Abstract
Mia40 and Erv1 execute a disulfide relay to import the small Tim proteins into the mitochondrial intermembrane space. Here, we have reconstituted the oxidative folding pathway in vitro with Tim13 as a substrate and determined the midpoint potentials of Mia40 and Tim13. Specifically, Mia40 served as a direct oxidant of Tim13, and Erv1 was required to reoxidize Mia40. During oxidation, four electrons were transferred from Tim13 with the insertion of two disulfide bonds in succession. The extent of Tim13 oxidation was directly dependent on Mia40 concentration and independent of Erv1 concentration. Characterization of the midpoint potentials showed that electrons flowed from Tim13 with a more negative midpoint potential of −310 mV via Mia40 with an intermediate midpoint potential of −290 mV to the C130-C133 pair of Erv1 with a positive midpoint potential of −150 mV. Intermediary complexes between Tim13-Mia40 and Mia40-Erv1 were trapped. Last, mutating C133 of the catalytic C130-C133 pair or C30 of the shuttle C30-C33 pair in Erv1 abolished oxidation of Tim13, whereas mutating the cysteines in the redox-active CPC motif, but not the structural disulfide linkages of the CX9C motif of Mia40, prevented Tim13 oxidation. Thus, we demonstrate that Mia40, Erv1, and oxygen are the minimal machinery for Tim13 oxidation.
INTRODUCTION
To import proteins from the cytosol, the mitochondrion contains translocons of the outer membrane (TOM) and inner membrane (TIM). Proteins with a typical N-terminal targeting sequence are imported via the TIM23 pathway, whereas polytopic inner membrane proteins use the TIM22 import pathway (Milenkovic et al., 2007; Neupert and Herrmann, 2007). In contrast, proteins imported into the intermembrane space use a variety of import pathways (Herrmann and Hell, 2005). Notably, a class of proteins that have conserved cysteine motifs, including the small Tim proteins with the twin CX3C motif and proteins with a twin CX9C motif, such as Cox17 and Cox19, use Mia40 and Erv1 (Chacinska et al., 2004; Allen et al., 2005; Mesecke et al., 2005; Rissler et al., 2005). The Mia40-Erv1 pathway represents the oxidative folding pathway of the intermembrane space (Koehler et al., 2006; Herrmann and Kohl, 2007; Hell, 2008; Stojanovski et al., 2008).
Substrates of the oxidative folding pathway consist of a subset of intermembrane space residents that contain disulfide bonds. These include the small Tim proteins Tim8, Tim9, Tim10, Tim12, and Tim13 that form juxtapositional disulfide bonds (Curran et al., 2002; Allen et al., 2003; Webb et al., 2006). A large set of CX9C proteins, including Mia40, Cox17, Cox19, and several of uncharacterized function, also possess thiol bonds (Nobrega et al., 2002; Arnesano et al., 2005; Gabriel et al., 2007; Banci et al., 2008; Hell, 2008). In addition, the proteins Sco1 and Sco2, which mediate cytochrome oxidase assembly, contain disulfide linkages in a single CX3C motif (Williams et al., 2005; Banci et al., 2007). The machinery responsible for mediating the oxidative folding of these substrates consists of the receptor Mia40 and the sulfhydryl oxidase Erv1 (Tokatlidis, 2005; Hell, 2008; Stojanovski et al., 2008). Electrons are shuttled from Erv1 to a variety of electron acceptors, including oxygen and cytochrome c (Bihlmaier et al., 2007; Dabir et al., 2007). Thus, the mitochondrial intermembrane space supports the assembly of disulfide bonds in proteins like the bacterial periplasm and the endoplasmic reticulum.
Mia40 acts as a receptor for the import of these cysteine-rich proteins and forms a transient disulfide bond with the substrate (Chacinska et al., 2004; Mesecke et al., 2005; Terziyska et al., 2005; Grumbt et al., 2007). Mia40 contains three disulfide bonds. Specifically, two disulfide bonds are formed by cysteines in the twin CX9C motif, which may function in a structural role; the third disulfide bond is formed by the CPC motif near the N terminus, which participates in the thiol exchange reaction (Grumbt et al., 2007). After binding, the substrate is released from Mia40 in a folded state, and the sulfhydryl oxidase Erv1 reoxidizes Mia40 (Allen et al., 2005; Mesecke et al., 2005; Rissler et al., 2005).
Erv1 has three redox-active centers (Coppock and Thorpe, 2006; Dabir et al., 2007). There are two pairs of redox-active cysteines at C30-C33 and C130-C133 that are separated by two amino acids (CX2C motif). The C130-C133 is located near the active site flavin and transfers electrons to FAD, whereas the C30-C33 pair, referred to as the “shuttle” disulfide motif for its role in mediating electron transfer from substrates to the active site, is located distal to the active site (Fass, 2008). Electrons are transferred from the substrate through the C30-C33 pair to C130-C133 and then to FAD (Fass, 2008). Subsequently, electrons from Erv1 are transferred via the flavin to a combination of electron acceptors that function synergistically (Farrell and Thorpe, 2005; Bihlmaier et al., 2007; Dabir et al., 2007). Erv1 forms a complex with cytochrome c (cyt c) and thus can transfer electrons directly to cyt c (Bihlmaier et al., 2007; Dabir et al., 2007). From cyt c, electrons are donated to the electron transport chain or to cytochrome c peroxidase (Ccp1) (Dabir et al., 2007). In addition, molecular oxygen accepts electrons from Erv1 to generate H2O2, which is in turn converted to water by Ccp1 (Farrell and Thorpe, 2005; Dabir et al., 2007). This oxidative folding pathway is therefore versatile at donating electrons to several terminal electron acceptors like the oxidative folding pathways in the bacterial periplasm (Bader et al., 1999).
That electrons can flow from a substrate to oxygen suggests that the midpoint potentials (Em) of the individual components are arranged from more negative to more positive values. The Em of a subset of the components has been determined. Specifically, the Em of substrates Tim9, Tim10, and Cox17 are −310, −320, and −340 mV, respectively (Lu and Woodburn, 2005; Voronova et al., 2007; Morgan and Lu, 2008). For the sulfhydryl oxidase Erv1, the Em for the active site C130-C133 pair, the shuttle C30-C33 pair, and the FAD are −150, −320, and −215 mV, respectively (Dabir et al., 2007). Electron acceptors cyt c and oxygen have an Em of +250 and +820 mV, respectively (Wallace, 1984).
Portions of this pathway have been reconstituted in vitro. For example, using the nonphysiological substrate dithiothreitol (DTT), electron transfer through Erv1 to oxygen and cyt c has been reconstituted (Farrell and Thorpe, 2005; Bihlmaier et al., 2007; Dabir et al., 2007). In addition, the transfer of disulfide bonds from Erv1 to Mia40 has been demonstrated previously (Grumbt et al., 2007). Here, we report in vitro reconstitution of the entire pathway from reduced Tim13 via Mia40 and Erv1 to oxygen with the subsequent generation of oxidized Tim13 and H2O2. The Em values of these components are generally aligned from more negative to more positive, supporting that this pathway is thermodynamically favorable for electron transfer. Thus, the oxidative folding of Tim13 can be modeled in vitro.
MATERIALS AND METHODS
Plasmids and Strains
Recombinant Erv1 was expressed and purified under native conditions as described previously (Dabir et al., 2007). The C-terminal region of Mia40 starting at amino acid 251 was amplified by PCR (primer pair 5′-GGAATTCTTAAGGTTTGGATTCCTC-3′ and 5′-CGGGATCCCGCTAAGCAATCTGAATCT-3′) from yeast stain GA74 and cloned into pET28a with an N-terminal 6xHIS-tag by using BamHI and EcoRI restriction sites. Mia40 was purified under native conditions using a similar protocol to that of Erv1 purification (Dabir et al., 2007). Cysteine point mutants of Mia40 and Erv1 were generated by site-directed mutagenesis using overlap extension and confirmed by nucleotide sequencing (Ho et al., 1989). Recombinant Tim13 was purified under denaturing conditions as described previously (Beverly et al., 2008) and was dialyzed into 20 mM Tris, pH 7.0, 150 mM KCl, and 1 mM EDTA. Proteins Tim13, Mia40, and Erv1 were detected with polyclonal antibodies and immunoblot analysis.
Redox Titrations
Redox titrations of disulfide/dithiol redox couples were performed as described previously (Krimm et al., 1998; Masuda et al., 2002; Dabir et al., 2007) by using either thiol labeling with monobromobimane (mBBr) or intrinsic tryptophan fluorescence. Redox equilibration was achieved in redox buffers consisting of defined mixtures of either reduced and oxidized dithiothreitol (DTT/DTTox) or glutathione (GSH/GSSG). DTT/DTTox was used for ambient redox potential (Eh) values from −230 to −400 mV, and GSH/GSSG for values more positive than −230 mV (Hirasawa et al., 2000).
More specifically, 100 μg of Tim13 was equilibrated in buffer containing 0.1 M HEPES, pH 7.0, and a DTT/DTTox ratio (total concentration of 2.0 or 5.0 mM) to generate ambient redox potentials of −240 to −400 mV. The samples were incubated for 2 h under aerobic conditions before incubation with mBBr for 30 min at 25°C. The protein was then precipitated by the addition of 10% trichloroacetic acid (TCA) and resuspended in 0.1 M Tris, pH 8.0, and 0.1% SDS. The fluorescence of the sample was measured on a Quant Master QM-4 spectrofluorometer from Photon Technology International (Lawrenceville, NJ), with an excitation wavelength of 380 nm and emission monitored at 475 nm.
Redox equilibrations for all Mia40 titrations were performed in the same manner. Thirty micrograms of Mia40 was equilibrated for 2 or 4 h in 0.1 M HEPES, pH 7.0, containing DTT or glutathione redox couples at a total concentration of 2.0 or 5.0 mM, generating ambient redox potentials of −100 to −400 mV under both aerobic and anaerobic conditions. Em values for Mia40 were independent of the redox equilibrium time and the total DTT or total GSH concentrations present in the redox equilibrium buffer (Setterdahl et al., 2003). Measurement of the midpoint potential using mBBr was performed as for Tim13 described above. To monitor the changes in the intrinsic tryptophan fluorescence after redox equilibration the fluorescence of the protein at 280 nm was immediately monitored by the spectrofluorimeter. For Mia40, differing results were obtained under ambient oxygen concentrations or under nitrogen atmosphere in the glove box, so the midpoint potential was based on the most reproducible method, which was intrinsic tryptophan fluorescence (Table 1). All Em values reported represent the average from at least three replicate titrations. The experimental errors in Em values estimated from the average deviations are all less than the inherent experimental uncertainties in redox titrations of approximately ±10 mV.
Table 1.
Midpoint potential measurements of Mia40
| Method | Status | Midpoint potential (mV) |
|---|---|---|
| mBBr titration | Anaerobic | −277 ± 7.5 |
| mBBr titration | Aerobic | −322 ± 2.5 |
| Intrinsic tryptophan fluorescence | Anaerobic | −289 ± 2.5 |
| Intrinsic tryptophan fluorescence | Aerobic | −289 ± 4.8 |
The raw data were normalized such that the fluorescent measurement at the most reducing redox potential was assigned a value of 100% reduced, and the fluorescent measurement at the most oxidizing redox potential was assigned a value of 0% reduced. A theoretical curve calculated from the Nernst equation at the respective reduction potentials was fit to the titration data for Mia40 and Tim13. The Mia40 data gave an excellent fit for a two-electron transfer, whereas the curve fit to the Tim13 data was solved for a four-electron transition. For titration of Tim13, curves for n = 2 (R2 = 0.9644) and n = 4 (R2 = 0.9923) electrons were included. The probability was higher (99.99%) that the data fit a curve for n = 4 electrons than n = 2 electrons (0.01%) as calculated using the informational theory approach Akaike's criterion using Prism (GraphPad Software, San Diego, CA; Akaike, 1974). If the difference in likelihood was not very large (i.e., 40 vs. 60%), either curve could have been likely.
Thiol Trapping Studies
For Tim13 samples after circular dichroism (CD) analysis, 1 μg of protein was modified by 20 mM maleimide-polyethylene glycol (PEG) 2K (Mal-PEG) (Figure 1C); addition of Mal-PEG increases the molecular mass by 5 kDa (Wu et al., 2000). Samples were resolved on 15% SDS gels and visualized by SYPRO Ruby stain using an FX ProPlus imager (Bio-Rad Laboratories, Hercules, CA).
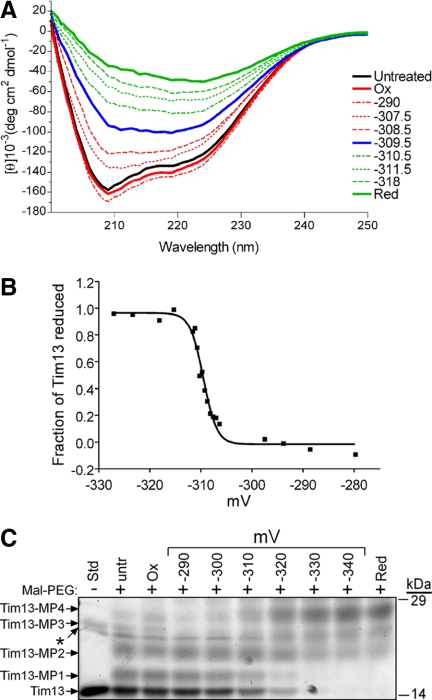
Figure 1.
The midpoint potential of Tim13 is −310 mV. The redox titration of the Tim13 dithiol/disulfide couple was performed in a DTT redox buffer at pH 7.0 under anaerobic conditions with a 20-h equilibration time. (A) The secondary structure of each sample was monitored by CD analysis at the indicated redox potentials. Control spectra for untreated, oxidized (Ox), and reduced (Red) Tim13 were included. (B) The fraction of reduced Tim13 in each sample from A was calculated from the differences in the CD spectra at 208 nm and plotted versus the midpoint potential of the sample. Data in all titrations were analyzed using nonlinear curve regression analysis, which yielded a sigmoidal curve. (C) Tim13 samples were incubated at the indicated redox potentials for 20 h followed by addition of Mal-PEG, analyzed by nonreducing SDS-PAGE, and detected by SYPRO Ruby staining. Up to four Mal-PEG equivalents attached to the four cysteine residues in Tim13, which is indicated by Tim13-MP1 through Tim13-MP4 (n = 5).
Mia40 was incubated in different redox buffers for 2 h at the indicated redox potentials (Figure 2C). Subsequently, Mia40 was precipitated using 10% TCA and then resuspended in sample buffer containing 20 mM 4-acetamido-4-maleimidylstilbene-2,2-disulfonic acid (AMS). The samples were resolved on a 6–16% Tris-Tricine gel under nonreducing conditions followed by SYPRO Ruby staining.
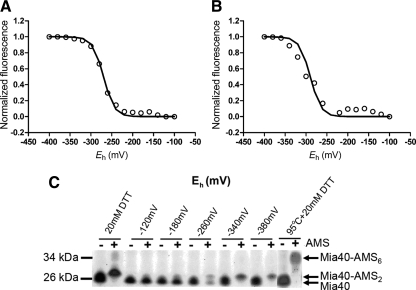
Figure 2.
The midpoint potential of Mia40 is approximately −290 mV. Redox titration of the dithiol/disulfide couple of Mia40 was performed in a DTT or glutathione redox buffer. Equilibration was achieved at 2 h at pH 7.0 under anaerobic conditions. Data in all titrations were fit to the Nernst equation for a two-electron carrier. (A) Redox titrations were performed with mBBr. The best fit to the mBBr fluorescence magnitude versus Eh value was obtained with an Em of −277 mV (n = 3). (B) Redox titrations were performed as in A and intrinsic tryptophan fluorescence was used to monitor the redox state of Mia40. The best fit to the data were for a single n = 2 component with Em = −289 mV (n = 3). (C) Mia40 samples were incubated in aerobic conditions at the indicated redox (Eh) values. Samples were treated with AMS, which adds a molecular mass of 0.5 kDa. As controls, reduced (20 mM DTT) and reduced-denatured (95°C + 20 mM DTT) samples were included, demonstrating the addition of two and six equivalents of AMS, respectively (n = 3).
Reconstitution Studies and Analysis of Trapped Mixed Disulfides
For the reconstitution assays, Tim13 was reduced in the presence of 5 mM DTT for 2 h. The DTT was removed by gel filtration using an NAP5 column, and Tim13 was eluted in 20 mM Tris-HCl, pH 7.0, 150 mM KCl, and 1 mM EDTA. In the reconstitution experiments, 15 μM reduced Tim13 was incubated with 0.5 μM Mia40 and/or Erv1, Erv1C133S, or Erv1C30S as indicated, for 4 h at 25°C in 20 mM Tris-HCl, pH 7.0, 150 mM KCl, and 1 mM EDTA. Concentrations of Mia40 and Erv1 and the length of incubation were varied per experiment as indicated. The redox state of Mia40 and Tim13 were determined by thiol trapping with AMS. For Tim13, 1 μg of sample was incubated with 20 mM AMS for 1 h at 37°C and then resolved by nonreducing SDS-polyacrylamide gel electrophoresis (PAGE). Tim13 was detected by immunoblot analysis. Quantitation was performed on a VersaDoc Imaging System model 5000 (Bio-Rad Laboratories) using Quantity One software (Bio-Rad Laboratories) and expressed as the fraction of monomeric Tim13 that is oxidized.
To detect the presence of mixed disulfide, 10 μM Mia40, Erv1, and reduced Tim13 were incubated at 30°C for 10 min. The reaction was halted by addition of 50 mM iodoacetimide (IAA). The proteins were then resolved on reducing and nonreducing SDS gels and detected by immunoblot analysis. As a control, similar amounts of each protein alone also were resolved on the same gel.
Analysis of Hydrogen Peroxide Levels
H2O2 levels in the reconstitution assays were measured using the Amplex Red Hydrogen Peroxide/Peroxidase Assay kit according to the manufacturer's protocol (Dabir et al., 2007; Invitrogen, Carlsbad, CA). In brief, Erv1 and Mia40 were mixed at the indicated concentrations with the Amplex Red/horseradish peroxidase reaction mix. The reaction was initiated by the addition of reduced Tim13. The Erv1-catalyzed reduction of O2 to H2O2 was measured by a FlexStation plate reader (Molecular Devices, Sunnyvale, CA) controlled via the SoftMax Pro software package (Molecular Devices) for data acquisition. Statistical analysis was performed using Prism software (GraphPad Software). The reaction was performed at a shorter time period than the reconstitution assays because the Amplex Red assay was very sensitive. Some background activity was detected in the assay, perhaps due to low levels of reductant from Tim13 purification. Therefore, we used the Amplex Red assay to verify that hydrogen peroxide was being produced in proportion to the oxidation of Tim13.
CD Studies
The redox equilibrations were performed in 20 mM Tris, pH 7.0, 150 mM KCl, and 1 mM EDTA. Tim13 (0.2 mg/ml) was equilibrated under anaerobic conditions for 20 h at the indicated redox potentials (Figure 1). The secondary structure content of recombinant proteins was detected using circular dichroism on a J-715 spectropolarimeter (Jasco, Tokyo, Japan) at 25°C. Each sample was analyzed with a scan speed of 50 nm/min, a bandwidth of 1.0 nm, and a wavelength interval of 0.5 nm. Four scans were averaged for each spectrum. The baseline correction option was used to subtract a buffer baseline. Spectra were recorded from 260 to 200 nm in a 1-mm pathlength cell, with protein concentrations of 0.1–0.2 mg/ml. Spectra were analyzed for secondary structure content by using a self-consistent method, the convex constraint algorithm for secondary structure prediction (Perczel et al., 1992; Sreerama and Woody, 1993; Greenfield, 2006), or both. For the folding studies in Figure 1A, control spectra in which Tim13 was reduced by incubation in 2 mM DTT, oxidized by incubation in 2 mM oxidized DTT, and untreated controls were included. Samples were equilibrated for 20 h. The change in the CD spectra at 208 nm was used to calculate the fraction of Tim13 reduced using the following equation:
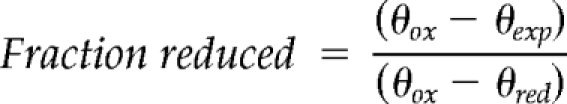 |
where θox is the molar ellipticity of the oxidized sample, θred is the molar ellipticity of the reduced sample, and θexp is the molar ellipticity of the experimental sample that is being calculated for each redox potential. We could not fit the Nernst equation to these data because it is an indirect measurement of electron transfer and a direct measure of changes in secondary structure content. Instead, nonlinear regression analysis was performed using Prism software (GraphPad Software). The midpoint from the generated curve is reported as the reduction potential for Tim13, which agrees with the midpoint potential calculated from mBBr redox titrations.
RESULTS
Tim13 Has a Midpoint Potential of −310 mV
Dissection of the specific steps in import of cysteine-rich proteins into the mitochondrial intermembrane space is difficult because Mia40 and Erv1 are essential proteins and mutations in key cysteine residues lead to inviability. However, mechanistic studies in the endoplasmic reticulum and bacterial periplasm have been completed by reconstitution experiments with excess amounts of substrate and catalytic amounts of oxidative folding components (Bader et al., 1998; Tu et al., 2000). With a similar goal of in vitro reconstitution of the oxidative folding pathway in the mitochondrion, the midpoint potentials of the individual components that were not characterized previously were determined. We measured the Em of Tim13 at equilibrium over a range of redox potentials by using mBBr titration (Dabir et al., 2007), CD studies (Curran et al., 2002), and thiol trapping (Curran et al., 2002; Dabir et al., 2007) in both anaerobic and aerobic conditions. Recombinant Tim13 was equilibrated in DTT/DTTox redox buffers to poise samples at Eh values ranging from −240 to −380 mV (Hirasawa et al., 1999; Dabir et al., 2007) and then incubated with mBBr to form a thiol-specific fluorescent covalent adduct (Dabir et al., 2007). The titration results were independent of the equilibration time and the anaerobic/aerobic state, indicating the likelihood of good redox equilibration between Tim13 and the ambient potential imposed by the redox buffers. The titration results were fit to curves for a two-electron and four-electron couple calculated from the Nernst equation. The titration gave an excellent fit to the Nernst equation for a four-electron redox couple and not a two-electron redox couple, suggesting that the two disulfide bonds formed simultaneously. A midpoint potential of −315 mV was calculated (Supplemental Figure S1).
When Tim13 was subject to CD studies, the recombinant protein displayed secondary structure content with α-helical properties (Figure 1A), as reported previously (Curran et al., 2002; Beverly et al., 2008). In the presence of oxidant (oxidized DTT), the secondary structure content of Tim13 was similar to that displayed by the untreated protein. When Tim13 was reduced by DTT treatment, the structure sustained conformational changes. This suggested that CD analysis could be used to determine the specific Em for Tim13. The critical midpoint potential was −310 mV; in reduction potentials more negative than −310 mV, Tim13 showed secondary structural content similar to Tim13 in the presence of oxidant; and in reduction potentials more positive than −310 mV, Tim13 showed secondary structural content similar to Tim13 in the presence of reductant (Figure 1A). The changes in CD spectra were used to calculate the fraction of Tim13 that was reduced. A nonlinear regression analysis on the resulting curve confirmed that the transition point was −310 mV (Figure 1B). Last, thiol-trapping with Mal-PEG was performed to confirm that the cysteine residues transitioned at this midpoint from a reduced to disulfide bonded state (Figure 1C). The addition of Mal-PEG to free cysteine residues increases the molecule weight by 5 kDa (Makmura et al., 2001). Tim13 was equilibrated at the indicated redox potentials and treated with Mal-PEG, followed by separation on nonreducing SDS-PAGE and detection with SYPRO Ruby staining. Indeed, at redox potentials more negative than −310 mV, approximately four Mal-PEG additions (designated Tim13-MP4) were detected, whereas at redox potentials more positive than −310 mV, the number of Mal-PEG additions decreased markedly to zero (Figure 1C). Together, Tim13 has one midpoint potential of −310 mV and four electrons were removed simultaneously in these equilibrium titration assays.
Mia40 Has a Midpoint Potential of Approximately −290 mV
We also determined the Em of Mia40 using mBBr titration and intrinsic tryptophan fluorescence titration under aerobic and anaerobic conditions. Mia40 has two CX9C motifs that assemble in structural disulfide bonds and one CPC motif that is seemingly redox active (Grumbt et al., 2007). With titration using mBBr or intrinsic tryptophan fluorescence in anaerobic or aerobic conditions, the analysis showed the presence of a single two-electron component (Figure 2, A and B). The average values for the Em were −277 and −289 mV for mBBr and intrinsic tryptophan fluorescence, respectively, as calculated by the Nernst equation. In contrast, when the Em was calculated in aerobic conditions, values of −322 and −289 mV were measured using titration methods with mBBr and intrinsic tryptophan fluorescence, respectively (Supplemental Figure S2). To confirm the redox potential titrations, the thiol status at the indicated redox potentials was monitored with the use of AMS, each AMS modification increases the molecular mass by 0.5 kDa) (Figure 2C). Mia40-AMS adducts were separated by nonreducing SDS-PAGE followed by detection with SYPRO Ruby staining. In control reactions, two AMS molecules were added when Mia40 was incubated in the presence of 20 mM DTT and six AMS molecules were added in the presence of 20 mM DTT and incubation at 95°C. Thus, full addition of AMS to the six cysteines was achieved only when the protein was completely unfolded by thermal denaturation (Grumbt et al., 2007). When Mia40 was incubated at redox potentials more reducing than −260 mV, two AMS equivalents were added; AMS addition was not detected at redox potentials more positive than −260 mV. Thus, the sum of several methods specify that the Em of Mia40 is approximately −290 mV (Table 1), placing it between that of Tim13 and the C130-C133 pair in the active site of Erv1. And of the six cysteine residues in Mia40, only two seem to be redox active.
The Combination of Erv1 and Mia40 Mediates Tim13 Oxidation In Vitro
To reconstitute the oxidative folding pathway with the minimal components, we expressed recombinant Erv1 and mutants C30S and C133S and characterized their sulfhydryl oxidase activity with the nonphysiologic substrate DTT (Supplemental Figure S3). Erv1 and the mutants C30S and C133S showed the same secondary structure content when analyzed by circular dichroism (Supplemental Figure S4). To generate a kinetic profile for Erv1 and C30S Erv1 mutant, both enzymes (1 μM) were incubated with DTT ranging from 0 to 400 μM. Enzymatic activity was measured using an Amplex Red substrate that is converted to fluorescently active resorufin in the presence of H2O2 (Dabir et al., 2007). The negative control for the H2O2 production assay was the enzymatically inactive C133S Erv1 mutant (1 μM) (Hofhaus et al., 2003). The velocity of H2O2 that was produced by the C133S Erv1 mutant was essentially zero, confirming that the mutation inactivates the enzyme. Erv1 and the C30S Erv1 mutant displayed a Vmax value of 1.18 and 0.68 pmol/s, respectively. In addition, the turnover numbers of 65 and 38 for Erv1 and the C30S Erv1 mutant were on par with the reported turnover numbers of 53 and 17 for Erv1 and the C30S Erv1 mutant (Hofhaus et al., 2003). Thus, the C30S Erv1 mutant showed some redox activity, indicating that the active site C130-C133 pair can partially oxidize the small substrate DTT and bypass the C30-C33 pair. Importantly, the recombinant Erv1 is an active sulfhydryl oxidase.
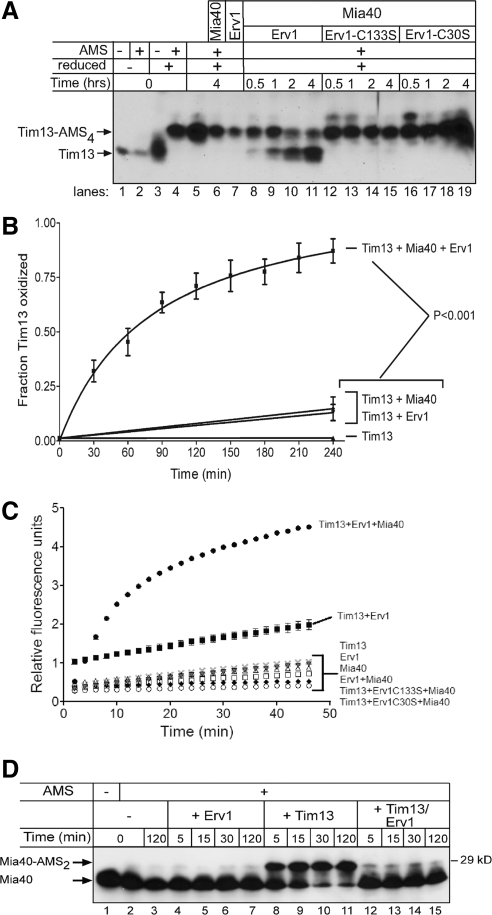
We reconstituted the oxidation of Tim13 by incubating 15 μM reduced Tim13 with 0.5 μM Mia40 and 0.5 μM Erv1 in an aerobic environment (Figure 3, A and B). At these concentrations, the substrate Tim13 is in excess and Mia40 and Erv1 are in catalytic concentrations, similar to systems reconstituted in the endoplasmic reticulum (Tu et al., 2000). Oxidation was monitored over a 4-h time course by the addition of AMS followed by nonreducing SDS-PAGE and immunoblot analysis with anti-Tim13; the absence of AMS addition indicated that Tim13 was oxidized. Tim13 was first reduced by treatment with DTT followed by gel filtration to remove the reductant. In a set of control reactions, AMS addition to the four cysteine residues of Tim13 occurred only after pretreatment of Tim13 with reductant (Figure 3A, lanes 1–4), confirming that the Tim13 thiols were reduced at the start of the assay. When Tim13 was incubated for 4 h alone or in the presence of Mia40 or Erv1, minor amounts of Tim13 were oxidized because AMS added to the majority of the Tim13 pool (Figure 3A, lanes 5–7, and B). This confirms that Tim13 did not oxidize spontaneously during the assay nor did Mia40 or Erv1 alone result in substantial oxidation of Tim13. However, when incubated with Erv1 and Mia40 together, Tim13 was oxidized as shown by the decreased AMS addition to the Tim13 pool over time (Figure 3A, lanes 8–11, and B). The amount of oxidized Tim13 was determined by quantitation using densitometry and the fraction of oxidized Tim13 (unmodified by AMS) was expressed as the percentage of total Tim13 (Figure 3B). Specifically, oxidized Tim13 was detected within 30 min after addition of both Mia40 and Erv1; and the fraction of oxidized Tim13 increased with the incubation time. Interestingly, an intermediate state with two AMS additions was not detected, indicating that all four cysteines were oxidized at once in this assay. Importantly, active Erv1 was required, because mutation of the shuttle (C30S) or active site (C133S) cysteine pair impaired Tim13 oxidation (Figure 3A, lanes 12–19). Therefore, the combination of both Erv1 and Mia40 is required for Tim13 oxidation.
Figure 3.
The combination of Mia40 and Erv1 directly oxidize Tim13 in vitro. (A) Reduced Tim13 (15 μM) was incubated with combinations of Mia40 (0.5 μM), Erv1 (0.5 μM), and mutant Erv1 (C133S or C30S, 0.5 μM) in a time course assay as indicated. Aliquots were removed at the indicated times and free thiols on Tim13 were blocked with AMS. Oxidized and reduced (Tim13-AMS4) Tim13 were detected by nonreducing SDS-PAGE followed by immunoblotting with antibodies against Tim13. (B) The amount of oxidized Tim13 was quantitated using VersaDoc and associated Quantity One software (Bio-Rad Laboratories) (p < 0.001; n = 4). (C) H2O2 production was monitored over a 50-min time period during Tim13 oxidation. H2O2 production was detected with the indicator Amplex Red and displayed as relative fluorescence units (n = 3). (D) As in A, Mia40 was incubated with Erv1, reduced Tim13, and the combination of reduced Tim13 and Erv1. Aliquots were removed at 5, 15, 30, and 120 min, and the Mia40 oxidation state was monitored by thiol trapping with AMS and immunoblotting with an antibody against Mia40. Note that the addition of two AMS molecules was detected (Mia40-AMS2; n = 3).
As Tim13 was oxidized, H2O2 was produced concomitantly with oxygen consumption by Erv1. We measured H2O2 production using the fluorometric assay (Figure 3C) (Dabir et al., 2007). As predicted, H2O2 production was minimal when Tim13 was incubated alone or in the presence of Mia40. The combination of Mia40 and Erv1 in the absence of Tim13 also led to negligible H2O2 production. Only when Tim13 was oxidized in the presence of both Mia40 and Erv1 did H2O2 production increase by 400%. Although the combination of Tim13 and Erv1 resulted in an increase in H2O2 production, oxidation of Tim13 was not detected by thiol-trapping (Figure 3A, lane 7). Finally, active Erv1 was required because the reactions with Erv1 mutants C30S and C133S produced negligible amounts of H2O2. Thus, the Amplex Red fluorometric assay is a reliable method to monitor the flow of electrons from Tim13 through Erv1.
We also measured the oxidation state of Mia40 in the reconstitution assay (Figure 3D). In the control reaction, Mia40 was oxidized at the start of the reaction (Figure 3D, lanes 1 and 2) and remained oxidized after incubating for 2 h (Figure 3D, lane 3), because AMS addition was not detected. When Mia40 was incubated with Erv1 (Figure 3D, lanes 4–7), Mia40 remained oxidized. However, Mia40 became reduced when incubated with reduced Tim13 alone (Figure 3D, lanes 8–11), as indicated by the addition of two AMS molecules. The addition of Erv1 to the combination of Tim13 and Mia40 resulted in reoxidation of Mia40 (Figure 3D, lanes 12–15). Thus, Mia40 oxidizes Tim13 directly and then Erv1 functions to reoxidize Mia40, completing the disulfide relay system.
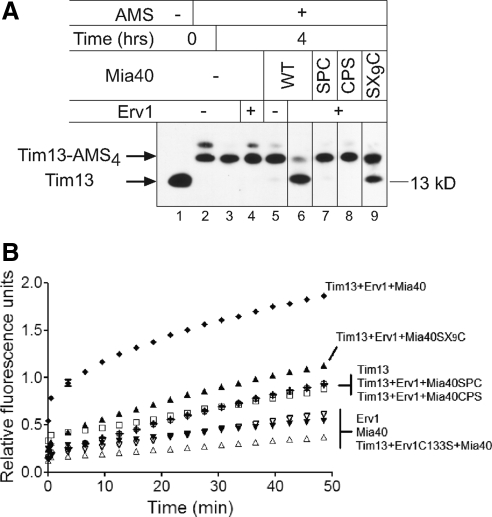
To test the requirement for the key cysteine residues of Mia40 in Tim13 oxidation, we sequentially mutated the first and second cysteines of the CPC motif to serine, generating SPC and CPS mutants, and the first cysteine of the CX9C motif, designated SX9C. From CD studies, the SPC and CPS mutants were folded similar to wild-type (wt) Mia40 (Supplemental Figure S4). In contrast, the SX9C mutant displayed less alpha-helical content than wt Mia40 as shown by the spectral differences at 222 nm (Supplemental Figure S4). These mutants were used in the oxidation assay as described in Figure 3A (Figure 4A). Whereas wt Mia40 in the presence of Erv1 oxidized Tim13, Mia40 mutants SPC and CPS in combination with Erv1 failed to oxidize Tim13. In contrast, the Mia40 mutant SX9C with Erv1 oxidized Tim13 but less efficiently compared with wt Mia40. The amount of H2O2 produced was significantly higher (p < 0.001) for the combination of Mia40 and Erv1 than the combination of Erv1 and the Mia40 mutants (Figure 4B). The cysteine residues in the CPC motif site are therefore redox active and essential for Tim13 oxidation.
Figure 4.
Mia40 is required for Tim13 oxidation in vitro. (A) As in Figure 3A, a time course of Tim13 oxidation was investigated at 4 h with Mia40 mutants (SPC, CPS, and SX9C). (B) As in Figure 3C, H2O2 production was monitored over a 50-min time period during Tim13 oxidation.
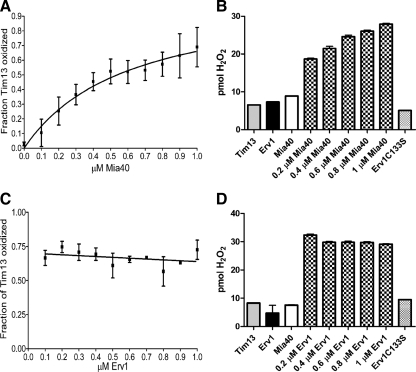
Dependence of Tim13 Oxidation on Mia40 Concentration
To pinpoint the dependence of the disulfide relay on Mia40 and Erv1, we tested whether Tim13 oxidation was dependent on Erv1 or Mia40 concentration. Specifically, the concentration of Tim13 (15 μM) and Erv1 (0.1 μM) was fixed, and the Mia40 concentration was varied over a range of 0–1 μM. The oxidation of Tim13 and the amount of H2O2 production was measured after a fixed period of time (Figure 5, A and B). As the concentration of Mia40 increased, the amount of Tim13 oxidation and of H2O2 production escalated. If the concentration of Erv1 was fixed in the assay at concentrations of 0.5 μM with varying Mia40 (0–1 μM), the oxidation curve for Tim13 was similar (data not shown). In reciprocal experiments in which Tim13 (15 μM) and Mia40 (1.0 μM) concentrations were fixed with varied Erv1 concentrations (0–1 μM), both the amount of Tim13 oxidized and H2O2 production remained constant (Figure 5, C and D). Based on the hypothesis of the disulfide relay, Mia40 seemingly oxidizes a small fraction of Tim13, presumably until Mia40 is completely reduced. Then, a catalytic amount of Erv1 subsequently reoxidizes Mia40 to allow further cycles of Tim13 oxidation. Therefore, the amount of Tim13 that becomes oxidized depends on the concentration of Mia40.
Figure 5.
Tim13 oxidation is dependent on Mia40 concentration. (A) The oxidation of 15 μM Tim13 was monitored as described in Figure 3, A and B, after 90 min in the presence of 0.1 μM Erv1 and 0–1.0 μM Mia40. (B) H2O2 production was monitored as in Figure 3C, after 35 minutes, in the presence of 15 μM Tim13, 1.0 μM Erv1, and 0–1.0 μM Mia40. (C) Tim13 oxidation was monitored as described in Figure 3, A and B, after 90 min in the presence of 1.0 μM Mia40 and 0.1–1.0 μM Erv1. (D) H2O2 production of was monitored as in Figure 3C, after 35 minutes, in the presence of 15 μM Tim13, 1.0 μM Mia40, and 0.2–1.0 μM Erv1 (n = 3).
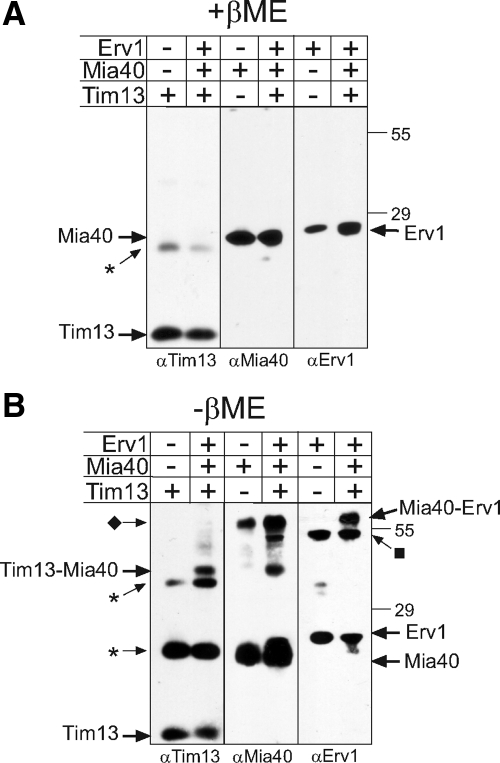
Tim13-Mia40 and Mia40-Erv1 Intermediates Are Formed during Tim13 Oxidation
Oxidation of Tim13 requires the generation of disulfide-bonded intermediates among Tim13, Mia40, and Erv1. To trap intermediates, equal concentrations of reduced Tim13 was incubated with Mia40 and Erv1, treated with IAA to block free thiols and subjected to nonreducing SDS-PAGE followed by immunoblotting for Tim13, Mia40, and Erv1 (Figure 6). As a control, the proteins were separated in the presence of β-mercaptoethanol (Figure 6A). A small fraction of the Tim13 formed multimers (indicated with the asterisk) that were not reduced by the presence of reductant. Mia40 and Erv1, however, were reduced and migrated as monomers.
Figure 6.
Mixed disulfides of Tim13-Mia40 and Mia40-Erv1 were detected in the presence of Tim13, Mia40, and Erv1. Equimolar amounts of recombinant Mia40, Erv1 and reduced Tim13 were incubated together for 10 min followed by treatment with IAA. The mix was separated on reducing (A) and nonreducing (B) SDS gels. Proteins were detected by immunoblotting with specific antibodies. As a control, a lane with the single protein is included. The asterisk denotes multimers of Tim13, the diamond marks a dimer of Mia40, and the square marks a dimer of Erv1.
In the absence of reductant, intermediates between Tim13-Mia40 and Mia40-Erv1 were detected by immunoblot analysis (Figure 6B). Again, nonspecific multimers of Tim13 (indicated by the asterisk) were formed. In addition, a dimer of Mia40 (indicated by ♦) and a dimer of Erv1 (indicated by ■) were identified. The intermediate between Mia40-Erv1 was difficult to detect because it migrates at a similar molecular mass as the Mia40 dimer, but given that the same band is detected with antibodies against Mia40 and Erv1 in the lanes where all three proteins were mixed, we interpret this as a Mia40-Erv1 intermediate. Neither a Tim13-Erv1 dimer nor a Tim13-Mia40-Erv1 trimer was identified. Together, this relay seems to consist of specific thiol linkages between Tim13-Mia40 and Mia40-Erv1.
DISCUSSION
The Midpoint Potentials of Tim13 and Mia40 Are Poised to Shuttle Electrons to Erv1
In this study, we present a detailed characterization of the redox behavior of Tim13 and Mia40, building on the previous study in which we examined the redox behavior of Erv1 (Dabir et al., 2007). Thus, we have filled in the gaps in the Em of the players of the disulfide relay and verified the direction of electron shuttling in this pathway. Like Tim9, Tim10, and Cox17 (Lu and Woodburn, 2005; Voronova et al., 2007; Banci et al., 2008; Morgan and Lu, 2008), the Em of Tim13 is very reducing at −310 mV (Figure 7). Interestingly, the equilibrium analysis (Figure 1) shows the presence of one transition of four electrons, indicating that Tim13 is either reduced or oxidized at once. This phenomena is shared by other substrates of this pathway as well such as Tim10 and Cox17 (Voronova et al., 2007; Banci et al., 2008). The CD analysis supports that this transition is accompanied by a structural change of “less structured” in the reduced state to “more structured” in the oxidized state. Unexpectedly, no evidence of a two electron transition for Tim13 was detected in our reconstitution experiments. Because the redox-active CPC motif of Mia40 is poised to receive only two electrons, two molecules of Mia40 may simultaneously bind to the substrate Tim13 or a combination of both Erv1 and Mia40 may directly oxidize Tim13 simultaneously. Alternatively, molecular oxygen could remove the second set of electrons from Tim13, but this does not seem likely, particularly in an anaerobic environment. Future studies will be required to analyze the coordinated removal of four electrons from Tim13.
Figure 7.
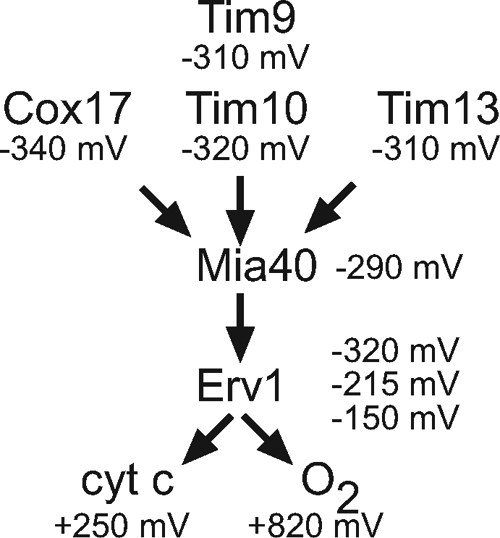
Schematic of electron flow during import of small Tim proteins in the mitochondrial intermembrane space. Electrons are transferred from substrates such as Tim10, Tim13, and Cox17 to Erv1 via Mia40. Electrons are then shuttled to cyt c and oxygen. See text for details.
In contrast, the Em of Mia40 was slightly more oxidizing at −290 mV. We used mBBr titration, intrinsic tryptophan fluorescence, and thiol trapping in anaerobic and aerobic conditions to investigate the redox status of Mia40. Although the measurements varied slightly (Table 1), the Em determined under anaerobic conditions seemed consistent (Figure 2). Given that Mia40 has six cysteine residues, up to three different Em were expected from the titration analysis. However, all methods consistently showed that only one inflection occurred that was accompanied by a two-electron transition when fit to the Nernst equation. This was specifically supported by the thiol trapping experiments with Mia40 (Figure 2C) in which two AMS molecules were added to Mia40 at potentials of −340 and −380 mV. Only when the protein was completely denatured by heat in the presence of reductant were six AMS molecules added. This suggests that the twin CX9C motif likely serves a structural role and that the CPC motif is involved in redox chemistry (Grumbt et al., 2007). It was interesting that the midpoint potential of human Mia40 was more oxidizing (−200 mV) (Banci et al., 2009), which might reflect a difference between mammalian and yeast proteins. Alternatively, the differing midpoint potentials may reflect differences in experimental conditions, especially as our study suggests that the observed midpoint potential can vary depending on experimental approach. Similarly, the redox titrations by Banci et al. (2009) were conducted with the glutathione redox couple that is poised for redox titrations more oxidizing than −230 mV (Hirasawa et al., 2000); midpoint potentials more reducing than −230 mV may not be measured as accurately with glutathione compared with DTT. Nevertheless, both studies place the midpoint potential of Mia40 between the substrates and Erv1.
From a previous study, we have determined the Em of Erv1 (Dabir et al., 2007). The C30-C33 pair has an Em of −320 mV and functions to shuttle electrons to the active site C130-C133 pair (Fass, 2008). The C130-C133 pair has an Em of −150 mV, whereas the FAD has a Em of −215 mV. Electron acceptors of Erv1, oxygen and cyt c, have more oxidative Em (Figure 7).
The Em are roughly sorted from more reducing for substrates to more oxidizing for Erv1 and cyt c, suggesting that shuttling of electrons from substrates to terminal electron acceptors is thermodynamically favored. Obviously, the Em of the C30-C33 pair of Erv1 suggests that it may be difficult to shuttle electrons from Mia40 (−290 mV) to the C30-C33 pair (−320 mV) and then to the active site cysteines (−150 mV) and FAD (−215 mV). However, the Em measurements are determined at thermodynamic equilibrium, and the Em of individual components within the context of the intermembrane space may vary, particularly in a kinetic scope when proteins are being imported and electrons are being transferred efficiently. Finally, the most positive Em of cyt c and oxygen may serve as a sink to draw electrons. Indeed, a hydrophobic oxygen channel in the related Erv2 sulfhydryl oxidase of the ER facilitates electron shuttling (Vitu et al., 2006).
Reconstitution of the Mia40-Erv1 Disulfide Relay
Although not as efficient as in organello import assays, the reconstitution assay with purified proteins allows us to investigate the details of this pathway and demonstrates clearly that catalytic amounts of Mia40 and Erv1 together are required to oxidize Tim13 efficiently. As shown in Figure 3D, Mia40 can directly oxidize Tim13, but Erv1 is needed to reoxidize Mia40. Our studies suggest that electrons flow from the substrate Tim13 through Mia40 to Erv1 and finally oxygen (Figure 7). To this end, intermediates between Tim13-Mia40 and Mia40-Erv1 were trapped, indicating that electrons likely flow through Mia40 to Erv1. Although Erv1 is seemingly not covalently bound to the Tim13-Mia40 intermediate, Erv1 dictates the redox state of Mia40 (Figure 3D). Also, the concentration of Mia40 determines the extent of Tim13 oxidation in the reconstitution assay. However, Erv1 is required for further rounds of import, because in its absence, Mia40 accumulates predominantly in the reduced form (Figure 3D) (Grumbt et al., 2007).
The CPC pair of Mia40 and both CX2C sites in Erv1 are critical for oxidation of Tim13. For Mia40, mutation of the first cysteine (SX9C) in the Mia40 twin CX9C motif slows, but does not abrogate, oxidation of Tim13. Because the SX9C mutation impairs folding of Mia40, the structure of the Mia40 core seems important for mediating thiol exchange reactions. For Erv1, both CX2C sites are essential for oxidation of Tim13, via recycling of Mia40, indicating that both sites are important in the disulfide relay. Based on the structure of Erv2, electrons most likely are shuttled through the distal C30-C33 pair to the active site C130-C133 and then the FAD (Fass, 2008). Even though the measured Em do not favor such a flow of electrons, our studies confirm that electrons most likely follow this pathway.
This work shows the reconstitution of the Mia40-Erv1 oxidative folding pathway in the mitochondrial intermembrane space. We have addressed specifically how the electrons are shuttled from substrate through components in this redox pathway. The in vitro reconstitution system is a novel approach particularly because the studies with Erv1 and Mia40 cysteine mutants cannot be done in vivo because the mutants are not viable. Establishing this assay also provides a framework to probe additional questions. Specifically, Mia40 has been the only substrate identified for Erv1. Given that Erv1 also may function in FeS cluster (Lange et al., 2001) and heme export (Dabir et al., 2007), it may have other substrates. In addition, it is not apparent how four electrons are removed simultaneously from Tim13 to Mia40, because an intermediate in which two electrons were first removed was not identified. It may be that such an intermediate is short-lived or an alternative mechanism may be used to remove the second set of electrons, with potential candidates including a second molecule of Mia40, Erv1, or molecular oxygen. Additional studies, designed to elucidate these pathways in greater detail, will be required.
Supplementary Material
ACKNOWLEDGMENTS
Establishment and equipment in the UCLA Mass Spectrometry and Proteomics Technology Center were supplied by a generous gift from the W. M. Keck Foundation. We thank F. Tsai for technical assistance and K. Beverly and M. Phillips for helpful discussions. This work was supported by U.S. Public Health Service National Research Service Award grant GM-07185 (to H.L.T.), U.S. Public Health Service National Research Service Award grant GM-008296 (to S.A.H.), United Mitochondrial Disease Foundation (to D.V.D.), UCLA Cota-Robles Fellowship (to S.A.N.), National Institutes of Health grant R01-GM-061721 (to C.M.K.), Muscular Dystrophy Association grant 022398 (to C.M.K.), and the American Heart Association grant 0640076N (to C.M.K.). C.M.K. is an Established Investigator of the American Heart Association.
Abbreviations used:
- AMS
4-acetamido-4-maleimidylstilbene-2,2-disulfonic acid
- CD
circular dichroism
- Em
midpoint potential
- IAA
iodoacetamide
- Mal-PEG
maleimide-PEG-2K
- mBBr
monobromobimane
- TIM
translocase of inner membrane
- TOM
translocase of inner membrane.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-10-1062) on May 28, 2009.
REFERENCES
- Akaike A new look at the statistical model identification. IEEE Trans. Automat. Control. 1974;19:716–723. [Google Scholar]
- Allen S., Balabanidou V., Sideris D. P., Lisowsky T., Tokatlidis K. Erv1 mediates the Mia40-dependent protein import pathway and provides a functional link to the respiratory chain by shuttling electrons to cytochrome c. J. Mol. Biol. 2005;353:937–944. doi: 10.1016/j.jmb.2005.08.049. [DOI] [PubMed] [Google Scholar]
- Allen S., Lu H., Thornton D., Tokatlidis K. Juxtaposition of the two distal “CX3C” motifs via intrachain disulfide bonding is essential for the folding of Tim10. J. Biol. Chem. 2003;278:38505–38513. doi: 10.1074/jbc.M306027200. [DOI] [PubMed] [Google Scholar]
- Arnesano F., Balatri E., Banci L., Bertini I., Winge D. R. Folding studies of Cox17 reveal an important interplay of cysteine oxidation and copper binding. Structure. 2005;13:713–722. doi: 10.1016/j.str.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Bader M., Muse W., Ballou D. P., Gassner C., Bardwell J. C. Oxidative protein folding is driven by the electron transport system. Cell. 1999;98:217–227. doi: 10.1016/s0092-8674(00)81016-8. [DOI] [PubMed] [Google Scholar]
- Bader M., Muse W., Zander T., Bardwell J. Reconstitution of a protein disulfide catalytic system. J. Biol. Chem. 1998;273:10302–10307. doi: 10.1074/jbc.273.17.10302. [DOI] [PubMed] [Google Scholar]
- Banci L., Bertini I., Cefaro C., Ciofi-Baffoni S., Gallo A., Martinelli M., Sideris D. P., Katrakili N., Tokatlidis K. MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat. Struct. Mol. Biol. 2009;16:198–206. doi: 10.1038/nsmb.1553. [DOI] [PubMed] [Google Scholar]
- Banci L., Bertini I., Ciofi-Baffoni S., Gerothanassis I. P., Leontari I., Martinelli M., Wang S. A structural characterization of human SCO2. Structure. 2007;15:1132–1140. doi: 10.1016/j.str.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Banci L., Bertini I., Ciofi-Baffoni S., Janicka A., Martinelli M., Kozlowski H., Palumaa P. A structural-dynamical characterization of human Cox17. J. Biol. Chem. 2008;283:7912–7920. doi: 10.1074/jbc.M708016200. [DOI] [PubMed] [Google Scholar]
- Beverly K. N., Sawaya M. R., Schmid E., Koehler C. M. The Tim8-Tim13 complex has multiple substrate binding sites and binds cooperatively to Tim23. J. Mol. Biol. 2008;382:1144–1156. doi: 10.1016/j.jmb.2008.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihlmaier K., Mesecke N., Terziyska N., Bien M., Hell K., Herrmann J. M. The disulfide relay system of mitochondria is connected to the respiratory chain. J. Cell Biol. 2007;179:389–395. doi: 10.1083/jcb.200707123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A., Pfannschmidt S., Wiedemann N., Kozjak V., Sanjuan Szklarz L. K., Schulze-Specking A., Truscott K. N., Guiard B., Meisinger C., Pfanner N. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 2004;23:3735–3746. doi: 10.1038/sj.emboj.7600389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppock D. L., Thorpe C. Multidomain flavin-dependent sulfhydryl oxidases. Antioxid. Redox. Signal. 2006;8:300–311. doi: 10.1089/ars.2006.8.300. [DOI] [PubMed] [Google Scholar]
- Curran S. P., Leuenberger D., Schmidt E., Koehler C. M. The role of the Tim8p-Tim13p complex in a conserved import pathway for mitochondrial polytopic inner membrane proteins. J. Cell Biol. 2002;158:1017–1027. doi: 10.1083/jcb.200205124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabir D. V., Leverich E. P., Kim S. K., Tsai F. D., Hirasawa M., Knaff D. B., Koehler C. M. A role for cytochrome c and cytochrome c peroxidase in electron shuttling from Erv1. EMBO J. 2007;26:4801–4811. doi: 10.1038/sj.emboj.7601909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell S. R., Thorpe C. Augmenter of liver regeneration: a flavin-dependent sulfhydryl oxidase with cytochrome c reductase activity. Biochemistry. 2005;44:1532–1541. doi: 10.1021/bi0479555. [DOI] [PubMed] [Google Scholar]
- Fass D. The Erv family of sulfhydryl oxidases. Biochim. Biophys. Acta. 2008;1783:557–566. doi: 10.1016/j.bbamcr.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Gabriel K., Milenkovic D., Chacinska A., Muller J., Guiard B., Pfanner N., Meisinger C. Novel mitochondrial intermembrane space proteins as substrates of the MIA import pathway. J. Mol. Biol. 2007;365:612–620. doi: 10.1016/j.jmb.2006.10.038. [DOI] [PubMed] [Google Scholar]
- Greenfield N. J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 2006;1:2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumbt B., Stroobant V., Terziyska N., Israel L., Hell K. Functional characterization of Mia40p, the central component of the disulfide relay system of the mitochondrial intermembrane space. J. Biol. Chem. 2007;282:37461–37470. doi: 10.1074/jbc.M707439200. [DOI] [PubMed] [Google Scholar]
- Hell K. The Erv1-Mia40 disulfide relay system in the intermembrane space of mitochondria. Biochim. Biophys. Acta. 2008;1783:601–609. doi: 10.1016/j.bbamcr.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Herrmann J. M., Hell K. Chopped, trapped or tacked–protein translocation into the IMS of mitochondria. Trends Biochem. Sci. 2005;30:205–211. doi: 10.1016/j.tibs.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Herrmann J. M., Kohl R. Catch me if you can! Oxidative protein trapping in the intermembrane space of mitochondria. J. Cell Biol. 2007;176:559–563. doi: 10.1083/jcb.200611060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa M., Ruelland E., Schepens I., Issakidis-Bourguet E., Miginiac-Maslow M., Knaff D. B. Oxidation-reduction properties of the regulatory disulfides of sorghum chloroplast nicotinamide adenine dinucleotide phosphate-malate dehydrogenase. Biochemistry. 2000;39:3344–3350. doi: 10.1021/bi9916731. [DOI] [PubMed] [Google Scholar]
- Hirasawa M., Schurmann P., Jacquot J. P., Manieri W., Jacquot P., Keryer E., Hartman F. C., Knaff D. B. Oxidation-reduction properties of chloroplast thioredoxins, ferredoxin:thioredoxin reductase, and thioredoxin f-regulated enzymes. Biochemistry. 1999;38:5200–5205. doi: 10.1021/bi982783v. [DOI] [PubMed] [Google Scholar]
- Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Hofhaus G., Lee J. E., Tews I., Rosenberg B., Lisowsky T. The N-terminal cysteine pair of yeast sulfhydryl oxidase Erv1p is essential for in vivo activity and interacts with the primary redox centre. Eur. J. Biochem. 2003;270:1528–1535. doi: 10.1046/j.1432-1033.2003.03519.x. [DOI] [PubMed] [Google Scholar]
- Koehler C. M., Beverly K. N., Leverich E. P. Redox pathways of the mitochondrion. Antioxid. Redox. Signal. 2006;8:813–822. doi: 10.1089/ars.2006.8.813. [DOI] [PubMed] [Google Scholar]
- Krimm I., Lemaire S., Ruelland E., Miginiac-Maslow M., Jaquot J. P., Hirasawa M., Knaff D. B., Lancelin J. M. The single mutation Trp35–>Ala in the 35–40 redox site of Chlamydomonas reinhardtii thioredoxin h affects its biochemical activity and the pH dependence of C36–C39 1H–13C NMR. Eur. J. Biochem. 1998;255:185–195. doi: 10.1046/j.1432-1327.1998.2550185.x. [DOI] [PubMed] [Google Scholar]
- Lange H., Lisowsky T., Gerber J., Muhlenhoff U., Kispal G., Lill R. An essential function of the mitochondrial sulfhydryl oxidase Erv1p/ALR in the maturation of cytosolic Fe/S proteins. EMBO Rep. 2001;2:715–720. doi: 10.1093/embo-reports/kve161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Woodburn J. Zinc binding stabilizes mitochondrial Tim10 in a reduced and import-competent state kinetically. J. Mol. Biol. 2005;353:897–910. doi: 10.1016/j.jmb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Makmura L., Hamann M., Areopagita A., Furuta S., Munoz A., Momand J. Development of a sensitive assay to detect reversibly oxidized protein cysteine sulfhydryl groups. Antioxid. Redox. Signal. 2001;3:1105–1118. doi: 10.1089/152308601317203611. [DOI] [PubMed] [Google Scholar]
- Masuda S., Dong C., Swem D., Setterdahl A. T., Knaff D. B., Bauer C. E. Repression of photosynthesis gene expression by formation of a disulfide bond in CrtJ. Proc. Natl. Acad. Sci. USA. 2002;99:7078–7083. doi: 10.1073/pnas.102013099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesecke N., Terziyska N., Kozany C., Baumann F., Neupert W., Hell K., Herrmann J. M. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121:1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Milenkovic D., Muller J., Stojanovski D., Pfanner N., Chacinska A. Diverse mechanisms and machineries for import of mitochondrial proteins. Biol. Chem. 2007;388:891–897. doi: 10.1515/BC.2007.097. [DOI] [PubMed] [Google Scholar]
- Morgan B., Lu H. Oxidative folding competes with mitochondrial import of the small Tim proteins. Biochem. J. 2008;411:115–122. doi: 10.1042/BJ20071476. [DOI] [PubMed] [Google Scholar]
- Neupert W., Herrmann J. M. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Nobrega M. P., Bandeira S. C., Beers J., Tzagoloff A. Characterization of COX19, a widely distributed gene required for expression of mitochondrial cytochrome oxidase. J. Biol. Chem. 2002;277:40206–40211. doi: 10.1074/jbc.M207348200. [DOI] [PubMed] [Google Scholar]
- Perczel A., Park K., Fasman G. D. Analysis of the circular dichroism spectrum of proteins using the convex constraint algorithm: a practical guide. Anal. Biochem. 1992;203:83–93. doi: 10.1016/0003-2697(92)90046-a. [DOI] [PubMed] [Google Scholar]
- Rissler M., Wiedemann N., Pfannschmidt S., Gabriel K., Guiard B., Pfanner N., Chacinska A. The essential mitochondrial protein Erv1 cooperates with Mia40 in biogenesis of intermembrane space proteins. J. Mol. Biol. 2005;353:485–492. doi: 10.1016/j.jmb.2005.08.051. [DOI] [PubMed] [Google Scholar]
- Setterdahl A. T., et al. Effect of pH on the oxidation-reduction properties of thioredoxins. Biochemistry. 2003;42:14877–14884. doi: 10.1021/bi0302088. [DOI] [PubMed] [Google Scholar]
- Sreerama N., Woody R. W. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem. 1993;209:32–44. doi: 10.1006/abio.1993.1079. [DOI] [PubMed] [Google Scholar]
- Stojanovski D., Muller J. M., Milenkovic D., Guiard B., Pfanner N., Chacinska A. The MIA system for protein import into the mitochondrial intermembrane space. Biochim. Biophys. Acta. 2008;1783:610–617. doi: 10.1016/j.bbamcr.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Terziyska N., Lutz T., Kozany C., Mokranjac D., Mesecke N., Neupert W., Herrmann J. M., Hell K. Mia40, a novel factor for protein import into the intermembrane space of mitochondria is able to bind metal ions. FEBS Lett. 2005;579:179–184. doi: 10.1016/j.febslet.2004.11.072. [DOI] [PubMed] [Google Scholar]
- Tokatlidis K. A disulfide relay system in mitochondria. Cell. 2005;121:965–967. doi: 10.1016/j.cell.2005.06.019. [DOI] [PubMed] [Google Scholar]
- Tu B. P., Ho-Schleyer S. C., Travers K. J., Weissman J. S. Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science. 2000;290:1571–1574. doi: 10.1126/science.290.5496.1571. [DOI] [PubMed] [Google Scholar]
- Vitu E., Bentzur M., Lisowsky T., Kaiser C. A., Fass D. Gain of function in an ERV/ALR sulfhydryl oxidase by molecular engineering of the shuttle disulfide. J. Mol. Biol. 2006;362:89–101. doi: 10.1016/j.jmb.2006.06.070. [DOI] [PubMed] [Google Scholar]
- Voronova A., Meyer-Klaucke W., Meyer T., Rompel A., Krebs B., Kazantseva J., Sillard R., Palumaa P. Oxidative switches in functioning of mammalian copper chaperone Cox17. Biochem. J. 2007;408:139–148. doi: 10.1042/BJ20070804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace C. J. The effect of complete or specific partial acetimidylation on the biological properties of cytochrome c and cytochrome c-T. Biochem. J. 1984;217:595–599. doi: 10.1042/bj2170595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb C. T., Gorman M. A., Lazarou M., Ryan M. T., Gulbis J. M. Crystal structure of the mitochondrial chaperone TIM9.10 reveals a six-bladed alpha-propeller. Mol. Cell. 2006;21:123–133. doi: 10.1016/j.molcel.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Williams J. C., Sue C., Banting G. S., Yang H., Glerum D. M., Hendrickson W. A., Schon E. A. Crystal structure of human SCO 1, implications for redox signaling by a mitochondrial cytochrome c oxidase “assembly” protein. J. Biol. Chem. 2005;280:15202–15211. doi: 10.1074/jbc.M410705200. [DOI] [PubMed] [Google Scholar]
- Wu H. H., Thomas J. A., Momand J. p53 protein oxidation in cultured cells in response to pyrrolidine dithiocarbamate: a novel method for relating the amount of p53 oxidation in vivo to the regulation of p53-responsive genes. Biochem. J. 2000;351:87–93. doi: 10.1042/0264-6021:3510087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.