Abstract
Background
Contamination of endoscopy equipment by Helicobacter pylori (H. pylori) frequently occurs after endoscopic examination of H. pylori-infected patients. In the hospital, manual pre-cleaning and soaking in glutaraldehyde is an important process to disinfect endoscopes. However, this might not be sufficient to remove H. pylori completely, and some glutaraldehyde-resistant bacteria might survive and be passed to the next patient undergoing endoscopic examination through unidentified mechanisms. We identified an Imp/OstA protein associated with glutaraldehyde resistance in a clinical strain, NTUH-C1, from our previous study. To better understand and manage the problem of glutaraldehyde resistance, we further investigated its mechanism.
Results
The minimal inhibitory concentrations (MICs) of glutaraldehyde andexpression of imp/ostA RNA in 11 clinical isolates from the National Taiwan University Hospital were determined. After glutaraldehyde treatment, RNA expression in the strains with the MICs of 4–10 μg/ml was higher than that in strains with the MICs of 1–3 μg/ml. We examined the full-genome expression of strain NTUH-S1 after glutaraldehyde treatment using a microarray and found that 40 genes were upregulated and 31 genes were downregulated. Among the upregulated genes, imp/ostA and msbA, two putative lipopolysaccharide biogenesis genes, were selected for further characterization. The sensitivity to glutaraldehyde or hydrophobic drugs increased in both of imp/ostA and msbA single mutants. The imp/ostA and msbA double mutant was also hypersensitive to these chemicals. The lipopolysaccharide contents decreased in individual imp/ostA and msbA mutants and dramatically reduced in the imp/ostA and msbA double mutant. Outer membrane permeability assay demonstrated that the imp/ostA and msbA double mutation resulted in the increase of outer membrane permeability. Ethidium bromide accumulation assay demonstrated that MsbA was involved in efflux of hydrophobic drugs.
Conclusion
The expression levels of imp/ostA and msbA were correlated with glutaraldehyde resistance in clinical isolates after glutaraldehyde treatment. Imp/OstA and MsbA play a synergistic role in hydrophobic drugs resistance and lipopolysaccharide biogenesis in H. pylori.
Background
Helicobacter pylori was first isolated from the gastric mucosa of a patient with gastritis and peptic ulceration by Marshall and Warren in 1982 [1]. It is an important human pathogen, responsible for type B gastritis and peptic ulcers. Furthermore, infection by H. pylori is a risk factor for gastric adenocarcinoma and for lymphoma in the mucosa-associated lymphoid tissue of the stomach in humans [2-5].
H. pylori is believed to be transmitted from person to person by oral-oral or oral-fecal routes [6]. However, another possible route involves transmission during endoscopic examination of patients because contamination of endoscopy equipment by H. pylori frequently occurs after endoscopic examination of H. pylori-infected patients [7-9]. Because H. pylori is prevalent in the population [10], it is important to prevent its transmission. In the hospital, manual pre-cleaning and soaking in glutaraldehyde is an important process used to disinfect endoscopes [7,11]. However, endoscopic disinfection might not be sufficient to remove H. pylori completely [12,13]. Some glutaraldehyde-resistant bacteria might survive and be passed to the next person undergoing endoscopic examination through unidentified mechanisms. Therefore, it is an important issue to clarify the mechanism of glutaraldehyde resistance.
In our previous study, we demonstrated that the Imp/OstA protein was associated with glutaraldehyde resistance in a clinical strain of H. pylori [14]. OstA (organic solvent tolerance) [15] has also been called imp (increased membrane permeability) [16], and was recently named lptD in Escherichia coli [17]. Imp/OstA exists widely in Gram-negative bacteria and participates in biogenesis of the cell envelope. It is an essential outer membrane protein in E. coli, depletion mutation of imp/ostA results in the formation of aberrant membranes [18]. Furthermore, Imp/OstA forms a complex with the RlpB lipoprotein and is responsible for lipopolysaccharide (LPS) assembly at the surface of the cell [17,19]. In addition, it mediates the transport of LPS to the surface in Neisseria meningitidis [20].
To further investigate the mechanism of glutaraldehyde resistance, we monitored the minimum inhibitory concentrations (MICs) and the expression of imp/ostA and Imp/OstA protein after glutaraldehyde treatment in 11 clinical isolates. Full-genome expression was also studied by microarray analysis; 40 genes were upregulated and 31 genes were downregulated in NTUH-S1 after glutaraldehyde treatment. Among the upregulated genes, msbA, was selected for further study. MsbA is an essential inner membrane protein in E. coli and a member of the ABC transporter superfamily of proteins [21]. MsbA produced in the Gram-positive organism Lactococcus lactis is capable of conferring drug resistance to the organism [22]. In addition, msbA is not essential in N. meningitidis and this organism can survive without LPS [23]. In E. coli, msbA was implicated in lipid A-core moiety flipping from the inner leaflet to outer leaflet of the inner membrane [24,25], and then Imp/RlpB protein complex was responsible for transport of LPS from the periplasm to the outer leaflet of the outer membrane [17]. Here we showed that imp/ostA and msbA might be synergistic in hydrophobic drugs resistance and LPS transport in H. pylori.
Methods
Chemicals
Glutaraldehyde was purchased from Electron Microscopy Sciences (Hatfield, PA). Chloramphenicol, erythromycin, kanamycin, novobiocin, rifampicin, ethidium bromide, and carbonyl cyanide m-chlorophenylhydrazone (CCCP) were purchased from Sigma Chemical Co (St Louis, MO).
Bacterial strains and culture conditions
Clinical isolates were collected from National Taiwan University Hospital (NTUH) as previously described [26]. H. pylori strains were grown on Columbia agar plates containing 5% sheep blood under microaerophilic conditions (5% O2, 10% CO2, and 85% N2) at 37°C. For microarray analysis, we selected a rapidly growing strain NTUH-S1 with a higher MIC (MIC = 6 μg/ml) to glutaraldehyde from a patient with gastritis to study gene expression. To screen for mutant strains, blood agar plates were supplemented with 4 μg/ml chloramphenicol or 10 μg/ml kanamycin. To screen for imp/ostA and msbA double deletion mutant or complementation strains, blood agar plates were supplemented with 4 μg/ml chloramphenicol and 10 μg/ml kanamycin.
Determination the MICs of glutaraldehyde and hydrophobic drugs in H. pylori
The MICs of glutaraldehyde and hydrophobic drugs (erythromycin, novobiocin, rifampicin, and ethidium bromide) were determined by the agar dilution method. Suspension of H. pylori was adjusted to 107 cells/ml. Five microliters of bacterial suspensions were spotted on blood agar plates supplemented with different concentrations of drugs. Results were observed after 72 h incubation under microaerophilic condition at 37°C.
RNA slot blot hybridization
Four strains with the MICs of 7–10 μg/ml glutaraldehyde (designed numbers 1~4), four with the MICs of 4–6 μg/ml glutaraldehyde (numbers 5~8), and three with the MICs of 1–3 μg/ml glutaraldehyde (numbers 9~11) were grown on Columbia blood agar plates for 48 h, and further passaged on Columbia blood agar plates or 0.5 μg/ml glutaraldehyde-containing blood agar plates for 48 h. Since 0.5 μg/ml was the half concentration of the minimum MIC for the 11 strains, we defined this as the induction concentration. Subsequently, RNA was extracted from the bacteria with or without glutaraldehyde treatment. Total RNA from each H. pylori clinical isolate was extracted as described previously [27]. Ten micrograms of total RNA was transferred onto a nylon membrane using a slot-blot system (Hoefer, Holliston, MA). The membrane was hybridized with DNA probes specific for 23S rRNA (0.9 kb PCR-amplified 23S rRNA-specific fragment using the forward primer: 5'-ATTGGAGGGAAGGCAAATCC-3' and the reverse primer: 5'-ATCACTATGACCGACTTTCG-3'), imp/ostA (0.8 kb PCR-amplified imp/ostA-specific fragment using the forward primer: 5'-CATTGATAACCCCATTTGGC-3' and the reverse primer: 5'-GCACATTCAAAGCGTTTTGC-3'), and msbA (0.8 kb PCR-amplified msbA-specific fragment using the forward primer: 5'-TAGCGTTAGTGGGGTTAGTC-3' and the reverse primer: 5'-ACACCCTTTGAGTGACAACG-3') labeled with DIG by PCR. Detection was performed with the DIG Luminescent Detection kit (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's instructions.
RNA isolation and quantitative real-time PCR
It takes 48 to 72 h to recover colonies when H. pylori were grown on blood agar plates. A previous report also detected consistent RNA expression levels changes of H. pylori after 48 h of growth on acidified blood agar plates [27]. H. pylori NTUH-S1 was grown on Columbia blood agar plates for 48 h, and further passaged on Columbia blood agar plates or 0.5 μg/ml glutaraldehyde-containing blood agar plates for 48 h. RNA was extracted by the QIAGEN RNeasy column purification kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Total RNA was quantified with a spectrophotometer and visualized on an ethidium bromide stained agarose gel. Total RNA served as a template for cDNA synthesis using the SuperScript II Reverse transcriptase (Invitrogen, Carlsbad, CA). Synthesis reactions were started with 1.5 μg total RNA per 20 μl reaction mixture. All reactions were normalized to the level of the 16S rRNA gene [28]. In real-time RT-PCR, amplification and detection of the cDNAs were monitored using the KAPA SYBR FAST qPCR kit (Kapabiosystems, Boston, MA) in an ABI 7900 thermocycler (Applied Biosystems, Carlsbad, CA). Gene-specific primers imp/ostA RT (F): 5'-TTTGTCTTTAGGGCTTTGGAATG-3', imp/ostA RT (R): 5'-GCACGAAGGAATTTTTAGATTGC-3' and 16S rRNA RT (F):5'-TGCGAAGTGGAGCCAATCTT-3', 16S rRNA RT (R): 5'-GGAACGTATTCACCGCAACA-3' were used for amplification of cDNAs in this experiment. For the imp/ostA gene, the calculated threshold cycle (Ct) was normalized to the Ct of the 16S rRNA gene from the same cDNA sample before the fold change was calculated using the ΔΔCt method as described previously [29].
Western blots analysis of cell extracts
Eleven strains (numbers 1~11, the same isolates as previously described in RNA slot blot hybridization experiments) were selected and grown on Columbia blood agar plates for 48 h, and further passaged on Columbia blood agar plates or 0.5 μg/ml glutaraldehyde-containing blood agar plates for 48 h. Bacteria were harvested by centrifugation. Cells were washed in phosphate-buffered saline (PBS), resuspended in lysis buffer (50 mM Tris-HCl, 500 mM NaCl, 0.1% SDS, 10% glycerol), and lysed by sonication. Total protein concentration was determined by using the Bio-Rad protein assay (Bio-Rad, Hercules, CA). Samples were loaded at identical protein concentrations from total cell extracts for Western blot analysis. SDS-PAGE was transferred to nitrocellulose for immunological detection. Membrane was blocked with 5% skimmed milk in TBS overnight at 4°C. Subsequently, membrane was incubated with anti-OstA polyclonal antibody [14] diluted 1:500 with 5% skimmed milk in TTBS (0.5% Tween-20) for 1 h at room temperature. Horseradish peroxidase-conjugated anti-rat IgG diluted 1:3000 with 5% skimmed milk in TTBS (0.5% Tween-20) was added and membrane was incubated for 1 h at room temperature. The membrane was washed three times with TTBS (0.5% Tween-20) between the incubation steps. Electrochemiluminescence (Amersham Biosciences, Fairfield, CT) was used for detection.
RNA isolation and microarray analysis of H. pylori NTUH-S1
H. pylori NTUH-S1 was grown on Columbia blood agar plates for 48 h and further passaged on Columbia blood agar plates or 3 μg/ml glutaraldehyde-containing blood agar plates for 48 h. RNA was extracted using the QIAGEN RNeasy column purification kit (Qiagen) according to the manufacturer's instructions.
cDNA was synthesized according to the SuperScript™ indirect cDNA Labeling System (Invitrogen). cDNA was then purified using the S.N.A.P column purification (Invitrogen) according to the manufacturer's instructions.
Aminoallyl dUTP-labeled cDNA was resuspended in 2 × coupling buffer and labeled with either Alexa Fluor 555 or 647 according to the manufacturer's protocol (Molecular Probes, Eugene, OR). Labeled cDNA was mixed together and purified by S.N.A.P column purification. Then, the labeled cDNA was concentrated with a Microcon YM-30 column (Millipore, Billerica, MA).
The Institute for Genomic Research (TIGR) provided a H. pylori whole-genome microarray. It consisted of 2,572 70-mer oligonucleotides, printed in quadruplicate and representing open reading frames from H. pylori 26695 and strain J99. Labeled cDNA was resuspended in filtered hybridization buffer (50% formamide, 5 × SSC, 0.1% sodium dodecyl sulfate, 0.1 M DTT, and 0.6 μg/ml salmon sperm DNA), denatured at 95°C for 5 min, and flicked for an additional minute. It was then denatured for another 5 min. The labeled probe was applied to the pre-hybridized microarray and placed in a hybridization chamber at 42°C for 16~20 h. Microarray scanning and analysis were performed on a scanner (GenePix 4000B with GenePix Pro 5.0 software; Axon, Foster City, CA).
Processed microarray data files have been deposited in the Center for Information Biology Gene Expression Database (CIBEX; http://cibex.nig.ac.jp) under accession number CBX86.
Construction of imp/ostA and msbA deletion mutants
The gene encoding Imp/OstA with the upstream and downstream 500 bp flanking region was amplified with the genomic DNA of wild-type NTUH-S1 by PCR. The forward primer was 5'-ATGCACTCTCCAAATTTAGA-3', and the reverse primer was 5'-GGGGCTAGGATAGGTTCTAA-3'. It was then cloned into a pGEM-T easy vector (Promega, Madison, WI).
The gene encoding MsbA with the upstream 458-bp and downstream 474-bp flanking regions was amplified with the genomic DNA of wild-type NTUH-S1. PCR was performed using the forward primer, 5'-ACGACAGGAAACCCTTTAGG-3' and the reverse primer was 5'-AGCGTAATAAACAGGCACGC-3'. It was also cloned into a pGEM-T easy vector (Promega).
The imp/ostA and msbA genes were deleted by inverse PCR, and a chloramphenicol-resistant cassette (a gift from Dr. D. E. Taylor, University of Alberta) with its coding region (from the 1-bp start codon to the 624-bp stop codon) was then cloned into the flanking regions to replace the full-length imp/ostA or msbA gene. This plasmid was natural transformed into the wild-type NTUH-S1 strain to generate deletion mutants. Chromosomal DNA of the transformants was checked by PCR with primers external and internal to the replacement site to verify the double-crossover event.
Complementation of imp/ostA and msbA
An imp/ostA complementation strain of NTUH-S1 was constructed as described previously [14]. The promoter site of msbA gene was predicted by using a tool available at the following website: http://www.fruitfly.org/seq_tools/promoter.html. The msbA gene containing the predicted promoter region (upstream 73 bp) was obtained by PCR using the forward primer: 5'-CCAATCGCTTTAAGCTG-3', and the reverse primer: 5'-TTAGCATTCTGTCAAACGCC-3'. Then the DNA fragment was cloned into the pGEM-T easy vector (Promega). The msbA gene with its promoter region was cut from the constructed pGEM-T easy vector and ligated into the NruI site of the shuttle vector pHel3 (plasmid pHel3 was a gift from Dr. R. Haas, Max-Planck-Institute für Biologie, Tübingen, Germany). The constructed shuttle vector was natural transformed into an msbA deletion mutant strain to generate the msbA complementation strain.
Construction of the imp/ostA and msbA double deletion mutant
The gene encoding MsbA with its upstream 458-bp and downstream 474-bp flanking region was cloned into the pGEM-T easy vector as described above. A kanamycin-resistant gene aphA-3 from Campylobacter jejuni was then cloned between the flanking regions to replace the full length msbA gene. This plasmid was natural transformed into the wild-type NTUH-S1 strain to generate the deletion mutant. Chromosomal DNA of the transformants was checked by PCR with primers external and internal to the replacement site to verify the double-crossover event. Then, chromosomal DNA from msbA deletion mutant strain (Kmr) was natural transformed into the imp/ostA deletion mutant to obtain a double deletion mutant strain. It was also confirmed by PCR with primers external and internal to the msbA gene replacement site.
Southern blotting
Approximately 5 μg of genomic DNA from H. pylori NTUH-S1 and the mutants was digested by Hind III and incubated at 37°C overnight for complete digestion. The digoxigenin-labeled imp/ostA and msbA probes (primers were the same as those described for slot blot) was generated by PCR. Hybridization and detection were performed with the DIG Luminescent Detection kit (Roche) according to the manufacturer's instructions.
Proteinase-K digested H. pylori
The procedure was followed as described previously [30]. H. pylori cells were collected and adjusted to a concentration of 2.5 × 109 cells/ml in PBS. Bacteria were boiled with 150 μl sample dye for 10 min at 100°C to disrupt the whole cells. Subsequently, the whole cell lysates were treated with proteinase K (Sigma) for 60 min at 60°C in a water bath. Then, 2.5 × 108 cells/ml were analyzed by 12% SDS-PAGE and stained with silver. The protein concentration of the 2.5 × 108 cells/ml was also determined by using the Bio-Rad protein assay (Bio-Rad) to serve as a loading control.
Immunoblots of LPS from H. pylori with anti-Lewis (Le) monoclonal antibody
H. pylori strains that have been screened serologically [31-33], and a previous study suggested that Asian isolates express predominantly type 1 (Lea, Leb) antigens compared to Western strains (predominantly expressing type 2 Lex and Ley determinants) [34]. We also primarily detected the Lewis antigen of NTUH-S1 with anti-Lea and anti-Leb antibody. Equivalent amounts of protein were loaded in each well and transferred to nitrocellulose for immunological detection with anti-Lea or anti-Leb monoclonal antibody (Seikagaku Corporation, Tokyo). For detection of Lewis antigen in proteinase K-digested whole cell lysates, nitrocellulose membrane was blocked with 5% skimmed milk in PBS for 1 h at room temperature. Subsequently, membrane was incubated with anti-Lea or anti-Leb antibody diluted 1:3000 with 5% skimmed milk in PBS overnight at 4°C. Horseradish peroxidase-conjugated anti-mouse IgG diluted 1:5000 with 5% skimmed milk in PBS was added and membrane was incubated for 1 h at room temperature. The membrane was washed three times with PBST (0.05% Tween-20) between the incubation steps. Electrochemiluminescence (Amersham Biosciences) was used for detection. Whole cells of the Lex and Ley antigen-expressing H. pylori 26695 strain [35] were used as a negative control in Western blots to ensure the specificity of the anti-Lea or anti-Leb antibody.
Measurement of outer membrane permeability by ethidium bromide
Outer membrane permeability can be measured by the fluorescence of the ethidium-polynucleotide complex in the cell because ethidium bromide displays approximately a 10-fold increase in fluorescence quantum yield upon binding to DNA [36]. The assay was modified as described previously [37]. Briefly,H. pylori were grown on Columbia blood agar plate for 48 h. Then, bacteria were pelleted and washed twice with ice-cold 50 mM potassium phosphate (pH 7.0) containing 5 mM MgSO4. Cells were resuspended in 1 ml of potassium phosphate buffer (pH 7.0) at an optical density (OD600) of 0.5 and incubated for 30 min at 37°C in the presence of 10 μM of CCCP to deplete cells of metabolic energy. Subsequently, cells were washed three times in ice-cold potassium phosphate (pH 7.0) containing 5 mM MgSO4 and loaded with 10 μg/ml ethidium bromide. At the 40-min time point, the increase in ethidium bromide fluorescence intensity was measured in Gemini XPS spectrofluorimeter (Molecular Devices, Sunnyvale, CA) at 30°C with the excitation wavelength set at 500 nm and the emission wavelength at 580 nm. Each measurement was repeated three times.
Ethidium bromide accumulation assay
The assay was modified as described previously [38]. Briefly,H. pylori were grown on Columbia blood agar plate for 48 h. Then, bacteria were pelleted and washed twice with ice-cold 50 mM potassium phosphate (pH 7.0) containing 5 mM MgSO4. Cells were resuspended in 1 ml of potassium phosphate buffer (pH 7.0) at an optical density (OD600) of 0.5. Cells were preloaded with 10 μg/ml ethidium bromide. At the 12-min time point, 10 μM of CCCP was added to the cells suspensions to assess energy-dependent efflux. CCCP was not added to the cells served as a control. The increase in ethidium bromide fluorescence intensity was measured in a Gemini XPS spectrofluorimeter at 30°C with excitation at 500 nm and emission at 580 nm. Each measurement was repeated three times.
Statistical analysis
For all experiments, a P value of < 0.05 was considered indicative of statistical significance, and all statistical analyses were determined with Student's t test.
Results
The MICs for glutaraldehyde in clinical isolates
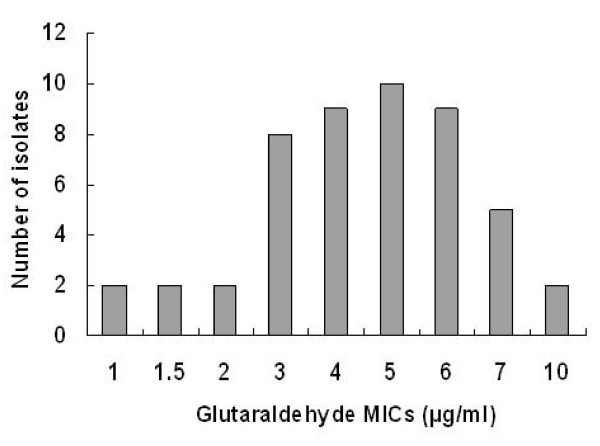
H. pylori strains were harvested during endoscopic examinations at National Taiwan University Hospital from 1991 to 2000 [39]. 49 clinical isolates were cultured successfully from stock and stored at -80°C. The patients from which these strains were isolated suffered from gastritis (15 strains), duodenal ulcer (16 strains), gastric ulcer (9 strains), mucosa-associated lymphoid tissue lymphoma (MALToma) (3 strains), and gastric cancer (6 strains). Subsequently, the MICs of glutaraldehyde were determined for these strains. The MICs of glutaraldehyde for most of the clinical isolates were the range of 3–6 μg/ml glutaraldehyde (Fig. 1). However, the diseases caused by the strains of H. pylori and the MICs of glutaraldehyde in these clinical isolates were not correlated (Table 1).
Figure 1.
The MICs of glutaraldehyde in clinical isolates from National Taiwan University Hospital.
Table 1.
The MICs of glutaraldehyde in clinical isolates during 1991–2000.
| Disease | Number of isolates | The MICs of glutaraldehyde in isolates (numbers) |
|---|---|---|
| Gastritis | 15 | 7 μg/ml (n = 2), 6 μg/ml (n = 1) 5 μg/ml (n = 3), 4 μg/ml (n = 4) 3 μg/ml (n = 5) |
| Duodenal ulcer | 16 | 10 μg/ml (n = 1), 7 μg/ml (n = 1) 6 μg/ml (n = 2), 5 μg/ml (n = 3) 4 μg/ml (n = 5), 2 μg/ml (n = 2) 1.5 μg/ml (n = 1), 1 μg/ml (n = 1) |
| Gastric ulcer | 9 | 10 μg/ml (n = 1), 7 μg/ml (n = 1) 6 μg/ml (n = 3), 5 μg/ml (n = 1) 3 μg/ml (n = 1), 1.5 μg/ml (n = 1) 1 μg/ml (n = 1) |
| MALToma | 3 | 7 μg/ml (n = 1), 5 μg/ml (n = 2) |
| Gastric cancer | 6 | 6 μg/ml (n = 3), 5 μg/ml (n = 1) 3 μg/ml (n = 2) |
The RNA and protein expression levels of imp/ostA in clinical isolates after glutaraldehyde treatment
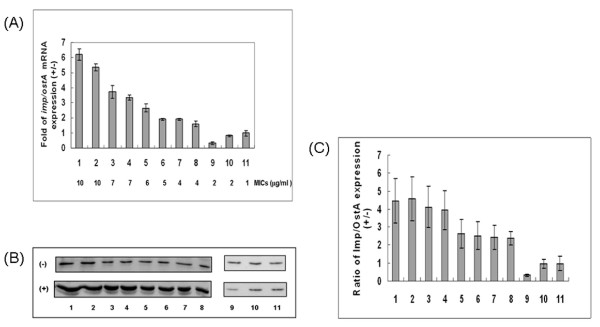
In our previous study [14], we found that the barrier function of the outer membrane against drugs might be decreased in the imp/ostA mutant strain, suggesting that glutaraldehyde may enter the mutant strain more rapidly than it enters the wild-type strain. Four strains with the MICs of 7–10 μg/ml (designed numbers 1~4), four with the MICs of 4–6 μg/ml (numbers 5~8), and three with the MICs of 1–3 μg/ml (numbers 9~11) were selected to clarify the correlation of imp/ostA expression with glutaraldehyde resistance. Subsequently, RNA was extracted from bacteria after 48 h with or without 0.5 μg/ml glutaraldehyde treatment. However, RNA expression of imp/ostA in strains without glutaraldehyde treatment was not detected by slot blot (data not shown). Therefore, we further examined RNA expression of imp/ostA by quantitative real-time PCR. The result indicated that RNA expression of imp/ostA induced by glutaraldehyde was higher in strains with the MICs of 4–10 μg/ml than that in strains with the MICs of 1–3 μg/ml (P= 0.001455) (Fig. 2A). Expression of Imp/OstA protein in these 11 strains after glutaraldehyde treatment was also examined (Fig. 2B). The intensity of protein expression in three independent experiments was analyzed by Image Quant 5.1, and the ratio of Imp/OstA protein expression in the 11 strains with and without glutaraldehyde treatment was calculated. The ratio of Imp/OstA expression induced by glutaraldehyde was higher for strains with the MICs of 4–10 μg/ml (numbers 1~8) than strains with the MICs of 1–3 μg/ml (numbers 9~11) (P = 6.1 × 10-5) (Fig. 2C). These results suggested that the expression of imp/ostA and Imp/OstA was involved in glutaraldehyde resistance in clinical isolates after glutaraldehyde treatment.
Figure 2.
The RNA and protein expression levels of imp/ostA in clinical isolates after glutaraldehyde treatment. (A) Quantitative real-time PCR analysis of the relative expression of imp/ostA mRNA after glutaraldehyde treatment in 11 clinical isolates. The MICs of the corresponding strains are shown in the lower portion of the figure. Each bar represents the relative expression after glutaraldehyde treatment. (B) Western blot analysis of Imp/OstA protein expression. (+) represents glutaraldehyde treatment; (-) represents no glutaraldehyde treatment. (C) The ratio of Imp/OstA protein expression with and without glutaraldehyde treatment. The results were from three independent experiments.
Full genome expression after glutaraldehyde treatment
We next examined the alterations in RNA expression in H. pylori NTUH-S1 induced by glutaraldehyde. After treatment with glutaraldehyde for 48 h, 40 genes were upregulated at least 2.5-fold, and 31 genes were downregulated at least 2.5-fold (see Additional File 1), compared to the untreated bacteria. The upregulated genes included imp/ostA, which was upregulated 9.218-fold. These results are in agreement with the quantitative real-time PCR data, showing that this gene was notably expressed after glutaraldehyde treatment. Another interesting finding was that glutaraldehyde upregulated another lipid transport gene msbA (HP1082) by 2.661-fold. Previous reports indicated that this subfamily of ABC transporters is involved in transport of many different substrates, such as peptides, lipids, hydrophobic drugs, polysaccharides, and proteins [40]. MsbA is a lipid flippase that transports the lipid A-core moiety from the inner to the outer leaflet of the inner membrane in E. coli [17,41]. Imp/OstA also participates in transport of LPS to the cell surface in E. coli [17] and N. meningitidis [20]. We proposed that MsbA might be correlated with LPS transport in H. pylori. The deficiency in a LPS biosynthesis gene could result in antibiotic susceptibility, especially for hydrophobic antibiotics [42-44]. Therefore, weregarded msbA as a suitable candidate for investigating glutaraldehyde or other hydrophobic drug transport in bacteria.
Reconfirmation of msbA expression in the clinical isolates by slot blots hybridization
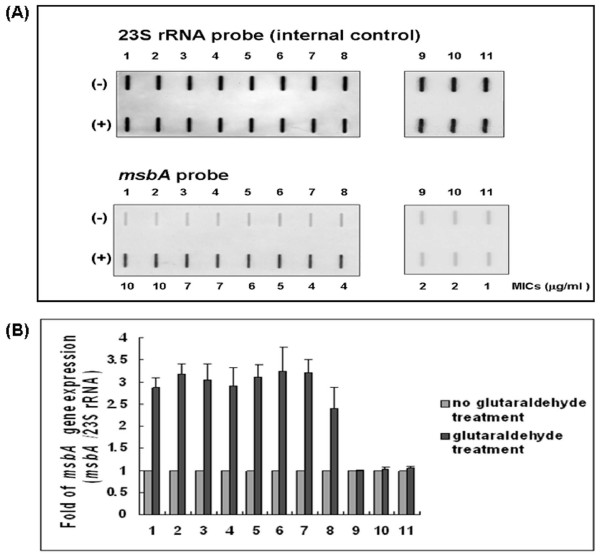
Microarray analysis demonstrated that msbA was upregulated by glutaraldehyde treatment, and the level of msbA expression in the clinical isolates after glutaraldehyde treatment was further determined by slot blot. RNA from the 11 strains used in the imp/ostA expression experiment (numbers 1~11) was extracted before or after glutaraldehyde treatment and hybridized with probes specific for 23S rRNA or msbA. The msbA transcripts were weakly detectable in the control without glutaraldehyde treatment; therefore, the RNA ratio (msbA/23S rRNA) without glutaraldehyde treatment was defined as 1, and the RNA ratio with glutaraldehyde treatment was calculated. The results confirmed the increased expression of msbA induced by glutaraldehyde (Fig. 3A). Furthermore, the level of msbA expression induced by glutaraldehyde was higher in strains with the MICs of 4–10 μg/ml than that in strains with the MICs of 1–3 μg/ml (P = 6.63 × 10-8) (Fig. 3B).
Figure 3.
The expression of msbA in 11 clinical isolates. (A) Slot blots analysis of msbA expression in 11 clinical isolates. Hybridization was performed with DIG probes specific for 23S rRNA and msbA. (+) represents glutaraldehyde treatment. (-) represents no glutaraldehyde treatment. (B) Bacteria were treated or not treated with glutaraldehyde by three independent experiments. The RNA ratio (msbA/23S rRNA) without glutaraldehyde treatment was defined as 1, and the RNA ratio with glutaraldehyde treatment was calculated.
Effect of imp/ostA on the transcription of msbA after glutaraldehyde treatment
The expression of both imp/ostA and msbA was increased in NTUH-S1 after glutaraldehyde treatment according to the results of the microarray analysis. To determine whether imp/ostA affects msbA gene expression after glutaraldehyde treatment and vice versa, RNA levels of imp/ostA and msbA in wild-type and mutant strains after 0.5 μg/ml glutaraldehyde treatment were analyzed by slot blot. The imp/ostA transcript was not affected in the msbA deletion mutant in comparison with the wild-type strain after glutaraldehyde treatment (Fig. 4). Likewise, the msbA transcript was not affected in the imp/ostA deletion mutant in comparison with the wild-type strain after glutaraldehyde treatment. This result indicated that imp/ostA and msbA were induced by glutaraldehyde through independent pathways.
Figure 4.
The effect of imp/ostA on the transcription of msbA after glutaraldehyde treatment and vice versa. Slot blots analysis of total RNA preparations of H. pylori NTUH-S1 wild-type and mutants after 0.5 μg/ml glutaraldehyde treatment for 48 h. Each well was loaded with 10 μg total bacterial RNA. The membrane was hybridized with DIG-labeled probes specific for H. pylori imp/ostA, msbA, and 23S rRNA.
The MICs of glutaraldehyde in isogenic mutants
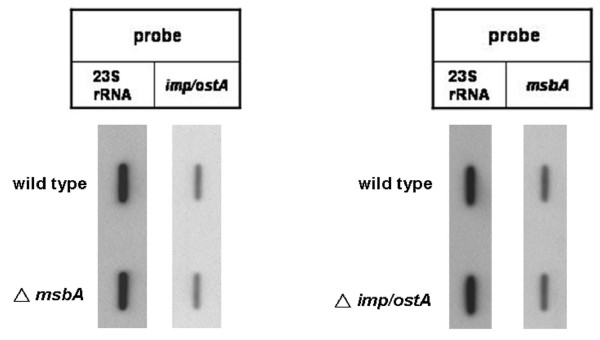
We had previously observed that the imp/ostA mutant became more sensitive to glutaraldehyde than wild-type strain [14]. Southern blot hybridizations were performed to confirm that imp/ostA or msbA were absent in the mutants (Fig. 5). We further investigated whether the sensitivities to glutaraldehyde ofisogenic msbA and an imp/ostA, msbA double mutants were altered. The MIC for the msbA single mutant (3.05 ± 0.27 μg/ml) was lower than for wild-type (5.45 ± 0.21 μg/ml) (wild-type vs.msbA single mutant, P = 2.84 × 10-7). For comparison, the MIC for the imp/ostA single mutant (1.40 ± 0.42 μg/ml) was also significantly lower than that of wild-type, as previously reported [14]. Furthermore, the MICs for imp/ostA and msbA double mutant (0.60 ± 0.14 μg/ml) was also significantly lower than that of wild-type and showed the most significant difference (P = 5.77 × 10-10). Complementation of the msbA mutation significantly restored the resistance to glutaraldehyde (Fig. 6A). These results suggested that imp/ostA and msbA were both involved in glutaraldehyde resistance, and the deficiency of these two genes in H. pylori led to hypersensitivity to glutaraldehyde.
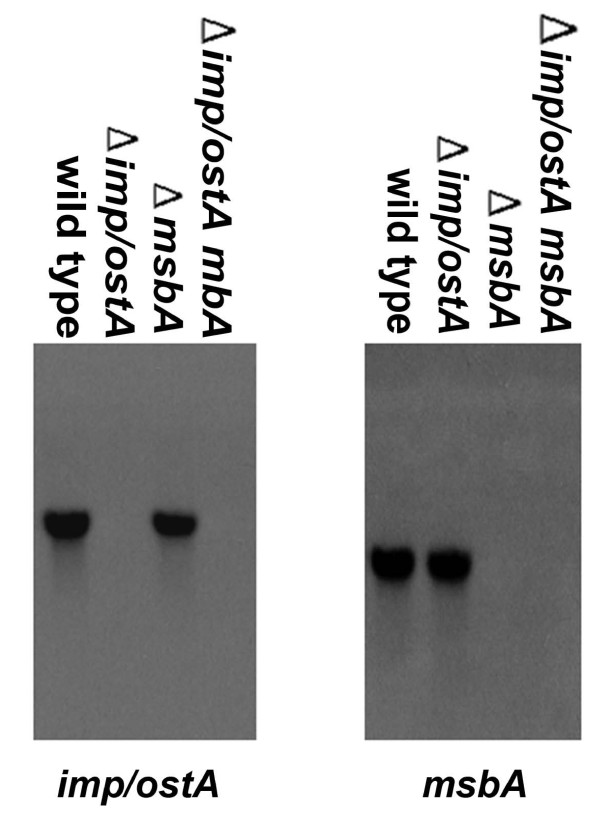
Figure 5.
Southern hybridization of Hind III-digested DNA from strains NTUH-S1 and mutants with imp/ostA (left) and msbA (right) probes. Approximately 5 μg of genomic DNA from H. pylori NTUH-S1 and the mutants was digested by Hind III. Hybridization and detection were performed with the DIG Luminescent Detection kit (Roche) according to the manufacturer's instructions.
Figure 6.
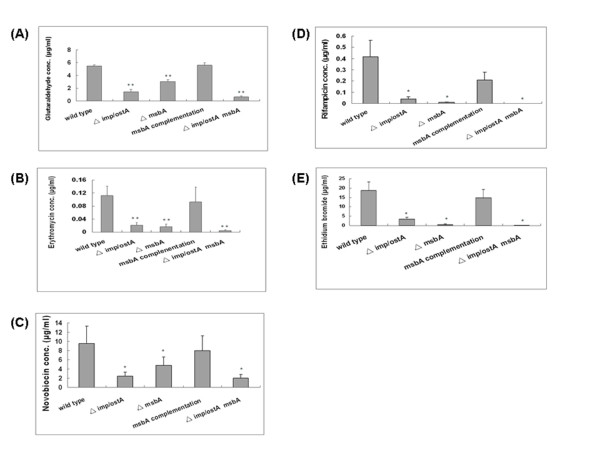
Determination of the MICs glutaraldehyde, erythromycin, novobiocin, rifampicin, and ethidium bromide in H. pylori and isogenic mutants (A-E). Experiments were repeated three to five times. *, P < 0.05 vs. wild-type, and **, P < 0.001 vs. wild-type. Error bars indicate the standard deviation.
The MICs of hydrophobic antibiotics in isogenic mutants
According to previous reports [41,45], MsbA interacts with multiple drugs, for example, multidrug resistance (MDR) substrates (doxorubicin, vinblastine, erythromycin, ethidium bromide) and non-MDR substrates (lipid A, Hoechst). In addition, MsbA increases resistance to erythromycin by 86-fold when it is expressed in L. lactis [22]. In contrast, expression of MsbA in Pseudomonas aeruginosa did not confer resistance to erythromycin, but introducing E. coli msbA into P. aeruginosa decreased the susceptibility of this bacterium to erythromycin by 4-fold [46]. Hence, we investigated whether the isogenic msbA mutant was more sensitive to erythromycin than wild-type H. pylori. The result showed that the MICs were 0.112 ± 0.029 μg/ml and 0.017 ± 0.008 μg/ml for wild-type and the msbA deletion mutant (wild-type vs. msbA deletion mutant, P= 0.00059, respectively). This indicated that MsbA participated in erythromycin resistance in H. pylori. In a previous study [14], it has been reported that the mutation of imp/ostA resulted in a lower MIC of erythromycin in H. pylori. In this study, deletion of both imp/ostA and msbA enhanced the susceptibility to erythromycin (P= 0.00055) (Fig. 6B).
Previous reports demonstrated that in Gram-negative bacteria, a deficiency of the LPS biosynthesis gene would result in antibiotic susceptibility, especially for hydrophobic antibiotics [42-44]. Therefore, we determined the MICs of two hydrophobic antibiotics, novobiocin and rifampicin, to investigate whether imp/ostA and msbA participated in resistance to hydrophobic antibiotics. Both imp/ostA and msbA single mutants were more sensitive to novobiocin and rifampicin than wild-type (Fig. 6C and 6D). These results indicated that imp/ostA and msbA are important for H. pylori resistence to hydrophobic antibiotics. The MIC of rifampicin for the imp/ostA and msbA double mutant (0.00037 ± 0.00013 μg/ml) decreased significantly.
In order to determine the transport route of hydrophobic drugs in bacteria, the hydrophobic compound ethidium bromide was used. In this way, the MIC of ethidium bromide for H. pylori was also examined. The result showed that the msbA mutant was more susceptible to ethidium bromide than the wild-type strain. This result suggests that MsbA might be involved in active efflux by H. pylori, as evidenced by an approximately 36-fold reduction in the MIC of the msbA mutant compared to the wild-type strain (Fig. 6E).
LPS production in H. pylori wild-type and isogenic mutants
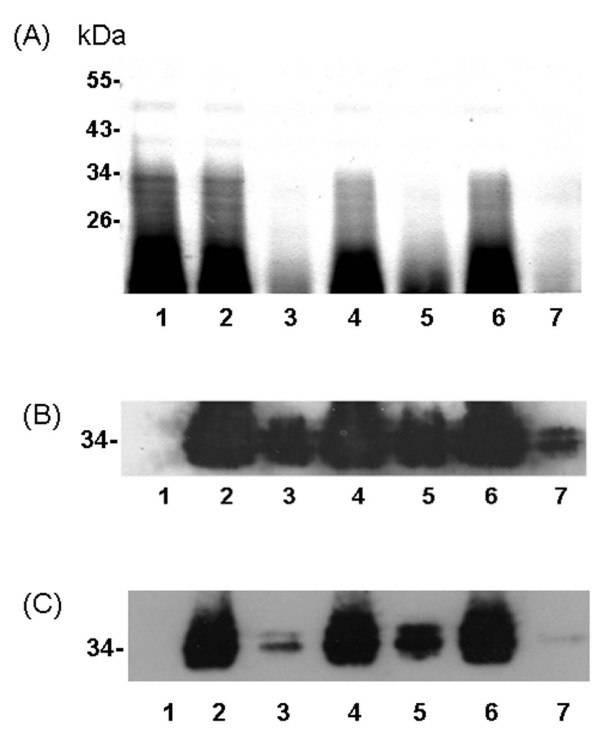
To investigate whether imp/ostA and msbA participated in LPS biogenesis, the equivalent amounts of proteinase K-digested whole cells were analyzed by silver staining (Fig. 7A). The total amount of LPS was drastically reduced in the imp/ostA single mutant compared with that in the wild-type strain (Fig. 7A, lane 3). This result indicated that imp/ostA participated in LPS biogenesis and is consistent with a similar finding in N. meningitidis [20]. Mutation of msbA decreased LPS production, although small amounts of LPS could be detected (Fig. 7A, lane 5). Furthermore, deletion of both imp/ostA and msbA severely reduced LPS production. The LPS in H. pylori was detected by using anti-Lea (Fig. 7B) or anti-Leb antibody (Fig. 7C). H. pylori 26695 strain expressing Lex and Ley but not Lea or Leb antigens was used as a negative control to ensure the specificity of the anti-Lea or anti-Leb antibody (Fig. 7B and 7C, lane 1). The data indicated that the NTUH-S1 strain expressed both the Lea and Leb antigens. In addition, the amounts of O-antigen (~34 kDa) in the imp/ostA or msbA single mutant were reduced, and it was especially reduced in the imp/ostA and msbA double mutant. The growth curves of the wild-type and mutant strains were also examined, and the growth rates of these mutants did not differ from that of the wild-type strain (data not shown). This result demonstrated that both imp/ostA and msbA were involved in the production of LPS.
Figure 7.
Silver-stained of proteinase-K digested whole cell lysate from H. pylori wild-type and isogenic mutants. (A) Lanes 1–7 were all loaded with 2.5 × 108 proteinase K-digested bacteria (~130 μg total protein). Lane 1, 26695; lane 2, wild-type; lane 3, imp/ostA single mutant strain; lane 4, imp/ostA complementation strain; lane 5, msbA single mutant strain; lane 6, msbA complementation strain; lane 7,imp/ostA and msbA double mutant strain. Molecular weights of the prestained markers are indicated. (B-C) Immunoblots of LPS from H. pylori with anti-Lea or anti-Leb monoclonal antibodies. (B) anti-Lea (1:3000) as the primary antibody and anti-mouse IgG (1:5000) as the secondary antibody, or (C) anti-Leb (1:3000) as the primary antibody and anti-mouse IgG (1:5000) as the secondary antibody.
Outer membrane permeability to ethidium bromide
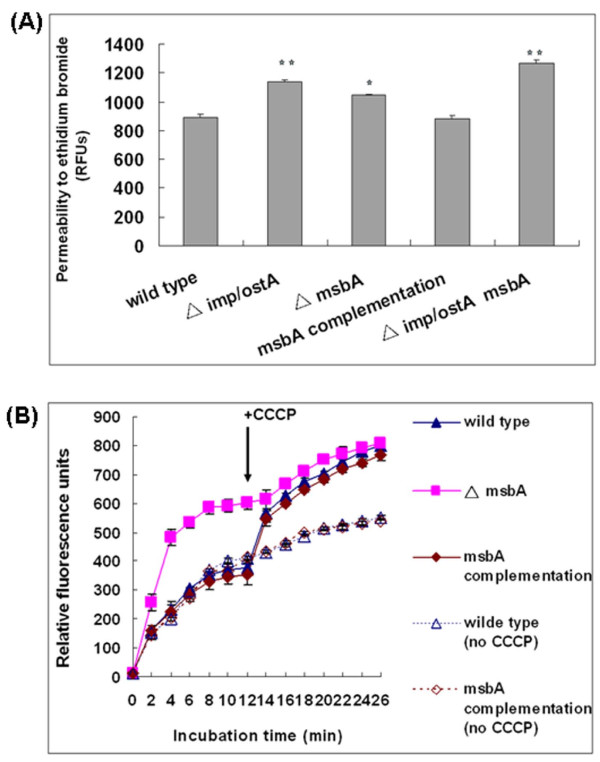
To investigate whether the permeability of the outer membrane was altered in the mutant strains, we measured the fluorescence intensity at a 40-min time point after addition of ethidium bromide and CCCP (Fig. 8A). The fluorescence intensity of the imp/ostA deletion mutant (1142.73 ± 12.38 relative fluorescence units [RFUs]) was higher than that of the wild-type (891.29 ± 20.62 RFUs, P = 0.0001). The fluorescence intensity of the msbA deletion mutant was also significantly higher than the wild-type (P = 0.00164). These results might due to the increase of outer membrane permeability when imp/ostA or msbA was mutated. Furthermore, the fluorescence intensity of the imp/ostA and msbA double mutant was also significantly higher than that of wild-type (P = 5.83 × 10-5). Therefore, the increased sensitivity to hydrophobic compounds conferred by imp/ostA and msbA mutations can be explained by the enhanced membrane permeability for the toxic substances moving in.
Figure 8.
Permeability and efflux of ethidium bromide. (A) Determination of the outer membrane permeability in H. pylori wild-type and isogenic mutants. Each measurement was repeated three times. *, P < 0.05 vs. wild-type, and **, P < 0.001 vs. wild-type. (B) Ethidium bromide accumulation assay. Cells were preloaded with 10 μg/ml ethidium bromide. At the 12-min time point, 10 μM of CCCP was added to the cells suspensions to assess energy-dependent efflux. CCCP was not added to the cells serving as controls (dotted lines). Each measurement was repeated three times.
The role of msbA in ethidium bromide efflux
As ethidium bromide is a hydrophobic aromatic compound, we used this compound to mimic glutaraldehyde or hydrophobic antibiotics moving in and efflux. The Ethidium bromide accumulation assay was performed to determine whether the msbA deletion mutant was more susceptible to glutaraldehyde or hydrophobic antibiotics due to the loss of an active efflux mechanism. The result showed that the msbA deletion mutant accumulated more amounts of ethidium bromide than wild-type (Fig. 8B). When CCCP was added to the cells containing ethidium bromide at 12 min, the accumulation of ethidium bromide increased in wild-type and reached to the level almost equal to that of msbA deletion mutant. This indicated that ethidium bromide was retained in the cells when efflux was blocked after the collapse of the cells' metabolic energy by CCCP. In contrast, CCCP had no significant effect on the level of ethidium bromide accumulated in the msbA deletion mutant. In addition, ethidium bromide accumulation in the msbA complementation strain reached a level almost equal to that of wild-type. CCCP was not added to wild-type or complementation strain containing ethidium bromide at 12 min served as a control. These data indicated that MsbA was involved in hydrophobic drug efflux and that it pumped out ethidium bromide in an energy-dependent process. We concluded that MsbA might pump out glutaraldehyde or hydrophobic antibiotics through an active efflux mechanism in H. pylori.
Discussion
We previously identified that imp/ostA was associated with glutaraldehyde resistance in a clinical H. pylori strain [14]. In order to further investigate the mechanism of glutaraldehyde resistance, the MICs and the levels of imp/ostA expression in clinical isolates were monitored. The result indicated that RNA and protein expression of imp/ostA induced by glutaraldehyde was higher in strains with the MICs of 4–10 μg/ml than that in strains with the MICs of 1–3 μg/ml. According to these results, we suggested that imp/ostA expression was correlated with glutaraldehyde resistance in H. pylori clinical isolates.
After treating NTUH-S1 with glutaraldehyde, 40 genes were found to be upregulated at least 2.5-fold by microarray analysis. For 14 of these genes, DNA or protein sequence alignment yielded no information about their function. The other genes could be divided into three groups: transporters, biosynthesis and metabolism genes, and motility and chemotaxis genes. Two genes were related to iron transport; nonheme iron-containing ferritin (HP0653, pfr), which participates in iron metabolism and in gastric colonization by H. pylori [47]; and an iron dicitrate ABC transporter (HP0889, fecD). Genes including aimF, bioC, ispB, NADH-flavin oxidoreductase (HP0642), and cytochrome c551 peroxidase (HP1461) were involved in biosynthesis and metabolism. Lastly, flagellar hook-associated protein 1 (HP1119, flgK) [48] and flagellar hook-associated protein 2 (HP0752, fliD) [49,50] were related to motility and chemotaxis. However, these genes might not be directly involved in resistance to glutaraldehyde, and their association with glutaraldehyde resistance needs further investigation. In addition, 31 genes were downregulated at least 2.5-fold after glutaraldehyde treatment. Several adjacent genes seemed to be co-regulated, which is indicative of operon structures. For example, HP0690-HP0693 [51] participated in fatty acid metabolism in the TCA cycle. HP0695-HP0696 [51] participated in hydantoin utilization. In addition, some genes are transcribed at different loci but are involved in outer-membrane composition, which included hopG, hofH, and homA. Lastly, two subunits of the 2-oxoglutarate oxidoreductase, oorB and oorD [52], are also involved in the TCA cycle for energy metabolism. The correlation between TCA cycle-related genes and glutaraldehyde resistance also needs to be investigated further.
Silver staining revealed that both imp/ostA and msbA participated in the biogenesis of LPS in H. pylori. Similarly mutation of the E. coli LPS biosynthesis gene, lpxA2, resulted in extreme susceptibility to antibiotics, especially hydrophobic antibiotics [42-44]. Therefore, mutation of the LPS biosynthesis genes, imp/ostA and msbA, might account for the reduction of the MICs for hydrophobic antibiotics.
In the beginning, we observed that the MICs of two glutaraldehyde-resistant strains were 10 μg/ml glutaraldehyde. In fact, this is the half concentration used in our hospital for disinfection during endoscopy. We proposed that some bacteria could survive at the low concentrations in the glutaraldehyde-treated endoscopic environment. According to the MICs tests, LPS analysis, outer membrane permeability assay, and ethidium bromide accumulation assay, the increased sensitivity to hydrophobic compounds conferred by mutations of imp/ostA and msbA can be explained by the defect in LPS production and increased outer membrane permeability. In addition, the increased sensitivity to hydrophobic compounds conferred by mutation of msbA might to the result of accumulation of chemicals that are not pumped out by the MsbA efflux pump. The combination of these effects of the imp/ostA and msbA would reduce the MICs of cells toward glutaraldehyde and hydrophobic antibiotics. These findings might help us to understand the mechanism of bacterial tolerance to chemical disinfectant and hydrophobic drugs.
Conclusion
The expression levels of imp/ostA and msbA were correlated with glutaraldehyde resistance in clinical isolates after glutaraldehyde treatment. Imp/OstA and MsbA play an important role in hydrophobic drugs resistance and LPS biogenesis in H. pylori.
Authors' contributions
HC, TL, and JW conceived and designed the experiments. HC carried out the experiments, analyzed the data, and drafted the manuscript. JY provided clinical isolate strains. TL and JW modified the manuscript. All the authors have read and approved the final manuscript.
Supplementary Material
microarray data. Genes were upregulated and downregulated after glutaraldehyde treatment by microarray analysis
Contributor Information
Hung-Chuan Chiu, Email: gaki98@yahoo.com.tw.
Tzu-Lung Lin, Email: f87445101@ntu.edu.tw.
Jyh-Chin Yang, Email: 81wendy@gmail.com.
Jin-Town Wang, Email: wangjt@ntu.edu.tw.
Acknowledgements
This work was supported by grants from the National Science Council, Taiwan.
References
- Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1(8390):1311–1315. doi: 10.1016/S0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- Blaser MJ. Hypotheses on the pathogenesis and natural history of Helicobacter pylori-induced inflammation. Gastroenterology. 1992;102(2):720–727. doi: 10.1016/0016-5085(92)90126-j. [DOI] [PubMed] [Google Scholar]
- Nomura A, Stemmermann GN, Chyou PH, Kato I, Perez-Perez GI, Blaser MJ. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N Engl J Med. 1991;325(16):1132–1136. doi: 10.1056/NEJM199110173251604. [DOI] [PubMed] [Google Scholar]
- Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, Sibley RK. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325(16):1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- Wotherspoon AC, Doglioni C, Diss TC, Pan L, Moschini A, de Boni M, Isaacson PG. Regression of primary low-grade B-cell gastric lymphoma of mucosa-associated lymphoid tissue type after eradication of Helicobacter pylori. Lancet. 1993;342(8871):575–577. doi: 10.1016/0140-6736(93)91409-F. [DOI] [PubMed] [Google Scholar]
- Cave DR. How is Helicobacter pylori transmitted? Gastroenterology. 1997;113(6 Suppl):S9–14. doi: 10.1016/s0016-5085(97)80004-2. [DOI] [PubMed] [Google Scholar]
- Kaczmarek RG, Moore RM Jr, McCrohan J, Goldmann DA, Reynolds C, Caquelin C, Israel E. Multi-state investigation of the actual disinfection/sterilization of endoscopes in health care facilities. Am J Med. 1992;92(3):257–261. doi: 10.1016/0002-9343(92)90074-L. [DOI] [PubMed] [Google Scholar]
- Fantry GT, Zheng QX, James SP. Conventional cleaning and disinfection techniques eliminate the risk of endoscopic transmission of Helicobacter pylori. Am J Gastroenterol. 1995;90(2):227–232. [PubMed] [Google Scholar]
- Langenberg W, Rauws EA, Oudbier JH, Tytgat GN. Patient-to-patient transmission of Campylobacter pylori infection by fiberoptic gastroduodenoscopy and biopsy. J Infect Dis. 1990;161(3):507–511. doi: 10.1093/infdis/161.3.507. [DOI] [PubMed] [Google Scholar]
- Graham DY, Malaty HM, Evans DG, Evans DJ Jr, Klein PD, Adam E. Epidemiology of Helicobacter pylori in an asymptomatic population in the United States. Effect of age, race, and socioeconomic status. Gastroenterology. 1991;100(6):1495–1501. doi: 10.1016/0016-5085(91)90644-z. [DOI] [PubMed] [Google Scholar]
- Reynolds CD, Rhinehart E, Dreyer P, Goldmann DA. Variability in reprocessing policies and procedures for flexible fiberoptic endoscopes in Massachusetts hospitals. Am J Infect Control. 1992;20(6):283–290. doi: 10.1016/S0196-6553(05)80231-7. [DOI] [PubMed] [Google Scholar]
- Nurnberg M, Schulz HJ, Ruden H, Vogt K. Do conventional cleaning and disinfection techniques avoid the risk of endoscopic Helicobacter pylori transmission? Endoscopy. 2003;35(4):295–299. doi: 10.1055/s-2003-38149. [DOI] [PubMed] [Google Scholar]
- Kaneko H, Mitsuma T, Kotera H, Uchida K, Furusawa A, Morise K. Are routine cleaning methods sufficient to remove Helicobacter pylori from endoscopic equipment? Endoscopy. 1993;25(6):435. doi: 10.1055/s-2007-1010358. [DOI] [PubMed] [Google Scholar]
- Chiu HC, Lin TL, Wang JT. Identification and characterization of an organic solvent tolerance gene in Helicobacter pylori. Helicobacter. 2007;12(1):74–81. doi: 10.1111/j.1523-5378.2007.00473.x. [DOI] [PubMed] [Google Scholar]
- Aono R, Negishi T, Aibe K, Inoue A, Horikoshi K. Mapping of organic solvent tolerance gene ostA in Escherichia coli K-12. Biosci Biotechnol Biochem. 1994;58(7):1231–1235. doi: 10.1271/bbb.58.1231. [DOI] [PubMed] [Google Scholar]
- Sampson BA, Misra R, Benson SA. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics. 1989;122(3):491–501. doi: 10.1093/genetics/122.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandeo P, Lau FK, Carpentieri A, De Castro C, Molinaro A, Deho G, Silhavy TJ, Polissi A. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J Bacteriol. 2008;190(13):4460–4469. doi: 10.1128/JB.00270-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun M, Silhavy TJ. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol Microbiol. 2002;45(5):1289–1302. doi: 10.1046/j.1365-2958.2002.03091.x. [DOI] [PubMed] [Google Scholar]
- Wu T, McCandlish AC, Gronenberg LS, Chng SS, Silhavy TJ, Kahne D. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2006;103(31):11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos MP, Tefsen B, Geurtsen J, Tommassen J. Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc Natl Acad Sci USA. 2004;101(25):9417–9422. doi: 10.1073/pnas.0402340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow M, Georgopoulos C. The essential Escherichia coli msbA gene, a multicopy suppressor of null mutations in the htrB gene, is related to the universally conserved family of ATP-dependent translocators. Mol Microbiol. 1993;7(1):69–79. doi: 10.1111/j.1365-2958.1993.tb01098.x. [DOI] [PubMed] [Google Scholar]
- Woebking B, Reuter G, Shilling RA, Velamakanni S, Shahi S, Venter H, Balakrishnan L, van Veen HW. Drug-lipid A interactions on the Escherichia coli ABC transporter MsbA. J Bacteriol. 2005;187(18):6363–6369. doi: 10.1128/JB.187.18.6363-6369.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefsen B, Bos MP, Beckers F, Tommassen J, de Cock H. MsbA is not required for phospholipid transport in Neisseria meningitidis. J Biol Chem. 2005;280(43):35961–35966. doi: 10.1074/jbc.M509026200. [DOI] [PubMed] [Google Scholar]
- Polissi A, Georgopoulos C. Mutational analysis and properties of the msbA gene of Escherichia coli, coding for an essential ABC family transporter. Mol Microbiol. 1996;20(6):1221–1233. doi: 10.1111/j.1365-2958.1996.tb02642.x. [DOI] [PubMed] [Google Scholar]
- Zhou Z, White KA, Polissi A, Georgopoulos C, Raetz CR. Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. J Biol Chem. 1998;273(20):12466–12475. doi: 10.1074/jbc.273.20.12466. [DOI] [PubMed] [Google Scholar]
- Hsieh PF, Yang JC, Lin JT, Wang JT. Molecular mechanisms of clarithromycin resistance in Helicobacter pylori. J Formos Med Assoc. 1998;97(7):445–452. [PubMed] [Google Scholar]
- Ang S, Lee CZ, Peck K, Sindici M, Matrubutham U, Gleeson MA, Wang JT. Acid-induced gene expression in Helicobacter pylori: study in genomic scale by microarray. Infect Immun. 2001;69(3):1679–1686. doi: 10.1128/IAI.69.3.1679-1686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockelmann U, Dorries HH, Ayuso-Gabella MN, Salgot de Marcay M, Tandoi V, Levantesi C, Masciopinto C, Van Houtte E, Szewzyk U, Wintgens T. Quantitative PCR monitoring of antibiotic resistance genes and bacterial pathogens in three European artificial groundwater recharge systems. Appl Environ Microbiol. 2009;75(1):154–163. doi: 10.1128/AEM.01649-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh PF, Lin TL, Lee CZ, Tsai SF, Wang JT. Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis. 2008;197(12):1717–1727. doi: 10.1086/588383. [DOI] [PubMed] [Google Scholar]
- Hitchcock PJ, Brown TM. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154(1):269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoons-Smit IM, Appelmelk BJ, Verboom T, Negrini R, Penner JL, Aspinall GO, Moran AP, Fei SF, Shi BS, Rudnica W. Typing of Helicobacter pylori with monoclonal antibodies against Lewis antigens in lipopolysaccharide. J Clin Microbiol. 1996;34(9):2196–2200. doi: 10.1128/jcm.34.9.2196-2200.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneghan MA, McCarthy CF, Moran AP. Relationship of blood group determinants on Helicobacter pylori lipopolysaccharide with host lewis phenotype and inflammatory response. Infect Immun. 2000;68(2):937–941. doi: 10.1128/IAI.68.2.937-941.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall DG, Hynes SO, Coleman DC, O'Morain CA, Smyth CJ, Moran AP. Lack of a relationship between Lewis antigen expression and cagA, CagA, vacA and VacA status of Irish Helicobacter pylori isolates. FEMS Immunol Med Microbiol. 1999;24(1):79–90. doi: 10.1111/j.1574-695X.1999.tb01268.x. [DOI] [PubMed] [Google Scholar]
- Moran AP. Lipopolysaccharide in bacterial chronic infection: insights from Helicobacter pylori lipopolysaccharide and lipid A. Int J Med Microbiol. 2007;297(5):307–319. doi: 10.1016/j.ijmm.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Wang G, Ge Z, Rasko DA, Taylor DE. Lewis antigens in Helicobacter pylori: biosynthesis and phase variation. Mol Microbiol. 2000;36(6):1187–1196. doi: 10.1046/j.1365-2958.2000.01934.x. [DOI] [PubMed] [Google Scholar]
- LePecq JB, Paoletti C. A fluorescent complex between ethidium bromide and nucleic acids. Physical-chemical characterization. J Mol Biol. 1967;27(1):87–106. doi: 10.1016/0022-2836(67)90353-1. [DOI] [PubMed] [Google Scholar]
- Giraud E, Cloeckaert A, Kerboeuf D, Chaslus-Dancla E. Evidence for active efflux as the primary mechanism of resistance to ciprofloxacin in Salmonella enterica serovar typhimurium. Antimicrob Agents Chemother. 2000;44(5):1223–1228. doi: 10.1128/AAC.44.5.1223-1228.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglierame P, Pasca MR, De Rossi E, Buroni S, Arrigo P, Manina G, Riccardi G. Efflux pump genes of the resistance-nodulation-division family in Burkholderia cenocepacia genome. BMC Microbiol. 2006;6:66. doi: 10.1186/1471-2180-6-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JT, Sheu JC, Lin JT, Wang TH, Wu MS. Direct DNA amplification and restriction pattern analysis of Helicobacter pylori in patients with duodenal ulcer and their families. J Infect Dis. 1993;168(6):1544–1548. doi: 10.1093/infdis/168.6.1544. [DOI] [PubMed] [Google Scholar]
- Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72(2):317–364. doi: 10.1128/MMBR.00031-07. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes CL, Ward A, Yu J, Chang G. The structures of MsbA: Insight into ABC transporter-mediated multidrug efflux. FEBS Lett. 2006;580(4):1042–1048. doi: 10.1016/j.febslet.2005.11.033. [DOI] [PubMed] [Google Scholar]
- Leive L, Telesetsky S, Coleman WG Jr, Carr D. Tetracyclines of various hydrophobicities as a probe for permeability of Escherichia coli outer membranes. Antimicrob Agents Chemother. 1984;25(5):539–544. doi: 10.1128/aac.25.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuorio R, Vaara M. The lipid A biosynthesis mutation lpxA2 of Escherichia coli results in drastic antibiotic supersusceptibility. Antimicrob Agents Chemother. 1992;36(4):826–829. doi: 10.1128/aac.36.4.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements JM, Coignard F, Johnson I, Chandler S, Palan S, Waller A, Wijkmans J, Hunter MG. Antibacterial activities and characterization of novel inhibitors of LpxC. Antimicrob Agents Chemother. 2002;46(6):1793–1799. doi: 10.1128/AAC.46.6.1793-1799.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G, Janvilisri T, Venter H, Shahi S, Balakrishnan L, van Veen HW. The ATP binding cassette multidrug transporter LmrA and lipid transporter MsbA have overlapping substrate specificities. J Biol Chem. 2003;278(37):35193–35198. doi: 10.1074/jbc.M306226200. [DOI] [PubMed] [Google Scholar]
- Ghanei H, Abeyrathne PD, Lam JS. Biochemical characterization of MsbA from Pseudomonas aeruginosa. J Biol Chem. 2007;282(37):26939–26947. doi: 10.1074/jbc.M702952200. [DOI] [PubMed] [Google Scholar]
- Waidner B, Greiner S, Odenbreit S, Kavermann H, Velayudhan J, Stahler F, Guhl J, Bisse E, van Vliet AH, Andrews SC. Essential role of ferritin Pfr in Helicobacter pylori iron metabolism and gastric colonization. Infect Immun. 2002;70(7):3923–3929. doi: 10.1128/IAI.70.7.3923-3929.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleumink-Pluym NM, Verschoor F, Gaastra W, Zeijst BA van der, Fry BN. A novel approach for the construction of a Campylobacter mutant library. Microbiology. 1999;145(Pt 8):2145–2151. doi: 10.1099/13500872-145-8-2145. [DOI] [PubMed] [Google Scholar]
- Kim JS, Chang JH, Chung SI, Yum JS. Molecular cloning and characterization of the Helicobacter pylori fliD gene, an essential factor in flagellar structure and motility. J Bacteriol. 1999;181(22):6969–6976. doi: 10.1128/jb.181.22.6969-6976.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KA, Karim N, Worku M, Penn CW, O'Toole PW. Helicobacter pylori flagellar hook-filament transition is controlled by a FliK functional homolog encoded by the gene HP0906. J Bacteriol. 2005;187(16):5742–5750. doi: 10.1128/JB.187.16.5742-5750.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflock M, Bathon M, Schar J, Muller S, Mollenkopf H, Meyer TF, Beier D. The orphan response regulator HP1021 of Helicobacter pylori regulates transcription of a gene cluster presumably involved in acetone metabolism. J Bacteriol. 2007;189(6):2339–2349. doi: 10.1128/JB.01827-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes NJ, Clayton CL, Chalk PA, Kelly DJ. Helicobacter pylori porCDAB and oorDABC genes encode distinct pyruvate:flavodoxin and 2-oxoglutarate:acceptor oxidoreductases which mediate electron transport to NADP. J Bacteriol. 1998;180(5):1119–1128. doi: 10.1128/jb.180.5.1119-1128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
microarray data. Genes were upregulated and downregulated after glutaraldehyde treatment by microarray analysis