Abstract
Histone levels are tightly regulated to prevent harmful effects such as genomic instability and hypersensitivity to DNA damaging agents due to the accumulation of these highly basic proteins when DNA replication slows down or stops. Although chromosomal histones are stable, excess (non-chromatin bound) histones are rapidly degraded in a Rad53 kinase dependent manner in Saccharomyces cerevisiae. Here we demonstrate that excess histones associate with Rad53 in vivo, appear to undergo modifications such as tyrosine phosphorylation and polyubiquitylation, before their proteolysis by the proteasome. We have identified the tyrosine 99 residue of histone H3 as being critical for the efficient ubiquitylation and degradation of this histone. We have also identified the E2 proteins Ubc4 and Ubc5, as well as the E3 ubiquitin ligase Tom1, as enzymes involved in the ubiquitylation of excess histones. Regulated histone proteolysis has major implications for the maintenance of epigenetic marks on chromatin, genomic stability and the packaging of sperm DNA.
Keywords: Histones, Rad53, Phosphorylation, Ubiquitylation, Proteasome, Ubc4, Ubc5, Tom1
In eukaryotic cells, DNA is wrapped around basic histone proteins to form a nucleoprotein filament called chromatin. Histones are essential for viability as they package DNA and regulate access to genetic information contained within DNA. However, the positively charged histones can also bind non-specifically to negatively charged nucleic acids and adversely affect processes that require access to DNA1. Hence, histone and DNA synthesis are coupled, with the bulk of histone synthesis occurring during S-phase where they are rapidly deposited onto replicated DNA by histone chaperones1. Absence of histone synthesis during DNA replication leads to inviability in yeast2, whereas moderate defects in histone expression adversely affect transcription and cell fitness3. Yeast cells lacking the histone chaperones CAF-1 (Chromatin Assembly Factor-1) and Asf1 (Anti Silencing Function 1) exhibit enhanced rates of gross chromosomal rearrangements4. In human cells, inhibiting histone expression during S-phase or inactivating CAF-1 triggers DNA damage and inviability5,6. Conversely, cells accumulate excess “free” histones when DNA synthesis decreases at the end of S-phase or when DNA synthesis is inhibited by genotoxic agents7–9. Even a slight stoichiometric excess of histones over DNA triggers chromatin aggregation and blocks transcription in vitro10. In yeast, elevated histone levels lead to enhanced DNA damage sensitivity9 and chromosome loss11, a form of genomic instability. Genomic instability is characterized by the acquisition of genome alterations and is associated with human cancers12. Overall, the aforementioned studies imply that improper histone stoichiometry or chromatin structures may contribute to genomic instability and tumorigenesis. Hence, cells have evolved numerous mechanisms to regulate histone levels. For example, DNA replication arrest elicits a downregulation of histone mRNAs via histone gene repression in yeast13 and degradation of histone mRNAs in higher eukaryotes14. We have previously shown that the basal kinase activity of the DNA damage checkpoint kinase Rad53 regulates histone protein levels in yeast9. Rad53 associates with excess histones and somehow targets them for degradation. Here, we have dissected the molecular pathway by which Rad53 triggers excess histone degradation.
Histones associated with Rad53 are phosphorylated and this modification is required for their efficient degradation
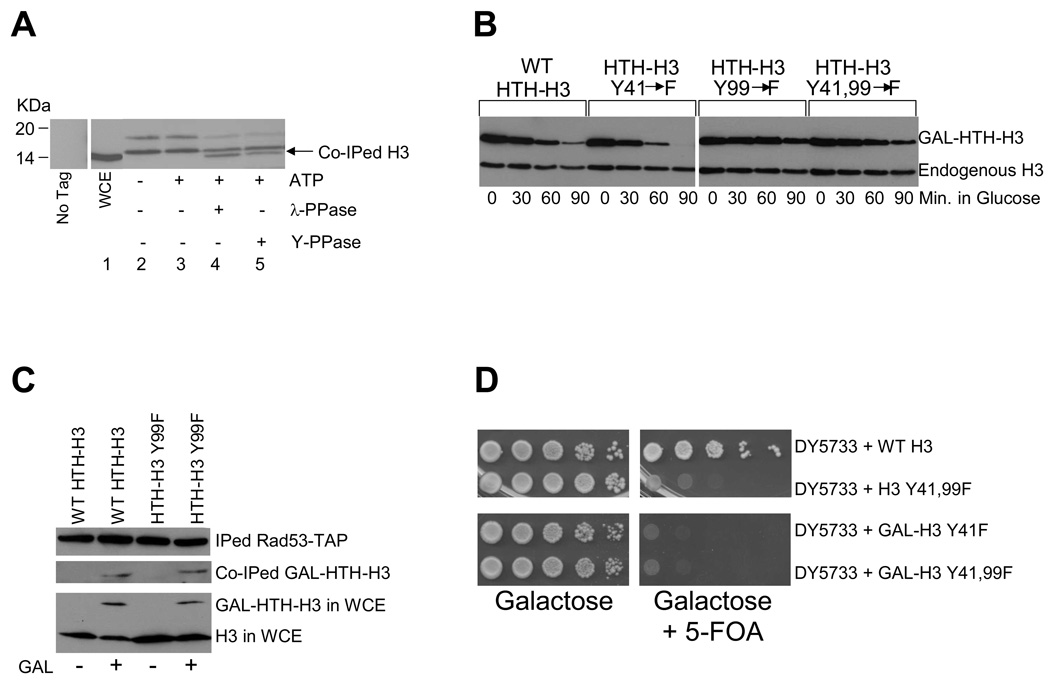
Since histones H3, H4 and H2B interact with the Rad53 kinase that can phosphorylate all core histones in vitro9, we investigated whether excess histones were being phosphorylated in vivo. We immunoprecipitated Rad53-TAP9 from yeast whole cell extracts (WCEs) and treated the immunoprecipitate with or without ATP and, subsequently, with either lambda phosphatase (λ-PPase, to dephosphorylate serines, threonines and tyrosines), or the YOP phosphatase15 (Y-PPase, to dephosphorylate tyrosines only). Histone H3 associated with Rad53-TAP was detected by Western blotting (Figure 1A) with the H3-C antibody9 raised against the C-terminus of H3 that is not posttranslationally modified. This antibody detects a single band for H3 in yeast WCE (lane 1) and does not detect anything in immunoprecipitates from an untagged strain (“No tag” in Figure 1A). Surprisingly, this antibody detects multiple bands in Rad53-TAP immunoprecipitates that migrate slower than the H3 band in yeast WCE (Figure 1A, lane 2), suggesting that most H3 associated with Rad53-TAP are posttranslationally modified. Incubation of the immunoprecipitate with ATP at 30°C did not affect the bands detected by the H3-C antibody (lane 3), although subsequent treatment with λ-PPase resulted in the regeneration of a band with mobility identical to the H3 band in WCE (compare the H3 band in lanes 1 and 4), concomitant with reduced intensity of slower migrating bands. Hence, H3 associated with Rad53 is likely to be phosphorylated, which affects its mobility during SDS-PAGE. Treatment with the Y–PPase also generated a band whose migration is identical to that of H3 in WCE (compare lanes 1 and 5), suggesting that H3 associated with Rad53 may be phosphorylated on tyrosines. Similar results were obtained for histone H4 associated with Rad53 (Supplemental Figure S1A). Interestingly, Rad53 is known to phosphorylate tyrosine in a co-precipitated yeast protein16. Although our data is consistent with Rad53 directly phosphorylating core histones in vivo, we cannot formally exclude the possibilities that Rad53 may phosphorylate some other target(s) needed for histone degradation, or that another kinase may be directly phosphorylating excess histones bound to Rad53 in vivo.
Figure 1. Histones associated with Rad53 are phosphorylated and this modification is required for efficient histone degradation.
(A) Histone H3 associated with Rad53 is phosphorylated. TAP-tagged Rad53 was immunoprecipitated using IgG-Sepharose beads as described in the Methods section. Rad53-TAP bound to beads was first equilibrated with lysis buffer lacking phosphatase inhibitors and then incubated with or without 1mM ATP for 1hour at 30°C, following which the ATP treated sample was divided into three equal aliquots. One aliquot was left untreated; the second was incubated with λ-phosphatase (λ-PPase), while the third was treated with YOP tyrosine specific phosphatase (Y-PPase) for an additional hour at 30°C. All samples were boiled in SDS-PAGE sample loading buffer and proteins were resolved through a SDS 4–12% polyacrylamide gradient gel, which was processed for Western blotting with the H3-C antibody9. The migration of histone H3 present in whole cell extract (WCE) is indicated by the arrow. The lane labeled “No Tag” shows a mock immunoprecipitation with IgG-Sepharose performed on a strain where Rad53 was untagged.
(B) Mutation of the tyrosine 99 phosphorylation site in histone H3 abolishes its degradation. Site-directed mutagenesis was used to mutate both the potential tyrosine (Y) phosphorylation sites in the H3 protein at positions 41 and 99 to phenylalanines (F), either individually (Y41F and Y99F), or in combination (Y41,99F). Histone degradation assays were carried out as described in the Methods section in wild type W303 cells bearing HTH-tagged wild type (WT) or mutant histone H3 plasmids.
(C) Mutation of the tyrosine 99 phosphorylation site on H3 does not preclude the recognition of this histone by Rad53. After a 2 hour galactose (GAL) induction of HTH-tagged wild-type and mutant H3 in asynchronous cells carrying the RAD53-TAP allele, WCE were prepared. Rad53-TAP was immunoprecipitated (IPed) from the WCEs and the co-immunoprecipitated (co-IPed) HTH tagged H3 was detected by Western blotting with HA antibodies. The relative amount of endogenous and HTH tagged histone H3 present in the WCE is depicted in the lower panel to demonstrate that roughly equal amounts of material was used for each immunoprecipitation reaction.
Tyrosine 99 residue of histone H3 is critical for efficient degradation of this histone
Histone phosphorylation has been implicated in several cellular processes17, but never in the degradation of histones themselves. Based on our data in Figure 1A, we hypothesized that phosphorylation of tyrosines in H3 may be important for its degradation. We used site-directed mutagenesis to mutate the two tyrosines (Y) present in H3 at positions 41 and 99 to phenylalanine (F) residues that are similar to tyrosines, but cannot be phosphorylated. Wild type and mutant H3 were tagged at their N-terminus with a HIS10-TEV-HA (HTH) epitope and expressed under the control of a galactose inducible promoter from a high copy plasmid (pYES2-HTH-HHT2)9. Degradation assays were carried out for HTH-tagged wild type and mutant H3 in wild type cells (W303) essentially as described previously9. The levels of chromatin-derived endogenous H3, which is not subject to degradation, remained constant in our assay (Figure 1B, lower band). In contrast, overexpressed HTH-tagged wild type and the Y41F mutant were degraded rapidly, with the Y41F mutant routinely degrading slightly faster than wild type H3 (Figure 1B, upper band). By 90 minutes, up to 80% of wild type and 100% of the Y41F mutant were degraded, compared to less than 20% of the Y99F mutant, which is similar to the defect observed in rad53 mutants9. Hence, phosphorylation of the H3 Y99 residue is required for its efficient degradation. Combining the two mutations (Y41,99F) led to an intermediate degradation phenotype compared to the individual Y41F and Y99F mutations. Both H3 mutants were expressed at levels similar to wild type H3 (Figure 1B, upper band), localized to the nucleus as judged by immunofluorescence microscopy and were present in the chromatin associated fraction (data not shown). Further, both the wild type and Y99F mutant HTH-H3 were incorporated equally well into the Rad53-histone complex (Figure 1C). We also obtained evidence that phosphorylation of internal residues in histone H4, including tyrosines, may be required for its efficient degradation (Supplemental Figure S1B, C, D). Mutation of H3 Y41, but not Y99 residue causes inviability18,19, possibly due to instability of the Y41 mutant (Figure 1B). However, plasmid-shuffle assays20 involving overexpression of either the unstable Y41F single, or the more stable Y41,99F double mutant as the only source of H3 did not result in viability (Supplemental Figure S2), suggesting that protein stability is not the essential role of H3 Y41.
Histones associated with Rad53 are ubiquitylated
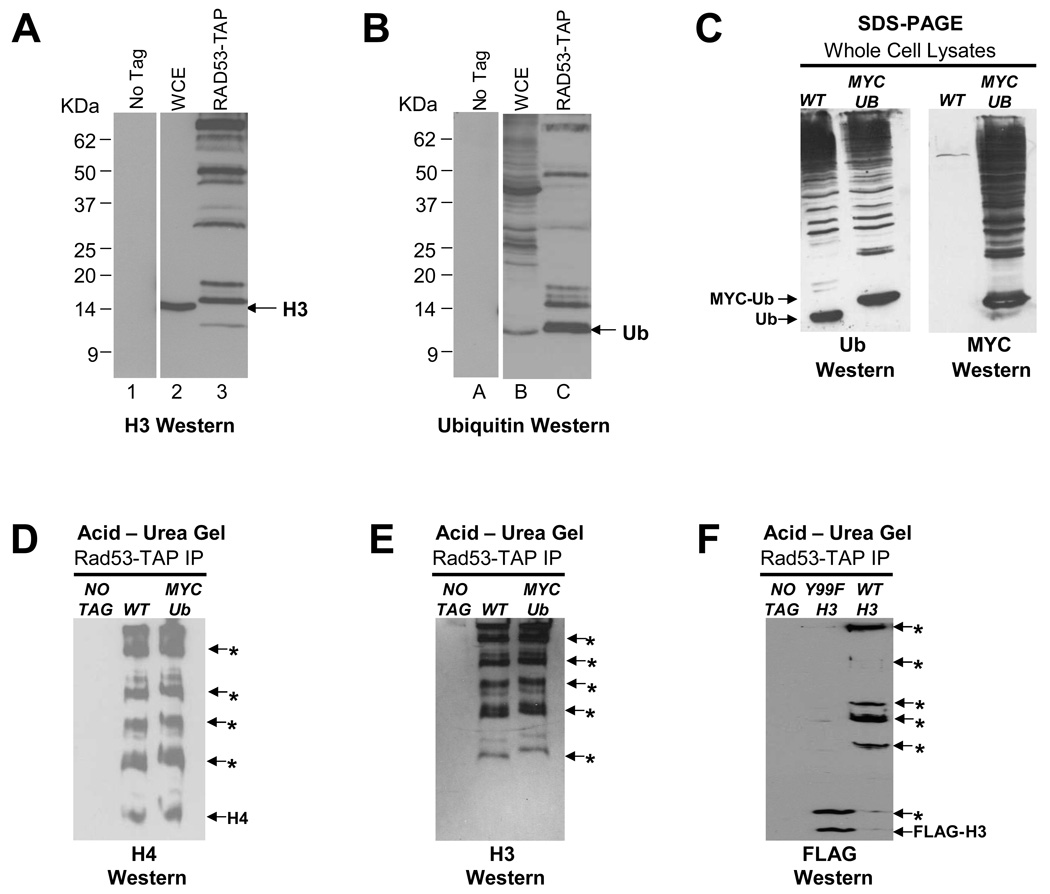
The ubiquitin-proteasome pathway21 allows cells to fine tune their metabolic processes by selectively degrading proteins via the covalent attachment of at least four molecules of the 76-residue ubiquitin to a lysine on the target protein through the sequential action of E1, E2 and E3 enzymes, resulting in a polyubiquitylated protein that is recognized and degraded by a multifunctional protease, the 26S proteasome. Histones incorporated into chromatin are extremely stable proteins22–24. This is likely important for maintaining histone modifications that contribute to stable gene silencing. However, excess free histones are degraded in a Rad53 dependent manner9, perhaps following their ubiquitylation. Analysis of Rad53-TAP immunoprecipitates by Western blotting with the H3-C antibody revealed multiple bands (Figure 2A, lane 3), some larger than 50kDa, which is several times the mass of histone H3 (15.4kDa) and cannot be explained by phosphorylation alone. Consistent with this, H3 species above 25kDa are not eliminated by phosphatase treatment (data not shown), suggesting that they could be ubiquitylated. Hence, we stripped the Western blot shown in Figure 2A and re-probed it with ubiquitin antibodies25. These antibodies detected free ubiquitin and several bands above it in Rad53-TAP immunoprecipitates (Figure 2B, lane C), some of which comigrate with bands detected by the H3-C antibody (Figure 2A, lane 3), implying that these bands correspond to multi- or poly-ubiquitylated histone H3. The ubiquitin antibody detects several bands that are not recognized by the H3-C antibody (Figure 2B versus 2A) and these may correspond to other ubiquitylated histones present in the Rad53-TAP complex. To confirm our results, we placed the HA and FLAG epitope tags on the N-terminus of endogenous H3 genes (HA-HHT1, FLAG-HHT2) and analyzed Rad53-TAP immunoprecipitates from these strains by Western blotting with HA antibodies, which detected multiple bands for HA-H3 only in the tagged strain (Supplemental Figure S3A; HA Western, lane 3). Importantly, many of the bands detected by HA antibodies are detected by ubiquitin antibodies as well, strongly suggesting that they correspond to ubiquitylated H3 (Supplemental Figure S3A; Ub Western, lane C).
Figure 2. Histones associated with Rad53 are ubiquitylated and the tyrosine 99 residue of histone H3 is critical for its ubiquitylation.
(A) Histone H3 associated with Rad53 is extensively modified. Rad53-TAP complexes were immunoprecipitated as described in Methods and analyzed by Western blotting with H3-C antibody. The migration of bulk histone H3 in WCE is indicated by the arrow. In Rad53 immunoprecipitates, multiple bands with slower mobility than the H3 band in WCE are detected, suggesting that histone H3 associated with Rad53 is heavily modified.
(B) Histone H3 associated with Rad53 appears to be ubiquitylated. The Western blot shown above in (A) was stripped and reprobed with an ubiquitin antibody. The migration of free ubiquitin (Ub) is indicated by the arrow.
(C) Characterization of a strain in which the only source of ubiquitin is His6-MYC tagged (MYC-Ub). WCEs prepared from wild type (WT) and MYC-Ub strains were resolved by SDS-PAGE and processed for Western blotting first with a ubiquitin (Ub) antibody (Ub Western) and, after stripping, with a MYC antibody (MYC Western).
(D) Histone H4 bound to Rad53 is either multi- or poly-ubiquitylated. Histone H4 present in Rad53-TAP immunoprecipitates was analyzed by acetic acid-urea (AU) gel electrophoresis followed by Western blotting with H4 antibodies. The mobility of unmodified histone H4 is indicated by “H4” while the putative ubiquitylated histone H4 species carrying increasing numbers of ubiquitin moieties are indicated by asterisks. The mobility of H4-MYC-Ub bands is slightly retarded in the lane containing Rad53-TAP immunoprecipitates from the MYC-Ub strain compared to those derived from the WT strain.
(E) Histone H3 bound to Rad53 is also either multi- or poly-ubiquitylated. Indicated samples were processed as described in (D) except that Western blotting was carried out to detect histone H3. Note that in this experiment all histone H3 appears to be modified.
(F) Tyrosine 99 residue of histone H3 is required for the efficient ubiquitylation of this histone. Strains carrying FLAG-tagged endogenous genes corresponding to either the wild type (WT) or Y99F mutant histone H3 were processed as described in (D) except that Western blotting was carried out using FLAG antibodies. Note the absence of high molecular mass FLAG-H3 species in the Y99F mutant.
Although polyubiquitylation of chromosomal histones has been observed previously26, 27, ubiquitylation of excess core histones in vivo has not been reported previously. Hence, we devised another strategy to prove that histones associated with Rad53 are ubiquitylated. We obtained a yeast strain lacking endogenous ubiquitin genes28 and carrying either wild type ubiquitin (Ub) or a 6His-MYC tagged ubiquitin (MYC-Ub) gene on a high copy plasmid as the only source of ubiquitin. The mobility of MYC-Ub is retarded compared to that of Ub due to additional mass of the epitope tag (Figure 2C, Ub Western) and a MYC antibody detects MYC-ubiquitylated proteins only in the MYC-Ub strain (Figure 2C, MYC Western). We then analyzed Rad53-TAP complexes immunoprecipitated from strains expressing Ub and MYC-Ub on an acetic acid-urea (AU) polyacrylamide gel29. Unlike SDS-PAGE, proteins are resolved on AU gels based on their mass and net charge, which allows only positively charged proteins, such as histones and their modified forms, to enter the gel29,30. We expected that the bands corresponding to ubiquitylated histones associated with Rad53 would exhibit slightly retarded AU gel mobility in samples derived from the MYC-Ub strain (due to the added mass and net negative charge of the 6His-MYC tag in ubiquitylated histones) when compared to samples from the Ub strain. In AU gels, a small, but clear retardation in the mobility of H4 bands was observed in Rad53-TAP immunoprecipitate from the MYC-Ub strain (Figure 2D), strongly suggesting that H4 is extensively ubiquitylated. We obtained identical results for histone H3 associated with Rad53 (Figure 2E). Diffuse bands corresponding to each ubiquitylated form of histone H3 and H4 are observed, possibly due to multiple posttranslational modifications (newly synthesized histones are extensively acetylated) within each major ubiquitylated/phosphorylated band, all of which have different AU gel mobilities due to charge differences.
We used epitope tagging of endogenous H3 genes instead of the MYC-Ub to obtain further evidence for histone ubiquitylation (Supplemental Figure 3B, C). Compared to untagged H3, a slight but clear retardation in mobility was imparted to tagged H3 species in Rad53-TAP immunoprecipitates resolved on AU gels (Supplemental Figure 3B, compare lanes 3 and 4). Importantly, the species recognized by H3-C antibodies are also detected by ubiquitin antibodies, thus confirming that these bands are ubiquitylated H3 (Supplemental Figure 3B, compare H3 Western to Ub Western).
Tyrosine 99 residue of histone H3 is critical for efficient ubiquitylation of this histone
We investigated if the H3 Y99F mutation that renders this protein resistant to degradation (Figure 1B) also prevents its efficient ubiquitylation. We resolved Rad53-TAP immunoprecipitated from cells expressing FLAG-tagged wild type or Y99F mutant H3 on AU gels. Western blotting using FLAG antibodies revealed that the majority of wild type H3 was present in high molecular mass species, consistent with extensive ubiquitylation (Figure 2F). However, the H3 Y99F mutant was predominantly present in unmodified or weakly modified forms of low molecular masses, suggesting that H3 Y99 phosphorylation may be critical for efficient ubiquitylation of excess H3.
Excess histones are degraded by the proteasome
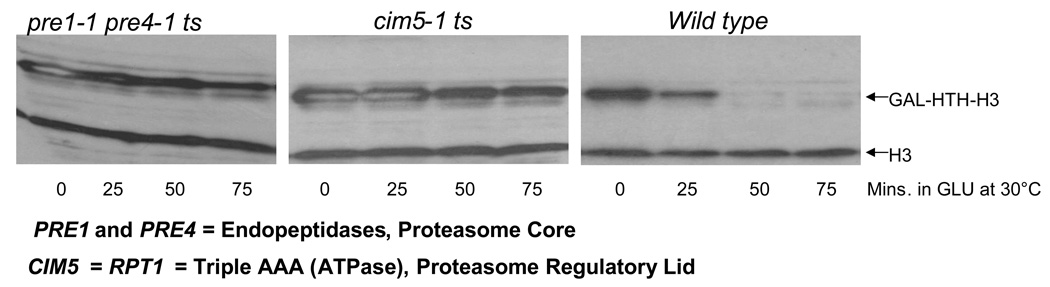
Next, we investigated whether proteasomes were involved in the degradation of excess histones by performing degradation assays for overexpressed HTH-tagged H3 in temperature sensitive (ts) proteasomal mutants. The pre1-1 pre4-1 ts strain carries mutations in the genes encoding two endopeptidase components of the proteasome core31, while the cim5-1 ts strain carries a mutation in the gene encoding Rpt1, an ATPase subunit of the proteasome lid32. Even at the semi-permissive temperature of 30°C, ts mutations in the proteasome inhibited degradation of HTH-H3 compared to wild type cells (Figure 3, upper band), suggesting that functional proteasomes are required for histone degradation.
Figure 3. Functional proteasomes are required for the degradation of excess histones.
Wild type and temperature sensitive (ts) proteasomal mutants carrying the pYES2-HTH-HHT2 plasmid9 encoding galactose inducible, HIS10-TEV-HA (HTH) tagged histone H3 were used to carry out histone degradation assay essentially as described in Methods, with minor changes. The assay was carried out at the semi-permissive temperature of 30°C for the ts strains and cells were harvested every 25 minutes after switching to glucose. The levels of HTH-tagged H3 and chromosomal histone H3 were quantitated by Western blotting using the H3-C antibody9.
Identification of E2 and E3 enzymes involved in degradation related histone ubiquitylation
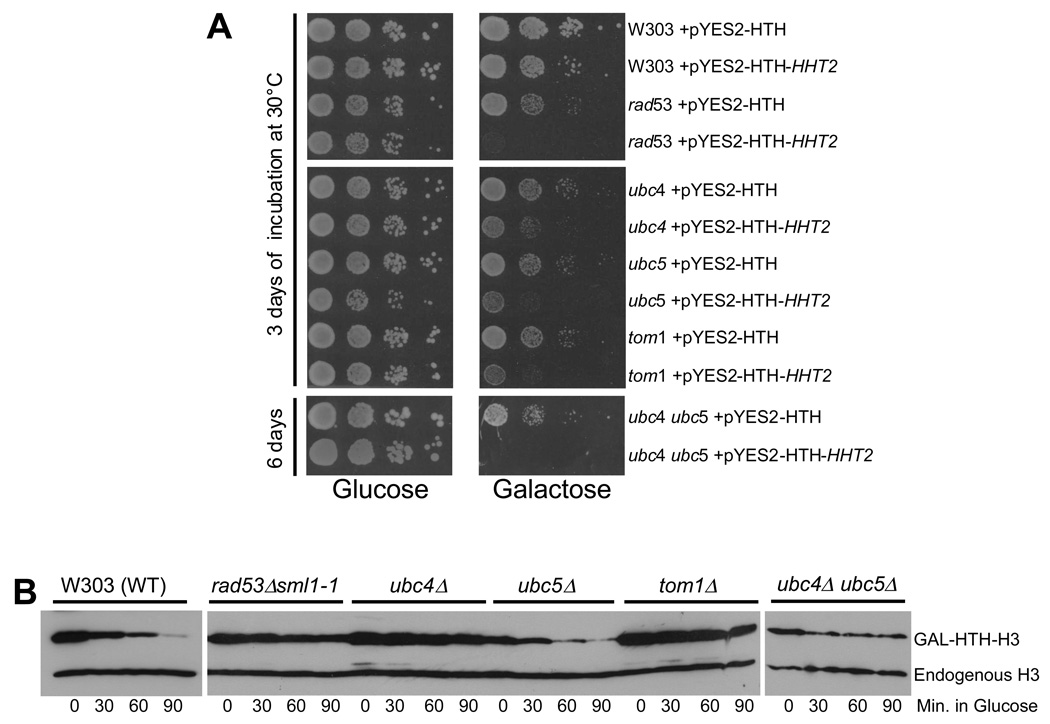
We next sought to identify the enzymes involved in ubiquitylating excess histones. Budding yeast has one E1 enzyme, Uba1, which is required for all ubiquitylation and is essential for viability33. There are thirteen E2 enzymes (Ubc1 to Ubc13), of which 11 are involved in ubiquitylation. Ubc9 is required for sumoylation and Ubc12 is needed for neddylation of proteins. We systematically screened strains carrying deletion or ts mutations of all the Ubc enzymes for sensitivity to histone H3 overexpression using a galactose inducible promoter, which identifies mutants defective in histone degradation by their poor growth on galactose media9. We found that ubc4 and ubc5 deletion strains were sensitive to histone overexpression, though not as sensitive as rad53 mutants (Figure 4A). Ubc4 and Ubc5 are 92% identical at the protein sequence level and function redundantly in protein degradation34. Hence, we constructed the ubc4 ubc5 double deletion strain, which proved as sensitive as rad53 mutants to histone overexpression (Figure 4A), implying that both Ubc4 and Ubc5 contribute to excess histone degradation. The remaining ubc mutants, including ubc2 (rad6), the E2 required for monoubiquitylation of histone H2B in vivo35, were insensitive to histone overexpression (Supplemental Figure S4), indicating that they are dispensable for histone degradation in vivo.
Figure 4. Identification of the putative E2 and E3 enzymes involved in the degradation related ubiquitylation of histones.
(A) Yeast strains lacking Ubc4, Ubc5 and Tom1 are sensitive to histone overexpression. Wild type W303 and congenic strains carrying deletions of genes encoding E2 or E3 enzymes were transformed with either a plasmid encoding galactose inducible, HIS10-TEV-HA (HTH) tagged histone H3 (pYES2-HTH-HHT2) or the empty vector (pYES2-HTH)9. The resulting transformants were grown overnight in minimal medium lacking uracil (to select for the plasmids) and containing 2% raffinose as carbon source. 10-fold serial dilutions of each strain were plated on minimal media minus uracil, with either glucose or galactose as carbon source. The plates were incubated for 3 days at 30°C, except for the slow growing ubc4 ubc5 double mutants which were incubated for 6 days. The rad53 deletion strain carries the sml1-1 null allele to suppress the lethality due to the loss of the essential function of Rad53.
(B) Disruption of the UBC4, UBC5 and TOM1 genes causes defects in the degradation of overexpressed histones. In vivo histone degradation assay with wild type (W303) and congenic mutant strains carrying the pYES2-HTH-HHT2 plasmid were performed as described in Methods. Cells were harvested every 30 minutes after switching to glucose. The levels of HTH-tagged H3 and chromosomal histone H3 were quantitated by Western blotting using the H3-C antibody9.
Yeast cells have many RING (Really Interesting New Gene) and HECT (Homologous to E6AP C-Terminus) domain containing proteins that are potential E3 ligases36, which makes the identification of any E3 ligase involved in histone degradation challenging. We screened yeast strains carrying deletions or ts mutations of all the known E3 enzymes in yeast for sensitivity to histone overexpression. Only cells carrying a deletion of the HECT domain ubiquitin ligase Tom1 were sensitive to histone overexpression (Figure 4A), whereas mutants corresponding to other known E3 ligases in yeast were insensitive (Supplemental Figure S5). This suggests that Tom1 is the likely E3 candidate, although we cannot rule out contribution from uncharacterized yeast E3 ligases in degradation related histone ubiquitylation.
ubc4-ubc5 and tom1 deletion mutants are defective in the degradation of excess histones
If UBC4-UBC5 and TOM1 are indeed involved in degradation related histone ubiquitylation, their mutants ought to be defective in histone degradation. We performed the histone degradation assay as described previously9 in Figure 1B to test this. Like the rad53 mutants, ubc4 and tom1 deletion strains were defective in degrading overexpressed HTH-H3 (Figure 4B). The ubc5 deletion strain degraded HTH-H3 as efficiently as wild type cells, while the ubc4 ubc5 double mutant was defective in histone degradation (Figure 4B). This is not surprising since Ubc4 and Ubc5 are functionally redundant34. Further, the requirement for Ubc5 in protein degradation is more critical in stationary phase cells34, whereas our degradation assays were performed with cells that had not reached stationary phase prior to G1 arrest. Ubc4 is more abundant than Ubc5 and they physically interact in vivo (Supplemental Figure S6A, B). Curiously, the Ubc4 protein is upregulated upon ubc5 deletion, whereas the Ubc5 protein is downregulated upon ubc4 deletion (Supplemental Figure S6B), which may further exacerbate the histone degradation defect in the ubc4 mutant. Not surprisingly, the ubc4 deletion strain, but not the ubc5 deletion mutant, exhibits a slight slow growth phenotype.
ubc4-ubc5 and tom1 deletion mutants accumulate high levels of endogenous histones bound to histone chaperones
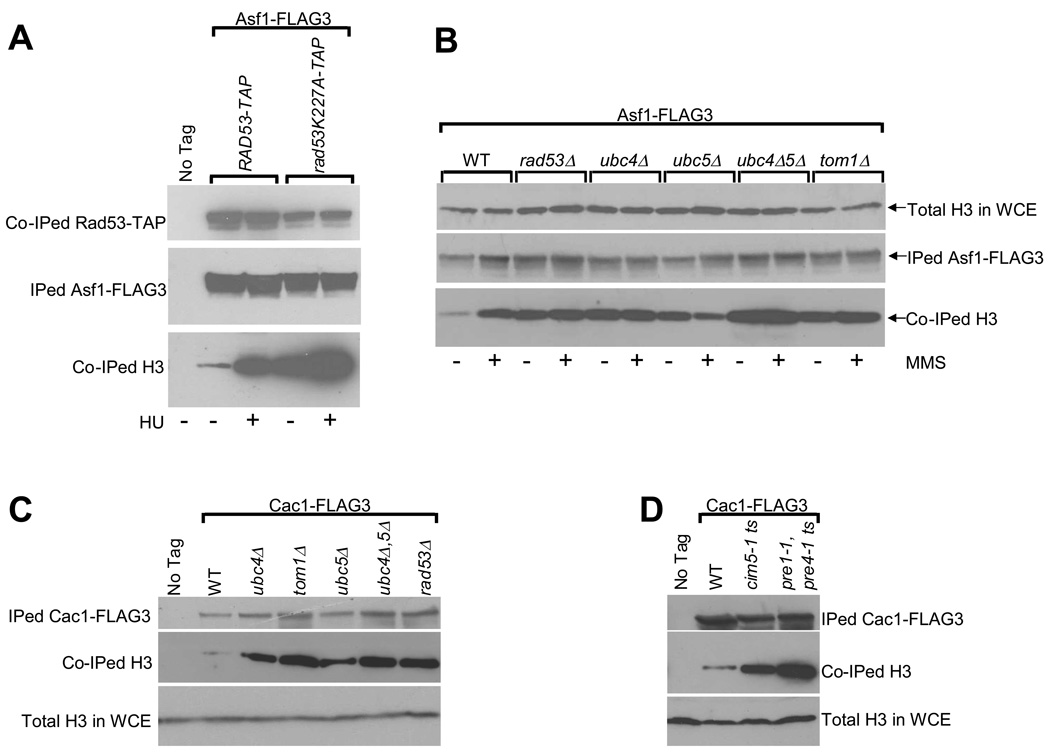
rad53 mutants defective in histone regulation accumulate high levels of endogenous histones on histone chaperones Cac1 and Hir29. Further, upon DNA damage or replication arrest, histone binding to chaperones increases dramatically. Here, we have investigated if mutants deficient in histone degradation accumulate high levels of endogenous histones on histone chaperone Asf137. Equal amounts of Asf1-FLAG3 were immunoprecipitated from exponentially growing cells carrying wild type RAD53-TAP or the “kinase dead” mutant38 rad53K227A-TAP. Compared to wild type cells, the rad53K227A mutant cells accumulated high levels of H3 on Asf1 both in the absence and presence of the replication inhibitor hydroxyurea (HU) (Figure 5A). We also immunoprecipitated Asf1-FLAG3 from wild type and E2/E3 mutant cells in the absence or presence of methyl methane sulfonate (MMS) induced alkylation damage that slows DNA synthesis39 and elicits excess histone accumulation 9. In the absence of MMS, small amounts of H3 were associated with Asf1-FLAG3 in wild type cells, which increased dramatically upon MMS treatment (Figure 5B). In contrast, rad53, ubc4, tom1 and, to a lesser extent, ubc5 deletion mutants showed high levels of H3 associated with Asf1-FLAG3 in the absence of DNA damage. No increase in histone binding to Asf1-FLAG3 was observed when these mutants were treated with MMS, suggesting that the histone binding capacity of Asf1 is saturated even in the absence of replication inhibition. We obtained identical results upon immunoprecipitating Cac1-FLAG3 where the E2 and E3 mutants showed higher levels of H3 associated with Cac1 (Figure 5C). Further, inhibition of proteasome function at 37°C in ts proteasomal mutants also resulted in the accumulation of H3 on Cac1-FLAG3 (Figure 5D).
Figure 5. Yeast cells lacking the factors involved in histone degradation accumulate excess endogenous histones bound to histone chaperones Asf1 and Cac1.
(A) Endogenous histone H3 accumulates on Asf1 in a rad53 “kinase dead” mutant38. Asynchronous cultures of the indicated strains carrying a FLAG3 epitope on the endogenous ASF1 gene were either left untreated, or treated with the DNA replication inhibitor hydroxyurea (HU) for 90 minutes. Cells were then harvested, WCEs prepared and Asf1-FLAG3 was immunoprecipitated (IPed) as described in Methods. The indicated IPed and co-immunoprecipitated (Co-IPed) proteins were detected by Western blotting. Note that although equal amounts of Asf1-FLAG3 were IPed in all the samples, the amount of co-IPed mutant rad53K227A-TAP protein was lower compared to the wild type Rad53-TAP due to the lower stability of the mutant protein.
(B) Deletion of the ubc4, ubc5 and tom1 genes results in an accumulation of excess endogenous histone H3 bound to Asf1. Asynchronous cultures of strains expressing the histone chaperone Asf1-FLAG3 were either left untreated or treated with the DNA alkylating agent methyl methane sulfonate (MMS, 0.033%) for 90 minutes and processed exactly as described above in (A). Total histone H3 levels in the WCEs are shown to demonstrate that roughly equal amounts of extracts were used for each immunoprecipitation reaction.
(C) Excess histone H3 accumulates on histone chaperone Cac1 upon the deletion of the ubc4, ubc5 and tom1 genes. The indicated strains carrying a FLAG3 epitope on the endogenous gene encoding Cac1 were harvested and processed as described above in (A) except that no treatment with genotoxic agents such as HU or MMS were carried out.
(D) Disruption of the proteasome function results in an accumulation of excess endogenous histone H3 bound to Cac1. Asynchronous cultures of the indicated temperature sensitive (ts) proteasome mutant strains expressing the histone chaperone Cac1-FLAG3 were grown overnight at 25°C and then shifted to the restrictive temperature of 37°C for 2.5 hours prior to harvesting the cells and processing them as described above in (C).
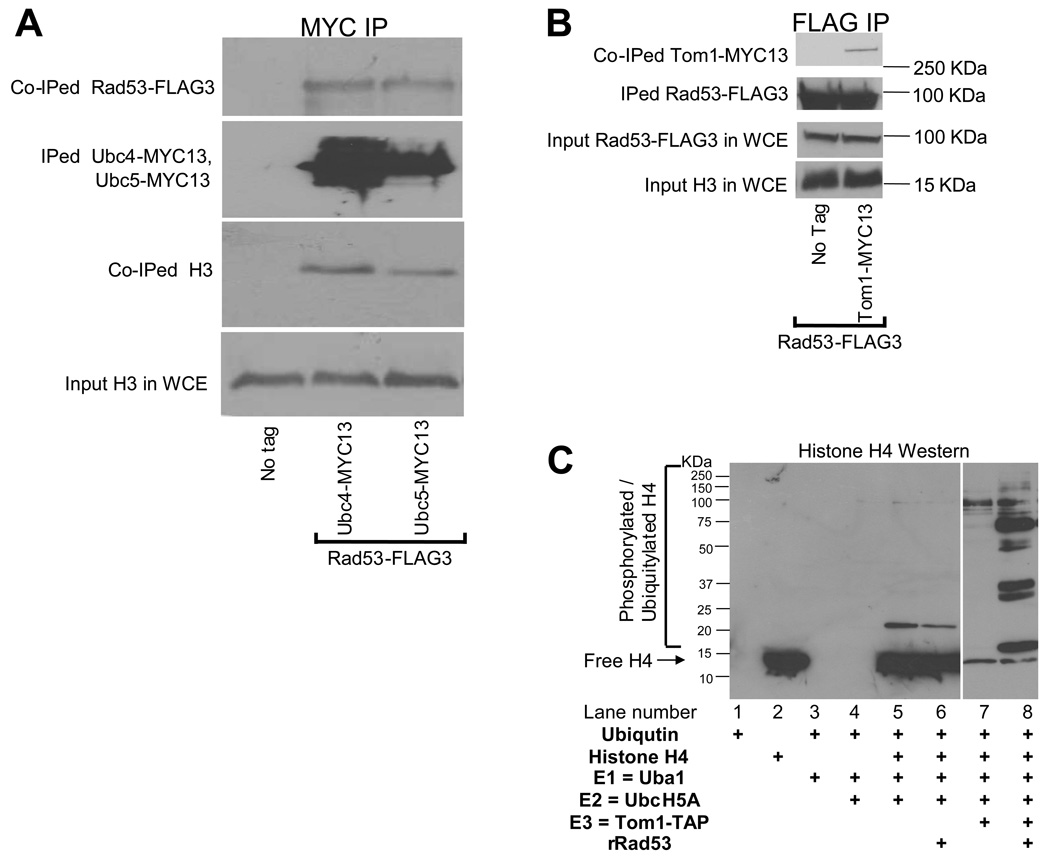
E2 and E3 enzymes involved in excess histones degradation interact with Rad53 in vivo and can ubiquitylate histones in vitro in a Rad53 dependent manner
Rad53 complexes contain free ubiquitin and modified histones (Figure 2B), suggesting that it may interact with the histone ubiquitylation machinery. To test for physical interactions among the factors involved in excess histone ubiquitylation, we tagged endogenous UBC4, UBC5 and TOM1 with the MYC13 epitope40 in wild type cells that were also expressing Rad53-FLAG3 from a plasmid (pRS316 RAD53 3xFLAG41). Then, we immunoprecipitated either the MYC-tagged proteins or Rad53-FLAG3 from WCEs and resolved them on SDS 4–12% gradient polyacrylamide gels. Western blotting with MYC, FLAG and H3-C antibodies was used to detect the respective tagged proteins (Figure 6A, B). Small amounts of histone H3 and Rad53-FLAG3 were co-immunoprecipitated with Ubc4-MYC13 and Ubc5-MYC13 (Figure 6A). We also clearly detected the presence of Tom1-Myc13 in Rad53-FLAG3 immunoprecipitates (Figure 6B). These results suggest that the enzymes involved in the Rad53-dependent ubiquitylation of excess histones do interact with each other, at least transiently. This observation encouraged us to reconstitute histone ubiquitylation in vitro using purified components. Our in vitro ubiquitylation reactions clearly demonstrate Rad53 and E2/E3 dependent H4 ubiquitylation (Figure 6C). However, mutations in these genes did not result in a reduction in the amount of ubiquitylated histones co-immunoprecipitated with Rad53 (data not shown). It is possible that ubiquitylation enzymes that do not normally act on histones in vivo are capable of non-specifically modifying histones associated with Rad53 under conditions of high effective local protein concentrations on sepharose beads during Rad53 immunoprecipitation.
Figure 6. Ubc4, Ubc5, Tom1 and Rad53 interact with each other in vivo and can ubiquitylate histones in vitro.
(A) The E2 enzymes Ubc4 and Ubc5 interact with Rad53 and histone H3. Asynchronous cultures of wild type (WT) strains expressing Rad53-FLAG3 and either Ubc4-MYC13 or Ubc5-MYC13 was harvested and WCEs prepared as described in Methods. Each WCE was immunoprecipitated (IPed) with MYC antibodies. The IPed material was resolved on a SDS 4–12% polyacrylamide gradient gel and processed for Western blotting to detect the IPed and co-IPed proteins. No protein was detected in the immunoprecipitated material from a WT strain lacking epitope tags (No tag). Total histone H3 levels in the WCEs are shown to demonstrate that roughly equal amounts of extracts were used for each immunoprecipitation reaction.
(B) Tom1 interacts with Rad53. Asynchronous cultures of the WT strain expressing either Rad53-FLAG3 alone or in combination with Tom1-MYC13 were harvested and processed as described above for (A). Rad53-FLAG3 was IPed with FLAG antibodies and processed for Western blotting to detect the IPed and co-IPed proteins. Total Rad53-FLAG3 and histone H3 levels in the WCEs are shown to demonstrate that roughly equal amounts of extracts were used for each immunoprecipitation reaction.
(C) In vitro ubiquitylation of histone H4 by E2 and E3 enzymes in a Rad53 dependent manner. Purified components were used to reconstitute the ubiquitylation of recombinant histone H4 in a Rad53 dependent manner in vitro as described in the Methods section and analyzed by Western blotting with H4 antibodies. Addition of E1 and E2 to histone H4 with or without Rad53 did not result in appreciable histone modifications (lanes 6 and 5 respectively) and neither did the addition of E3 in the absence of Rad53 (lane 7). Only the addition of Rad53 to a mixture of histone H4, ubiquitin, E1, E2 and E3 resulted in a dramatic stimulation of histone modifications, which are likely to be a mixture of ubiquitylation and phosphorylation (lane 8).
Discussion
In this study, we demonstrated that Rad53 triggers excess histone degradation via a phosphorylation/ubiquitylation/proteasome dependent pathway (Supplemental Figure S7). A freezer mill for preparing frozen WCEs under liquid nitrogen temperatures (−196°C) proved crucial for detecting extensive ubiquitylation of excess histones. Only unmodified histones were detected in Rad53-TAP complexes purified from WCEs prepared using a French press or a bead beater, despite the presence of protease inhibitors. Further, histones carrying modifications that signal their degradation occur only in association with Rad53 and their half-life is likely very short. Hence, it is hardly surprising that extensively ubiquitylated histones were not detected previously, despite the use of comprehensive mass spectrometric approaches that identified many ubiquitylated proteins42,43, including the known H2B monoubiquitylation35. Importantly, these considerations do not imply that excess histone degradation is rare, but merely indicate that extensively phosphorylated/ubiquitylated histones are only present as a very small fraction of total histones and cannot be detected simply by Western blotting of WCEs.
Our data suggests that the H3 Y99 residue is important for efficient degradation of this histone (Figure 1B), possibly upon phosphorylation (Figure 1A). At least some tyrosine residues on histones are buried within the nucleosome44,45 and these may only be accessible to a kinase in the context of excess non-chromosomal histones. The H3 Y99 residue is inaccessible in the nucleosome (Supplemental Figure S8), but may be available for phosphorylation in excess H3 not associated with chromatin. In vivo, newly synthesized histones associate as H2A-H2B or H3-H4 heterodimers for incorporation into chromatin37,46. Interestingly, the H3 Y99 residue is at the H3-H4 heterodimer interface and exhibits stacking interactions with H4 F61 residue, which contributes significantly towards stabilizing the heterodimer. An intriguing possibility is that phosphorylation of H3 Y99 may preclude or disrupt H3-H4 heterodimerization and thus prevent its deposition on chromatin, with degradation being its fate by default.
Phosphorylated tyrosines may not be the only marks that target degradation of histones other than H3. Indeed, we found that mutation of either all the tyrosine or serine/threonine residues alone did not compromise degradation of excess histone H4 (data not shown). In contrast, a mutant H4 lacking all putative phosphorylation sites (except serine 1, which is a stabilizing residue according to the N-end rule for protein degradation47) is degraded with slower kinetics than wild type H4 (Supplemental Figure S1B, C), despite the fact that it is efficiently recognized by Rad53 (Supplemental Figure S1D). This suggests that phosphorylation of serine, threonine and tyrosine residues contribute to excess H4 degradation.
In this study we provided evidence that extensively ubiquitylated forms of histones are degraded in vivo (Figure 2B, 2D and 2E; Supplemental Figure S3), although our current data cannot distinguish between monoubiquitylation of multiple lysines versus polyubiquitylation of one or more lysines on histones. Nevertheless, we believe that excess histones associated with Rad53 are polyubiquitylated, since polyubiquitin chains consisting of at least four ubiquitins linked via lysine 48 are needed for substrate presentation to proteasomes48. Although this currently represents a considerable technical challenge (owing to the limited amounts of excess histones and their complex modifications), mass spectrometric analysis of histones associated with Rad53 will be necessary to ascertain the exact structure of their ubiquitylation and identify key ubiquitylation sites that signal excess histone degradation.
We identified Ubc4 and Ubc5 as partially redundant E2 enzymes, and Tom1 as the E3 ligase involved in excess histone degradation (Figure 4, Figure 5 and Figure 6). Tom1 has been implicated in diverse cellular processes including mRNA export, transcriptional regulation and maintenance of nuclear structure49,50. Further, Npi46, a protein that binds to the histone H2B nuclear localization signal (NLS), suppresses tom1 mutations upon overexpression51. More interestingly, Ubc4-Ubc5 and Tom1-related enzymes are implicated in histone degradation during vertebrate spermiogenesis. To package the haploid genome within the narrow sperm head, histones are first replaced by transition proteins and then by protamines, while the displaced histones are ubiquitylated and proteolyzed26,52. A testis-specific isoform of UBC4 is induced during late stages of spermiogenesis in rat53,54. Remarkably, an ubiquitin ligase homologous to Tom1, known as LASU1 or E3Histone, was purified from bovine testis and functions with UBC4 to ubiquitylate all four core histones in vitro55. This strongly hints at evolutionary conservation of enzymes involved in histone degradation and based on our work, it will be interesting to determine whether histone phosphorylation by the Rad53 homolog CHK2 (or the highly related CHK1), or other kinases, is prerequisite to their degradation during spermiogenesis.
Excess histones increase the incidence of chromosome loss11 and sensitize cells to genotoxic agents9. Consistent with this, we find that ubc4 ubc5 and tom1 mutants are more sensitive to DNA damaging agents (Supplemental Figure S9) and exhibit higher rates of chromosome loss compared to wild type cells (Supplemental Figure S10). Interestingly, proteasomes are recruited to DNA double strand breaks (DSBs) and although proteasomal substrates relevant for DSB repair are not known56, histones are lost from the vicinity of a DSB57. Our results raise the exciting possibility that chromosomal histones may be a substrate for the proteasome at DSBs and this may depend upon Rad53, Ubc4-Ubc5 and Tom1. It will also be of interest to decipher and compare the molecular framework of Rad53-independent histone degradation pathways that eliminate mis-incorporated centromeric histone variant CENP-A from ectopic loci58,59, and histones covalently attached to DNA by oxidative stress60. In summary, in addition to their far-reaching implications for many DNA metabolic processes1, the principles that govern the degradation of excess histones uncovered here may also apply to other biological systems that necessitate regulated histone degradation.
METHODS
Yeast strains, plasmids and Western blotting
Yeast strains are listed in Supplemental Table 1 in the Supplemental Information section provided with the electronic version of the manuscript. The 2μ based, URA3 marker and GAL1 promoter carrying pYES2 vector (Invitrogen) was used to sub-clone the HIS10-TEV-HA (HTH) epitope tag downstream of the GAL1 promoter between the HindIII and KpnI restriction sites in the Multiple Cloning Site (MCS) to generate the empty vector (pYES2-HTH) used in our studies. Histone H3 encoded by the HHT2 gene was then inserted downstream of the HTH tag between the EcoRI and XbaI restriction sites of the MCS to generate the H3 overexpression plasmid pYES2-HTH-HHT2 (previously referred to as 2μ-URA3-GAL-HIS10-TEV-HHT29). Our antibodies and Western blotting procedure have been described in detail elsewhere9.
Immunoprecipitation
For routine immunoprecipitation of tagged proteins, cells were harvested from 1-liter cultures at ~2×107 cells/ml. For Rad53-TAP immunoprecipitation, unless indicated otherwise, 4L cultures were used for each lane on a Western Blot, with the exception of the experiments shown in Figure 1C and Supplemental Figure S1D, where 0.5L cultures were used. Whole-cell extracts (WCEs) were prepared by grinding cells in liquid nitrogen in a SPEX CertiPrep 6850 Freezer Mill in 20ml lysis buffer (per liter of yeast culture) containing protease, phosphatase and proteasome inhibitors (20mM Hepes-KOH pH 7.5, 110mM potassium acetate, 10% glycerol, 0.1% Tween-20, 1mM sodium vanadate, 50mM sodium fluoride, 50mM sodium β-glycerophosphate, 10mM sodium butyrate, 10mM 2-mercaptoethanol, 2X Roche protease inhibitor cocktail, 10µM MG-132 proteasome inhibitor). Extracts from equal amounts of cells were incubated overnight at 4°C with FLAG M2 antibody resin (Sigma), 9E10 antibody sepharose beads, 3F10 antibody resin (Roche) or IgG sepharose (Amersham Biosciences) to immunoprecipitate FLAG, MYC, HA and TAP- tagged proteins respectively. The immunoprecipitated material was resolved on a 4–12% polyacrylamide gradient gel (BioRad) and processed for Western blotting as described previously9 using specific antibodies to detect the presence of FLAG (M2 from Sigma), MYC (4A6 from Millipore), HA (HA.11 from Covance) or TAP (TAP from Open Biosystems) tagged immunoprecipitated and co-immunoprecipitated proteins.
In vivo histone degradation assay
Degradation assays were carried out for overexpressed tagged histones in the indicated strains essentially as described previously9. Briefly, cells synchronized in G1 with α-factor were treated with galactose (GAL) for 90 minutes to induce the tagged histone, after which glucose (GLU) was added to repress the galactose promoter in the continued presence of α-factor. Aliquots of cells were harvested at the indicated time intervals after the addition of glucose and frozen in liquid nitrogen. Whole cell lysates were prepared as described previously61 and resolved on SDS-18% polyacrylamide gels. The levels of endogenous H3, as well as HTH tagged H3, were quantitated by Western blotting with histone H3-C antibodies as described previously9. In Supplemental Figure S1B, the levels of H4-HA3 were quantitated using HA antibodies while endogenous chromosomal H4 was detected using polyclonal H4 antibodies9.
In vitro ubiquitylation assay
Purified components that were either commercially available or prepared in our laboratory were used to reconstitute the ubiquitylation of recombinant histone H4 in a Rad53 dependent manner in vitro. Reactions were carried out in 25mM Tris-HCl pH8.0, 125mM NaCl, 2mM MgCl2, 50µM DTT, 2mM ATP, 1µM ubiquitin aldehyde (inhibitor of deubiquitylases) and 10µM MG132 (proteasomal inhibitor). Where indicated, the reaction was supplemented with the following: 1µg recombinant human histone H4 (New England Biolabs), 1µg recombinant 6-His-tagged human ubiquitin, 0.05µM E1 enzyme Uba1 from yeast, 0.5µM E2 enzyme recombinant GST-UbcH5A (human homolog of yeast Ubc4 which exhibits 81% identity and 94% similarity and can complement the loss of Ubc4 from yeast62) (all from Boston Biochem), 50ng of recombinant 6His-tagged Rad53 and 50ng of Tom1-TAP purified from yeast. Reactions were carried out in a total volume of 15µl and incubated for 1h at 30°C, following which 5µl of 4x SDS-PAGE loading buffer was added and the ubiquitylated proteins and the entire reaction was resolved on a 18% SDS-polyacrylamide gel. The gel was processed for Western Blotting using a H4 antibody described previously9.
Supplementary Material
Acknowledgements
We wish to thank Drs. John Diffley, Dan Finley, Mark Hochstrasser, Stefan Jentsch, Noel Lowndes, Carl Mann, Hiroshi Masumoto, Mary Ann Osley, Rodney Rothstein, David Stern, David Stillman, Jesper Svejstrup, Alain Verreault and Yanchang Wang for strains and reagents. We thank Drs. Alain Verreault and David Brown for critical reading of this manuscript. We also thank Dr. Munah Abdul-Rauf for construction of the pYES2-HTH and pYES2-HTH-HHT2 plasmids, as well as the H4 mutants used in the supplemental data. We thank undergraduate students Shari Eckley and Melanie Gonzalez for technical assistance with the harvesting of many liters of yeast cultures for our experiments. Research in AG’s laboratory is supported by a Bankhead-Coley Cancer Research Program grant (07BN-02) from the Florida Department of Health, while research in JP's laboratory is funded by a NIH grant (R15GM079678-01).
References
- 1.Gunjan A, Paik J, Verreault A. The emergence of regulated histone proteolysis. Curr. Opin. Genet. Dev. 2006;16:112–118. doi: 10.1016/j.gde.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 2.Han M, Chang M, Kim UJ, Grunstein M. Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication, and transcription. Cell. 1987;48:589–597. doi: 10.1016/0092-8674(87)90237-6. [DOI] [PubMed] [Google Scholar]
- 3.Eriksson PR, Mendiratta G, McLaughlin NB, Wolfsberg TG, Marino-Ramirez L, Pompa TA, Jainerin M, Landsman D, Shen CH, Clark DJ. Global regulation by the yeast Spt10 protein is mediated through chromatin structure and the histone upstream activating sequence elements. Mol. Cell. Biol. 2005;25:9127–9137. doi: 10.1128/MCB.25.20.9127-9137.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Myung K, Pennaneach V, Kats ES, Kolodner RD. Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc. Natl. Acad. Sci. USA. 2003;100:6640–6645. doi: 10.1073/pnas.1232239100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson DM, Ye X, Hall C, Santos H, Ma T, Kao GD, Yen TJ, Harper JW, Adams PD. Coupling of DNA synthesis and histone synthesis in S phase independent of cyclin/cdk2 activity. Mol. Cell. Biol. 2002;22:7459–7472. doi: 10.1128/MCB.22.21.7459-7472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye X, Franco AA, Santos H, Nelson DM, Kaufman PD, Adams PD. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol. Cell. 2003;11:341–351. doi: 10.1016/s1097-2765(03)00037-6. [DOI] [PubMed] [Google Scholar]
- 7.Bonner WM, Wu RS, Panusz HT, Muneses C. Kinetics of accumulation and depletion of soluble newly synthesized histone in the reciprocal regulation of histone and DNA synthesis. Biochemistry. 1988;27:6542–6550. doi: 10.1021/bi00417a052. [DOI] [PubMed] [Google Scholar]
- 8.Groth A, Ray-Gallet D, Quivy JP, Lukas J, Bartek J, Almouzni G. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol. Cell. 2005;17:301–311. doi: 10.1016/j.molcel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 9.Gunjan A, Verreault A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell. 2003;115:537–549. doi: 10.1016/s0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- 10.Steger DJ, Workman JL. Transcriptional analysis of purified histone acetyltransferase complexes. Methods. 1999;19:410–416. doi: 10.1006/meth.1999.0877. [DOI] [PubMed] [Google Scholar]
- 11.Meeks-Wagner D, Hartwell LH. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell. 1986;44:43–52. doi: 10.1016/0092-8674(86)90483-6. [DOI] [PubMed] [Google Scholar]
- 12.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 13.Osley MA. The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- 14.Kaygun H, Marzluff WF. Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol. Cell. Biol. 2005;25:6879–6888. doi: 10.1128/MCB.25.16.6879-6888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan KL, Dixon JE. Protein tyrosine phosphatase activity of an essential virulence determinant in Yersinia. Science. 1990;249:553–556. doi: 10.1126/science.2166336. [DOI] [PubMed] [Google Scholar]
- 16.Stern DF, Zheng P, Beidler DR, Zerillo C. Spk1, a new kinase from Saccharomyces cerevisiae, phosphorylates proteins on serine, threonine, and tyrosine. Moll. Cell. Biol. 1991;11:987–1001. doi: 10.1128/mcb.11.2.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 18.Dai J, Hyland EM, Yuan DS, Huang H, Bader JS, Boeke JD. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell. 2008;134:1066–1078. doi: 10.1016/j.cell.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakanishi S, Sanderson BW, Delventhal KM, Bradford WD, Staehling-Hampton K, Shilatifard A. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat. Struct. Mol. Biol. 2008;15:881–888. doi: 10.1038/nsmb.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wittschieben BO, Fellows J, Du W, Stillman DJ, Svejstrup JQ. Overlapping roles for the histone acetyltransferase activities of SAGA and elongator in vivo. EMBO J. 2000;19:3060–3068. doi: 10.1093/emboj/19.12.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hershko A, Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 22.Commerford SL, Carsten AL, Cronkite EP. Histone turnover within nonproliferating cells. Proc. Natl. Acad. Sci. USA. 1982;79:1163–1165. doi: 10.1073/pnas.79.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsvetkov S, Ivanova E, Djondjurov L. Metabolic behaviors of the core histones in proliferating Friend cells. Exp. Cell. Res. 1989;180:94–105. doi: 10.1016/0014-4827(89)90215-2. [DOI] [PubMed] [Google Scholar]
- 24.Wunsch AM, Lough J. Histones synthesized at different stages of myogenesis are differentially degraded in myotube cells. J. Cell Physiol. 1989;141:97–102. doi: 10.1002/jcp.1041410115. [DOI] [PubMed] [Google Scholar]
- 25.Haas AL, Bright PM. The immunochemical detection and quantitation of intracellular ubiquitin-protein conjugates. J. Biol. Chem. 1985;260:12464–12473. [PubMed] [Google Scholar]
- 26.Chen HY, Sun JM, Zhang Y, Davie JR, Meistrich ML. Ubiquitination of histone H3 in elongating spermatids of rat testes. J. Biol. Chem. 1998;273:13165–13169. doi: 10.1074/jbc.273.21.13165. [DOI] [PubMed] [Google Scholar]
- 27.Geng F, Tansey WP. Polyubiquitylation of histone H2B. Mol. Biol. Cell. 2008;19:3616–3624. doi: 10.1091/mbc.E08-01-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spence J, Gali RR, Dittmar G, Sherman F, Karin M, Finley D. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell. 2000;102:67–76. doi: 10.1016/s0092-8674(00)00011-8. [DOI] [PubMed] [Google Scholar]
- 29.Davie JR. Peptide mapping of basic proteins by proteolysis in acetic acid/urea-minislab polyacrylamide gels. Anal. Biochem. 1985;144:522–526. doi: 10.1016/0003-2697(85)90149-6. [DOI] [PubMed] [Google Scholar]
- 30.Li W, Nagaraja S, Delcuve GP, Hendzel MJ, Davie JR. Effects of histone acetylation, ubiquitination and variants on nucleosome stability. Biochem. J. 1993;296:737–744. doi: 10.1042/bj2960737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hilt W, Enenkel C, Gruhler A, Singer T, Wolf DH. The PRE4 gene codes for a subunit of the yeast proteasome necessary for peptidylglutamyl-peptide-hydrolyzing activity. Mutations link the proteasome to stress- and ubiquitin-dependent proteolysis. J. Biol. Chem. 1993;268:3479–3486. [PubMed] [Google Scholar]
- 32.Ghislain M, Udvardy A, Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- 33.McGrath JP, Jentsch S, Varshavsky A. UBA1: an essential yeast gene encoding ubiquitin-activating enzyme. EMBO J. 1991;10:227–236. doi: 10.1002/j.1460-2075.1991.tb07940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seufert W, Jentsch S. Ubiquitin-conjugating enzymes Ubc4 and Ubc5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 1990;9:543–550. doi: 10.1002/j.1460-2075.1990.tb08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 36.Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays Biochem. 2005;41:15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- 37.English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Di Fiore PP, Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paulovich AG, Hartwell LH. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 40.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae . Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 41.Lee SJ, Schwartz MF, Duong JK, Stern DF. Rad53 phosphorylation site clusters are important for Rad53 regulation and signaling. Mol. Cell. Biol. 2003;23:6300–6314. doi: 10.1128/MCB.23.17.6300-6314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayor T, Lipford JR, Graumann J, Smith GT, Deshaies RJ. Analysis of polyubiquitin conjugates reveals that the Rpn10 substrate receptor contributes to the turnover of multiple proteasome targets. Mol. Cell. Proteomics. 2005;4:741–751. doi: 10.1074/mcp.M400220-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat. Biotech. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 44.Santisteban MS, Arents G, Moudrianakis EN, Smith MM. Histone octamer function in vivo: mutations in the dimer-tetramer interfaces disrupt both gene activation and repression. EMBO J. 1997;16:2493–2506. doi: 10.1093/emboj/16.9.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White CL, Suto RK, Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 47.Varshavsky A. The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Utsugi T, Hirata A, Sekiguchi Y, Sasaki T, Toh-e A, Kikuchi Y. Yeast tom1 mutant exhibits pleiotropic defects in nuclear division, maintenance of nuclear structure and nucleocytoplasmic transport at high temperatures. Gene. 1999;234:285–295. doi: 10.1016/s0378-1119(99)00197-3. [DOI] [PubMed] [Google Scholar]
- 50.Saleh A, Collart M, Martens JA, Genereaux J, Allard S, Cote J, Brandl CJ. TOM1p, a yeast hect-domain protein which mediates transcriptional regulation through the ADA/SAGA coactivator complexes. J. Mol. Biol. 1998;282:933–946. doi: 10.1006/jmbi.1998.2036. [DOI] [PubMed] [Google Scholar]
- 51.Shan X, Xue Z, Mélèse T. Yeast NPI46 encodes a novel prolyl cis-trans isomerase that is located in the nucleolus. J. Cell Biol. 1994;126:853–862. doi: 10.1083/jcb.126.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meistrich ML. Boca Raton: CRC Press; 1989. Histones and other basic nuclear proteins. [Google Scholar]
- 53.Rajapurohitam V, Morales CR, El-Alfy M, Lefrançois S, Bedard N, Wing SS. Activation of a UBC4-dependent pathway of ubiquitin conjugation during postnatal development of the rat testis. Dev. Biol. 1999;212:217–228. doi: 10.1006/dbio.1999.9342. [DOI] [PubMed] [Google Scholar]
- 54.Wing SS, Bedard N, Morales C, Hingamp P, Trasler J. A novel rat homolog of the Saccharomyces cerevisiae ubiquitin-conjugating enzymes Ubc4 and Ubc5 with distinct biochemical features is induced during spermatogenesis. Mol. Cell. Biol. 1996;16:4064–4072. doi: 10.1128/mcb.16.8.4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Z, Oughtred R, Wing SS. Characterization of E3Histone, a novel testis ubiquitin protein ligase which ubiquitinates histones. Mol. Cell. Biol. 2005;25:2819–2831. doi: 10.1128/MCB.25.7.2819-2831.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krogan NJ, Lam MH, Fillingham J, Keogh MC, Gebbia M, Li J, Datta N, Cagney G, Buratowski S, Emili A, Greenblatt JF. Proteasome involvement in the repair of DNA double-strand breaks. Mol. Cell. 2004;16:1027–1034. doi: 10.1016/j.molcel.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 57.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Collins KA, Furuyama S, Biggins S. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 2004;14:1968–1972. doi: 10.1016/j.cub.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 59.Moreno-Moreno O, Torras-Llort M, Azorin F. Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila, to centromeres. Nucleic Acids Res. 2006;34:6247–6255. doi: 10.1093/nar/gkl902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ullrich O, Reinheckel T, Sitte N, Hass R, Grune T, Davies KJ. Poly-ADP ribose polymerase activates nuclear proteasome to degrade oxidatively damaged histones. Proc. Natl. Acad. Sci. USA. 1999;96:6223–6228. doi: 10.1073/pnas.96.11.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 62.Matuschewski K, Hauser HP, Treier M, Jentsch S. Identification of a novel family of ubiquitin-conjugating enzymes with distinct amino-terminal extensions. J. Biol. Chem. 1996;271:2789–2794. doi: 10.1074/jbc.271.5.2789. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.