Abstract
Passive immunotherapy (PI) is being explored as a potential therapeutic against Alzheimer’s disease. The most promising antibodies (Abs) used in PI target the EFRH motif of the Aβ N-terminus. The monoclonal anti-Aβ Ab PFA1 recognizes the EFRH epitope of Aβ. PFA1 has a high affinity for Aβ fibrils and protofibrils (0.1 nM), as well as good affinity for Aβ monomers (20 nM). However, PFA1 binds the toxic N-terminally modified pyro-glutamate peptide pyro-Glu3-Aβ with a 77-fold loss in affinity compared to the WT Aβ(1–8). Furthermore, our earlier work illustrated PFA1’s potential for cross-reactivity. The receptor tyrosine kinase Ror2, which plays a role in skeletal and bone formation, possesses the EFRH sequence. PFA1 Fab binds the Ror2(518–525) peptide sequence REEFRHEA with a 3-fold enhancement over WT Aβ(1–8). In this work, the crystal structures of the hybridoma-derived PFA1 Fab in complex with pyro-Glu3-Aβ peptide and with a cross-reacting peptide from Ror2 have been determined at resolutions of 1.95 and 2.7 Å, respectively. As with wild type Aβ, these peptides bind to the Fab via a combination of charge- and shape-complementarity, hydrogen bonding, and hydrophobic interactions. Comparison of the structures of the four peptides Aβ(1–8), Grip1, pyro-Glu3-Aβ and Ror2 in complex with PFA-1 show that the greatest conformational flexibility occurs at residues 2–3 and 8 of the peptide. These structures provide a molecular basis of the specificity tolerance of PFA1 and its ability to recognize Aβ N-terminal heterogeneity. The structures provide clues to improving mAb specificity and affinity for pyro-Glutamate Aβ.
Keywords: Alzheimer’s disease, passive immunotherapy, pyroglutamate, amyloid beta, Crystal structure
The deposition of amyloid aggregates of the amyloid-beta (Aβ) peptide is a hallmark of the underlying pathology of Alzheimer’s disease (AD). The Aβ peptides (1– 40) and (1–42) are degradation products of the amyloid precursor protein (APP) resulting from the sequential proteolysis by the secretases, β and γ, respectively (1). With two extra C-terminal residues, Aβ (1–42) is considered the more toxic species found in amyloid deposits of both humans and in animal models of AD. The direct immunization of patients with Aβ fibrils in a phase II clinical trial was halted due to the development of meningoencephalitis in 6% of the patients. Passive immunotherapy (PI) where the humanized IgGs are intravenously administered is potentially a safer treatment. At least three mechanisms for the effectiveness of Aβ immunotherapy in AD models have been proposed. Two mechanisms, microglial activation (2, 3) and catalytic dissolution (4), require that the antibody enter the brain. Another mechanism, sometimes called the peripheral sink hypothesis, does not have this requirement. In the peripheral sink hypothesis (5), Abs bind to Aβ in the blood and shift the distribution of Aβ between the brain and the peripheral circulatory system, leading to a net efflux of Aβ from the CNS to the peripheral circulatory system where it is degraded (5). All three mechanisms require Ab binding to Aβ in either monomer or aggregated forms. Since it is unclear which of these mechanisms contributes to AD therapy, it can be argued that the most appropriate Abs for therapy are those capable of recognizing all assembly forms of Aβ peptides.
In a recent study, we demonstrated the molecular basis of passive immunotherapy by determining the first X-ray crystal structures of the N-terminal epitope of Aβ(1–8) (sequence DAEFRHDS) bound to the monoclonal antibodies (mAbs) PFA1 and PFA2 (6). Briefly, the essential binding motif on Aβ is the EFRH sequence at positions 3–6; this motif is recognized by the Fabs' complementarity determining regions, notably a WWDDD region within heavy chain CDR2 in both PFA1 and PFA2.
PFA1 and PFA2 mAbs bind Aβ monomers, protofibrils, and fibrils with nM affinities (6), and hence show promise as potential immunotherapy agents. Besides universal binding to all known physical forms of Aβ(1–40) and Aβ(1–42), however, a desired antibody or antibodies for treating AD should theoretically exhibit certain properties in their binding profile. First, there are a variety of chemical degradation products of Aβ in vivo, some of which are of equal or greater concern-, vis a vis aggregate formation and toxicity than the intact Aβ sequence. Secondly, since the mAb's binding epitope is a relatively short sequence, it is possible that similar or identical sequences might exist elsewhere in the proteome, and exhibit cross-reactivity to the mAb with potential toxic side effects. Thus, the ideal Ab for PI would exhibit good specificity for all toxic and aggregating forms of Aβ while at the same time avoiding high-affinity interactions with other proteins.
The analysis of neuritic and vascular plaques shows that there is substantial N-terminal heterogeneity of Aβ. For example, the Aβ peptide can have its D residues at positions 1 and 7 racemized or isomerized, and there are pyro-glutaminyl (pyro-Glu) forms at residue 3 and 11 (7–10). In fact, there is evidence to suggest the major component of AD senile plaques is the N-terminal modified Aβ beginning at E3, which is post-translationally modified to an N-terminal pyro-Glu(11–15). In some cases pyro-Glu forms of AD at residues 3 (pyro-Glu3-Aβ(3–8)) and 11 (pyro-Glu11-Aβ), constitute more than 50% of the Aβ in neuritic plaques (11). Interestingly, relative to age-matched controls, AD sufferers have an abundance of pyro-Glu-Aβ in their neuritic amyloid deposits, suggesting that pyro-Glu-Aβ may play an important role in the onset of AD.
Supporting evidence for a role for pyro-Glu-Aβ peptides in AD include: (a) the pyro-Glu residue stabilizes the amyloid peptides against degradation by aminopeptidases (16, 17); (b) pyro-Glu forms of Aβ appear in early stages of the disease (18); (c) pyro-Glu-modified peptides form aggregates 250-fold faster than WT Aβ (19); (d) pyro-Glu-Aβ peptides display an enhanced cytotoxicity compared to WT Aβ (9, 14); (e) based on in vitro studies, pyro-Glu-modified Aβ peptides are potential seeding species for Aβ aggregate formation in vivo (20, 21), and (f) recently, inhibitors of the glutaminyl cyclase was shown to reduce plaque burden dramatically and improve cognition in AD mice (22). Unfortunately, given pyro-Glu3-Aβ's potential importance in disease etiology, we found that PFA1 Fab binds pyro-Glu3-Aβ with a much reduced affinity (77-fold difference in Kd) when compared to Aβ(1–8) (6). We would like to explore the structural basis of this loss of affinity.
The shortness of the Aβ sequence epitope also raises possible specificity issues. As a preliminary investigation of the specificity of PFA1 and PFA2 with respect to the human genome, the WT Aβ(2–7) peptide AEFRHD and mutants derived from proteins listed in (6), AKFRHD, AEIRHD, AEFRSD, and REEFRHEA, were synthesized and their affinity for the two Fab fragments was determined by surface plasmon resonance (SPR) (Table 1). AEIRHD and AEFRSD showed no measurable binding to PFA1 and PFA2. However, we found that PFA1-Fab binds to AKFRHD (a sequence found in glutamate receptor interacting protein 1 (GRIP1)) with an affinity 28 times lower than to AEFRHD (6). Perhaps more significantly, the peptide sequence REEFRHEA, found in the cytosolic tyrosine kinase domain of human receptor-related neurotrophic tyrosine kinase (Ror2(518–525)), actually binds to PFA1 and PFA2 with approximately twice the affinity of the WT Aβ(2–7) peptide AEFRHD. Interestingly, Ror2 plays a role in bone formation (23), while Grip1 is responsible for maintaining the plasticity of α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate (AMPA) glutamate receptors by PDZ domain interactions (24, 25).
Table 1.
Alignment of peptide sequences relevant to this work and Kd measurements for peptide binding to the PFA1 Fab taken from (6).
| Protein/Peptide | Sequence | Kd (nM) Measurements from SPR |
|---|---|---|
| Aβ(1–40) WT (“WT”) Aβ(2–7) WT |
DAEFRHDS… AEFRHD |
39.0 60 |
| Pyro-Glu3-Aβ(3–8) | pEFRHDS | 3000 |
| Ror2(518–525) | REEFRHEA | 24 |
| Grip1(110–115) | AKFRHD | 3400 |
| Position 4 mutant | AEIRHD | No binding |
| Position 6 mutant | AEFRSD | No binding |
Here we report the crystal structures of PFA1 complexed with pyro-Glu3-Aβ(3–8) and Ror2(518–525). These structures provide the molecular details that should make possible the optimization of mAb structure to appropriately adjust its binding affinities for these peptides.
Materials and Methods
Protein production and purification
The hybridoma production of PFA1 and its purification and Fab generation were described previously(6).
Crystallization, data collection and processing
Crystal screening was performed with the PEGs Suite (QIAGEN) using the PHOENIX crystallization robot (Art Robbins Instruments). The PFA1-Ror2(518–525) complex and the PFA1-pyro-Glu3-Aβ(3–8) complex were crystallized using the sitting drop vapor diffusion method. Co-crystals of the PFA1-Ror2(518–525) peptide complex appeared in a solution containing 0.2 M LiCl and 20% w/v PEG3350 without a buffer. The co-crystals of the PFA1-pyro-Glu3-Aβ(3–8) complex were grown in a solution consisting of 0.1 M Tris-HCl, pH 8.5 and 25% w/v PEG2000-MME. For X-ray data collection, all crystals were harvested into a cryo-protectant solution. For the PFA1-Ror2(518–525) co-crystals, this consisted of 0.2 M LiCl, 25% PEG3350, and 20% glycerol. The PFA1-Pyro-Glu3-Aβ(3–8) co-crystals were cryo-protected using 0.1 M Tris-HCl, pH 8.5, 28% PEG2000-MME, and 20% glycerol. Crystals were flash-frozen in liquid nitrogen. All diffraction data were collected at the Advanced Photon Source, beamline 14BMC (BIOCARS-CAT) with a Q315 detector. The oscillation range was 0–250° in 0.5° increments for the PFA1-Ror2(518– 525) co-crystals and was 0–190° in 0.5° increments for the PFA1-Pyro-Glu3-Aβ(3–8) co-crystals. Data for both the co-crystals were indexed, integrated, and scaled using the HKL2000 package (26). Although the data for PFA1 complexed with pyro-Glu is 92% complete at 2.2 Å resolution, the data can be processed to 1.95 Å resolution with greater than 50% completeness in the last shell. The PFA1-Ror2 complex diffracted to 2.7 Å resolution with 85% completeness. The lack of >90% completeness can be attributed to the large blind spot region encountered in the P1 space group.
Molecular replacement and structure refinement
The structures were solved by molecular replacement using the constant (CL+CH1) and variable portions of PFA1 (PDB code: 1IPU) separately using MOLREP (27). Protein rebuilding, including water-picking, was performed using Coot (28). Refinement was conducted using Refmac5 (29) and the structures were validated with SFCHECK (30), PROCHECK (31), and MOLPROBITY (32). As reported by PROCHECK, 99.7% and 99.5% of the residues lie in the allowed regions of the Ramachandran plot for PFA1-Pyro-Glu3-Aβ(3–8) and the PFA1-Ror2(518–525) complex structures, respectively. Data collection and refinement statistics appear in Table 2. Calculation of buried surface area was carried out using AREAIMOL (33, 34) with a 1.4 Å probe radius.
Table 2.
Crystallographic data collection and refinement parameters
| PFA1- Ror2(518–525) |
PFA1-PYRO- Glu3-Aβ(3–8) |
|
|---|---|---|
| Data collection | ||
| Space group | P1 | P1 |
| Cell dimensions | ||
| a, b, c (Å) | 41.6 | 41.7 |
| 42.6 | 42.7 | |
| 58.7 | 58.6 | |
| α, β, γ (°) | 96.1 | 96.1 |
| 93.6 | 93.8 | |
| 91.7 | 91.7 | |
| Resolution (Å) | 30-2.7 | 35-1.9 |
| (last shell) | (2.8-2.7) | (2.02-1.95) |
| Rsym | 0.083(0.222) | 0.049(0.284) |
| I / σI | 12.8(2.8) | 17.5(2.2) |
| Completeness | 85.0(49.9) | 86.4(54.2) |
| (%) | ||
| Redundancy | 2.3(1.6) | 1.8(1.5) |
| Refinement | ||
| Resolution (Å) | 2.7 | 1.9 |
| No. reflections | 8780 | 23906 |
| TLS groups | 5 | 0 |
| Rwork / Rfree % | 17.7/24.8 | 20.4/25.6 |
| No. atoms | ||
| Protein(non-H) | 3421 | 3495 |
| Ligand/ion | 0 | 6 (glycerol) |
| Water | 23 | 113 |
| B-factors (Å2) | 32 | 35 |
| Bound peptide | 40 | 52 |
| Ligand/ion | None | 41 (glycerol) |
| Water | 30 | 40 |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.010 | 0.010 |
| Bond angles (°) | 1.2 | 1.3 |
| Ramachandran | 99.5% | 99.7% |
| allowed region | ||
| PDB code | 3EYU | 3EYS |
Results
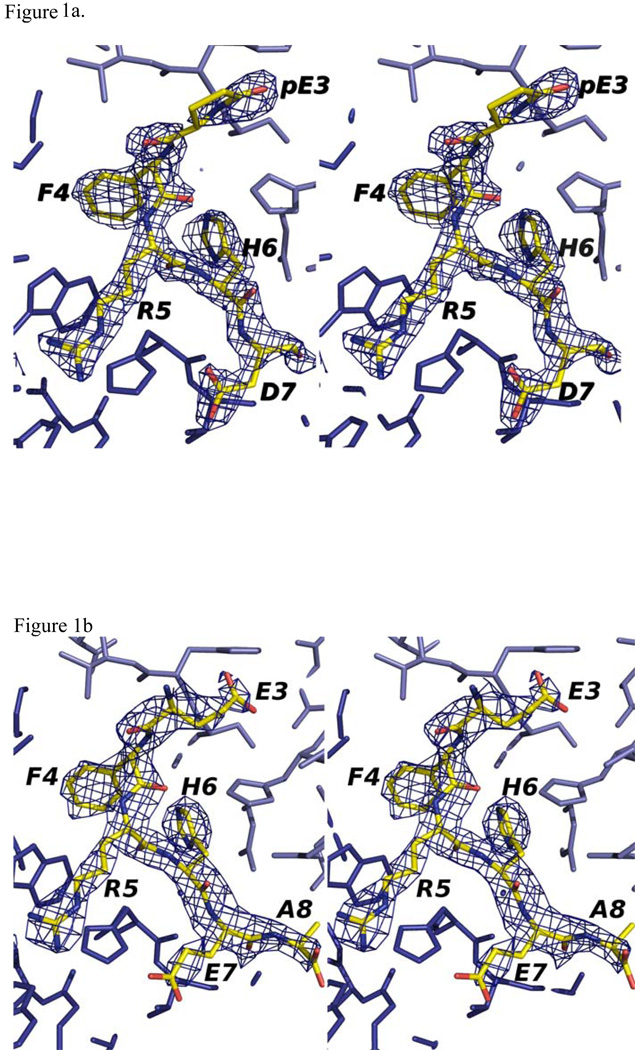
To explore the binding mode of pyro-Glu3-F-R-H-D-S (pyro-Glu3-Aβ(3–8)) peptide to the mAb PFA1, as well as the potential for cross-reactivity with the protein Ror2, we determined the three-dimensional X-ray structures of the two binary co-complexes PFA1-Fab-pyro-Glu3-Aβ(3–8) and PFA1-Fab-Ror2(518–525) to 1.9 Å and 2.7 Å, respectively (see Table 2 for peptide sequences). Both structures have reasonable refinement statistics as shown in Table 2, and they are isomorphous to the apo PFA1 structure (6). The 2Fo-Fc difference Fourier electron density map clearly defines the first five residues of the pyro-Glu3-Aβ(3–8) and the EFRHEA sequence of the Ror2(518–525) of the two structures (Figure 1 and Table 1), and the electron density for the complementarity determining regions (CDRs) is readily traced. The antigen-binding site, which lies in a cleft between the light- and heavy-chain variable domains, is formed by four of the six CDRs: CDR-L1 (QSIVHSNGNTY), CDR-L3 (FQGSHVPLTF), CDR-H2 (IW-WDDDR), and CDR-H3 (VRRAHTTVLGDWFAY). The residues in bold-face type directly interact with the antigen.
Figure 1.
Stereo view of 2Fo-Fc maps of PFA1 binding to the pyro-Glu3-Aβ(3–8) peptide: (a) and the Ror2(518–525), (b). The maps were contoured at 1.1 σ (within 2 Å of the peptide). Numbering scheme is that of Aβ(1–8) WT (see Table 1).
Overall, the pyro-Glu3-Aβ(3–8) peptide adopts a slightly extended coil conformation that is stabilized by hydrogen bonds, van der Waals, and ion-pair interactions (see Figure 2A). The crucial mAb-binding epitope is the 3EFRH6 at the N-terminus of Aβ, which we previously showed to be extremely sensitive to mutations. Mutating any single residue in this epitope to alanine essentially abolishes binding (6). The (FR) of the 3EFRH6 motif makes interactions with a WWDDD motif belonging to CDR-H2, similar to that observed in the Aβ WT complex.
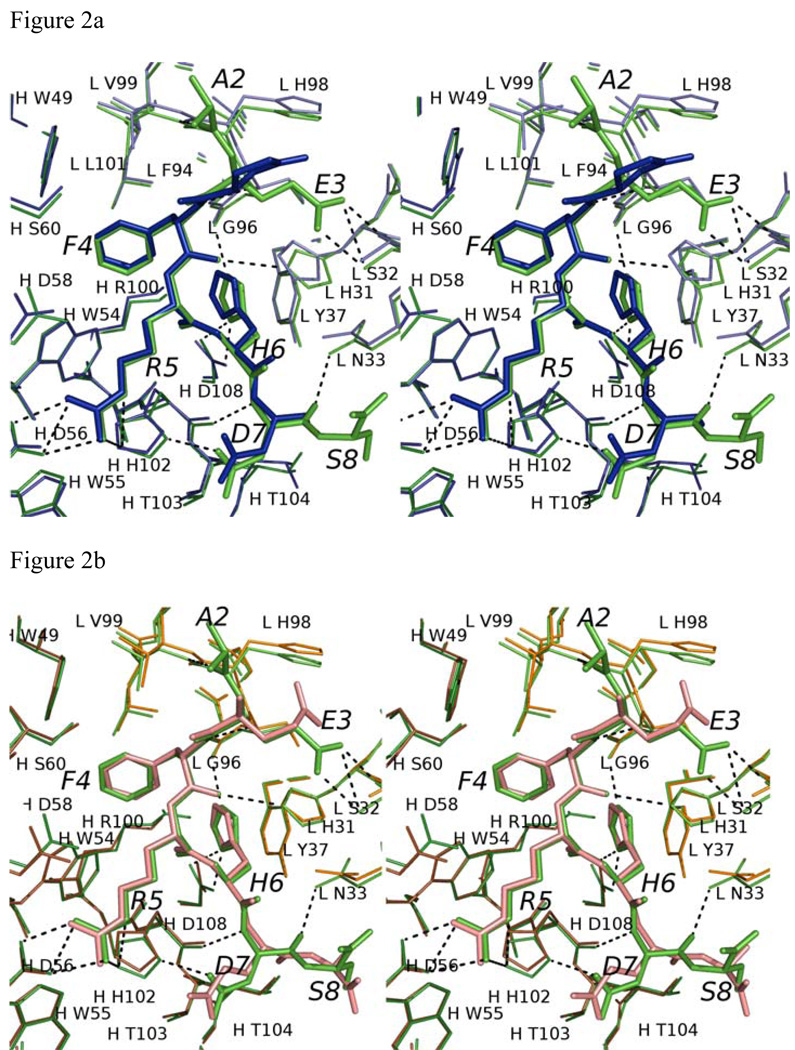
Figure 2.
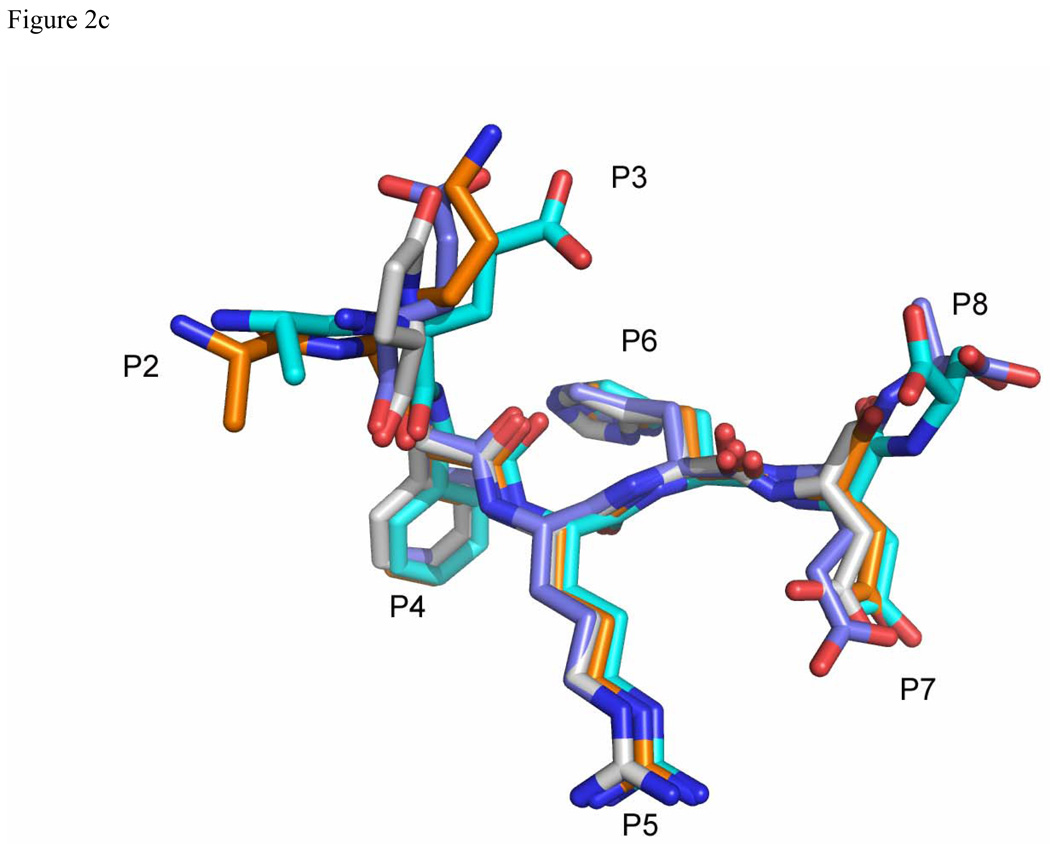
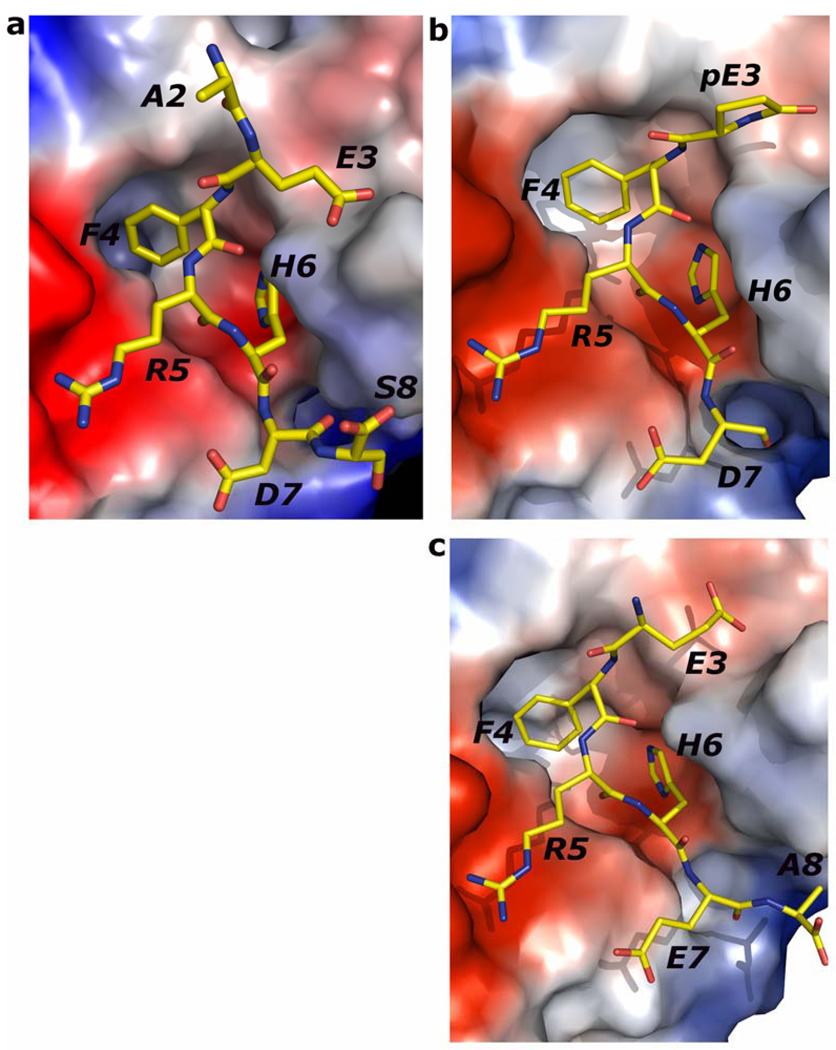
Comparison of Aβ and related peptide structures. Stereo view of the overlay of Aβ(1–8) WT peptide (green) with (a) pyro-peptide (blue), (b) Ror2(518–525) (pink). The PFA1 residues are drawn with thinner bonds and similarly color-coded, save that the light chain is shown in a paler shade and the heavy chain is shown in a darker shade. The numbering scheme is that of Aβ(1–8) WT. The WWDDD motif appears in the lower left corner of (a) and (b); D57 points away from the bound peptide and does not bind to it. (c) Superposition of Aβ-related peptide structures demonstrating coupled position changes (“cross-talk”) of residues 3 and 8: Aβ(1–8) (cyan), Grip1(110–115) (orange), pyro-Glu3 Aβ (white) and Ror2(518–525) (purple) bound to PFA1. The side-chains are marked P2–P8, where P stands for the peptide position.
The Glu to pyro-Glu substitution at position 3 has a drastic effect on mAb binding (6). However, the Cα RMS deviation for residues 3–8 between the WT Aβ and pyro-Glu3-Aβ(3–8) is only 0.24 Å. This is not surprising as the main-chain and side-chain conformations between the two peptides are extremely conserved beyond residue 3 (see Figure 2A). In fact, the main-chain conformation of all the peptide complexes determined by our laboratory is extremely conserved as reflected by their small RMS deviations (see Table 3 and Figure 2C). The greatest structural difference between WT Aβ(1–8) and pyro-Glu3-Aβ(3–8) is found at position 3. Interestingly, the pyro-glutaminyl ring occupies overlapping space with the E3 side-chain of WT Aβ (see Figure 2A). Consequently, this places the pyro-Glu3 amide nitrogen within hydrogen bonding distance of the carbonyl of S97, an interaction that the WT Aβ cannot make. However, while the carbonyl oxygen of pyro-Glu3 occupies a position merely 1.9 Å away from that of the Oε1 atom of the unmodified Aβ E3 side-chain, this is still too far away to form compensatory hydrogen bonds; the nearest hydrogen bonding donor to the carbonyl oxygen atom of pyro-Glu3 is 4 Å away. The carboxylate side chain of E3 in WT Aβ forms two hydrogen bonds to the amide of the main-chain and the hydroxyl group of LC S32. The distance between the amide of LC S32 and the Oε1 of E3 is 2.6 Å, while the distance between the hydroxyl of S32 and the Oε2 of E3 is 2.8 Å. In the PFA1-pyro-Glu3-Aβ(3–8) structure, pyro-Glu3 does not form these two important hydrogen bonds. Instead, the oxygen that is attached to the pyro-glutaminyl ring is almost 4.7 Å away from the amide nitrogen of S32. Furthermore, E3 makes some long-range (< 5 Å) ion-pair interactions with the LC H98 side chain. A complete list of pyro-Glu3 interactions with PFA1 is compared with those of the WT Aβ(1–8) in SD Table 1A–B.
Table 3.
Root Mean Square Differences (RMSDs) for the CDRs of five PFA1 structures.
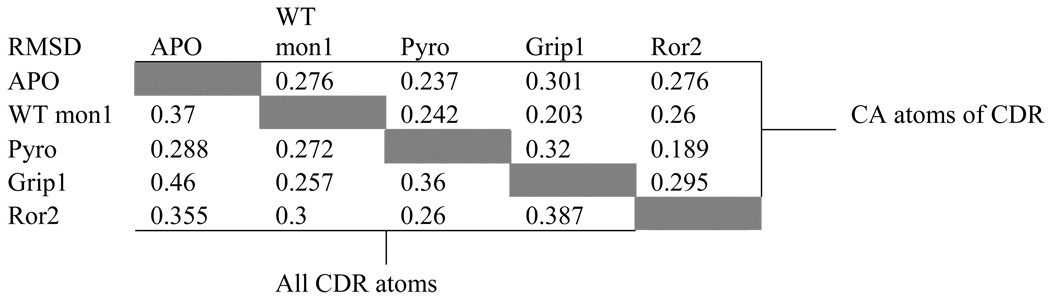 |
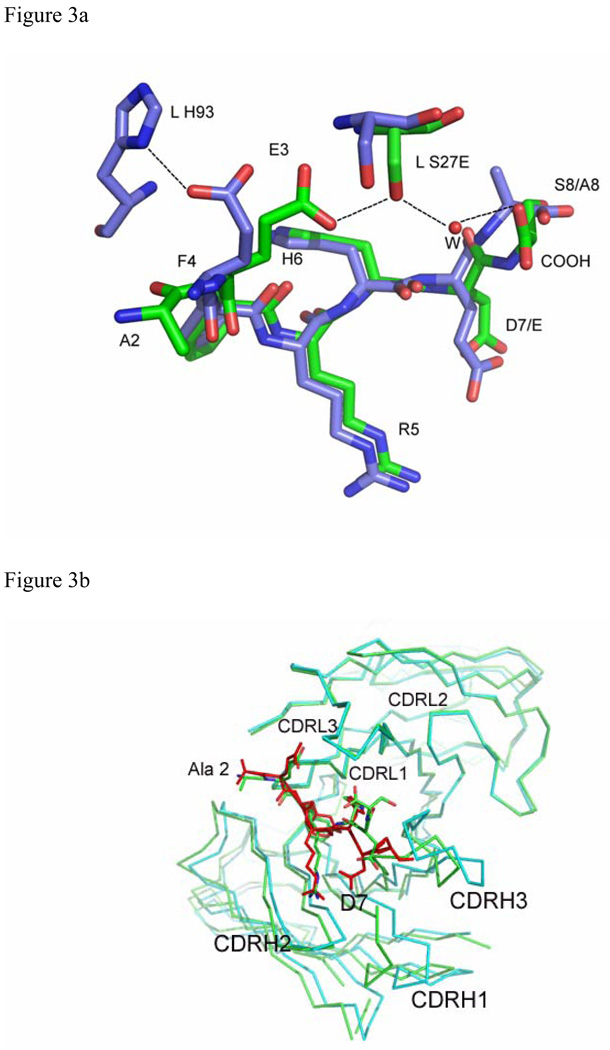
Although there are sequence differences at four positions between WT Aβ(1–8) and the Ror2(518–525) (Table 1), the Ror2(518–525) binds PFA1 similarly to the WT Aβ(1–8). The RMSD between the Cα atoms is 0.3 Å (Table. 3). The greatest differences between the two peptide structures occur at E3, at position 7 where the WT Aβ residue D7 is substituted by E in Ror2(518–525), and at the C-terminus, where S8 of the WT Aβ is replaced by A in Ror2(518–525) (Table 1 and Figure. 2B and Figure 3A). The E3 side-chain adopts a different conformation in the WT Aβ; the carboxylate side-chain hydrogen bonds with the hydroxyl of LC S27E, while in Ror2(518–525) the E3 side-chain hydrogen-bonds with the imidazole side-chain of LC H93. Interestingly, the differing E3 interactions between the two peptides appear to result from the S→A substitution at position 8 in Ror2(518–525) that leads to a chain of events spanning nearly an 8 Å distance. First, the substitution causes a main-chain conformational difference that places the carboxyl terminus of Ror2(518–525) further from LC S27E, abrogating the formation of a water-mediated second sphere hydrogen bond between the COOH and LC S27E observed in the WT Aβ complex (see Figure 3A). Second, the Ror2(518–525)’s COOH group is positioned such that it is unable to stabilize the aforementioned water molecule. Indeed, we do not observe electron density for a water molecule near this position in the PFA1- Ror2(518–525) complex structure. Third, the absence of the water molecule prevents the formation of the hydrogen bond between the water and E3 that is observed in WT Aβ, resulting in side-chain conformational flexibility of Ror2(518–525)’s E3 which finds a new hydrogen bond partner in LC H93. These observations suggest that there is cross-talk between the binding sites 3 and 8 (Figure. 2B and Figure 3A). Also, the substitution of D to E at position 7 results in a slight change in the main-chain conformation between WT Aβ and Ror2(518–525) (Figure 2B). This, combined with the larger E side-chain of Ror2(518–525), positions its carboxyl group further away from the imidazole hydrogen bond donor of HC H102, resulting in a longer hydrogen bond (3.4 Å for PFA1-Ror2(518–525) versus 2.6 Å for PFA1-Aβ(1–8)) between D7 of the bound peptide and H-H102 of PFA1. However, compared to the WT Aβ, the larger E7 side-chain of Ror2(518–525) makes a much shorter intramolecular ion-pair interaction with the guanidinium group of R5 (4.7 Å versus 5.7 Å) belonging to the EFRH motif. A complete list of the Ror2(518–525) peptide’s interactions with PFA1 is compared with those of the WT Aβ(1–8) in SD Table 1B–C. PFA1 binds Ror2(518–525) with an approximately 3-fold improved affinity compared to WT Aβ.
Figure 3.
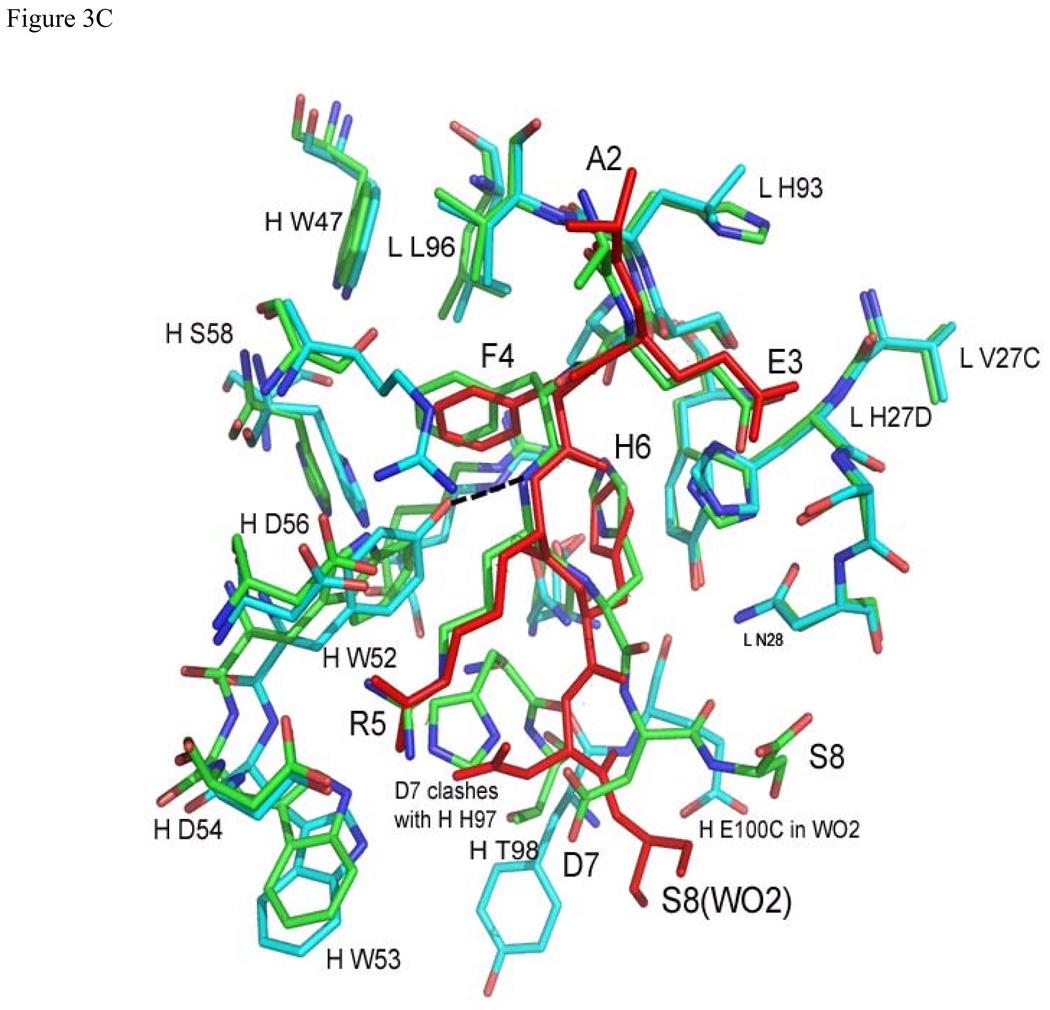
Superposition of Aβ and related structures: (a) Aβ(1–8) (green) and Ror2(518–525) (purple) bound to PFA1 (PFA1 residues are drawn in the same colors corresponding to each peptide), (b) Superposition of PFA1 (CDRs and peptide drawn in green) and WO2 (CDRs cyan, and peptide drawn in red) CDRs and (c) Comparison of PFA1-Aβ(1–8) (peptide and Fab residues are in green) and WO2-Aβ(1–16) (peptide drawn in red and the Fab residues are in cyan) binding sites. One hydrogen bond is shown in dashed lines between HC Y52 of WO2 and the amide of R3 of the Aβ(1–16) to highlight the CDR sequence difference at HC Y52. The Fab residues are labeled according to the PFA1 sequence and numbered according to the Kabat convention (http://www.biochem.ucl.ac.uk/~martin/abs/GeneralInfo.html).
DISCUSSION
To understand why the pyro-Glu modification lowers affinity to PFA1, it is helpful to review the core binding for the WT Aβ peptide. The E3 side-chain of the Aβ peptide makes important hydrogen bonds with the antibody, its only charge-complementarity is with LC H31's side chain, which has an average ~10% charge at physiological pH, so the mAb does not exploit charge-complementarity to significantly stabilize E3 (Figure 2A). The Aβ peptide residues F4, R5, and H6 make more specific interactions, however. F4 sits in a complementarily shaped hydrophobic pocket made up of heavy chain residues W47, H50, W52, and F100(E) (Kabat numbering) and light chain residuesV94 and L96 (Figure 4A). Arginine 5 fits neatly into a shallow cavity consisting of heavy chain residues D54 and D56 for charge complementarity, with residue H97 providing pi-overlap for the guanidinium group and with residue W52 providing a hydrophobic patch near the aliphatic atoms of R5's side chain. Finally, H6 points into a complementary pocket, with pi-overlap provided by light chain residue Y32. H6 forms hydrogen bonds to light chain residue G91 and to heavy chain residue D100(C); light chain residue H27(D) also contributes to the shape of this cavity. The full impact of the pyro-Glu3 modification to PFA1 binding can be observed when comparing electrostatic binding surfaces (Figure 4A–B). Binding energy of protein-protein or protein-ligand can be expressed as a linear combination of the differences in polar and apolar solvent-accessible surface areas between those states (35).The pyro-Glu3-Aβ(3–8) peptide buries 432.9 Å2 of the antibody surface when binding to PFA1, which is some 60 Å2 less than that of the WT Aβ. The difference in SA is ultimately attributable to the reduced length of the pyro-Glu3-Aβ(3–8) peptide (6 residues) compared to WT Aβ (8 residues). The PFA1-pyro-Glu3-Aβ(3–8) co-crystal structure reveals that the loss of binding of pyro-Glu3-Aβ to PFA1 is in part due to the loss of crucial hydrogen bonds and long-range ion-pair interactions with E3. It is also due to the loss of buried SA caused by the shorter pyro-Glu3-Aβ(3–8) peptide compared to WT Aβ peptide.
Figure 4.
Electrostatics of binding: (a) The electrostatic potential surface of PFA1 with bound Aβ(1–8)peptide, (b) with bound pyro-Glu3-peptide and (c) Ror2(518–525). Blue represents positive charge, red indicates negative charge, and the uncharged portions are shown in white. Each of the peptides is drawn with carbon (yellow), nitrogen (blue), and oxygen (red). The numbering scheme is that of Aβ(1–8) WT.
As noted before (19), the following factors may contribute to pyro-Glu-Aβ peptides' acceleration of fibril forming kinetics. First, the pyro-Glu(3–40)-Aβ lacks a charged N-terminal group, making it more hydrophobic (depicted in Figure 4). Second, it has been noted that the capped N-terminus resulting from cyclization reduces Aβ's propensity to form intramolecular antiparallel β-sheets (19). This is particularly important, as fiber diffraction and solid-state NMR data have shown the fibrils are composed of intermolecular, in-register, parallel β-sheets formed by hydrogen bonding between Aβ monomers (36, 37). Moreover, the introduction of a lactam modification that restrains rotation of the peptide bond promotes fibrilogenesis by favoring a conformation prone to aggregation (38). Finally, unlike other N-terminal modifications, which are extremely unstable, the pyro-Glu residue stabilizes the Aβ peptide against degradation by aminopeptidases (16, 17). Hence, its stability, hydrophobicity, and structure make it a perfect seed for fibrilogenesis, and thus a good target for mAb therapy.
Recently, two other related structures have been reported (39, 40). The first structure is of Aβ(17–36) in complex with an affibody adopting a β-conformation (39). However, as the N-terminus of Aβ is absent in this structure, we are unable to do a comparison with our PFA1-Aβ(1–8) complex. The second structure is that of Aβ(1–16) in complex with the Fab fragment of the mAb WO2 (40). Although residues 1–16 of Aβ were used in the co-crystallization experiment, only residues 1–8 of Aβ were observed in the electron density maps. Comparison of the PFA1-Aβ(1–8) with the WO2-Fab-Aβ(1–16) structure reveals that the greatest conformational differences between the two Fabs occur at the two CDRs H2 and H3 that interact with residues 7–8 of the Aβ peptide (see Figures. 3b–c). The Aβ(1–8) peptide conformations bound to PFA1 and WO2 are extremely well conserved between residues 1–6 (see Figure 3c). The FR residues of the EFRH motif in the WO2Fab-Aβ(1–16) structure, as in our PFA1-Aβ(1–8) structure, interacts with the CDRH2 residues 52–56; in PFA1-Aβ(1–8), this segment comprises a WWDDD motif. This motif is conserved in WO2, except that in the WO2 mAb, HC W52 is substituted by Y. The side-chain hydroxyl of HC Y52 in WO2 makes a hydrogen-bond with the main-chain amide of R5 of Aβ, an interaction not observed in the PFA1-Aβ(1–8) complex. Residues D7 and S8 of the Aβ(1–8) peptide show the greatest main-chain conformational difference between the PFA1-Aβ(1–8) and the WO1Fab-Aβ(1–16) structure. One reason for this can be attributed to the previously mentioned chemical and conformational difference at CDRH3 between the two structures − we observe D7 of Aβ(1–8) in the WO1 complex occupying a sterically overlapping position with the imidazole side-chain of HC H97 of PFA1 (Figures. 3b–c). For these reasons, the Fab contact surface of Aβ(7–8) differs between the two complexes. In the WO2-Fab-Aβ(1–16) complex, residues 7–8 contact HC Y100B; in PFA1, they interact instead with HC T98 and HC E100C. The E100C does not have a spatially overlapping residue in PFA1 due to the conformational differences in loop CDRH3 between the two mAbs. Other substitutions between the PFA1 and WO2 include: HC S58→R and LC H93→L.
The EFRH motif of the Aβ N-terminus (comprising of residues 3–6) is the dominant binding epitope for PFA1 and for the recently reported IgG, WO2. The examination of the Ror2(518–525)-PFA1 complex reveals that the Fab's enhanced affinity for Ror2(518–525) when compared to Aβ is likely due to an increased number of electrostatic interactions as well as more favorable intramolecular interactions, possibly stabilizing the peptide conformation. Although, the Ror2(518–525) sequence is found in the (predicted) cytosolic portion of the protein, Tampellini et al have shown evidence for anti-Aβ mAb internalization(41, 42). Hence, its enhanced affinity for PFA1 does strikingly illustrate the risk of cross-reactivity that stems from the short EFRH epitope. Based on our four PFA1-peptide complex structures (Table 1), the greatest variation in the peptide conformations occurs at the N-terminus (positions 2 and 3) and at position 8 at the C-terminus, and less so at position 7 (Figure 2C). Structurally, the FRH main-chain and side-chain conformations are extremely conserved. Hence, to ensure binding to N-terminal variants of Aβ such as pyro-Glu3-Aβ while improving the specificity of PFA1-derived mAbs toward the N-terminus of Aβ, mutations to PFA1 will have to concentrate on optimizing interactions at subsites 7 and beyond. Two such possibilities that we have modeled are to mutate HC 98 to K, or to mutate HC 100 (Kabat numbering) from V to R or K, introducing charge-complementarity to enhance binding to D7 of Aβ. To enhance affinity to the N-terminus of the pyro-Glu3-Aβ peptide, mutating mAb residue LC S27E→Q would put that side chain in a position to form a hydrogen bond with the lactam carbonyl oxygen atom of peptide residue pyro-Glu3. While these mutations might not help distinguish Aβ from Ror2 as such (Ror2 is E7 at that position), they could nonetheless address a general problem with most anti Aβ mAbs: the short length of immunodominant EFRH binding epitope.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the members of the BMC beamline at BioCARS at APS for assistance with data collection. We thank Dr. Brad Bennett and Jay Prendergast for proofreading this manuscript. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract No. W-31-109-Eng-38. Use of the BioCARS Sector 14 was supported by the National Institutes of Health, National Center for Research Resources, under grant number RR007707.
This work was supported by NIH grant numbers NIH NS46356-04A1 and NIH R01AG018416.
The abbreviations used are
- Aβ
amyloid beta
- AD
Alzheimer’s disease
- PFA1
Protofibril antibody 1
- PFA2
Protofibril antibody 2
- SPR
Surface plasmon resonance
- GRIP1
glutamate receptor interacting protein 1
- Ror2
receptor-related neurotrophic tyrosine kinase
- (AMPA)′
α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate
- PEG3350
polyethylene glycol, average molecular weight 3350
Footnotes
The atomic coordinates have been deposited with RCSB ID 3EYU (PFA1-Ror2(518–525)) and 3EYS (PFA1-pyro-Glu3-Aβ(3–8)).
Supporting Information Available: The Table SD1 contains hydrogen bonding data for the PFA1-pyro-Glu3-Aβ and PFA1-Ror2(518–525) structures. This material is available free of charge via the Internet at http://pubs.acs.org
REFERENCES
- 1.Haass C, Koo EH, Mellon A, Hung AY, Selkoe DJ. Targeting of cell-surface beta-amyloid precursor protein to lysosomes: alternative processing into amyloid-bearing fragments. Nature. 1992;357:500–503. doi: 10.1038/357500a0. [DOI] [PubMed] [Google Scholar]
- 2.Schenk D, Barbour R, Dunn W, Gordon G, Grajeda H, Guido T, Hu K, Huang J, Johnson-Wood K, Khan K, Kholodenko D, Lee M, Liao Z, Lieberburg I, Motter R, Mutter L, Soriano F, Shopp G, Vasquez N, Vandevert C, Walker S, Wogulis M, Yednock T, Games D, Seubert P. Immunization with amyloid-beta attenuates Alzheimer-disease-like pathology in the PDAPP mouse. Nature. 1999;400:173–177. doi: 10.1038/22124. [DOI] [PubMed] [Google Scholar]
- 3.Bard F, Cannon C, Barbour R, Burke RL, Games D, Grajeda H, Guido T, Hu K, Huang J, Johnson-Wood K, Khan K, Kholodenko D, Lee M, Lieberburg I, Motter R, Nguyen M, Soriano F, Vasquez N, Weiss K, Welch B, Seubert P, Schenk D, Yednock T. Peripherally administered antibodies against amyloid beta-peptide enter the central nervous system and reduce pathology in a mouse model of Alzheimer disease. Nat Med. 2000;6:916–919. doi: 10.1038/78682. [DOI] [PubMed] [Google Scholar]
- 4.Solomon B, Koppel R, Frankel D, Hanan-Aharon E. Disaggregation of Alzheimer beta-amyloid by site-directed mAb. Proc Natl Acad Sci U S A. 1997;94:4109–4112. doi: 10.1073/pnas.94.8.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeMattos RB, Bales KR, Cummins DJ, Dodart JC, Paul SM, Holtzman DM. Peripheral anti-A beta antibody alters CNS and plasma A beta clearance and decreases brain A beta burden in a mouse model of Alzheimer's disease. Proc Natl Acad Sci U S A. 2001;98:8850–8855. doi: 10.1073/pnas.151261398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardberg AS, Dice LT, Ou S, Rich RL, Helmbrecht E, Ko J, Wetzel R, Myszka DG, Patterson PH, Dealwis C. Molecular basis for passive immunotherapy of Alzheimer's disease. Proc Natl Acad Sci U S A. 2007;104:15659–15664. doi: 10.1073/pnas.0705888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuo YM, Emmerling MR, Woods AS, Cotter RJ, Roher AE. Isolation, chemical characterization, and quantitation of A beta 3-pyroglutamyl peptide from neuritic plaques and vascular amyloid deposits. Biochem Biophys Res Commun. 1997;237:188–191. doi: 10.1006/bbrc.1997.7083. [DOI] [PubMed] [Google Scholar]
- 8.Roher AE, Lowenson JD, Clarke S, Wolkow C, Wang R, Cotter RJ, Reardon IM, Zurcher-Neely HA, Heinrikson RL, Ball MJ, et al. Structural alterations in the peptide backbone of beta-amyloid core protein may account for its deposition and stability in Alzheimer's disease. J Biol Chem. 1993;268:3072–3083. [PubMed] [Google Scholar]
- 9.He W, Barrow CJ. The A beta 3-pyroglutamyl and 11-pyroglutamyl peptides found in senile plaque have greater beta-sheet forming and aggregation propensities in vitro than full-length A beta. Biochemistry. 1999;38:10871–10877. doi: 10.1021/bi990563r. [DOI] [PubMed] [Google Scholar]
- 10.Sergeant N, Bombois S, Ghestem A, Drobecq H, Kostanjevecki V, Missiaen C, Wattez A, David JP, Vanmechelen E, Sergheraert C, Delacourte A. Truncated beta-amyloid peptide species in pre-clinical Alzheimer's disease as new targets for the vaccination approach. J Neurochem. 2003;85:1581–1591. doi: 10.1046/j.1471-4159.2003.01818.x. [DOI] [PubMed] [Google Scholar]
- 11.Saido TC, Iwatsubo T, Mann DM, Shimada H, Ihara Y, Kawashima S. Dominant and differential deposition of distinct beta-amyloid peptide species, A beta N3(pE), in senile plaques. Neuron. 1995;14:457–466. doi: 10.1016/0896-6273(95)90301-1. [DOI] [PubMed] [Google Scholar]
- 12.Russo C, Schettini G, Saido TC, Hulette C, Lippa C, Lannfelt L, Ghetti B, Gambetti P, Tabaton M, Teller JK. Presenilin-1 mutations in Alzheimer's disease. Nature. 2000;405:531–532. doi: 10.1038/35014735. [DOI] [PubMed] [Google Scholar]
- 13.Russo C, Salis S, Dolcini V, Venezia V, Song XH, Teller JK, Schettini G. Amino-terminal modification and tyrosine phosphorylation of [corrected] carboxy-terminal fragments of the amyloid precursor protein in Alzheimer's disease and Down's syndrome brain. Neurobiol Dis. 2001;8:173–180. doi: 10.1006/nbdi.2000.0357. [DOI] [PubMed] [Google Scholar]
- 14.Hosoda R, Saido TC, Otvos L, Jr, Arai T, Mann DM, Lee VM, Trojanowski JQ, Iwatsubo T. Quantification of modified amyloid beta peptides in Alzheimer disease and Down syndrome brains. J Neuropathol Exp Neurol. 1998;57:1089–1095. doi: 10.1097/00005072-199811000-00012. [DOI] [PubMed] [Google Scholar]
- 15.Saido TC, Yamao-Harigaya W, Iwatsubo T, Kawashima S. Amino- and carboxyl-terminal heterogeneity of beta-amyloid peptides deposited in human brain. Neurosci Lett. 1996;215:173–176. doi: 10.1016/0304-3940(96)12970-0. [DOI] [PubMed] [Google Scholar]
- 16.Kraus M, Bienert M, Krause E. Hydrogen exchange studies on Alzheimer's amyloid-beta peptides by mass spectrometry using matrix-assisted laser desorption/ionization and electrospray ionization. Rapid Commun Mass Spectrom. 2003;17:222–228. doi: 10.1002/rcm.901. [DOI] [PubMed] [Google Scholar]
- 17.Piccini A, Russo C, Gliozzi A, Relini A, Vitali A, Borghi R, Giliberto L, Armirotti A, D'Arrigo C, Bachi A, Cattaneo A, Canale C, Torrassa S, Saido TC, Markesbery W, Gambetti P, Tabaton M. beta-amyloid is different in normal aging and in Alzheimer disease. J Biol Chem. 2005;280:34186–34192. doi: 10.1074/jbc.M501694200. [DOI] [PubMed] [Google Scholar]
- 18.Vanderstichele H, De Meyer G, Andreasen N, Kostanjevecki V, Wallin A, Olsson A, Blennow K, Vanmechelen E. Amino-truncated beta-amyloid42 peptides in cerebrospinal fluid and prediction of progression of mild cognitive impairment. Clin Chem. 2005;51:1650–1660. doi: 10.1373/clinchem.2005.051201. [DOI] [PubMed] [Google Scholar]
- 19.Schilling S, Lauber T, Schaupp M, Manhart S, Scheel E, Bohm G, Demuth HU. On the seeding and oligomerization of pGlu-amyloid peptides (in vitro) Biochemistry. 2006;45:12393–12399. doi: 10.1021/bi0612667. [DOI] [PubMed] [Google Scholar]
- 20.Tekirian TL, Yang AY, Glabe C, Geddes JW. Toxicity of pyroglutaminated amyloid beta-peptides 3(pE)-40 and -42 is similar to that of A beta1-40 and -42. J Neurochem. 1999;73:1584–1589. doi: 10.1046/j.1471-4159.1999.0731584.x. [DOI] [PubMed] [Google Scholar]
- 21.Geddes JW, Tekirian TL, Mattson MP. N-terminus truncated beta-amyloid peptides and C-terminus truncated secreted forms of amyloid precursor protein: distinct roles in the pathogenesis of Alzheimer's disease. Neurobiol Aging. 1999;20:75–79. doi: 10.1016/s0197-4580(99)00012-3. discussion 87. [DOI] [PubMed] [Google Scholar]
- 22.Schilling S, Zeitschel U, Hoffmann T, Heiser U, Francke M, Kehlen A, Holzer M, Hutter-Paier B, Prokesch M, Windisch M, Jagla W, Schlenzig D, Lindner C, Rudolph T, Reuter G, Cynis H, Montag D, Demuth HU, Rossner S. Glutaminyl cyclase inhibition attenuates pyroglutamate Abeta and Alzheimer's disease-like pathology. Nat Med. 2008;14:1106–1111. doi: 10.1038/nm.1872. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Ross JF, Bodine PV, Billiard J. Homo-dimerization of Ror2 Tyrosine Kinase Receptor Induces 14 3-3{beta} Phosphorylation and Promotes Osteoblast Differentiation and Bone Formation. Mol Endocrinol. 2007 doi: 10.1210/me.2007-0323. [DOI] [PubMed] [Google Scholar]
- 24.Burette A, Wyszynski M, Valtschanoff JG, Sheng M, Weinberg RJ. Characterization of glutamate receptor interacting protein-immunopositive neurons in cerebellum and cerebral cortex of the albino rat. J Comp Neurol. 1999;411:601–612. [PubMed] [Google Scholar]
- 25.Wyszynski M, Valtschanoff JG, Naisbitt S, Dunah AW, Kim E, Standaert DG, Weinberg R, Sheng M. Association of AMPA receptors with a subset of glutamate receptor-interacting protein in vivo. J Neurosci. 1999;19:6528–6537. doi: 10.1523/JNEUROSCI.19-15-06528.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW, Sweet RM, editors. Macromolecular Crystallography, Part A. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 27.Zhu X, Heine A, Monnat F, Houk KN, Janda KD, Wilson IA. Structural basis for antibody catalysis of a cationic cyclization reaction. J Mol Biol. 2003;329:69–83. doi: 10.1016/s0022-2836(03)00406-6. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 30.Vaguine AA, Richelle J, Wodak SJ. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr D Biol Crystallogr. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 31.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK:a program to check the stereochemical quality of protein structures. J. Appl Cryst. 1993;26:283–291. [Google Scholar]
- 32.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collaborative Computational Project. N The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 34.Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 35.Murphy KP, Xie D, Garcia KC, Amzel LM, Freire E. Structural energetics of peptide recognition: angiotensin II/antibody binding. Proteins. 1993;15:113–120. doi: 10.1002/prot.340150203. [DOI] [PubMed] [Google Scholar]
- 36.Blake CC, Serpell LC, Sunde M, Sandgren O, Lundgren E. A molecular model of the amyloid fibril. Ciba Found Symp 199. 1996:6–15. doi: 10.1002/9780470514924.ch2. discussion 15–21,40-16. [DOI] [PubMed] [Google Scholar]
- 37.Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer's beta-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 38.Sciarretta KL, Gordon DJ, Petkova AT, Tycko R, Meredith SC. Abeta40-Lactam(D23/K28) models a conformation highly favorable for nucleation of amyloid. Biochemistry. 2005;44:6003–6014. doi: 10.1021/bi0474867. [DOI] [PubMed] [Google Scholar]
- 39.Hoyer W, Gronwall C, Jonsson A, Stahl S, Hard T. Stabilization of a beta-hairpin in monomeric Alzheimer's amyloid-beta peptide inhibits amyloid formation. Proc Natl Acad Sci U S A. 2008;105:5099–5104. doi: 10.1073/pnas.0711731105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miles LA, Wun KS, Crespi GA, Fodero-Tavoletti MT, Galatis D, Bagley CJ, Beyreuther K, Masters CL, Cappai R, McKinstry WJ, Barnham KJ, Parker MW. Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope. J Mol Biol. 2008;377:181–192. doi: 10.1016/j.jmb.2007.12.036. [DOI] [PubMed] [Google Scholar]
- 41.Tampellini D, Magrane J, Takahashi RH, Li F, Lin MT, Almeida CG, Gouras GK. Internalized antibodies to the Abeta domain of APP reduce neuronal Abeta and protect against synaptic alterations. J Biol Chem. 2007;282:18895–18906. doi: 10.1074/jbc.M700373200. [DOI] [PubMed] [Google Scholar]
- 42.Arbel M, Solomon B. Immunotherapy for Alzheimer's disease: attacking amyloid-beta from the inside. Trends Immunol. 2007;28:511–513. doi: 10.1016/j.it.2007.09.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.