Abstract
Control of cell cycle progression by stress-activated protein kinases (SAPKs) is essential for cell adaptation to extracellular stimuli. Exposure of yeast to osmostress activates the Hog1 SAPK, which modulates cell cycle progression at G1 and G2 by the phosphorylation of elements of the cell cycle machinery, such as Sic1 and Hsl1, and by down-regulation of G1 and G2 cyclins. Here, we show that upon stress, Hog1 also modulates S phase progression. The control of S phase is independent of the S phase DNA damage checkpoint and of the previously characterized Hog1 cell cycle targets Sic1 and Hsl1. Hog1 uses at least two distinct mechanisms in its control over S phase progression. At early S phase, the SAPK prevents firing of replication origins by delaying the accumulation of the S phase cyclins Clb5 and Clb6. In addition, Hog1 prevents S phase progression when activated later in S phase or cells containing a genetic bypass for cyclin-dependent kinase activity. Hog1 interacts with components of the replication complex and delays phosphorylation of the Dpb2 subunit of the DNA polymerase. The two mechanisms of Hog1 action lead to delayed firing of origins and prolonged replication, respectively. The Hog1-dependent delay of replication could be important to allow Hog1 to induce gene expression before replication.
INTRODUCTION
Activation of stress-activated protein kinases (SAPKs) is essential for proper cell adaptation to extracellular stimuli (Kyriakis and Avruch, 2001). In the budding yeast Saccharomyces cerevisiae, the presence of high osmolarity in the extracellular environment results in the activation of the p38-related stress-activated Hog1 kinase. Activation of Hog1 is essential for cell survival in response to high osmolarity, because the SAPK elicits an extensive program required for cell adaptation that includes regulation of gene expression, translation, and cell cycle progression (de Nadal et al., 2002; Hohmann, 2002). The presence of stress critically affects progression through the cell cycle, and cells must modulate cell cycle to allow for proper cellular adaptation. Activation of the Hog1 SAPK mediates a transient cell cycle arrest at both G1 and G2 phases of the cell cycle (Alexander et al., 2001; Yaakov et al., 2003; Escote et al., 2004; Clotet et al., 2006; Clotet and Posas, 2007).
Under normal growth conditions, control of cell cycle entry at START is exerted mainly by the activity of the cyclin-dependent kinase (CDK) Cln3-Cdc28, which promotes transcription of Cln1 and Cln2 cyclins upon cell growth. S phase is initiated when Clb5/6-Cdc28 is activated. The activity of this complex depends on the levels of Clb5/6 cyclins and the CDK inhibitor Sic1. Sic1 inactivates Clb5/6-Cdc28 complexes in late G1, and progression into S phase therefore requires degradation of Sic1. The control exerted by Hog1 at G1 requires the down-regulation of CLN1 and CLN2 expression as well as the direct phosphorylation and stabilization of the CDK-inhibitor protein Sic1 (Escote et al., 2004; Zapater et al., 2005). This combinatorial mechanism results in Sic1 stabilization and inhibition of cell cycle progression to prevent premature entry into S phase without proper adaptation to osmostress. Cells lacking Sic1 or containing a Sic1 allele mutated in the Hog1 phosphorylation site are unable to arrest at G1 upon Hog1 activation and enter S phase without being properly adapted, rendering them osmosensitive (Escote et al., 2004; Zapater et al., 2005).
Hog1 regulation of progression into mitosis also displays some similarities to its control over G1. The mitogen-activated protein kinase (MAPK) Hog1 mediates cell cycle arrest by the down-regulation of the mitotic cyclin Clb2 as well as by promoting the stabilization of the Swe1 protein kinase via Hsl1 phosphorylation (Clotet et al., 2006). In addition, it seems that Hog1 is important to modulate exit from mitosis, although the exact mechanism remains unclear (Reiser et al., 2006). Thus, Hog1 controls progression through multiple stages of the cell cycle.
The replication of the S. cerevisiae genome is a massive task in which many proteins are involved. Initiation of replication from replication origins (ARSs) is temporally controlled throughout S phase. The assembly of the replication complex (RC) at origins of replication is a highly ordered process beginning with the assembly of the prereplicative complex (pre-RC) from late mitosis to G1. In S phase, the pre-RC is converted into a fully assembled preinitiation complex and origins of replication are fired. This process is fully dependent on S-CDK (Cdc28-Clb5/6) and Dbf4-dependent kinase (DDK) (Cdc7-Dbf4) activity. Sld2 and Sld3 are the minimal set of essential S-CDK substrates for the onset of replication (Tanaka et al., 2007; Zegerman and Diffley, 2007). The role of S-CDK in the initiation of replication is circumvented in a strain in which these phosphorylation events have been bypassed by genetic means (Zegerman and Diffley, 2007). DDK acts locally and is essential for the firing of individual origins of replication. Accordingly, inactivation of temperature-sensitive alleles, such as cdc7ts4, prevents further replication origin firing in cells that have already entered S phase (Bousset and Diffley, 1998; Donaldson et al., 1998a). Although DDK is known to phosphorylate multiple subunits of the minichromosome maintenance (MCM) complex both in vitro and in vivo, the role of these phosphorylation events is not entirely understood except for the phosphorylation of Mcm4 by DDK (Bell and Dutta, 2002; Sheu and Stillman, 2006).
The DNA replication process can be disrupted by the presence of DNA lesions. This interferes with normal fork progression and leads to activation of the S phase DNA checkpoint. The Mec1 kinase of the ataxia telangiectasia mutated-related protein/ataxia telangiectasia mutated family is recruited to the stalled forks and leads to the autophosphorylation of the Rad53 effector kinase in coordination with the adaptor proteins Rad9 and Mrc1 (Longhese et al., 2003; Bartek and Lukas, 2007). This checkpoint elicits a response that includes the stabilization of replication forks and the inhibition of firing at late origins (Tercero and Diffley, 2001). Rad53 is the predominant effector kinase and is indispensable for both of these responses.
Here, we show that in addition to the control of G1 and G2 phases of the cell cycle, the MAPK Hog1 is also able to modulate S phase progression. The MAPK exerts its effect independently of the S phase DNA checkpoint as well as of Sic1 and Swe1. Hog1 controls S phase by delaying the expression of S phase cyclins Clb5 and Clb6 as well as by controlling replication when cells are stressed in late S phase, via a mechanism independent of the delay on Clb5 and Clb6 accumulation. Thus, the Hog1 MAPK regulates multiple stages of the cell cycle to enable cells to adapt before progressing through essential cell cycle transitions.
MATERIALS AND METHODS
Yeast Strains and Plasmids
Strains used were as follows: TM141 (MATa ura3 leu2 trp1 his3) and its derivatives (sln1-ts4), (sln1-ts4 hog1::LEU2), (sln1-ts4 sic1::KanMX), and (sln1-ts4 swe1::KanMX), and W303 (MATa, his3 leu2 trp1 ura3 ade2 can1) and its derivative (sic1-HA3::KanMX). Wild-type (wt) strain BY4741 (MATa his3-1 leu2-0 met15-0 ura3-0), its derivate (sic1::KanMX bar1::Hyg), and various tandem affinity purification (TAP)-tagged strains used were also from the BY4741 full TAP-fusion collection. Hemagglutinin (HA) tags were amplified using pCYC59 as a template and integrated at the C terminus of the RAD53, SLD2, or DPB2 alleles of wt BY4741 cells or TM141 sln1ts4 and its hog1 derivative. The W303-1b strain (MATa cdc7-4, MATa ade2-1, ura3-1, his3-11, 15 trp1-1, leu2-3, can100) was kindly provided by D. Quintana (Department of Biochemistry and Molecular Biology, School of Medicine, Universitat Autonoma de Barcelona, Barcelona, Spain) and crossed with W303-1a (MATa ade2-1, ura3-1, his3-11, 15 trp1-1, leu2-3, can100, hog1::LEU). The resulting spores were analyzed, and MATa strains carrying the cdc7-ts4 allele alone or combined with the hog1::LEU were isolated and used in this study. The YJT72 (sml1Δ) and YJT75 (rad53Δ sml1Δ) strains were also provided by David Quintana and have been described previously (Tercero and Diffley, 2001; Gunjan and Verreault, 2003). The yeast strain with the full bypass system was kindly provided by Dr. J.F.X. Diffley (Cancer Research UK London Research Institute, Clare Hall Laboratories, London, United Kingdom) y2067 sld2Δ::LEU2, trp1::PSLD2-sld2(T84D)::TRP1, sld3-600, 609, 622A-dpb11 (253-764)::KanMx, ura3::PGAL1-sic1ΔNT-myc::URA3 (Zegerman and Diffley, 2007).
The pRS426TEG1 and pRS426TEG1-Hog1 to express glutathione transferase (GST) and GST-Hog1 in yeast were described previously (Alepuz et al., 2003). The PBS2DD allele contains two mutations that substitute both phosphorylation sites required for Pbs2 activation to aspartic acid (Ser514 and Thr518).
Cell Synchronization, Cell Growth, and Fluorescence-activated Cell Sorting (FACS) Analyses
Cell synchronization in G1 was accomplished by treatment of exponentially growing cells (OD = 0.7) with 40 μg/ml α-factor pheromone for 3 h. For release from α-factor, cells were washed twice in fresh medium. For hydroxyurea (HU) synchronization, cultures were treated with 200 mM HU for 1 h directly after release from α-factor. Cells were liberated of HU by washing them twice in fresh medium. All time courses were carried out at 25°C, except for when temperature-sensitive mutants were shifted to their nonpermissive temperature of 37°C.
For flow cytometry analyses, cells were fixed in 70% ethanol, washed with 50 mM Na-citrate, treated with 0.1 mg/ml RNase A at 37°C overnight, stained with 4 μg/ml propidium iodide, delicately sonicated to disrupt cell aggregates, and analyzed in a FACScan flow cytometer (BD Biosciences, San Jose, CA). Ten thousand cells were analyzed for each sample. WinMDI 2.9 software (http://en.bio-soft.net/other/WinMDI.html; J. Trotter, Scripps Research Institute, La Jolla, CA) was used to attain FACS profiles.
Western Blot Analyses
For detecting gel mobility shifts of Rad53, Sld2, and Dpb2, the protein samples were resolved on maxigels of 7% acrylamide. Antibodies used were α-HA extracted from 12CA5 hybridoma and Peroxidase Anti-Peroxidase (PAP) soluble complex (Sigma-Aldrich, St. Louis, MO) to detect TAP, α-GST (27457701; APBiotech, Piscataway, NJ), α-Dbf4 yA-16 (Santa Cruz Biotechnology, Santa Cruz, CA), and α-Hog1 yC-20 (Santa Cruz Biotechnology). For Western blot analysis, trichloroacetic acid protein extracts were used as described in Bell et al. (2001).
Protein Extraction and Coimmunoprecipitation Binding Experiments
Protein from cells was extracted in buffer A + NaCl (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 15 mM EDTA, 15 mM EGTA, 0.1% Triton X-100, and 2 mM dithiothreitol supplemented with protease and phosphatase inhibitors). Cells were lysed twice with glass beads for 30 s in a FastPrep system (MP Biomedicals, Irvine, CA), and the lysates were cleared with a 5-min centrifugation at 13,000 × g. To analyze the association of Hog1 with components of the replication complex, protein was extracted from osmotically challenged HU-synchronized cells expressing GST or GST-Hog1 and specific chromosomally TAP-tagged proteins. One milligram of protein extract was incubated with 50 μl of glutathione-Sepharose 4B beads overnight at 4°C. The beads were washed extensively with buffer A + NaCl, resuspended in loading buffer, and resolved by SDS-polyacrylamide gel electrophoresis (PAGE).
Northern Blot Analysis
Total mRNA was obtained using standard procedures and expression of specific genes were probed using labeled polymerase chain reaction (PCR) fragments containing the open reading frame of RNR1 (1.7 kbp), GRE2 (1.0 kbp), HSP12 (0.3 kbp), or RPL28 (0.45 kbp).
Two-dimensional (2D) Electrophoresis and Hybridization
Total DNA from 100 ml of mid-log phase cells was isolated according to Allers and Lichten (2000). DNA was restricted with EcoRV and HindIII. First dimensions were run at room temperature for 20 h at 40 V in 0.4% agarose gels in 1× Tris borate-EDTA (TBE). Second dimensions were run at 4°C for 12 h at 140 V in 1.1% gels in 1× TBE + 0.3 μgr/ml ethidium bromide recirculating buffer. DNA was transferred onto a Hybond-XL membrane (GE Healthcare, Chalfont St. Giles, Buckinghamshire, United Kingdom) and hybridized with a specific probe for ARS305 obtained by PCR. Membranes were exposed to a Typhoon 8600 (GE Healthcare) and analyzed using the imaging software supplied.
RESULTS
Activation of Hog1 Provokes a Transient Cell Cycle Arrest at S Phase
Cells transiently arrest at G1 when osmotically stressed after release from pheromone arrest (Escote et al., 2004). In addition, we found that when cells were subjected to stress after crossing START, they were unable to arrest before S phase but instead progressed slowly into G2. Thus, we decided to study whether Hog1 could control cell cycle progression during S phase. Yeast cells were synchronized using α-factor, released into fresh media, and then subjected to osmostress (0.4 M NaCl) at different times after their release. As shown in Figure 1a, cells stressed 20 min after release into fresh media arrested ∼40 min in G1 before proceeding into S. However, when cells were stressed at minute 30, they entered S phase but took a longer time to complete S phase. Interestingly, these cells completed S phase with kinetics similar to those in cells that transiently arrested at G1, indicating that cells encountered a similar delay before reaching G2 (Figure 1a). Similar results were observed when cells were subjected to 0.8 M sorbitol instead of 0.4 M NaCl (Supplemental Figure 3). Thus, our results suggest that cells stressed after START delay cell cycle progression during S phase.
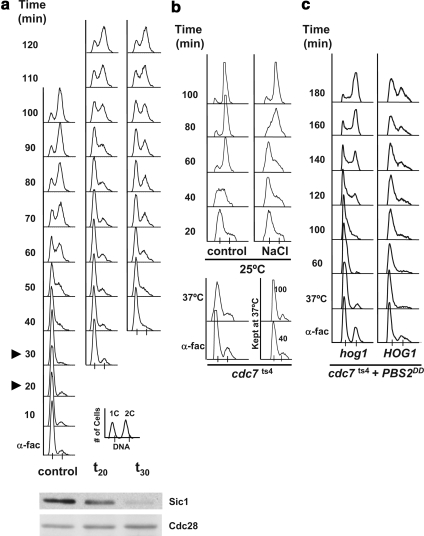
Figure 1.
HOG activation induces an S phase delay. (a) Cells osmostressed in S phase exhibit a distinct cell cycle delay than that in cells stressed in G1. Cells growing exponentially in YPD at 25°C were synchronized in G1 phase with α-factor for 3 h (α-fac), washed of α-factor, and released into fresh YPD medium (control) or subjected to 0.4 M NaCl 20 min (t20) or 30 (t30) min subsequent to release from α-factor, as indicated by the arrowheads in the control cells. Samples were taken every 10 min and assessed for total DNA content by FACS analysis. A key for the FACS profiles is presented at the bottom right corner. A Western blot monitoring Sic1 levels is shown underneath. There the left lane shows the amount of Sic1 just after release from alpha factor, the middle panel shows the amount of Sic1 at time 20 after release, and in the right panel the amount present at time 30. Cdc28 was used as a loading control for the Western blot. (b) Cells delay when osmostressed during S phase. An exponential culture of cdc7ts4 cells grown at 25°C was incubated with α-factor for 3 h (α-fac), washed free of α-factor, and immediately shifted to the nonpermissive temperature of 37°C for 1 h (37°C) to synchronize the cells in S phase. The culture was then shifted back to 25°C (control) and half of the cells were subjected to 0.4 M NaCl (NaCl). Part of the culture was maintained at 37°C throughout the experiment, and samples were taken after 40 and 100 min (kept at 37°C, bottom right-hand side). (c) S phase delay is dependent on HOG1. cdc7ts4 and the isogenic hog1Δ mutant harboring a plasmid bearing the Pbs2DD constitutively active MAPKK under the GAL1 promoter were grown and synchronized as described in b. Cells were grown with raffinose and galactose was added upon shifting the cells to 37°C. Experiments were performed at least twice, and representative results are shown.
To further characterize the effects of osmostress on S phase progression, a distinct experimental system was used using a temperature-sensitive CDC7 allele (cdc7ts4). At restrictive temperature, these cells arrest at the beginning of S phase because they cannot fire replication origins (see Introduction). cdc7ts4 cells were first arrested in G1 with α-factor and then shifted to nonpermissive temperature. Incubation of cdc7ts4 cells at nonpermissive temperature directly after release from α-factor resulted in cells efficiently synchronized in early S phase. Cells were then subsequently released at permissive temperature into media with or without NaCl (Figure 1b). As a control, a fraction of the cells was kept at restrictive temperature throughout the entire experiment and as expected, these cells were unable to progress through S phase. As shown in Figure 1b, whereas the control cells completed replication ∼60 min after their release, the osmostressed cells took ∼100 min to reach a similar profile. Therefore, cells delay progression through the S phase in response to osmostress.
To establish the involvement of Hog1 in the S phase delay upon stress, we analyzed the effect of sustained activation of the HOG pathway on S phase progression. We have shown previously that activation of the Hog1 MAPK via expression of PBS2DD or via inactivation of the Sln1 osmosensor leads to G1 and G2 arrest (Maeda et al., 1994; Escote et al., 2004; Clotet et al., 2006). cdc7ts4 or cdc7ts4 hog1 strains containing a plasmid expressing the PBS2DD allele under the control of the GAL1 promoter were grown at the nonpermissive temperature (37°C) and then released to 25°C in the presence of galactose. Expression of PBS2DD resulted in a strong S phase delay in cells containing wild-type HOG1 compared with hog1 cells (Figure 1c). Similar results were obtained after release from HU arrest when Hog1 activation was induced by inactivation of Sln1 or in response to osmostress (Figure 2). The HU-synchronized cells were faithfully synchronized in S phase with no detectable Sic1, with high Clb5 protein levels as well as RNR1 mRNA (Figure 2b). Correspondingly, when hog1-deficient cells were subjected to osmostress they were unable to delay S phase upon stress (Supplemental Figure 1). Therefore, activation of Hog1 leads to a transient S phase cell cycle delay.
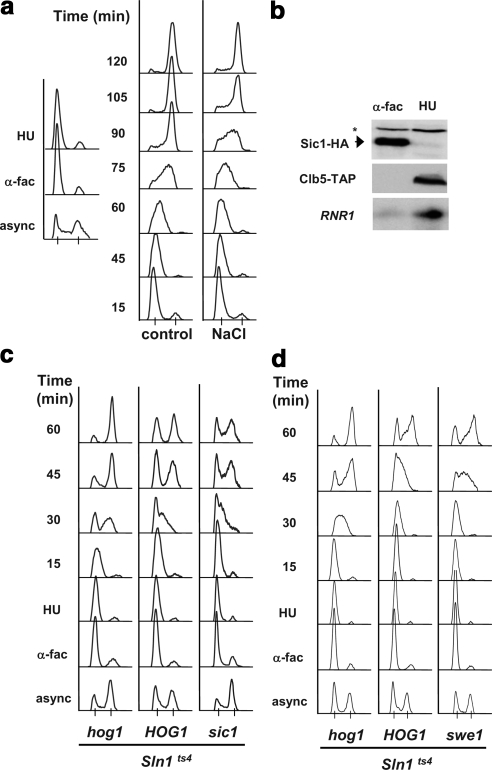
Figure 2.
Hog1 induces an S phase delay independently of the Sic1 and Swe1 Hog1 targets. (a) Osmostressed cells are delayed in S phase progression. Wild-type cells were grown exponentially (async.) and then incubated with α-factor for 3 h (α-fac), washed free of the α-factor, and incubated with 200 mM HU for 1 h (left-hand side). The cells were then washed and liberated of HU and allowed to progress through the cell cycle at 25°C in YPD medium (control) or YPD supplemented with 0.4 M NaCl (NaCl). Samples were analyzed by FACS. (b) HU treated cells are synchronized in S phase. Protein and RNA samples were extracted from α-fac and HU-synchronized cells. C-Terminally tagged Sic1-HA and Clb5-TAP levels were measured by Western blot. The asterisk indicates a nonspecific band. Northern blot analysis was performed with a probe directed at RNR1 mRNA. (c) S phase delay is independent of SIC1 but depends on HOG1. Exponential cultures of sln1ts4 and the isogenic hog1 or sic1 mutants were grown at the permissive temperature of 25°C in which they were synchronized first with α-factor and then with HU as described in a. The cells were then washed free of HU at 37°C. Samples were analyzed by FACS. (d) S phase delay is independent of Swe1. sln1ts4, sln1ts4 hog1, and sln1ts4 Swe1 cells analyzed as described in c. Experiments were performed at least twice, and representative results are shown.
The Control of S Phase by Hog1 Does Not Require Sic1 or Swe1
The well established role of the cell cycle inhibitors Sic1 and Swe1 in the corresponding Hog1-induced G1 and G2 cell cycle arrests prompted us to test their involvement in the S phase delay as well. We have demonstrated previously that the inactivation of Sln1 at G1 lead to cell cycle arrest mediated by Sic1, whereas inactivation of Sln1 at G2 resulted in G2 arrest mediated by Swe1. Thus, sln1ts4, sln1ts4 hog1 (which are refractory to the upstream activation of the pathway), and sln1ts4 sic1 or sln1ts4 swe1 cells were synchronized in S phase with HU. Cells were released from HU at the nonpermissive temperature and their S phase delay upon Hog1 activation was analyzed. As shown in Figure 2c, induction of Hog1 activity resulted in cells that transiently arrested at S phase as compared with sln1ts4 hog1 cells. It is worth noting that sln1ts4 hog1 cells bearing a plasmid that contained a catalytically inactive HOG1 allele (hog1knn) behaved as hog1-deficient cells, indicating that it is the catalytic activity of Hog1 and not solely the presence of the protein that leads to S phase delay (Supplemental Figure 5). Furthermore, sic1 cells arrested at S phase to the same degree as wild-type cells (HOG1). Therefore, as anticipated by the absence of Sic1 protein during S phase, deletion of SIC1 did not prevent S phase delay upon Hog1 activation. Swe1, although classically characterized as a cell cycle inhibitor at the G2/M transition, has also been implicated in the DNA damage response (Liu and Wang, 2006). Nevertheless, when Hog1 was activated in swe1 cells, cells delayed in S phase as efficiently as wild-type cells (Figure 2d). Thus, neither Sic1 nor Swe1 are involved in the S phase delay mediated by Hog1.
S Phase Delay Does Not Depend on the S Phase DNA Checkpoint
Cells can arrest replication in response to DNA damage and under replication stress conditions. The S phase DNA checkpoint plays a key role in protecting cells during replication. Therefore, we tested whether this pathway was involved in the ability of Hog1 to delay replicating cells. The S phase DNA checkpoint converges mainly into a single effector kinase, Rad53. This protein undergoes massive autophosphorylation upon activation resulting in reduced migration on SDS PAGE. Thus, we tested whether Rad53 was activated upon osmostress by monitoring Rad53 hyperphosphorylation. To carry out this experiment, cells could not be synchronized with HU, because this would directly lead to the phosphorylation of Rad53. In fact, HU treatment served as a positive control to visualize the reduced mobility of activated Rad53 (Figure 3a, HU). To bypass this obstacle, cells were synchronized in G1 with α-factor, released, and then allowed to cycle for an additional 20 or 40 min to let them enter S phase. The cultures were then subjected to osmotic stress (0.4 M NaCl), and activation of Rad53 was monitored during the subsequent hour. Notably, an S phase delay was exerted by the HOG pathway under the very same conditions (Figure 1a). However, no hyperphosphorylation of Rad53 was detected (Figure 3a). Similar results were obtained when cells were exposed to a more severe (1M NaCl) osmostress (data not shown). These results suggest that osmostress does not lead to activation of Rad53.
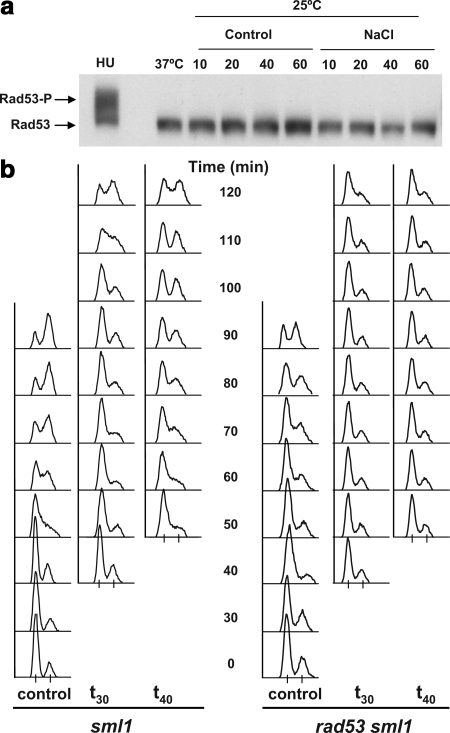
Figure 3.
The Rad53 DNA damage checkpoint is not involved in the Hog1 induced S phase delay. (a) The effector kinase Rad53 is not hyper-phosphorylated in response to osmostress. Exponentially growing cells with endogenously HA-tagged Rad53 were synchronized with α-fac for 3 h. Cells were washed free of the α-factor and stressed with 0.4 M NaCl either 20 min (left, t20) or 40 min (right, t40) after α-factor release. Samples were taken immediately before NaCl addition (0) and 10, 20, 40, and 60 min after it. Protein was extracted, separated on 7% gel, and Western blot analysis was performed with α-HA antibodies. Part of the culture was incubated with HU for 1 h immediately after release from α-factor (HU). Nonphosphorylated Rad53 is indicated by the bottom arrow and slower migrating phosphorylated Rad53 forms are present above in the HU sample (P-Rad53-HA). (b) Osmostressed rad53 cells delay in S phase. sml1 (left-hand side) and sml1 rad53 cells (right-hand side) were grown, synchronized with α-factor, and released as described in a. Cultures were subsequently osmostressed with 0.4 M NaCl 30 min (t30) or 40 min (t40) subsequent to α-factor release. Samples were collected and analyzed by FACS. The progression of a nonstressed culture is shown (control). Experiments were performed at least twice, and representative results are shown.
To rule out the involvement of Rad53 in the Hog1-induced S phase delay, we tested whether cells lacking RAD53 were able to delay upon osmostress. Attesting to the vital role of Rad53 in the cell, the deletion of this allele is inviable. However, deletion of the SML1 gene suppresses this lethality (Zhao et al., 1998). Thus, rad53 sml1 cells could be analyzed for their capacity to delay S phase in osmostress. The parental sml1 single mutant was utilized as a control in parallel. rad53Δ cells are hypersensitive to HU treatment (Gunjan and Verreault, 2003), and so S phase cells were obtained as described above. As shown in Figure 3b, rad53 sml1 cells delayed in S phase (Figure 3b; data not shown). In fact, they delayed more than the control sml1 cells when exposed to NaCl. This, as well as the limited α-factor arrest, is most probably to their being sick, as evident by comparing the control series of the sml1 and rad53 sml1 strains. Together, these results demonstrate that Hog1 does not induce S phase delay via the S phase DNA damage pathway.
Hog1 Mediates a Strong Down-Regulation of the S Phase Cyclins Clb5 and Clb6 upon Stress
The Hog1 MAPK down-regulates Cln1, Cln2, and Clb2 cyclin levels to control G1 and G2 progression (see Introduction). Thus, we analyzed whether Clb5, Clb6, and Dbf4 levels were altered upon osmostress during S phase. Exponentially growing sic1 cells with endogenously C-terminal TAP-tagged Clb5 or Clb6 were synchronized in α-factor for 3 h and released into fresh YPD medium. All cells used in these experiments were deleted of SIC1 to ensure that no Sic1-mediated G1 arrest would be exerted by Hog1. Cells were grown in YPD (control) or osmostressed with 0.4 M NaCl 30 min after release from α-factor (t30). Samples were analyzed by SDS-PAGE and probed with α-TAP antibodies to detect endogenous Clb5-TAP and Clb6-TAP levels (top of each panel). As shown in Figure 4, a and b, Clb5 and Clb6 begin to show up 30 min after release, just as cells are entering S phase (also see Figure 1a). It was exactly at this point that the cells were osmostressed. Clb5 and Clb6 accumulation was clearly delayed in response to osmostress. In contrast, expression of stress responsive genes such as GRE2 or HSP12 was not affected during S phase (Supplemental Figure 2).
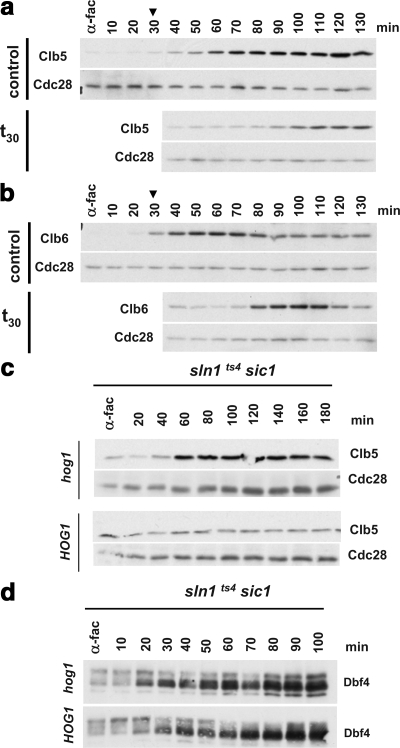
Figure 4.
Clb5 and Clb6 protein accumulation is delayed in response to osmotic shock. (a and b) Clb5 and Clb6 protein expression are delayed in osmostressed cells. Exponentially growing sic1 cells with endogenously TAP-tagged Clb5 (a) or Clb6 (b) were synchronized in α-factor for 3 h and released into fresh YPD medium. Cells were either not osmostressed (control) or stressed with 0.4 M NaCl 30 min (t30) after α-factor release as indicated by the arrowhead. Samples were analyzed by SDS-PAGE and probed with α-TAP antibodies to detect endogenous Clb5-TAP and Clb6-TAP levels (top part of each panel). Blots were then stripped and probed with α-PSTAIR to detect Cdc28 protein levels (bottom of each panel). (c) Clb5 protein accumulation during S phase is delayed in a Hog1-dependent manner. TAP-Clb5 tagged sln1ts4 sic1Δ with the wild-type HOG1 allele (bottom) or a hog1Δ deletion (top) were grown, synchronized with α-fac, and released into fresh medium. The cultures were shifted to 37°C after their release from α-fac. Samples were analyzed for Clb5 and Cdc28 levels as described in a and b. Experiments were performed at least twice, and representative results are shown. (d) Hog1 activation does not delay the Dbf4 protein accumulation during S-phase. sln1ts4 sic1 cells containing the wild type HOG1 allele (top) or hog1 deletion (bottom) were grown, synchronized with α-fac, and released into fresh medium and shifted to 37°C. Samples were analyzed by SDS-PAGE and blotted and probed with specific polyclonal antibodies to detect the endogenous levels of the Dbf4.
We then tested whether the delay of Clb5 accumulation upon osmostress was dependent on Hog1 activation. Thus, TAP-Clb5 tagged sln1ts4 sic1 and sln1ts4 hog1 sic1 cells were synchronized with α-factor and then released at the nonpermissive temperature. As shown in Figure 4c, whereas cells unable to activate the HOG pathway (hog1) showed a strong increase of Clb5 60 min after release at 37°C, cells with activated Hog1 were unable to induce Clb5 accumulation. These results demonstrate that indeed Hog1 is preventing the accumulation of Clb5 levels upon osmostress. In contrast, under the same experimental conditions, the levels of Dbf4 did not alter when the MAPK was activated (Figure 4d).
The Control of S Phase Also Occurs at High Clb5 Levels
Cells synchronized in S Phase by virtue of the cdc7ts4 allele or HU treatment still delayed in response to osmostress after their release (Figures 1 and 2) despite possessing high Clb5 levels (Figure 2b and Supplemental Figure 3). Thus, Hog1 is able to regulate yeast progression through the S phase of the cell cycle even in the presence of high levels of Clb5. Importantly, the maximal Clb5 levels did not decline when HU-arrested cells were osmostressed after release (data not shown). Therefore, other means to delay S phase progression should exists in addition to the delay on Clb5,6 accumulation. To directly analyze this hypothesis, we took advantage of a genetic tool that enables cells to progress into S phase without S-CDK activity. It has been shown that Sld2 and Sld3 are the minimal and essential set of S-CDK substrates for the initiation of DNA replication (Tanaka et al., 2007; Zegerman and Diffley, 2007). The essential S-CDK requirement in the initiation of DNA replication is bypassed in a strain in which the phosphorylation of Sld2 and Sld3 are simulated by genetic means (Zegerman and Diffley, 2007). In this strain, mutant forms of Sld2 and Sld3 mimic the formation of the trimeric complex Sld2-Sld3-Dpb11. As a consequence, these cells can initiate DNA replication even in the absence of S-CDK activity when the inhibitor Sic1 is overexpressed.
Based on our evidence that Hog1 also regulates S phase progression at a stage subsequent to the initiation of Clb5,6 protein synthesis, it would be expected that the bypass strain, which does not require neither Clb5 nor Clb6 to promote replication (Zegerman and Diffley, 2007), should also arrest in response to osmostress. Yeast cells containing Sld2T84D and Sld3T600-609-622A-Dpb11(253-764) and expressing stabilized Sic1 (Sic1ΔN) under the control of GAL1 promoter were synchronized at G1 with α-factor and then released into fresh media containing either glucose or in galactose. Cells were exposed to osmostress (0.4 M NaCl) 30 min after the α-factor release, or allowed to progress under normal conditions (control). As shown in Figure 5, control cells progressed through S phase after release on galactose, demonstrating that the S-CDK bypass system functioned. However, these cells delayed in S phase in the presence of osmostress showing that even cells that do not require S-CDK activity to replicate delay in response to osmostress. Similar results were obtained with S-CDK bypass cells synchronized with HU (data not shown). Together, in addition to down-regulating Clb5,6 levels, Hog1 also mediates S phase delay independently of S-CDK activity.
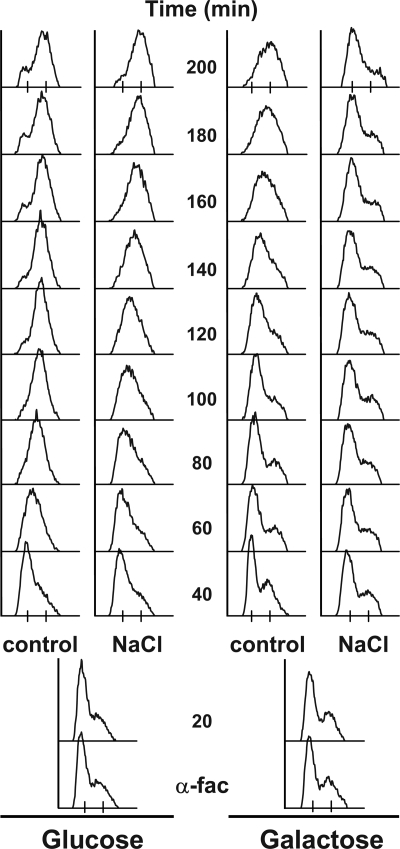
Figure 5.
Hog1 can induce an S phase delay independent of S-CDK activity. Cells of the S-CDK bypass system (Zegerman and Diffley, 2007) capable of initiating replication without Cdc28-Clb5,6 activity were synchronized in α-factor for 3 h (α-fac) in either glucose (left) or in galactose, in which Sic1ΔNT is expressed and Cdc28-Clb5/6 activity is inhibited. Cells were liberated of α-factor into fresh YPD (control). Half of the culture was exposed to 0.4 M NaCl 20 min after release from α-factor (NaCl).
Hog1 Associates with Components of the Basic Replication Machinery
Hog1 binds to DNA-associated proteins to regulate transcription (Alepuz et al., 2001; Proft et al., 2006). To gain further insight into the mechanism by which Hog1 inflicts an delay on replicating cells independently of Clb5,6 delay, we analyzed the capacity of the MAPK to interact with various components of the replication machinery. HU-synchronized cells with the endogenously expressed TAP-tagged protein of interest and bearing GST-tagged Hog1 or a control GST-expressing plasmid were subjected to osmostress (0.4 M NaCl; 10 min). Cells were lysed and the TAP-tagged protein was probed for after incubation with glutathione beads and extensive washes. Representative proteins of DNA polymerase ε and of each of the major subcomplexes of the replication machinery were tested for their capacity to coprecipitate with Hog1; Orc2 for the origin recognition complex (ORC) complex, Mcm4 for the MCM complex, Cdc45 for its complex, and Psf2 for GINS. The Dpb2 subunit of the replicative polymerase ε and DDK Cdc7 kinase were also analyzed (Kelly and Brown, 2000; Bell and Dutta, 2002).
Each of the tested TAP-tagged proteins was equally expressed in the cells harboring the GST versus the GST-Hog1–expressing plasmid (α-TAP, total). In addition, GST and GST-Hog1 were efficiently precipitated in all samples (bottom α-GST panel). As shown in Figure 6, for almost all proteins tested, a specific interaction was observed between GST-Hog1 and the replication-based protein. A striking exception is Orc2. The inability of the ORC complex to interact with Hog1 was retested and confirmed with TAP-Orc1 (data not shown). Also, an interaction between Hog1 and replication complex proteins was reproduced when the TAP tag was replaced with an HA tag in the coimmunoprecipitation analysis (data not shown). Thus, Hog1 associates with the replication complex machinery. In good agreement, a portion of endogenously expressed Hog1 from osmostressed HU-synchronized cells coelutes from a gel filtration column with a very high molecular weight fraction that includes Dpb2 and Cdc45 (data not shown).
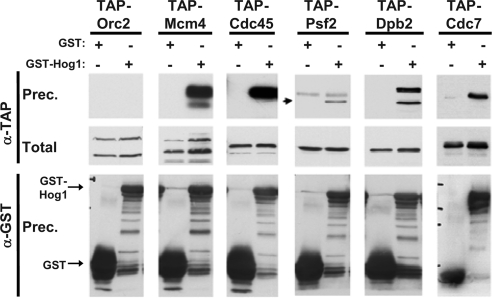
Figure 6.
Hog1 interacts in vivo with various proteins of the replication complexes. Coimmunoprecipitation experiments were individually performed with the indicated TAP-tagged strains (Orc2, Mcm4, Cdc45, Psf2, Dpb2, or Cdc7). Each strain was transformed with either a control vector expressing GST or a vector expressing GST-Hog1. Cultures were treated with 200 mM HU for 2 h, osmostressed with 0.4 M NaCl for 10 min and then collected. One milligram of protein extracts was incubated with glutathione beads, extensively washed, eluted, and analyzed by SDS-PAGE. The resulting blots were probed with α-GST antibody to detect the immunoprecipitated GST or GST-Hog1 (α-GST, prec.) and α-TAP antibody to detect the coimmunoprecipitated proteins (α-TAP, prec.). Twenty micrograms of total protein extracts was run in parallel and probed with α-TAP (Total). Experiments were performed at least twice, and representative results are shown.
The interaction of Hog1 with the basic replicative machinery led us test whether there was a molecular target for the MAPK within the complex. We therefore attempted to identify a direct substrate phosphorylated by Hog1 within the replication machinery by in vitro phosphorylation assays. More than 25 proteins involved in replication were individually tested and none of them was found to be phosphorylated by the MAPK (data not shown).
Hog1 Promotes S Phase Delay after Sld2 Phosphorylation and before Phosphorylation of Dpb2
The stepwise recruitment of proteins to the replication complex and its concurrent conversion from a prereplication complex into a fully functional active complex is accompanied by various molecular events. Two of these include the phosphorylation of the Sld2 and the Dpb2 proteins (Masumoto et al., 2002; Kesti et al., 2004). In brief, the hyperphosphorylation of Sld2 by S-CDK coordinates the recruitment of replicative polymerases to the replication complex and is essential for the replication of chromosomal DNA. The Dpb2 subunit of the DNA polymerase ε is also phosphorylated by CDK. Importantly, these two spatially and temporarily distinct events can serve as biochemical markers for the qualitative state of the replication complex. Indeed, Sld2 phosphorylation has been shown to precede Dpb2 phosphorylation in vivo (Masumoto et al., 2002). Thus, a defined timeframe for the action of Hog1 on the replication process could be established.
To test whether Hog1 acts before or succeeding the phosphorylation of Sld2 by CDK, Sld2 phosphorylation kinetics were followed in cells in which the HOG pathway was activated 20 min after release from α-factor. As in previous experiments, all strains used in these experiments were deleted of SIC1. As observed in Figure 7a, a gradual hyperphosphorylation of Sld2 is observed upon release of the G1-arrested cells. All of the Sld2 protein is hyperphosphorylated within 30 min. Notably, this is also the case in osmostressed cells. In addition, in both cultures, the maximal phosphorylation levels were obtained by minute 70, at which point its phosphorylation began to decrease in the nonstressed cells. Maximal Sld2 phosphorylation was still maintained in the osmostressed culture through to the end of the experiment. This is most probably indicative of the longer S phase these cells posses due to the Hog1 induced delay. Indeed, these osmostressed cells portrayed the expected FACS delay (data not shown). These results reveal that although the cells delay upon osmostress, the Sld2 phosphorylation pattern is identical. Together with our previous observations that HU synchronized cells and the S-CDK bypass cells delayed when osmostressed in S phase, it seems clear that Hog1 does not act before or directly on this key initial event of the replication process but rather must act subsequent to it.
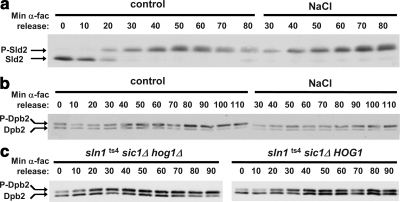
Figure 7.
Hog1 acts subsequent to Sld2 phosphorylation and before Dpb2 phosphorylation. (a) Sld2 phosphorylation kinetics are identical in nonstress and osmostress conditions. Sic1 cells with endogenously tagged Sld2-HA were synchronized with α-fac for 3 h and released into fresh medium (control). Half of the culture was exposed to 0.4 M NaCl 20 min after release from α-factor (NaCl). Protein extracts were prepared, separated on 7% SDS-PAGE, and probed with α-HA antibodies to detect Sld2-HA. The slower migrating phosphorylated form of Sld2 (P-Sld2) and the nonphosphorylated form (Sld2) are indicated. (b) Dpb2 phosphorylation is delayed in osmostressed cells. sic1 mutant cells with endogenously tagged Dpb2-HA were grown and assayed as described in a. (c) The delayed Dpb2 phosphorylation kinetics depends on Hog1. Dpb2-HA tagged sln1ts4 sic1 with wild-type HOG1 (right) or a hog1 deletion (left) were grown, synchronized with α-fac and released into fresh medium as described in a. The cultures were shifted to 37°C 20 min after their release from α-fac. Dpb2 phosphorylation was followed as described above. Experiments were performed at least twice, and representative results are shown.
As a next stage of the biochemical dissection of the replication process, the phosphorylation pattern of Dpb2 was analyzed. Dpb2 phosphorylation is believed to contribute to the processivity of DNA polymerase ε or to its affinity toward the replication complex proteins (Kesti et al., 2004). Unlike the phosphorylation of Sld2, it is not essential for the replication process. Indeed, this is reflected by the extent of phosphorylated protein in α-factor cells versus HU arrested cells. Although the entire population of Sld2 shifts to the hyperphosphorylated form in replicating cells, Dpb2 is already very significantly phosphorylated in G1 (∼50%; average of multiple experiments) and is augmented to a maximum of ∼70% in HU-treated cells (data not shown). This is also the maximal Dpb2 phosphorylation detected after prolonged release of the cells from G1 arrest (see below). Thus, although not as prominent as Sld2 phosphorylation, Dpb2 phosphorylation is still clearly measurable and indicative of the replication state. As shown in Figure 7b, Dpb2 was gradually phosphorylated in nonstressed cells throughout the course of the experiment. In contrast, Dpb2 phosphorylation only very slightly increased and then maintained a low stable level in the osmostressed cells. Thus, unlike the case with Sld2, Dpb2 phosphorylation pattern is altered in osmostressed cells.
To test the specific role of Hog1 in this phenomenon, Dpb2 phosphorylation kinetics was also followed in the sln1ts4 system. Understandably, the overall kinetics and timing of Dpb2 phosphorylation in the two experimental setups are distinct due to the different experimental conditions. In this system, Dpb2 was gradually phosphorylated in the hog1 cells until it reached maximum phosphorylation levels at minute 50–60. In contrast, in the parental cells in which the HOG pathway was effectively activated as confirmed by FACS analysis (data not shown), Dpb2 phosphorylation remained minimal throughout the experiment (Figure 7c). Together, unlike the case with Sld2, Hog1 activation alters the phosphorylation dynamics of the Dpb2 polymerase subunit. This suggests that Hog1 exerts its effect, at least in part, in the modulation of S phase progression between these two phosphorylation events.
Replication Is Altered in Response to Osmostress
To further study the effect of osmostress in S phase, we predicted that endogenous chromosomal replication should show reduced firing efficiency, prolonged replication, or both. We therefore tested the efficiency of firing and duration of the replication process for the early origin ARS305 by 2D-gel analysis. Initially, exponentially growing cells (sic1 bar1) growing at 25°C were synchronized in α-factor for 3 h and released into fresh medium (control) or subjected to 0.4 M NaCl (NaCl) 15 min after release. DNA was isolated and probed for ARS305 replication by 2D electrophoresis. As shown in Figure 8b, the appearance of replication bubbles was delayed in the presence of stress compared with control cells. In addition, replication lasted longer in cells subjected to stress albeit having fired a bit later. Thus, firing of replication is delayed and the replication process seems to last longer in the presence of stress.
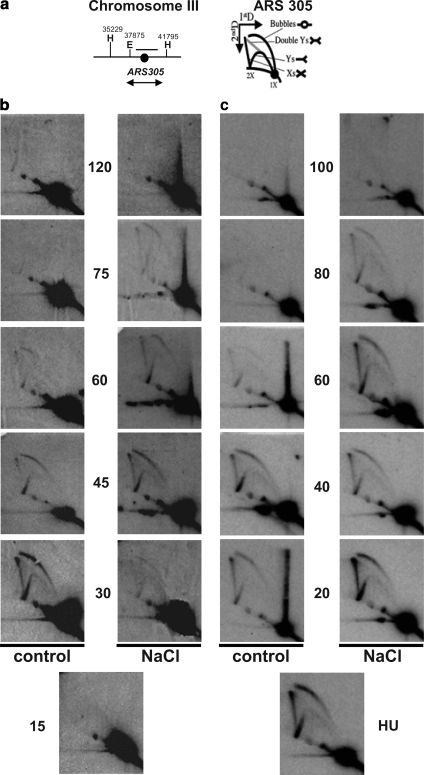
Figure 8.
Replication is altered in osmostressed cells. (a) Schemes of the chromosome III region around ARS305 analyzed (left) and of the migration pattern of the Y-, bubble-, double Y-, and X-shaped replication intermediates in 2D-gel electrophoresis (right). The region used as a probe in the hybridization experiments is indicated (b) Replication firing is delayed in cells osmostressed after α-fac synchronization. Exponentially growing sic1 bar1 cells growing at 25°C were synchronized in α-fac for 3 h and liberated into fresh medium (control). Half of the culture was exposed to 0.4 M NaCl 15 min after release from α-factor (NaCl). 2D-gel analysis of EcoRV–HindIII restriction fragment covering ARS305 is shown. (c) Cells osmostressed after HU synchronization take longer to replicate. Cells as described in a were synchronized for 1 h in 200 mM HU directly after washing away the α-fac. The culture was then liberated of HU into fresh medium (control) or medium supplemented with 0.4 M NaCl (NaCl). 2D-gel analysis was performed as described in a.
In addition to regulating S phase initiation, the MAPK is also capable of slowing down S phase progression when cells were stressed at late S phase. Thus, even cells synchronized with HU in which the early origins have already fired, are expected to take longer to replicate when stressed. sic1 bar1 cells growing at 25°C were synchronized in α-factor for 3 h and then released for 1 h into 200 mM HU. The S phase synchronized culture was then freed into fresh medium (control) or medium supplemented with 0.4 M NaCl (NaCl). As expected, the early origin ARS305 had already fired in the presence of HU, as can be seen by the appearance of replication bubbles (Figure 8c). More importantly, the replication process lasted longer in cells subjected to stress than in control cells, as observed with both the bubbles and the Y-arcs of the replication forks where initiated at either ARS305 or another ARS, respectively. Together, both the timing of replication firing and the duration of the replication process are affected in response to osmostress.
DISCUSSION
Regulation of cell cycle progression by external stimuli requires complex regulatory mechanisms. Indeed, in response to osmostress, both lower and higher eukaryotic cells activate signaling pathways that control several key elements of the cell cycle machinery to prevent cell cycle progression without proper adaptation to stress (Sheikh-Hamad and Gustin, 2004). In yeast, activation of the Hog1 MAPK signaling pathway results in the control of G1, G2, and possibly mitosis in response to osmostress (Clotet and Posas, 2007). Here, we show that the Hog1 MAPK pathway also modulates S phase progression upon osmostress.
Neither Sic1 nor Swe1, previously well characterized cell cycle regulators targeted by the MAPK, are involved in S phase control, as expected by their known roles in cell cycle control upon osmostress. More striking is the observation that the S phase DNA checkpoint is not involved in the replication delay provoked by osmotic stress. The lack of detectable Rad53 hyperphosphorylation in response to osmostress and the fact that rad53 cells delayed at S phase as efficiently as wild-type cells indicate that Hog1 must modulate S phase progression by different means. This is an interesting observation as the DNA damage S phase checkpoint is such a central and robust pathway in response to a wide range of stimuli that impinges on the DNA or its replication (Branzei and Foiani, 2006; Harrison and Haber, 2006).
Activation of Hog1 during G1 and G2 mediates cell cycle arrest by targeting specific cell cycle inhibitors in combination with the down-regulation of G1 and G2 cyclins (i.e., Cln1 and Cln2 as well as Clb2). In response to osmostress, there is a strong delay in the accumulation of both cyclins. The resulting reduction in S-CDK activity might mediate the delay at early S phase. Correspondingly, cells stressed before the firing of an early origin such as ARS305 showed a clear delay in firing. It is known that the control of the S-CDK activity (Cdc28-Clb5,6) is critical in the G1-S transition for the firing of DNA replication (Donaldson et al., 1998a; Nougarede et al., 2000). However, this does not exclude that S-CDK activity is required throughout the whole duration of the S phase for efficient firing of all replication origins. In fact, although Clb6 is degraded shortly after initiation of DNA replication, Clb5 accumulates throughout the S phase and is not degraded until G2-M. Accordingly, the rate of DNA replication is unaffected in a clb6 strain, whereas a clb5 strain has a much longer S phase due to deficient late origin firing (Donaldson et al., 1998b). Therefore, the continuous presence of Clb5 during S phase might be critical for the correct firing of the full set of replication origins. Hog1 activation clearly affects Clb5 accumulation in response to stress. Thus, it is likely that the reduction in the levels of Clb5 upon osmostress affects both origin firing and, at least partially, could also explain the slow rate of S phase progression.
It is worth noting that cells synchronized at different points of the S phase, by virtue of a cdc7ts strain or the presence of HU, still delayed when subjected to osmotic stress, albeit already possessing high Clb5 levels. More importantly, cells that have been genetically modified and do not require S-CDK activity to replicate, delay in response to osmostress. Thus, several lines of experimental evidence indicate that an alternative mechanism in addition to inhibition of the Clbs must be exerted by the MAPK. The Hog1 MAPK plays a key role in the regulation of transcription and does so by the direct interaction with the basic transcription machinery (Alepuz et al., 2003; de Nadal et al., 2004; Zapater et al., 2007). The MAPK also seems to be part of the basic replicative machinery. Thus, it is likely that there could be a molecular target on the complex for the MAPK. Although we tested several proteins within the complex by in vitro phosphorylation assays, we were not able to identify any potential target for the MAPK. This negative result cannot exclude the existence of a target for Hog1 in the replication complex, because the MAPK could exert its effect through an intermediate protein. Alternatively, it could be that we have missed the target due to the sensitivity of our in vitro assays or the fact that we have not been able to analyze all the components that form the replicative machinery. We believe, however, that such a substrate might exist because the catalytically deficient mutant of Hog1 was unable to provoke an S phase delay.
The analyses of the kinetics of phosphorylation of both Sld2 and Dpb2 have defined that the activity of the MAPK should be exerted mechanistically after Sld2 but before Dpb2 phosphorylation events. This is consistent with the observation that in a strain containing a bypass system that circumvents Sld2 phosphorylation by S-CDK, osmostress leads to cell cycle delay. Furthermore, cells in which replication has fired (after release from HU) exhibited prolonged replication in the presence of stress as seen in the 2D-gel analyses. These analyses also support the idea that a component of the replication complex must be regulated. Interestingly, in contrast to the rest of the replication proteins assayed, no interaction was detected between the Hog1 and the ORC subcomplex. A marked difference between ORC and the remainder of the proteins tested is that the former remains constitutively bound to the origin of replication, whereas the remainder progress with the dynamic replication fork (Bell and Dutta, 2002). This raises the intriguing possibility that Hog1 interacts with the replication complex specifically when it is actively replicating. This notion is also consistent with Hog1 acting on a component of the active replication complex.
Together, our data suggest that, upon stress, two mechanisms are involved in the S phase delay; the inhibition of Clb5 and Clb6 cyclins and an additional component, most likely involved in the replication process per se. This dual control of progression might be of critical relevance as the regulation of G1 and G2 phases of the cell cycle is also mediated by the delay of cyclins and cell cycle regulators upon osmostress. Notably, the two mechanisms are not mutually exclusive. These may be exerted jointly by Hog1 in the same cell, resulting in the simultaneous action on both origins which have fired (early) and origins that have not yet fired (late).
One possible reason for cells to delay when replication and osmostress coincide is the need to transcriptionally adapt to osmotic changes. A major adaptive response to osmostress is the control of gene expression by Hog1. It is therefore conceivable that cells must deal with the possibility that a significant amount of transcription might coincide with the initiating or ongoing replication which might lead to transcription associated recombination (Aguilera, 2002; Aguilera and Gomez-Gonzalez, 2008). Hence, these two major dynamic complexes, replication and transcription, could feasibly interfere with each other. Indeed, there are >300 genes transcriptionally induced by Hog1 and ∼400 origins of replication. We measured whether cells are competent to transcribe stress genes during S phase and found that their expression is as efficient as in G1 (Supplemental Figure 2). Thus, delaying replication in response to osmostress might be important to avoid the collision of the two essential machineries that is known to lead to genomic instability (Aguilera, 2002).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to M. A. Adrover and Dr. M. Aldea for helpful discussions and suggestions; Drs. J.F.X. Diffley and D. Quintana for strains; Dr. D. O. Morgan for critical reading of the manuscript; Dr. E. de Nadal for constant help and support; and M. L. Rodriguez and L. Subirana for excellent technical assistance. G. Y. was a recipient of European Molecular Biology Organization short-term fellowship ASTF 144.00-05. This work was supported by UNICELLSYS from the European Community's seventh framework program (FP7, agreement 201142) (to F. P.); the Spanish Ministry of Science and Innovation (grant BFU2006-00984) [to F. P.] and grant BFU2006-05260 [to A. A.)]; and Junta de Andalucía (CVI-624) (to A. A.) through contract ERAS-CT-2003-980409 of the European Commission, DG Research, FP6 as part of a EURYI scheme award (www.esf.org/euryi); and support from the Fundación Marcelino Botín (to F. P.) and Consolider Ingenio 2010 program (grant CSD2007-0015) of the Spanish Government (to F. P. and A. A.). F. P.'s research is supported by “ICREA Acadèmia” for excellence in research (Generalitat de Catalunya).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-02-0129) on May 28, 2009.
REFERENCES
- Aguilera A. The connection between transcription and genomic instability. EMBO J. 2002;21:195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A., Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- Alepuz P. M., de Nadal E., Zapater M., Ammerer G., Posas F. Osmostress-induced transcription by Hot1 depends on a Hog1-mediated recruitment of the RNA Pol II. EMBO J. 2003;22:2433–2442. doi: 10.1093/emboj/cdg243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alepuz P. M., Jovanovic A., Reiser V., Ammerer G. Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol. Cell. 2001;7:767–777. doi: 10.1016/s1097-2765(01)00221-0. [DOI] [PubMed] [Google Scholar]
- Alexander M. R., Tyers M., Perret M., Craig B. M., Fang K. S., Gustin M. C. Regulation of cell cycle progression by Swe1p and Hog1p following hypertonic stress. Mol. Biol. Cell. 2001;12:53–62. doi: 10.1091/mbc.12.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T., Lichten M. A method for preparing genomic DNA that restrains branch migration of Holliday junctions. Nucleic Acids Res. 2000;28:e6. doi: 10.1093/nar/28.2.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J., Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 2007;19:238–245. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Bell M., Capone R., Pashtan I., Levitzki A., Engelberg D. Isolation of hyperactive mutants of the MAPK p38/Hog1 that are independent of MAPK kinase activation. J. Biol. Chem. 2001;276:25351–25358. doi: 10.1074/jbc.M101818200. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Bousset K., Diffley J. F. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 1998;12:480–490. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., Foiani M. The Rad53 signal transduction pathway: replication fork stabilization, DNA repair, and adaptation. Exp. Cell Res. 2006;312:2654–2659. doi: 10.1016/j.yexcr.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Clotet J., Escote X., Adrover M. A., Yaakov G., Gari E., Aldea M., de Nadal E., Posas F. Phosphorylation of Hsl1 by Hog1 leads to a G2 arrest essential for cell survival at high osmolarity. EMBO J. 2006;25:2338–2346. doi: 10.1038/sj.emboj.7601095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clotet J., Posas F. Control of cell cycle in response to osmostress: lessons from yeast. Methods Enzymol. 2007;428:63–76. doi: 10.1016/S0076-6879(07)28004-8. [DOI] [PubMed] [Google Scholar]
- de Nadal E., Alepuz P. M., Posas F. Dealing with osmostress through MAP kinase activation. EMBO Rep. 2002;3:735–740. doi: 10.1093/embo-reports/kvf158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nadal E., Zapater M., Alepuz P. M., Sumoy L., Mas G., Posas F. The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature. 2004;427:370–374. doi: 10.1038/nature02258. [DOI] [PubMed] [Google Scholar]
- Donaldson A. D., Fangman W. L., Brewer B. J. Cdc7 is required throughout the yeast S phase to activate replication origins. Genes Dev. 1998a;12:491–501. doi: 10.1101/gad.12.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson A. D., Raghuraman M. K., Friedman K. L., Cross F. R., Brewer B. J., Fangman W. L. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol. Cell. 1998b;2:173–182. doi: 10.1016/s1097-2765(00)80127-6. [DOI] [PubMed] [Google Scholar]
- Escote X., Zapater M., Clotet J., Posas F. Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat. Cell Biol. 2004;6:997–1002. doi: 10.1038/ncb1174. [DOI] [PubMed] [Google Scholar]
- Gunjan A., Verreault A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell. 2003;115:537–549. doi: 10.1016/s0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- Harrison J. C., Haber J. E. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T. J., Brown G. W. Regulation of chromosome replication. Annu. Rev. Biochem. 2000;69:829–880. doi: 10.1146/annurev.biochem.69.1.829. [DOI] [PubMed] [Google Scholar]
- Kesti T., McDonald W. H., Yates J. R., III, Wittenberg C. Cell cycle-dependent phosphorylation of the DNA polymerase epsilon subunit, Dpb2, by the Cdc28 cyclin-dependent protein kinase. J. Biol. Chem. 2004;279:14245–14255. doi: 10.1074/jbc.M313289200. [DOI] [PubMed] [Google Scholar]
- Kyriakis J. M., Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 2001;81:807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- Liu H., Wang Y. The function and regulation of budding yeast Swe1 in response to interrupted DNA synthesis. Mol. Biol. Cell. 2006;17:2746–2756. doi: 10.1091/mbc.E05-11-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese M. P., Clerici M., Lucchini G. The S-phase checkpoint and its regulation in Saccharomyces cerevisiae. Mutat. Res. 2003;532:41–58. doi: 10.1016/j.mrfmmm.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Maeda T., Wurgler-Murphy S. M., Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- Masumoto H., Muramatsu S., Kamimura Y., Araki H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature. 2002;415:651–655. doi: 10.1038/nature713. [DOI] [PubMed] [Google Scholar]
- Nougarede R., Della S. F., Zarzov P., Schwob E. Hierarchy of S-phase-promoting factors: yeast Dbf4-Cdc7 kinase requires prior S-phase cyclin-dependent kinase activation. Mol. Cell. Biol. 2000;20:3795–3806. doi: 10.1128/mcb.20.11.3795-3806.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft M., Mas G., de Nadal E., Vendrell A., Noriega N., Struhl K., Posas F. The stress-activated hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Mol. Cell. 2006;23:241–250. doi: 10.1016/j.molcel.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Reiser V., D'Aquino K. E., Ee L. S., Amon A. The stress-activated mitogen-activated protein kinase signaling cascade promotes exit from mitosis. Mol. Biol. Cell. 2006;17:3136–3146. doi: 10.1091/mbc.E05-12-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh-Hamad D., Gustin M. C. MAP kinases and the adaptive response to hypertonicity: functional preservation from yeast to mammals. Am. J. Physiol. Renal Physiol. 2004;287:F1102–F1110. doi: 10.1152/ajprenal.00225.2004. [DOI] [PubMed] [Google Scholar]
- Sheu Y. J., Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol. Cell. 2006;24:101–113. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Umemori T., Hirai K., Muramatsu S., Kamimura Y., Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- Tercero J. A., Diffley J. F. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- Yaakov G., Bell M., Hohmann S., Engelberg D. Combination of two activating mutations in one HOG1 gene forms hyperactive enzymes that induce growth arrest. Mol. Cell. Biol. 2003;23:4826–4840. doi: 10.1128/MCB.23.14.4826-4840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapater M., Clotet J., Escote X., Posas F. Control of cell cycle progression by the stress-activated Hog1 MAPK. Cell Cycle. 2005;4:6–7. doi: 10.4161/cc.4.1.1344. [DOI] [PubMed] [Google Scholar]
- Zapater M., Sohrmann M., Peter M., Posas F., de Nadal E. Selective requirement for SAGA in Hog1-mediated gene expression depending on the severity of the external osmostress conditions. Mol. Cell. Biol. 2007;27:3900–3910. doi: 10.1128/MCB.00089-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P., Diffley J. F. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- Zhao X., Muller E. G., Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.