Abstract
Rationale: Tobacco smoking is responsible for 85% of all lung cancers. To further our understanding of the molecular pathogenesis of lung cancer, we determined whether smoking history leads to the emergence of specific genomic alterations found in non–small cell lung cancer (NSCLC).
Objectives: To identify gene copy number alterations in NSCLCs associated with smoking history or DNA repair capacity.
Methods: Seventy-five NSCLCs were selected for this study from patients with current, none, or past smoking history, including pack year information. Tissue sections were microdissected, and DNA was extracted, purified, and labeled by random priming before hybridization onto bacterial artificial chromosome (BAC) arrays. Normalized ratios were correlated with smoking history and DNA repair capacity was measured by an in vitro lymphocyte assay in the same patients.
Measurements and Main Results: We identified smoking-related genomic signatures in NSCLCs that could be predicted with an overall 74% accuracy. Lung tumors arising from current-smokers had the greatest number of copy number alterations. The genomic regions most significantly associated with smoking were located within 60 regions and were functionally associated with genes controlling the M phase of the cell cycle, the segregation of chromosomes, and the methylation of DNA. Verification of the data is provided from data in the public domain and by quantitative real-time polymerase chain reaction. The associations between genomic abnormalities and DNA repair capacity did not reach statistical significance.
Conclusions: These findings indicate that smoking history leaves a specific genomic signature in the DNA of lung tumors and suggest that these alterations may reflect new molecular pathways to cancer development.
Keywords: array comparative genomic hybridization, tobacco, profile, microarray
AT A GLANCE COMMENTARY
Scientific Knowledge on the Subject
Lung tumors that develop in smokers and nonsmokers have similar clinical behavior and yet result from different injury to the airways. It remains to be determined whether tobacco smoking is responsible for the development of a specific genomic signature that is related to tumorigenesis.
What This Study Adds to the Field
Lung cancers from smokers carry a genomic signature that is distinct from that of never-smokers. The genes associated with these genomic regions of aberration are involved in tobacco-related molecular pathways of tumorigenesis and may represent new targets for chemoprevention.
Lung cancer remains the leading cause of cancer mortality for both men and women in the United States (1). Greater than 85% of all lung cancers are attributed to cigarette smoking; however, only a fraction of long-term cigarette smokers develop lung cancer, suggesting a role for interindividual variation in genetic susceptibility for lung tumorigenesis. Lung cancer also develops through a multistage process in a background of increasing genomic instability (2). Therefore, elucidating the molecular determinants responsible for the development of lung cancer and identifying intermediate biomarkers associated with malignant progression remain important challenges.
Over the last 10 years, we have learned that the somatic molecular alterations in cancers yield signatures that can be used for subclassification (3–5) and that they provide information relevant to predicting patient survival (3, 6), risk of recurrence (7), and response to therapy (4, 8). Nevertheless, non–small cell lung cancer (NSCLC) is still typically managed as a single major entity using similar preventive, diagnostic, and therapeutic approaches. Cigarette smoking contributes to the accumulation of genetic alterations in lung cancer (9–11). Therefore, it is critical to elucidate whether phenotypically similar tumors arising among ever-smokers and lifetime never-smokers acquire specific molecular abnormalities that could further elucidate lung tumorigenesis.
Genetic alterations in NSCLC have been recently measured by array comparative genomic hybridization (12–15). Specific areas of amplification and deletion distinguish squamous from adenocarcinoma of the lung. To better understand how tobacco smoking participates in the molecular pathogenesis of lung cancer, we used array comparative genomic hybridization (CGH) to discover specific patterns of genomic alterations found in NSCLC that might be related to tobacco smoking history. We evaluated whether these somatic patterns of genomic abnormalities were associated with molecular pathways including DNA repair capacity as measured by in vitro peripheral lymphocyte assays of the same patients.
METHODS
Patient Population and Tumor Samples
Frozen samples from 75 resected lung tumors, 32 squamous carcinomas, and 43 adenocarcinoma of the lung, were selected from the pathology archives from the University of Texas M. D. Anderson Cancer Center between 2000 and 2002. Detailed questionnaire data derived from personal interviews were available on all study subjects, including demographic characteristics and smoking history. The clinical and pathological characteristics of the patients are summarized in Table 1. Group 1 consisted of 30 current-smokers (CS) at the time of diagnosis or those who quit less than a year before the diagnosis of lung cancer. Group 2 consisted of 22 former smokers (FS) who quit smoking between 2 and 22 years before diagnosis of lung cancer. Group 3 included 17 lifetime never-smokers (NS; fewer than 100 cigarettes in their lifetime) and 6 long-time quitters (LTQ) who had quit smoking between 23 and 56 years before diagnosis. This cut-off was selected based on the overall distribution of smoking history in the population. Hematoxylin and eosin stained paraffin sections from all tumors were reviewed and tumor-containing areas were circled by our pathologist (I.W.) to identify regions containing greater than 70% tumor cells. The tumor tissues were then microdissected from adjacent twenty-micron methyl green stained tissue sections under a dissecting microscope. The study was approved by the local Institutional Review Boards at Vanderbilt University and the M. D. Anderson Cancer Center.
TABLE 1.
PATIENT CHARACTERISTICS
| Current-smokers | Former-smokers | Nonsmokers/Long-term Quitters | P value* | |
|---|---|---|---|---|
| Patients, n (%) | 30 (40) | 22 (29) | 23 (31) | |
| Age, mean (SD) | 63 (9) | 64 (9) | 68 (12) | 0.131 |
| Sex, n (%) | 0.735 | |||
| Female | 15 (50) | 11 (50) | 14 (61) | |
| Male | 15 (50) | 11 (50) | 9 (39) | |
| PKY, mean (SD) | 60 (50) | 51 (34) | 4 (13) | <0.001 |
| Histology, n (%) | 0.013 | |||
| Adenocarcinoma | 19 (63) | 7 (32) | 17 (74) | |
| Squamous | 11 (37) | 15 (68) | 6 (26) | |
| Stage, n (%) | 0.259 | |||
| I | 16 (53) | 10 (45) | 12 (52) | |
| II | 7 (23) | 9 (41) | 3 (13) | |
| III | 6 (20) | 3 (14) | 8 (35) | |
| IV | 1 (3) | — | — | |
| Ethnicity, n (%) | 0.595 | |||
| Caucasian | 27 (90) | 21 (95) | 20 (87) | |
| Hispanic | — | — | 1 (4) | |
| African | 3 (10) | 1 (5) | 1 (4) | |
| Korean | — | — | 1 (4) | |
| DRC, mean (SD) | 8.23 (2.87) | 8.09 (2.32) | 8.06 (2.41) | 0.99 |
| BIC strand breaks, mean (SD) | 0.77 (0.37) | 0.64 (0.27) | 0.61 (0.27) | 0.352 |
Definition of abbreviations: BIC = bleomycin induced chromatin; DRC = DNA repair capacity; PKY = pack years.
Kruskal-Wallis test for groups with continuous outcomes; Fisher's exact test for categorical outcomes.
DNA repair capacity had been previously assayed in these patients by an in vitro lymphocyte culture-based host cell reactivation assay. The host cell reactivation assay measures the activity of a reactivated reporter gene in cells transfected with benzo(a)diol epoxide–treated plasmids (16). Because a single unrepaired DNA adduct can effectively block transcription (17), any activity reflects the ability of the host cells to remove benzo(a)diol epoxide–induced adducts from the plasmids. Mutagen sensitivity is measured by quantifying the chromatid breaks induced by an in vitro mutagen challenge (in this instance, bleomycin) in short-term lymphocyte cultures and reported as the mean number of breaks per cell (18).
DNA Extraction and Array Comparative Genomic Hybridization Analyses
DNA was extracted, purified, and labeled by random priming (12) before hybridization onto a 2,464 bacterial artificial chromosome (BAC) clone array obtained through the University of California, San Francisco (UCSF) array comparative genomic hybridization (CGH) core laboratory (12, 19–22). One μg of tumor and sex-matched control DNA from a pool of normal peripheral blood monocytes (Promega, Madison, WI, Catalog number is G1471/G1521) were differentially labeled and hybridized onto a 2,464 BAC clones array CGH slide (http://cancer.ucsf.edu/array/services.php#humanBAC). Array CGH hybridizations were performed as described elsewhere (23). Approximately 500 ng of each test and reference probe were coprecipitated with 50 μg of human Cot-1 DNA (Invitrogen, Carlsbad, CA) and resuspended in 20 μl of hybridization mix [50% formamide, 10% dextran sulfate, 2 × sodium chloride/sodium citrate (SSC), 2% sodium dodecyl sulfate (SDS)]. Probes were denatured at 72°C for 7 minutes, incubated at 37°C for 45 minutes, applied to the array slide inside a rubber cement dam and incubated at 37°C in a humidified chamber overnight. Slides were washed and dried before analysis.
Array CGH Data Normalization
Hybridization signals for each array element consisting of red to green intensity ratio of median values obtained by Gene Pix 4.0 (Axon Instruments Inc., Foster City, CA) were analyzed using the snap CGH package in R 2.5.0 (http://www.r-project.org). The data were first filtered by flag and R square ≤ 0.81 (the correlation of the CY3 labeled tumor specimen pixel intensities versus CY5 labeled control DNA). The background was corrected by a log linear interpolation method (24). Within-array normalization was obtained by print-tip lowess methods and between-array normalization with quantile normalization. All data were log2 transformed and lung tissue samples from nine individuals with smoking history but without evidence of malignancy were used to compare tumor classes versus normal signals and to determine a statistical cut-off for amplification and/or deletion (±2 SD). Our report of microarray experiments conforms to Minimum Information about a Microarray Experiment (MIAME) guidelines. The raw data from the experiments is deposited in a publicly accessible database (http://www.vicc.org/biostatistics/supp.php).
Quantitative Real-time Polymerase Chain Reaction
Gene copy number was determined by quantitative real-time polymerase chain reaction (PCR). The method was described by Wang and Velculescu (25). Briefly, real-time PCR was performed on an iQ5-Cycler (Bio-Rad, Hercules, CA) using SYBR Green Supermix (Bio-Rad). Primers for each gene analyzed were designed by PerlPrimer (http://perlprimer.sourceforge.net/). We used the following for PCR amplification protocol: one cycle of 95.0° C for 15 minutes, followed by 45 cycles of 95.0° C for 15 seconds, 59.0° C for 30 seconds and 72.0° C for 30 seconds. FAM5B 5′-CAAAGATAATCTAAGCCCTCACC-3′ forward primer, 5′-TTAGTTGTAGCCTCCCTGTG-3′ reverse primer; MCM2 5′-ATCAACATCCACAACCTCTC-3′ forward, 5′-AGAAACAAACAGTCATGCCAG-3′ reverse; NSD1 5′-ACCTGTCATCAAGCATATCCA-3′ forward, 5′-TTTAGACCATCCACTTTCCCA-3′ reverse; CTSB 5′-TTGAAGTCTACTCTGATGGG-3′ forward, 5′-CGAGAAGTTAAGATGAAGTCC-3′ reverse.
Statistical Analysis
Statistical analyses focused on establishing patterns of recurrent copy number abnormality by smoking status and identification of BAC clones closely associated with the groups. The analysis followed the following steps:
1. Selection of copy number (CN) abnormalities in BAC clones between the study groups.
The selection was based on Wilcoxon rank-sum test, two-sample t test, Kolomogorov-Smirnov test, significance analysis of microarrays (SAM) (26), and weighted gene analysis (WGA) (27). The cut-off points were determined based on SAM >4, WGA >2 and false discovery rate (FDR) <0.05 for Wilcoxon rank-sum test, two-sample t test and Kolomogorov-Smirnov test. The BAC was on the final selection list if it met at least one of the five selection criteria above.
2. Class prediction model.
The weighted flexible compound covariate method (WFCCM) (28–30) was used in the class-prediction model based on the selected BACs to determine whether the genomic patterns could be used to classify tissue samples into two classes (current- versus former-smokers). We estimated the misclassification rate using the leave-one-out cross-validation (LOOCV) class prediction method based on the WFCCM. We used LOOCV to estimate the accuracy of the prediction model. LOOCV uses a single observation as the test set and the remaining observations as the training set. A prediction model involving statistical processes (1) and (2) was built from the training set and used to predict the class of a single-observation test set. The predicted class was compared with the true class to estimate accuracy. The process was repeated n times, where n is the number of samples. During the LOOCV, n different models were created and each one was used to predict the class of the omitted sample. At the end, the n accuracies from LOOCV were averaged to provide an estimate of the accuracy of the prediction model using all n samples.
3. Cluster analysis.
The agglomerative hierarchical clustering algorithm (28) was applied to examine similarity in samples across array elements and to investigate the pattern among the statistically significant discriminator features as well as disease status using M. Eisen's software (31)
4. Data visualization.
Rank-based visualization method of microarray clusters allows users to easily see patterns of trend associated with specific variable (32) that are not apparent in traditional visualizations, and it is more robust to noise. Clustering algorithms that use a rank-based distance metric will group together variables based on their pattern of expression, which can result in clusters that look very nonuniform when traditionally displayed.
Spearman correlation was used to assess the association between BAC copy number and the clinical factors with continuous measurement such as DNA repair capacity (DRC), bleomycin induced chromatin (BIC), pack year history and number of cigarettes smoked per day. The selection of important BACs was based on the cut-off of the FDR < 0.05. Ordinary regression was used to detect whether there was a linear trend between genomic alterations and smoking status (CS, FS and NS or LTQ) with a P value < 0.0005 as the selection cut-off of important BACs.
RESULTS
Patient Characteristics
Characteristics of the patients studied are summarized in Table 1. The mean age of the cases was 63 years for CS, 64 years for FS and 68 years for NS or LTQ. Fifty three percent were female. Women were overrepresented (61%) in the NS group. Seventy-two patients (87%) were non-Hispanic whites, one Hispanic, five African Americans and one was Korean American. Thirty-two patients presented with adenocarcinoma of the lung and 43 with squamous carcinoma of the lung. There was no significant smoking history difference between histological subgroups. The average smoking intensity for CS was 60 pack years (PKY) (number of packs of cigarettes smoked per day × number of years) compared with 51 for FS and 4.5 for NS/LTQ. Seventy-six percent of the tumors were early stage (I or II). Sixty-one percent of patients were alive after 3 years of follow-up. There were no significant difference in DRC, and BIC strand breaks between the smoking groups. NS exhibited the lowest DRC (8.06%) and BIC (0.61), compared with CS (8.23% and 0.77, respectively).
Copy Number Alterations in NSCLCs
Thirty-two squamous cell carcinoma samples, forty-three adenocarcinomas of the lung samples, and nine normal samples were analyzed by array CGH. To confirm the reproducibility of the platform reported in the literature, we tested the variability of the copy number assessment within triplicate analyses of two lung tumors. Copy number measurements across the array were found to have a coefficient of variance of less than 4%. In the analysis of between-group variability, we normalized the data to assays obtained from nine histologically normal lung samples obtained from patients undergoing anatomic resection of lung cancer. The greatest variability in gene copy number was observed in tumors derived from patients with a current smoking history (see Table E1 in the online supplement). The genomic profile of a typical squamous carcinoma is shown in Figure E1 (see online supplement). Significantly more genomic alterations were found among NSCLC from CS (14.9%) than among FS (12.7%) and NS/LTQ (12.0%), P < 0.001. All of the chromosomes were involved with alterations. The most frequent copy number alterations among the three groups are available in the online dataset and presented in supplement Figure E2. In addition, the histologies could be predicted based on 63, 81, and 28 BAC clones with an overall 83, 75, and 78% accuracy for the following comparisons: adenocarcinoma versus squamous carcinoma, adenocarcinoma versus normal, and squamous cell carcinoma versus normal, respectively (Table 2). These results were based on leave one out cross-validation analysis (33).
TABLE 2.
PERFORMANCE OF A PREDICTION MODEL BASED ON SMOKING-RELATED GENOMIC SIGNATURES AND HISTOLOGICAL SUBTYPES
| Accuracy | Specificity | Sensitivity | Classifiers, n | |
|---|---|---|---|---|
| Smoking (CS vs. NS/LTQ) | 74 (62,85) | 77 (62,92) | 70 (51,88) | 35 |
| Smoking (CS vs. FS) | 62 (48,75) | 60 (42,78) | 64 (44,84) | 85 |
| Smoking (FS vs. NS/LTQ) | 45 (30,60) | 45 (23,67) | 45 (25,66) | 81 |
| Smoking (CS vs. NS/LTQ) within Adenocarcinoma | 78 (64,91) | 74 (68,100) | 71 (49,92) | 123 |
| Smoking (CS vs. NS/LTQ) within Squamous | 41 (18,65) | 64 (35,92) | 0 (0,0) | 8 |
| Adenocarcinoma vs. Squamous | 83 (70,97) | 82 (59,100) | 84 (68,100) | 63 |
| Normal vs. Adenocarcinoma | 75 (63,87) | 89 (68,100) | 72 (59,85) | 81 |
| Normal vs. Squamous | 78 (65,91) | 100 (100,100) | 72 (56,87) | 28 |
Definition of abbreviations: CS = current-smokers; FS = former-smokers; LTQ = long-time quitters; NS = never-smokers.
Values are presented as percent (95% confidence interval).
In an effort to compare the frequency and location of the observed copy number changes in all NSCLC to published data, we defined amplification or deletion regions if the log2 ratio was outside of the mean (9 normal samples) ±2 SD. If amplification or deletion frequency for a BAC clone was equal to or greater than 25% of the normal value, this BAC clone was defined as amplification or deletion. The fluorescent in situ hybridization (FISH) bands mapping to these BAC clones, copy number variation (CNV) regions, were used to compare with published datasets. Comparing our CNV regions from all samples (including both adenocarcinoma and squamous) to Table 1 in the article by Zhao and colleagues (13), 5 of 5 regions of deletions were found, whereas 10 of 11 (91%) amplified regions were overlapping. Comparing our CNV regions (based on adenocarcinoma samples only) to the supplementary Table 2 in the article by Weir and colleagues (15), there was an excellent overlap of the regions of interest, since 16 regions of deletion and 10 regions of amplification were also found in our dataset.
NSCLC Genomic Signature Related to Smoking Status
The genomic signature related to smoking status was obtained in three ways. First we looked for BAC clones associated with smoking status. Second, we looked for BAC clones associated with a smoking trend in NS/LTQ, FS, and CS; and third, we looked for BAC clones associated with PKY.
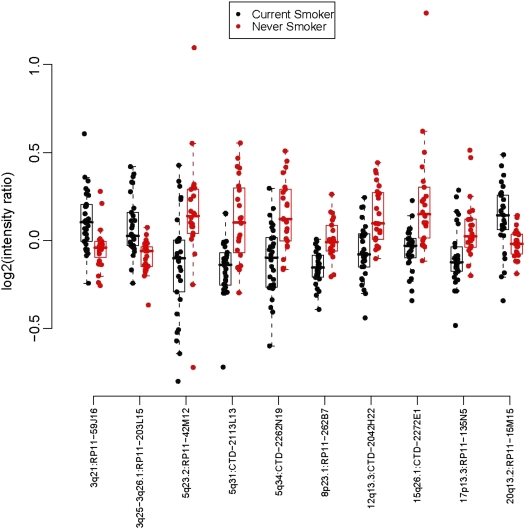
The selection of copy number abnormalities among BAC clones tested between the lung tumors of patients with different smoking status (CS, FS and NS/LTQ) was based on the analysis as described above. To classify the genomic signatures of tumors by smoking status, we used a class-prediction model based on the selected BACs to determine whether the genomic patterns could be used to classify tissue samples between classes. The agglomerative hierarchical clustering algorithm presented in Figure 1 shows similarity in samples across array elements and determines patterns among the statistically significant discriminator features among the smoking groups independently from the histological subgroup. Our prediction model classified lung tumors of CS from those of NS/LTQ or from those of FS with accuracies of 74 and 62% based on 35 and 85 BAC clones, respectively (Table 2). In contrast, we were not able to distinguish NSCLC genomic signatures of FS from NS/LTQ. The data presented are the results of an estimated misclassification rate using the LOOCV class prediction method (classifier selection repeated with each training set prior to the classification of LOOCV (33). From this iterative process in comparing CS with NS/LTQ, we identified 10 BAC clones at genomic locations 3q21, 3q25-3q26, 5q23.2, 5q31, 5q34, 8p23.1, 12q13.3, 15q26.1, 17p13.3 and 20q13.2 that overlap each of the hundreds of cross-validation tests. Those are presented in Figure 2. We also determined our ability to obtain such a signature among adenocarcinomas and obtained a similar accuracy of 78% (Table 2). For squamous carcinoma of the lung, however, and using the same statistical cut-offs, no BAC clones were found to be significantly associated with smoking status.
Figure 1.
Genomic signature of lung cancers according to smoking status: current-smokers, never-smokers, or long-term quitters. On the y axis, the brown label represents the current-smokers group and the blue the never-smokers or long-term quitters. S = squamous carcinoma; A = adenocarcinoma.
Figure 2.
Scatter box plot of median values for copy number (log2) for ten classifiers between current-smokers and never-smokers or long-term quitters.
We additionally looked for BAC clones associated with a trend of smoking status (from NS/LTQ to FS and CS). We found 32 genomic locations strongly associated with this trend (P < 0.0005 as cut-off points), 10 of which were in the list of 35 from the covariate model in the analysis based on smoking status. Finally, we searched for clones associated with smoking history defined as PKY. An additional 6 BAC clones were also found to be associated with smoking PKY and number of cigarettes smoked per day.
The final number of 60 genomic locations associated with smoking (see Table E2 in the online supplement) was used in subsequent analyses to identify candidate genes associated with cigarette smoking. For each of the 60 BAC clones, we used the UCSC online genome browser (http://www.genome.ucsc.edu) to output the reference sequence genes located within an arbitrary 300,000 genomic distance, fixed on the centroid of the BAC. This arbitrary genomic distance for each BAC was chosen because of the absence of full sequencing information on all BACs. This process returned 330 genes (see Table E2 in the online supplement). Twenty-five percent of 60 genomic alterations occurred at known fragile sites (34).
Normally Occurring Copy Number Alterations among Selected Clones
To determine whether any of the smoking-related 60 genomic alterations could be the result of natural copy number variation we compared our results to a study on naturally occurring copy number alterations by Richard Redon (35) (see http://www.nature.com/nature/journal/v444/n7118/full/nature05329.html). We cross-referenced our 2,464 clones to Redon's list of clones that naturally exhibit copy number variations. We found 13 of our 60 selected genomic regions with known naturally occurring copy number variation. Only 3 of the 13 regions of CNV (RP11–123E5, RP11–198G24, RP11–203L17) varied in the same direction (gain or loss) in our dataset and may therefore carry lower significance.
Verification of Copy Number Alterations Associated with Smoking History
We verified the copy number alterations related to smoking in two ways. First, we compared our data to those published by others and second, by quantitative real-time PCR. We compared our array CGH data with the data recently published by Weir and colleagues (15) where five main regions of aberrations were associated with smoking history in adenocarcinoma of the lung. Of the five regions, four were found also altered in our dataset and in the same direction. Specifically these were regions on 7q, 7p, 10q and 16p (see Table E2 in the online supplement). When we tested clones associated with a smoking trend, three of five regions were also identified on 7p, 7q, and 16p.
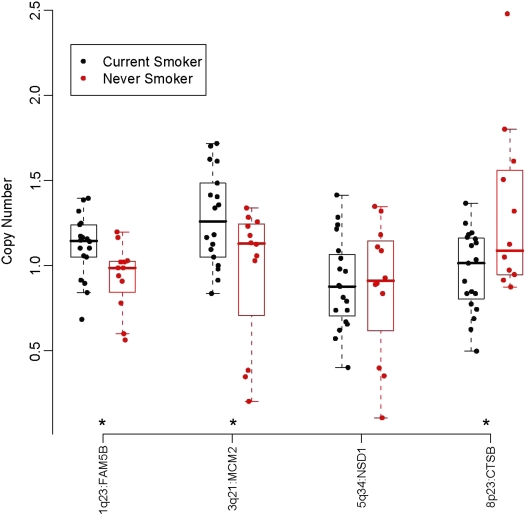
We then selected four other genomic regions strongly associated with cigarette-smoking history representative of our data for quantitative real-time PCR verification. We chose these four regions based on statistical significance and biological implications and tested the copy number of genes within these regions, specifically, FAM5B (1q23), MCM2 (3q21), NSD1 (5q34) and CTSB (8p23.1). Three of these regions were related to the smoking-status analysis and one region to the smoking-trend analysis (FAM5B). Quantitative real-time PCR was performed on the DNA of tumors tested by array CGH (CS, n = 19 and NS/LTQ, n = 12) and we demonstrated in three out of four a significant change in copy number following the same direction as the array CGH data (Figure 3).
Figure 3.
Scatter box plot of median values for copy number for four classifiers between current-smokers and never-smokers or long-term quitters obtained by quantitative real time polymerase chain reaction. Asterisks indicate significant difference between groups, P < 0.05.
KEGG and GO Pathway Analysis of Smoking-Related Genes
To determine whether our smoking-related genes were enriched for some biological processes, cellular components, or molecular pathways and output gene ontology information on our genes of interest, we used the WebGestalt program (36, 37). Using our 2,464 BAC array gene universe as a reference, including 7,300 known genes, we found that our gene set is enriched for the cell cycle pathway in the Kyoto Encyclopedia of Genes and Genomes (KEGG), a knowledge base for systematic analysis of gene functions in terms of the networks of genes and molecules (38). This analysis revealed a strong node in genes related to cell cycle function (CDC25C; CDC23; CDK2; YWHAG; MCM2; ANAPC4; CCNB1; CDK7). The gene ontology annotation available on WebGestalt was provided by the Stanford Source database (39, 40). The analysis in the Gene Oncology (GO) database revealed significant enriched population (P < 0.05) 1) in genes related to the M phase of the cell cycle (CDC25C; CDC23; CDK2; PAFAH1B1; SPBC24; SMC4; YWHAG; ANAPC4; CDCA5; CCNB1); 2) in genes related to chromosome organization and biogenesis (a process that results in the formation, arrangement of constituent parts, or disassembly of eukaryotic chromosomes) (BRD8; SMARCC2; SMG6; TERF2IP; TRIM23; NSD1; JMJD1B; SMARCA4; CARM1; SMC4; MCM2; ARD1A; CDCA5; CENPH; WHSC1); and 3) in genes involved in the transfer of methyl groups (NSD1; CARM1; AMT; WHSC1).
Genomic Signature Associated with DNA Repair Capacity and Polymorphism of Genes Known to Be Associated with Risk of Development of Lung Cancer
There was no association between genomic abnormalities by array CGH, DRC assay, or BIC strand breaks obtained from the peripheral blood of matched individuals. The Spearman correlations between DRC and copy number among BAC clones ranges from −0.47 to 0.57 with FDR adjusted P values ranging from 0.999 to 1. The Spearman correlations between BIC strand breaks and copy number among BAC clones range from −0.49 to 0.45 with FDR adjusted P values ranging from 0.707 to 1.
DISCUSSION
Genomic alterations found in lung cancer may relate to the pathogenesis of this disease and specifically elucidate how smoking history leads to specific patterns of aberrations. Here, we focused on the discovery of genomic signatures found in NSCLC as they relate to smoking history. We found an increase in copy number alterations in tumors from current-smokers. We identified specific patterns of genomic abnormalities that active smoking confers during lung cancer development.
Genetic alterations such as mutations (9), aneuploidy, and gene copy number alterations (deletions and amplifications) (12, 41–43) have long been recognized in lung cancer. Recently, attempts have been made to look at the genome in comprehensive ways to identify tumors with common groups of genetic features that might provide biological or clinical guidance beyond traditional classification by light microscopy. Single nucleotide polymorphism arrays have been developed that are able to analyze loss or gain of genetic material at very high resolution (13, 44), and cancer genome resequencing efforts are likely to uncover common mutations (45). From genomic analyses, relatively small differences have been observed between squamous and adenocarcinoma of the lung. Main differences were described on chromosome 3q and include the p63 gene (14, 46, 47). Minimal chromosomal regions of alterations are now under investigation in the context of mechanistic studies of lung tumorigenesis.
Although the airway epithelium of patients with and without lung cancer may be particularly useful in assessing disease elsewhere in the airways or risk of transformation (48, 49) it provides limited information as to how different lung cancers develop their specific signatures. The signatures we have detected may yield new insights in how cigarette exposure may select or apply the necessary pressure to select copy number alteration in specific regions of the genome. In fact, these signatures may reflect new pathways to cancer development or that may occur later in the process and be a product of selection of genomic alterations related to smoking. Experimental data will be needed to confirm the role of these signatures in tumor development.
From our pathway analysis, the genes found in regions showing association between smoking status and significant copy number differences were implicated in DNA replication, and chromosomal segregation such as SMC4, ANAPC4, BML, SPC24, CARM1 and MCM2. The copy number alterations of these genes controlling chromosomal stability may have resulted in a less stable genome in smokers, a hypothesis that will need to be tested prospectively. These data are consistent with the literature on genotoxicity of tobacco smoke that is known to induce DNA strand breaks, aneuploidy, and mutations in germ cells (50, 51).
The smoking-related signature also affects copy number changes in a series of genes associated with DNA methyltransferase activity, including genes such as NSD1, CARM1, and WHSC1 involved in histone methyltransferase activity, and AMT an aminomethyltransferase. Histones may be methylated and thereby allow the recruitment of regulatory proteins (52, 53). Specific methylated residues confer gene activation within euchromatin (54). The alterations found in these tumors suggest an association between histone methylation, chromatin structure, and development of lung cancer in smokers. Further investigation will be required to explore the role of tobacco smoking in regulating lung carcinogenesis.
Cancer is thought to be clonal in nature, but because of the related genomic instability within tumor cells we can expect great variability of DNA abnormalities between cells within a cancer (55). Yet among these lung tumors, some signatures seem to persist after tumor development throughout their progression and their histological differentiation. The fact that smoking history leads to specific genomic alterations across tumor types also suggests a specific pathogenesis with genomic alteration resulting from a series of dysregulation in the DNA repair mechanism and chromosomal segregation that needs to be validated.
Previously we have shown that suboptimal DNA repair capacity, as measured by the host cell reactivation assay, is associated with up to a two-fold statistically significant increased risk of lung cancer (56, 57). Likewise, the in vitro mutagen sensitivity assay quantifies chromatid breaks induced by bleomycin as an indirect reflection of repair ability. Higher bleomycin sensitivity is associated with 1.6 to 1.9-fold lung cancer risks with evidence of a dose–response relationship (56, 58). The absence of association between copy number alterations and DNA repair capacity and genomic signatures suggests that the processes are independent from one another. Possible explanations could be that the sample size of our study may not be large enough to have the power to detect a significant difference between smokers and never smokers, that the repair capacity was measured in a surrogate tissue (peripheral lymphocytes) and not in the target tissue, and that repair capacity may be a marker of cancer risk and may not impact the genomic regions selected by the cancer related to this phenotype.
In summary, we identified patterns of genomic abnormalities associated with smoking exposure. These signatures may reflect new pathways affected by smoking that lead to lung cancer development. This initial study will need to be validated in an independent set of tumors, but identification of specific genetic abnormalities related to smoking history may allow for the identification of key genes in individuals at high risk who may be targeted for early detection and prevention strategies.
Supplementary Material
Acknowledgments
We thank Jay Snoddy and the Vanderbilt Bioinformatics Resource Center for their assistance in data analysis and Zsuzsanna Adam for her technical support with microarray preparation.
Supported by CA102353, lung SPORE CA90949, lung SPORE CA70907, CA55769 and the Damon Runyon Cancer Research Foundation (Ci-# 19-03 to P.P.M.).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.200801-142OC on September 5, 2008
Conflict of Interest Statement: None of the authors have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin 2007;57:43–66. [DOI] [PubMed] [Google Scholar]
- 2.Hittelman WN. Genetic instability in epithelial tissues at risk for cancer. Ann N Y Acad Sci 2001;952:1–12. [DOI] [PubMed] [Google Scholar]
- 3.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med 2002;8:816–824. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. Classification of human lung carcinomas by mrna expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA 2001;98:13790–13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayes DN, Monti S, Parmigiani G, Gilks CB, Naoki K, Bhattacharjee A, Socinski MA, Perou C, Meyerson M. Gene expression profiling reveals reproducible human lung adenocarcinoma subtypes in multiple independent patient cohorts. J Clin Oncol 2006;24:5079–5090. [DOI] [PubMed] [Google Scholar]
- 6.Lu Y, Lemon W, Liu PY, Yi Y, Morrison C, Yang P, Sun Z, Szoke J, Gerald WL, Watson M, et al. A gene expression signature predicts survival of patients with stage 1 non–small cell lung cancer. PLoS Med 2006;3:e467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xi L, Lyons-Weiler J, Coello MC, Huang X, Gooding WE, Luketich JD, Godfrey TE. Prediction of lymph node metastasis by analysis of gene expression profiles in primary lung adenocarcinomas. Clin Cancer Res 2005;11:4128–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Potti A, Mukherjee S, Petersen R, Dressman HK, Bild A, Koontz J, Kratzke R, Watson MA, Kelley M, Ginsburg GS, et al. A genomic strategy to refine prognosis in early-stage non–small-cell lung cancer. N Engl J Med 2006;355:570–580. [DOI] [PubMed] [Google Scholar]
- 9.Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in p53. Science 1996;274:430–432. [DOI] [PubMed] [Google Scholar]
- 10.Wiencke JK, Thurston SW, Kelsey KT, Varkonyi A, Wain JC, Mark EJ, Christiani DC. Early age at smoking initiation and tobacco carcinogen DNA damage in the lung. J Natl Cancer Inst 1999;91:614–619. [DOI] [PubMed] [Google Scholar]
- 11.Sanchez-Cespedes M, Ahrendt SA, Piantadosi S, Rosell R, Monzo M, Wu L, Westra WH, Yang SC, Jen J, Sidransky D. Chromosomal alterations in lung adenocarcinoma from smokers and nonsmokers. Cancer Res 2001;61:1309–1313. [PubMed] [Google Scholar]
- 12.Massion PP, Kuo WL, Stokoe D, Olshen AB, Treseler PA, Chin K, Chen C, Polikoff D, Jain AN, Pinkel D, et al. Genomic copy number analysis of non–small cell lung cancer using array comparative genomic hybridization: implications of the phosphatidylinositol 3-kinase pathway. Cancer Res 2002;62:3636–3640. [PubMed] [Google Scholar]
- 13.Zhao X, Weir BA, LaFramboise T, Lin M, Beroukhim R, Garraway L, Beheshti J, Lee JC, Naoki K, Richards WG, et al. Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Res 2005;65:5561–5570. [DOI] [PubMed] [Google Scholar]
- 14.Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y, Khatry DB, Protopopov A, You MJ, Aguirre AJ, et al. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci USA 2005;102:9625–9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature 2007;450:893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Athas WF, Hedayati MA, Matanoski GM, Farmer ER, Grossman L. Development and field-test validation of an assay for DNA repair in circulating human lymphocytes. Cancer Res 1991;51:5786–5793. [PubMed] [Google Scholar]
- 17.Koch KS, Fletcher RG, Grond MP, Inyang AI, Lu XP, Brenner DA, Leffert HL. Inactivation of plasmid reporter gene expression by one benzo(a)pyrene diol-epoxide DNA adduct in adult rat hepatocytes. Cancer Res 1993;53:2279–2286. [PubMed] [Google Scholar]
- 18.Hsu TC, Johnston DA, Cherry LM, Ramkissoon D, Schantz SP, Jessup JM, Winn RJ, Shirley L, Furlong C. Sensitivity to genotoxic effects of bleomycin in humans: possible relationship to environmental carcinogenesis. Int J Cancer 1989;43:403–409. [DOI] [PubMed] [Google Scholar]
- 19.Mosquera JM, Perner S, Demichelis F, Kim R, Hofer MD, Mertz KD, Paris PL, Simko J, Collins C, Bismar TA, et al. Morphological features of tmprss2-erg gene fusion prostate cancer. J Pathol 2007;212:91–101. [DOI] [PubMed] [Google Scholar]
- 20.Krzywinski M, Bosdet I, Mathewson C, Wye N, Brebner J, Chiu R, Corbett R, Field M, Lee D, Pugh T, et al. A BAC clone fingerprinting approach to the detection of human genome rearrangements. Genome Biol 2007;8:R224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albertson DG. Profiling breast cancer by array CGH. Breast Cancer Res Treat 2003;78:289–298. [DOI] [PubMed] [Google Scholar]
- 22.Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, Hamilton G, Hindle AK, Huey B, Kimura K, et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet 2001;29:263–264. [DOI] [PubMed] [Google Scholar]
- 23.Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet 1998;20:207–211. [DOI] [PubMed] [Google Scholar]
- 24.Edwards D. Non-linear normalization and background correction in one-channel CDNA microarray studies. Bioinformatics 2003;19:825–833. [DOI] [PubMed] [Google Scholar]
- 25.Wang TL, Maierhofer C, Speicher MR, Lengauer C, Vogelstein B, Kinzler KW, Velculescu VE. Digital karyotyping. Proc Natl Acad Sci USA 2002;99:16156–16161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001;98:5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi OP, et al. Gene-expression profiles in hereditary breast cancer. N Engl J Med 2001;344:539–548. [DOI] [PubMed] [Google Scholar]
- 28.Yamagata N, Shyr Y, Yanagisawa K, Edgerton M, Dang TP, Gonzalez A, Nadaf S, Larsen P, Roberts JR, Nesbitt JC, et al. A training-testing approach to the molecular classification of resected non–small cell lung cancer. Clin Cancer Res 2003;9:4695–4704. [PubMed] [Google Scholar]
- 29.Tukey JW. Tightening the clinical trial. Control Clin Trials 1993;14:266–285. [DOI] [PubMed] [Google Scholar]
- 30.Shyr Y, Kim K. Weighted flexible compound covariate method for classifying microarray data. In: Berrar D, editor. A practical approach to microarray data analysis. New York: Kluwer Academic; 2003. p. 186–200.
- 31.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 1998;95:14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hibbs MA, Dirksen NC, Li K, Troyanskaya OG. Visualization methods for statistical analysis of microarray clusters. BMC Bioinformatics 2005;6:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon R, Radmacher MD, Dobbin K, McShane LM. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J Natl Cancer Inst 2003;95:14–18. [DOI] [PubMed] [Google Scholar]
- 34.Buttel I, Fechter A, Schwab M. Common fragile sites and cancer: targeted cloning by insertional mutagenesis. Ann N Y Acad Sci 2004;1028:14–27. [DOI] [PubMed] [Google Scholar]
- 35.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. Global variation in copy number in the human genome. Nature 2006;444:444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirov SA, Peng X, Baker E, Schmoyer D, Zhang B, Snoddy J. Genekeydb: a lightweight, gene-centric, relational database to support data mining environments. BMC Bioinformatics 2005;6:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang B, Kirov S, Snoddy J. Webgestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 2005;33 (Web Server Issue):W741–W748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ermolaeva O, Rastogi M, Pruitt KD, Schuler GD, Bittner ML, Chen Y, Simon R, Meltzer P, Trent JM, Boguski MS. Data management and analysis for gene expression arrays. Nat Genet 1998;20:19–23. [DOI] [PubMed] [Google Scholar]
- 39.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. The gene ontology (GO) database and informatics resource. Nucleic Acids Res 2004;32:D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camon E, Magrane M, Barrell D, Lee V, Dimmer E, Maslen J, Binns D, Harte N, Lopez R, Apweiler R. The gene ontology annotation (GOA) database: sharing knowledge in uniprot with gene ontology. Nucleic Acids Res 2004;32:D262–D266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Racz A, Brass N, Hofer M, Sybrecht GW, Remberger K, Meese EU. Gene amplification at chromosome 1pter-p33 including the genes pax7 and eno1 in squamous cell lung carcinoma. Int J Oncol 2000;17:67–73. [DOI] [PubMed] [Google Scholar]
- 42.Testa JR, Liu Z, Feder M, Bell DW, Balsara B, Cheng JQ, Taguchi T. Advances in the analysis of chromosome alterations in human lung carcinomas. Cancer Genet Cytogenet 1997;95:20–32. [DOI] [PubMed] [Google Scholar]
- 43.Gray JW, Collins C. Genome changes and gene expression in human solid tumors. Carcinogenesis 2000;21:443–452. [DOI] [PubMed] [Google Scholar]
- 44.Zhao X, Li C, Paez JG, Chin K, Janne PA, Chen TH, Girard L, Minna J, Christiani D, Leo C, et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res 2004;64:3060–3071. [DOI] [PubMed] [Google Scholar]
- 45.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002;3:415–428. [DOI] [PubMed] [Google Scholar]
- 46.Massion PP, Taflan PM, Jamshedur Rahman SM, Yildiz P, Shyr Y, Edgerton ME, Westfall MD, Roberts JR, Pietenpol JA, Carbone DP, et al. Significance of p63 amplification and overexpression in lung cancer development and prognosis. Cancer Res 2003;63:7113–7121. [PubMed] [Google Scholar]
- 47.Garnis C, Lockwood WW, Vucic E, Ge Y, Girard L, Minna JD, Gazdar AF, Lam S, MacAulay C, Lam WL. High resolution analysis of non–small cell lung cancer cell lines by whole genome tiling path array CGH. Int J Cancer 2006;118:1556–1564. [DOI] [PubMed] [Google Scholar]
- 48.Spira A, Beane J, Shah V, Liu G, Schembri F, Yang X, Palma J, Brody JS. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci USA 2004;101:10143–10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spira A, Beane JE, Shah V, Steiling K, Liu G, Schembri F, Gilman S, Dumas YM, Calner P, Sebastiani P, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med 2007;13:361–366. [DOI] [PubMed] [Google Scholar]
- 50.DeMarini DM. Genotoxicity of tobacco smoke and tobacco smoke condensate: a review. Mutat Res 2004;567:447–474. [DOI] [PubMed] [Google Scholar]
- 51.Varella-Garcia M, Chen L, Powell RL, Hirsch FR, Kennedy TC, Keith R, Miller YE, Mitchell JD, Franklin WA. Spectral karyotyping detects chromosome damage in bronchial cells of smokers and patients with cancer. Am J Respir Crit Care Med 2007;176:505–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bannister AJ, Kouzarides T. Reversing histone methylation. Nature 2005;436:1103–1106. [DOI] [PubMed] [Google Scholar]
- 53.Gosden RG, Feinberg AP. Genetics and epigenetics—nature's pen-and-pencil set. N Engl J Med 2007;356:731–733. [DOI] [PubMed] [Google Scholar]
- 54.Shames DS, Girard L, Gao B, Sato M, Lewis CM, Shivapurkar N, Jiang A, Perou CM, Kim YH, Pollack JR, et al. A genome-wide screen for promoter methylation in lung cancer identifies novel methylation markers for multiple malignancies. PLoS Med 2006;3:e486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen JJ, Peck K, Hong TM, Yang SC, Sher YP, Shih JY, Wu R, Cheng JL, Roffler SR, Wu CW, et al. Global analysis of gene expression in invasion by a lung cancer model. Cancer Res 2001;61:5223–5230. [PubMed] [Google Scholar]
- 56.Spitz MR, Wei Q, Dong Q, Amos CI, Wu X. Genetic susceptibility to lung cancer: the role of DNA damage and repair. Cancer Epidemiol Biomarkers Prev 2003;12:689–698. [PubMed] [Google Scholar]
- 57.Wei Q, Cheng L, Amos CI, Wang LE, Guo Z, Hong WK, Spitz MR. Repair of tobacco carcinogen-induced DNA adducts and lung cancer risk: a molecular epidemiologic study. J Natl Cancer Inst 2000;92:1764–1772. [DOI] [PubMed] [Google Scholar]
- 58.Wu X, Lin J, Etzel CJ, Dong Q, Gorlova OY, Zhang Q, Amos CI, Spitz MR. Interplay between mutagen sensitivity and epidemiological factors in modulating lung cancer risk. Int J Cancer 2007;120:2687–2695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.