Abstract
Oxidative protein folding in Gram-negative bacteria results in the formation of disulfide bonds between pairs of cysteine residues. This is a multistep process in which the dithiol-disulfide oxidoreductase enzyme, DsbA, plays a central role. The structure of DsbA comprises an all helical domain of unknown function and a thioredoxin domain, where active site cysteines shuttle between an oxidized, substrate-bound, reduced form and a DsbB-bound form, where DsbB is a membrane protein that reoxidizes DsbA. Most DsbA enzymes interact with a wide variety of reduced substrates and show little specificity. However, a number of DsbA enzymes have now been identified that have narrow substrate repertoires and appear to interact specifically with a smaller number of substrates. The transient nature of the DsbA-substrate complex has hampered our understanding of the factors that govern the interaction of DsbA enzymes with their substrates. Here we report the crystal structure of a complex between Escherichia coli DsbA and a peptide with a sequence derived from a substrate. The binding site identified in the DsbA-peptide complex was distinct from that observed for DsbB in the DsbA-DsbB complex. The structure revealed details of the DsbA-peptide interaction and suggested a mechanism by which DsbA can simultaneously show broad specificity for substrates yet exhibit specificity for DsbB. This mode of binding was supported by solution nuclear magnetic resonance data as well as functional data, which demonstrated that the substrate specificity of DsbA could be modified via changes at the binding interface identified in the structure of the complex.
Introduction
The formation of disulfide bonds is a critical step in the correct folding and stability of many secreted proteins. In Gram-negative bacteria, disulfide bond formation occurs in the periplasm and is catalyzed by enzymes of the Dsb family. The Dsb family contains several members, which mediate different aspects of disulfide bond formation and isomerization (1). DsbA is the enzyme that is primarily responsible for the formation of disulfide bonds in newly synthesized substrate proteins (Fig. 1). In this reaction, oxidized DsbA reacts with a substrate protein to generate a mixed disulfide intermediate. This covalent reaction intermediate is rapidly resolved to release the oxidized substrate and reduced DsbA. Reduced DsbA is in turn reoxidized by the inner membrane protein DsbB (2).
FIGURE 1.
The catalytic cycle of DsbA. a, oxidized DsbA reacts with a variety of substrate proteins to generate a mixed disulfide DsbA-substrate complex (b). The complex is rapidly resolved with release of oxidized substrate and reduced DsbA (c). Reduced DsbA reacts specifically with membrane-bound DsbB to form a mixed disulfide DsbA-DsbB complex (d), which results in reoxidation of DsbA and reduction of DsbB.
Bacteria lacking a functional DsbA display pleiotropic phenotypes, since the folding of a large number of disulfide bond-containing proteins is disrupted. Many of these are secreted proteins, such as toxins and surface proteins, that contribute to bacterial virulence (3). For example, DsbA is required for the formation of a functional type III secretion system in many bacteria, including Pseudomonas aeruginosa (4, 5), Shigella flexneri (6), Salmonella enterica serovar Typhimurium (7), and Yersinia pestis (8); dsbA− Vibrio cholerae are unable to secrete cholera toxin (9); dsbA− strains of E. coli exhibit reduced levels of β-lactamase activity (10) and are hypersensitive to benzyl penicillin, dithiothreitol (11), and some divalent metal cations (12). Furthermore, DsbA has been shown to be necessary for intracellular survival of S. flexneri (13) and P. aeruginosa (4), and DsbA is required for virulence of S. enterica in a mouse infection model (7). Each of these phenotypes has been attributed to the lack of disulfide formation in protein substrates of DsbA. Thus, there has been considerable interest in the structural basis of DsbA activity and selectivity and its role in bacterial virulence.
The structures of DsbA from E. coli (EcDsbA) and V. cholerae (VcDsbA) have been solved in both their reduced and oxidized forms (14–18). Each contains a thioredoxin (TRX)5 domain, a common structural fold of thiol-disulfide oxidoreductases (19), and an inserted helical domain (14). DsbA enzymes contain a single pair of redox-active cysteines in a CXXC motif (Cys30-Pro31-His32-Cys33 in EcDsbA) and a cis-proline residue (Pro151 in EcDsbA) that is adjacent in the three-dimensional fold but distant in the sequence; both are conserved features of the redox-active TRX fold (19). EcDsbA and VcDsbA share relatively low sequence similarity (<40%), yet retain conserved surface features around their active sites, which have been proposed to form the substrate-binding site (20). These include a hydrophobic patch and a hydrophobic groove, which flank the active site of the enzyme. Within the hydrophobic groove is the cis-proline residue that is a conserved feature of the TRX fold and in DsbA is found in a loop at the end of a long helix, which links the α-helical and TRX domains. The structure of EcDsbA has been determined in complex with DsbB (21), which revealed that a periplasmic loop of DsbB is bound within the hydrophobic groove of DsbA (21). The hydrophobic groove has also been widely assumed to be the substrate-binding site of DsbA, although the transient nature of DsbA-substrate complexes has hindered their structural characterization.
Many DsbA enzymes display broad substrate specificity. In E. coli, for example, it has been estimated that DsbA may oxidize hundreds of different substrate proteins (22). More recently, a number of DsbA enzymes have been described that display narrower substrate specificity, and the structures of several of these have been reported. These include a DsbA enzyme from Neisseria meningitidis (NmDsbA3) (23) as well as DsbA from the Gram-positive organism Staphylococcus aureus (SaDsbA) (24) and a second DsbA enzyme (DsbL) (25) that is present in some uropathogenic strains of E. coli (25). Functional characterization has revealed that both SaDsbA and NmDsbA3 can only partially complement dsbA− E. coli (24, 26), suggesting that both have a narrower substrate repertoire than EcDsbA, whereas biochemical analysis has revealed that NmDsbA3 is a substrate for EcDsbB (23) and that SaDsbA is not (24). DsbL has been shown to partially restore motility to dsbA− E. coli in a DsbA complementation assay, but biochemical analysis revealed that it does not show DsbA-like activity in standard oxidoreductase assays, such as RNase refolding and insulin reduction (25).
Despite their functional differences, all of the characterized DsbA enzymes share a similar tertiary structure, and minimal changes have been observed between the reported structures of reduced and oxidized forms of the protein. There are, however, some differences in the sequences around the active sites that may contribute to the observed differences in activity between these DsbA enzymes. For example, in most DsbA enzymes, there is a valine residue preceding the conserved cis-proline (VcP). In both SaDsbA and NmDsbA3, the residue preceding the cis-proline is a threonine (TcP). The TcP motif is also found in Gram-negative disulfide isomerase proteins (DsbC and DsbG). It has previously been demonstrated that mutations in the cis-proline loop of DsbC and DsbG can affect the substrate specificity of these enzymes (27), and more recently, it has been demonstrated that this residue is important in dictating the function of many TRX fold proteins (28). Although DsbL maintains the VcP sequence, structures have revealed that SaDsbA and DsbL lack the hydrophobic surface features surrounding their active sites, whereas NmDsbA3 presents a similar hydrophobic surface around the active site but adopts a different conformation in the loops linking the two domains, which are adjacent to the active site.
These findings suggest that there are subtle differences in the processes that mediate recognition of substrates by oxidized DsbA and recognition of DsbB by reduced DsbA. This observation is supported by analysis of N. meningitidis, which express three chromosomally encoded DsbA enzymes (29, 30). Two of these Neisserial DsbA enzymes (NmDsbA1 and NmDsbA2) share 73% sequence identity (29) and maintain the canonical DsbA active site sequence (Cys-Pro-His-Cys). However, complementation of dsbA− E. coli with NmDsbA1 or NmDsbA2 produced transformants with different phenotypes (29), suggesting that these enzymes are capable of discriminating between specific substrates (29, 30). However, both NmDsbA1 and NmDsbA2 were shown to complement the DTT sensitivity of dsbA− E. coli, suggesting that although they differ in their specificity for oxidizing substrate proteins (29), both are functional oxidoreductases, and are capable of being reoxidized by EcDsbB.
Structural details of how DsbA recognizes its substrates have proved difficult to obtain, since the covalent reaction intermediates between DsbA and its substrates are relatively short lived. Although a mutant DsbA protein has been identified that stabilizes the mixed disulfide intermediates (31), there are currently no structural details of any DsbA-substrate complex. Herein we report the structure of a covalent DsbA-peptide complex refined to a resolution of 1.9 Å. It has previously been found that reduced peptides and proteins are oxidized with similar kinetics (32–34), suggesting that peptides are suitable models of the reduced unfolded proteins that are the substrates of DsbA. Our structure reveals that the peptide binds to DsbA at a surface formed by residues at the interface between the α-helical and TRX domains and not within the hydrophobic groove that is the binding site for DsbB. Phenotypic assays were undertaken using chimeric proteins containing the α-domains of Neisserial DsbA enzymes grafted into the TRX domain of EcDsbA that introduce mutations at the domain interface and cis-proline loop that form the peptide binding site while maintaining the hydrophobic groove that is the binding site of DsbB. These revealed that the chimeras were functional oxidoreductase enzymes but that the phenotypes observed in the assays were modulated by changes at the peptide binding site identified in our structures of the complex. Taken together, these data suggest that the interface formed between the TRX and α-helical domains of DsbA is an important determinant of specificity in the reaction between oxidized DsbA and its substrates and that DsbA activity may be modulated through modifications to the nature of this surface and changes in the cis-proline loop.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Standard molecular biology techniques were utilized throughout. The gene encoding EcDsbA was cloned into the expression vector pTrc99A (35), which was used to transform E. coli BL21 DE3 Codon Plus cells (Stratagene). Bacterial cultures were grown at 37 °C until A600 reached 0.6, when expression was induced with the addition of isopropyl 1-thio-β-d-galactopyranoside (1 mm), and the cultures were grown for a further 5 h. Periplasmic extracts were obtained by resuspending the cell pellet in 20 mm Tris, pH 8.0, 150 mm NaCl containing polymyxin B sulfate (1 mg ml−1) for 2 h at 4 °C. DsbA was purified as described previously, reduced with Tris(2-carboxyethyl)phosphine-HCl at a 1:10 molar ratio, buffer-exchanged into 10 mm HEPES (pH 7.8), 0.1 mm EDTA, and concentrated to 20 mg ml−1 for crystallization. Uniformly 15N and 13C/15N isotope-labeled protein was produced according to the method of Marley et al. (36) and purified as described above. Samples for NMR contained uniformly 13C/15N EcDsbA (300 μm in 20 mm HEPES, pH 7.0, 150 mm NaCl, 10% D2O). Homogeneity of the purified proteins was assessed by SDS-PAGE (silver staining) and by electrospray ionization mass spectrometry on a Micromass Platform II liquid chromatography/quadrupole mass spectrometry system (Manchester, UK). Protein concentration was determined by UV-visible spectrophotometry using a molar extinction coefficient at 280 nm of 23,045 cm−1 m−1.
Kinetics of Disulfide Oxidation
Disulfide oxidase activity was measured by following the change in the intrinsic fluorescence of EcDsbA upon oxidation of a peptide derived from the mature sequence of the S. flexneri autotransporter protein SigA. Synthesis of the N-terminally acetylated sequence, Ac-G-N(Trt)-N(Trt)-N(Trt)-C(Trt)-P-I-P-F-L-C(Trt)-Q(Trt)-K(t-butoxycarbonyl)-D(t-butoxy)-RESIN was performed on Rink Resin under standard solid phase peptide synthesis conditions. Cleavage and deprotection was carried out using 95% trifluoroacetic acid from which the peptide was recovered using a standard sequence of ether precipitation, dissolution in 50% aqueous acetonitrile, and freeze drying. The product was subjected to semipreparative HPLC purification, allowing the isolation of the peptide in high purity (m/z 1604.5 MH+). The kinetics of oxidation of the SigA peptide by EcDsbA was determined in phosphate-buffered saline (pH 7.2), using an Applied Photophysics SF.18MV stopped-flow apparatus, thermostatted at 30 °C. Peptide oxidation was followed by monitoring the change in intrinsic fluorescence of EcDsbA upon reduction at wavelengths of >320 nm (λex = 295 nm). Oxidation reaction mixtures contained EcDbsA (1–10 μm) to which the SigA peptide was added over a range of concentrations (1–25 μm). The initial rates of the oxidation reactions were determined from the linear portion of the fluorescence data, and these were used to determine the apparent second order rate constant for the reaction as described previously (34).
Peptide Synthesis
Synthesis of the N-terminally acetylated sequence from SigA, Ac-P-I-P-F-L-Hse(Trt)-Q(Trt)-K(t-butoxycarbonyl)-D(t-butoxy)-RESIN, was performed on Rink Resin under standard solid phase peptide synthesis conditions. The peptide was cleaved and deprotected with 95% trifluoroacetic acid recovered as described above and purified using semipreparative HPLC, yielding the peptide in high purity (m/z 1100 MH+). The strategy for formation of the covalent complex followed to a large extent the method described by Couprie et al. (37). For synthesis of the bromopeptide, the homoserine sequence was assembled on resin as above, trityl protecting groups were removed by five cycles of treatment with 1:5:94 trifluoroacetic acid/triisopropylsilane/dichloromethane, and the exposed alcohol was brominated by extended treatment with CBr4/Ph3P in tetrahydrofuran. After extensive washing, final cleavage and deprotection was carried out using 95% trifluoroacetic acid. The brominated product was efficiently recovered after trifluoroacetic acid cleavage by ether precipitation, dissolution in 50% aqueous acetonitrile, and freeze drying. The product was stable to semipreparative HPLC purification, allowing the isolation of the peptide in high purity (m/z 1162 MH+) and good yield (20 mg). The homoserine-containing peptide (m/z 1100 MH+) was a minor component from the cleavage reaction. The bromopeptides were found to be stable to storage and not readily hydrolyzed under the aqueous conditions of complex formation with the protein.
Crystallization
Crystals of reduced EcDsbA were grown at 4 °C using the hanging drop technique. Crystals grew within 3 days from equal volumes of protein solution and a reservoir solution containing 0.2 m ammonium acetate, 20% (w/v) polyethlylene glycol 4000, and 0.1 m sodium citrate (pH 5.8). Crystals were soaked overnight at 4 °C in solution containing 0.2 m ammonium acetate, 28% (w/v) polyethlylene glycol 4000, 10% (v/v) glycerol, and 0.1 m sodium citrate (pH 5.8) supplemented with 3.5 mm bromopeptide, 3.5 mm Tris(2-carboxyethyl)phosphine-HCl, and 0.1 mm EDTA. Soaked crystals were then transferred into a cryoprotectant solution containing 0.2 m ammonium acetate, 28% (w/v) polyethlylene glycol 4000, 10% (v/v) glycerol, and 0.1 m sodium citrate (pH 5.8) and flash-frozen prior to data collection.
Structure Determination and Refinement
Data were collected at the Advanced Photo Source (Argonne National Laboratory, Argonne, IL) on beamline IMCA-CAT-17-ID-B at 100 K and were processed and scaled using the HKL suite. The crystals belong to space group P212121, and the unit cell dimensions were consistent with 4 molecules/asymmetric unit (see Table 1). The structure of DsbA in complex with the SigA peptide was determined by molecular replacement using a previously determined structure of reduced DsbA (Protein Data Bank accession code 1a2l) as the search model. The model was built manually using the program O and refined to 1.9 Å resolution using the CCP4 program suite (38). Each of the four DsbA molecules was built independently, and no NCS restraints were used during the refinement. The quality of the model was determined by monitoring the Rwork and Rfree values. In the final model 93.6% of residues were found within the most favored regions of the Ramachandran plot, with the remaining 6.4% in the allowed regions. Statistics for the final structures are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics
Values in parenthesis are for the highest resolution shell.
| DsbA-peptide complex | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 85.4, 87.9, 112.6 |
| α, β, γ (degrees) | 90.0, 90.0, 90.0 |
| Resolution (Å) | 1.9 |
| Rmerge | 0.109 (0.793) |
| I/σ (I) | 22.1 (2.5) |
| Completeness (%) | 99.8 (99.7) |
| Redundancy | 7.1 (6.6) |
| Refinement | |
| Resolution (Å) | 1.9 |
| No. of reflections | 66227 |
| Rwork/Rfree | 21.7/25.2 |
| No. of atoms | |
| Protein | 5805 |
| Ligand/ion | 117 |
| Water | 488 |
| B-factors | |
| Protein | 24.48 |
| Ligand/ion | 37.41 |
| Water | 28.84 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (degrees) | 1.02 |
NMR Spectroscopy
NMR experiments were carried out on a Varian Unity 600-MHz spectrometer equipped with a single axis gradient triple resonance cryoprobe. Standard triple resonance experiments were employed to confirm the published assignments for EcDsbA (16). Data were processed using NMRPipe (39) and were analyzed using the program SPARKY. Overall weighted average chemical shift changes (Δavg) were calculated for all residues, using the equation (40),
where δH and δN denote the changes in chemical shift for amide proton and nitrogen resonances, respectively, upon the addition of the SigA homoserine-containing peptide (Ac-PIPFL-Hse-QKD-NH2) (1 mm). The assigned 1H and 15N resonances for the apo- and peptide-complex of EcDsbA are presented in supplemental Table 1. Residues that underwent the greatest changes in chemical shift were mapped onto the crystal structure of the complex to identify the binding location of the peptide.
Construction of Chimeric DsbA Proteins
Chimeric dsbA genes were created that contained the EcDsbA TRX domain and the NmDsbA1 or NmDsbA2 α-domains. This was achieved through three-way splice overlap extension PCR (41). Each of the domains was amplified in the first round of PCR with primers having 6-bp overhangs of sequence complementary to the adjacent fragment to which it was to anneal. The second round of PCR allowed the 12-bp overlaps to anneal to form the chimeric dsbA gene template that was then amplified. Expression plasmids were constructed by cloning the appropriate gene into the expression plasmid pTrc99A (35).
Complementation of dsbA− E. coli
The E. coli JCB strains were kindly provided by J. C. Bardwell (Harvard Medical School) (10), and transformations of E. coli strain JCB571 were performed chemically (42). Transformants were selected on LB agar containing appropriate antibiotics and confirmed by colony PCR.
Phenotypic Assays
Oxidoreductase activity assays were undertaken for each expression construct by testing the DTT sensitivity of transformed cells (43). Bacteria were grown in LB to midlog phase when the cell density was determined spectrophotometrically, and the cultures were standardized to a concentration of 2 × 108 cells/ml. 10-Fold serial dilutions were prepared, and an aliquot (5 μl) of each concentration was deposited onto LB-agar plates with and without DTT (15 mm). After drying, the plates were incubated at 37 °C for 18 h.
Motility assays were performed essentially as described (44) on soft LB-agar (0.4%). Bacteria were grown in LB to midlog phase when the cell density was determined spectrophotometrically, and the cultures were standardized to a concentration of 2 × 108 cells/ml. Samples (5 μl) were drop-inoculated onto the soft agar and incubated at 37 °C overnight to allow swarming.
Conscore
A simple conservation scheme (45) was used to calculate amino acid conservation in confirmed DsbA substrates. The substrates used in the analysis are listed in supplemental Table 2. Five residues on either side of cysteine residues in known substrates of EcDsbA were investigated. Conservative substitution was defined as belonging to one of the following seven groups: 1) hydrophobic (alanine, valine, leucine, and isoleucine), 2) aromatic hydrophobic (tryptophan, tyrosine, and phenylalanine), 3) basic (arginine, histidine, and lysine), 4) acidic (glutamic acid and aspartic acid), 5) neutral polar (serine, methionine, threonine, cysteine, asparagine, and glutamine), 6) glycine, and 7) proline. 45 cysteine-containing sequences from proteins that have been shown to be substrates of EcDsbA were analyzed to determine the degree of conservation in residues surrounding the cysteine.
RESULTS
Disulfide Oxidase Activity
Disulfide oxidase activity was determined in vitro by measuring the rate of oxidation of a model substrate peptide. The secreted domain of the S. flexneri autotransporter protein SigA contains a single pair of cysteine residues (569CPIPFLC575), which are oxidized to form a disulfide in the mature protein. The sequence around the two cysteines (Ac-NNNCPIPFLCQKD-NH2) was synthesized and used as a model substrate peptide. Oxidation of the peptide by EcDsbA (which is identical in sequence to S. flexneri DsbA) was monitored by stopped-flow fluorescence. In the course of the assay, oxidation of the peptide substrate resulted in concomitant reduction of the DsbA enzyme. The intrinsic fluorescence of the reduced form of EcDsbA is significantly higher than that of oxidized EcDsbA (46), which enabled the progress of the reaction to be followed by monitoring the increase in fluorescence (Fig. 2). Analysis of the initial rates indicated that the reaction was first order with respect to peptide and EcDsbA and proceeded with a second order rate constant of k = 1.4 × 106 m−1 s−1. This is similar to the derived rate constants that have been reported previously for the reaction of DsbA enzymes with peptide and protein substrates (32–34) and indicated that the SigA peptide was efficiently oxidized by EcDsbA.
FIGURE 2.
Oxidation of the SigA peptide by DsbA. Oxidase activity was determined by monitoring the oxidation of SigA peptide substrate. The kinetics of the oxidation reactions were determined using stopped-flow fluorescence by monitoring the increase in fluorescence upon reduction of EcDsbA (1 μm) in the assay. Progress curves for oxidation of the peptide substrate are shown at increasing peptide concentrations (0, 2, 5, and 10 μm). The rate of oxidation increases with increasing peptide concentration.
Structure Determination of the DsbA-Peptide Complex
In order to characterize substrate binding to DsbA, the crystal structure of a covalent complex between DsbA and the SigA peptide was determined. The covalent complex was stabilized by replacing the intermolecular disulfide with a more stable isosteric thioether bond (37) to prevent product release. To form the complex, a nine-residue peptide encompassing SigA residues Pro570–Asp578 was synthesized with a homoserine residue (Hse) in place of the native cysteine (Ac-P1IPFL-Hse-QKD9-NH2). The hydroxyl of the homoserine was substituted with bromine to generate the homobromoalanine analogue, which was reacted with Cys30 of EcDsbA to form a covalent thioether complex between the peptide and EcDsbA. The isosteric thioether bond is more stable than the corresponding disulfide, and hence the thioether-linked peptide is essentially bound irreversibly to EcDsbA. The peptide was synthesized with an acetylated N terminus and an amidated C terminus to avoid introduction of charges at the termini of the peptide, which would not be present in the context of the intact SigA protein. Diffraction quality crystals of the complex were formed by soaking the homobromoalanine-SigA-peptide into preformed crystals of reduced EcDsbA. Analysis of the crystals by SDS-PAGE revealed that the peptide had formed a complex with ∼50% of the DsbA in the crystal, as estimated from the intensity of the respective bands on SDS-polyacrylamide gels (Fig. 3a) The structure of the complex was solved by molecular replacement and refined to a resolution of 1.9 Å (Rwork = 21.7%, Rfree = 25.2%) (Table 1).
FIGURE 3.
Analysis of the DsbA-peptide crystal. a, analysis by SDS-PAGE (lane ii) revealed that approximately half of the EcDsbA in the crystal had formed a covalent complex with the peptide. The peptide-DsbA complex (lane iii) was clearly resolved from free EcDsbA (lane iv) under the conditions used to run the gel. (Molecular weight markers are shown in lane i.) b, ribbon diagram of EcDsbA (Protein Data Bank code 3DKS, chain C). Each of the four molecules in the asymmetric unit adopted a typical DsbA fold comprising a thioredoxin (blue) and α-helical (magenta) domains and the insertion points (orange). The sulfur atoms of the active site cysteine residues are shown in yellow CPK representation.
Description of the Structure
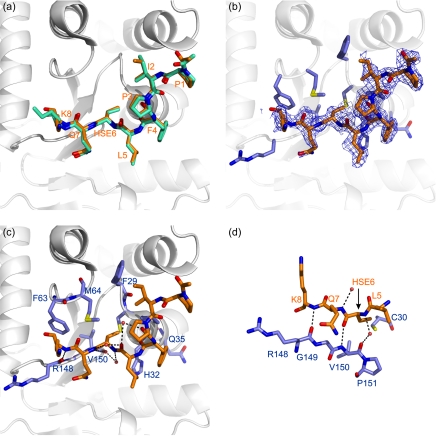
Each of the four EcDsbA molecules within the asymmetric unit adopted a typical DsbA fold (Fig. 3b), wherein the active site C30PHC33 sequence of EcDsbA is positioned at the end of helix α1 within the TRX domain of the protein. The TRX domain contains an inserted α-helical domain. The two domains are linked via a loop between strand β3 of the TRX domain and helix α2 in the helical domain (residues 62–66) and through a loop at the end of the long helix α6 (residues 129–144) (14). The four EcDsbA molecules within the asymmetric unit were highly similar to one another and superimposed over the backbone atoms for residues 2–184 with overall root mean square (r.m.s.) deviation of <0.5 Å. They are also similar to previously reported structures of reduced (Protein Data Bank code 1A2L) and oxidized (Protein Data Bank code 1FVK) EcDsbA and superimpose over the backbone atoms for residues 2–184 with overall r.m.s. deviation of <0.5 Å. Of the four EcDsbA molecules in the asymmetric unit, two (chains C and D) were covalently linked to a SigA peptide (chains E and F, respectively), whereas the other two DsbA molecules (chains A and B) were present in their reduced form. The covalent complex was formed via a thioether bond between the Cγ in the homoserine of the SigA peptide and Sγ in Cys30 of EcDsbA. Analysis of the structures revealed that binding of the peptide to the first EcDsbA molecule blocked the equivalent binding site on the adjacent EcDsbA, accounting for the observation that only half of the EcDsbA molecules in the crystal were present as a complex. The peptides in each of the two EcDsbA-peptide complexes were highly similar and had an r.m.s. deviation of 0.21 Å over eight Cα atoms (Fig. 4a). The side chains of the glutamine at position 7 and lysine at position 8 were mobile in both peptide complexes, as determined by the lack of prominent electron density in the 2Fo − Fc map (Fig. 4b). The aspartate at position 9 was disordered and not modeled in either complex.
FIGURE 4.
The substrate binding site of DsbA. a, comparison of the peptide conformation in the two models of the EcDsbA-peptide complex (Protein Data Bank code 3DKS). Peptide chains E and F are shown in orange and green stick representation, respectively, and the residues are labeled in orange. The EcDsbA molecule to which peptide E is covalently attached (chain C) is shown in a schematic diagram. b, omit (2Fo − Fc) electron density map (contoured at 0.8 σ) for the peptide (chain E). EcDsbA residues in contact with the peptide are shown in a blue stick representation. c, polar interactions observed in the complex are shown as black dotted lines between the interacting residues. Water atoms are shown as red spheres. The EcDsbA-peptide complex (chains C and E) is shown in the same orientation as above, and EcDsbA residues are labeled in blue. d, the peptide forms an antiparallel interaction with residues in the cis-proline loop of EcDsbA. Polar interactions are shown as black dotted lines between the peptide (in orange) and EcDsbA (in blue).
DsbA-Peptide Interaction
Analysis of the structure of the covalent EcDsbA-SigA-peptide complex revealed that the peptide was bound at the interface of the TRX and α-helical domains of DsbA, with contacts being made to both domains. Details of the DsbA-peptide complex are presented in Fig. 4. The intermolecular thioether bond displayed an architecture similar to that of a right-handed disulfide bond (47) with dihedral angles defined by CαC30-CβC30-SγC30-CγHse, CβC30-SγC30-CγHse-CβHse, and SγC30-CγHse-CβHse-CαHse of 76°/81°, 92°/100°, and 147°/154° in the two intermolecular complexes of the asymmetric unit, respectively. This is a conformation similar to that of the mixed disulfide that is observed in a TRX-substrate complex (48), indicating that the thioether provides a suitable isosteric replacement of the disulfide.
In addition to the covalent linkage between the peptide and EcDsbA, substrate binding was stabilized by hydrogen bonding and van der Waals interactions (Fig. 4, c and d), and the binding interface was almost identical in each complex. Hse6 and Lys8 from SigA formed hydrogen bonds with Val150 and Arg148 in the cis-proline loop of EcDsbA, respectively, such that the loop and substrate peptide were arranged in an antiparallel fashion, and a water-mediated hydrogen bond was formed between the amide proton of Hse6 and the carbonyl oxygen of Val150 (Fig. 4d). In addition to the hydrogen-bonding interactions, Phe29 of EcDsbA packed against Ile2 of the peptide, whereas Pro31, His32, and Gln35 of DsbA made contacts to Phe4 of SigA. Hse6 of the peptide contacted Met64 of DsbA, and Gln7 and Lys8 made contacts with Phe63, Arg148, and Gly149. The binding interface in the DsbA-peptide complex has a total buried surface area of 760 and 795 Å2 in the two structures, respectively, and a surface complementarity of 74% as calculated in CCP4 (49). There were very slight differences in the packing interactions in the two DsbA-substrate complexes present in the asymmetric unit of the unit cell as a result of small changes in the orientation of the side chain of Ile2 and the mobility of Lys8 (Fig. 4a). In addition, there were slight differences in the side chain orientations of Phe29 and Met64 in the two molecules of EcDsbA that were covalently attached to the peptide. Notwithstanding these small differences, the 10 residues of EcDsbA that contact the peptide in the complex could be superimposed over their Cα atoms with an r.m.s. deviation of 0.12 Å, and the H-bonding interactions were conserved in the two complexes, indicating that the mode of interaction is similar. In both cases, residues around the active site (29FCPH32) together with those in the loop connecting the TRX and α-helical domains (Phe63 and Met64) and residues in the loop preceding the cis-Pro151 (Arg148, Gly149, and Val150) formed >95% of the binding surface area.
Characterization of DsbA-Peptide Binding
Given that only two of the EcDsbA molecules out of the four present in the asymmetric unit were observed to bind the peptide, there was concern that the mode of peptide binding was a crystallographic artifact. In order to address this concern, the binding location of the SigA peptide was determined by measuring chemical shift perturbations in 1H-15N HSQC NMR experiments on uniformly 15N-labeled EcDsbA in the absence and presence of the peptide (Fig. 5). The chemical shifts of resonances in HSQC spectra are exquisitely sensitive to changes in the environment of the amide groups, and measurement of chemical shift perturbation is widely used as a means to identify the location of ligand binding. A peptide containing homoserine in place of cysteine was employed to prevent formation of a covalent complex and to allow identification of residues whose resonances were perturbed in the context of a noncovalent complex between the peptide and EcDsbA. Analysis of the spectra revealed that many of the most significant perturbations were observed for residues that formed a continuous surface at the interface between the α-helical and TRX domains (Fig. 5a). The location of the perturbed chemical shifts in the NMR spectra was consistent with the mode of binding observed in the x-ray structure and suggests that the structure represents a relevant model of the biological complex. In contrast, many of the residues in the hydrophobic groove that is the binding site for the periplasmic loop of DsbB were observed in the 15N HSQC spectrum of EcDsbA but were not significantly perturbed upon the addition of the peptide. This suggests that the SigA peptide binds in a location that is distinct from the binding site observed in the crystal structure of the DsbA-DsbB complex (21). An interesting feature of NMR data was that no peak was observed in the 15N HSQC spectrum for His32 either in the presence or absence of peptide, which is consistent with this residue undergoing conformational exchange, which has previously been suggested to be important in substrate recognition and catalysis in DsbA enzymes (50).
FIGURE 5.
Analysis of EcDsbA-SigA peptide interaction by NMR spectroscopy. a, EcDsbA residues for which chemical shift perturbations are observed upon the addition of the peptide are colored in magenta on the crystal structure of the EcDsbA-peptide complex (Protein Data Bank code 3DKS, chain C; the peptide 3DKS, chain E, is in orange stick format). His32, which is not observed in either HSQC spectrum is colored in cyan. A continuous surface is formed by residues that form the EcDsbA-peptide interface in the crystal. b, 15N HSQC spectrum of EcDsbA in the absence (magenta) and presence (cyan) of SigA peptide. The expansion demonstrates perturbations that are observed for some residues (e.g. Phe29 and Cys33), whereas other residues (e.g. Gly6 and Asp86) are unaffected.
Comparison of DsbA-Substrate and DsbA-DsbB Complex
Comparison of our structure with the EcDsbA-EcDsbB complex structure revealed that the conformation of EcDsbA was similar in both cases (Fig. 6). The two EcDsbA molecules (Protein Data Bank codes 3DKSD and 2HI7A) superimposed with an r.m.s. deviation of 0.59 Å over 179 Cα atoms. Both the SigA peptide and the periplasmic loop of EcDsbB interacted with 148RGV150 in the cis-proline loop of EcDsbA; however, the side chains of residues 99PFA101 from the DsbB loop were buried in the hydrophobic groove, where they interacted with Pro151, Pro163, Gln164, Thr168, Met171, and Phe174 of EcDsbA (Fig. 6b). Thus, although the general mode of interaction with DsbA is maintained, DsbB appears to make additional interactions within the hydrophobic groove on the surface of DsbA. These result in the higher buried surface area observed in the DsbA-DsbB complex (1340 Å2) (21), which may also account for the greater specificity of the DsbA-DsbB interaction.
FIGURE 6.
Comparison of the complexes of substrate-DsbA and DsbB-DsbA (Protein Data Bank code 2HI7). a, surface view of the interaction between EcDsbA and SigA peptide. The surface of EcDsbA is shown with the SigA peptide in an orange stick representation. The position of His32 on the surface is highlighted in magenta. b, structure of the DsbA-DsbB complex. The surface of EcDsbA is shown with the bound Cys104 loop in a green stick representation, and the position of His32 is highlighted. c, superposition of the two complexes reveals that His32 in the complex with SigA peptide (magenta) adopts a different rotameric state compared with that observed in the complex with DsbB.
Despite their general similarity, there were some differences observed between the DsbA molecules in the DsbA-substrate and DsbA-DsbB complexes. A notable difference was the orientation of the side chain of His32 (Fig. 6c). In the complex with peptide (Protein Data Bank code 3DKS chain C), H32 adopted a gauche+ conformation (χ1 = 82°), where its side chain lay across the face of the hydrophobic groove and blocked access to the peptide substrate. In the complex with DsbB (Protein Data Bank code 2HI7), His32 adopted a trans conformation (χ1 = −177°), which allowed access to the hydrophobic groove of DsbA and where its side chain made van der Waals interactions with Ala102-Thr103 in the DsbB loop. In the original crystal structure of oxidized EcDsbA (14), there were two molecules in the asymmetric unit. These two molecules of oxidized EcDsbA differed in the orientation of the His32 side chain (in the structure of oxidized EcDsbA, Protein Data Bank code 1FVK, χ1 = 160 and 72°, respectively). His32 has been shown to destabilize the oxidized form of EcDsbA (50), and it has been suggested previously that movement of this residue may be required to allow substrate access to the active site (50).
Functional Analysis of DsbA
To determine the role of the substrate-binding residues of DsbA in defining specificity, we constructed chimeric proteins comprising the TRX domain of EcDsbA and the α-domain of either NmDsbA1 or NmDsbA2. These are referred to as EcTDNmDsbA1α and EcTDNmDsbA2α, respectively. Alignment of the sequences of EcDsbA, NmDsbA1, and NmDsbA2 (Fig. 7a) revealed that within the chimeric DsbA enzymes, the loop 148RGV150 in EcDsbA is replaced by DGT and SGT in EcTDNmDsbA1α and EcTDNmDsbA2α, respectively. In addition, residues Phe63-Met64 of EcDsbA are replaced with WQ and WG in EcTDNmDsbA1α and EcTDNmDsbA2α, respectively. In contrast, the residues that form the hydrophobic groove in EcDsbA are retained in the chimeras.
FIGURE 7.
Phenotypic analysis of chimeric DsbA. a, primary structure of EcDsbA, NmDsbA1, and NmDsbA2. Residues comprising the thioredoxin domain of EcDsbA are underlined. Residues that make contacts to the peptide substrate in the crystal structure of the complex are highlighted (*). b–e, phenotypic assays on dsbA− E. coli (JCB571) complemented with different DsbA proteins. b, JCB571 alone (lane 3), JCB571 complemented with EcDsbA in pTrc99A as a positive control (lane 4), and JCB571 complemented with EcTDNmDsbA1α (lane 1) or EcTDNmDsbA2α (lane 2) were able to grow on LB agar without DTT, indicating that each of the cell lines was viable. c, the same cell lines were tested for growth on LB agar containing 15 mm DTT and 1 mm isopropyl 1-thio-β-d-galactopyranoside to induce DsbA expression. Each of the cell lines that had been complemented with the DsbA proteins grew, whereas JCB571 did not, indicating that each construct expressed an active DsbA. d, neither JCB571 (zone 2) nor JCB571 complemented with EcTDNmDsbA1α (zone 3) was motile. In contrast, the positive control (JCB571 complemented with EcDsbA) (zone 1) was motile. e, both the positive control strain (zone 1) and JCB571 complemented with EcTDNmDsbA2α (zone 3) were motile, whereas JCB571 (zone 2) was not.
In order to test the effect of these mutations, phenotypic analysis was performed using the dsbA− E. coli strain JCB571 complemented with EcDsbA, EcTDNmDsbA1α, or EcTDNmDsbA2α. First, the sensitivity to DTT of each of the transformants was determined. Strains of dsbA− E. coli, such as JCB571, are sensitive to DTT and are unable to grow on LB agar containing DTT (Fig. 7, b and c). Transformation of JCB571 with either of the chimeric proteins or EcDsbA restored resistance to DTT. This suggested that each construct was capable of expressing a functional DsbA enzyme and that the expressed DsbA was able to be reoxidized in the periplasm.
DsbA is also required for the correct folding and stability of a component of the flagellar motor of E. coli (FlgI) (51). In the absence of DsbA, the P-ring component of the flagellar motor is not formed, which renders dsbA− E. coli, such as JCB571, nonmotile. The phenotypic assays revealed that cells transformed with either EcDsbA or EcTDNmDsbA2α were motile, whereas those transformed with EcTDNmDsbA1α were not (Fig. 7, d and e). This is in contrast to observations with the intact Neisserial DsbA enzymes, wherein NmDsbA1 restored motility to dsbA− E. coli, whereas NmDsbA2 did not (29). However, when the five residues immediately preceding the cis-proline in EcTDNmDsbA1α (QIDGT) were replaced with the corresponding sequence from EcDsbA (QLRGV), the resulting chimeric protein was capable of both conferring resistance to DTT and restoring motility in JCB571 (Table 2). Similarly, replacing the five residues preceding the cis-proline in EcTDNmDsbA2α (QISGT) with the corresponding sequence from EcDsbA (QLRGV), the resulting chimeric protein was capable of both conferring resistance to DTT and restoring motility in JCB571.
TABLE 2.
Phenotypic analysis of DsbA chimeras
All dsbA genes were cloned into the low copy vector pHSG576 and assessed for their ability to restore E. coli dsbA mutant strain JCB571 resistance to DTT and motility.
| dsbA | DTT resistance | Motility |
|---|---|---|
| JCB570 (wild type) | Yes | Yes |
| JCB571 (dsbA−) | No | No |
| JCB571 transformed with | ||
| EcDsbA | Yes | Yes |
| EcTDNMDsbA1α | Yes | No |
| EcTDNMDsbA1α (QLRGV) | Yes | Yes |
| EcTDNMDsbA2α | Yes | Yes |
| EcTDNMDsbA1α (QLRGV) | Yes | Yes |
Despite the relatively high conservation within the sequences of NmDsbA1 and NmDsbA2, our analysis indicates that changes introduced by constructing the chimeric proteins are sufficient to alter the substrate specificity, but they do not destroy the oxidoreductase activity of the proteins or, apparently their ability to be reoxidized by DsbB. These data suggest that neither the TRX domain nor the α-domain is solely responsible for conferring substrate specificity and are consistent with the structural details, which reveal that binding takes place at a surface that encompasses the interface formed between the two domains. Further, they suggest that substrate recognition by oxidized DsbA is different in some respects from the interaction of reduced DsbA with DsbB, such that changes in substrate specificity can be achieved without apparent loss of recognition of DsbB.
DISCUSSION
We have determined a high resolution crystal structure of EcDsbA in complex with a nine-residue peptide derived from the autotransporter protein SigA of S. flexneri. This structure revealed that the peptide bound outside the hydrophobic groove of EcDsbA and contacted residues at the interface between the TRX and α-domains. The complex was stabilized by main-chain hydrogen bonds between substrate and residues in the cis-proline loop of the TRX domain, such that the two peptide chains were arranged in an antiparallel fashion. The mode of binding observed in the peptide-DsbA complex displayed some similarity to that of a trapped substrate-thioredoxin complex (Protein Data Bank entry 2IWT) (48). In both cases, the complex was stabilized by backbone-to-backbone hydrogen bonds between the substrate and residues in the cis-proline loop of the TRX domain, and the loop and peptide were arranged in an antiparallel fashion. The heavy atoms of the residues of the active site (XCXX(C/S)) and the residues in the loop immediately preceding the conserved cis-proline residue for the two complexes (R148GVP in EcDsbA and E88AMP in thioredoxin) superimposed with an r.m.s. deviation value of 0.52 Å. Furthermore, the buried surface areas observed for the DsbA-substrate complex (760 and 795 Å2) were similar to that in the thioredoxin-substrate complex (760 Å2) (48). The presence of mostly main-chain interactions as well as the relatively low interacting surface in the EcDsbA-SigA complex is consistent with the broad substrate specificity observed with most DsbA enzymes and the diverse sequences of potential DsbA substrates (31). Calorimetric studies have previously demonstrated that the interactions between DsbA and its substrate proteins are relatively weak (37), which suggested that small changes in the active site of the enzyme may be sufficient to alter substrate specificity. This is supported by previous mutational data, whereby it was demonstrated that a V150G mutation in EcDsbA was defective in complex formation with substrate proteins (31). A similar mode of binding has now been observed for a number of TRX domain-containing enzymes, including thioredoxin (48, 52, 53), glutaredoxin (54), glutathione transferase (55) as well as the mixed disulfide complex of DsbC-DsbD (56). This mode of interaction is emerging as a general feature of substrate recognition in TRX domain-substrate complexes (48).
However, comparison of the current structure and that of the EcDsbA-EcDsbB complex, suggests that there are subtle differences in the manner in which reduced and oxidized EcDsbA bind their respective substrates. Thus, EcDsbB bound within the hydrophobic groove of reduced EcDsbA, whereas the SigA peptide substrate did not. Comparison of the primary structure of proteins that have been identified as substrate proteins for EcDsbA (Table 3) reveals that there is no conserved sequence of hydrophobic residues in the substrate proteins that would presumably be required for favorable interaction within the hydrophobic groove. In fact, for those proteins that have been confirmed to be substrates of EcDsbA, there is no sequence conservation in the residues flanking the cysteine (Table 3).
TABLE 3.
Sequence conservation in DsbA substrates
Forty-five sequences of confirmed substrates of DsbA were compared to assess sequence conservation around the cysteine residue. Analysis of the sequences revealed that there was no conservation beyond the cysteine residue itself, as determined using Conscore.
| Conservative mutations | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Hydrophobic, small (AVLI) | 17 | 9 | 13 | 8 | 13 | 0 | 15 | 15 | 11 | 5 | 7 |
| Hydrophobic, aromatic (FYW) | 2 | 8 | 3 | 7 | 3 | 0 | 3 | 4 | 4 | 5 | 2 |
| Acidic (DE) | 6 | 5 | 6 | 5 | 7 | 0 | 5 | 4 | 2 | 7 | 9 |
| Basic (RHK) | 7 | 4 | 5 | 6 | 1 | 0 | 5 | 6 | 6 | 8 | 4 |
| Neutral polar (SMTCNQ) | 10 | 15 | 9 | 14 | 18 | 45 | 14 | 9 | 13 | 18 | 15 |
| Glycine | 2 | 3 | 6 | 4 | 2 | 0 | 2 | 1 | 5 | 1 | 2 |
| Proline | 1 | 1 | 3 | 1 | 1 | 0 | 1 | 6 | 4 | 1 | 5 |
| Percentage of conserved mutations | 40.5 | 36.6 | 36.1 | 35.0 | 42.9 | 100.0 | 35.7 | 39.5 | 36.1 | 41.9 | 40.5 |
As a consequence of the different location of binding, the SigA peptide did not contact the conserved cis-Pro151 within the hydrophobic groove. It has previously been demonstrated that the mutation P151T in EcDsbA results in the accumulation of DsbA-substrate complexes (31) in the periplasm of E. coli. This suggests that the P151T mutant is able to both recognize and form a mixed disulfide with a range of substrate proteins, but is defective in the step of the reaction pathway where the complex is resolved with the release of oxidized substrate and reduced DsbA (Fig. 1). The structure and activity of EcDsbA in which the cis-proline was mutated to alanine has previously been reported (57). The activity of EcDsbAP151A, as determined from its ability to oxidize alkaline phosphatase in a cell-based assay, was significantly lower than that of wild type EcDsbA. However, EcDsbB-mediated reoxidation of EcDsbAP151A was not impaired. In the structure of EcDsbAP151A, the conformation of the active site residues 30CPHC33 was unchanged, whereas the positions of residues in the cis-proline loop were significantly perturbed. These findings suggest that the residues in the loops surrounding the active site may be of greater importance in substrate recognition and binding by DsbA than the hydrophobic groove that is the binding site for DsbB.
Structures have recently been reported for SaDsbA (24) and NmDsbA3 (23), both of which are oxidoreductase enzymes that, in contrast to EcDsbA, display a limited substrate repertoire as well as E. coli DsbL, which is thought to have specificity for the enzyme arylsulfate sulfotransferase (25). Comparison of each of these structures with that of EcDsbA reveals significant differences around the regions identified as the peptide binding surface in the current study. Notably, SaDsbA and NmDsbA3 have a similar TcP sequence in the cis-proline loop. In the case of SaDsbA, mutation of this sequence to VcP, which is more common in Gram-negative DsbA enzymes, enhanced the oxidoreductase activity as measured in an insulin reduction assay (24). Thus, it appears that the sequence of the cis-proline loop may be important in dictating the substrate binding of DsbA, as has previously been demonstrated with other TRX domain proteins (27, 28). Although DsbL retains the VcP sequence in the cis-proline loop, it lacks the hydrophobic surface features that surround the active site in EcDsbA and VcDsbA, which supports the notion that these are important for substrate binding and specificity.
Although the structures of EcDsbA are very similar throughout the catalytic cycle, differences have been observed in the relative orientation of the TRX and α-helical domains in several structures of DsbA (17, 18). These are present both for different DsbA molecules in the same crystal structure, between DsbA structures solved by different groups, and between the different forms of DsbA. This has led to the suggestion that dynamics may play a role in the catalytic activity of DsbA (20). Analysis of the dynamics in reduced and oxidized forms of VcDsbA has revealed the presence of interdomain motions in the reduced form of the protein (18), which result in an opening of the hydrophobic groove. Redox-dependent conformational changes have previously been demonstrated to result in the opening of a cavity in the TRX domain-containing protein ResA (58), which contributes to substrate specificity. In the case of DsbA, the conformations of the different redox states are similar, but changes in the dynamics appear to allow the reduced form of DsbA to access a more open conformation, which may facilitate the interaction with the DsbB loop and enable the formation of a mixed disulfide between Cys104 of EcDsbB and Cys30 of EcDsbA.
The structure of the EcDsbA-peptide complex presented herein provides an insight into the specificity observed within the catalytic cycle of this enzyme. In conjunction with the description of the structure of a EcDsbA-EcDsbB complex (21) and differences in the dynamics of the different redox states of the VcDsbA enzyme (18), a clearer picture is beginning to emerge of the factors regulating the catalytic cycle of DsbA. Thus, oxidized DsbA appears to bind its substrates through a mechanism that is common to many TRX family oxidoreductases via the formation of backbone hydrogen bonds to a loop immediately prior to the conserved cis-proline residue of the TRX fold. Subtle changes in the composition of this loop and the residues that surround it appear to influence substrate specificity. The relatively higher specificity of reduced DsbA for DsbB results from additional interactions of DsbB within the hydrophobic groove on the surface of DsbA, which may become more accessible as a result of interdomain motions that are present in the reduced DsbA.
Supplementary Material
Acknowledgments
We greatly appreciate the gift of strains JCB570 and JCB571 from Professor James Bardwell (University of Michigan, Ann Arbor, MI). We thank the BIOCARS staff at the Advanced Photon Source for assistance with data collection. NMR data were analyzed using the program SPARKY.6
This work was supported in part by Australian Research Council (ARC) Grant LP0455508 and National Health and Medical Research Council (NHMRC) Grant 455860.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4 and Tables 1 and 2.
The atomic coordinates and structure factors (code 3DKS) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
6 T. D. Goddard and D. G. Kneller, unpublished data.
- TRX
- thioredoxin
- Trt
- triphenylmethyl
- cP
- cis-proline
- DTT
- dithiothreitol
- HPLC
- high pressure liquid chromatography
- Hse
- homoserine residue
- r.m.s.
- root mean square.
REFERENCES
- 1.Collet J. F., Bardwell J. C. ( 2002) Mol. Microbiol. 44, 1– 8 [DOI] [PubMed] [Google Scholar]
- 2.Bardwell J. C., Lee J. O., Jander G., Martin N., Belin D., Beckwith J. ( 1993) Proc. Natl. Acad. Sci. U. S. A. 90, 1038– 1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heras B., Shouldice S. R., Totsika M., Scanlon M. J., Schembri M. A., Martin J. L. ( 2009) Nat. Rev. Microbiol. 7, 215– 225 [DOI] [PubMed] [Google Scholar]
- 4.Ha U. H., Wang Y., Jin S. ( 2003) Infect. Immun. 71, 1590– 1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dacheux D., Epaulard O., de Groot A., Guery B., Leberre R., Attree I., Polack B., Toussaint B. ( 2002) Infect. Immun. 70, 3973– 3977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watarai M., Tobe T., Yoshikawa M., Sasakawa C. ( 1995) Proc. Natl. Acad. Sci. U. S. A. 92, 4927– 4931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miki T., Okada N., Danbara H. ( 2004) J. Biol. Chem. 279, 34631– 34642 [DOI] [PubMed] [Google Scholar]
- 8.Jackson M. W., Plano G. V. ( 1999) J. Bacteriol. 181, 5126– 5130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peek J. A., Taylor R. K. ( 1992) Proc. Natl. Acad. Sci. U. S. A. 89, 6210– 6214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bardwell J. C., McGovern K., Beckwith J. ( 1991) Cell 67, 581– 589 [DOI] [PubMed] [Google Scholar]
- 11.Missiakas D., Georgopoulos C., Raina S. ( 1993) Proc. Natl. Acad. Sci. U. S. A. 90, 7084– 7088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stafford S. J., Humphreys D. P., Lund P. A. ( 1999) FEMS Microbiol. Lett. 174, 179– 184 [DOI] [PubMed] [Google Scholar]
- 13.Yu J. ( 1998) Infect. Immun. 66, 3909– 3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin J. L., Bardwell J. C., Kuriyan J. ( 1993) Nature 365, 464– 468 [DOI] [PubMed] [Google Scholar]
- 15.Hu S. H., Peek J. A., Rattigan E., Taylor R. K., Martin J. L. ( 1997) J. Mol. Biol. 268, 137– 146 [DOI] [PubMed] [Google Scholar]
- 16.Schirra H. J., Renner C., Czisch M., Huber-Wunderlich M., Holak T. A., Glockshuber R. ( 1998) Biochemistry 37, 6263– 6276 [DOI] [PubMed] [Google Scholar]
- 17.Guddat L. W., Bardwell J. C., Martin J. L. ( 1998) Structure 6, 757– 767 [DOI] [PubMed] [Google Scholar]
- 18.Horne J., d'Auvergne E. J., Coles M., Velkov T., Chin Y., Charman W. N., Prankerd R., Gooley P. R., Scanlon M. J. ( 2007) J. Mol. Biol. 371, 703– 716 [DOI] [PubMed] [Google Scholar]
- 19.Martin J. L. ( 1995) Structure 3, 245– 250 [DOI] [PubMed] [Google Scholar]
- 20.Guddat L. W., Bardwell J. C., Zander T., Martin J. L. ( 1997) Protein Sci. 6, 1148– 1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inaba K., Murakami S., Suzuki M., Nakagawa A., Yamashita E., Okada K., Ito K. ( 2006) Cell 127, 789– 801 [DOI] [PubMed] [Google Scholar]
- 22.Dutton R. J., Boyd D., Berkmen M., Beckwith J. ( 2008) Proc. Natl. Acad. Sci. U. S. A. 105, 11933– 11938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vivian J. P., Scoullar J., Robertson A. L., Bottomley S. P., Horne J., Chin Y., Wielens J., Thompson P. E., Velkov T., Piek S., Byres E., Beddoe T., Wilce M. C., Kahler C. M., Rossjohn J., Scanlon M. J. ( 2008) J. Biol. Chem. 283, 32452– 32461 [DOI] [PubMed] [Google Scholar]
- 24.Heras B., Kurz M., Jarrott R., Shouldice S. R., Frei P., Robin G., Cemazar M., Thöny-Meyer L., Glockshuber R., Martin J. L. ( 2008) J. Biol. Chem. 283, 4261– 4271 [DOI] [PubMed] [Google Scholar]
- 25.Grimshaw J. P., Stirnimann C. U., Brozzo M. S., Malojcic G., Grütter M. G., Capitani G., Glockshuber R. ( 2008) J. Mol. Biol. 380, 667– 680 [DOI] [PubMed] [Google Scholar]
- 26.Dumoulin A., Grauschopf U., Bischoff M., Thöny-Meyer L., Berger-Bächi B. ( 2005) Arch. Microbiol. 184, 117– 128 [DOI] [PubMed] [Google Scholar]
- 27.Hiniker A., Ren G., Heras B., Zheng Y., Laurinec S., Jobson R. W., Stuckey J. A., Martin J. L., Bardwell J. C. ( 2007) Proc. Natl. Acad. Sci. U. S. A. 104, 11670– 11675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren G., Stephan D., Xu Z., Zheng Y., Tang D., Harrison R. S., Kurz M., Jarrott R., Shouldice S. R., Hiniker A., Martin J. L., Heras B., Bardwell J. C. ( 2009) J. Biol. Chem. 284, 10150– 10159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sinha S., Langford P. R., Kroll J. S. ( 2004) Microbiology 150, 2993– 3000 [DOI] [PubMed] [Google Scholar]
- 30.Tinsley C. R., Voulhoux R., Beretti J. L., Tommassen J., Nassif X. ( 2004) J. Biol. Chem. 279, 27078– 27087 [DOI] [PubMed] [Google Scholar]
- 31.Kadokura H., Tian H., Zander T., Bardwell J. C., Beckwith J. ( 2004) Science 303, 534– 537 [DOI] [PubMed] [Google Scholar]
- 32.Darby N. J., Creighton T. E. ( 1995) Biochemistry 34, 3576– 3587 [DOI] [PubMed] [Google Scholar]
- 33.Ruddock L. W., Hirst T. R., Freedman R. B. ( 1996) Biochem. J. 315, 1001– 1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skórko-Glonek J., Sobiecka-Szkatuła A., Lipiñska B. ( 2006) Acta Biochim. Pol. 53, 585– 589 [PubMed] [Google Scholar]
- 35.Amann E., Ochs B., Abel K. J. ( 1988) Gene 69, 301– 315 [DOI] [PubMed] [Google Scholar]
- 36.Marley J., Lu M., Bracken C. ( 2001) J. Biomol. NMR 20, 71– 75 [DOI] [PubMed] [Google Scholar]
- 37.Couprie J., Vinci F., Dugave C., Quéméneur E., Moutiez M. ( 2000) Biochemistry 39, 6732– 6742 [DOI] [PubMed] [Google Scholar]
- 38.Collaborative Crystallography Project 4 ( 1994) Acta Crystallogr. Sect. D 50, 760– 76315299374 [Google Scholar]
- 39.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. ( 1995) J. Biomol. NMR 6, 277– 293 [DOI] [PubMed] [Google Scholar]
- 40.Ayed A., Mulder F. A., Yi G. S., Lu Y., Kay L. E., Arrowsmith C. H. ( 2001) Nat. Struct. Biol. 8, 756– 760 [DOI] [PubMed] [Google Scholar]
- 41.Warrens A. N., Jones M. D., Lechler R. I. ( 1997) Gene 186, 29– 35 [DOI] [PubMed] [Google Scholar]
- 42.Chung C. T., Miller R. H. ( 1988) Nucleic Acids Res. 16, 3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sardesai A. A., Genevaux P., Schwager F., Ang D., Georgopoulos C. ( 2003) EMBO J. 22, 1461– 1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macnab R. M. ( 1986) Methods Enzymol. 125, 563– 581 [DOI] [PubMed] [Google Scholar]
- 45.Wielens J., Crosby I. T., Chalmers D. K. ( 2005) J. Comput. Aided Mol. Des. 19, 301– 317 [DOI] [PubMed] [Google Scholar]
- 46.Bader M., Muse W., Zander T., Bardwell J. ( 1998) J. Biol. Chem. 273, 10302– 10307 [DOI] [PubMed] [Google Scholar]
- 47.Petersen M. T., Jonson P. H., Petersen S. B. ( 1999) Protein Eng. 12, 535– 548 [DOI] [PubMed] [Google Scholar]
- 48.Maeda K., Hägglund P., Finnie C., Svensson B., Henriksen A. ( 2006) Structure 14, 1701– 1710 [DOI] [PubMed] [Google Scholar]
- 49.Lawrence M. C., Colman P. M. ( 1993) J. Mol. Biol. 234, 946– 950 [DOI] [PubMed] [Google Scholar]
- 50.Guddat L. W., Bardwell J. C., Glockshuber R., Huber-Wunderlich M., Zander T., Martin J. L. ( 1997) Protein Sci. 6, 1893– 1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hizukuri Y., Yakushi T., Kawagishi I., Homma M. ( 2006) J. Bacteriol. 188, 4190– 4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qin J., Clore G. M., Kennedy W. M., Huth J. R., Gronenborn A. M. ( 1995) Structure 3, 289– 297 [DOI] [PubMed] [Google Scholar]
- 53.Qin J., Clore G. M., Kennedy W. P., Kuszewski J., Gronenborn A. M. ( 1996) Structure 4, 613– 620 [DOI] [PubMed] [Google Scholar]
- 54.Nordstrand K., Åslund F., Holmgren A., Otting G., Berndt K. D. ( 1999) J. Mol. Biol. 286, 541– 552 [DOI] [PubMed] [Google Scholar]
- 55.Ladner J. E., Parsons J. F., Rife C. L., Gilliland G. L., Armstrong R. N. ( 2004) Biochemistry 43, 352– 361 [DOI] [PubMed] [Google Scholar]
- 56.Haebel P. W., Goldstone D., Katzen F., Beckwith J., Metcalf P. ( 2002) EMBO J. 21, 4774– 4784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Charbonnier J. B., Belin P., Moutiez M., Stura E. A., Quéméneur E. ( 1999) Protein Sci. 8, 96– 105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Colbert C. L., Wu Q., Erbel P. J., Gardner K. H., Deisenhofer J. ( 2006) Proc. Natl. Acad. Sci. U. S. A. 103, 4410– 4415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.