Abstract
A novel approach for boosting antimicrobial peptides through end tagging with hydrophobic oligopeptide stretches is demonstrated. Focusing on two peptides derived from kininogen, GKHKNKGKKNGKHNGWK (GKH17) and HKHGHGHGKHKNKGKKN (HKH17), tagging resulted in enhanced killing of Gram-positive Staphylococcus aureus, Gram-negative Escherichia coli, and fungal Candida albicans. Microbicidal potency increased with tag length, also in plasma, and was larger for Trp and Phe stretches than for aliphatic ones. The enhanced microbicidal effects correlated to a higher degree of bacterial wall rupture. Analogously, tagging promoted peptide binding to model phospholipid membranes and liposome rupture, particularly for anionic and cholesterol-void membranes. Tagged peptides displayed low toxicity, particularly in the presence of serum, and resisted degradation by human leukocyte elastase and by staphylococcal aureolysin and V8 proteinase. The biological relevance of these findings was demonstrated ex vivo and in vivo in porcine S. aureus skin infection models. The generality of end tagging for facile boosting of antimicrobial peptides without the need for post-synthesis modification was also demonstrated.
Introduction
Because of increasing bacterial resistance, there is a need for new types of antibiotics (1). Antimicrobial peptides (AMPs)2 are interesting in this context because they provide rapid and broad spectrum response toward both Gram-negative and Gram-positive bacteria, as well as fungi (2–8), at seemingly limited resistance development (6). Although there is increasing evidence that AMPs influence bacteria in a multitude of ways (5), as well as a considerable diversity among the 900 AMPs presently known, bacterial wall rupture plays a key role in the bactericidal action of most AMPs. From a therapeutic perspective, AMPs should ideally rupture bacterial walls specifically, leaving (human) eukaryotic cells unaffected. Approaches for reaching this include quantitative structure-activity relationship in combination with directed amino acid modifications (9–12) and identification of selective AMPs generated through infection-related proteolysis of endogenous proteins (13–15).
From a mechanistic perspective, high AMP adsorption is a prerequisite for potent membrane disruption, which scales with the amount of AMP bound to the lipid membrane (16–19). Although high AMP adsorption can sometimes be reached by highly charged and hydrophilic AMPs, Staphylococcus aureus and other important pathogens have a relatively low electrostatic surface potential, which may be reduced or even reversed, e.g. by l-lysine modification of phosphatidylglycerol, d-alanine modification of cell wall teichoic acid, and aminoarabinose modifications in LPS, reducing AMP binding to Gram-positive and Gram-negative bacteria, respectively (6). Furthermore, electrostatic AMP binding is salt-sensitive, and bactericidal potency of such peptides at high ionic strength is limited. Given this and inspired by lipopeptides (20–23) and other amphiphilic structures (24), we identified end tagging of AMPs with hydrophobic amino acid stretches as an interesting approach to achieve high AMP adsorption and potency, also at high ionic strength and against bacteria of low electrostatic charge density. Because cholesterol should preclude membrane tag insertion (25), selectivity between bacteria and eukaryotic cells could also be expected. In the present investigation, this concept was investigated with a focus on previously identified highly charged AMPs derived from kininogen, interesting from a therapeutic perspective because of their ability to withstand infection-related proteolysis (14), another key defense and resistance mechanism of bacteria against AMPs (6).
EXPERIMENTAL PROCEDURES
Peptides
High quality peptides were synthesized by Biopeptide Co. (San Diego, CA), with the exception of LL-37, which was obtained from Innovagen AB (Lund, Sweden). The purity (>95%) of these peptides was confirmed by mass spectral analysis (MALDI-TOF Voyager). Peptides for the initial screening were from Sigma-Genosys (Sigma-Aldrich), generated by a peptide synthesis platform (PEPscreen®; Custom Peptide Libraries, Sigma Genosys) with a yield of ∼1–6 mg. MALDI-TOF mass spectrometry was performed on these peptides, and the average crude purity of the peptides was found to be 60–70%. Prior to biological testing, the peptides were diluted in H2O (5 mm stock) and stored at −20 °C. This stock solution was used for the subsequent experiments.
Microorganisms
Escherichia coli ATCC 25922, S. aureus ATCC 29213, and Candida albicans ATCC 90028 were from the Department of Clinical Bacteriology of Lund University Hospital. Additional S. aureus clinical isolates were obtained from patients with skin infections. S. aureus molecular typing was performed using ADSRRS (amplification of DNA fragments surrounding rare restriction sites) fingerprinting analysis as described previously (26).
Radial Diffusion Assay
Essentially as described earlier (27, 28), bacteria were grown to mid-logarithmic phase in 10 ml of full-strength (3% w/v) trypticase soy broth (TSB) (Becton-Dickinson). The microorganisms were then washed once with 10 mm Tris, pH 7.4. Subsequently, 4 × 106 CFU were added to 15 ml of the underlay agarose gel, consisting of 0.03% (w/v) TSB, 1% (w/v) low electroendosmosis type agarose (Sigma-Aldrich) and 0.02% (v/v) Tween 20 (Sigma-Aldrich). The underlay was poured into a Ø 144-mm Petri dish. After agarose solidification, 4-mm-diameter wells were punched, and 6 μl of peptide solution of required concentration was added to each well. The plates were incubated at 37 °C for 3 h to allow peptide diffusion. The underlay gel was then covered with 15 ml of molten overlay (6% TSB and 1% low electroendosmosis type agarose in distilled H2O). Antimicrobial activity of a peptide was visualized as a zone of clearing around each well after 18–24 h of incubation at 37 °C.
Minimal Inhibitory Concentration Assay
MIC assay was carried out by a microtiter broth dilution method as described previously (11, 29) with slight modifications. For preparation of refined media, 100 ml of Luria Bertani broth was prepared in 10 mm Tris, pH 7.4, and applied to a column packed with 40 ml of DEAE Sephacel (9013-34-7; Sigma-Aldrich). The column was previously rinsed with a volume two times the bed volume using water, 2 m NaCl, 0.1 m NaOH, water, and 70% ethanol, respectively, and finally equilibrated in 10 mm Tris (pH 7.4) before use. The medium was passed through the column twice and finally sterilized by filtration (Millex GP filter unit, 0.22 μm) and stored at 8 °C until use. For determination of MIC, the peptides were dissolved in 10 mm Tris, pH 7.4, at a concentration 10 times higher than the required range by serial dilutions from a stock solution. Ten μl of each concentration was added to each corresponding well of a 96-well microtiter plate (polypropylene; Costar Corp., Cambridge, MA). Bacteria grown overnight in 3% TSB were rinsed with Tris, pH 7.4, and diluted in refined LB medium to a concentration of ∼1 × 105 CFU/ml. Ninety μl of bacterial solution in the refined LB medium was added to each well containing the test peptides. The plate was incubated at 37 °C overnight. MIC was taken as the concentration at which >90% of growth inhibition was observed.
Protease Sensitivity Assay
Peptides (1 μg) were incubated at 37 °C with S. aureus aureolysin (0.1 μg, 25,000 units/mg), S. aureus V8 proteinase (0.1 μg, 2000 milliunits), both from BioCol μGmbH (Potsdam, Germany), or neutrophil elastase (0.4 μg, 29 units/mg; Calbiochem (La Jolla, CA)) in a total volume of 30 μl for 3 h. The materials were analyzed on 16.5% precast SDS-PAGE Tris-Tricine gels (Bio-Rad) and analyzed after staining with Coomassie Blue R-250.
MTT Assay
Sterile filtered MTT (Sigma-Aldrich) solution (5 mg/ml in PBS) was stored protected from light at −20 °C until usage. HaCaT keratinocytes, 3000 cells/well, were seeded in 96-well plates and grown in keratinocyte-SFM/BPE-rEGF medium to confluence. Keratinocyte-SFM/BPE-rEGF medium alone or keratinocyte-SFM supplemented with 20% serum was added, followed by peptide addition to 60 μm. After incubation overnight, 20 μl of the MTT solution was added to each well, and the plates were incubated for 1 h in CO2 at 37 °C. The MTT-containing medium was then removed by aspiration. The blue formazan product generated was dissolved by the addition of 100 μl of Me2SO/well, gently swirling the plates for 10 min at room temperature. The absorbance was monitored at 550 nm. The results given represent mean values from triplicate measurements.
Lactate Dehydrogenase (LDH) Assay
HaCaT keratinocytes were grown in 96-well plates (3000 cells/well) in SFM supplemented with bovine pituitary extract and recombinant EGF (BPE-rEGF) (Invitrogen) to confluency. The medium was then removed, and the peptides (at 60 μm, diluted in 200 μl of serum-free keratinocyte medium/BPE-rEGF or keratinocyte serum-free medium supplemented with 20% human serum) were added. The LDH-based TOX-7 kit (Sigma-Aldrich) was used for quantification of LDH release from the cells. The results represent mean values from triplicate measurements and are given as fractional LDH release relative to the positive control consisting of 1% Triton X-100 (yielding 100% LDH release).
Hemolysis Assay
EDTA-blood was centrifuged at 800 × g for 10 min, after which plasma and buffy coat were removed. The erythrocytes were washed three times and resuspended in PBS, pH 7.4, to get a 5% suspension. For experiments in 50% blood, EDTA blood was diluted (1:1) with PBS. The cells were incubated with end-over-end rotation for 1 h at 37 °C in the presence of peptides (60 μm). 2% Triton X-100 (Sigma-Aldrich) served as a positive control. The samples were then centrifuged at 800 × g for 10 min. Hemoglobin release was monitored by absorbance at 540 nm and is expressed as a percentage of Triton X-100-induced hemolysis. The results given represent mean values from triplicate measurements.
Transmission Electron Microscopy
S. aureus ATCC 29213 bacteria were grown in Todd-Hewitt medium at 37 °C to mid-logarithmic phase. The bacteria were washed in buffer (10 mm Tris, pH 7.4, 0.15 m NaCl, 5 mm glucose) and resuspended in the same buffer. Peptides at 30 μm were incubated with S. aureus ATCC 29213 (1 × 108 bacteria) for 2 h in a total volume of 50 ml in 10 mm Tris, pH 7.4, 5 mm glucose. Ten μl of the bacterial suspensions were then adsorbed onto carbon-coated copper grids for 1 min, washed briefly with two drops of water, and negatively stained by two drops of 0.75% uranyl formate. The grids were rendered hydrophilic by glow discharge in low pressure residual air. Specimens were observed in a Jeol JEM 1230 electron microscope operated at 60 kV accelerating voltage, and the images were recorded with a Gatan Multiscan 791 CCD camera.
Fluorescence Microscopy
For monitoring of membrane permeabilization using the impermeant probe fluorescein isothiocyanate (Sigma-Aldrich), S. aureus ATCC 29213 bacteria were grown to mid-logarithmic phase in TSB medium. The bacteria were washed and resuspended in buffer (10 mm Tris, pH 7.4, 0.15 m NaCl, 5 mm glucose) to yield a suspension of 1 × 107 CFU/ml. 100 μl of the bacterial suspension was incubated with 30 μm of the respective peptides at 30 °C for 30 min. The microorganisms were then immobilized on poly-l-lysine-coated glass slides by incubation for 45 min at 30 °C, followed by addition onto the slides of 200 μl of fluorescein isothiocyanate (6 μg/ml) in buffer and incubation for 30 min at 30 °C. The slides were washed, and bacteria were fixed by incubation, first on ice for 15 min and then in room temperature for 45 min in 4% paraformaldehyde. The glass slides were subsequently mounted on slides using Prolong Gold antifade reagent mounting medium (Invitrogen). For fluorescence analysis, bacteria were visualized using a Nikon Eclipse TE300 (Nikon) inverted fluorescence microscope equipped with a Hamamatsu C4742-95 cooled CCD camera (Hamamatsu) and a Plan Apochromat ×100 objective (Olympus). Differential interference contrast (Nomarski) imaging was used for visualization of the microbes themselves.
Viable Count Analysis
S. aureus ATCC 29213 bacteria were grown to mid-logarithmic phase in Todd-Hewitt medium. Following this, bacteria (50 ml; 2 × 106 CFU/ml) were incubated, at 37 °C for 2 h, with GKH17, HKH17, their tagged variant peptides, or LL-37 (at 0.1, 0.5, 1.0, 5, 10, 50, and 100 μm) in 10 mm Tris, 0.15 m NaCl, with or without 20% human citrate plasma. To quantify bactericidal activity, serial dilutions of the incubation mixtures were plated on Todd-Hewitt agar, followed by incubation at 37 °C overnight, and the number of CFU was determined. 100% survival was defined as total survival of bacteria in the same buffer and under the same condition in the absence of peptide.
Antibacterial Effects ex Vivo and in Vivo
For evaluation of antibacterial effects on pig skin in vivo, female SPF pigs (cross-bred of Danish Country and Yorkshire, 30–35 kg) were used. On either side of the back, 10 treatment areas of 40 × 40 mm were marked. For inoculation, S. aureus ATCC 29213 were inoculated on each area. Bacteria (1 × 106 CFU) were diluted in 50 μl of 10 mm Tris, pH 7.4. This inoculum was distributed by a sterile spatula over the specified skin surface. Approximately 15 min after the inoculation, the GKH17-WWW-containing polyethylene glycol/water formulation (PEG400/PEG3350/water 54/36/10 w/w) or control formulation without peptide was applied. The areas were then covered with a polyurethane film dressing (Tegaderm®) (3M, St. Paul, MN) and fixed by Fixomul® (Smith & Nephew Wound Management, Hull, UK). The dressings and the Fixomul® were retained by a Vetflex® bandage. Four hours later, the animal was anesthetized again and sacrificed by pentobarbital injection. CFU of S. aureus was determined according to the Kligman method (30). Sterile plastic wells were placed in the center of the inoculated areas, and 1 ml of 10 mm phosphate buffer, pH 7.4, 0.05% Triton X-100 was added. The skin was scrubbed back and forth for 25 cycles using a sterile blunted, stainless-steel spatula. The liquid was recovered, and the procedure was repeated once. The two samples were pooled, and CFU was determined after appropriate dilutions. For evaluating AMPs ex vivo, a pig skin model was used as described previously (31) but with modifications. Defatted pig hides were first washed with water and then 70% ethanol. They were then destubbed with disposable razors and 8 × 8-cm pieces were cut, sealed in plastic wrap, and frozen at −20 °C. Before use, the skin samples were thawed and then washed with ethanol (70%) and water. To separate the inoculation areas, sterilized tubings (polyethylene, quality 9.6 m, Nalgene® VWR 228-0170) were cut into ∼10-mm lengths and glued onto the skin samples (cyanoacrylate glue; Henkel, Düsseldorf, Germany). The skin was infected by adding 1 × 106 CFU of an overnight culture of S. aureus ATCC 29213 in a total volume of 10 μl. After an incubation time of 4 h at 37 °C, peptide-containing solutions (0.5 and 1 mm in 100 μl) or the above formulations of GKH17-WWW and as well as ∼25 mg of control formulation (PEG400/PEG3350/water (54/36/10 w/w) without peptide) were applied. (The formulation without peptide had no bactericidal effect on either S. aureus or E. coli.) Bacterial sampling was then performed after an incubation time of 2 h by washing the reaction chambers twice with 250 μl of 10 mm Na2HPO4, pH 7.4, 0.05% Triton X-100, supplemented with 0.1% dextran sulfate (molecular mass, 500 kDa; Sigma-Aldrich), added to block peptide activity during sampling.
Liposome Preparation and Leakage Assay
The liposomes investigated were either zwitterionic (DOPC/cholesterol 60/40 mol/mol or DOPC without cholesterol) or anionic (DOPE/DOPG 75/25 mol/mol). DOPG (monosodium salt), DOPC, and DOPE were all from Avanti Polar Lipids and of >99% purity, whereas cholesterol (>99% purity) was from Sigma-Aldrich. Because of the long, symmetric, and unsaturated acyl chains of these phospholipids, several methodological advantages are reached. In particular, membrane cohesion is good, which facilitates very stable, unilamellar, and largely defect-free liposomes (observed from cryo-TEM) and well defined supported lipid bilayers (observed by ellipsometry and atomic force microscopy), allowing detailed values on leakage and adsorption density to be obtained. The lipid mixtures were dissolved in chloroform, after which solvent was removed by evaporation under vacuum overnight. Subsequently, 10 mm Tris buffer, pH 7.4, was added together with 0.1 m carboxyfluorescein (CF) (Sigma). After hydration, the lipid mixture was subjected to eight freeze-thaw cycles consisting of freezing in liquid nitrogen and heating to 60 °C. Unilamellar liposomes of about Ø140 nm were generated by multiple extrusions through polycarbonate filters (pore size, 100 nm) mounted in a LipoFast miniextruder (Avestin, Ottawa, Canada) at 22 °C. Untrapped CF was removed by two subsequent gel filtrations (Sephadex G-50; GE Healthcare) at 22 °C, with Tris buffer as eluent. CF release from the liposomes was determined by monitoring the emitted fluorescence at 520 nm from a liposome dispersion (10 μm lipid in 10 mm Tris, pH 7.4). An absolute leakage scale was obtained by disrupting the liposomes at the end of each experiment through the addition of 0.8 mm Triton X-100 (Sigma-Aldrich). A SPEX fluorolog 1650 0.22-m double spectrometer (SPEX Industries) was used for the liposome leakage assay and also for monitoring Trp absorption spectra of GKH17-WWW in Tris buffer in the absence and the presence of liposomes under the conditions described above. The measurements were performed in triplicate at 37 °C.
Dynamic Light Scattering
The mean hydrodynamic diameter of GKH17 and GKH17-WWW was determined at a peptide concentration of 10 μm in Tris buffer, pH 7.4, by dynamic light scattering at a 173° scattering angle, using a Zetasizer Nano ZS (Malvern Instruments, Malvern, UK).
CD Spectroscopy
CD spectra were measured by a Jasco J-810 spectropolarimeter. The measurements were performed in triplicate at 37 °C in a 10-mm quartz cuvette under stirring with a peptide concentration of 10 μm. The effect on peptide secondary structure of liposomes at a lipid concentration of 100 μm was monitored in the range 200–250 nm. To account for instrumental differences between measurements, the background value (detected at 250 nm, where no peptide signal is present) was subtracted. Signals from the bulk solution were also corrected for.
Ellipsometry
Peptide adsorption to supported lipid bilayers was studied in situ by null ellipsometry, using an Optrel Multiskop (Optrel, Kleinmachnow, Germany) equipped with a 100 milliwatt argon laser. All of the measurements were carried out at 532 nm and an angle of incidence of 67.66° in a 5-ml cuvette while stirring (300 rpm). Both the principles of null ellipsometry and the procedures used have been described extensively before (32). In brief, by monitoring the change in the state of polarization of light reflected at a surface in the absence and the presence of an adsorbed layer, the mean refractive index (n) and thickness (d) of the adsorbed layer can be obtained. From the thickness and refractive index the adsorbed amount (Γ) was calculated according to Ref. 33 by the following equation,
 |
where dn/dc is the refractive index increment (0.154 cm3/g) (34), and n0 is the refractive index of the bulk solution. Corrections were routinely done for changes in bulk refractive index caused by changes in temperature and excess electrolyte concentration.
Zwitterionic bilayers were deposited by co-adsorption from a mixed micellar solution containing 60/40 mol/mol DOPC/cholesterol and n-dodecyl-β-d-maltoside (≥98% purity; Sigma-Aldrich), as described in detail previously (34). In brief, the mixed micellar solution was formed by the addition of 19 mm n-dodecyl-β-d-maltoside in water to DOPC/cholesterol dry lipid films, followed by stirring overnight, yielding a solution containing 97.3 mol% n-dodecyl-β-d-maltoside, 1.6 mol% DOPC and 1.1 mol% cholesterol. This micellar solution was added to the cuvette at 25 °C, and the following adsorption was monitored as a function of time. When adsorption had stabilized, rinsing with Milli-Q water at 5 ml/min was initiated to remove mixed micelles from solution and surfactant from the substrate. By repeating this procedure and subsequently lowering the concentration of the micellar solution, stable and densely packed bilayers were formed, with structural characteristics similar to those of bulk lamellar structures of the lipids (34). Because subbilayer and patchy adsorption resulted from the above mixed micelle approach in the case of the anionic lipid mixture, supported lipid bilayers were generated from liposome adsorption in this case. DOPE/DOPG liposomes (75/25 mol/mol) were prepared as described above, but the dried lipid films resuspended in Tris buffer only with no CF present. To avoid adsorption of peptide directly at the silica substrate through any defects of the supported lipid layer, poly-l-lysine (molecular mass, 170 kDa; Sigma-Aldrich) was preadsorbed from water prior to lipid addition to an amount of 0.045 ± 0.01 mg/m2, followed by removal of nonadsorbed poly-l-lysine by rinsing with water at 5 ml/min for 20 min. Water in the cuvette was then replaced by buffer containing also 150 mm NaCl, which was followed by addition of liposomes in buffer at a lipid concentration of 20 μm and subsequently by rinsing with buffer (5 ml/min for 15 min) when the liposome adsorption had stabilized. The final layer formed had structural characteristics (thickness, 40 ± 10 Å; mean refractive index, 1.47 ± 0.026) similar to those of the zwitterionic bilayers, which suggests that a layer fairly close to a complete bilayer is formed. Again, the bilayer build-up was performed at 25 °C. After lipid bilayer formation, temperature was raised, and the cuvette content was replaced by buffer at a rate of 5 ml/min over a period of 30 min. After stabilization for 40 min, the peptide was added to a concentration of 0.01 μm, followed by three subsequent peptide additions to 0.1, 0.5, and 1 μm, in all cases monitoring the adsorption for 1 h. All of the measurements were made at least in duplicate.
Slot-Blot Assay
LPS binding ability of the peptides were examined by slot-blot assay. Peptides (1, 2, and 5 μg) were bound to nitrocellulose membrane (Hybond-C; GE Healthcare), presoaked in PBS, by vacuum. The membranes were then blocked by 2 weight % bovine serum albumin in PBS, pH 7.4, for 1 h at room temperature and subsequently incubated with 125I-labeled LPS (40 μg/ml; 0.13 × 106 cpm/μg) for 1 h at room temperature in PBS. After LPS binding, the membranes were washed three times, 10 min each time, in PBS and visualized for radioactivity on Bas 2000 radioimaging system (Fuji).
Statistics
The values are reported as the means ± standard deviation of the means. To determine significance, analysis of variance with ANOVA (SigmaStat; SPSS Inc., Chicago, IL), followed by post hoc testing using the Holm-Sidak method, or Student's t test, were used as indicated in the figure legends, where n denotes the number of independent experiments. Significance was accepted at p < 0.05.
RESULTS
Microbicidal Potency and Proteolytic Stability
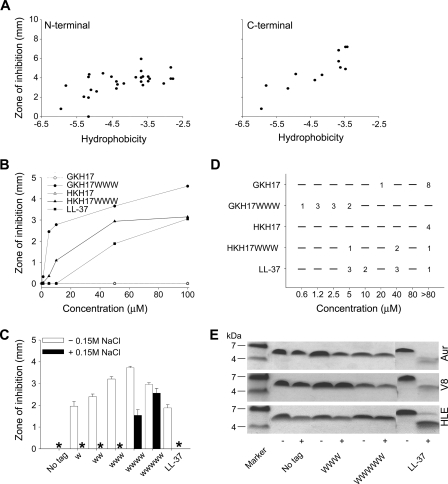
Focusing on two kininogen-derived peptides, GKHKNKGKKNGKHNGWK (GKH17) and HKHGHGHGKHKNKGKKN (HKH17) (14), hydrophobic end tags were investigated concerning effects of the chemical nature, length, and position of the tags on bactericidal and cytotoxic properties of these peptides. As can be seen in Fig. 1A, tagging of GKH17 in both C and N termini resulted in enhanced bactericidal effect against Gram-positive S. aureus, an effect that increased with the overall hydrophobicity (35–37) of the tag. Similar effects were obtained with Gram-negative E. coli, as well as the fungus C. albicans (supplemental Table S1). Trp and Phe tags emerge as particularly potent, although a spread can be noted, both between microbial isolates and between N- and C-terminal tagging (supplemental Fig. S1). Although the initial screening reported in Fig. 1A was done in a PEPscreen format, all of the following experiments were performed for a smaller number of peptides of high (>95%) purity. As above, aliphatic LLL modifications resulted in substantially weaker antimicrobial effect than the corresponding WWW modifications (results not shown). Additionally, the bactericidal effect, given by the size of the inhibition zone, increased with the number of Trp residues in the hydrophobic tag and was significantly larger than that of the benchmark peptide LL-37 even for a tag consisting of two Trps only. For tags consisting of three Trps or more, peptides significantly more potent than LL-37 were obtained (Fig. 1, B and C). Trp tags also yielded increased activity at physiological salt conditions, particularly noted for the longer tag lengths (Fig. 1C and supplemental Fig. S1). As shown in Fig. 1B, the potentiating effect of the hydrophobic tag can be observed at all peptide concentrations. Extending these investigations also to MIC for a number of S. aureus (both ATCC and clinical) isolates, Fig. 1D shows that WWW tagging of GKH17 and HKH17 resulted in a broad reduction in MIC values. This was particularly noted for GKH17-WWW, demonstrating MICs significantly lower than those for the benchmark peptide LL-37. Quantitatively, untagged GKH17 has a MIC of >80 μm for eight of the nine S. aureus isolates investigated, and of 20 μm for the remaining isolate. For GKH17-WWW, on the other hand, one of these clinical isolates shows a MIC of 0.6 μm, three isolates show a MIC of 1.2 μm, three isolates show a MIC of 2.5 μm, and two isolates show a MIC of 5 μm. As can be seen in Fig. 1E, both GKH17 and GKH17-WWW displayed good stability against proteolysis by human leukocyte elastase, as well as staphylococcal aureolysin and V8 proteinase. Similar results were obtained for HKH17 (results not shown). In contrast, and in agreement with previous findings (38), LL-37 undergoes substantial degradation by all these enzymes. From this it is clear that the stability displayed by GKH17 and HKH17 against proteolytic degradation by HLE, V8, and aureolysin is maintained also after WWW tagging.
FIGURE 1.
Antibacterial activities and protease effects of peptides. A, antimicrobial activity as assessed by RDA against S. aureus of peptides with either N-terminal (left panel) or C-terminal (right panel) hydrophobic modifications. For determination of activity, bacteria (4 × 106 CFU/ml) were inoculated in 0.1% TSB-agarose gel, and 4-mm wells were punched. In each well, 6 μl of peptide (at 100 μm) was loaded. The zones of clearance correspond to the inhibitory effect of each peptide after incubation at 37 °C for 18–24 h (mean values, n = 3). The activity is plotted versus peptide hydrophobicity (Kyte and Doolittle scale). (The correlation coefficient (R) and the coefficient of determination (R2) in Fig. 1A are 0.583 and 0.340, respectively, for the N-terminal modifications, and 0.881 and 0.776, respectively, for the C-terminal modifications.) B, dose-dependent activity of the selected peptides GKH17 and HKH17 and WWW-tagged variants against S. aureus (4 × 106 CFU) in RDA. (The difference between the original peptides GKH17, HKH17, and their tagged variants is significant (p < 0.05, two-way ANOVA).) C, effect of tag length on GKH17 peptide activity (at 50 μm) against S. aureus ATCC 29213 as assessed by RDA in the presence and absence of 0.15 m NaCl. * denotes no detectable clearance zone. (In absence of salt the difference between all tagged peptides, and no tag is significant. In presence of 0.15 m NaCl, the difference between the WWW and WWWWW peptides versus no tag is significant (p < 0.05, one-way ANOVA).) D, MIC of indicated peptides against different S. aureus isolates. E, protease sensitivity of peptides. The indicated peptides were incubated with (+) or without (−) the S. aureus enzymes aureolysin (Aur), V8 proteinase (V8), or human leukocyte elastase (HLE) and analyzed by SDS-PAGE (16.5% Tris-Tricine gels).
Eukaryotic Cell Responses
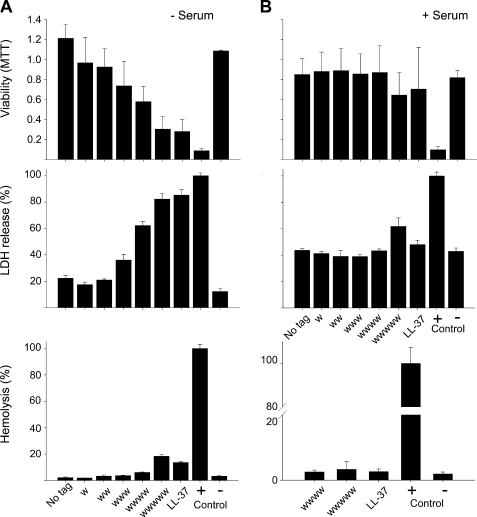
Peptide-induced eukaryotic cell permeabilization was investigated with three different assays to get a broader picture on the potential toxic effects of these peptides. As can be seen in Fig. 2, hydrophobic tagging increased peptide permeabilization of both erythrocytes and human keratinocytes (HaCat) somewhat for short end tags, an effect increasing substantially with the length of the hydrophobic tag. Quantitatively, the permeabilization in buffer was quite low for tags up to WWW and significantly lower than that of endogenous LL-37. For the longest tag investigated, WWWWW, permeabilization was comparable with or lower than that of LL-37. The toxicity was further reduced and reached very low levels for both the tagged peptides and LL-37 in the presence of serum.
FIGURE 2.
Activities of peptides against eukaryotic cells. A, effects of peptides on HaCaT cells and erythrocytes in absence of human serum. The MTT assay (top panel) was used to measure viability of HaCaT keratinocytes in the presence of GKH17 peptides with variable Trp tagging. In the assay, MTT is modified into a dye, blue formazan, by enzymes associated with metabolic activity. The absorbance of the dye was measured at 550 nm. The cell permeabilizing effects of the indicated peptides (middle panel) were measured by the LDH-based TOX-7 kit. Hemolytic effects (bottom panel) of the indicated GKH17 variant peptides were investigated, and the corresponding data for LL-37 are included for comparison. The cells were incubated with the peptides at 60 μm, and 2% Triton X-100 served as positive control. The absorbance of hemoglobin release was measured at 540 nm and is expressed as a percentage of Triton X-100-induced hemolysis (note scale of y axis). MTT, the difference between the WWW and longer variant peptides versus no tag is significant (p < 0.05, one-way ANOVA). LDH, the difference between the WWW and longer variant peptides versus no tag is significant (p < 0.05, one-way ANOVA). Hemolysis, the difference between the WWWWW variant versus no tag is significant (p < 0.05, one-way ANOVA). B, effects of GKH17 and Trp-modified variants in the presence of human serum. Identical experiments as in A were performed in presence of human 20% serum. For assessment of hemolysis in presence of serum, only those peptides displaying significant hemolytic activities in absence of serum were analyzed. MTT, there is no significant difference between the peptide variants. LDH, the difference between the WWWWW variant peptide versus no tag is significant (p < 0.05, one-way ANOVA). Hemolysis, there is no significant difference between the peptide variants.
Permeabilization of Bacteria and Liposomes
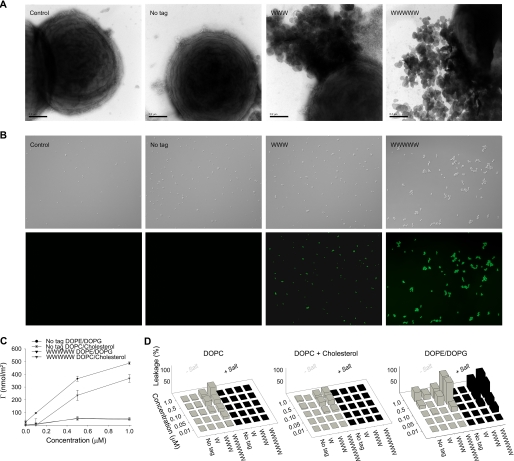
To learn more about the mechanism behind these observations, a series of experiments was performed. As can be seen in Fig. 3 (A and B), tagging promoted binding of GKH17 to bacteria and subsequent permeabilization. Supplemental Fig. S2 shows that the peptide remained largely disordered on membrane binding and that Trp tags are located in the proximity of the membrane polar headgroup region. Although permeabilization by nontagged GKH17 was essentially nonobservable, it increased strongly with the length of the hydrophobic tag (Fig. 3B). In parallel, tagged GKH17 caused release of intracellular material of bacteria, similar to LL-37, quantitatively increasing with the length of the hydrophobic tag (Fig. 3A). In agreement with these findings, the binding of GKH17-WWW was higher than that of GKH17 at model DOPE/DOPE lipid membranes (Fig. 3C), and peptide-induced leakage from liposomes prepared from the same lipids was dramatically enhanced for GKH17-WWW compared with GKH17 (Fig. 3D). Also in parallel to bacterial killing, salt resistance of the tagged peptides with respect to liposome leakage induction increased with the length of the hydrophobic tag. In contrast to the anionic and cholesterol-free DOPE/DOPG (“bacterial”) membrane, that mimicking eukaryotic cell membranes (DOPC/cholesterol) displayed lower peptide binding, also for the Trp-tagged peptides, as well as lower liposome leakage induction. In addition to lipid membrane effects, Trp tagging facilitated binding to LPS (supplemental Fig. S2), an effect that can be completely reversed through the addition of the anionic competitor heparin (results not shown).
FIGURE 3.
Peptide-mediated permeabilization of S. aureus bacteria and liposomes. A, S. aureus ATCC 29213 was incubated with GKH17 and the indicated hydrophobically modified variants (all at 30 μm) and analyzed with electron microscopy (negative staining). The scale bars correspond to 0.5 μm. B, bacterial viability after incubation with peptides. S. aureus ATCC 29213 was incubated with GKH17 or Trp-tagged GKH17 peptides (all at 30 μm) in buffer at physiological salt (0.15 m NaCl) for 2 h at 37 °C. The upper images in each row are Nomarski Differential Interference Contrast images, whereas the lower images show fluorescein isothiocyanate fluorescence of bacteria. C, peptide adsorption to supported lipid bilayers composed of DOPC/cholesterol (60/40 mol/mol; zwitterionic) and DOPE/DOPG (75/25 mol/mol; anionic). (There is statistical difference between control GKH17 and GKH17-WWWWW, as well as between DOPE/DOPG and DOPC/cholesterol for GKH17-WWWWW (p < 0.05, Student's t test).) D, effects of peptides on liposomes in the presence and absence of 0.15 m NaCl. The membrane-permeabilizing effect was recorded by measuring fluorescence release of carboxyfluorescein from liposomes. The left panel shows DOPC (zwitterionic) liposomes, the center panel shows DOPC/cholesterol (60/40 mol/mol; zwitterionic) liposomes, and the right panel shows DOPE/DOPG (75/25 mol/mol; anionic) liposomes.
Results ex Vivo and in Vivo in Porcine Skin Infection Models
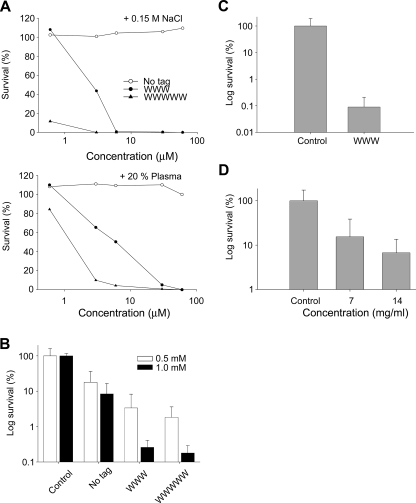
To demonstrate the biological relevance of these effects, a series of experiments were performed. Fig. 4A shows results on bacterial killing as a function of peptide concentration, both at physiological ionic strength and in the presence of human plasma. As demonstrated, the potentiating effect of the hydrophobic tag remains under these conditions, with GKH17-WWW and GKH17-WWWWW being effective against S. aureus also in the presence of plasma. Furthermore, in an ex vivo porcine skin model, both GKH17-WWW and GKH17-WWWWW showed efficient dose-dependent reduction of S. aureus, with a slightly more efficient bactericidal effect demonstrated for the latter peptide (Fig. 4, B and C). In parallel, efficient and dose-dependent reduction of S. aureus was demonstrated in vivo using a porcine skin infection model, although the presence of competing factors, e.g. related to bacterial regrowth caused by incomplete eradication, and the presence of scavengers of peptide action necessitate higher peptide concentrations than those used in vitro (Fig. 4D).
FIGURE 4.
Activities of peptides at physiological conditions, ex vivo, and in vivo. A, antibacterial effects of the indicated peptides against S. aureus in viable count assay. 2 × 106 CFU/ml of bacteria were incubated in 50 μl with the indicated peptides at 0.06–60 μm in 10 mm Tris, pH 7.4, 0.15 m NaCl (upper panel) or the same buffer containing 20% human plasma (lower panel). (The difference between the original peptide and its tagged variants is significant, p < 0.05, two-way ANOVA). B, activities of peptides in an ex vivo skin infection model. Pig skin was inoculated with S. aureus, and peptides at the indicated concentrations were added after an incubation time of 4 h. Bacteria were collected, and CFU was determined (mean values are presented; n = 6). Note the logarithmic scale on the y axis. (There is a statistically significant difference (p < 0.001 two-way ANOVA) between the tagged peptides versus control as well as the native peptide (no tag) at the two doses.) C, inoculation similar as in B, but pig skin was treated with GKH17 and GKH17-WWW (2.8 mm) in a polyethylene glycol formulation (PEG400/PEG3350/water 54/36/10). (There is a statistically significant difference between control treatment and WWW (p < 0.001, Student's t test).) D, treatment of S. aureus infected pig skin in vivo. Treatment with GKH17-WWW at two concentrations in a polyethylene glycol formulation (PEG400/PEG3350/water 54/36/10) and with formulation in the absence of peptide (2.8 and 5.6 mm). (There is a statistically significant difference between control treatment and the two concentrations of WWW (p < 0.05, one-way ANOVA).)
Concept Generalization
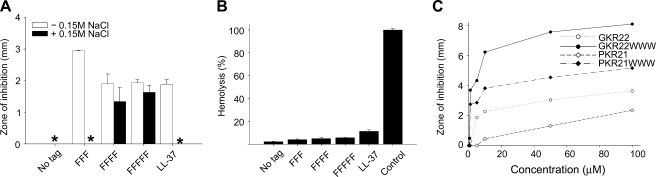
Although much of the results presented above have focused on Trp tagging of GKH17, the concept is applicable also to other hydrophobic tags and other polar and highly charged AMPs. Thus, results similar to those for Trp were obtained with Phe tags, showing increasing potency and salt resistance with increasing tag length (Fig. 5A and supplemental Fig. S1). As with Trp, Phe-tagged GKH17 displays very limited toxicity, lower than that of LL-37, particularly in the presence of serum (Fig. 5B and supplemental Fig. S1). Additionally, Fig. 5C and supplemental Fig. S1 show that a similar bactericidal effects can be obtained with other peptides as well, demonstrated for GKR22 (GKRKKKGKGLGKKRDPCLRKYK) and PKR 21 (PKRKKKGGKNGKNRRNRKKKN), originating from HB-EGF and amphiregulin, respectively (15). Similar results were found also for QTP22 (QPTRRPRPGTGPGRRPRPRPRP), derived from PRELP (39), for which QTP22-WWW displayed much higher bactericidal potency than the untagged QTP22 (results not shown). In contrast, hydrophobic end tagging is not applicable to nonbactericidal peptides. As can be seen in supplemental Fig. S3, neither the untagged S10 (decaserine) and D10 (decaaspartic acid) homopeptides (uncharged and negatively charged, respectively) nor their tagged WWW variants displayed any bactericidal effect on E. coli or S. aureus nor hemolysis above that of the negative control. Neither does the WWW tripeptide tag in the absence of cationic template peptide display any bactericidal effect against E. coli or S. aureus or cause hemolysis above that of the negative control. Thus, it is the combined effect of Trp tagging and the bactericidal properties of the template AMP that underlies activity boosting.
FIGURE 5.
Generalization of the concept of end tagging by hydrophobic amino acid stretches. A, antimicrobial activity as assessed by RDA against S. aureus of the indicated GKH17 peptides (at 50 μm) in the absence (open bars) or presence (black bars) of 0.15 m NaCl (mean values are presented, n = 3). * denotes no detectable clearance zone. B, analysis of hemolytic effects of peptides and comparison with LL-37. The cells were incubated with peptides at 60 μm, and 2% Triton X-100 served as positive control. The absorbance of hemoglobin release was measured at 550 nm and is expressed as a percentage of Triton X-100-induced hemolysis (mean values are presented, n = 3). C, hydrophobic modification of antimicrobial peptides from HB-EGF (GKR22) and amphiregulin (PKR21) increases their bactericidal activity. Antimicrobial activity of the indicated peptides was assessed by RDA against S. aureus at the indicated concentration (mean values are presented, n = 3).
Supplemental Data
PEPscreen RDA data on effect of tag composition and location, RDA data on effect of tag length for E. coli, data on cell toxicity for Phe-tagged peptides, RDA data on GKR22 (-WWW) and PKR21 (-WWW) for E. coli, CD spectra for GKH17 and its Trp-tagged variants, Trp fluorescence spectra, dynamic light scattering data, as well as peptide structures, hydrophobicities, and tabulated RDA data for the initial peptide screening, are available as supplemental data.
DISCUSSION
This report presents a powerful and generally applicable, practical, and tunable platform based on hydrophobic end modifications of AMPs. Although such modifications can be designed in a number of ways, end tagging by hydrophobic amino acid stretches is an attractive alternative because it allows the primary AMP sequence to be retained at the same time as efficient but selective membrane anchoring is achieved. End tagging also does not affect proteolytic stability of the tagged AMPs detrimentally, a factor of importance for bactericidal potency on S. aureus, Pseudomonas aeruginosa, and other bacteria that secrete AMP-degrading proteases (6). A number of mechanisms by which AMPs induce membrane defects have been observed. For some peptides, e.g. melittin, alamethicin, magainin 2, and gramicidin A (5, 7, 40, 41), transmembrane structures have been reported, sometimes coupled to peptide self-assembly, although there is some uncertainty on some of these systems (42). Not rarely, such structures are associated with an ordered secondary structure in the membrane-bound peptide. For disordered and highly charged peptides, including the ones studied here, membrane disruption is obtained by other mechanisms, e.g. induction of a negative curvature strain, membrane thinning, or local packing defects associated with peptide localization primarily in the phospholipid polar headgroup region (5, 16–19, 43). For the latter type of AMPs, defect formation scales with the amount of peptide adsorbed at the lipid membrane, hence reaching a high peptide adsorption at the membrane is a prerequisite for AMP potency (16–19). For mechanisms based on peptide adsorption in the proximity of the polar headgroup region of the phospholipid membrane, a high peptide net charge facilitates membrane rupture. Efficient AMPs fulfill both of these criteria, so that a high concentration of electric charges is located at the membrane interface. Although electrostatic interactions provide a driving force for adsorption of positively charged AMPs to highly negatively charged bacterial membranes, these are screened at high ionic strength, resulting in partial or complete loss in bactericidal capacity, e.g. at physiological conditions. Given this, and the ability of S. aureus and other important pathogens to reduce their surface charge density, hydrophobic end tagging of AMPs may constitute a strategy for improving bactericidal potency of AMPs. Although a number of hydrophobic amino acids may be used as end tags, Trp- and Phe-based ones seem to be particularly potent. These bulky and polarizable residues have an affinity to interfaces and are frequently located in the proximity of the polar headgroup region in phospholipid membranes (44–49). In the case of antimicrobial peptides, examples of this have been provided, e.g. by Glukhov et al. (44), who found sequence-dependent penetration of KKKKKKAAXAAWAAXAA (X being Trp or Phe) of 2.5–8 Å. Similarly, Li et al. (46) investigated aurein 1.2 analogs and found the Phe residues in these peptides to penetrate 2–5 Å below the polar headgroup region. Through this interaction with the phospholipid membrane, Trp/Phe residues are able to insert into the membrane, acting as an anchor for the peptide (48, 50) and resulting in increased bactericidal effects (49, 51, 52) and salt resistance (48). In contrast to these previous investigations, which report on position-specific single tryptophan substitutions in AMPs, however, the present work describes a generally applicable approach for boosting potency of essentially all (highly charged) AMPs without interfering with the AMP sequence. Thus, various hydrophobic end tags can be used (Trp and Phe being most potent), resulting in efficiency boosting, which is largely independent of AMP sequence, whereas the end tags themselves, or end-tagged non-AMPs, do not display any bactericidal (nor toxic) effects. In contrast, previous results have shown that peptides based on overall hydrophobic interactions are less selective between bacterial and eukaryotic lipid membranes, resulting in enhanced cytotoxicity of hydrophobic AMPs. For example, peptides composed of Leu and Lys only, with a high hydrophobicity/charge ratio (e.g. KLLKLLLKLLKLLLKLLLKLLK) are even more hemolytic than bee venom melittin (4). In comparison, the peptide end tags investigated here contribute to AMP selectivity between bacterial and eukaryotic membranes. Thus, bacterial membranes are void of cholesterol and rich in anionic phospholipids such as phosphatidyl glycerol, diphosphatidylglycerol, and phosphatidyl serine, whereas eukaryotic membranes contain cholesterol and are dominated by zwitterionic phospholipids (e.g. phosphatidyl choline). (Although minute lipid component exchange between bacteria and eukaryotic cells in contact during infection cannot be completely excluded, cholesterol content in bacteria membranes is expected to be negligible also under these circumstances.) For a hydrophobic residue to be able to penetrate into the phospholipid membrane, it must overcome the cohesive energy of the latter (53). As cholesterol condenses the phospholipid bilayer and increases its elasticity drastically (25), end-tagged peptides become sensitive to cholesterol. Particularly for bulky groups such as Trp and Phe, which tend to locate in an orientation parallel to the lipid membrane surface and thus require substantial area expansion (46), tag insertion into membranes containing cholesterol becomes an energetically costly process. Consequently, the adsorption of tagged peptides is low to cholesterol-containing membranes, as is membrane-induced leakage induction of the corresponding liposomes. This difference in membrane interactions in the presence of absence of cholesterol, together with that caused by the charge difference between anionic (ζ ≈ −30 mV) and zwitterionic (ζ ≈ −10 mV) (17) membranes, most likely contributes to the selectivity observed with the presently investigated peptides. For Gram-negative bacteria, this phospholipid membrane selectivity is probably accompanied by selectivity originating from LPS binding, which is significantly enhanced through the Trp tagging (supplemental Fig. S2).
Clearly, the end-tagged peptides resemble lipopeptides (20–23). The latter consist of a linear or a cyclic peptide sequence of either positive or negative charge, with a hydrophobic moiety, e.g. a fatty acid, covalently attached, frequently to the N terminus of the peptide. Lipopeptides, such as polymyxin B and colistin, are potent against particularly Gram-negative bacteria, although somewhat less so against Gram-positive ones. In parallel, however, these peptides display substantial toxicity, which has restricted their use to local applications, and to severe indications for which other antibiotics are ineffective, e.g. multiresistant P. aeruginosa infections in cystic fibrosis (20). As for antimicrobial peptides, one of the main targets of lipopeptides is the bacterial membrane(s). Through insertion into the membrane (in the case of daptomycin mediated by Ca2+ ions (21)), lipopeptides introduce defects, leading to membrane depolarization, reduced ability for ATP synthesis and many other effects (20). Particularly closely related to AMPs end-tagged by hydrophobic amino acids stretches are previously investigated cationic peptides conjugated to cholesterol or fatty acids (20–23). In analogy to the present findings, elongation of the acyl chain in such lipopeptides resulted in increased membrane disruption in model PG/PE membranes, as well as activity against bacteria and fungi (20). This may be related to an increased membrane insertion capacity for longer acyl moieties but may also be an effect of the longer acyl chains promoting lipopeptide oligomerization in a manner similar to that reported by Avrahami and Shai (22). Interestingly, oligomerization seems to play a key role for these lipopeptides (as for negatively charged daptomycin (21)), whereas in our case, the Trp fluorescence spectra show no sign of aggregation for WWW-modified peptides (supplemental Fig. S2). Thus, the high λmax obtained (360 nm) shows that tryptophan is in contact with the ambient aqueous buffer solution (54). Micelles and other hydrophobically driven self-assemblies, on the other hand, are characterized by very low water content in the self-assembly interior by thermodynamic necessity (24). In addition, dynamic light scattering of GKH17-WWW shows a narrow peak corresponding to a hydrodynamic diameter of 2 nm, similar to that for untagged GKH17 (supplemental Fig. S2). Together, these results show that possible self-assemblies formed in this system (if any) are very small. Thus, aggregation seems not to be required for membrane rupture for these peptides. In analogy to surfactants containing aromatic tails (24), the bulky and polarizable nature of Trp and Phe is expected to preclude self-assembly, resulting in low aggregation numbers, high critical aggregation concentrations, and fast oligomer dissociation kinetics, limiting the relative importance of the aggregated state for membrane insertion and defect formation. Additionally, opposite to several of the previously investigated lipopeptides (e.g. magainin variants), which display helix induction both on interaction with phospholipid membranes and on conjugation of the peptide to the fatty acid moiety, secondary structure induction seems to play a limited role in the action of the presently investigated peptides. Thus, Trp tagging does not induce any ordered secondary structure in GKH17, both the tagged and the nontagged peptides remaining disordered both in bulk solution and on interaction with anionic liposomes.
As demonstrated, end tagging by hydrophobic amino acid stretches is a facile and flexible approach of general applicability, by which AMP boosting is obtained for a range of hydrophobic amino acids. It is also applicable for a spectrum of microorganisms, here demonstrated for Gram-positive and Gram-negative bacteria, as well as for fungi. Through tag composition and/or length, potency and toxicity can be tuned, e.g. depending on whether the peptide is to be applied in an environment containing serum or not or depending on the relative need for microbicidal potency and limited toxicity. This flexibility is attractive from a therapeutic perspective, because it allows AMPs to be tagged to fit the conditions of the indication at hand. Particularly for AMPs not sensitive to infection-related proteolysis, the finding that hydrophobic tagging may be achieved without affecting stability against proteolytic degradation also opens up applications characterized by high proteolytic activity, such as infected wounds, eye infections, and cystic fibrosis. Importantly, hydrophobic tagging may be applied to a broad range of AMPs, particularly polar and highly charged ones. In fact, the approach may possibly also be employed to enhance biological peptide activities in a broader perspective. Although dealing with receptor interactions rather than antimicrobial effects, Ember et al. (55) found that hydrophobic tagging increased the biological potency of short peptide stretches originating from complement factor 3a. In line with the present findings, peptide potency increased with the number of terminal Trp residues, Trp being more efficient than Ile in increasing peptide biological activity. Although membrane interactions played a role, Trp tagging notably promoted specific peptide binding to the complement factor 3a receptor. End tagging by hydrophobic amino acid stretches may thus potentially offer a way to also promote more specific peptide docking and functions.
Supplementary Material
Acknowledgments
We thank Lotta Wahlberg and Maria Baumgarten for expert technical assistance.
This work was supported by Swedish Research Council Projects 521-2006-2784 and 621-2003-4022, the Royal Physiographic Society in Lund, the Welander-Finsen, Söderberg, Schyberg, Crafoord, Alfred Österlund, and Kock Foundations, and the Swedish Government Funds for Clinical Research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S3.
- AMP
- antimicrobial peptide
- CFU
- colony-forming unit(s)
- CF
- carboxyfluorescein
- RDA
- radial diffusion assay
- TSB
- trypticase soy broth
- LPS
- lipopolysaccharide
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- MIC
- minimal inhibitory concentration(s)
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- LDH
- lactate dehydrogenase
- SFM
- serum-free medium
- EGF
- epidermal growth factor
- PBS
- phosphate-buffered saline
- PEG
- polyethylene glycol
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phoshocholine
- DOPE
- 1,2-dioleoyl-sn-glycero-3-phoshoetanolamine
- DOPG
- 1,2-dioleoyl-sn-glycero-3-phosphoglycerol
- ANOVA
- analysis of variance.
REFERENCES
- 1.French G. L. ( 2005) Adv. Drug Deliv. Rev. 57, 1514– 1527 [DOI] [PubMed] [Google Scholar]
- 2.Zasloff M. ( 2002) Nature 415, 389– 395 [DOI] [PubMed] [Google Scholar]
- 3.Marr A. K., Gooderham W. J., Hancock R. E. ( 2006) Curr. Opin. Pharmacol. 6, 468– 472 [DOI] [PubMed] [Google Scholar]
- 4.Tossi A., Sandri L., Giangaspero A. ( 2000) Biopolymers 55, 4– 30 [DOI] [PubMed] [Google Scholar]
- 5.Brogden K. A. ( 2005) Nat. Rev. Microbiol. 3, 238– 250 [DOI] [PubMed] [Google Scholar]
- 6.Nizet V. ( 2006) Curr. Issues Mol. Biol. 8, 11– 26 [PubMed] [Google Scholar]
- 7.Huang H. W. ( 2006) Biochim. Biophys. Acta 1758, 1292– 1302 [DOI] [PubMed] [Google Scholar]
- 8.Hancock R. E., Sahl H. G. ( 2006) Nat. Biotechnol. 24, 1551– 1557 [DOI] [PubMed] [Google Scholar]
- 9.Zelezetsky I., Tossi A. ( 2006) Biochim. Biophys. Acta 1758, 1436– 1449 [DOI] [PubMed] [Google Scholar]
- 10.Bhonsle J. B., Venugopal D., Huddler D. P., Magill A. J., Hicks R. P. ( 2007) J. Med. Chem. 50, 6545– 6553 [DOI] [PubMed] [Google Scholar]
- 11.Pasupuleti M., Walse B., Svensson B., Malmsten M., Schmidtchen A. ( 2008) Biochemistry 47, 9057– 9070 [DOI] [PubMed] [Google Scholar]
- 12.Jenssen H., Fjell C. D., Cherkasov A., Hancock R. E. ( 2008) J. Pept. Sci. 14, 110– 114 [DOI] [PubMed] [Google Scholar]
- 13.Nordahl E. A., Rydengård V., Nyberg P., Nitsche D. P., Mörgelin M., Malmsten M., Björck L., Schmidtchen A. ( 2004) Proc. Natl. Acad. Sci. U. S. A. 101, 16879– 16884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nordahl E. A., Rydengård V., Mörgelin M., Schmidtchen A. ( 2005) J. Biol. Chem. 280, 34832– 34839 [DOI] [PubMed] [Google Scholar]
- 15.Malmsten M., Davoudi M., Walse B., Rydengård V., Pasupuleti M., Mörgelin M., Schmidtchen A. ( 2007) Growth Factors 25, 60– 70 [DOI] [PubMed] [Google Scholar]
- 16.Ringstad L., Andersson Nordahl E., Schmidtchen A., Malmsten M. ( 2007) Biophys. J. 92, 87– 98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ringstad L., Kacprzyk L., Schmidtchen A., Malmsten M. ( 2007) Biochim. Biophys. Acta 1768, 715– 727 [DOI] [PubMed] [Google Scholar]
- 18.Ringstad L., Protopapa E., Lindholm-Sethson B., Schmidtchen A., Nelson A., Malmsten M. ( 2008) Langmuir 24, 208– 216 [DOI] [PubMed] [Google Scholar]
- 19.Ringstad L., Schmidtchen A., Malmsten M. ( 2006) Langmuir 22, 5042– 5050 [DOI] [PubMed] [Google Scholar]
- 20.Jerala R. ( 2007) Expert Opin. Investig. Drugs 16, 1159– 1169 [DOI] [PubMed] [Google Scholar]
- 21.Straus S. K., Hancock R. E. ( 2006) Biochim. Biophys. Acta 1758, 1215– 1223 [DOI] [PubMed] [Google Scholar]
- 22.Avrahami D., Shai Y. ( 2002) Biochemistry 41, 2254– 2263 [DOI] [PubMed] [Google Scholar]
- 23.Avrahami D., Shai Y. ( 2004) J. Biol. Chem. 279, 12277– 12285 [DOI] [PubMed] [Google Scholar]
- 24.Malmsten M. ( 2003) Surfactants and Polymers in Drug Delivery, 1st Ed., Marcel Dekker, New York [Google Scholar]
- 25.Mouritsen O. G., Zuckermann M. J. ( 2004) Lipids 39, 1101– 1113 [DOI] [PubMed] [Google Scholar]
- 26.Masny A., Plucienniczak A. ( 2001) BioTechniques 31, 930– 936 [DOI] [PubMed] [Google Scholar]
- 27.Lehrer R. I., Rosenman M., Harwig S. S., Jackson R., Eisenhauer P. ( 1991) J. Immunol. Methods 137, 167– 173 [DOI] [PubMed] [Google Scholar]
- 28.Andersson E., Rydengård V., Sonesson A., Mörgelin M., Björck L., Schmidtchen A. ( 2004) Eur. J. Biochem. 271, 1219– 1226 [DOI] [PubMed] [Google Scholar]
- 29.Wu M., Hancock R. E. ( 1999) J. Biol. Chem. 274, 29– 35 [DOI] [PubMed] [Google Scholar]
- 30.Williamson P., Kligman A. M. ( 1965) J. Investig. Dermatol. 45, 498– 503 [DOI] [PubMed] [Google Scholar]
- 31.McDonnell G., Haines K., Klein D., Rippon M., Walmsley R., Pretzer D. ( 1999) J. Microbiol. Methods 35, 31– 35 [DOI] [PubMed] [Google Scholar]
- 32.Malmsten M. ( 1994) J. Colloid Interface Sci. 166, 333– 342 [Google Scholar]
- 33.De Feijter J. A., Benjamins J., Veer F. A. ( 1978) Biopolymers 17, 1759– 1772 [Google Scholar]
- 34.Tiberg F., Harwigsson I., Malmsten M. ( 2000) Eur. Biophys. J. 29, 196– 203 [DOI] [PubMed] [Google Scholar]
- 35.Kyte J., Doolittle R. F. ( 1982) J. Mol. Biol. 157, 105– 132 [DOI] [PubMed] [Google Scholar]
- 36.Tossi A., Sandri L., Giangaspero A. ( 2002) in Peptides 2002: Proceedings of the Twenty-seventh European Peptide Symposium, pp. 416– 417, Edizioni Ziino, Napoli, Italy [Google Scholar]
- 37.Eisenberg D., Schwarz E., Komaromy M., Wall R. ( 1984) J. Mol. Biol. 179, 125– 142 [DOI] [PubMed] [Google Scholar]
- 38.Schmidtchen A., Frick I. M., Andersson E., Tapper H., Björck L. ( 2002) Mol. Microbiol. 46, 157– 168 [DOI] [PubMed] [Google Scholar]
- 39.Malmsten M., Davoudi M., Schmidtchen A. ( 2006) Matrix Biol. 25, 294– 300 [DOI] [PubMed] [Google Scholar]
- 40.Strömstedt A. A., Wessman P., Ringstad L., Edwards K., Malmsten M. ( 2007) J. Colloid Interface Sci. 311, 59– 69 [DOI] [PubMed] [Google Scholar]
- 41.Ramamoorthy A., Thennarasu S., Lee D. K., Tan A., Maloy L. ( 2006) Biophys. J. 91, 206– 216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bechinger B. ( 1999) Biochim. Biophys. Acta 1462, 157– 183 [DOI] [PubMed] [Google Scholar]
- 43.Chen F. Y., Lee M. T., Huang H. W. ( 2003) Biophys. J. 84, 3751– 3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glukhov E., Stark M., Burrows L. L., Deber C. M. ( 2005) J. Biol. Chem. 280, 33960– 33967 [DOI] [PubMed] [Google Scholar]
- 45.Haney E. F., Lau F., Vogel H. J. ( 2007) Biochim. Biophys. Acta 1768, 2355– 2364 [DOI] [PubMed] [Google Scholar]
- 46.Li X., Li Y., Peterkofsky A., Wang G. ( 2006) Biochim. Biophys. Acta 1758, 1203– 1214 [DOI] [PubMed] [Google Scholar]
- 47.de Planque M. R., Kruijtzer J. A., Liskamp R. M., Marsh D., Greathouse D. V., Koeppe R. E., 2nd, de Kruijff B., Killian J. A. ( 1999) J. Biol. Chem. 274, 20839– 20846 [DOI] [PubMed] [Google Scholar]
- 48.Deslouches B., Phadke S. M., Lazarevic V., Cascio M., Islam K., Montelaro R. C., Mietzner T. A. ( 2005) Antimicrob. Agents Chemother. 49, 316– 322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McInturff J. E., Wang S. J., Machleidt T., Lin T. R., Oren A., Hertz C. J., Krutzik S. R., Hart S., Zeh K., Anderson D. H., Gallo R. L., Modlin R. L., Kim J. ( 2005) J. Investig. Dermatol. 125, 256– 263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strömstedt A. A., Pasupuleti M., Schmidtchen A., Malmsten M. ( 2009) Antimicrob. Agents Chemother. 53, 593– 602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fimland G., Eijsink V. G., Nissen-Meyer J. ( 2002) Biochemistry 41, 9508– 9515 [DOI] [PubMed] [Google Scholar]
- 52.Wei S. Y., Wu J. M., Kuo Y. Y., Chen H. L., Yip B. S., Tzeng S. R., Cheng J. W. ( 2006) J. Bacteriol. 188, 328– 334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ishitsuka Y., Pham D. S., Waring A. J., Lehrer R. I., Lee K. Y. ( 2006) Biochim. Biophys. Acta 1758, 1450– 1460 [DOI] [PubMed] [Google Scholar]
- 54.Lotte K., Plessow R., Brockhinke A. ( 2004) Photochem. Photobiol. Sci. 3, 348– 359 [DOI] [PubMed] [Google Scholar]
- 55.Ember J. A., Johansen N. L., Hugli T. E. ( 1991) Biochemistry 30, 3603– 3612 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.