Abstract
Purpose of review:
Severe congenital neutropenia has been a well-known haematological condition for over 50 years. Over this long period of time, the variable genetic etiology and associated sequelae of the disease have been ascertained, and successful treatment strategies developed. Over the last 2 years, however, new studies have added greatly to our understanding of the molecular basis of the disease, details of which are presented in this review.
Recent findings:
Recent studies have elucidated a role for the unfolded protein response in mediating the pathogenic effects of ELA2 mutations, the most common mutation in SCN as well as cyclic neutropenia. Genetic lesions in HAX1 have also been identified in the original Kostmann pedigree representing the autosomal recessive form of SCN. An emerging theme is the convergence of these and other genetic lesions underlying SCN in enhancing neutrophil apoptosis. Other studies have revealed the importance of multiple independent mutations in these and other genes in SCN. Finally, the key role for STAT5 in mediating the effects of G-CSFR truncation mutations in the development of MDS/AML following SCN has been elucidated.
Summary:
As the full spectrum of molecular mutations causing neutropenia emerges it is becoming possible to differentiate patients into sub-types with different prognoses, for whom tailored therapies are indicated.
Keywords: Severe congenital neutropenia, ELA2, CSF3R, HAX1, GFI1, WASp, neutrophil elastase, G-CSF receptor
INTRODUCTION
Severe congenital neutropenia (SCN) represents a heterogeneous disease, with autosomal recessive, autosomal dominant, sporadic and X-linked forms. The majority of patients present with life-threatening infections during the first 6 months of life, due to extremely low numbers of circulating neutrophils [1]. Treatment with pharmacological doses of granulocyte colony-stimulating factor (G-CSF) has proven to be effective in restoring the neutrophil count in the majority of SCN patients, with a concomitant reduction in infection-related events [2, 3]. However, some SCN patients remain unresponsive [3]. Moreover, surviving SCN patients remain at high risk of developing myelodysplastic syndrome (MDS) and/or acute myeloid leukemia (AML) [4, 5*, 6**].
REVIEW TEXT
Neutropenias represent a series of potentially life-threatening disorders characterised by a reduction in circulating neutrophils. Since neutrophils play a major role in host defense against bacteria, neutropenia patients suffer from frequent episodes of opportunistic bacterial infections [7**]. Severe congenital neutropenia (SCN) is a heterogeneous group of disorders characterized by a severe decrease in the number of blood neutrophils (<0.5×109/l), and a maturation arrest of bone marrow progenitor cells mainly at the promyelocyte/myeloid stage [5*, 7**]. Although SCN was originally described as an autosomal recessive disorder in Swedish families, this form is now recognized as a separate syndrome, Kostmann's neutropenia, which produces even lower neutrophil counts (<0.2×109/l) [8*]. More commonly, SCN occurs as a sporadic and autosomal dominant disorder, and as a feature of several other inherited disorders. Most SCN patients are successfully treated by G-CSF therapy, although around 10% are unresponsive. However, a major clinical concern for SCN patients remains their increased risk of developing myelodysplastic syndrome (MDS) and/or acute myeloid leukemia (AML) with poor prognosis for survival [5*, 6**].
Previous studies have shown that constitutive mutations in the ELA2 gene (encoding neutrophil elastase) are found in the majority of SCN patients [9, 10*] and cause neutropenia [11]. Other important observations are the finding of acquired mutations in the CSF3R gene (encoding the G-CSF receptor, G-CSFR) in the majority of patients transforming to MDS/AML [12**], and constitutive (alternate) mutations in the same gene leading to poor responsiveness to G-CSF [13**]. This review describes recent studies that have furthered our understanding of each of these mutations, as well as other mutations responsible for other variants of this disease.
Molecular basis of disease
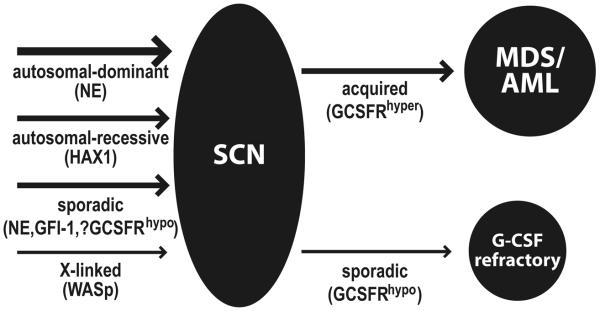
A number of genes have now been identified that appear to contribute to the etiology of SCN or its associated sequelae (Figure 1).
Figure 1. Mutations in severe congenital neutropenia.
Model for the involvement of mutations in severe congenital neutropenia (SCN). Mutations underlying the different forms of SCN are indicated on the left hand side, while mutations associated with predisposition of these patients to MDS/AML, or refractoriness to G-CSF treatment are shown on the right hand side.
Neutrophil elastase (NE)
Neutrophil elastase, encoded by the ELA2 gene, is serine protease produced at the promyelocyte stage of neutrophilic differentiation and stored within the primary granules of mature neutrophils [14]. Over 50 mutations in ELA2 have been found in patients with autosomal dominant and sporadic forms of SCN, as well as in cyclic neutropenia [10*]. While it has been hypothesized that defective enzyme activity or inappropriate localization may represent the mechanism of pathogenesis for the various mutations [10*], more recent studies argue that NE mutations elicit the unfolded protein response (UPR), which increases the transcription of chaperone-encoding, endoplasmic reticulum-associated protein degradation (ERAD), and pro-apoptotic genes, which ultimately leads to apoptosis [15, 16*].
Granulocyte colony-stimulating factor receptor (G-CSFR)
The G-CSF-R, encoded by the CSF3R gene, plays a crucial role in the production and function of neutrophilic granulocytes, being able to stimulate the proliferation, differentiation and survival of cells along the neutrophilic lineage, activate the functions of mature neutrophils, as well as mobilize various precursor cells [17*]. Two classes of CSF3R mutations have been associated with SCN, with quite different roles [5*, 13**].
Acquired mutations in the CSF3R gene have been identified in around 20-30% of SCN [17*, 18]. These mutations produce C-terminally truncated hyper-responsive forms of the receptor (G-CSFRhyper), which act in a dominant-negative manner to enhance proliferation at the expense of maturation [13**, 19]. The role of G-CSFRhyper mutations in neutropenia appears to be relatively modest [5*, 13**]. However, SCN patients carrying G-CSFRhyper mutations show a strong predisposition to both MDS and AML, where they appear to represent an early step in leukemogenesis [5*, 12**]. Recent studies suggest that the pathogenic properties of G-CSFRhyper mutation are largely due to the enhanced Stat5 activation they elicit [20, 21**], which appears to provide a selective advantage HSCs expressing this mutation [21**, 22*].
Constitutive mutations in the CSF3R gene, leading to hypo-responsive forms of the receptor (G-CSFRhypo), have been reported in several SCN patients who were unable to respond to normal G-CSF therapy [23]. Again, these mutations – which perturb the extracellular domain – act in a dominant manner over wild-type receptors, probably by disrupting normal ligand binding [13**]. While G-CSFRhypo mutations have not formally been shown to cause SCN, this remains likely, but such mutations are certainly responsible for refractoriness to G-CSF treatment observed in these cases.
HAX1
HAX1 is a ubiquitously-expressed mitochondrial protein, which functions as an anti-apoptotic protein, in a manner similar to bcl-2 with which it has weak homology [24**]. Mutations in HAX1 have been reported in cases of autosomal-recessive SCN (as described in the original Kostmann pedigree) [24**], with some mutations also producing neurological disorders [25*, 26*]. In each case, the genetic lesions serve to inactivate the HAX1 protein, leading to a loss of mitochondrial membrane potential, release of proapoptotic proteins and subsequent apoptosis of neutrophils [24**].
Wiskott-Aldrich syndrome protein (WASp)
WASp is exclusively expressed in hematopoietic cell, where it plays a regulatory key role actin polymerization involved in cell signaling, cell-cell interactions and cell motility. Patients with X-linked SCN have been reported with activating mutations in WASp leading to a constitutively-active form of the protein, and unregulated actin polymerization [27, 28]. Concomitant defects in mitosis and cytokinesis lead to decreased proliferation and increased apoptosis in myeloid progenitors [29**].
Growth factor-independent protein 1 (GFI1)
GFI1 is a zinc finger protein which appears to function as a transcriptional repressor [30]. Inactivating mutations in this protein have been reported in a small number of SCN patients [31]. Two distinct mechanisms have been proposed for its action, based on the two genes identified to be up-regulated once the repressive effects of GFI1 have been alleviated by mutation: (i) upregulation of NE [31] leading to induction of the unfolded protein response and hence apoptosis [7**]; (ii) upregulation of C/EBPε leading to induction of CSF-1 expression and lineage switching to the macrophage lineage [32].
Other proteins
Many cases of SCN exist for which no underlying molecular cause have been identified, although several of the above candidates have been excluded, making it likely that other mutations also contribute to neutropenia. In one such case, CD40 ligand deficiency has been suggested as a possible cause [33*]. Moreover, there are several disorders which exhibit neutropenia as part of a broader spectrum of disease spectrum, the molecular basis of which have been determined. These include mutations in the Rab27 protein, a small GTPase, in Griscelli syndrome type 2 [34], the MAPBPIP scaffolding protein in so-called ‘p14 deficiency’ [35], the AP3B1 adapter protein in Hermansky-Pudlak syndrome type 2 [36] and the CHS1/LYST protein in Chediak-Higashi syndrome [37*].
Key themes
Emerging from the most recent studies are some consistent themes, which serve as a framework for future work.
Convergence of mutations at the biological level
Many neutropenia-associated mutations converge to disrupt the delicate developmental pathway required to form these protease-packed cells [38*]. Defects in protein trafficking [10*], as well as the molecular defects underpinning p14 deficiency [35], Griscelli syndrome type 2 [34], Hermansky-Pudlak syndrome type 2 [36] and Chediak-Higashi syndrome [37*] appear to cause a similar outocme. Recently, the unfolded protein response has both associated with mutations in both NE [15, 16*] and GFI1 [7**]. Several of the SCN-related mutations result in increased apoptosis, including those in NE [15, 16*], WASp [29**], G-CSFRhypo [39], HAX1 [24**], and potentially GFI1 [7**], suggesting that mistakes in trafficking and the unfolded protein response are trigger events for initiating premature cell death. This hypothesis has a number of implications for therapy. Firstly, such a scenario would imply that the key role of G-CSF therapy in SCN is as a survival factor for neutrophils, rather than simply stimulating the production of neutrophils (as is its role in the treatment of other forms of neutropenia). Secondly, it is possible that other agents that enhance neutrophil survival might be effective therapeutic agents. Thirdly, the expansion of acquired G-CSFRhyper mutant clones might be due to enhanced clonal survival rather than exclusively an enhanced proliferative advantage.
Co-operation between mutations
Another common theme from recent studies is the presence of combinations of the above mentioned mutations in neutropenia. For example, one of the original Kostman family possessed an NE mutation and another member had an acquired G-CSFRhyper mutation, presumably on the background of an HAX1 mutation [40]. Similarly, combinations of NE and acquired G-CSFRhyper mutations have been reported [5*], while we recently reported a patient with constitutive NE and G-CSFRhypo mutations, who acquired sequential G-CSFRhyper mutations [41*]. Other cases have been reported with multiple G-CSFRhyper mutations [5*], as well as with multiple NE mutations [42*]. In the case of constitutive mutations, such combinations are presumably the result of chance, although there remains a possibility that the mutations synergise in some way, particularly those that converge at a similar biological level. In the case of the acquired G-CSFRhyper mutations, it is possible that the presence of an altered myeloid compartment and G-CSF therapy in neutropenic patients creates an millieau favourable for the expansion of clones possessing such a mutation [13**]. Indeed, such acquired G-CSFRhyper mutations may partially rescue neutropenia caused by NE and/or G-CSFRhypo mutations [41*, 43].
Treatment strategies
Treatment with G-CSF is effective in the majority of SCN [3]. However, alternative therapies are needed, particularly for patients who are refractory to G-CSF treatment, and those acquiring truncating G-CSFRhyper mutations on G-CSF treatment due to concerns about possible contribution of G-CSF to progression to MDS/AML. One approach would be to improve the reduced neutrophil survival common in neutropenia. While there are several strategies for doing this, including the inhibition of NE, one key therapeutic target is STAT5 that we and others have shown to be a key mediator of survival in neutrophilic granulocyte [41*, 44*]. Various strategies can be brought to bear to target this molecule [45*]. Indeed, we have successfully used corticosteroids to enhance Stat5 activation and survival in vitro with successful application in the treatment of neutropenia [39], and more recently showed that constitutively-active Stat5 could improve survival in a cell model of granulopoiesis [41*]. Undoubtedly, other aspects of the cell survival machinery could also be targeted.
CONCLUSIONS
Recent studies have elucidated several genetic and molecular perturbations leading to severe congenital neutropenia. They provide important new insights into both normal and pathogenic myelopoiesis. Diagnosis and classification based of this new genetic, molecular and cellular information affords the opportunity to develop tailored, and potentially new, therapeutic strategies, and much improved care for SCN patients.
Acknowledgments
This work was supported by funds from the Deakin University Molecular Medicine and Nutrition Research Cluster, Geelong Victoria, Australia and a grant from the National Institutes of Health, NIAID, R24AI49393, Bethesda, Maryland, USA.
REFERENCES
- 1.Welte K, Zeidler C, Dale DC. Severe congenital neutropenia. Semin. Hematol. 2006;43:189–195. doi: 10.1053/j.seminhematol.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 2.Dale DC, Bonilla MA, Davis MW, et al. A randomized controlled phase III trial of recombinant human granulocyte colony-stimulating factor (Filgrastim) for treatment of severe chronic neutropenia. Blood. 1993;81:2496–2502. [PMC free article] [PubMed] [Google Scholar]
- 3.Dale DC, Bolyard AA, Schwinzer BG, et al. The severe congenital neutropenia international registry: 10-year follow-up report. Support Cancer Ther. 2006;3:220–231. doi: 10.3816/SCT.2006.n.020. [DOI] [PubMed] [Google Scholar]
- 4.Rosenberg PS, Alter BP, Bolyard AA, et al. Severe Congenital Neutropenia International Registry. The incidence of leukemia and mortality from sepsis in patients with severe congenital neutropenia receiving long-term G-CSF therapy. Blood. 2006;107:4628–4635. doi: 10.1182/blood-2005-11-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5*.Germeshausen M, Ballmaier M, Welte K. Incidence of CSF3R mutations in severe congenital neutropenia and relevance for leukemogenesis: results of a long-term survey. Blood. 2007;109:93–99. doi: 10.1182/blood-2006-02-004275. Detailed analysis of G-CSFR mutations in SCN, including their role in leukemogenesis. [DOI] [PubMed] [Google Scholar]
- 6**.Rosenberg PS, Alter BP, Link DC, et al. Neutrophil elastase mutations and risk of leukaemia in severe congenital neutropenia. Br. J. Haematol. 2008;140:210–213. doi: 10.1111/j.1365-2141.2007.06897.x. Definitive study on the role of ELA2 mutations in leukemogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7**.Berliner N. Lessons from congenital neutropenia: 50 years of progress in understanding myelopoiesis. Blood. 2008;111:5427–5432. doi: 10.1182/blood-2007-10-077396. Seminal history of research into congenital neutropenia, providing an overview of the key clinical and molecular findings and how these have added to our understanding of normal myelopoiesis. [DOI] [PubMed] [Google Scholar]
- 8*.Carlsson G, Melin M, Dahl N, et al. Kostmann syndrome or infantile genetic agranulocytosis, part two: understanding the underlying genetic defects in severe congenital neutropenia. Acta Paediatr. 2007;96:813–819. doi: 10.1111/j.1651-2227.2007.00274.x. First detailed analysis of the genetic defects in Kostmann syndrome. While failing to identify the underlying cause, additional mutations in neutrophil elastase and GCSFR were identified. [DOI] [PubMed] [Google Scholar]
- 9.Dale DC, Person RE, Bolyard AA, et al. Mutations in the gene encoding neutrophil elastase in congenital and cyclic neutropenia. Blood. 2000;96:2317–2322. [PubMed] [Google Scholar]
- 10*.Horwitz MS, Duan Z, Korkmaz B, et al. Neutrophil elastase in cyclic and severe congenital neutropenia. Blood. 2007;109:1817–1824. doi: 10.1182/blood-2006-08-019166. Comprehensive review of neutrophil elastase mutations and their role in the pathogenesis of cyclic and severe congenital neutropenia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boxer LA, Stein S, Buckley D, et al. Strong evidence for autosomal dominant inheritance of severe congenital neutropenia associated with ELA2 mutations. J. Paediatr. 2006;148:633–636. doi: 10.1016/j.jpeds.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 12**.Touw IP, Bontenbal M. Granulocyte colony-stimulating factor: key (f)actor or innocent bystander in the development of secondary myeloid malignancy. J. Natl. Cancer Inst. 2007;99:183–186. doi: 10.1093/jnci/djk057. Definitive articulation of the case ‘for’ and ‘against’ truncating GCSFR mutations playing a role in secondary myeloid malignancy, with the conclusion that they likely do make an important contribution. [DOI] [PubMed] [Google Scholar]
- 13**.Ward AC. The role of the granulocyte colony-stimulating factor receptor (G-CSF-R) in disease. Front. Biosci. 2007;12:608–618. doi: 10.2741/2086. Comprehensive review of G-CSFR mutations, their molecular consequences and relative contribution to the pathogenesis of SCN and associated disorders. [DOI] [PubMed] [Google Scholar]
- 14.Korkmaz B, Moreau T, Gauthier F. Neutrophil elastase, proteinase 3 and cathepsin G: physiochemical properties, activity and physiopathological functions. Biochimie. 2008;90:227–242. doi: 10.1016/j.biochi.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 15.Kollner I, Sodeik B, Schreek S, et al. Mutations in neutrophil elastase causing congenital neutropenia lead to cytoplasmic protein accumulation and induction of the unfolded protein response. Blood. 2006;108:493–500. doi: 10.1182/blood-2005-11-4689. [DOI] [PubMed] [Google Scholar]
- 16*.Grenda DS, Murakami M, Ghatak J, et al. Mutations of the ELA2 gene found in patients with severe congenital neutropenia induce the unfolded response and cellular apoptosis. Blood. 2007;110:4179–4187. doi: 10.1182/blood-2006-11-057299. *Confirmatory study of the role of the ‘unfolded protein response’ in congenital neutropenia caused by neutrophil elastase mutations, providing key mechanistic detail of how apoptosis is induced by this process. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17*.Touw IP, van de Geijn GJ. Granulocyte colony-stimulating factor and its receptor in normal myeloid cell development, leukemia and related blood cell disorders. Front. Biosci. 2007;12:800–815. doi: 10.2741/2103. Insightful review of the role of G-CSF and its receptor in normal development, and how this is altered in disease, including leukemia. [DOI] [PubMed] [Google Scholar]
- 18.Dong F, Brynes RK, Tidow N, et al. Mutations in the gene for the granulocyte colony-stimulating-factor receptor in patients with acute myeloid leukemia preceded by severe congenital neutropenia. N. Engl. J. Med. 1995;333:487–493. doi: 10.1056/NEJM199508243330804. [DOI] [PubMed] [Google Scholar]
- 19.Ward AC, van Aesch YM, Schelen AM, Touw IP. Defective internalization and sustained activation of truncated granulocyte colony-stimulating factor receptor found in severe congenital neutropenia/acute myeloid leukemia. Blood. 1999;93:447–458. [PubMed] [Google Scholar]
- 20.Gits J, van Leeuwen D, Carroll HP, et al. Multiple pathways contribute to the hyperproliferative responses from truncated granulocyte colony-stimulating factor receptors. Leukemia. 2006;20:2111–2118. doi: 10.1038/sj.leu.2404448. [DOI] [PubMed] [Google Scholar]
- 21**.Liu F, Kunter G, Krem MM, et al. Csf3r mutations in mice confer a strong clonal HSC advantage via activation of Stat5. J. Clin. Invest. 2008;118:946–955. doi: 10.1172/JCI32704. Elegant study demonstrating in vivo that truncating GCSFR mutations provide a growth advantage to hematopoietic stem cell populations, which is mediating through Stat5 activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22*.Germeshausen M, Skokowa J, Ballmaier M, et al. G-CSF receptor mutations in patients with congenital neutropenia. Curr. Opin. Hematol. 2008;15:332–337. doi: 10.1097/MOH.0b013e328303b9f6. Useful review of GCSFR mutations in congenital neutropenia. [DOI] [PubMed] [Google Scholar]
- 23.Ward AC, van Aesch YM, Gits J, et al. Novel point mutation in the extracellular domain of the granulocyte colony-stimulating factor (G-CSF) receptor in a case of severe congenital neutropenia hyporesponsive to G-CSF treatment. J. Exp. Med. 1999;190:497–507. doi: 10.1084/jem.190.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24**.Klein C, Grudzien M, Appaswamy G, et al. HAX1 deficiency causes autosomal recessive severe congenital neutropenia (Kostmann disease) Nature Genet. 2007;39:86–92. doi: 10.1038/ng1940. First study identifying and characterizing HAX1 mutations in the etiology of classical autosomal recessive congenital neutropenia (Kostmann syndrome)elicitied by mutations in neutrophil elastase. [DOI] [PubMed] [Google Scholar]
- 25*.Germeshausen M, Grudzien M, Zeidler C, et al. Novel HAX1 mutations in patients with severe congenital neutropenia reveal isoform-dependent genotype-phenotype association. Blood. 2008;111:4954–4957. doi: 10.1182/blood-2007-11-120667. Describes the elucidation of distinct mutant HAX1 isoforms with differentially associated with neurological symptoms associated with this form of congenital neutropenia. [DOI] [PubMed] [Google Scholar]
- 26*.Carlsson G, Van't Hooft I, Melin M, et al. Central nervous system involvement in severe congenital neutropenia: neurological and neuropsychological abnormalities associated with specific HAX1 mutations. J. Intern. Med. May 29;2008 doi: 10.1111/j.1365-2796.2008.01982.x. Epub. *Confirmatory study of the role of distinct HAX1 mutations in cases of congenital neutropenia with associated neurological symptoms. [DOI] [PubMed] [Google Scholar]
- 27.Devriendt K, Kim AS, Mathijs G, et al. Constitutively activating mutation in WASP causes X-linked severe congenital neutropenia. Nat. Genet. 2001;27:313–317. doi: 10.1038/85886. [DOI] [PubMed] [Google Scholar]
- 28.Ancliff PJ, Blundell MP, Cory GO, et al. Two novel activating mutations in the Wiskott-Aldrich syndrome protein result in congenital neutropenia. Blood. 2006;108:2182–2189. doi: 10.1182/blood-2006-01-010249. [DOI] [PubMed] [Google Scholar]
- 29**.Moulding DA, Blundell MP, Spiller DG, et al. Unregulated actin polymerization by WASp causes defects of mitosis and cytokinesis in X-linked neutropenia. J. Exp. Med. 2007;204:2213–2224. doi: 10.1084/jem.20062324. Definitive study identifying the molecular perturbation caused by constitutively-active forms of WASp associated with X-linked neutropenia, implicating mitotic and other defects in inducing neutrophil apoptosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hock H, Orkin SH. Zinc finger transcription factor Gfi1: versatile regulator of lymphocytes, neutrophils and hematopoietic stem cells. Curr. Opin. Hematol. 2006;13:1–6. doi: 10.1097/01.moh.0000190111.85284.8f. [DOI] [PubMed] [Google Scholar]
- 31.Person RE, Li FQ, Duan Z, et al. Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat. Genet. 2003;34:308–12. doi: 10.1038/ng1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhuang D, Qiu Y, Kogan SC, Dong F. Increased CCAAT enhancer binding protein e (C/EBPe) expression and premature apoptosis in myeloid cells expressing Gfi-1 N382S mutant associated with severe congenital neutropenia. J. Biol. Chem. 2006;281:10745–10750. doi: 10.1074/jbc.M510924200. [DOI] [PubMed] [Google Scholar]
- 33*.Rezaei N, Aghamohammadi A, Ramyar A, et al. Severe congenital neutropenia or hyper-IgM syndrome? A novel mutation of CD40 ligand in a patient with severe neutropenia. Int. Arch. Allergy Immunol. 2008;147:255–259. doi: 10.1159/000142050. Short report identifying a CD40 ligand mutation associated in a severely neutropenic patient, and hypothesizing a potential mechanistic link. [DOI] [PubMed] [Google Scholar]
- 34.Menasche G, Feldmann J, Houdusse A, et al. Biochemical and functional characterization of Rab27a mutations in Griscelli syndrome patients. Blood. 2003;101:2736–2742. doi: 10.1182/blood-2002-09-2789. [DOI] [PubMed] [Google Scholar]
- 35**.Bohn G, Allroth A, Brandes G, et al. A novel primary immunodeficiency syndrome caused by deficiency of the endosomal adaptor protein p14. Nature Med. 2007;13:38–45. doi: 10.1038/nm1528. First report identifying mutations in the endosomal adaptor protein p14 in cases of primary immunodeficiency. [DOI] [PubMed] [Google Scholar]
- 36.Jung J, Bohn G, Allroth A, et al. Identification of a homozygous deletion in the AP3B1 gene causing Hermansky-Pudlak syndrome, type 2. Blood. 2006;108:362–369. doi: 10.1182/blood-2005-11-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37*.Kaplan J, De Domenico I, Ward DM. Chediak-Higashi syndrome. Curr. Opin. Hematol. 2008;15:22–29. doi: 10.1097/MOH.0b013e3282f2bcce. Useful review of the clinical and molecular features of Chediak-Higashi syndrome. [DOI] [PubMed] [Google Scholar]
- 38*.Dale DC, Boxer LA, Liles WC. The phagocytes: neutrophils and monocytes. Blood. 2008;112:935–945. doi: 10.1182/blood-2007-12-077917. Definitive contemporary guide to phagocytes. [DOI] [PubMed] [Google Scholar]
- 39.Dror Y, Ward AC, Touw IP, Freedman MH. Combined corticosteroid/granulocyte colony-stimulating factor (G-CSF) therapy in the treatment of severe congenital neutropenia unresponsive to G-CSF: Activated glucocorticoid receptors synergize with G-CSF signals. Exp. Hematol. 2000;28:1381–9. doi: 10.1016/s0301-472x(00)00544-0. [DOI] [PubMed] [Google Scholar]
- 40.Carlsson G, Aprikyan AA, Ericson KG, et al. Neutrophil elastase and granulocyte colony-stimulating factor receptor mutation analyses and leukemia evolution in severe congenital neutropenia pateints belonging to the original Kostmann family in northern Sweden. Haematologica. 2006;91:589–595. [PubMed] [Google Scholar]
- 41*.Ward AC, Gits J, Majeed F, et al. Functional interaction between mutations in the granulocyte colony-stimulating factor receptor in severe congenital neutropenia. Br. J. Haematol. 2008;142:653–656. doi: 10.1111/j.1365-2141.2008.07224.x. Short report elucidating multiple mutations in a patient with severe congenital neutropenia hyporesponsive to G-CSF, including constitutive neutrophil elastase and GCSFRhypo mutations and sequential GCSFR truncating mutations, the latter hypothesized to partially ‘rescue’ the GCSFRhypo mutation. [DOI] [PubMed] [Google Scholar]
- 42*.Salipante SJ, Benson KF, Luty J, et al. Double de novo mutations of ELA2 in cyclic and severe congenital neutropenia. Hum. Mutat. 2007;28:874–881. doi: 10.1002/humu.20529. Study identifying double mutations in the gene encoding neutrophil elastase in different forms of neutropenia, demonstrating both mutations occurred de novo. [DOI] [PubMed] [Google Scholar]
- 43.Aprikyan AA, Kutyavin T, Stein S, et al. Cellular and molecular abnormalities in severe congenital neutropenia predisposing to leukemia. Exp. Hematol. 2003;31:372–381. doi: 10.1016/s0301-472x(03)00048-1. [DOI] [PubMed] [Google Scholar]
- 44*.Fievez L, Desmet C, Henry E, et al. STAT5 is an ambivalent regulator of neutrophil homeostasis. PLoS ONE. 2007;2:e727. doi: 10.1371/journal.pone.0000727. Study demonstrating differential roles for Stat5 in neutrophil production, potentially explaining some aspects of SCN pathogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45*.Lewis RS, Ward AC. Stat5 as a marker of leukemia. Expert Rev. Mol. Diag. 2008;8:73–82. doi: 10.1586/14737159.8.1.73. Short review focused on the association of Stat5 with leukemia (including SCN-associated), identifying it as both a marker of the disease as well as a potential therapeutic target. [DOI] [PubMed] [Google Scholar]