Abstract
Abiotic stresses, such as drought and salinity, lead to crop growth damage and a decrease in crop yields. Stomata control CO2 uptake and optimize water use efficiency, thereby playing crucial roles in abiotic stress tolerance. Hydrogen peroxide (H2O2) is an important signal molecule that induces stomatal closure. However, the molecular pathway that regulates the H2O2 level in guard cells remains largely unknown. Here, we clone and characterize DST (drought and salt tolerance)—a previously unknown zinc finger transcription factor that negatively regulates stomatal closure by direct modulation of genes related to H2O2 homeostasis—and identify a novel pathway for the signal transduction of DST-mediated H2O2-induced stomatal closure. Loss of DST function increases stomatal closure and reduces stomatal density, consequently resulting in enhanced drought and salt tolerance in rice. These findings provide an interesting insight into the mechanism of stomata-regulated abiotic stress tolerance, and an important genetic engineering approach for improving abiotic stress tolerance in crops.
Keywords: Rice, zinc finger protein, drought tolerance, salt tolerance, stomatal aperture, hydrogen peroxide
Drought and salinity are two main abiotic stresses negatively affecting plant growth and seed production. Plants respond to and adapt to these stresses in order to survive. The understanding of plant responses to stresses in physiology, genetics, and molecular biology will be greatly helpful in improving the tolerance of plants to abiotic stresses through genetic engineering. Stomatal pores that are located in the epidermis of plant leaves control the uptake of CO2 for photosynthesis and the water loss during transpiration, and play a crucial role in abiotic stress tolerance (Schroeder et al. 2001a; Hetherington and Woodward 2003). Under drought stress, abscisic acid (ABA)-mediated stomatal closure is a mechanism that plants use to adapt to water deficiency (Schroeder et al. 2001b; Zhu 2002; Fan et al. 2004). Reactive oxygen species (ROS) including hydrogen peroxide (H2O2) that are widely generated under stress have been proposed to function as second messengers in ABA signaling in guard cells (McAinsh et al. 1996; Pei et al. 2000; Kohler et al. 2003; Bright et al. 2006). Early work has shown that H2O2 can induce stomata closure (McAinsh et al. 1996; Zhang et al. 2001). In guard cells, ABA-stimulated ROS accumulation activates plasma membrane calcium channels and triggers stomatal closure (Hamilton et al. 2000; Pei et al. 2000).
ABA-induced ROS are mainly generated by NADPH oxidase in guard cells (Kwak et al. 2003). Mutation of two genes encoding subunits of an NADPH oxidase (AtrbohD and AtrbohF) impairs ABA-induced stomatal closing (Kwak et al. 2003). To cope with increased levels of ROS, plants have evolved at least four kinds of ROS scavenging enzymes including catalase, superoxide dismutase, ascorbate peroxidase, and glutathione peroxidase. It has been shown that H2O2 is scavenged by glutathione peroxidase in the guard cells (Miao et al. 2006). So far, the genes that regulate the expression of H2O2 scavenging enzymes have not been identified, and the mechanism of regulation of H2O2 levels, especially in the stomata, is largely unknown.
Previous studies have shown that the transcriptional factors play important roles in plant tolerance to abiotic stresses (Stockinger et al. 1997; Liu et al. 1998; Haake et al. 2002; Yamaguchi-Shinozaki and Shinozaki 2006). The Cys-2/His-2-type (C2H2) zinc finger protein—also called a classical or TFIIIA-type zinc finger—that was first found in Xenopus oocytes (Miller et al. 1985) is an important class of eukaryotic transcription factors. Several plant members of this family have various regulatory roles in stress responses and developmental processes (Kim et al. 2001; Sakamoto et al. 2004; de Lorenzo et al. 2007; Xu et al. 2007). However, it is still not known whether the C2H2-type zinc finger protein is involved in regulating stomatal aperture, although previous studies have shown that several transcription factors have been associated with stomata movement (Cominelli et al. 2005; Liang et al. 2005; Hu et al. 2006). In this study, we characterized a novel C2H2 zinc finger transcription factor, DST (drought and salt tolerance), that controls stomatal aperture under drought and salt stress in rice. DST contributes to stomata movement via regulation of genes involved in ROS homeostasis. These findings will shed light on the process of stomatal movement in plants and on the engineering of drought and salt tolerance in crops.
Results
Isolation of dst mutant
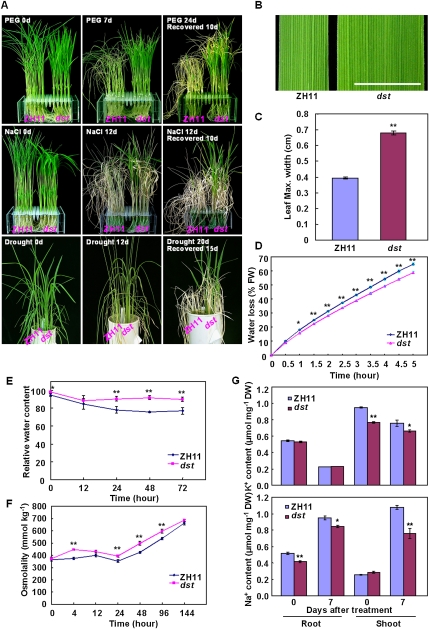
We performed a large-scale screen for the mutants to identify genetic loci that affect drought and salt tolerance in rice and isolated a dst mutant line. When treated with 20% PEG4000 (simulation of drought stress), the dst mutant exhibited less severe wilting than wild-type (ZH11) plants (Fig. 1A, top panel). Furthermore, ∼70% of dst mutant plants survived under 140 mM NaCl treatment, but nearly all of the wild-type plants wilted (Fig. 1A; Supplemental Fig. S1A). The dst mutant exhibited better recovery than wild-type plants from both drought and salt stresses (Fig. 1A). To accurately evaluate the effect of the DST mutation on drought tolerance, we performed a soil drought experiment in deep polyvinyl chloride (PVC) pipes. The dst mutant plants showed stronger drought tolerance than the wild-type plants (Fig. 1A, bottom panel). These results indicated that mutation of the DST locus significantly improves drought and salt tolerance. There were no significant morphological alterations in dst mutants, except for a markedly wider leaf width (Fig. 1B,C). Most agronomic traits were unchanged in the mutants, with the exception of the panicle number per plant and the main panicle length (Supplemental Fig. S2).
Figure 1.
The dst mutant shows tolerance to drought and salt stress. (A) PEG4000, NaCl, and drought stress treatment of wild-type plants Zhonghua11 (ZH11) and the dst mutant. Twenty-day-old plants were treated with 20% PEG4000 for 7 d (top panel) or 140 mM NaCl for 12 d (middle panel), and then recovered as indicated. (Bottom panel) Thirty days after transplantation, plants were subjected to drought stress for 12 or 20 d and then allowed to recover for 15 d. (B) Leaf of ZH11 and dst mutant. Bar, 0.5 cm. (C) Quantitative measurement of the maximum leaf width of ZH11 and dst mutant (n = 20). (D) Water loss of ZH11 and dst mutant. For each repeat, 15 fully expanded leaves of 25-d-old plants were used in a triplicate experiment (n = 3). (E) Relative water content of ZH11 and dst mutant treated with 18% PEG4000 using the fully expanded leaves of 20-d-old plants (n = 9). (F) Osmolality measurement in ZH11 and dst mutant under 18% PEG4000 treatment using the fully expanded leaves of 25-d-old plants (n = 8). (G) Na+ and K+ contents in the roots and shoots of ZH11 and dst mutant plants under non-salt stress and 100 mM NaCl treatment for 7 d (n = 3). Data in C–G are presented as mean ± SEM. (*) P ≤ 0.05; (**) P ≤ 0.01, Student's t-test.
Physiological analysis of dst mutant
To elucidate the physiological mechanism of drought and salt tolerance in the dst mutant, we first investigated the water status in wild-type and dst mutants. Under dehydration stress, the dst mutant lost less water and maintained higher water content than wild-type plants (Fig. 1D,E). The dst mutant also maintained higher osmolality during drought treatment (Fig. 1F). These results indicate that the enhanced drought tolerance of the dst mutant is due to an increased ability to maintain water. Finally, we compared the K+ and Na+ content in the roots and shoots in response to NaCl stress. There was no significant difference in K+ content between the roots of the wild type and dst mutant; however, K+ content was lower in the shoots of dst mutants under both normal and salt stress conditions (Fig. 1G). NaCl treatment resulted in a lower accumulation of Na+ in the roots and shoots of dst mutant plants (Fig. 1G), indicating that the enhanced salt tolerance of dst mutant might be due to a reduction of Na+ toxicity. These results demonstrate that the DST mutation leads to enhancement of drought and salt tolerance in rice plants.
Stomatal movement changes in the dst mutant due to H2O2 accumulation
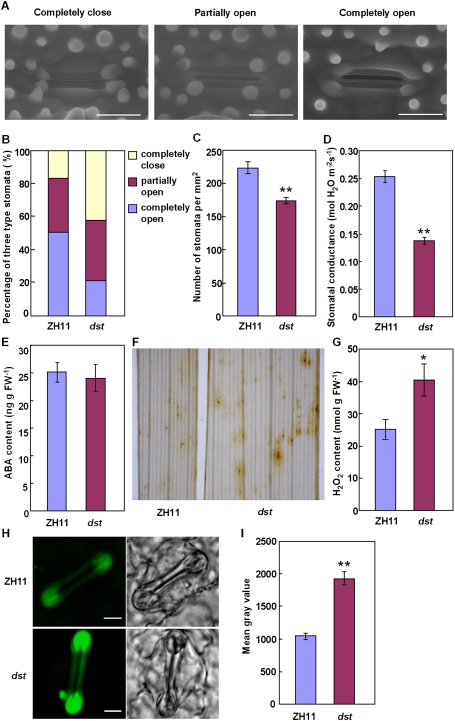
Because stomata are involved in responses to abiotic stresses (Hetherington and Woodward 2003), we examined the stomatal status of dst mutant and wild-type plants. The results showed that 42.4% of stomata were completely closed in the dst mutant plants, but only 16.7% were completely closed in the wild-type plants; also, only 21.2% of stomata were completely open in the dst mutants, but 50.8% were completely open in wild-type plants (Fig. 2A,B). The percentage of partially open stomata was similar in dst mutant and wild-type plants (32.5%–36.4%). These results showed that stomatal movement is greatly affected in dst mutants. A significant decrease in stomatal density was also found in dst mutant plants (Fig. 2C), whereas there was no significant difference in guard cell size between dst mutant and wild-type plants (data not shown). Meanwhile, the stomatal conductance was lower in dst mutant plants than that of wild-type plants (Fig. 2D). We further investigated the stomatal status of the dst mutant under stress conditions, and found that the stomatal conductance of the dst mutant was significantly lower than that of the wild-type plants not only in the normal growth condition, but also in the salt stress condition (Supplemental Fig. S3). These results demonstrate that the enhanced drought tolerance displayed by the dst mutant is primarily due to increased stomatal closure and decreased stomatal density, which prevent water loss, and that the enhanced salt tolerance of the dst mutant might be the result of a low transpiration rate that reduces Na+ transport from roots to shoots.
Figure 2.
H2O2 accumulation induces stomata closing in the dst mutant. (A) Environmental scanning electron microscopy images of three levels of stomatal opening. Bar, 10 μm. (B) The percentage of three levels of stomatal opening in ZH11 and dst mutant (n = 132 stomata for ZH11; n = 126 stomata for dst). (C) Stomatal density of the middle leaves of ZH11 and dst mutant (n = 10). Three random scopes were used in each repeat (356.20 μm × 356.20 μm in each scope). (D) Stomatal conductance of ZH11 and dst mutant (n = 12). (E) ABA content of ZH11 and dst mutant (n = 5 repeats, 16 plants in each repeat). (F) DAB staining in the seedling leaves of ZH11 (left) and dst mutant (right). (G) Quantitative measurement of H2O2 in the seedling leaves of ZH11 and dst mutant (n = 4 repeats, 16 plants in each repeat). (H) Examination of H2O2 production in the guard cell of ZH11 and dst mutant plant with H2DCFDA. Bar, 5 μm. (I) Quantitative analysis of H2O2 production in the guard cell of ZH11 and dst mutant plant (n = 104 stomata for ZH11; n = 99 stomata for dst). Data are presented as mean ± SEM in (C–E, G, and I). (*) P ≤ 0.05; (**) P ≤ 0.01, Student's t-test.
The phytohormone ABA induces stomatal closure (Schroeder et al. 2001b; Zhu 2002). However, measurement of endogenous ABA content revealed no significant difference between dst mutant and wild-type plants (Fig. 2E). Therefore, the increased stomatal closure of the dst mutant is not the result of ABA accumulation. Since H2O2 induces leaf stomatal closure (McAinsh et al. 1996; Hamilton et al. 2000; Apel and Hirt 2004; Bright et al. 2006), we examined H2O2 content in rice leaves and found a higher accumulation in dst mutant plants (Fig. 2F,G). Furthermore, by using a fluorescence dye, 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA), we found that dst mutant plants accumulated more H2O2 in the guard cells than those of wild-type plants (Fig. 2H,I). These results suggested that accumulation of H2O2 in guard cells probably underlies the increased stomatal closure in dst mutant plants.
DST encodes a novel zinc finger protein of unknown function
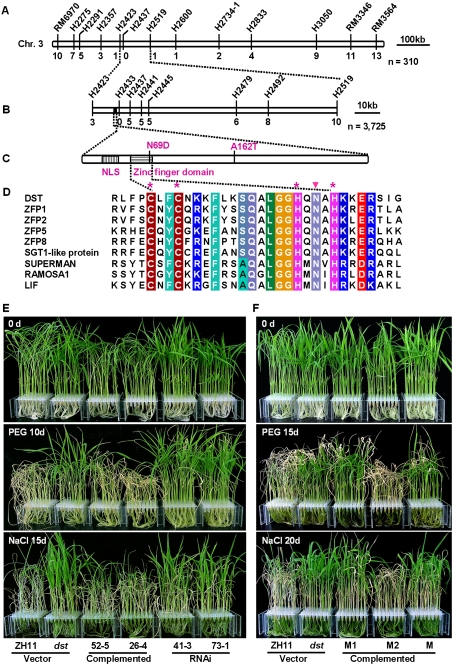
A map-based cloning approach was used to isolate the DST gene. Since the wide leaf is a typical and visible phenotype of the dst mutant (Fig. 1B), it was adopted as the standard dst phenotype. All F1 progeny showed a narrow leaf phenotype similar to that of wild-type plants (data not shown). In the F2 population, narrow and wide leaf plants segregated in a 3:1 ratio (241 narrow vs. 63 wide; χ2 = 2.96 < χ20.05 = 3.84; P > 0.05), demonstrating that the wide leaf phenotype of the dst mutant is controlled by a single recessive nuclear gene. The DST locus was initially mapped to the long arm of chromosome 3 between the markers H2423 and H2519 (Fig. 3A; Supplemental Table S1), and was subsequently fine-mapped to a 14-kb region between H2423 and H2437 (Fig. 3B). Only one candidate gene, which encodes a 301-residue polypeptide with unknown function, localized in this region (Fig. 3C). Protein sequence analysis showed that DST contains a nuclear localization signal (NLS) peptide and a single C2H2-type zinc finger motif at the N terminus (Fig. 3C,D). A BLAST search found only one copy of DST in the rice genome. A comparison of the nucleotide sequences of the gene in dst mutant and wild-type plants revealed that two nucleotide substitutions lead to two amino acid substitutions: asparagine (N69) to aspartic acid (D) and alanine (A162) to threonine (T) (Fig. 3C), of which the N69 is a conserved residue of the zinc finger motif.
Figure 3.
Map-based cloning of DST and complementation test. (A) The DST locus was mapped to chromosome 3 between markers H2423 and H2519 using 310 homozygous F2 plants. (B) Fine mapping of the DST locus. The DST locus was narrowed to a 14-kb region between markers H2423 and H2437. Numbers below the horizontal line in A and B are the number of recombinants. (C) The protein structure of DST. Two nucleotide substitutions in dst lead to an asparagine-to-aspartic acid change at amino acid 69 and an alanine-to-threonine change at amino acid 162. (D) Amino acid sequence alignments of the zinc finger domain. Identical and similar residues are displayed in colored boxes. The asterisk marks the cysteine and histidine in the C2H2-type zinc finger domain. The solid triangle denotes the point mutation in the zinc finger domain. GenBank accession numbers are ZFP1 (NP_178188), ZFP2 (NP_200560), ZFP5 (NP_172518), ZFP8 (NP_181725), SGT1-like protein (BAD73077), SUPERMAN (AAC49116), RAMOSA1 (AAY17040), and LIF (BAB58897). (E) Drought and salt treatment of T2 transgenic lines including vector control, complementation, and RNAi. Twenty-day-old plants (top) were treated with 18% PEG4000 for 10 d (middle) or 100 mM NaCl for 15 d (bottom). (F) Drought and salt treatment of T2 transgenic lines with point mutation constructs. Twenty-day-old plants (top) were treated with 18% PEG4000 for 15 d (middle) or 100 mM NaCl for 20 d (bottom). M1, M2, and M indicate transgenic plants with N69D, A162T, and both point mutation constructs, respectively.
A complementation test was carried out to confirm the mapping result. Transfer of a 4.632-kb wild-type DNA fragment containing the DST promoter region and the entire ORF into dst mutants conferred phenotypes similar to those of wild-type plants, including narrow leaf shape, sensitivity to drought and salt stresses, stomatal opening level, and H2O2 content (Fig. 3E; Supplemental Fig. S1A,B,D,E), whereas a knockout of DST using RNAi technology produced the dst phenotypes (Fig. 3E; Supplemental Fig. S1A–E). These results demonstrate that DST is indeed responsible for the phenotypic changes found in dst mutants, and that DST is a negative regulator of drought and salt tolerance.
We constructed N69D (M1) and A162T (M2) mutants to evaluate the role of the two substitutions (Fig. 3C). These two constructs and the dst (M, containing the two mutations) were each transformed into the dst mutant. Only those transformants expressing the M2 construct complemented dst phenotypes (Fig. 3F). Therefore, the mutation of N69, but not A162 is primarily responsible for the phenotypes of the dst mutant.
Subcellular localization and expression pattern of DST
Protein structure prediction showed that DST has a putative NLS (Fig. 3C). We transiently expressed the DST-GFP fusion protein in onion epidermal cells and confirmed that the fusion protein specifically localizes to the nucleus (Fig. 4A). The fluorescence of DST-GFP was also detected exclusively in the nucleus in transgenic rice plants that were transformed with a DST-GFP fusion construct under the control of the cauliflower mosaic virus (CaMV) 35S promoter (Supplemental Fig. S4). The tissue expression pattern of DST was determined by transforming rice plants with a β-glucuronidase (GUS) construct driven by the DST promoter. Strong GUS signals were detected in vascular tissues of the roots, internodes, and leaf sheath (Fig. 4B–H). In the leaves, the GUS signal was also detected in the stomata apparatus (Fig. 4I,J). To confirm the expression of DST in the stomata apparatus, we generated transgenic rice plants that were transformed with a DST-GFP fusion construct under the control of the DST native promoter. Confocal microscopic analysis showed that the fluorescence of DST-GFP was detected in the guard cells and the subsidiary cells (Fig. 4K; Supplemental Fig. S5), which was consistent with the role of DST in stomata regulation.
Figure 4.
Subcellular localization and expression pattern of DST. (A) Subcellular localization of DST. GFP and the DST-GFP fusion gene under the control of the CaMV 35S promoter were expressed transiently in onion epidermal cells. The DST-GFP fusion protein was specifically expressed in the nucleus. (B–J) DST promoter-GUS expression patterns in transgenic rice plants. GUS expression was observed in the vascular system of roots (B–D), nodes (E,F), leaf sheathes (G,H), and leaves (I). (J) GUS expression was also observed in the stomata. The arrow indicates the stoma. Bars: C,D,J, 100 μm. (K) The stoma of the transgenic rice plants that were transformed with a DST-GFP fusion construct under the control of DST native promoter. The GFP fluorescence was detected in the guard cells and the subsidiary cells. Bar, 5 μm. (L) Relative expression levels of DST by quantitative real-time PCR in the shoots of plants treated with 18% PEG4000 and 100 mM NaCl. Data are presented as mean ± SEM (n = 3).
We also tested whether the expression of DST is regulated by drought and salt stresses. Real-time PCR analysis showed that DST is down-regulated rapidly after 0.5 h of drought or salt treatment, but that the expression resumed continuously after 24 h of treatment (Fig. 4L). These data are consistent with its role in regulating stomatal movement under drought and salt stresses. To exclude the possibility that DST is regulated by a circadian rhythm, we analyzed the rhythmic profiles of DST and found that DST is not regulated significantly by a circadian rhythm (Supplemental Fig. S6).
DST is a transcription factor with transactivational activity
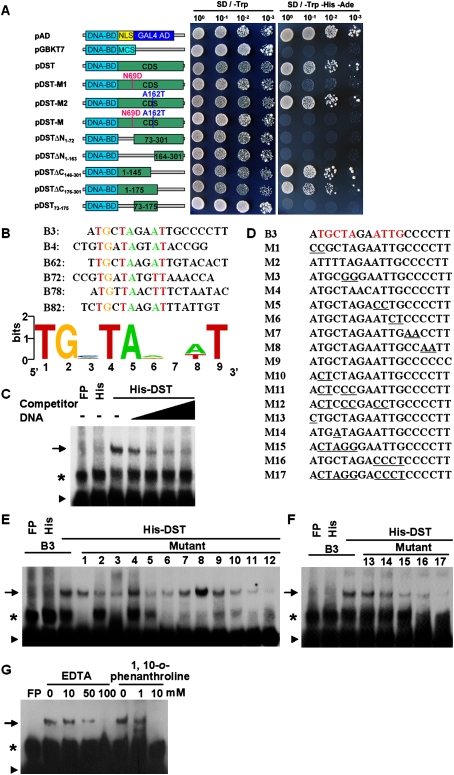
We performed a transcriptional activation assay and found that expression of the DST-BD (GAL4-binding domain) fusion protein in yeast resulted in high reporter gene expression (Fig. 5A), which revealed that DST has strong activity as a transcription activator. Assessment of the two amino acid substitutions in the dst mutant showed that the N69D substitution decreased the level of transcriptional activation, while the A162T substitution did not alter the activation level. These results indicated that the N69 of DST is required for transcriptional activation. Interestingly, when the two amino acid substitutions were combined, activation was lower than that of the N69D mutation alone (Fig. 5A), possibly because the two amino acid substitutions modify DST protein configuration. Further quantitative assays of the β-galactosidase activity confirmed these results (Supplemental Fig. S7). Deletion analysis shows that the first 72 amino acids, containing the zinc finger motif, are required for transcriptional activation, and that the activation domain is localized in the first 145 N-terminal residues (Fig. 5A).
Figure 5.
DST has transactivation activity and DNA-binding functions. (A) Schematic diagrams of various constructs used for transactivation activity assay. (DNA-BD) GAL4 DNA-binding domain; (GAL4 AD) GAL4 activation domain; (NLS) NLS sequence; (MCS) multiple cloning site; (CDS) coding sequence of DST. (B) Alignment of representative sequences identified in bacterial one-hybrid experiment. Conserved nucleotides are shown in color. Sequence logo was generated using Weblogo (see the Materials and Methods for detail). (C) EMSA showing that the His-DST fusion protein, not His alone, can bind the B3 sequence. Competitive binding analysis was performed with increasing amounts of unlabeled probe. The amount of competitive unlabeled probe in lanes 4–7 was 10, 50, 100, and 200 times of that of the labeled probe, respectively. (D) B3 sequence and its various mutants. (E,F) EMSA with the same amount of wild-type B3 probe or mutant probes. (G) Two metal chelators, EDTA or 1,10-o-phenanthroline, effectively abolish the binding of DST to B3 probe, suggesting that the zinc finger motif of DST might be the DNA-binding domain. The arrow, asterisk, and triangle in C and E–G indicate the up-shifted bands, nonspecific binding, and free probe, respectively. (FP) Free protein.
DST binds to the TGCTANNATTG element by the zinc finger domain
We used a bacterial one-hybrid system (Meng et al. 2005) to identify the DNA-binding sequence of DST. Among the sequences identified, six sequences had a conserved TGNTANN(A/T)T sequence (Fig. 5B). Electrophoretic mobility shift assay (EMSA) showed that the His-DST fusion protein, but not His alone, could bind to a B3 sequence that contains this conserved sequence, whereas an excess unlabeled B3 sequence effectively reduced or abolished the His-DST binding to the labeled B3 sequence (Fig. 5C). Meanwhile, the His-DST protein could bind to the B3 sequence in a dosage-dependent manner (Supplemental Fig. S8A). These results demonstrated that DST binds specifically to this sequence. Individual mutations of TG (M10), TA (M3), and AT (M5), and simultaneous mutations (M11 and M12) in the TGNTANNAT sequence markedly reduced or abolished the binding of His-DST (Fig. 5D,E). The mutants (M4, M7, M8, M9 and M13) with the conserved nucleotides did not significantly reduce the binding of His-DST (Fig. 5D–F). However, other mutants (M6 and M14) markedly reduced the binding of His-DST, showing that TG and C are also required for DST binding (Fig. 5D–F). Taken together, these results showed that DST binds to the TGCTANNATTG sequence, which is a novel cis-element and named DBS (DST-binding sequence). This was further demonstrated by those mutations (M15, M16, and M17) in this core sequence that completely abolished the binding of His-DST (Fig. 5D–F). Interestingly, the M8 mutant, which produced new ATTG core nucleotides, increased the binding of His-DST (Fig. 5E; Supplemental Table S2). We also tested whether the mutant DST protein in dst mutants could bind to DBS, and found that the mutant form of DST protein could bind to DBS as the wild-type protein did (Supplemental Fig. S8B–D).
Based on previous studies (Wolfe et al. 2000; Sakamoto et al. 2004; Lin et al. 2007), we assumed that the zinc finger motif of DST might be the DNA-binding domain. The preincubation of the His-DST fusion protein with two metal chelators—EDTA or 1,10-o-phenanthroline—effectively reduced or abolished the DNA-binding ability of His-DST, suggesting that DST binds to the DBS element by the N-terminal zinc finger motif (Fig. 5G). This suggestion was further confirmed by EMSA using a truncate DST protein with deletion of the zinc finger domain (Supplemental Fig. S8B–D).
DST targets to genes related to H2O2 homeostasis
Expression profile analysis using the Affymetrix Rice Genome Genechip was used to identify DST target genes. We found that among ∼50,000 rice genes, the transcription levels of 60 genes were altered significantly (more than threefold change) in dst mutant plants (Supplemental Table S3). A high percentage (60%) of these 60 genes had lower expression levels in dst mutants (Supplemental Table S3), and mutation of the DST protein in dst mutants lost its transactivation activity (Fig. 5A), indicating that DST might promote expression of these genes. It is noteworthy that at least five of the down-regulated genes encode proteins related to ROS homeostasis: two peroxidases (24 precursor, 12 precursor; EC1.11.1.7), glutathione S-transferase (OsGSTU2; EC2.5.1.18), and two cytochrome P450s (71D10, 94A2; EC1.14.-.-) (Jakoby 1978; Marrs 1996; Noctor and Foyer 1998; Asada 1999; Rodriguez Milla et al. 2003; Mittler et al. 2004; Passardi et al. 2004, 2005; Campos et al. 2005; Moller et al. 2007). The expression of these genes was greatly down-regulated in dst mutant plants (Supplemental Table S3), four of which were confirmed by RT–PCR (Supplemental Fig. S9).
We subsequently searched the promoter regions of four genes and found two or three DBSs in each promoter (Supplemental Fig. S10). The promoter sequences of two genes were used for EMSA, showing that DST can bind to the promoter sequences (Fig. 6A). Furthermore, a chromatin immunoprecipitation (ChIP) assay using wild-type plants showed that the anti-DST antibody specifically precipitated promoter sequences of all four genes from the cells of wild-type plants (Fig. 6B), indicating that DST binds directly to the promoters of these genes in vivo. Noticeably, in these five genes, peroxidase 24 precursor may directly scavenge H2O2. Peroxidase activity assay confirmed that peroxidase 24 precursor catalyzes the reduction of H2O2 (Fig. 6C,D). Real-time PCR analysis showed that the expression level of peroxidase 24 precursor was higher in the guard cell protoplast (GCP)-enriched sample compared with the mesophyll cell protoplast (MCP)-enriched sample (Fig. 6E). This result indicated that peroxidase 24 precursor was expressed in guard cells, which was consistent with the expression of DST in the guard cells. Furthermore, peroxidase 24 precursor was not only down-regulated in the leaf, but also in the GCP of the dst mutant (Fig. 6F). Meanwhile, under salt and drought treatment, peroxidase 24 precursor was down-regulated and then resumed (Fig. 6G), which was similar to the expression pattern of DST. These results indicated that DST might directly regulate the expression of peroxidase 24 precursor under salt and drought stresses. Collectively, we propose that accumulation of H2O2 in the dst mutant (Fig. 2F–I) may be the result of down-regulation of the genes related to H2O2 homeostasis.
Figure 6.
DST targets to genes related to H2O2 homeostasis. (A) EMSA shows that His-DST fusion protein, but not His alone, can bind to the promoter sequences of Peroxidase 24 precursor and Cytochrome P450 71D10. The 50-bp or 52-bp promoter sequences containing DBS were synthesized, biotin-labeled, and used as probes in EMSA. The arrow and triangle indicate the up-shifted bands and free probe, respectively. (B) ChIP assay using the wild-type seedlings shows that DST binds to promoters of four genes related to H2O2 homeostasis in vivo. Immunoprecipitation was performed with anti-DST antibody (Anti-DST) or without antibody (no Ab). (C) Time-course analysis of HRP, peroxidase 24 precursor fusion protein (6xHis-PRX), and 6xHis protein in reduction of H2O2. One milliunit per milliliter HRP, 1.34 μg of 6xHis-PRX, and 2.14 μg of 6xHis protein were used in the reaction with excess 2 mM H2O2. HRP and 6xHis were used as positive control and negative control, respectively. Data are presented as mean ± SEM (n = 3 for HRP and n = 10 for 6xHis-PRX and 6xHis). (D) Relative enzyme activity of 6xHis and 6xHis-PRX when incubated with 2 mM H2O2 for 60 min. Data are presented as mean ± SEM (n = 10). (**) P ≤ 0.01, Student's t-test. (E) Comparison of the expression level of peroxidase 24 precursor in the MCP and GCP of ZH11. Data are presented as mean ± SEM (n = 3). (**) P ≤ 0.01, Student's t-test. (F) Relative expression levels of peroxidase 24 precursor in the MCP, the GCP, and the leaf of ZH11 or dst mutant. Data are presented as mean ± SEM (n = 3). (*) P ≤ 0.05; (**) P ≤ 0.01. Student's t-test. (G) Relative expression levels of peroxidase 24 precursor under the treatment with 18% PEG4000 or 100 mM NaCl. Data are presented as mean ± SEM (n = 3).
Discussion
Generally, C2H2-type zinc finger proteins contain more than one zinc finger motif for DNA binding in the N terminus and separable transcriptional activation (or repression) domains in the C terminus (Wolfe et al. 2000). The zinc finger motif is thought to recognize and bind its target DNA sequence, but has never been shown to be required for transcriptional activity (Wolfe et al. 2000; Sakamoto et al. 2004; Lin et al. 2007). However, here we surprisingly found that the single zinc finger motif of DST is required not only for its DNA-binding activity but also its transcriptional activity, indicating that DST is a novel transcription factor. It brings us brand-new knowledge and a new understanding about the zinc finger transcription factor.
H2O2 plays an important role in mediating ABA-induced stomatal closure (McAinsh et al. 1996; Pei et al. 2000; Kohler et al. 2003; Bright et al. 2006). In Arabidopsis guard cells, H2O2 was scavenged by glutathione peroxidase (Miao et al. 2006). Mutation of Arabidopsis thaliana glutathione peroxidase 3 (ATGPX3) leads to the accumulation of H2O2 in guard cells. However, direct upstream regulators of genes related to H2O2 homeostasis, such as ATGPX3, have not been identified. Here, we showed that rice peroxidase 24 precursor has peroxidase activity to scavenge H2O2 (Fig. 6C,D). Meanwhile, EMSA and ChIP results showed that DST targets to the promoter sequence of peroxidase 24 precursor (Fig. 6A,B). Therefore, DST is, to our knowledge, the first known direct upstream regulator of the genes related to H2O2 homeostasis in guard cells. Mutation of DST results in the down-regulation of peroxidase 24 precursor (a scavenger of H2O2), might lead to the accumulation of H2O2 in guard cells and trigger stomatal closure, and enhances drought and salt tolerance. Recently, Arabidopsis mutants lacking either or both a cytosolic and chloroplastic ascorbate peroxidase, which were responsible for H2O2 removal, were found to be more tolerant to salinity stress (Miller et al. 2007). Interestingly, two ROS scavengers (glutathione S-transferase GSTU6 and peroxidase 16 precursor) were found to be up-regulated in the dst mutant (Supplemental Table S3), and they might contribute to the removal of excess ROS because high levels of ROS have been shown to be toxic (Apel and Hirt 2004).
In the last decade, many investigators have made progress in illuminating the regulatory pathways involved in abiotic stress tolerance (Zhu 2002; Shinozaki et al. 2003; Yamaguchi-Shinozaki and Shinozaki 2006). For example, dehydration-responsive element (DRE)/DRE-binding protein (DREB) pathways (Stockinger et al. 1997; Liu et al. 1998); the ABA-responsive element (ABRE)/ABRE-binding protein ABI5 pathway (Choi et al. 2000; Finkelstein and Lynch 2000; Lopez-Molina and Chua 2000; Uno et al. 2000); and the MYB recognition sequence (MYBRS)/MYB transcription factor pathway (Abe et al. 1997, 2003). The cis-acting and trans-acting regulatory elements in these pathways have been precisely analyzed at the molecular level (Yamaguchi-Shinozaki and Shinozaki 2005). DBS is a novel cis-acting regulatory element that was found in the promoter sequences of the genes related to ROS homeostasis. DST binds to DBS and regulates the expression of ROS homeostasis-related genes. In the initial stage of dehydration stress, the expression of DST was repressed, resulting in the down-regulation of peroxidase 24 precursor—whose promoter contains DBS—giving rise to H2O2 accumulation and promoting stomatal closure.
ABA is an important regulatory component of plant responses to abiotic stresses such as drought and salinity. Many genes that are induced by drought and salt treatment also are induced, or not, in response to exogenous ABA treatment, suggesting that there are ABA-dependent or ABA-independent stress response pathways (Zhu 2002; Yamaguchi-Shinozaki and Shinozaki 2006). Treatment of exogenous ABA did not significantly affect the expression of DST, and there were no significant differences between stomatal openings among wild type, the dst mutant, the complementation line, and the RNAi line under ABA treatment (Supplemental Fig. S11A,B), indicating that the pathway that DST was involved in was ABA-independent.
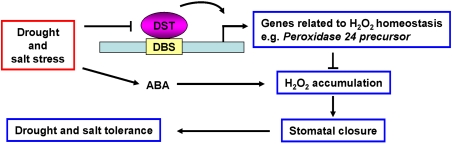
Guard cells or stomatal apparatus cells are important cell types for dehydration tolerance. Although the transcriptional network in these cells is quite important in understanding plant response mechanisms to dehydration and for improvement of dehydration tolerance of crops, progress in this area has been relatively limited. The DST regulatory pathway revealed in this study will lead to better understanding of the transcription network in the stomatal apparatus. In addition, DST also has pleiotropic effects—at least on stomatal density and leaf width—implying that DST also is involved in regulating the development of tissues or organs. How DST regulates stomatal density and leaf width is an intriguing question to be answered in future studies. Based on our data, we summarized a model to explain the potential role of DST in the regulation of stomatal status and abiotic stress tolerance (Fig. 7).
Figure 7.
A proposed model for the role of DST in regulation of abiotic stress tolerance. DST binds directly to the DBS element in the promoter of genes related to H2O2 homeostasis and activates their transcription, thereby inhibiting H2O2 accumulation. Inhibition of H2O2 accumulation influences stomatal closure and, ultimately, abiotic stress tolerance. However, the pathway for regulation of stomatal density remains to be elucidated in future studies. Under drought and salt stress, the expression of DST was repressed, resulting in the down-regulation of genes related to H2O2 homeostasis, such as peroxidase 24 precursor, whose promoter contains DBS, giving rise to H2O2 accumulation and promoting stomatal closure, and finally enhancing drought and salt tolerance. This pathway is ABA-independent, and is different from the ABA-induced H2O2 accumulation pathway in controlling stomatal closure.
Engineering of the stomatal response to reduce water loss would be a powerful tool for the enhancement of drought tolerance in crops (Schroeder et al. 2001b). Down-regulation of DST function, which constitutively reduces the stomatal opening and stomatal density, would allow engineering of crop plants (such as rice) to reduce water loss, enhance drought tolerance, and reduce water requirements in agricultural regions with marginal fresh water availability. Moreover, the down-regulation of DST does not affect rice grain yield (Supplemental Fig. S2F); so although the DST mutation results in a smaller stomatal opening and a lower stomatal density (Fig. 2A–C), it is sufficient for CO2 influx and growth. This characteristic shows the strong potential value of DST in abiotic stress tolerance engineering. Therefore, our findings not only provide new insights into the molecular mechanisms underlying abiotic stress tolerance in plants, but also facilitate molecular breeding efforts to improve drought and salt tolerance in staple crops and the conservation of water resources in agricultural regions where fresh water is scarce.
Materials and methods
Plant material and growth conditions
The wild-type plant Zhonghua 11 (ZH11) is a drought- and salt-sensitive japonica variety. We screened >270,000 ethyl methanesulfonate (EMS)-mutagenized M2 rice seedlings (∼9000 lines) treated with 140 mM NaCl. In the first round of screening, >10 lines were isolated as putative salt tolerance mutant plants. Among these mutant lines, one mutant maintained the highest ability to survive in the treatment of NaCl. When treated with 20% PEG4000 (simulation of drought stress), this mutant exhibited less severe wilting than wild-type (ZH11) plants. This mutant was named as dst. The dst mutant was backcrossed to the wild-type and then a dst mutant line after propagation for at least five generations was used for further analysis. Seeds were placed for 1 wk or more at 42°C to break any possible dormancy, soaked in the water at room temperature for 3 d, and then germinated for 1 d at 37°C. The most uniformly germinated seeds were sown in a 96-well plate from which the bottom was removed. The plate was floated on water for 1 d at 37°C in the dark to encourage root growth, before removal to a growth chamber with a 13-h light (24°C)/11-h dark (20°C) photoperiod. Five days later, the seedlings were cultured with Yoshida's culture solution with or without 1% hygromycin (for the selection of transgenic plants). For salt treatment, 20-d-old seedlings were transferred to culture solution containing 100 mM or 140 mM NaCl. For PEG treatment, 20-d-old seedlings were transferred to culture solution containing 18% or 20% (w/v) PEG4000. For soil drought experiments in PVC pipes, 25-d-old seedlings cultured in the growth chamber were transplanted to PVC pipes and cultured in the greenhouse at 24°C–30°C and 50%–60% relative humidity. Thirty days after transplantation, water was removed from the bottom of the PVC pipes for dehydration treatment.
Diaminobenzidine (DAB) staining and quantitative measurement of H2O2
H2O2 was detected by DAB staining as described previously (ThordalChristensen et al. 1997). The fully expanded leaves of 20-d-old wild-type and dst plants were detached and vacuum-infiltrated with the DAB solution. The sampled leaves were placed in a growth chamber for 8 h at 28°C and cleared in boiling ethanol (96%) for 10 min before photographing. A measurement of H2O2 production was performed by extracting H2O2 from leaves according to a previously described method (Rao et al. 2000) and quantifying the molecule with an Amplex Red Hydrogen/Peroxidase Assay Kit (Molecular Probes) according to the manufacturer's instructions.
Detection of H2O2 production in guard cells
H2O2 production in guard cells was detected using H2DCFDA (Molecular Probes) as described previously (Zhang et al. 2001) with modifications. Leaves of 45- to 50-d rice seedlings were immerged in 0.01% Tween-20 and vacuum-infiltrated for 5 min. After rinsing twice with distilled water, leaves were incubated in 2% (w/v) cellulase RS (Yakult Honsha) for 5 h at 40°C without shaking to facilitate peeling off the epidermal strips. The epidermal strips were peeled off from leaves using tissue forceps. After washing with loading buffer (10 mM Tris-HCl, 50 mM KCl at pH 7.2), the epidermal strips were incubated in staining buffer (loading buffer containing 50 μM H2DCFDA) for 10 min at room temperature in the dark. The epidermal strips were washed with distilled water to remove the excess dye. The fluorescence was examined using a confocal laser-scanning microscope (Carl Zeiss, LAM510) with the following settings: excitation, 488 nm; emission, 525 nm; and power, 5%. All confocal images were taken under identical conditions: 12 bits; scanning speed, 7; frame, 1024 × 1024; pinhole, 300; and detector gain, 750. For quantifying the gray value (0–4095) in the guard cell, the guard cell region was selected, and the mean gray value was recorded. For dst and wild-type plants, nine independent experiments were performed, and >10 stoma were selected randomly for analysis in each experiment.
Imaging of rice stomata and measurement of stomatal conductance
Leaves of 25-d-old plants were detached and immediately fixed by liquid nitrogen. The stomatal pictures were obtained using an environmental scanning electron microscopy (XT30 ESEM-TMP, Philips). For the stomatal conductance measurement, seedlings were cultured in a growth chamber for 25 d, and then transplanted to soil in a greenhouse under natural growth conditions. At tillering stage (∼35 d after transplant), the second fully expanded leaf, counting from the top of plant, was used for stomatal conductance measurement with a portable gas analysis system (LI-COR 6400, LI-COR, Inc.). The measurement was performed under the following conditions: a constant water concentration of 18.85 ± 0.02 mmol mol−1, a constant temperature of 28.80°C ± 0.02°C, and a constant CO2 concentration of 350.20 ± 0.02 μL L−1.
Bacterial one-hybrid analysis and EMSA
Bacterial one-hybrid analysis was performed as described previously (Meng et al. 2005) with some modifications. The full-length coding sequence of DST was PCR-amplified with primers containing BstXI and BamHI sites, and then cloned into the AvrII–BamHI sites of pH1B1 (Supplemental Table S2). High-efficiency electrocompetent DH10B was substituted for XL1-blue. The 3-amino-1,2,4-triazole (3-AT) selection plate contained 3 mM or 4 mM 3-AT (Sigma). Twenty clones grown on 3 mM or 4 mM 3-AT selection plates were selected randomly and sequenced. Fifteen unique sequences were analyzed using the Gibbs Motif Sampler (http://bayesweb.wadsworth.org/cgi-bin/gibbs.12.pl?data_type=DNA). Among them, six sequences containing a conserved sequence were identified. The sequence logo was generated using Weblogo 3 (http://weblogo.berkeley.edu/logo.cgi). The plasmids pB1H1 (Addgene plasmid 12611) and pH3U3-mcs (Addgene plasmid 12609) and bacteria strain US0hisB-pyrF- (Addgene plasmid 12614) were obtained from Professor Scot Wolfe through Addgene (http://www.addgene.org).
For EMSA, 6xHis and 6xHis-DST recombinant fusion proteins were expressed in the Escherichia coli BL21 (DE3) strain and purified using Ni-NTA resin (Qiagen). The complementary pairs of biotin-labeled oligonucleotides were obtained by PCR amplification using 5′-biotin-labeled primers. The PCR products were purified by electroelution with a GeBAflex tube (Gene Bio-Application Ltd.) according to the manufacturer's instructions. EMSA was performed with the Light Shift Chemiluminescent EMSA Kit (Pierce) according to the instructions. The binding reactions—containing 10 mM Tris (pH 7.5), 50 mM KCl, 1 mM DTT, 2.5% glycerol, 0.05% NP-40, 5 mM MgCl2, 0.5 mM EDTA, 5 ng/μL poly (dI•dC), 1 μg 6xHis-DST recombinant fusion protein, and 100 fmol biotin-labeled DNA—were kept for 30 min at room temperature before loading buffer was added. For metal chelation experiments, His-DST fusion protein was preincubated with 1,10-o-phenanthroline or EDTA for 30 min before mixing with the labeled probes. Gel electrophoresis was performed on a 10% native polyacrylamide gel (79:1 acryl/bis). After blotting on a positively charged nylon membrane (Amersham), the DNA was linked using a UV light cross-linker instrument equipped with 254-nm bulbs for 0.8 min exposure. The primer sequences are listed in Supplemental Table S2.
Preparation of His-DST fusion protein and DST antibody
To generate His-DST expression plasmid, the pDST plasmid was digested with EcoRI and SalI to release the ORF of DST, which was then inserted into the EcoRI–SalI site of pET-32a (Novagen). To construct the truncate DST expression plasmid (His-DST-ΔZF), a 687-base-pair (bp) coding sequence of DST was PCR-amplified with primers containing BamHI and EcoRI sites, and then cloned into the BamHI–EcoRI sites of pET-32a (Supplemental Table S2). The His-DST-M protein construct was generated by digesting the plasmid pDST-M with EcoRI and SalI to release the mutation form of the ORF of DST, and then inserted into the EcoRI–SalI site of pET-32a. The His-DST expression plasmid was transformed into the bacterial strain BL21 (DE3). The transformed cells were cultured at 37°C until the OD600 of the cell culture was 0.5, and then induced with 1 mM IPTG (isopropyl-1-thio-β-D-galactopyranoside) for 36 h at 12°C. For extraction of native fusion protein, the cultured bacteria cells were lysed by using a high-pressure cell crusher and the fusion proteins were purified with Ni-NTA resin (Qiagen).
For preparation of DST antibody, >5 mg of purified His-DST protein were used to immune rabbit at 3-wk intervals. DST-specific antibodies were purified from the crude rabbit antiserum using MBP-DST fusion protein. The MBP-DST plasmid was constructed by digesting the pDST plasmid with HindIII and SalI to release the ORF of DST, which was then inserted into the HindIII–SalI site of pMAL-c2X (New England Biolabs). The MBP-DST fusion protein was prepared as described above and purified with anti-MBP magnetic beads (New England Biolabs) according to the manufacturer's instructions.
Preparation of 6xHis-PRX fusion protein and proxidase activity assay
To generate the 6xHis-PRX, the ORF of peroxidase 24 precursor was PCR-amplified with the primers listed in Supplemental Table S2. The PCR products were digested with EcoRI and HindIII, and inserted into the EcoRI–HindIII site of pET-32a (Novagen). The 6xHis-PRX expression plasmid was transformed into the bacterial strain BL21 (DE3) pLysS. The transformed cells were cultured at 37°C until the OD600 of the cell culture was 0.5, and then induced with 1 mM IPTG for 36 h at 12°C. For extraction of native fusion protein, the cultured bacteria cells were lysed by using a high-pressure cell crusher and the fusion proteins were purified with Ni-NTA resin (Qiagen) according to the manufacturer's instructions.
The peroxidase activity assay was performed using an Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Molecular Probes) according to the manufacturer's instructions. About 1 μg of 6xHis-PRX fusion protein was incubated with Amplex Red reaction mixture at 30°C. Absorbance at 560 nm was measured in a SpectraMax 190 (Molecular Devices) every 1 min for 60 min. Values were subtracted to the background absorbance. About 2 μg of 6xHis protein and 100 μU/mL horseradish peroxidase (HRP) were used as negative and positive controls. For generation of a standard curve, 0, 25, 37.5, 50, 62.5, 75, 87.5, and 100 μU/mL HRP were incubated with Amplex Red reaction mixture for 60 min at 30°C. After subtracting to the background absorbance, the values were used to generate a linear regression equation. The relative activity of 6xHis-PRX and 6xHis protein was calculated with equal mole protein according to the linear regression equation.
Other experimental methods are described in the Supplemental Material.
Accession numbers
Raw microarray data have been deposited in Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) with GEO Series accession number GSE11175. The GenBank accession number for DST sequences is GQ178286.
Acknowledgments
We thank Professors D.-Q. Xu and Z.-Z. Piao and Dr. Y. Chen for technical assistances; Professor Scot Wolfe for providing the bacterial one-hybrid system; and Arizona Genomics Institute for providing the BAC clone. This work was supported by grants from the National Natural Science Foundation of China (30730058 and 30821004), the Ministry of Science and Technology of China (2006CB100100 and 2006AA10A102), the Ministry of Agriculture of China (2009ZX08009-067B), the Chinese Academy of Sciences (KSCX2-YW-N-011), and the Shanghai Science and Technology Development Fund (09DJ1400503).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1812409.
Supplemental material is available at http://www.genesdev.org.
References
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K, Hirt H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Asada K. The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:601–639. doi: 10.1146/annurev.arplant.50.1.601. [DOI] [PubMed] [Google Scholar]
- Bright J, Desikan R, Hancock JT, Weir IS, Neill SJ. ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 2006;45:113–122. doi: 10.1111/j.1365-313X.2005.02615.x. [DOI] [PubMed] [Google Scholar]
- Campos EG, Jesuino RS, Dantas Ada S, Brigido Mde M, Felipe MS. Oxidative stress response in Paracoccidioides brasiliensis. Genet Mol Res. 2005;4:409–429. [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY. ABFs, a family of ABA-responsive element binding factors. J Biol Chem. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol. 2005;15:1196–1200. doi: 10.1016/j.cub.2005.05.048. [DOI] [PubMed] [Google Scholar]
- de Lorenzo L, Merchan F, Blanchet S, Megias M, Frugier F, Crespi M, Sousa C. Differential expression of the TFIIIA regulatory pathway in response to salt stress between Medicago truncatula genotypes. Plant Physiol. 2007;145:1521–1532. doi: 10.1104/pp.107.106146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan LM, Zhao Z, Assmann SM. Guard cells: A dynamic signaling model. Curr Opin Plant Biol. 2004;7:537–546. doi: 10.1016/j.pbi.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell. 2000;12:599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 2002;130:639–648. doi: 10.1104/pp.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton DW, Hills A, Kohler B, Blatt MR. Ca2+ channels at the plasma membrane of stomatal guard cells are activated by hyperpolarization and abscisic acid. Proc Natl Acad Sci. 2000;97:4967–4972. doi: 10.1073/pnas.080068897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington AM, Woodward FI. The role of stomata in sensing and driving environmental change. Nature. 2003;424:901–908. doi: 10.1038/nature01843. [DOI] [PubMed] [Google Scholar]
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci. 2006;103:12987–12992. doi: 10.1073/pnas.0604882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby WB. The glutathione S-transferases: A group of multifunctional detoxification proteins. Adv Enzymol Relat Areas Mol Biol. 1978;46:383–414. doi: 10.1002/9780470122914.ch6. [DOI] [PubMed] [Google Scholar]
- Kim JC, Lee SH, Cheong YH, Yoo CM, Lee SI, Chun HJ, Yun DJ, Hong JC, Lee SY, Lim CO, et al. A novel cold-inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plants. Plant J. 2001;25:247–259. doi: 10.1046/j.1365-313x.2001.00947.x. [DOI] [PubMed] [Google Scholar]
- Kohler B, Hills A, Blatt MR. Control of guard cell ion channels by hydrogen peroxide and abscisic acid indicates their action through alternate signaling pathways. Plant Physiol. 2003;131:385–388. doi: 10.1104/pp.016014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JD, Schroeder JI. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003;22:2623–2633. doi: 10.1093/emboj/cdg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang YK, Dubos C, Dodd IC, Holroyd GH, Hetherington AM, Campbell MM. AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr Biol. 2005;15:1201–1206. doi: 10.1016/j.cub.2005.06.041. [DOI] [PubMed] [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H. Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science. 2007;318:1302–1305. doi: 10.1126/science.1146281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Chua NH. A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 2000;41:541–547. doi: 10.1093/pcp/41.5.541. [DOI] [PubMed] [Google Scholar]
- Marrs KA. The functions and regulation of glutathione S-transferases in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:127–158. doi: 10.1146/annurev.arplant.47.1.127. [DOI] [PubMed] [Google Scholar]
- McAinsh MR, Clayton H, Mansfield TA, Hetherington AM. Changes in stomatal behavior and guard cell cytosolic free calcium in response to oxidative stress. Plant Physiol. 1996;111:1031–1042. doi: 10.1104/pp.111.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Brodsky MH, Wolfe SA. A bacterial one-hybrid system for determining the DNA-binding specificity of transcription factors. Nat Biotechnol. 2005;23:988–994. doi: 10.1038/nbt1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Lv D, Wang P, Wang XC, Chen J, Miao C, Song CP. An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell. 2006;18:2749–2766. doi: 10.1105/tpc.106.044230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Rizhsky L, Hegie A, Koussevitzky S, Mittler R. Double mutants deficient in cytosolic and thylakoid ascorbate peroxidase reveal a complex mode of interaction between reactive oxygen species, plant development, and response to abiotic stresses. Plant Physiol. 2007;144:1777–1785. doi: 10.1104/pp.107.101436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Moller IM, Jensen PE, Hansson A. Oxidative modifications to cellular components in plants. Annu Rev Plant Biol. 2007;58:459–481. doi: 10.1146/annurev.arplant.58.032806.103946. [DOI] [PubMed] [Google Scholar]
- Noctor G, Foyer CH. Ascorbate and glutathione: Keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- Passardi F, Longet D, Penel C, Dunand C. The class III peroxidase multigenic family in rice and its evolution in land plants. Phytochemistry. 2004;65:1879–1893. doi: 10.1016/j.phytochem.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Passardi F, Cosio C, Penel C, Dunand C. Peroxidases have more functions than a Swiss army knife. Plant Cell Rep. 2005;24:255–265. doi: 10.1007/s00299-005-0972-6. [DOI] [PubMed] [Google Scholar]
- Pei ZM, Murata Y, Benning G, Thomine S, Klusener B, Allen GJ, Grill E, Schroeder JI. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature. 2000;406:731–734. doi: 10.1038/35021067. [DOI] [PubMed] [Google Scholar]
- Rao MV, Lee H, Creelman RA, Mullet JE, Davis KR. Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell. 2000;12:1633–1646. doi: 10.1105/tpc.12.9.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez Milla MA, Maurer A, Rodriguez Huete A, Gustafson JP. Glutathione peroxidase genes in Arabidopsis are ubiquitous and regulated by abiotic stresses through diverse signaling pathways. Plant J. 2003;36:602–615. doi: 10.1046/j.1365-313x.2003.01901.x. [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 2004;136:2734–2746. doi: 10.1104/pp.104.046599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D. Guard cell signal transduction. Annu Rev Plant Physiol Plant Mol Biol. 2001a;52:627–658. doi: 10.1146/annurev.arplant.52.1.627. [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Kwak JM, Allen GJ. Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature. 2001b;410:327–330. doi: 10.1038/35066500. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol. 2003;6:410–417. doi: 10.1016/s1369-5266(03)00092-x. [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ThordalChristensen H, Zhang ZG, Wei YD, Collinge DB. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 1997;11:1187–1194. [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys(2)His(2) zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- Xu S, Wang X, Chen J. Zinc finger protein 1 (ThZF1) from salt cress (Thellungiella halophila) is a Cys-2/His-2-type transcription factor involved in drought and salt stress. Plant Cell Rep. 2007;26:497–506. doi: 10.1007/s00299-006-0248-9. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005;10:88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang L, Dong F, Gao J, Galbraith DW, Song CP. Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol. 2001;126:1438–1448. doi: 10.1104/pp.126.4.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]