. Arrows in the upper panel indicate the PPARγ protein and 45-kDa fragment.
Increase in the PPARγ Fragment Induced by TNFα Does Not Require New Protein Synthesis
A Pparγ mRNA, γORF4, which is generated by alternative splicing has been reported recently by Sabatino and colleagues (43). The γORF4 transcript results in the synthesis of a truncated PPARγ protein containing the first 273 amino acids of the mature PPARγ but lacking the entire ligand-binding domain. Thus, one possibility is that the 45-kDa PPARγ immunoreactive band is a translation product of an alternatively spliced Pparγ mRNA expressed upon TNFα stimulation. To test this possibility 3T3-L1 adipocytes were preincubated with the protein synthesis inhibitor cycloheximide (5 μg/ml) and then treated for 6 h with 50 ng/ml TNFα. Cells were then harvested and cell lysate samples immunoblotted for PPARγ protein. Inhibition of protein synthesis with cycloheximide did not significantly block the ability of TNFα to induce the production of the 45-kDa PPARγ fragment (data not shown) suggesting that this product is a result of post-translational truncation of existing PPARγ protein.
Reductions in PPARγ Protein Levels by TNFα Involve the Proteasome Degradation Pathway
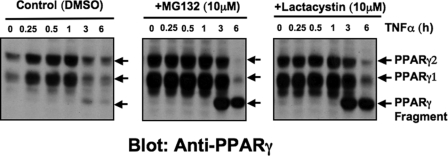
We and other groups have shown that PPARγ degradation rates are remarkably rapid as measured in the presence of cycloheximide (t½ = 2 h) (44, 45). Furthermore, treatment of cultured adipocytes with the proteasome inhibitor MG132 increases the protein levels, suggesting that the proteasome degradation pathway regulates PPARγ protein levels in 3T3-L1 adipocytes (32). To test whether the 45-kDa fragment seen in cells stimulated with TNFα is a product of proteasomal degradation of PPARγ in 3T3-L1 adipocytes, we treated cells with TNFα plus proteasome inhibitors. As depicted in Fig. 2, a marked enhancement of PPARγ protein levels was observed in the presence of the proteasome inhibitors MG132 and lactacystin. Moreover, addition of the proteasome inhibitors partially prevented PPARγ protein degradation mediated by TNFα treatment of adipocytes for 3 h (Fig. 2). However, the formation of the 45-kDa PPARγ fragment was not prevented by these proteasome inhibitors, rather, accumulation of the fragment seemed to be enhanced by inhibition of proteasome function. Thus, these results suggest that suppression of the PPARγ protein level through TNFα may also involve proteasome-mediated protein degradation of this nuclear receptor. Generation of the 45-kDa fragment does not require activity of the proteasomal pathway, but ultimate degradation of the fragment may involve the proteasome.
FIGURE 2.
Reductions in PPARγ protein levels mediated by TNFα involve the proteasome degradation pathway. 3T3-L1 adipocytes were treated for 20 min with 1% dimethyl sulfoxide (DMSO) (control), 10 μm MG132, or 10 μm lactacystin and then incubated in the presence or absence of 50 ng/ml TNFα for the indicated periods of time. Cells were then harvested and PPARγ protein analyzed by Western blot using a carboxyl-terminal anti-PPARγ monoclonal antibody. Depicted Western blots are representative of three different experiments. Arrows indicate PPARγ proteins and the 45-kDa fragment.
TNFα-induced Cleavage of PPARγ Is Caspase-dependent in Intact Cells
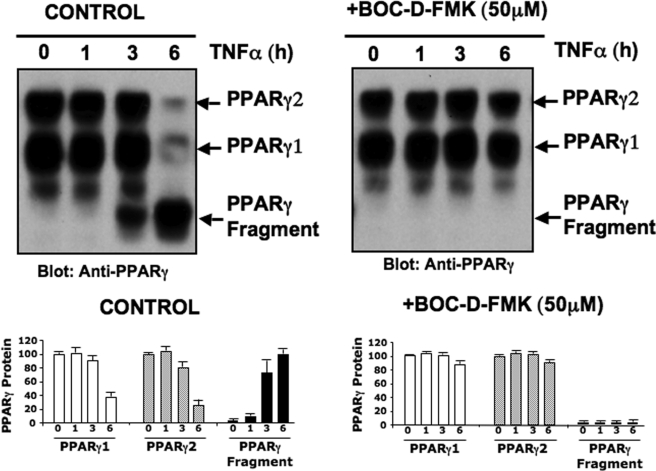
TNFα signaling leads to activation of caspase cascades in adipocytes and cleavage of several signaling molecules (35). To identify potential caspase cleavage sites in the PPARγ protein sequence, we utilized the web-based tool GraBCas (46). Several putative caspase consensus cleavage sites in the PPARγ protein sequence were identified as shown in Table 1. These observations prompted us to examine whether caspase activities are involved in PPARγ cleavage elicited by TNFα treatment of cultured adipocytes. To determine whether PPARγ was cleaved by caspases, 3T3-L1 adipocytes were preincubated with the broad-spectrum caspase inhibitor Boc-d-FMK and then stimulated with TNFα for up to 6 h (Fig. 3). Lysates from the harvested cells were then immunoblotted with anti-PPARγ monoclonal antibody. Consistent with the results depicted in Figs. 1 and 2, treatment with TNFα for 6 h reduced full-length PPARγ protein levels and induced formation of a 45-kDa fragment. However, the TNFα-mediated decrease in PPARγ protein and appearance of the 45-kDa fragment was abrogated by this broad-spectrum caspase inhibitor (Fig. 3, right panels). This result strongly indicates that TNFα receptor signaling in fat cells induces cleavage of PPARγ by caspases and/or caspase-like proteases. They also suggest that caspase-mediated PPARγ proteolysis may be part of the mechanism whereby TNFα suppresses PPARγ expression levels in adipocytes.
TABLE 1.
Putative caspase cleavage sites in PPARγ protein
Potential caspase cleavage sites in the amino acid sequence of PPARγ2 as identified by the web-based program GraBCas.
| Amino acid | Amino acid sequence | Caspase |
|---|---|---|
| 49 | SSVD↓L | 3, 7 |
| 69 | TTVD↓F | 3, 6, 7, 8 |
| 228 | ISSD↓I | 2 |
| 230 | SDID↓Q | 6, 7, 8 |
| 238 | ESAD↓L | 2, 3, 4 |
| 341 | DLND↓Q | 2 |
| 408 | LELD↓D | 1, 3, 4, 5, 6, 8, 9 |
| 411 | DDSD↓L | 2, 8 |
| 490 | TETD↓M | 1, 2, 3, 4, 5, 6, 7, 8, 9 |
FIGURE 3.
TNFα-induced cleavage of PPARγ is caspase-dependent. Upper panel, caspase activity is required for TNFα-mediated cleavage of the PPARγ protein and 45-kDa PPARγ fragment formation. 3T3-L1 adipocytes were treated in the presence of 10 μm MG132 and for 20 min with 1% dimethyl sulfoxide (control) or 50 μm of the general caspases inhibitor Boc-d-FMK. Cells were then treated or not with 50 ng/ml TNFα for the indicated periods of time. Cells were then harvested and PPARγ protein analyzed by Western blot, using a carboxyl-terminal anti-PPARγ monoclonal antibody. Depicted Western blots are representative of five different experiments. Lower panels show quantification of PPARγ protein levels by densitometry analysis. Data are representative of five independent experiments. Arrows in the upper panel indicate the PPARγ protein and 45-kDa fragment.
Caspases-3, -6, and -8 Are Required for TNFα-induced PPARγ Cleavage in Vivo
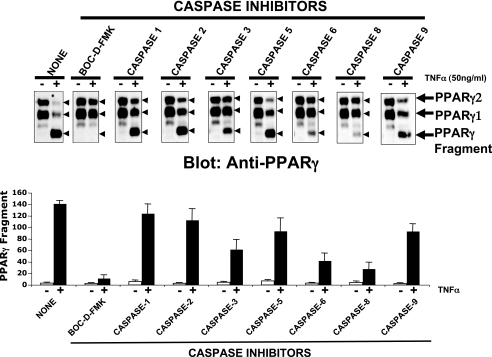
Because a general caspase inhibitor prevented the TNFα-induced degradation and cleavage of PPARγ protein (Fig. 3), we next examined the involvement of particular caspases in this process. Thus, a similar experiment was performed with inhibitors of specific caspases. Adipocytes were treated with a panel of caspase inhibitors, then exposed to TNFα for 6 h and lysates were subjected to immunoblot analysis with anti-PPARγ antibody. Consistent with the results depicted in Fig. 3, the general caspase inhibitor Boc-d-FMK completely inhibited the decline of PPARγ protein and appearance of the 45-kDa PPARγ fragment (Fig. 4, upper panel). The caspase-8 inhibitor (Z-IETD-FMK) blocked both TNFα-induced PPARγ degradation and 45-kDa PPARγ fragment formation to nearly the same extent as the broad-spectrum caspase inhibitor Boc-d-FMK (Fig. 4). Among the remaining compounds tested, inhibitors of caspases-1, -2, -5, and -9 had essentially no effect. Only the caspase-6 (Z-VEID-FMK) and caspase-3 (Z-DEVD-FMK) inhibitors partially blocked the decline of PPARγ protein and the cleavage process. These results indicate that caspases-3, -6, and -8 activities are required for TNFα-induced PPARγ cleavage in vivo. They (47) also suggest that TNFα activation of the caspase-8 → caspase-3 → caspase-6 cascade is necessary for down-regulation of the PPARγ protein in cultured adipocytes stimulated with this cytokine.
FIGURE 4.
Caspases-3, -6, and -8 are required for TNFα-induced cleavage of PPARγ. Upper panels, 3T3-L1 adipocytes were treated with 10 μm MG132 and during the first 20 min, with 1% dimethyl sulfoxide (NONE), 50 μm Boc-d-FMK, or 50 μm of the indicated caspase inhibitors and then treated (+) or not (−) with 50 ng/ml TNFα for 6 h. Cells were then harvested and PPARγ protein analyzed by Western blot, using the carboxyl-terminal anti-PPARγ monoclonal antibody E-8. Depicted Western blots are representative of four different experiments. Arrows indicate PPARγ proteins and the 45-kDa fragment. Lower panels show quantification of PPARγ protein levels by densitometry analysis. Data are means of four independent experiments and S.E.
In Vitro Cleavage of PPARγ with Recombinant Caspase-3 and Caspase-6
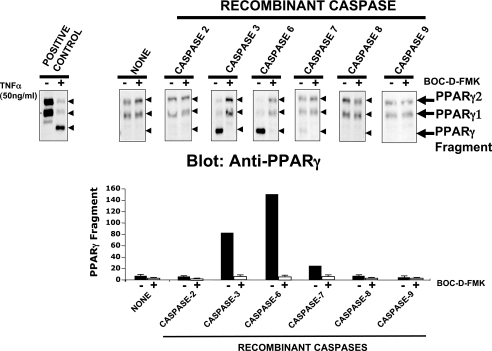
The results depicted in Fig. 4, suggesting that caspase-3, -6, and -8 are required for PPARγ protein degradation induced by TNFα, prompted us to investigate whether PPARγ proteins are directly targeted by the activity of these caspases. Nuclear fractions from 3T3-L1 adipocytes were isolated and incubated with recombinant caspases 1 (data not shown), and caspase-2, -3, -6, -7, -8, and -9, in the presence or absence of Boc-d-FMK. As depicted in Fig. 5, PPARγ proteins were cleaved in vitro only by recombinant caspase-3 and -6, as shown by the disappearance of the full-length PPARγ band and the generation of a 45-kDa PPARγ fragment. As expected, PPARγ protein cleavage by recombinant caspase-3 and -6 was abrogated in the presence of the general caspase inhibitor Boc-d-FMK. Interestingly, recombinant caspase-3 and -6 generated 45-kDa PPARγ fragments in vitro, similar to the 45-kDa fragment induced by TNFα in vivo (Figs. 1–4). Treatment with recombinant caspase-8 was unable to cleave PPARγ in vitro although inhibition of caspase-8 blocked PPARγ protein cleavage in cells (Fig. 4). Taken together, these results shown that in vitro treatment of PPARγ with recombinant caspase-3 and -6 generated a 45-kDa cleavage product, consistent with that seen in adipocytes stimulated with TNFα. They also reinforce the notion that PPARγ protein is directly targeted by caspase-3 and -6 during TNFα signaling in adipose cells in a pathway indirectly involving caspase-8.
FIGURE 5.
Recombinant caspase-3 and -6, but not caspase-8, cleave PPARγ. Upper panels, caspases-3 and -6 mediate cleavage of PPARγ protein and generate the 45-kDa PPARγ fragment. Isolated nuclear subcellular fractions from 3T3-L1 adipocytes were incubated for 12 h with (+) or without (−) 50 μm Boc-d-FMK and in the absence (NONE) or presence of the indicated recombinant caspase proteins. Nuclear fractions were then resolved by SDS-PAGE and PPARγ protein analyzed by Western blot, using a carboxyl-terminal anti-PPARγ monoclonal antibody. Depicted Western blots are representative of three different experiments. The left, upper panel depicts a positive control where lysate from cells treated (+) or not (−) with 50 ng/ml TNFα for 6 h were resolved by SDS-PAGE and immunobloted with anti-PPARγ antibody. Lower panels show quantification of PPARγ protein levels by densitometry analysis. Data shown are mean ± S.E. of three independent experiments. Arrows and arrowheads in the upper panel indicate the PPARγ protein and 45-kDa fragment.
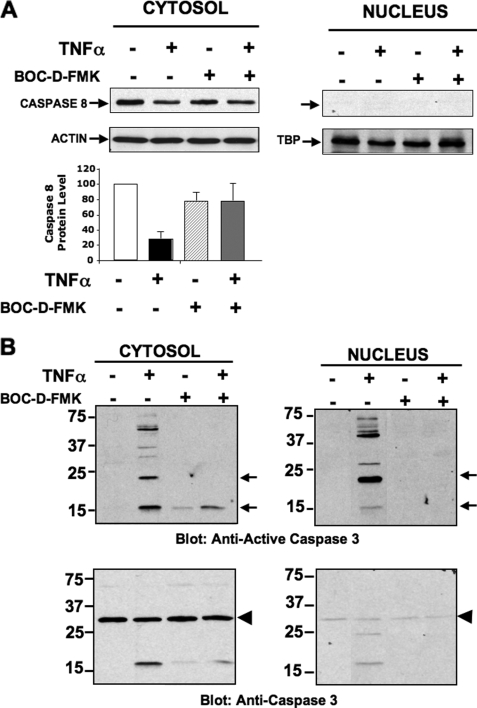
Active Caspase-3 and Caspase-6 Are Translocated into Adipocyte Nuclei Upon TNFα Stimulation
Given the predominantly nuclear localization of PPARγ protein in adipocytes (44, 48, 49), we next examined whether TNFα treatment induces active caspases to be translocated into the nucleus where they could target PPARγ protein. Thus cytosolic and nuclear subcellular fractions from 3T3-L1 adipocytes, treated or not with TNFα, were isolated and then subjected to immunoblot analysis with anti-procaspase and anti-active caspase antibodies to examine the subcellular distribution of active caspases in response to TNFα. As shown in Fig. 6A, full-length caspase-8 was present in the cytosolic, but not nuclear fraction. Based on previous results, treatment with TNFα was expected to induce cleavage and activation of full-length caspase-8. Although the antibody used does not detect the activated cleaved caspase-8, we interpret the decrease in full-length caspase-8 protein levels in the cytosolic fractions (Fig. 6A, upper and lower panel) as indicative of cleavage and activation (50). This result suggests that stimulation of cultured adipocytes with TNFα induces activation of caspase-8 in the cytosolic compartment, but not the nucleus. We further examined whether other caspases downstream to caspase-8, such as caspase-3 and -6, predicted to be the activities that directly target PPARγ, were also activated under these conditions and translocated to the nucleus. Thus, nuclear and cytosolic fractions from adipocytes stimulated or not with TNFα were immunobloted for pro-caspase-3 and active caspase-3. As depicted in Fig. 6B (lower panel), pro-caspase-3 was present mostly in the cytosolic fraction, with minimal detection in the nuclear fraction. Remarkably, treatment of 3T3-L1 adipocytes with TNFα for 6 h caused a dramatic increase in active caspase-3 (i.e. caspase-3-p17 subunit) associated with the nuclear subcellular fraction (Fig. 6B, upper panel). Moreover, incubation of 3T3-L1 adipocytes with the general caspase inhibitor Boc-d-FMK before treatment of cells with TNFα blocked recruitment of active caspase-3 into the nucleus (Fig. 6B, upper panel), consistent with activation and translocation of caspase-3 being dependent on activity of an upstream caspase, most likely caspase-8. Similarly, stimulation of cultured adipocytes with TNFα also caused a significant recruitment of active caspase-6 into the nucleus (data not shown). Thus, these results are consistent with the hypothesis that recruitment of active caspase-3 and -6 into the nucleus of TNFα-stimulated adipose cells mediates cleavage of the PPARγ protein.
FIGURE 6.
TNFα induces nuclear translocation of active caspase-3. A, upper panels, immunoblotting of pro-caspase-8 in 3T3-L1 adipocytes cells treated or not with 50 ng/ml TNF-α plus MG132 in the absence or presence of 50 μm Boc-d-FMK, as indicated. Nuclear and cytosolic subcellular fractions were then prepared and subjected to SDS-PAGE and immunoblotting with anti-pro-caspase-8 antibody. Cytosolic and nuclear fractions were also used for detection of actin and TATA-binding protein (loading controls) by immunoblot. Lower panel depicts quantification of pro-caspase-8 protein levels. Data are representative of four determinations ± S.E. B, immunoblotting of active caspase-3 (upper panel), and pro-caspase-3 (lower panel) in 3T3-L1 adipocytes cells treated or not with 50 ng/ml TNF-α plus MG132 and in the absence or presence of 50 μm Boc-d-FMK, as indicated. Nuclear and cytosolic subcellular fractions were then prepared and subjected to SDS-PAGE and immunoblotting with anti-active caspase-3 or anti-pro-caspase-3 antibodies. Data are representative of three independent experiments.
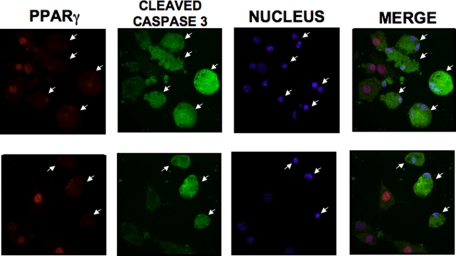
TNFα-mediated Caspase-3 and Caspase-6 Activation Disrupts Nuclear Localization of PPARγ in Cultured Adipocytes
We next investigated whether caspase-dependent cleavage of the N-terminal region of PPARγ upon TNFα stimulation affects the subcellular localization of PPARγ. To examine whether caspase-dependent cleavage of PPARγ induced by TNFα has an impact on its subcellular distribution, we treated 3T3-L1 adipocytes with or without TNFα for 6 h, similar to the experiments described in Figs. 1–4. Adipocytes were then fixed and stained with anti-active caspase-3 and anti-PPARγ antibodies, followed by immunofluorescence microscopy analysis to visualize the subcellular distribution of endogenous PPARγ protein in cells displaying activation of caspase-3. As shown in Fig. 7, TNFα induced a remarkable dispersion of PPARγ protein from the nuclear region of 3T3-L1 adipocytes exclusively in cells in which active caspase-3 was also detected. In these cells PPARγ was detected throughout the cytosolic compartment. Nuclei remained intact under these conditions as evidenced by the persistence of nuclear staining (Fig. 7) as well as continued nuclear localization of the general transcription factor TATA-binding protein (data not shown), illustrating specific relocation of PPARγ. Overall, this result supports the hypothesis that in TNFα-stimulated adipose cells, the caspase-mediated cleavage of PPARγ protein at its N-terminal end promotes nuclear export and cytosolic degradation of this transcriptional factor. This export to the cytoplasm likely leads to loss of its activation function in the nucleus.
FIGURE 7.
TNFα-mediated nuclear translocation of active caspase-3 disrupts nuclear localization of PPARγ protein. Differentiated 3T3-L1 adipocytes were treated with 50 ng/ml TNFα in the presence of 10 μm MG132. After 6 h, cells were fixed, permeabilized, and stained with monoclonal anti-PPARγ (clone E-8), polyclonal anti-active caspase-3 antibodies, and Hoechst 33342 trihydrochloride trihydrate, to visualize endogenous PPARγ and active caspase-3 proteins and nucleus. Most of the PPARγ protein (red) was localized within the nucleus (blue) of cultured adipocytes with no detection of caspase-3 activation. However, PPARγ protein was quite dispersed from the nuclei of cells displaying caspase-3 activation (green), indicated by the arrows. Depicted immunofluorescence microscopic images are representative of three experiments.
DISCUSSION
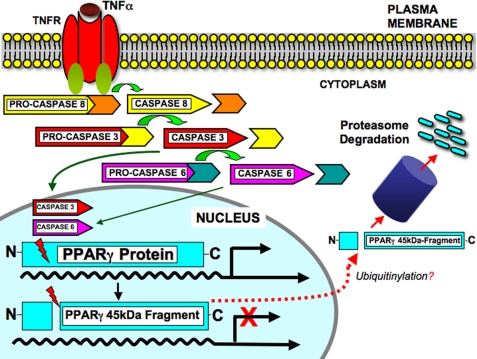
The inflammatory state associated with excess adipose tissue in obesity is characterized by the production of cytokines including TNFα, which may adversely affect the capability of fat tissue to sequester excess lipids. This contributes to impaired insulin responsiveness of other tissues. We have described here a newly recognized mechanism by which TNFα may impair adipocyte function by decreasing the levels and activity of the key transcriptional activator PPARγ. Our data support a model summarized in Fig. 8, in which TNF receptor signaling in adipocytes activates caspase-8 in the cytosol. Caspase-8 in turn promotes the cleavage and activation of caspases-3 and -6. The latter activated caspases are translocated to the adipocyte nucleus where they directly target PPARγ resulting in the cytosolic accumulation and ultimate degradation of a 45-kDa PPARγ cleavage product (Fig. 8). The size of this fragment as well as the predicted specificity of caspases for the cleavage site (Table 1) are consistent with cleavage at Asp69. This is likely to result in loss of PPARγ transcriptional activation by several means: 1) disruption of the N-terminal activation domain by the probable cleavage site, likely impairing transcription activation; 2) loss of nuclear localization of the cleaved PPARγ fragment; and 3) ultimate degradation of the cleaved PPARγ fragment via a proteasomal pathway.
FIGURE 8.
Model of caspase-dependent PPARγ protein cleavage. Cell-surface TNFα receptor signaling induces the activation of caspase-3, -6, and -8. In turn, activated caspases-3 and-6 are translocated to the adipocyte nucleus (green arrows) where they cleave PPARγ most likely at the identified consensus site TTVDF69 (Table 1) to generate a COOH-terminal 45-kDa fragment. Cleaved PPARγ protein is translocated to the cytoplasm (dashed red arrow) and degraded via a proteasome-mediated process, thus down-regulating PPARγ functions. Moreover, following PPARγ protein cleavage, ubiquitinylation might also occur.
Our results are largely consistent with a study published recently by He and colleagues (36). They report caspase-dependent cleavage of PPARγ resulting in the accumulation of a 45-kDa cleavage product in response to TNFα. In that study, specific inhibition of caspase-1, but not caspase-3, -6, or -8, was able to prevent PPARγ cleavage in response to TNFα. The reasons for this discrepancy between the results published by He et al. (36) and our data are not clear. However, in their report, caspase cleavage and production of the 45-kDa fragment was only observed under conditions of translation inhibition by cycloheximide in addition to TNFα treatment. In contrast, we observe PPARγ fragment generation irrespective of the presence of cycloheximide. Thus, it is possible that activation of distinct caspase pathways might result from these differing treatments. For instance, it has been established that cycloheximide sensitizes many types of cells to TNFα-induced apoptosis, mainly due to its ability to block de novo synthesis of the endogenous caspase inhibitors, such as the cellular FLICE-inhibitory protein (c-FLIP) and members of the inhibitors of apoptosis protein family. These proteins are up-regulated by TNFα-receptor via the NFκB pathway and function through direct interactions to inhibit the activity of several caspases, including caspase-3, caspase-7, caspase-8, and caspase-9 (51, 52). Hence, upon inhibition of protein synthesis by cycloheximide treatment, suppression in the levels of these caspase inhibitors could trigger a marked activation of different sets of caspases in adipocytes. In turn, highly active caspase activities due to the absence of their endogenous inhibitors could target the PPARγ protein in adipocytes.
Another possible explanation for the differences between the present results and those of He and colleagues (36) is the effect of cycloheximide on PPARγ protein turnover. Cycloheximide treatment reveals a remarkably rapid PPARγ degradation in adipocytes (44, 45). It is possible that under these conditions of disruption in protein synthesis and rapid PPARγ protein turnover, this transcriptional factor could be targeted to a different set of caspases.
However, data presented here including inhibitor studies in cells as well as direct in vitro cleavage by recombinant caspases-3 and -6 strongly suggest that the caspase-3, -6, and -8 cascade is involved. In any case we concur with He et al. (36) that caspase-mediated cleavage of PPARγ modulates its levels and function in response to TNFα, and it will be of interest to determine the precise pathway and its relevance in vivo.
TNFα has been implicated in suppressing adipose cell metabolism through down-regulation of PPARγ function. The present work adds to an emerging picture of this regulation occurring at multiple levels. Transcripitional regulation of PPARγ has been long recognized (reviewed in Ref. 53). Rapid turnover of adipocyte PPARγ as well as control at the translational level have more recently been described by us and others (44, 45, 54). Furthermore, regulation of PPARγ transcription activation activity through modulation of coactivator or corepressor activities has also been noted (53). Collectively these findings indicate a complex and multifactorial regulation of PPARγ in response to inflammatory cytokines like TNFα. The extent of the contribution of these various pathways to impairment of adipocyte function and resulting dysfunction in lipid metabolism, insulin sensitivity, and glucose homeostasis is yet to be determined. The description here of caspase pathways involved in PPARγ down-regulation adds another potential link between obesity-associated inflammation and insulin resistance.
Acknowledgments
We thank Andrea C. Hubbard for excellent technical assistance at the initial stage of this work. We also thank Dr. Joseph V. Virbasius for valuable discussions. We also appreciate the technical help of Dr. Paul Furcinitti, from the Imaging Core Facility of the University of Massachusetts Medical School and the Diabetes and Endocrinology Research Center of the University of Massachusetts Medical School (supported by Grant DK325220).
This work was supported, in whole or in part, by National Institutes of Health Grants DK30898 and DK030638 (to M. P. C.).
- FFAs
- free fatty acids
- TNFα
- tumor necrosis factor α
- PPARγ
- peroxisome proliferator-activated receptor γ
- C/EBP
- CCAAT/enhancer-binding protein
- NFκB
- nuclear factor κB
- Boc-d-FMK
- t-butoxycarbonyl-Asp(O-Me)-fluoromethyl ketone
- MG132
- Z-Leu-Leu-Leu-CHO
- Z
- benzyloxycarbonyl.
REFERENCES
- 1.Guilherme A., Virbasius J. V., Puri V., Czech M. P. ( 2008) Nat. Rev. Mol. Cell Biol. 9, 367– 377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sims E. A., Danforth E., Jr., Horton E. S., Bray G. A., Glennon J. A., Salans L. B. ( 1973) Recent Prog. Horm. Res. 29, 457– 496 [DOI] [PubMed] [Google Scholar]
- 3.Freidenberg G. R., Reichart D., Olefsky J. M., Henry R. R. ( 1988) J. Clin. Investig. 82, 1398– 1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Unger R. H. ( 2002) Annu. Rev. Med. 53, 319– 336 [DOI] [PubMed] [Google Scholar]
- 5.Savage D. B., Petersen K. F., Shulman G. I. ( 2007) Physiol. Rev. 87, 507– 520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boden G. ( 1997) Diabetes 46, 3– 10 [PubMed] [Google Scholar]
- 7.Kelley D. E., Mokan M., Simoneau J. A., Mandarino L. J. ( 1993) J. Clin. Investig. 92, 91– 98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santomauro A. T., Boden G., Silva M. E., Rocha D. M., Santos R. F., Ursich M. J., Strassmann P. G., Wajchenberg B. L. ( 1999) Diabetes 48, 1836– 1841 [DOI] [PubMed] [Google Scholar]
- 9.Oakes N. D., Bell K. S., Furler S. M., Camilleri S., Saha A. K., Ruderman N. B., Chisholm D. J., Kraegen E. W. ( 1997) Diabetes 46, 2022– 2028 [DOI] [PubMed] [Google Scholar]
- 10.Puri V., Ranjit S., Konda S., Nicoloro S. M., Straubhaar J., Chawla A., Chouinard M., Lin C., Burkart A., Corvera S., Perugini R. A., Czech M. P. ( 2008) Proc. Natl. Acad. Sci. U. S. A. 105, 7833– 7838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nordström E. A., Rydén M., Backlund E. C., Dahlman I., Kaaman M., Blomqvist L., Cannon B., Nedergaard J., Arner P. ( 2005) Diabetes 54, 1726– 1734 [DOI] [PubMed] [Google Scholar]
- 12.Farmer S. R. ( 2006) Cell Metab. 4, 263– 273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosen E. D., MacDougald O. A. ( 2006) Nat. Rev. Mol. Cell Biol. 7, 885– 896 [DOI] [PubMed] [Google Scholar]
- 14.Rosen E. D., Spiegelman B. M. ( 2006) Nature 444, 847– 853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chawla A., Schwarz E. J., Dimaculangan D. D., Lazar M. A. ( 1994) Endocrinology 135, 798– 800 [DOI] [PubMed] [Google Scholar]
- 16.Farmer S. R. ( 2005) Int. J. Obes. 29, Suppl. 1, S13– 16 [DOI] [PubMed] [Google Scholar]
- 17.Lazar M. A. ( 2005) Biochimie 87, 9– 13 [DOI] [PubMed] [Google Scholar]
- 18.Rosen E. D. ( 2005) Prostaglandins Leukot. Essent. Fatty Acids 73, 31– 34 [DOI] [PubMed] [Google Scholar]
- 19.Tontonoz P., Hu E., Graves R. A., Budavari A. I., Spiegelman B. M. ( 1994) Genes Dev. 8, 1224– 1234 [DOI] [PubMed] [Google Scholar]
- 20.Tontonoz P., Hu E., Devine J., Beale E. G., Spiegelman B. M. ( 1995) Mol. Cell. Biol. 15, 351– 357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tontonoz P., Graves R. A., Budavari A. I., Erdjument-Bromage H., Lui M., Hu E., Tempst P., Spiegelman B. M. ( 1994) Nucleic Acids Res. 22, 5628– 5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu E., Tontonoz P., Spiegelman B. M. ( 1995) Proc. Natl. Acad. Sci. U. S. A. 92, 9856– 9860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tontonoz P., Hu E., Spiegelman B. M. ( 1994) Cell 79, 1147– 1156 [DOI] [PubMed] [Google Scholar]
- 24.Jones J. R., Barrick C., Kim K. A., Lindner J., Blondeau B., Fujimoto Y., Shiota M., Kesterson R. A., Kahn B. B., Magnuson M. A. ( 2005) Proc. Natl. Acad. Sci. U. S. A. 102, 6207– 6212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lehmann J. M., Moore L. B., Smith-Oliver T. A., Wilkison W. O., Willson T. M., Kliewer S. A. ( 1995) J. Biol. Chem. 270, 12953– 12956 [DOI] [PubMed] [Google Scholar]
- 26.Wilson-Fritch L., Nicoloro S., Chouinard M., Lazar M. A., Chui P. C., Leszyk J., Straubhaar J., Czech M. P., Corvera S. ( 2004) J. Clin. Investig. 114, 1281– 1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson-Fritch L., Burkart A., Bell G., Mendelson K., Leszyk J., Nicoloro S., Czech M., Corvera S. ( 2003) Mol. Cell. Biol. 23, 1085– 1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wellen K. E., Hotamisligil G. S. ( 2003) J. Clin. Investig. 112, 1785– 1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephens J. M., Lee J., Pilch P. F. ( 1997) J. Biol. Chem. 272, 971– 976 [DOI] [PubMed] [Google Scholar]
- 30.Zhang B., Berger J., Hu E., Szalkowski D., White-Carrington S., Spiegelman B. M., Moller D. E. ( 1996) Mol. Endocrinol. 10, 1457– 1466 [DOI] [PubMed] [Google Scholar]
- 31.Ruan H., Hacohen N., Golub T. R., Van Parijs L., Lodish H. F. ( 2002) Diabetes 51, 1319– 1336 [DOI] [PubMed] [Google Scholar]
- 32.Hauser S., Adelmant G., Sarraf P., Wright H. M., Mueller E., Spiegelman B. M. ( 2000) J. Biol. Chem. 275, 18527– 18533 [DOI] [PubMed] [Google Scholar]
- 33.Shao D., Rangwala S. M., Bailey S. T., Krakow S. L., Reginato M. J., Lazar M. A. ( 1998) Nature 396, 377– 380 [DOI] [PubMed] [Google Scholar]
- 34.Diradourian C., Girard J., Pegorier J. P. ( 2005) Biochimie 87, 33– 38 [DOI] [PubMed] [Google Scholar]
- 35.Medina E. A., Afsari R. R., Ravid T., Castillo S. S., Erickson K. L., Goldkorn T. ( 2005) Endocrinology 146, 2726– 2735 [DOI] [PubMed] [Google Scholar]
- 36.He F., Doucet J. A., Stephens J. M. ( 2008) Obesity 16, 1735– 1741 [DOI] [PubMed] [Google Scholar]
- 37.Tang X., Powelka A. M., Soriano N. A., Czech M. P., Guilherme A. ( 2005) J. Biol. Chem. 280, 22523– 22529 [DOI] [PubMed] [Google Scholar]
- 38.Swick A. G., Blake M. C., Kahn J. W., Azizkhan J. C. ( 1989) Nucleic Acids Res. 17, 9291– 9304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dignam J. D., Lebovitz R. M., Roeder R. G. ( 1983) Nucleic Acids Res. 11, 1475– 1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacDougald O. A., Cornelius P., Lin F. T., Chen S. S., Lane M. D. ( 1994) J. Biol. Chem. 269, 19041– 19047 [PubMed] [Google Scholar]
- 41.Tang X., Guilherme A., Chakladar A., Powelka A. M., Konda S., Virbasius J. V., Nicoloro S. M., Straubhaar J., Czech M. P. ( 2006) Proc. Natl. Acad. Sci. U. S. A. 103, 2087– 2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hotamisligil G. S., Murray D. L., Choy L. N., Spiegelman B. M. ( 1994) Proc. Natl. Acad. Sci. U. S. A. 91, 4854– 4858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sabatino L., Casamassimi A., Peluso G., Barone M. V., Capaccio D., Migliore C., Bonelli P., Pedicini A., Febbraro A., Ciccodicola A., Colantuoni V. ( 2005) J. Biol. Chem. 280, 26517– 26525 [DOI] [PubMed] [Google Scholar]
- 44.Floyd Z. E., Stephens J. M. ( 2002) J. Biol. Chem. 277, 4062– 4068 [DOI] [PubMed] [Google Scholar]
- 45.Christianson J. L., Nicoloro S., Straubhaar J., Czech M. P. ( 2008) J. Biol. Chem. 283, 2906– 2916 [DOI] [PubMed] [Google Scholar]
- 46.Backes C., Kuentzer J., Lenhof H. P., Comtesse N., Meese E. ( 2005) Nucleic Acids Res. 33, W208– W213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chowdhury I., Tharakan B., Bhat G. K. ( 2008) Comp. Biochem. Physiol. B Biochem. Mol. Biol. 151, 10– 27 [DOI] [PubMed] [Google Scholar]
- 48.Akiyama T. E., Baumann C. T., Sakai S., Hager G. L., Gonzalez F. J. ( 2002) Mol. Endocrinol. 16, 707– 721 [DOI] [PubMed] [Google Scholar]
- 49.Berger J., Patel H. V., Woods J., Hayes N. S., Parent S. A., Clemas J., Leibowitz M. D., Elbrecht A., Rachubinski R. A., Capone J. P., Moller D. E. ( 2000) Mol. Cell. Endocrinol. 162, 57– 67 [DOI] [PubMed] [Google Scholar]
- 50.Nisoli E., Cardile A., Bulbarelli A., Tedesco L., Bracale R., Cozzi V., Morroni M., Cinti S., Valerio A., Carruba M. O. ( 2006) Cell Death Differ. 13, 2154– 2156 [DOI] [PubMed] [Google Scholar]
- 51.Budihardjo I., Oliver H., Lutter M., Luo X., Wang X. ( 1999) Annu. Rev. Cell Dev. Biol. 15, 269– 290 [DOI] [PubMed] [Google Scholar]
- 52.Kreuz S., Siegmund D., Scheurich P., Wajant H. ( 2001) Mol. Cell. Biol. 21, 3964– 3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye J. ( 2008) Biochem. Biophys. Res. Commun. 374, 405– 408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Puri V., Virbasius J. V., Guilherme A., Czech M. P. ( 2008) Acta Physiol. 192, 103– 115 [DOI] [PMC free article] [PubMed] [Google Scholar]