Abstract
CFTR chloride channels are encoded by the gene mutated in patients with cystic fibrosis. These channels belong to the superfamily of ABC transporter ATPases. ATP-driven conformational changes, which in other ABC proteins fuel uphill substrate transport across cellular membranes, in CFTR open and close a gate to allow transmembrane flow of anions down their electrochemical gradient. New structural and biochemical information from prokaryotic ABC proteins and functional information from CFTR channels has led to a unifying mechanism explaining those ATP-driven conformational changes.
The all-too-common deadly disease of young people, cystic fibrosis (CF), is caused by failure of a chloride-ion channel with the unwieldy name of CFTR, standing for cystic fibrosis transmembrane conductance regulator. Anion flow through this channel is needed for normal function of epithelia such as those that line airways and the intestinal tract and ducts in the pancreas, testes and sweat glands. Without anion flow, water movement slows, and dehydrated mucus clogs ducts and collects in the lung where it fosters ultimately lethal bacterial infections. The amino-acid sequence of CFTR immediately identified it as a member of the superfamily of ATP-binding cassette (ABC) transporter ATPases. Among the thousands of ABC family members, only CFTR is an ion channel. High-resolution electrophysiological studies of single CFTR channels therefore offer a unique window into mechanisms of ABC transporter function. And recent structural and biochemical advances in the study of bacterial ABC proteins have shed light on how opening and closing of the CFTR chloride channel are regulated.
Here we bring together the latest information on CFTR-channel function and on prokaryotic ABC-transporter structure to provide an up-to-the-minute view of what CFTR channels normally do, what a CFTR channel might look like and how its anion pathway is opened and closed. The gate that regulates chloride-ion flow through the membrane-spanning pore in CFTR is thought to be opened when ATP binds to CFTR’s two cytoplasmic nucleotide-binding domains (NBD1 and NBD2) and, acting as molecular glue, holds them together. Hydrolysis of one of the ATPs disrupts the NBD1–NBD2 interaction and closes the channel gate, terminating anion flow. Kinase-mediated phosphorylation of a cytoplasmic regulatory domain in CFTR is needed for successful transmission of NBD events to the channel gate.
Evolutionary conservation of structural components of the NBD1–NBD2 interface makes it probable that a similar cycle of ATP binding and hydrolysis drives function in most ABC family members. Even so, important differences between NBD1 and NBD2 in CFTR seem to have evolved to allow only one of the bound ATPs to be hydrolysed while the other remains unaltered during numerous gating cycles. Similar differences in the NBDs are found in CFTR’s closest relatives in the human genome. These are the multidrug-resistance-related proteins (MRPs), which are ATP-driven efflux pumps, and the sulphonylurea receptors (SURs), which associate with inward rectifier potassium-ion channels to form nucleotide-sensitive potassium conductance (see the review in this issue by Nichols, p. 470). So mechanisms inferred for CFTR are likely to apply also to MRP and SUR.
Cystic fibrosis and CFTR
In 1989, when the gene mutated in CF patients was identified by positional cloning1,2, the expectation was that it would encode a chloride-ion channel. This was because CF epithelia behaved as though they were impermeable to chloride3, and chloride channels could be activated by cAMP-dependent protein kinase (PKA) in normal but not in CF airway epithelium4,5. So when the mutant gene was found to encode an ABC transporter homologue, with several consensus sites for phosphorylation by PKA, its product was suggested to regulate chloride channels by pumping out a cytoplasmic inhibitor: hence the name, CFTR2. But there were more surprises. CFTR itself was shown to be a chloride channel, directly activated by phosphorylation by PKA (Fig. 1). This channel allowed chloride ions to flow equally readily in either direction, though at modest rates (conductance ≤10 pS; for example, see ref. 6). However, the regulated channels previously studied in airway epithelia4,5 conducted chloride ions rapidly (conductance ≥30 pS) and preferentially into the cell, and so were not CFTR channels. How, and even whether, the function of these and other channels and transporters is modulated by interaction with CFTR is still unsettled7,8.
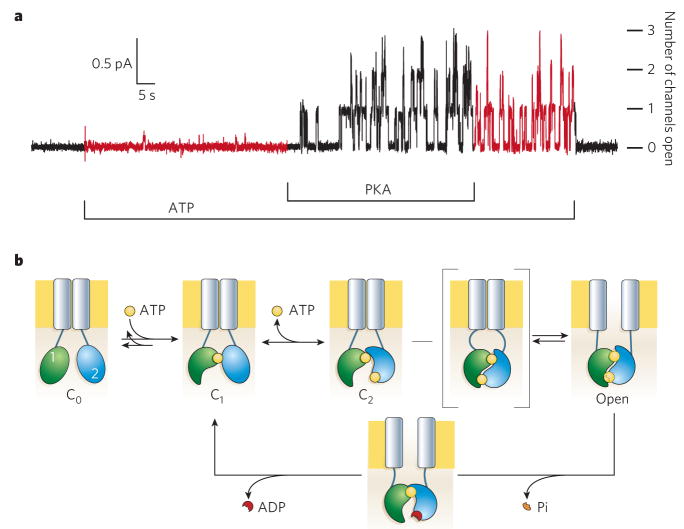
Figure 1. Opening and closing of CFTR channels.
a, CFTR’s regulatory (R) domain must be phosphorylated by PKA before ATP is able to support channel opening. The recording shows chloride current flow through individual CFTR channels upon opening. Endogenous membrane-attached phosphatases partly dephosphorylate the R domain on PKA withdrawal, reducing the probability of finding a channel open. But channels do not stop opening until ATP is removed; numbers of simultaneously open channels are indicated at the right. b, Present structural interpretation of ATP-dependent gating cycle of phosphorylated CFTR channels. The R domain is omitted. ATP (yellow) remains tightly bound to NBD1 (green) Walker motifs for several minutes, during which time many closed–open–closed gating cycles occur. ATP binding to NBD2 (blue) is followed by a slow channel opening step (C2-to-Open) that proceeds through a transition state (square brackets) in which the intramolecular NBD1–NBD2 tight heterodimer is formed but the transmembrane pore (grey rectangles) has not yet opened. The relatively stable open state becomes destabilized by hydrolysis of the ATP bound at the NBD2 composite catalytic site and loss of the hydrolysis product, inorganic phosphate (Pi). The ensuing disruption of the tight dimer interface leads to channel closure.
Although two-thirds of all CF disease cases can be attributed to a single mutation in CFTR, deletion of Phe 508 in NBD1, over 1,000 CF-disease-associated mutations of this 1,480-amino-acid protein have been logged (http://www.genet.sickkids.on.ca/cftr/). ΔF508-CFTR protein fails to mature properly and is tagged for degradation9. But cell incubation at room, rather than blood, temperature leads to maturation, trafficking and insertion of ΔF508 CFTR into the surface membrane, where it functions nearly as well as wild type. Much effort is therefore being directed to identifying small molecules that might chaperone ΔF508 to the cell surface in CF patients. Other CF-causing mutations lead to truncated protein or to full-length channels with reduced chloride-transport capacity because they either spend a smaller fraction of time open, or when open, conduct chloride ions more slowly than normal. Most disease-causing mutations have yet to be functionally characterized.
CF is a pleiotropic disease affecting all exocrine epithelia. Morbidity and mortality are largely due to chronic bacterial infection and inflammation in the lung. Inhaled DNase can ameliorate thickening of airway mucus by aggregated DNA from dead cells. Although the underlying pathophysiology of CF is still debated10, the simplest hypothesis is that impaired anion movement is the primary cause. In the handful of cases examined, severity of disease seemed related to impairment of channel function. Therapeutic efforts are therefore centred on restoring the activity of mutant CFTR or enhancing alternative pathways for anion flow. Gene therapy trials to introduce CFTR DNA into airway epithelial cells began over a decade ago, but difficulties with gene delivery and expression11 have led to refocusing of efforts to rescue ΔF508 CFTR.
Overview of CFTR structure and function
As ABC proteins occupy up to 5% of each prokaryotic genome sequenced, there are thousands of examples of them, although few have been studied in any depth. They conform to a modular architecture, thought to comprise two transmembrane domains (TMDs), with typically six membrane-spanning α-helices each, and two cytoplasmic NBDs. Individual bacterial genes often encode single domains or fused pairs of domains (TMD–TMD, NBD–NBD or TMD–NBD), but three-quarters of the 48 human ABC genes (assigned to seven subfamilies) encode full-length ABC proteins12. In the case of CFTR, a unique regulatory (R) domain, the target of PKA phosphorylation, links two homologous halves, giving the overall domain organization TMD1–NBD1–R–TMD2–NBD2 (ref. 2).
A common assertion, rarely tested, is that a functional ABC protein contains two TMDs and two NBDs, as found in crystal structures of the bacterial ABC transporters MsbA13 and BtuCD14 and in a low-resolution structure of the multidrug transporter P-gp from two-dimensional (2D) cryo-electron crystallography15. For CFTR, this means a single polypeptide, and functional measurements have addressed this question. Two CFTR polypeptides can associate16, at least through their carboxy-terminal PDZ-domain-binding motifs17. However, co-expression of two kinds of CFTR channels with distinct functional characteristics produced no hybrid channels18, and cysteine-modification tests showed that a single introduced cysteine contributed only once to a CFTR pore19. So each CFTR channel seems to be constructed from just one CFTR polypeptide.
It seems that negatively charged ions, such as chloride, accumulate near positively charged regions at each end of the pore in CFTR’s trans-membrane domain20,21 and flow through the pore after the opening of its gate by events initiated by ATP binding to the cytoplasmic NBDs (Figs 1, 2; N-terminal NBD1, green; C-terminal NBD2, blue). Before events at CFTR’s NBDs can trigger gating of the pore, the R domain must be phosphorylated by PKA22 (Fig. 1a). Once phosphorylated, CFTR channels need ATP to open normally; they close within 1 s of ATP withdrawal (Fig. 1a) and tend to stay shut until ATP is restored. A simple scheme linking these events to the pore gate (Fig. 1b) can account for most of the experimental evidence, although many details remain to be clarified. Once ATP binds to homologous nucleotide-interacting motifs in the two NBDs, these domains are thought to approach each other closely, sandwiching two ATPs in the NBD1–NBD2 interface. Upon this intramolecular heterodimer-like interaction, a signal would be transmitted through cytoplasmic-linking domains to open the gate in the transmembrane domain (Fig. 1b, grey). That channel-opening signal would be sustained until hydrolysis of one of the ATPs leads to disruption of the NBD1–NBD2 interface and separation of the NBDs. Loss of the signal allows the channel gate to close, terminating anion flow until ATP again binds to the NBDs.
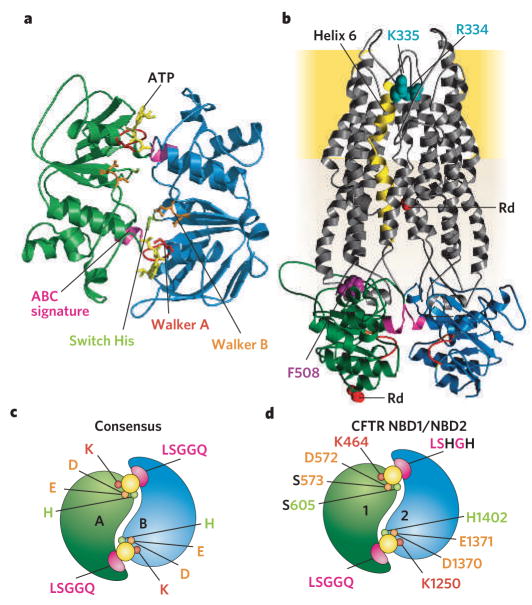
Figure 2. Possible structure and organization of domains in CFTR.
a, Homology model of CFTR NBD heterodimer (NBD1 in green, NBD2 in blue) based on several ATP (yellow) bound NBD crystals, including human CFTR NBD1 F508A31 and dimeric MJ079625, MalK26 and HlyB27. b, Highly speculative homology model of CFTR including its two TMDs (grey) modelled on bacterial MsbA structures13. Each TMD comprises cytoplasmic segments that link membrane-embedded α-helices (one directly) to the NBDs (colour-coded as in a). Membrane-spanning helix 6 (gold) has been implicated in forming part of the anion-permeation pathway, and positively charged residues (blue-green) in it are thought to recruit anions into an outer vestibule. In CF patients, deletion of Phe 508 (purple), at the interface between NBD1 and TMD1 causes post-translational misfolding and degradation9. c, d, Illustrations contrasting NBD-homodimer (c; for example, MJo796 and MalK) consensus catalytic site residues (for example, MJ0796 aand MalK) and expected CFTR NBD1–NBD2 heterodimer (d) with only one consensus catalytic site.
Nucleotide-binding domains
High-resolution structures are not yet available for any full-length eukaryotic ABC protein but have been obtained not only for the prokaryotic MsbA and BtuCD entire transporters but also for several NBDs (for example, see refs 23–27). The NBD structures all reveal the same basic fold, with highly conserved sequence motifs positioned to interact with bound ATP (Fig. 2a).
In an ATP-bound NBD monomer23, the phosphate chain of the nucleotide is cradled by a canonical P-loop, or Walker28 A sequence (red, Fig. 2). This lies at the N terminus of a helix adjacent to a parallel β-sheet tipped by loops (one including the Walker B sequence (orange, Fig. 2), another including a ‘switch’ His (light green, Fig. 2)) that contact the γ phosphate of ATP in an F1-ATPase-like ATP-binding core. On the other side of that helix, an extended loop arising from the middle of an ABC-specific anti-parallel β-region contains an aromatic residue against which the nucleotide base stacks. Evident lack of nucleotide interaction with the conserved ‘ABC signature’ sequence (consensus LSGGQ; pink, Fig. 2), some 15 Å distant in the so-called α-helical domain (also containing Phe 508; Fig. 2b), was initially puzzling. The puzzle was solved by biochemical demonstration of nucleotide-induced dimerization of bacterial NBDs29, and by crystal structures that showed rotationally symmetric ‘head-to-tail’ NBD homodimers25–27 (compare with Fig. 2a). These nucleotide-bound NBD dimers enclosed two ATP molecules in interfacial composite sites, each comprising ATP-interacting motifs from the ‘head’ of one monomer and signature sequence residues from the ‘tail’ of the other (Fig. 2a, c).
Recent crystal structures30–32 of NBD1 of CFTR show that it, too, conforms to the established NBD architecture, although its ABC-specific β-sheet includes a long, inserted loop, not found in other NBDs, and whose shape and position are still unclear. Assuming that NBD1 and NBD2 in CFTR approach each other in the way that bacterial NBDs do in the homodimeric crystals (as illustrated in Fig. 2c, d) allows construction of speculative homology models of CFTR (Fig. 2a, b), based on the structures of prokaryotic NBD dimers and of MsbA. These structural diagrams serve to illustrate the relative positioning of the transmembrane domains, cytoplasmic linking segments and NBDs, and concur with a low-resolution map33 from cryo-electron crystallography of 2D crystals of CFTR. Much remains uncertain, however, including the precise structure of the TMDs, where homology between MsbA and CFTR is low. The structure and disposition of the R domain, lacking in other ABC proteins, is completely unknown and so is omitted from Fig. 2b (it must somehow connect the red spacefill residues, marked Rd).
A key distinction between homodimeric interactions seen so far in crystal structures of bacterial ABC proteins and the proposed NBD1–NBD2 interaction in CFTR is the asymmetry of the two expected interfacial nucleotide-binding sites, echoing the mere 27% identity between CFTR’s NBD1 and NBD2. In contrast to the two essentially identical catalytic sites found in bacterial NBD homodimers24–27, each with its full complement of conserved consensus residues (Fig. 2c), the expected NBD1–NBD2 ‘heterodimeric’ arrangement comprises only one consensus catalytic site (Fig. 2d). We call that site the ‘NBD2 composite’ site, as NBD2 contributes its nucleotide-binding residues of the Walker motifs, the residues that hold ATP in ATP-bound NBD monomers. The other interfacial site, the ‘NBD1 composite’ site, includes non-canonical residues from NBD1 Walker B (Ser instead of Glu) and switch (Ser instead of His) motifs, as well as from the NBD2 signature sequence (LSHGH instead of LSGGQ). Not surprisingly, as we will see below, evidence suggests that only the NBD2 composite site is catalytically competent.
The CFTR pore
Where is the pore in CFTR? Few systematic mutational studies have been attempted, making the residues that contribute to CFTR’s anion pore uncertain. The focus of most attention has been the sixth putative membrane-spanning helix in TMD1 (gold helix, Fig. 2b), whose cytoplasmic extension connects TMD1 to NBD1. As befits a contributor to an anion-selective pore, transmembrane helix 6 is predicted to contain at least three basic residues, Arg 334 and Lys 335 (blue-green spacefill, Fig. 2b) towards the extracellular end and Arg 347 four helical turns closer to the cytoplasm. Although initially proposed to face the pore34, Arg 347 was subsequently shown35 to form a salt bridge to an Asp in transmembrane helix 8. A number of approaches have led to the conclusion that the positive charges at Arg 334 and Lys 335 promote entry of anions into an outer vestibule, and that the side chain of Thr338, one helical turn in, faces the pore. These include measurements of the ease of entering the pore and of passing through it, for a variety of monovalent anions, and measurements of the influence of covalent modification of introduced cysteines (reviewed in ref. 20).
Vast spans of the TMDs have not yet received attention (for example, helices 7 to 10), and effects on anion conduction of mutations at selected positions in helices 1 to 5 and 11 are not yet fully understood20,21. Although the symmetrical arrangement of membrane-spanning helices in the paired TMDs of homodimeric ABC proteins, such as MsbA and BtuCD, imbues CFTR models based on them with comparable twofold symmetry (see Fig. 2b), a lot more work is needed before the true disposition of transmembrane helices around CFTR’s anion-selective pore can be specified.
Like most other chloride channels (see the review in this issue by Miller, p. 484), CFTR selects relatively poorly among small monovalent anions. Halide and pseudohalide anions larger than chloride, which escape their water shell more easily, also enter the channel more easily than chloride20. Their lower avidity for water also keeps larger anions in the pore longer, allowing them to impair chloride-ion flow by competition20. These characteristics imply a fairly featureless interior of the CFTR pore where no region makes very intimate contact with permeating chloride ions20. This contrasts with the close embrace of potassium ions by rings of backbone carbonyl oxygens in the selectivity filter of potassium channels. Whereas potassium channels need to discriminate between similarly sized sodium and potassium ions, CFTR channels face the simpler task of distinguishing chloride (and probably bicarbonate) from larger anions such as phosphate and sulphate. Loss of regulated conduction of bicarbonate10 by CFTR channels may contribute to both airway and pancreatic-duct disease in CF.
Phosphorylation of the R domain
Gating of the CFTR channel is strictly dependent on R-domain phosphorylation by PKA. CFTR channel activity increases at least 100-fold upon phosphorylation36 (from an open probability of <0.003 for non-phosphorylated CFTR channels in saturating [ATP]). The R domain comprises the ~200 residues linking NBD1 to TMD2 and contains multiple consensus sites for phosphorylation by PKA; up to six have been found phosphorylated in cells37–39 and up to eight in isolated CFTR40. Among other kinases, both Ca2+-dependent and independent isoforms of protein kinase C (PKC) can phosphorylate R-domain sites22,38,41. Although CFTR phosphorylation by PKC itself causes only modest channel activation22,41, it potentiates22, and might even be a prerequisite for42,43, subsequent channel activation by PKA.
The unphosphorylated R domain exerts an inhibitory influence on CFTR channel gating that can be relieved by phosphorylating it, or deleting it44,45. Thus, at saturating [ATP], the open probability of CFTR channels cut in half and lacking the R domain is ~60% of that of fully phosphorylated CFTR channels similarly cut in half but retaining the entire CFTR sequence, including the R domain45 (which all testifies to the nature of CFTR’s modular ABC-protein architecture). So loss of inhibition by the unphosphorylated R domain can account for over half of the more than two orders of magnitude increase in open probability of intact wild-type CFTR channels upon phosphorylation36. Any additional stimulatory effect of the phosphorylated R domain46 is therefore probably small (less than twofold).
Just how R-domain phosphorylation controls channel gating is not clear. No other ABC protein contains an R domain, and its sequence, though conserved across species47, shows no homology to other proteins. Its shape and location in the CFTR channel are therefore unknown. CFTR-channel activity changes gradually as increasing numbers of target serines are mutated37,48 or as increasing PKA concentrations incrementally phosphorylate wild-type CFTR39,43. Results like these have fostered notions of redundancy among serines in a structureless R domain, and of action by accumulated negative charge37,49. But this is unlikely to be the whole story. For instance, purified R-domain protein undergoes discernible conformational changes upon phosphorylation39,50. In addition, two of the most readily phosphorylated serines inhibit channel activity when phosphorylated, despite undoubted introduction of negative charge39,51. Although direct introduction of negative charge by substituting aspartate52 or glutamate53 for six to eight serines does result in partly active channels, their maximal activity is 2–3-fold lower than that of phosphorylated wild-type CFTR. And circular dichroism spectra of the purified R domain indicated that the introduced acidic residues imperfectly mimic phosphoserines54. Also, a role for a conformational change of the R domain is suggested by the finding that the prolyl-isomerase cyclophilin A stimulated wild-type CFTR channel activity, but not that of mutants lacking three conserved R-domain prolines55.
The R domain does not seem to prevent ATP from binding at the NBDs or hydrolysis, because phosphorylation is reported not to alter photolabelling by 8-azido-ATP56–58. Nor can inhibition of gating in the unphosphorylated channel be attributed to the serine-containing nonconserved structural elements in and around CFTR’s NBD130–32, say by preventing NBD1–NBD2 interaction. This is because excision of neither segment vitiated the strict dependence of channel gating on phosphorylation by PKA36.
By contrast, simply severing CFTR where the R domain connects to TMD2 partly disrupts inhibition of gating in unphosphorylated channels45,59. So the closed conformation of the pore might be stabilized (relative to the open conformation) by an action on the TMDs of the unphosphorylated R domain that requires its covalent attachment to transmembrane helix 7? That interaction could be relieved by R-domain phosphorylation — perhaps through its association with some other part of the channel — and the free energy difference between open- and closed-state conformations thereby lowered sufficiently to enable ATP-mediated closed-to-open transitions. Physical association of the R domain with CFTR’s N terminus might be required for channel activation by PKA60 and might be promoted by R-domain phosphorylation61.
CFTR channel gating by ATP
The location of the gate that prevents ion flow through closed CFTR channels is not known. But the machinery attached to the strings that pull the gate open and closed has recently come into view. Opening and closing of the CFTR pore are now believed to be linked to formation and disruption of a tight NBD1–NBD2 complex, driven by ATP binding and hydrolysis events (Fig. 1b). Phosphorylated CFTR channels can be opened by a broad range of nucleoside triphosphates, including ATP, GTP, ITP, UTP, CTP, AMP-CPP62 and 8-azido-ATP58. But they are opened only inefficiently by millimolar concentrations of poorly hydrolysable analogues such as AMP-PNP, AMP-PCP or ATP-γS53,62–64, and not at all by ADP62. The frequency of CFTR-channel opening increases with ATP concentration along a Michaelis curve and is half maximal at about 50 μM ATP (Fig. 3a). At saturating concentrations of ATP, a channel waits in the closed state for a second or so (depending on the temperature) before opening. This means that binding of ATP limits channel opening at low concentrations, but at saturating ATP concentrations some other slow step controls how quickly the channel can open once ATP has bound.
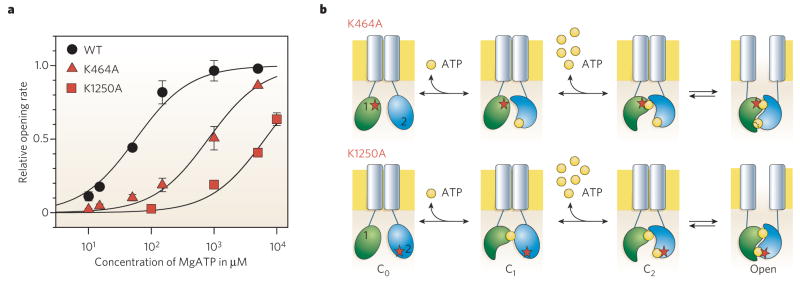
Figure 3. The conserved Walker A lysine is critical for ATP binding in each NBD.
a, Dependence on cytoplasmic-surface ATP concentration of rate of channel opening for wild-type CFTR (WT) and for CFTR channels with a single mutation of the NBD1 (K464) or NBD2 (K1250) Walker A lysine. Either mutation lowers the apparent affinity for ATP-dependent channel opening, implying that ATP normally binds at both sites before a channel opens. b, Diagram illustrating the simplest interpretation of results in a. At lower [ATP], channel opening is limited by low success rate of ATP binding to the composite site that harbours the mutation (red star) of the conserved lysine. ATP first binds to the non-mutant site, that is to the NBD2 site (blue) in K464A channels (upper row), but to the NDB1 site (green) in K1250A channels (lower row). When at higher concentrations, ATP also occupies the mutant site, the NBDs dimerize and the channel opens.
ATP binding and hydrolysis
Where does this rate-limiting binding of ATP occur? Mutagenesis experiments (Fig. 3) to alter the Walker motifs that contact the bound ATP in NBD crystals (Fig. 2a) confirm that, as expected, the ATP acts at CFTR’s NBDs. The fact that neutralization of the Walker A Lys at either the NBD1 or the NBD2 composite site severely impairs channel opening at low ATP concentrations (Fig. 3, but compare it with ref. 65) implies that ATP binding at either site can be made rate limiting. This suggests that ATP normally occupies both sites before a channel opens64,66 (but compare with ref. 67).
Photolabelling of CFTR with 8-azido nucleotide, followed by separation of NBD1- and NBD2-containing fragments, confirmed binding to both NBD1 and NBD2. But only 8-azido nucleotide at NBD1 could survive washes at 0 °C before crosslinking the nucleotide with UV irradiation57,58,68. NBD1 was photolabelled to a similar extent with 8-azido-[α-32P]-ATP or 8-azido-[γ-32P]ATP57,58, even after prolonged washes at 30° C before UV irradiation58, whereas, even without washing, NBD2 could never be photolabelled with 8-azido-[γ32P]ATP57. This all suggests that hydrolysis of 8-azido-ATP is rapid at the NBD2 composite catalytic site57 but negligibly slow at the NBD1 composite site, even at 30 °C58. As loss of 8-azido-ATP from NBD1 at 30 °C before photolabel-ling takes minutes, but opening and closing of CFTR channels in ATP takes seconds (Fig. 1a; also in 8-azido-ATP58), nucleotide-dependent gating would seem to be controlled predominantly by events at the NBD2 composite catalytic site58.
But how are nucleotide comings and goings at the NBD2 composite site linked to channel opening and closing? The principal consequence of mutations at the key catalytic NBD2 Walker A Lys and Walker B Asp and Glu residues (K1250, D1370 and E1371 in Fig. 2d), at saturating concentrations of ATP, is a marked increase in the time for which channels remain open (Fig. 4b). This is most easily seen from the markedly delayed step-like closure of individual mutant channels after sudden withdrawal of ATP (Fig. 4b) compared with the rapid closure of wild-type CFTR channels (Figs 1a, 4a). As biochemical measurements confirm that homologous mutations abrogate ATP hydrolysis in other ABC proteins (reviewed in refs 69, 70), and mutation K1250A has been shown to abolish ATPase activity of CFTR71, the implication is that hydrolysis of the ATP bound at the NBD2 composite catalytic site occurs before normal rapid channel closure64,72,73. The idea that ATP hydrolysis sets the timing of normal CFTR-channel closure is further supported by the strong dependence of channel closing rate on Mg2+ concentration74 and on temperature (activation enthalpy, ~70 kJ mol−1)45,75. It is also supported by the finding that closure is markedly delayed in the presence of ATP plus the poorly hydrolysable analogue AMP-PNP63,76 or of ATP plus the inorganic phosphate analogue orthovanadate58,76,77, which forms a stable complex with the hydrolysis product ADP after dissociation of the γ-phosphate (for example, see ref. 78).
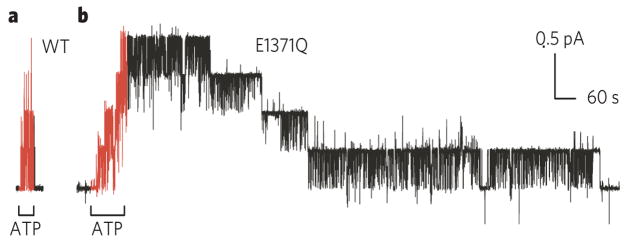
Figure 4. ATP hydrolysis prompts channel closure.
Comparison of speed of closing of wild-type CFTR (a) and of mutant CFTR bearing the single mutation E1371Q (b), of the conserved Walker B glutamate. This glutamate is required for ATP hydrolysis, but not for binding of ATP29,69,70,81. The greatly delayed closing of all four E1371Q channels (b), compared with the four WT CFTR channels (a), after removal of ATP supports the conclusion that ATP hydrolysis at the NBD2 composite catalytic site controls normal channel closing63–66,72–77,80, and that the channel open-burst state corresponds to NBD dimerization22,29,69,70,80.
NBD dimerization
So if hydrolysis of the NBD2-site ATP controls the timing of channel closing, what normally triggers its opening? The slow step that limits the speed of channel opening at saturating nucleotide concentration is sensitive to the structure around the γ-phosphate and, for AMP-PNP, AMP-PCP or ATP-γS, that step occurs at ~4% of the maximal rate seen with ATP64. The step is also dependent on Mg2+ and, at saturating ATP concentrations, in the absence of Mg2+ ions, occurs at only <2% of the maximal rate seen in their presence74. The slow opening step is also highly temperature sensitive (activation enthalpy ≥100 kJ mol−1)75,79. These characteristics suggested that the process that usually opens CFTR channels is likely to be formation of an ATP-driven tight NBD1–NBD2 heterodimeric interaction80 (Figs 1b and 2d), which is analogous to the homodimeric interaction (Figs 2a, c, 5a) found in biochemical and structural studies of bacterial NBDs21–24,29,69,70,81.
One clue that the CFTR-channel open state corresponds to the heterodimerized NBD1–NBD2 conformation comes from the ~1,000-fold stabilization of the open state (Fig. 4b) caused by the mutation E1371Q of the Walker B Glu (the possible catalytic base29,81). The homologous Gluto-Gln mutation in bacterial NBDs was required to produce the highly stable ATP-bound homodimers needed for crystallization25,29. Even with that hydrolysis-impairing mutation, stable dimers were not obtained with ADP29, which is consistent with a cycle of ATP-induced NBD dimerization and hydrolysis-induced dimer disruption24. Opening of a CFTR channel can then be viewed as being triggered by a pincer-like drawing together of ATP-bound NBD1 and ATP-bound NBD2, as inferred from the relative positions of dimerized bacterial NBDs in the presence and absence of ATP26,69 (Fig. 5a, b). Channel closing would follow hydrolysis of the ATP at the NBD2 composite site, loss of the liberated phosphate and subsequent disruption of the heterodimeric NBD1–NBD2 interaction, as illustrated in Fig. 1b.
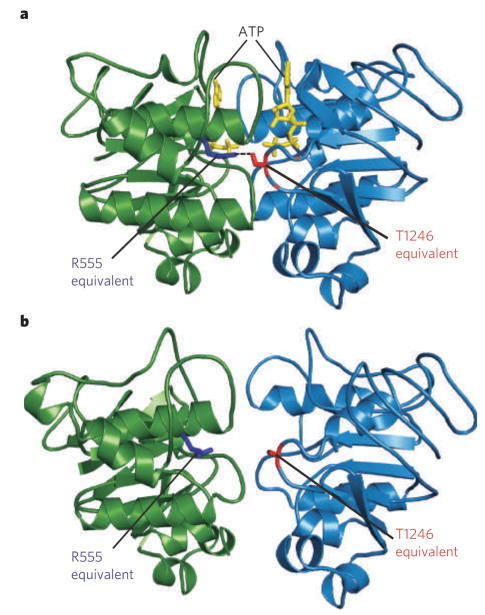
Figure 5. Structures of the MalK homodimer.
The MalK homodimer was crystallized with (a) and without (b) ATP (yellow). The structures illustrate pincer-like motion of MalK that closes the catalytic interface upon binding of two ATP molecules26. The residues corresponding to CFTR R555 (blue) and T1246 (red) form a hydrogen bond (a, dotted line) linking Walker A of one subunit with the signature sequence of the other, only in the ATP-bound crystal (a).
Direct evidence for a head-to-tail NBD1–NBD2 interaction in functioning CFTR channels comes from two sources. First, target cysteines introduced in pairs, one in NBD1 and the other in NBD2, can be crosslinked by short bifunctional thiol-specific reagents only when, for example, one cysteine is in a Walker motif and the other is in a signature sequence, and not when both are in Walker motifs or both are in signature sequences82. Second, measurements of the energetics of channel gating revealed that a residue near the NBD1 signature sequence and another in the NBD2 Walker A motif make an energetically favourable interaction when the channels are open, or in the transition state (Fig. 1b) between closed and open states. But these residues do not interact when the channels are closed, either before or after (states C1 and C2, Fig. 1b) binding the second ATP, which is thought to trigger channel opening80. The sign and magnitude of that coupling energy are consistent with formation of a hydrogen bond between these interacting residues as the channel opens. The corresponding two residues (labelled ‘R555 equivalent’ and ‘T1246 equivalent’ in Fig. 5b) in bacterial NBD homodimers are indeed linked by a hydrogen bond in the ATP-bound, tightly dimerized conformation25–27 (Fig. 5a) but not in the nucleotide-free, ‘open dimer’ conformation26 (Fig. 5b). This strongly supports a cyclic, dynamic restructuring of the NBD1–NBD2 heterodimeric interface during the CFTR-channel gating cycle.
A bonus of the large complement of ABC-protein genes in prokaryotic genomes is that over 20,000 NBD sequences are available (http://www.sanger.ac.uk/cgi-bin/Pfam/getacc?PF00005). Statistical analysis83 of these sequences suggests that the two residues that hydrogen bond across the ATP-bound NBD dimer interface (that is, the ‘R555 equivalent’ and ‘T1246 equivalent’ residues) were subject to evolutionary pressure as a pair, rather than individually. They seem to have co-evolved to retain the ability to form a hydrogen bond that precisely spaces their a-carbon atoms in the dimer (as in Fig. 5a). Analysis of the distributions of potential hydrogen-bond donor (R555 equivalent) and acceptor (T1246 equivalent) side chains shows that the two positions have changed in concert so that a shorter acceptor is more frequently paired with a longer donor and vice versa80. The broad conservation of NBD-sequence motifs throughout prokaryotic and eukaryotic ABC proteins, and specific conservation of the structural underpinnings of the hydrogen bond found to undergo dynamic changes during the gating cycle of CFTR channels, implies that such NBD dimerization is crucial to the function of most ABC proteins.
Outlook
Although progress is being made, much remains to be learned about CFTR, including the structures and locations of the anion pore in the transmembrane domain, of its gate and of the R domain. It would also be good to know how graded R-domain phosphorylation controls communication between the NBDs and the gate. The transduction pathway by which conformational rearrangements at the NBDs are transmitted through the protein to the transmembrane domains is not known for any ABC protein. This question could be addressed by further examination of the extensive evolutionary record for ABC proteins. Statistical analysis of sequence alignments may reveal sets of positions at which amino-acid distributions are correlated. Thermodynamic mutant-cycle analysis in CFTR80 could then be used to test whether statistical coupling reflects conservation of a network of energetically coupled residues that mediates transmission of allosteric signals. It may be possible to follow how energetic coupling between side-chain pairs changes, both as a function of their location in the protein and as a function of time during CFTR’s channel-gating cycle.
Sequence analysis suggests that CFTR’s asymmetric catalytic interface, with one active and one dead composite site, may be characteristic of all ABC-C subfamily members (CFTR, MRPs and SURs) but not of ABC proteins in general. The structural and mechanistic detail relevant to ABC-C proteins that can be gleaned from studies of symmetric, homodimeric ABC proteins is therefore limited. Determination of the structure of any comparably asymmetric ABC protein would greatly advance our understanding of how the ABC-C proteins CFTR, MRP and SUR work.
Much also remains to be learned about the physiological roles and functions of CFTR — for example, the identities of other proteins with which CFTR interacts (reviewed in refs 7, 8), whether CFTR functions as an ATP-driven transporter84 and whether, and under what conditions, CFTR behaves like an adenylate kinase85,86.
Present treatment of cystic fibrosis is essentially symptomatic, and although average life expectancy for CF patients is now just over 30 years, there is as yet no cure. Development of procedures for delivery of wild-type CFTR-encoding DNA using viral and non-viral (such as liposome or nanoparticle) agents or of functional CFTR protein (by means of liposomes) to affected cells continues apace11. Because of the many hurdles to be overcome, parallel efforts to harness high-throughput methods to find small molecules that would allow maturation of ΔF508 CFTR or enhance activity of other mutant CFTR channels are important. Because of the continuous turnover of epithelia, repetition of all these treatments will be required. On the other side of the coin, small molecule inhibitors87 will be used in the near future against CFTR channels in the intestinal tract to combat excessive fluid loss, not least in travellers’ diarrhoea.
Acknowledgments
This review and our research on CFTR were supported by grants from the NIH and Fogarty International Center (to D.C.G). We dedicate this review to our late colleague Benjamin Angel, MA, MB BS (3rd December 1978 to 2nd October 2005), whose courage and grace in the face of CF continue to inspire us.
Footnotes
Author Information Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
References
- 1.Rommens JM, et al. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- 2.Riordan JR, et al. Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 3.Quinton PM. Chloride impermeability in cystic fibrosis. Nature. 1983;301:421–422. doi: 10.1038/301421a0. [DOI] [PubMed] [Google Scholar]
- 4.Schoumacher RA, et al. Phosphorylation fails to activate chloride channels from cystic fibrosis airway cells. Nature. 1987;330:752–754. doi: 10.1038/330752a0. [DOI] [PubMed] [Google Scholar]
- 5.Li M, et al. Cyclic AMP-dependent protein kinase opens chloride channels in normal but not cystic fibrosis airway epithelium. Nature. 1988;331:358–360. doi: 10.1038/331358a0. [DOI] [PubMed] [Google Scholar]
- 6.Bear CE, et al. Purification and functional reconstitution of the cystic fibrosis transmembrane conductance regulator (CFTR) Cell. 1992;68:809–818. doi: 10.1016/0092-8674(92)90155-6. [DOI] [PubMed] [Google Scholar]
- 7.Altschuler Y, Hodson C, Milgram SL. The apical compartment: trafficking pathways, regulators and scaffolding proteins. Curr Opin Cell Biol. 2003;15:423–429. doi: 10.1016/s0955-0674(03)00084-x. [DOI] [PubMed] [Google Scholar]
- 8.Li C, Naren AP. Macromolecular complexes of cystic fibrosis transmembrane conductance regulator and its interacting partners. Pharmacol Ther. 2005;108:208–223. doi: 10.1016/j.pharmthera.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Du K, Sharma M, Lukacs GL. The DeltaF508 cystic fibrosis mutation impairs domain-domain interactions and arrests post-translational folding of CFTR. Nature Struct Mol Biol. 2005;12:17–25. doi: 10.1038/nsmb882. [DOI] [PubMed] [Google Scholar]
- 10.Wine JJ. Acid in the airways. Focus on ‘Hyperacidity of secreted fluid from submucosal glands in early cystic fibrosis’. Am J Physiol Cell Physiol. 2006;290:C669–C671. doi: 10.1152/ajpcell.00525.2005. [DOI] [PubMed] [Google Scholar]
- 11.Davies JC, Alton EW. Airway gene therapy. Adv Genet. 2005;54:291–314. doi: 10.1016/S0065-2660(05)54012-4. [DOI] [PubMed] [Google Scholar]
- 12.Dean M, Rzhetsky A, Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001;11:1156–1166. doi: 10.1101/gr.184901. [DOI] [PubMed] [Google Scholar]
- 13.Reyes CL, Chang G. Structure of the ABC transporter MsbA in complex with ADP. vanadate and lipopolysaccharide. Science. 2005;308:1028–1031. doi: 10.1126/science.1107733. [DOI] [PubMed] [Google Scholar]
- 14.Locher KP, Lee AT, Rees DC. The E. coli BtuCD structure: A framework for ABC transporter architecture and mechanism. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- 15.Rosenberg MF, Callaghan R, Modok S, Higgins CF, Ford RC. Three-dimensional structure of P-glycoprotein: the transmembrane regions adopt an asymmetric configuration in the nucleotide-bound state. J Biol Chem. 2005;280:2857–2862. doi: 10.1074/jbc.M410296200. [DOI] [PubMed] [Google Scholar]
- 16.Ramjeesingh M, Kidd JF, Huan LJ, Wang Y, Bear CE. Dimeric cystic fibrosis transmembrane conductance regulator exists in the plasma membrane. Biochem J. 2003;374:793–797. doi: 10.1042/BJ20030683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raghuram V, Mak DD, Foskett JK. Regulation of cystic fibrosis transmembrane conductance regulator single-channel gating by bivalent PDZ-domain-mediated interaction. Proc Natl Acad Sci USA. 2001;98:1300–1305. doi: 10.1073/pnas.031538898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen JH, Chang XB, Aleksandrov AA, Riordan JR. CFTR is a misnomer: Biochemical and functional evidence. J Membr Biol. 2002;188:55–71. doi: 10.1007/s00232-001-0174-2. [DOI] [PubMed] [Google Scholar]
- 19.Zhang ZR, et al. Determination of the functional unit of the cystic fibrosis transmembrane conductance regulator chloride channel: One polypeptide forms one pore. J Biol Chem. 2005;280:458–468. doi: 10.1074/jbc.M409626200. [DOI] [PubMed] [Google Scholar]
- 20.Dawson DC, Liu X, Zhang Z, McCarty NA. In: Cystic Fibrosis Transmembrane Conductance Regulator. Kirk KL, Dawson DC, editors. Kluwer/Plenum; New York: 2003. pp. 1–34. [Google Scholar]
- 21.Linsdell P. Mechanism of chloride permeation in the cystic fibrosis transmembrane conductance regulator chloride channel. Exp Physiol. 2006;99:123–129. doi: 10.1113/expphysiol.2005.031757. [DOI] [PubMed] [Google Scholar]
- 22.Tabcharani JA, Chang XB, Riordan JR, Hanrahan JW. Phosphorylation-regulated Cl− channel in CHO cells stably expressing the cystic fibrosis gene. Nature. 1991;352:628–631. doi: 10.1038/352628a0. [DOI] [PubMed] [Google Scholar]
- 23.Hung LW, et al. Crystal structure of the ATP-binding subunit of an ABC transporter. Nature. 1998;396:703–707. doi: 10.1038/25393. [DOI] [PubMed] [Google Scholar]
- 24.Hopfner KP, et al. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 25.Smith PC, et al. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol Cell. 2002;10:139–149. doi: 10.1016/s1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J, Lu G, Lin J, Davidson AL, Quiocho FA. A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol Cell. 2003;12:651–661. doi: 10.1016/j.molcel.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 27.Zaitseva J, Jenewein S, Jumpertz T, Holland IB, Schmitt L. H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. EMBO J. 2005;24:1901–1910. doi: 10.1038/sj.emboj.7600657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moody JE, Millen L, Binns D, Hunt JF, Thomas PJ. Cooperative, ATP-dependent association of the nucleotide-binding cassettes during the catalytic cycle of ATP-binding cassette transporters. J Biol Chem. 2002;277:21111–21114. doi: 10.1074/jbc.C200228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis HA, et al. Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J. 2004;23:282–293. doi: 10.1038/sj.emboj.7600040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis HA, et al. Impact of the deltaF508 mutation in first nucleotide-binding domain of human cystic fibrosis transmembrane conductance regulator on domain folding and structure. J Biol Chem. 2005;280:1346–1353. doi: 10.1074/jbc.M410968200. [DOI] [PubMed] [Google Scholar]
- 32.Thibodeau PH, Brautigam CA, Machius M, Thomas PJ. Side chain and backbone contributions of Phe508 to CFTR folding. Nature Struct Mol Biol. 2005;12:10–16. doi: 10.1038/nsmb881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenberg MF, et al. Purification and crystallization of the cystic fibrosis transmembrane conductance regulator (CFTR) J Biol Chem. 2004;279:39051–39057. doi: 10.1074/jbc.M407434200. [DOI] [PubMed] [Google Scholar]
- 34.Tabcharani JA, et al. Multi-ion pore behaviour in the CFTR chloride channel. Nature. 1993;366:79–82. doi: 10.1038/366079a0. [DOI] [PubMed] [Google Scholar]
- 35.Cotten JF, Welsh MJ. Cystic fibrosis-associated mutations at arginine 347 alter the pore architecture of CFTR. Evidence for disruption of a salt bridge. J Biol Chem. 1999;274:5429–5435. doi: 10.1074/jbc.274.9.5429. [DOI] [PubMed] [Google Scholar]
- 36.Csanády L, Chan KW, Nairn AC, Gadsby DC. Functional roles of nonconserved structural segments in CFTR’s NH2-terminal nucleotide binding domain. J Gen Physiol. 2005;125:43–55. doi: 10.1085/jgp.200409174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng SH, et al. Phosphorylation of the R domain by cAMP-dependent protein kinase regulates the CFTR chloride channel. Cell. 1991;66:1027–1036. doi: 10.1016/0092-8674(91)90446-6. [DOI] [PubMed] [Google Scholar]
- 38.Picciotto M, Cohn J, Bertuzzi G, Greengard P, Nairn AC. Phosphorylation of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 1992;267:12742–12752. [PubMed] [Google Scholar]
- 39.Csanády L, et al. Preferential phosphorylation of R-domain Serine 768 dampens activation of CFTR channels by PKA. J Gen Physiol. 2005;125:171–186. doi: 10.1085/jgp.200409076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neville DCA, et al. Evidence for phosphorylation of serine 753 in CFTR using a novel metal-ion affinity resin and matrix-assisted laser desorption mass spectrometry. Protein Sci. 1997;6:2436–2445. doi: 10.1002/pro.5560061117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berger HA, Travis SM, Welsh MJ. Regulation of the cystic fibrosis transmembrane conductance regulator Cl− channel by specific protein kinases and protein phosphatases. J Biol Chem. 1993;268:2037–2047. [PubMed] [Google Scholar]
- 42.Jia Y, Mathews CJ, Hanrahan JW. Phosphorylation by protein kinase C is required for acute activation of cystic fibrosis transmembrane conductance regulator by protein kinase A. J Biol Chem. 1997;272:4978–4984. doi: 10.1074/jbc.272.8.4978. [DOI] [PubMed] [Google Scholar]
- 43.Chappe V, et al. Phosphorylation of protein kinase C sites in NBD1 and the R domain control CFTR channel activation by PKA. J Physiol. 2003;548:39–52. doi: 10.1113/jphysiol.2002.035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rich DP, et al. Effect of deleting the R domain on CFTR-generated chloride channels. Science. 1991;253:205–207. doi: 10.1126/science.1712985. [DOI] [PubMed] [Google Scholar]
- 45.Csanády L, et al. Severed channels probe regulation of gating of CFTR by its cytoplasmic domains. J Gen Physiol. 2000;116:477–500. doi: 10.1085/jgp.116.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winter MC, Welsh MJ. Stimulation of CFTR activity by its phosphorylated R domain. Nature. 1997;389:294–296. doi: 10.1038/38514. [DOI] [PubMed] [Google Scholar]
- 47.Gadsby DC, Nairn AC. Control of CFTR channel gating by phosphorylation and nucleotide hydrolysis. Physiol Rev. 1999;79:S77–S107. doi: 10.1152/physrev.1999.79.1.S77. [DOI] [PubMed] [Google Scholar]
- 48.Chang XB, et al. Protein kinase A(PKA) still activates CFTR chloride channel after mutagenesis of all 10 PKA consensus phosphorylation sites. J Biol Chem. 1993;268:11304–11311. [PubMed] [Google Scholar]
- 49.Ostedgaard LS, Baldursson O, Vermeer DW, Welsh MJ, Robertson AD. A functional R domain from cystic fibrosis transmembrane conductance regulator is predominantly unstructured in solution. Proc Natl Acad Sci USA. 2000;97:5657–5662. doi: 10.1073/pnas.100588797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dulhanty AM, Riordan JR. Phosphorylation by cAMP-dependent protein kinase causes a conformational change in the R domain of the cystic fibrosis transmembrane conductance regulator. Biochemistry. 1994;33:4072–4079. doi: 10.1021/bi00179a036. [DOI] [PubMed] [Google Scholar]
- 51.Wilkinson DJ, et al. CFTR activation: additive effects of stimulatory and inhibitory phosphorylation sites in the R domain. Am J Physiol. 1997;273:L127–L133. doi: 10.1152/ajplung.1997.273.1.L127. [DOI] [PubMed] [Google Scholar]
- 52.Rich DR, et al. Regulation of the cystic fibrosis transmembrane conductance regulator Cl channel by negative charge in the R domain. J Biol Chem. 1993;268:20259–20267. [PubMed] [Google Scholar]
- 53.Aleksandrov AA, Chang X, Aleksandrov L, Riordan JR. The non-hydrolytic pathway of cystic fibrosis transmembrane conductance regulator ion channel gating. J Physiol. 2000;528:259–265. doi: 10.1111/j.1469-7793.2000.00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dulhanty AM, Chang XB, Riordan JR. Mutation of potential phosphorylation sites in the recombinant R domain of the cystic fibrosis transmembrane conductance regulator has significant effects on domain conformation. Biochem Biophys Res Commun. 1995;206:207–214. doi: 10.1006/bbrc.1995.1029. [DOI] [PubMed] [Google Scholar]
- 55.Xie J, Zhao J, Davis PB, Ma J. Conformation, independent of charge, in the R domain affects cystic fibrosis transmembrane conductance regulator channel openings. Biophys J. 2000;78:1293–1305. doi: 10.1016/S0006-3495(00)76685-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Travis SM, Carson MR, Ries DR, Welsh MJ. Interaction of nucleotides with membrane-associated cystic fibrosis transmembrane conductance regulator. J Biol Chem. 1993;268:15336–15339. [PubMed] [Google Scholar]
- 57.Aleksandrov L, Aleksandrov AA, Chang XB, Riordan JR. The first nucleotide binding domain of cystic fibrosis transmembrane conductance regulator is a site of stable nucleotide interaction, whereas the second is a site of rapid turnover. J Biol Chem. 2002;277:15419–15425. doi: 10.1074/jbc.M111713200. [DOI] [PubMed] [Google Scholar]
- 58.Basso C, Vergani P, Nairn AC, Gadsby DC. Prolonged nonhydrolytic interaction of nucleotide with CFTR’s NH2-terminal nucleotide binding domain and its role in channel gating. J Gen Physiol. 2003;122:333–348. doi: 10.1085/jgp.200308798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ostedgaard LS, Rich DP, DeBerg LG, Welsh MJ. Association of domains within the cystic fibrosis transmembrane conductance regulator. Biochemistry. 1997;36:1287–1294. doi: 10.1021/bi962174s. [DOI] [PubMed] [Google Scholar]
- 60.Naren AP, et al. CFTR chloride channel regulation by an interdomain interaction. Science. 1999;286:544–548. doi: 10.1126/science.286.5439.544. [DOI] [PubMed] [Google Scholar]
- 61.Chappe V, Irvine T, Liao J, Evagelidis A, Hanrahan JW. Phosphorylation of CFTR by PKA promotes binding of the regulatory domain. EMBO J. 2005;24:2730–2740. doi: 10.1038/sj.emboj.7600747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anderson MP, et al. Nucleoside triphosphates are required to open the CFTR chloride channel. Cell. 1991;67:775–784. doi: 10.1016/0092-8674(91)90072-7. [DOI] [PubMed] [Google Scholar]
- 63.Hwang TC, Nagel GA, Nairn AC, Gadsby DC. Regulation of the gating of CFTR Cl channels by phosphorylation and ATP hydrolysis. Proc Natl Acad Sci USA. 1994;91:4698–4702. doi: 10.1073/pnas.91.11.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vergani P, Nairn AC, Gadsby DC. On the mechanism of MgATP-dependent gating of CFTR Cl− channels. J Gen Physiol. 2003;121:17–36. doi: 10.1085/jgp.20028673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Powe ACJ, Al-Nakkash L, Li M, Hwang TC. Mutation of Walker-A lysine 464 in cystic fibrosis transmembrane conductance regulator reveals functional interaction between its nucleotide-binding domains. J Physiol. 2002;539:333–346. doi: 10.1113/jphysiol.2001.013162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berger AL, Ikuma M, Welsh MJ. Normal gating of CFTR requires ATP binding to both nucleotide-binding domains and hydrolysis at the second nucleotide-binding domain. Proc Natl Acad Sci USA. 2005;102:455–460. doi: 10.1073/pnas.0408575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bompadre SG. CFTR gating II: effects of nucleotide binding on the stability of open states. J Gen Physiol. 2005;125:377–394. doi: 10.1085/jgp.200409228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Szabó K, Szakács G, Hegedus T, Sarkadi B. Nucleotide occlusion in the human cystic fibrosis transmembrane conductance regulator. J Biol Chem. 1999;274:12209–12212. doi: 10.1074/jbc.274.18.12209. [DOI] [PubMed] [Google Scholar]
- 69.Davidson AL, Chen J. ATP-binding cassette transporters in bacteria. Annu Rev Biochem. 2004;73:241–268. doi: 10.1146/annurev.biochem.73.011303.073626. [DOI] [PubMed] [Google Scholar]
- 70.Higgins CF, Linton KJ. The ATP switch model for ABC transporters. Nature Struct Mol Biol. 2004;11:918–926. doi: 10.1038/nsmb836. [DOI] [PubMed] [Google Scholar]
- 71.Ramjeesingh M, et al. Walker mutations reveal loose relationship between catalytic and channel-gating activities of purified CFTR (cystic fibrosis transmembrane conductance regulator) Biochemistry. 1999;38:1463–1468. doi: 10.1021/bi982243y. [DOI] [PubMed] [Google Scholar]
- 72.Gunderson KL, Kopito RR. Conformational states of CFTR associated with channel gating: the role ATP binding and hydrolysis. Cell. 1995;82:231–239. doi: 10.1016/0092-8674(95)90310-0. [DOI] [PubMed] [Google Scholar]
- 73.Carson MR, Travis SM, Welsh MJ. The two nucleotide-binding domains of cystic fibrosis transmembrane conductance regulator (CFTR) have distinct functions in controlling channel activity. J Biol Chem. 1995;270:1711–1717. doi: 10.1074/jbc.270.4.1711. [DOI] [PubMed] [Google Scholar]
- 74.Dousmanis AG, Nairn AC, Gadsby DC. Distinct Mg(2+)-dependent steps rate limit opening and closing of a single CFTR Cl(−) channel. J Gen Physiol. 2002;119:545–559. doi: 10.1085/jgp.20028594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mathews CJ, Tabcharani JA, Hanrahan JW. The CFTR chloride channel: nucleotide interactions and temperature-dependent gating. J Membr Biol. 1998;163:55–66. doi: 10.1007/s002329900370. [DOI] [PubMed] [Google Scholar]
- 76.Gunderson KL, Kopito RR. Effects of pyrophosphate and nucleotide analogs suggest a role for ATP hydrolysis in cystic fibrosis transmembrane regulator channel gating. J Biol Chem. 1994;269:19349–19353. [PubMed] [Google Scholar]
- 77.Baukrowitz T, Hwang TC, Nairn AC, Gadsby DC. Coupling of CFTR Cl channel gating to an ATP hydrolysis cycle. Neuron. 1994;12:473–482. doi: 10.1016/0896-6273(94)90206-2. [DOI] [PubMed] [Google Scholar]
- 78.Urbatsch IL, Sankaran B, Weber J, Senior AE. P-glycoprotein is stably inhibited by vanadate-induced trapping of nucleotide at a single catalytic site. J Biol Chem. 1995;270:19383–19390. doi: 10.1074/jbc.270.33.19383. [DOI] [PubMed] [Google Scholar]
- 79.Aleksandrov AA, Riordan JR. Regulation of CFTR ion channel gating by MgATP. FEBS Lett. 1998;431:97–101. doi: 10.1016/s0014-5793(98)00713-3. [DOI] [PubMed] [Google Scholar]
- 80.Vergani P, Lockless SW, Nairn AC, Gadsby DC. CFTR channel opening by ATP-driven tight dimerization of its nucleotide-binding domains. Nature. 2005;433:876–880. doi: 10.1038/nature03313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tombline G, Bartholomew LA, Urbatsch IL, Senior AE. Combined mutation of catalytic glutamate residues in the two nucleotide binding domains of P-glycoprotein generates a conformation that binds ATP and ADP tightly. J Biol Chem. 2004;279:31212–31220. doi: 10.1074/jbc.M404689200. [DOI] [PubMed] [Google Scholar]
- 82.Mense M, Nairn AC, Gadsby DC. CFTR chloride channel activation in Xenopus oocytes by forskolin/IBMX promotes formation of a Rad50-like NBD1/NBD2 dimer. Biophys J. 2006;90:310a. [Google Scholar]
- 83.Lockless SW, Ranganathan R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- 84.Kogan I, et al. CFTR directly mediates nucleotide-regulated glutathione flux. EMBO J. 2003;22:1981–1989. doi: 10.1093/emboj/cdg194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Randak CO, Welsh MJ. Adenylate kinase activity in ABC transporters. J Biol Chem. 2005;280:34385–34388. doi: 10.1074/jbc.R500009200. [DOI] [PubMed] [Google Scholar]
- 86.Gross CH, et al. Nucleotide binding domains of cystic fibrosis transmembrane conductance regulator, an ABC-transporter, catalyze adenylate kinase activity but not ATP hydrolysis. J Biol Chem. doi: 10.1074/jbc.M511113200. in the press. [DOI] [PubMed] [Google Scholar]
- 87.Muanprasat C, et al. Discovery of glycine hydrazide pore-occluding CFTR inhibitors: mechanism, structure-activity analysis, and in vivo efficacy. J Gen Physiol. 2004;124:125–137. doi: 10.1085/jgp.200409059. [DOI] [PMC free article] [PubMed] [Google Scholar]