Abstract
As in centuries past, the main weapon against human malaria infections continues to be intervention with drugs, despite the widespread and increasing frequency of parasite populations that are resistant to one or more of the available compounds. This is a particular problem with the lethal species of parasite, Plasmodium falciparum, which claims some two million lives per year as well as causing enormous social and economic problems. Amongst the antimalarial drugs currently in clinical use, the antifolates have the best defined molecular targets, namely the enzymes dihydrofolate reductase (DHFR) and dihydropteroate synthase (DHPS), which function in the folate metabolic pathway. The products of this pathway, reduced folate cofactors, are essential for DNA synthesis and the metabolism of certain amino acids. Moreover, their formation and interconversions involve a number of other enzymes that have not as yet been exploited as drug targets. Antifolates are of major importance as they currently represent the only inexpensive regime for combating chloroquine-resistant malaria, and are now first-line drugs in a number of African countries. Aspects of our understanding of this pathway and antifolate drug resistance are reviewed here, with a particular emphasis on approaches to analysing the details of, and balance between, folate biosynthesis by the parasite and salvage of pre-formed folate from exogenous sources.
Keywords: Antifolate drugs, Drug resistance, Folate metabolism, Folate salvage, Gene disruption, Metabolic labelling
1. Introduction to the folate pathway
Folate metabolism is critically important to the viability of malaria parasites and the pathway has been targeted in both treatment and prophylaxis of the disease for over half a century, following closely on the discovery and introduction of antifolate drugs to combat bacterial infections. The most widely used antimalarial drugs of this type include pyrimethamine (PYR), proguanil, sulfadoxine (SDX) and dapsone, which have long provided chemotherapy at a price affordable by poorer nations and have been especially important as resistance to the principal antimalarial drug, chloroquine, has inexorably grown (Sibley et al., 2001). However, despite the long history, considerable importance and impact of these drugs, we still lack a detailed understanding of the underlying metabolism in the parasite. In other organisms, the pathways in which folate cofactors are involved include production of purines and pyrimidines for DNA replication as well as the synthesis and/or catabolism of several amino acids (Met, Gly, Ser, Glu, His). However, the repertoire of folate-dependent reactions that the malaria parasite can carry out is not yet fully defined. For example, whereas we know that it is dependent upon an external supply of purines from its host, other possible ‘missing reactions’ are more conjectural.
Among the different species of malaria parasites, most work has been done on that which is lethal to humans, Plasmodium falciparum, a pathogen that continues to exact a death toll of some two million a year and imposes a massive burden of morbidity and economic hardship on populations throughout the tropics and sub-tropics (Greenwood and Mutabingwa, 2002). Several approaches are in principle possible to study the folate pathway, although technical difficulties in working with such parasites are an important factor in designing experiments. Classical biochemistry is hampered by the limited amounts of both enzymes and substrates that can be obtained from in vitro cultures of P. falciparum, compounded by the instability of many of the molecules involved, while genetic studies along traditional lines have been confined to the very small number of controlled sexual crosses that it has been possible to carry out between defined clones of the parasite (Walliker et al., 1987; Wellems et al., 1990). The easiest route has been to build up a picture of the pathway via identification and analysis of relevant genes in the first instance, using more developed studies on other organisms as a guide, followed where possible with expression and characterisation of the recombinant gene products. However, as described below, approaches involving metabolic labelling with radioactive precursors have also been undertaken, as has a limited degree of genetic manipulation of the parasite to probe folate pathway gene function.
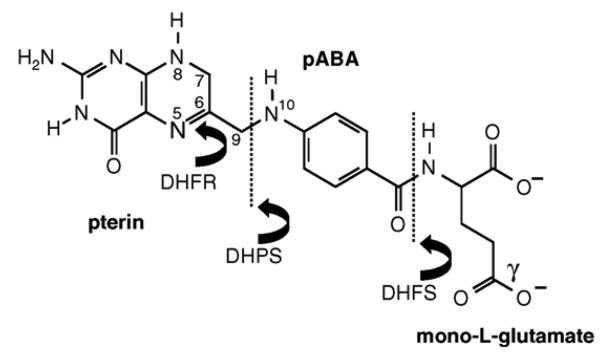
The purpose of the folate pathway is to provide cofactors that are essential for the metabolic events mentioned above, which involve the transfer of one-carbon (C1) units. Thus, in the case of DNA synthesis, folate in the form of 5,10-methylenetetrahydrofolate (methyleneTHF) is necessary to provide the methyl group that converts dUMP to dTMP, whose triphosphate derivative is used by DNA polymerase to add ‘T’ nucleotides to growing DNA chains. It should be noted that the term folate is used generically to cover the metabolites in the cell carrying the canonical folate moiety, but it should be borne in mind that a number of closely related derivatives are actually involved, varying in the degree of oxidation of the pterin ring, the nature of the substituents on the 5 and 10 positions of the molecule, and the number of glutamate residues attached to the para-aminobenzoate moiety that links the pterin ring to these glutamates (see Fig. 1).
Fig. 1.
Structure of dihydrofolate (DHF) indicating its construction from the pterin, pABA and l-glutamate moieties. The enzyme DHPS links pABA to an activated pterin ring to give dihydropteroate, which is converted to dihydrofolate by addition of a single l-glutamate residue by DHFS. The 5,6 double bond in DHF is reduced by DHFR to give tetrahydrofolate (THF); all active folate cofactors are found in this fully reduced state. They are also subject to polyglutamation by the FPGS activity, whereby additional glutamate residues are linked via peptide bond formation at the gamma position. Important atomic positions on the dihydropteroate moiety are numbered. For enzyme abbreviations, see the legend to Fig. 2.
2. Principal components of the pathway in P. falciparum
The schema in Fig. 2 depicts the enzymes that conventionally constitute the main folate pathway, as identified in other microorganisms, covering the biosynthesis from GTP of the basic folate moiety, dihydrofolate (DHF), as well as reduction of the latter to the biologically active, fully reduced forms of tetrahydrofolate (THF). The production of each molecule of dTMP results in the oxidation of the THF molecule to DHF, which must be recycled by dihydrofolate reductase (DHFR) back to the THF form. Biosynthesis of the folate moiety itself is mediated by five enzyme activities, while interconversion of folate among the various forms required for C1 transfer reactions is principally carried out by a further four activities. One of the latter, folylpolyglutamate synthase (FPGS), converts reduced folates to their polyglutamated forms, in which extra glutamate residues are conjugated to the single glutamate whose addition to dihydropteroate produces DHF. It has been shown in other systems that this process is critical for the cellular retention of folates as well as enhancing their affinity for other folate-dependent enzymes (Krumdieck et al., 1992).
Fig. 2.
Principal enzymes and substrates of the folate pathway involved in formation of tetrahydrofolate (THF) and its utilisation in the thymidylate cycle. Other enzymes involved in the interconversion of tetrahydrofolate forms modified at the N5 and N10 positions of the pteroate moiety are not shown. Salvaged pre-formed folate feeds into the pathway, commonly in the form of folic acid or folinic acid (from culture medium) or 5-MeTHF (from host plasma), and modified appropriately. pABA can also be salvaged from the above sources or synthesised de novo from the shikimate pathway. Note (1), the triphosphate group is removed before the DHNA step, thought to be by non-enzymatic loss of pyrophosphate and subsequent removal of the final phosphate by a non-specific phosphatase activity. Note (2), polyglutamated forms are the preferred substrates for folate pathway enzymes and the predominant forms within the cell. It is not known whether newly synthesised folate in P. falciparum is polyglutamated at the DHF or THF stage (or both). For simplicity, it is shown here as occurring at the THF stage (large brackets), but omitted from substrate names in the thymidylate cycle. Activities targeted by the components of Fansidar, pyrimethamine (PYR) and sulfadoxine (SDX), are indicated. GTPC, GTP cyclohydrolase I (EC 3.5.4.16); DHNA, dihydroneopterin aldolase (EC 4.1.2.25); PPPK, hydroxymethyldihydropterin pyrophosphokinase (EC 2.7.6.3); DHPS, dihydropteroate synthase (EC 2.5.1.15); DHFS, dihydrofolate synthase (EC 6.3.2.12); DHFR, dihydrofolate reductase (EC 1.5.1.3); FPGS, folylpolyglutamate synthase (EC 6.3.2.17); SHMT, serine hydroxymethyltransferase (EC 2.1.2.1); TS, thymidylate synthase (EC 2.1.1.45).
Starting with the cloning of the dihydrofolate reductase-thymidylate synthase (dhfr-ts) gene in 1987 (Bzik et al., 1987), genes encoding all of these enzymic functions had been putatively or conclusively identified either before the P. falciparum genome sequencing project began in 1996, in the case of dhfr-ts and the hydroxymethylpterin pyrophosphokinase-dihydropteroate synthase (pppk-dhps) gene (Brooks et al., 1994; Triglia and Cowman, 1994), or before it was completed in 2002, in the case of the GTP cyclohydrolase I (gtpc) (Lee et al., 2001), the dihydrofolate synthase-folylpolyglutamate synthase (dhfs-fpgs) (Lee et al., 2001; Salcedo et al., 2001) and the serine hydroxymethyltransferase (shmt) genes (Alfadhli and Rathod, 2000; Lee et al., 2001), with the notable exception of the dihydroneopterin aldolase (dhna) gene. It was anticipated that publication of the completed genome sequence (Gardner et al., 2002) would clear up this loose end, but no candidate was identified from the predicted protein sequences of the ~5300 putative open reading frames. Despite considerable efforts in attempting to pinpoint this gene, both experimentally and using sequence- and structure-based search programs, we have as yet been unable to identify a plausible candidate. At present the possibilities are that this protein, which is poorly conserved among other organisms, is so divergent that it is unrecognisable even using very sophisticated bioinformatics tools, that its gene is fragmented into too many small exons for ORF prediction programs to cope with, or that it is genuinely absent from the parasite. If the last of these, the necessary step of removing the aldol group from 7,8-dihydroneopterin before addition of the pyrophosphate moiety to the resulting hydroxyl group would have to be carried out by a different, non-orthologous protein, or, also a formal possibility, by a non-protein-based mechanism. In parasite systems, it is not uncommon for a host function to be subverted by the parasite to its own ends, but in this case humans, incapable of making their own folates, lack a DHNA homologue. One possibility is that the PPPK enzyme that follows DHNA in the conventional catalytic sequence also has this ability, but no functional studies have yet been reported for this domain of the bifunctional PPPK-DHPS protein. It is interesting to note however, that the P. falciparum PPPK domain includes two long insertions of ~90 amino acids each (Brooks et al., 1994; Triglia and Cowman, 1994) that are not found in any of the known PPPK molecules from bacterial or non-apicomplexan eukaryotic sources. The first is highly conserved in length and sequence in other Plasmodium species, whereas the second is only about 40 amino acids long in the latter, albeit well conserved where it aligns with the much longer P. falciparum sequence. The orthologue from the closely related apicomplexan parasite Toxoplasma gondii has similar but shorter insertions (50 and 12 amino acids, respectively) in the equivalent positions (Pashley et al., 1997). Another candidate that would keep DHNA in the family of conventional folate pathway enzymes would be the N-terminal region of the GTP cyclohydrolase I (GTPC) molecule upstream of all of the residues known to participate in the catalytic function. This N-terminal domain is much extended compared to that found in the orthologues of other organisms (Lee et al., 2001); for example, the plasmodial molecules exceed the human protein by lengths ranging from 25 (P. chabaudi) to 201 (P. knowlesi) amino acids (see GenBank entries AF486639, AF486640, AY582138, AY604168, AF043557, AY458431). Interestingly however, while the GTPC molecules from the primate and human malaria parasites display reasonable sequence complexity in this region, we found those from the rodent malarias (especially Plasmodium yoelii nigeriensis and P. yoelii yoelii) to be based to a greater or lesser degree on imperfect repeats of a pentapeptide motif. Overall, the degree of conservation among the various Plasmodium species, both at the primary sequence and predicted secondary structure levels, is very low in this part of the molecule, strongly suggesting that the latter harbours no extra catalytic activity, although it may play some kind of species-specific regulatory and/or targeting role.
Of the gene products in Fig. 2 other than DHNA, the predicted activities of all but GTPC have been verified experimentally, either by standard enzyme assays performed on recombinant proteins produced from the various cloned genes (e.g. Sirawaraporn et al., 1990; Triglia et al., 1997; Shallom et al., 1999; Alfadhli and Rathod, 2000), or by creating Escherichia coli or Saccharomyces cerevisiae mutants deficient in the relevant activity and restoring prototrophy by transforming in the candidate P. falciparum gene (Hall et al., 1991; Wooden et al., 1997; Shallom et al., 1999; Salcedo et al., 2001). The identification of the gtpc gene thus rests at present on its having the only open reading frame (ORF) in the genome whose predicted protein sequence shows very strong similarity to the C-proximal, catalytic domain of known enzymes from other organisms. Outside of the schema of Fig. 2, other enzymes feed into the folate pathway or depend upon folate cofactors, and, while not detailed here, we may note that there are further gaps compared to other organisms that conceptual translation of the complete genome sequence has not obviously filled.
3. Drug resistance in vitro and in vivo
DHFR is the target of the antifolate drugs PYR and cycloguanil (the active metabolite of the pro-drug proguanil), while the sulphur-based drugs (‘sulfa drugs’) such as SDX and dapsone inhibit DHPS. The role of a small number of point mutations in the dhfr domain in generating drug resistance first became apparent in the late 1980s (Cowman et al., 1988; Peterson et al., 1988; Snewin et al., 1989), followed in 1994 by a similar discovery for dhps (Brooks et al., 1994; Triglia and Cowman, 1994), and has been well documented in subsequent reviews (e.g. Hyde, 1990, 2002; Plowe et al., 1998; Sibley et al., 2001). A question that has long been associated with these studies is whether the mutations observed in DHFR and DHPS account fully for resistance levels seen in vitro and in vivo. This is difficult to investigate systematically as the drugs targeting these enzymes act synergistically (see Section 6) and the different combinations of dhps and dhfr haplotypes seen in individual samples in the field currently exceeds 30 (Plowe et al., 1997; Wang et al., 1997a). In natural infections, it would also be expected that host factors play a significant part in the eventual outcome of drug intervention, and it has often been observed that parasites resistant to drugs in vitro by virtue of their mutational status can be cleared by some patients, emphasising the important role of host immunity in this process (Sibley et al., 2001; Djimde et al., 2003). Conversely, clinical drug failures where parasites are typed as wild-type or only moderately resistant to antifolates presumably result from poor compliance or differences in metabolic clearance rates. Studies in vitro indicate that the concentration of folate-related molecules in the medium and in the host blood cells can have an important influence on the ability of a drug to inhibit parasites, especially in the case of the sulfa drugs, such as SDX (Wang et al., 1997c), discussed further below in connection with folate salvage. Also relevant is the several-fold stimulation of DHFR and TS activities induced by challenge with antifolate drugs that is postulated to occur by a mechanism involving the derepression of mRNA translation, and is independent of the mutational status of the dhfr-ts gene (Nirmalan et al., 2004b). Moreover, it cannot be excluded that other gene products contribute in at least some cases to antifolate drug resistance. This is suggested by the recent discovery of sulfa-resistant isolates of P. falciparum that are still wild-type in dhps (Mberu et al., 2002), and has been directly demonstrated in the rodent parasite P. chabaudi, where quantitative trait locus analysis of a genetic cross between clones with different resistance patterns to PYR, SDX and PYR/SDX indicates the influence of one or more genes other than dhfr and dhps on the observed levels of resistance in cross progeny (Hayton et al., 2002).
An important aspect of resistance to antifolates in vivo is the selection of resistant populations from parasites in the presence of sub-lethal amounts of a drug. It has been shown that inhibitors such as PYR and SDX that are cleared relatively slowly from the body are more likely to encourage selection than drugs with short half-lives (Watkins et al., 1997; Nzila et al., 2000b; Hastings et al., 2002). Attempts have been made to predict how this phenomenon will play out for Fansidar (the clinical formulation of PYR and SDX), depending upon the initial dhfr and dhps genotypes of the infecting parasites, and what the clinical outcome is likely to be, taking into account the range of drug clearance rates observed in different patients (Watkins et al., 1997; Wang et al., 1999; Hastings et al., 2002). A new combination of antifolates, lapudrin (chlorproguanil) and dapsone, which have much shorter half-lives than PYR and SDX, is currently being deployed (as ‘LapDap’) in East Africa. This potent combination (Nzila-Mounda et al., 1998) is able to clear Fansidar-resistant parasites and it is hoped that resistance to it will arise much more slowly than has occurred with Fansidar itself, despite ostensibly targeting the same enzymes (Mutabingwa et al., 2001; Sulo et al., 2002). Such resistance may be further delayed by the addition of an artemisinin-based component, which targets a quite different parasite function (or functions), and such artemisinin combination treatments (ACT) are currently being intensively explored as a general means of maintaining existing drugs as useful therapeutic tools for much longer periods than would be the case if they were deployed individually (Price and Nosten, 2001; Winstanley et al., 2002).
4. Two routes to folate metabolites:salvage and biosynthesis
As indicated above, most microorganisms are able to synthesise the folates they need from the simple precursors GTP, p-aminobenzoic acid (pABA) and l-glutamate, which are assembled into the canonical folate structure shown in Fig. 1. Higher organisms such as ourselves are unable to do this, so we depend on dietary intake of pre-formed folate as an essential nutrient. P. falciparum has the ability to exploit both of these routes (Milhous et al., 1985; Krungkrai et al., 1989; Wang et al., 1997b, 1999), utilising folate either provided in culture medium in vitro or salvaged from the host plasma in vivo on the one hand, or converting the above precursors de novo into folate derivatives on the other (Fig. 2). This is at once a feature of considerable interest as well as a complicating factor for experiments that attempt to unravel the finer details of folate metabolism.
A central question, with important implications for the development and validation of new drugs targeting the folate pathway, concerns the relative importance of the biosynthetic pathway compared to folate salvage from the human host in vivo and the interplay between them. Crucially, we have little knowledge of the needs of the parasite to use each of these routes, nor of its flexibility in altering their balance in adverse circumstances, such as sulfa-drug inhibition of DHPS in the de novo pathway, or depleted exogenous folate levels in the host plasma as a result of nutritional deficiency. Thus, folate is available in the human host plasma principally as 5-methyltetrahydrofolate (MeTHF), but at a low concentration, averaging around 10–15 nM in adults (Baker et al., 1994). However, in cases of malnutrition, this can fall to ~10-fold less than normal (Nelson et al., 2003). Inhibition studies of the DHPS activity with high levels of SDX indicate that in vitro, blockage of folate biosynthesis can apparently be bypassed via the salvage route, suggesting that biosynthesis may be dispensable, although there are significant differences among strains in the efficiency with which salvaged folate is utilised (Wang et al., 1997b) (see below). In the field, however, there is a strong correlation between SDX-resistant forms of the parasite carrying mutations in the dhps gene and the prevalence of Fansidar usage (Wang et al., 1997a; Plowe et al., 1998; Nzila et al., 2000a; Sibley et al., 2001), suggesting that in normal infections of the human host, folate salvage cannot completely satisfy the parasite's requirements, a conclusion in accord with recent data from genetically modified parasites (see Section 5).
4.1. Genetic analysis of folate salvage
One approach to investigating salvage that we adopted was to ask whether different strains of the parasite could utilise exogenous folate to the same degree. This has largely taken the form of two types of investigation. The first was to exploit the observation that, among the small number of sexual crosses that have been performed with P. falciparum (Walliker et al., 1987; Wellems et al., 1990), one parental pair, the cloned lines HB3 and Dd2, show a quite marked difference in phenotype with respect to salvage. This is most easily observed in the way these parasites respond to exogenous folic acid when in the presence of high concentrations of SDX, which inhibits growth by suppression of de novo synthesis of folate. Addition of low levels of folic acid to Dd2 (~10 nM) in these circumstances restores growth to levels similar to those in the absence of drug. In contrast, addition of quite high concentrations of folic acid to HB3 (>200 nM) alleviates the growth inhibition to only a small degree, suggesting that this strain is much less able to use the folate provided to bypass the SDX block on biosynthesis. To investigate the genetic basis of this phenotypic difference, we followed the pattern of folic acid utilisation in a large number of cloned progeny lines resulting from the cross. Initial data (Wang et al., 1997b) pointed to a locus on chromosome 4, the home of the dhfr-ts gene, but a more accurate assignment awaited completion of the DNA sequence of this chromosome (Hall et al., 2002). Knowing then the precise positions of the polymorphic markers that flagged crossover points in the recombinant progeny (Su et al., 1999), we were able to map the critical locus to a relatively small stretch of DNA (48.6 kb) that encompassed dhfr-ts and seven other genes (Wang et al., 2004a). Although we cannot yet formally exclude all of these neighbouring genes, there are seemingly no promising candidates for folate processing proteins among them, six of which encode hypothetical proteins with no obvious counterparts in other organisms, and the seventh a putative regulator of protein kinase A that appears not to be expressed in the asexual parasite stages used in our studies. We therefore adopted the working hypothesis that the dhfr-ts flanking sequence or its coding sequence is intimately linked with the differences in folate utilisation that we observed among the parents and progeny of the HB3-Dd2 cross, as well as in other parasite lines. Although we found an interesting dimorphism in the 5′ untranslated region of Dd2 and HB3 mRNA in the form of a directly repeated 256 bp sequence, present in two copies in Dd2, but only one in HB3 (Nirmalan et al., 2002), this did not correlate in the cross progeny or other parasite lines with either the folate utilisation phenotype or levels of mRNA and protein. Moreover, no other significant differences were found between HB3 and Dd2 in the entire ~2.5 kb intergenic region between the dhfr-ts coding sequence and the next upstream ORF. In fact, the phenotype correlated only with the coding sequence of the dhfr domain, leading to the suggestion that it arises from a particular property of the single mutant (S108N) form of this gene product, which is not evident in the case of the wild-type sequence nor, perhaps unexpectedly, when further mutations at positions 51 and 59 accompany the S108N alteration at position 108 (as in Dd2) (Wang et al., 2004a).
4.2. Biochemical analysis of folate salvage and biosynthesis
To complement the above genetic analysis, a second approach was to supply radioactive folates to parasites and follow the fate of their derivatives on HPLC as they are utilised via the salvage pathway, and to compare this with conversion of radioactive pABA to folate end-products via de novo synthesis (Wang et al., 2004a). Earlier results using this technology (Krungkrai et al., 1989) showed clearly that exogenous folic acid was indeed taken up by the parasite and then converted to polyglutamated end-products, this latter step expected from studies on other organisms. We have used this approach to compare HB3 and Dd2, as well as to compare stably transfected parasites carrying a defective gene of interest with the unmanipulated host (see Section 5). In HPLC separations, it is difficult to differentiate among folates that have different oxidation states of the pterin ring and/or variation in the substituents at the 5 and 10 positions. However, it is more straightforward to resolve folates differing in their level of polyglutamation, which has a more profound influence on their retention times (Selhub, 1989). It has been reported (Krungkrai et al., 1989) that the predominant polyglutamated form of folate in P. falciparum is the pentaglutamate, and we in turn have seen no evidence for folate end-products extending beyond this level of modification.
As the conventional sequence of events in other microorganisms is the reduction of folate to the THF level followed by addition of the polyglutamyl tail (Green et al., 1996), we used progression towards the highest degree of polyglutamation over a given time as a measure of the facility with which a particular precursor was utilised.
For both the HB3 and Dd2 parasite lines, as well as the FCR3 line used in our transfection studies (Section 5), the de novo biosynthetic route using the pABA provided appeared more efficient than the salvage of pre-formed folate, especially for HB3 and Dd2, as judged by their ability to convert precursors to polyglutamated end-products in a given period. Moreover, for all of the parasite lines we have tested to date, conversion of salvaged folic acid was much less facile than seen for the fully reduced folinic acid (5-formyltetrahydrofolic acid). This is a probable indication that reduction of the pterin ring from its fully oxidised form represents a significant hurdle to either uptake or processing beyond the monoglutamate stage. However, we consistently found that labelling with folic acid compared to folinic acid was less efficient for HB3 than Dd2 by a factor of ca. 2–3-fold. Although a relatively small factor, this may be a crucial contributor to relative survival in our folate utilisation experiments when the parasites become dependent upon the salvage route following blockage of biosynthesis by SDX.
4.3. How is exogenous folic acid utilised?
The proven ability of P. falciparum to utilise folic acid necessitates the action of a folate reductase (FR) activity or a pteridine reductase akin to that found in trypanosomatids (Robello et al., 1997; Luba et al., 1998). However, no obvious homologue of the latter is found in Plasmodium, consistent with the fact that wild-type malaria parasites are highly sensitive to DHFR inhibitors whereas trypanosomatids are not, as the pteridine reductase activity in the latter also has the ability to reduce DHF, bypassing the block on DHFR. From our cross data and from biochemical considerations, the primary candidate for providing a FR activity would be DHFR-TS. Thus, DHFR molecules from widely different organisms exhibit a slow-acting low-level FR activity (Futterman and Silverman, 1957; Coward et al., 1974; Baccanari et al., 1975). In mammals, ratios of Vmax for FR relative to DHFR at physiological pH range from ~0.01 to 0.04-fold, with Km values for folic acid correspondingly ~8 to 20-fold higher than for DHF (Coward et al., 1974). Although a FR activity has not been reported in Plasmodium, its likely low level, as evidenced by the inefficiency with which oxidised folic acid is utilised relative to folinic acid, would require an extremely sensitive assay, and a definitive identification might be difficult to achieve. However, if parasite DHFR-TS does have FR activity, it is likely to be differentially affected by the various PYR-resistance mutations. This is the case for the DHFR activity itself, where for its normal substrate, DHF, the singly mutant DHFR-TS (S108N) from HB3 has a Km ~12-fold higher than the wild-type and ~3-fold higher than the double mutant (S108N, N51I), as measured both for the native enzymes extracted directly from parasites (Chen et al., 1987) and for the full-length recombinant equivalents of these DHFRTS variants expressed in E. coli (Sirawaraporn et al., 1990). Moreover, reported turnover numbers (kcat/Km) for the native enzymes are 15.2 μM−1 s−1 for wild-type, 4.0 μM−1 s−1 for the double mutant, but only 1.6 μM−1 s−1 for singly mutant HB3 (Chen et al., 1987). Thus, the DHFR activity of HB3 appears to be particularly adversely affected by the S108N alteration, although a direct comparison with triply mutant Dd2 cannot be made as equivalent data for the full-length DHFR-TS from this line have not been reported. It should also be noted that studies on the isolated DHFR domains carrying the various altered amino acids do not support this trend (Sirawaraporn et al., 1997a,b). If true for the native DHFR-TS enzymes, however, passage of folate around the thymidylate cycle to produce dTMP would also be predicted to be less efficient for HB3-type parasites, whatever its initial oxidation state. Thus, a combination of relatively poor FR and DHFR activities in HB3 compared to other variants might then be problematic under conditions of SDX inhibition where folic acid is the sole source of salvageable material, and could perhaps explain the marked differences in survival that we see when HB3 and Dd2 are compared in the above assay (Wang et al., 1997b). A sub-optimal DHFR activity with reduced fitness may also explain the apparent under-representation of such singly mutant strains, compared to wild-type and higher mutants, that is often observed amongst field isolates of P. falciparum. For example, in >120 samples reported as having single dhfr genotypes from a number of regions where antifolates were deployed, but wild-type parasites were still common, the overall relative abundances of the variant forms were 42% wild type, 3% single (S108N) mutants, 26% double mutants and 28% triple mutants (Reeder et al., 1996; Wang et al., 1997a). These data may indicate that mutations over and above the key S108N alteration act largely as compensatory changes in the molecular structure to ensure that the parasite can continue to process the natural substrates to an adequate degree, while significantly reducing the binding affinity of the drug.
4.4. Utilisation of pABA
Analogous to the situation with folate, which can be acquired via the salvage route or by de novo biosynthesis, is that of the pABA required in the latter pathway. Thus, exogenous pABA can be taken up from standard medium in vitro or from host plasma in vivo, where it is estimated to be present in widely varying concentrations over the range of ~0.1 to 6 μM (Schapira et al., 1986). However, genes and enzymes of the shikimate pathway have been identified in P. falciparum (Roberts et al., 1998; Triglia and Cowman, 1999; Fitzpatrick et al., 2001; Gardner et al., 2002), from which pABA can also be synthesised de novo via chorismate. Again, the question arises as to the relative dependency of the parasite on these two potential sources. Compounds that bind to 5-enolpyruvyl shikimate 3-phosphate synthase in the shikimate pathway inhibit growth of P. falciparum, and this can be relieved by providing exogenous pABA (Roberts et al., 1998). Moreover, mutant parasites that are auxotrophic for pABA have been produced by gamma-irradiation, where it was deduced that a pABA synthase activity had been disabled (McConkey et al., 1994). These experiments suggest that this pathway does produce pABA in metabolically significant concentrations. Conversely, however, the rodent parasite P. yoelii has been shown to be incapable of surviving without exogenous pABA, despite the presence of the shikimate pathway (Kicska et al., 2003). We are currently attempting to disable the relevant genes of this pathway in P. falciparum to better assess its importance as a source of pABA, but it may well be that, similar to the situation with folate itself, the wild-type parasite requires input from both the endogenous and exogenous routes. Indeed, in the latter case, experimental evidence suggests that either pre-formed folate or pABA will serve as an adequate supplement for normal development, although growth rates tend to be somewhat slower in pABA-only medium.
5. Gene disruption to study function
A potentially powerful approach to further investigating the functions of the individual folate pathway genes is to use parasite transfection to knock out or modulate their activities. Such transfectants can then be subjected to experiments of the type described above, to shed additional light on synergy, folate synthesis and processing, drug inhibition, etc. Although technically challenging because of an extremely low frequency of integration of the modifying sequence (O'Donnell et al., 2001; Duraisingh et al., 2002), this general approach has been successfully used to investigate, for example, candidate parasite genes involved in cytoadhesion (Crabb et al., 1997a), cellular invasion (Triglia et al., 2000), resistance to quinoline drugs (Reed et al., 2000; Waller et al., 2003) and sexual development (Kongkasuriyachai et al., 2004). However, to apply this technique to the folate pathway, we required a system that did not depend upon the resistance to the antifolate drugs PYR or WR99210 (Fidock and Wellems, 1997) as the selectable marker for successful transfection. Such resistance has been used in most reported transfection experiments to date and is conferred by an integrated mutant dhfr gene from Toxoplasma gondii or Homo sapiens (e.g. Crabb et al., 1997b; Fidock et al., 1998). As expression of these genes will strongly perturb folate pools within the parasite, we developed an alternative system exploiting resistance to the antibiotics blasticidin S (BS) and geneticin (G418) as markers, previously used to select transient transfectants of P. falciparum (Ben Mamoun et al., 1999). In this system, integrants carrying the sequence used for gene modification are positively selected with these drugs, which are not known to be active against enzymes of the folate pathway (Wang et al., 2002). We targeted initially the dhps domain of pppk-dhps, which as described above, constitutes a critical component of the biosynthetic pathway, with a view to investigating further the importance of folate biosynthesis in the parasite compared to the salvage of pre-formed folate. P. falciparum is a haploid organism throughout its erythrocytic life cycle and our hypothesis was that, in vitro, transfection mutants with a disruption in the single pppk-dhps gene should be viable if an adequate amount of folate could be salvaged and properly processed from the culture medium, in which it is provided at high concentration. This was tested using a series of constructs that varied in the degree to which the wild-type sequence had been altered, to allow for the possibility that complete elimination of DHPS activity might be lethal, regardless of exogenous folate supplementation (Wang et al., 2004a). This indeed appeared to be the case as repeated transfections with either of two replacement dhps genes truncated in different positions towards the 3′ end yielded no detectable integrants after extensive periods of selection, even in the presence of reduced folate in the form of folinic acid, which as described above, is a more readily utilised substrate than folic acid. As both truncations were likely to result in complete disablement of the dhps gene product, this indicated that a total loss of activity could not, in fact, be tolerated by the parasite. In contrast, constructs transfected in parallel that reconstituted the full-length gene reproducibly yielded viable lines with the correct integration after a short period of G418 selection. One of these had been mutated in two active-site residues within a key, highly conserved motif near the C-terminus of the DHPS domain, where RVHDV (residues 686–690) was altered to QVQDV. This integrant still retained a very low level of DHPS activity, as demonstrated by its ability to convert radioactive pABA to folate end-products at levels just detectable using HPLC. However, although viable with folate supplementation, this mutant lost the ability to survive when reliant on conversion of pABA as the sole source of folate.
The conclusion from these experiments, that some DHPS activity must be maintained for parasite viability, albeit at a low level, was in apparent conflict with the earlier studies described above (Wang et al., 1997b,c), which showed that parasite lines like Dd2 could grow at a normal rate in high concentrations of SDX if exogenous folate was present. However, this assumes that the SDX present in such cultures shuts down DHPS activity completely. One resolution of this apparent paradox would be that DHPS activity is required in more than one compartment in the cell, and that folate salvaged exogenously cannot fulfil the requirements of all compartments, at least one of which would be dependent upon provision of DHF synthesised de novo. It would also follow that the DHPS in such a compartment would necessarily be less susceptible to SDX inhibition than DHPS located in the cytoplasm. Possible compartments could include the mitochondrion, where folate biosynthesis occurs in a range of organisms, including yeast, plants and mammals (DeSouza et al., 2000; Ravanel et al., 2001), the apicoplast, an organelle related to chloroplasts where folates are also found in plants (Gambonnet et al., 2001) and the nucleus, for the last of which we have preliminary immunofluorescence and EM data indicating the presence of folate pathway enzymes (see Section 7). Another scenario could be that, whereas the exogenous folate might satisfy the parasite's requirements for C1 transfer reactions, a residual DHPS activity is still needed to help counteract a potentially toxic build-up of folate breakdown products. Thus, in an older study using radiolabelled precursors, it was estimated that >50% of the folate salvaged by the parasite was split into pABG and pterin-6-CHO (Krungkrai et al., 1989). Further productive usage of these components would ultimately require DHPS to reform the 9–10 bond of the folate structure by rejoining pABG to appropriately modified pterin moieties. However, we found that growth of the transfectant with the heavily compromised DHPS was comparable to that of the host over many cycles when either exogenous folic acid or folinic acid was provided, suggesting that DHPS does not have a major role in the provision of functional folates from salvaged material that has subsequently been broken down. Consistent with this, we saw no evidence of radiolabelled catabolic intermediates in parasite extracts. This suggests that cleavage of salvaged folate does not, in fact, occur to an appreciable extent in vivo before it is utilised or stored, but we cannot yet exclude a role for DHPS in dealing with any unwanted side-products.
Another important conclusion can be drawn from the metabolic labelling studies using radioactive pABA, folic acid or folinic acid precursors. Significantly, in the case of both folic and folinic acid, the dhps-disabled mutant was not only more highly labelled than the unmodified host FCB, but also yielded a higher ratio of polyglutamated to monoglutamated labelled products. This is likely to reflect the fact that this mutant, having a minimal ability to produce folate de novo, is now reliant on deriving a much greater proportion of the required flux into polyglutamated end-products from the salvage pathway. Whatever the relative importance of the biosynthetic and salvage pathways in natural isolates, the picture thus seems to be emerging that both routes are required by the parasite, and that it can compensate to a certain degree if one or other of these routes is compromised. We have shown this in parasites where the DHPS activity has been drastically reduced and are currently investigating the converse situation where salvageable folate is either not provided or its uptake inhibited. That salvage cannot be dispensed with entirely is suggested by the simple observation that parasites starved of folate supplementation are very difficult to maintain in culture for any length of time, although, as noted previously, provision of exogenous pABA can serve as a reasonably effective substitute.
6. Synergy of antifolate drugs
An important property of antifolate combinations such as Fansidar is the considerable synergy displayed by their constituent drugs. This has been directly responsible for the continuing therapeutic utility of SDX and PYR, despite the appearance and widespread occurrence of parasites resistant to the individual components. Given the key role of synergy in clinical efficacy, it is important to understand its molecular basis. Commonly though, theories of synergy assume sequential blockade in a linear pathway (Burchall, 1977), but as well as synthesising THF de novo from DHF produced via DHPS and DHFS, DHFR is involved in the cyclical pathway of thymidylate synthesis, reducing the DHF produced by TS each time the latter converts a molecule of dUMP to dTMP (Fig. 2). Moreover, the existence of the salvage pathway described above further complicates the picture, because as we have seen, reducing the supply of folate via de novo synthesis can be compensated for by increasing flux through the salvage pathway, although eliminating it entirely appears to result in non-viability (Wang et al., 2004a).
Comparisons of transfected parasites deficient in DHPS activity with host FCB in their responses to PYR and SDX proved that synergy is absolutely dependent upon DHPS activity (Wang et al., 2004a), ruling out one hypothesis that it was due to simultaneous action of SDX and PYR on DHFR (Chulay et al., 1984). This is consistent with studies using purified DHFR from P. falciparum that showed negligible synergy at physiological drug levels (Wang et al., 1999).
Alternative scenarios include interference of PYR with exogenous folate utilisation, considerably increasing the reliance of the parasite on the biosynthetic pathway, and thus its susceptibility to sulfa drugs (Sims et al., 1999; Wang et al., 1999), and the conversion of SDX by DHPS to sulfa-pterin adducts that inhibit activity further downstream, possibly at DHFR as previously speculated (Chulay et al., 1984). Such adducts, currently being investigated in P. falciparum (Patel et al., 2004), are found to be inhibitory to several folate pathway enzymes of E. coli, but not to DHFR (Roland et al., 1979). They also inhibit the growth of yeast, but their target has not yet been identified (Patel et al., 2003, 2004). If adduct formation does have a significantly detrimental effect on parasite metabolism, then the observed sulfa-drug resistant mutants may be selected on the basis that they produce lower concentrations of such toxic molecules, rather than merely reduce competition between pABA and sulfa-drug binding to DHPS.
Interestingly, the transfected line defective in DHPS activity is considerably more sensitive to PYR alone than the host (Wang et al., 2004b). This could reflect the fact that once parasites have lost most of their ability to make their own folate de novo, pools of DHF are likely to be diminished and thus compete less effectively with the drug for binding to DHFR, despite some of this deficit apparently being offset by increased flux through the salvage pathway. This is complementary to the observation that inhibition of folate uptake with the anti-gout drug probenecid increases sensitivity to antifolate inhibitors (Nzila et al., 2003), and again emphasises the likely importance to P. falciparum of both arms of the pathway.
7. Conclusions and some future perspectives
Despite the fact that the folate pathway inhibitors are amongst the oldest synthetic antimalarials, and that widespread resistance to Fansidar now exists amongst populations of P. falciparum, this metabolic pathway is one of the very few in the parasite's repertoire that is a clinically proven target. This, combined with the fact that only two of the component enzymes have been targeted to date, means that it is clearly important to thoroughly evaluate other members of the pathway as potential novel targets—not only the enzymes described here, but also those outside the scope of this article that utilise folate-based cofactors. Such evaluation needs to be considered in the context of the precise function(s) of the gene products and the interplay between the biosynthetic and salvage routes in providing the requisite folate cofactors. While a clearer picture of this pathway is beginning to emerge, there are still fundamental questions to be answered. For example, we do not know the mechanisms by which exogenous folates are transported into the infected red blood cell and then into the parasite itself. Moreover, we have no clear idea of the intracellular location(s) of the folate pathway enzymes nor of the extent of their co-localisation or possible assembly into supramolecular complexes. As indicated above, biochemical studies hint at a possible compartmentation, but direct visualisation is required to establish a true picture. To this end, we have raised polyclonal antibodies to recombinant versions of all of the enzymes for which we have cloned genes and are using these in confocal microscopy studies. Preliminary data indicate that patterns of distribution are complex and dynamic, are not confined to the cytoplasm, despite a lack of known targeting sequences in any of the proteins, and vary across the different stages of the erythrocytic life cycle (Read and Hyde, unpublished data).
At the structural level, the recent solution of the X-ray structure for DHFR-TS from P. falciparum (Yuvaniyama et al., 2003) represents a major step forward in understanding in detail the mechanism of action of the two component activities and how the well-characterised mutations in the DHFR domain lead to resistance to anti-DHFR inhibitors like PYR and cycloguanil, and why experimental compounds such as WR99210 (Kinyanjui et al., 1999) should bind so tightly to all mutant forms of DHFR. Importantly, this opens the door to rational design of less toxic versions of potent molecules like WR99210, and of inhibitors that can overcome the loss of binding suffered by PYR and cycloguanil by avoiding unfavourable contacts with the mutant amino-acid residues. Proof of principle for this type of design has already been demonstrated using 3D models of DHFR-TS based on determined structures of orthologues from other organisms to produce effective variants of PYR (McKie et al., 1998; Sardarian et al., 2003). However, the acquisition of the plasmodial X-ray structure allows this type of approach to be confidently taken to a new level, not only for the DHFR domain, but also for TS, and already, new types of antifolate compounds with good inhibitory properties designed on the basis of the crystallographic results are being reported (Rastelli et al., 2003). It is likely that the PPPK-DHPS molecule will be the next in the pathway to have its 3D structure solved, and this will represent the first molecule of the bifunctional type found in protozoa and plants to be so characterised. Such data provide a major impetus for new ideas and experiments to identify and develop effective inhibitors of the parasite. Where there are human counterparts, as for GTPC (used for reduced biopterin synthesis in the nitric oxide pathway, rather than folate synthesis), DHFR, TS, SHMT and FPGS, comparison of host and parasite 3D structures will allow the design of molecules with much higher affinities for the parasite version of the enzyme, rather than depending upon empirical discovery of differential binding properties, as was the case for PYR and cycloguanil.
Whereas structural studies of the different enzymes will be invaluable for understanding catalytic and resistance mechanisms, and permitting rational design and improvement of drugs, we have as yet virtually no knowledge of the factors regulating these activities in the folate pathway. No transcription factors are known that operate on the genes encoding the enzymes, and we know very little about feedback mechanisms. Some interesting insights have however been gained. Thus, it has been shown that, similar to the mammalian system, the DHFR-TS protein of P. falciparum is capable of binding to its own mRNA and suppressing its translation (Zhang and Rathod, 2002), although there are conflicting data as to the ability of antifolate drugs to perturb and relieve this suppression (Zhang and Rathod, 2002; Nirmalan et al., 2004b). This specific binding ability also appears to extend to the gene sequence itself (Rathod, personal communication), and we have preliminary immunofluorescence and EM data indicating that the DHFR-TS protein can be found in the nucleus. This raises the fascinating possibility, for this key folate pathway enzyme at least, that it is capable of directly regulating its own production both at the transcriptional and translational levels.
The generally low level of expression of the folate pathway enzymes means that we have been unable as yet to identify any members on silver-stained 2D gels of parasite lysates, but we and others (Florens et al., 2002; Lasonder et al., 2002) have identified PPPK-DHPS, DHFS-FPGS, DHFR-TS and SHMT after 2D liquid chromatography separation of extracts followed by mass spectrometry, although the identification of DHFS-FPGS was tenuously based upon only a single tryptic peptide (Lasonder et al., 2002). However, these powerful technical advances open the way for studying how levels of these and other proteins are altered by external perturbations such as drug treatment of the parasites or alteration in the availability of folate metabolites, and will complement studies of these factors at the transcriptional level (Nirmalan et al., 2002). In anticipation of such studies, we have recently developed a quantitative proteomics method that can be applied to essentially all proteins of P. falciparum, whereby cultures are labelled with heavy-isotope-containing isoleucine and compared to parallel cultures grown in the presence of normal isoleucine (Nirmalan et al., 2004a). Proteins from experimental and control cultures can then be accurately quantified after mixing at the cell stage and processing of this mixture for measurement of light and heavy peptide counterparts in a single scan on the mass spectrometer. Quantitative measurements of the relevant metabolites as well as the enzymes involved will ultimately be essential to enable flux analysis of the folate system to be undertaken, and thus to gain a more complete understanding of this complex pathway and the mechanisms of resistance that the parasite can deploy to combat chemical intervention.
Acknowledgement
Work described above from the author's laboratory was principally supported by grants 056845 and 073896 from the Wellcome Trust, UK.
References
- Alfadhli S, Rathod PK. Gene organization of a Plasmodium falciparum serine hydroxymethyltransferase and its functional expression in Escherichia coli. Mol. Biochem. Parasitol. 2000;110:283–291. doi: 10.1016/s0166-6851(00)00282-6. [DOI] [PubMed] [Google Scholar]
- Baccanari D, Phillips A, Smith S, Sinski D, Burchall J. Purification and properties of Escherichia coli dihydrofolate reductase. Biochemistry. 1975;14:5267–5273. doi: 10.1021/bi00695a006. [DOI] [PubMed] [Google Scholar]
- Baker H, Tenhove W, Baker E, Frank O. Vitamin activities in human portal, hepatic and femoral blood after vitamin ingestion. Int. J. Vitam. Nutr. Res. 1994;64:60–67. [PubMed] [Google Scholar]
- Ben Mamoun C, Gluzman IY, Goyard S, Beverley SM, Goldberg DE. A set of independent selectable markers for transfection of the human malaria parasite Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 1999;96:8716–8720. doi: 10.1073/pnas.96.15.8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DR, Wang P, Read M, Watkins WM, Sims PFG, Hyde JE. Sequence variation of the hydroxymethyldihydropterin pyrophosphokinase—dihydropteroate synthase gene in lines of the human malaria parasite, Plasmodium falciparum, with differing resistance to sulfadoxine. Eur. J. Biochem. 1994;224:397–405. doi: 10.1111/j.1432-1033.1994.00397.x. [DOI] [PubMed] [Google Scholar]
- Burchall JJ. Synergism between trimethoprim and sulfamethoxazole. Science. 1977;197:1300–1301. [PubMed] [Google Scholar]
- Bzik DJ, Li WB, Horii T, Inselburg J. Molecular cloning and sequence analysis of the Plasmodium falciparum dihydrofolate reductase-thymidylate synthase gene. Proc. Natl. Acad. Sci. U.S.A. 1987;84:8360–8364. doi: 10.1073/pnas.84.23.8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GX, Mueller C, Wendlinger M, Zolg JW. Kinetic and molecular properties of the dihydrofolate reductase from pyrimethamine-sensitive and pyrimethamine-resistant clones of the human malaria parasite Plasmodium falciparum. Mol. Pharmacol. 1987;31:430–437. [PubMed] [Google Scholar]
- Chulay JD, Watkins WM, Sixsmith DG. Synergistic antimalarial activity of pyrimethamine and sulfadoxine against Plasmodium falciparum in vitro. Am. J. Trop. Med. Hyg. 1984;33:325–330. doi: 10.4269/ajtmh.1984.33.325. [DOI] [PubMed] [Google Scholar]
- Coward JK, Parameswaran KN, Cashmore AR, Bertino JR. 7,8-Dihydropteroyl oligo-gamma-l-glutamates: synthesis and kinetic studies with purified dihydrofolate reductase from mammalian sources. Biochemistry. 1974;13:3899–3903. doi: 10.1021/bi00716a013. [DOI] [PubMed] [Google Scholar]
- Cowman AF, Morry MJ, Biggs BA, Cross GAM, Foote SJ. Amino-acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 1988;85:9109–9113. doi: 10.1073/pnas.85.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb BS, Cooke BM, Reeder JC, Waller RF, Caruana SR, Davern KM, Wickham ME, Brown GV, Coppel RL, Cowman AF. Targeted gene disruption shows that knobs enable malaria-infected red cells to cytoadhere under physiological shear stress. Cell. 1997a;89:287–296. doi: 10.1016/s0092-8674(00)80207-x. [DOI] [PubMed] [Google Scholar]
- Crabb BS, Triglia T, Waterkeyn JG, Cowman AF. Stable transgene expression in Plasmodium falciparum. Mol. Biochem. Parasitol. 1997b;90:131–144. doi: 10.1016/s0166-6851(97)00143-6. [DOI] [PubMed] [Google Scholar]
- DeSouza L, Shen Y, Bognar AL. Disruption of cytoplasmic and mitochondrial folylpolyglutamate synthetase activity in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 2000;376:299–312. doi: 10.1006/abbi.2000.1741. [DOI] [PubMed] [Google Scholar]
- Djimde AA, Doumbo OK, Traore O, Guindo AB, Kayentao K, Diourte Y, Niare-Doumbo S, Coulibaly D, Kone AK, et al. Clearance of drug-resistant parasites as a model for protective immunity in Plasmodium falciparum malaria. Am. J. Trop. Med. Hyg. 2003;69:558–563. [PubMed] [Google Scholar]
- Duraisingh MT, Triglia T, Cowman AF. Negative selection of Plasmodium falciparum reveals targeted gene deletion by double crossover recombination. Int. J. Parasit. 2002;32:81–89. doi: 10.1016/s0020-7519(01)00345-9. [DOI] [PubMed] [Google Scholar]
- Fidock DA, Nomura T, Wellems TE. Cycloguanil and its parent compound proguanil demonstrate distinct activities against Plasmodium falciparum malaria parasites transformed with human dihydrofolate reductase. Mol. Pharmacol. 1998;54:1140–1147. doi: 10.1124/mol.54.6.1140. [DOI] [PubMed] [Google Scholar]
- Fidock DA, Wellems TE. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc. Natl. Acad. Sci. U.S.A. 1997;94:10931–10936. doi: 10.1073/pnas.94.20.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick T, Ricken S, Lanzer M, Amrhein N, Macheroux P, Kappes B. Subcellular localization and characterization of chorismate synthase in the apicomplexan Plasmodium falciparum. Mol. Microbiol. 2001;40:65–75. doi: 10.1046/j.1365-2958.2001.02366.x. [DOI] [PubMed] [Google Scholar]
- Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, Haynes JD, Moch JK, Muster N, Sacci JB, et al. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419:520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- Futterman S, Silverman M. The “inactivation” of folic acid by liver. J. Biol. Chem. 1957;224:31–40. [PubMed] [Google Scholar]
- Gambonnet B, Jabrin S, Ravanel S, Karan M, Douce R, Rebeille F. Folate distribution during higher plant development. J. Sci. Food Agric. 2001;81:835–841. [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JM, Nichols BP, Matthews RG. Folate biosynthesis, reduction, and polyglutamation. In: Neidhardt FC, editor. Escherichia coli and Salmonella. ASM Press; Washington, DC: 1996. pp. 665–673. [Google Scholar]
- Greenwood B, Mutabingwa T. Malaria. Nature. 2002;415:670–672. doi: 10.1038/415670a. [DOI] [PubMed] [Google Scholar]
- Hall N, Pain A, Berriman M, Churcher C, Harris B, Harris D, Mungall K, Bowman S, Atkin R, et al. Sequence of Plasmodium falciparum chromosomes 1, 3–9 and 13. Nature. 2002;419:527–531. doi: 10.1038/nature01095. [DOI] [PubMed] [Google Scholar]
- Hall SJ, Sims PFG, Hyde JE. Functional expression of the dihydrofolate reductase and thymidylate synthetase activities of the human malaria parasite Plasmodium falciparum in Escherichia coli. Mol. Biochem. Parasitol. 1991;45:317–330. doi: 10.1016/0166-6851(91)90100-k. [DOI] [PubMed] [Google Scholar]
- Hastings IM, Watkins WM, White NJ. The evolution of drug-resistant malaria: the role of drug elimination half-life. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2002;357:505–519. doi: 10.1098/rstb.2001.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayton K, Ranford-Cartwright LC, Walliker D. Sulfadoxine-pyrimethamine resistance in the rodent malaria parasite Plasmodium chabaudi. Antimicrob. Agents Chemother. 2002;46:2482–2489. doi: 10.1128/AAC.46.8.2482-2489.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde JE. The dihydrofolate reductase-thymidylate synthetase gene in the drug resistance of malaria parasites. Pharmacol. Therap. 1990;48:45–59. doi: 10.1016/0163-7258(90)90017-v. [DOI] [PubMed] [Google Scholar]
- Hyde JE. Mechanisms of resistance of Plasmodium falciparum to antimalarial drugs. Microbes Infect. 2002;4:165–174. doi: 10.1016/s1286-4579(01)01524-6. [DOI] [PubMed] [Google Scholar]
- Kicska GA, Ting LM, Schramm VL, Kim K. Effect of dietary p-aminobenzoic acid on murine Plasmodium yoelii infection. J. Inf. Dis. 2003;188:1776–1781. doi: 10.1086/379373. [DOI] [PubMed] [Google Scholar]
- Kinyanjui SM, Mberu EK, Winstanley PA, Jacobus DP, Watkins WM. The antimalarial triazine WR99210 and the prodrug PS-15: folate reversal of in vitro activity against Plasmodium falciparum and a non-antifolate mode of action of the prodrug. Am. J. Trop. Med. Hyg. 1999;60:943–947. doi: 10.4269/ajtmh.1999.60.943. [DOI] [PubMed] [Google Scholar]
- Kongkasuriyachai D, Fujioka H, Kumar N. Functional analysis of Plasmodium falciparum parasitophorous vacuole membrane protein (Pfs16) during gametocytogenesis and gametogenesis by targeted gene disruption. Mol. Biochem. Parasitol. 2004;133:275–285. doi: 10.1016/j.molbiopara.2003.10.014. [DOI] [PubMed] [Google Scholar]
- Krumdieck CL, Eto I, Baggott JE. Regulatory role of oxidized and reduced pteroylpolyglutamates. Ann. N.Y. Acad. Sci. 1992;669:44–58. doi: 10.1111/j.1749-6632.1992.tb17088.x. [DOI] [PubMed] [Google Scholar]
- Krungkrai J, Webster HK, Yuthavong Y. De novo and salvage biosynthesis of pteroylpentaglutamates in the human malaria parasite, Plasmodium falciparum. Mol. Biochem. Parasitol. 1989;32:25–37. doi: 10.1016/0166-6851(89)90126-6. [DOI] [PubMed] [Google Scholar]
- Lasonder E, Ishihama Y, Andersen JS, Vermunt AMW, Pain A, Sauerwein RW, Eling WMC, Hall N, Waters AP, Stun-nenberg HG, Mann M. Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature. 2002;419:537–542. doi: 10.1038/nature01111. [DOI] [PubMed] [Google Scholar]
- Lee CS, Salcedo E, Wang Q, Wang P, Sims PFG, Hyde JE. Characterization of three genes encoding enzymes of the folate biosynthetic pathway in Plasmodium falciparum. Parasitology. 2001;122:1–13. doi: 10.1017/s0031182000006946. [DOI] [PubMed] [Google Scholar]
- Luba J, Nare B, Liang PH, Anderson KS, Beverley SM, Hardy LW. Leishmania major pteridine reductase 1 belongs to the short chain dehydrogenase family: stereochemical and kinetic evidence. Biochemistry. 1998;37:4093–4104. doi: 10.1021/bi972693a. [DOI] [PubMed] [Google Scholar]
- Mberu EK, Nzila AM, Nduati E, Ross A, Monks SM, Kokwaro GO, Watkins WM, Sibley CH. Plasmodium falciparum: in vitro activity of sulfadoxine and dapsone in field isolates from Kenya: point mutations in dihydropteroate synthase may not be the only determinants in sulfa resistance. Exp. Parasitol. 2002;101:90–96. doi: 10.1016/s0014-4894(02)00108-x. [DOI] [PubMed] [Google Scholar]
- McConkey GA, Ittarat I, Meshnick SR, McCutchan TF. Auxotrophs of Plasmodium falciparum dependent on p-aminobenzoic acid for growth. Proc. Natl. Acad. Sci. U.S.A. 1994;91:4244–4248. doi: 10.1073/pnas.91.10.4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKie JH, Douglas KT, Chan C, Roser SA, Yates R, Read M, Hyde JE, Dascombe MJ, Yuthavong Y, Sirawaraporn W. Rational drug design approach for overcoming drug resistance: application to pyrimethamine resistance in malaria. J. Med. Chem. 1998;41:1367–1370. doi: 10.1021/jm970845u. [DOI] [PubMed] [Google Scholar]
- Milhous WK, Weatherly NF, Bowdre JH, Desjardins RE. In vitro activities of and mechanisms of resistance to antifol antimalarial drugs. Antimicrob. Agents Chemother. 1985;27:525–530. doi: 10.1128/aac.27.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutabingwa T, Nzila A, Mberu E, Nduati E, Winstanley P, Hills E, Watkins W. Chlorproguanil-dapsone for treatment of drug-resistant falciparum malaria in Tanzania. Lancet. 2001;358:1218–1223. doi: 10.1016/S0140-6736(01)06344-9. [DOI] [PubMed] [Google Scholar]
- Nelson BC, Pfeiffer CM, Margolis SA, Nelson CP. Affinity extraction combined with stable isotope dilution LC/MS for the determination of 5-methyltetrahydrofolate in human plasma. Anal. Biochem. 2003;313:117–127. doi: 10.1016/s0003-2697(02)00531-6. [DOI] [PubMed] [Google Scholar]
- Nirmalan N, Sims PFG, Hyde JE. Quantitative proteomics of the human malaria parasite Plasmodium falciparum and its application to studies of development and inhibition. Mol. Microbiol. 2004a;52:1187–1199. doi: 10.1111/j.1365-2958.2004.04049.x. [DOI] [PubMed] [Google Scholar]
- Nirmalan N, Sims PFG, Hyde JE. Translational up-regulation of antifolate drug targets in the human malaria parasite Plasmodium falciparum upon challenge with inhibitors. Mol. Biochem. Parasitol. 2004b;136:63–70. doi: 10.1016/j.molbiopara.2004.02.013. [DOI] [PubMed] [Google Scholar]
- Nirmalan N, Wang P, Sims PFG, Hyde JE. Transcriptional analysis of genes encoding enzymes of the folate pathway in the human malaria parasite Plasmodium falciparum. Mol. Microbiol. 2002;46:179–190. doi: 10.1046/j.1365-2958.2002.03148.x. [DOI] [PubMed] [Google Scholar]
- Nzila A, Mberu E, Bray P, Kokwaro G, Winstanley P, Marsh K, Ward S. Chemosensitization of Plasmodium falciparum by probenecid in vitro. Antimicrob. Agents Chemother. 2003;47:2108–2112. doi: 10.1128/AAC.47.7.2108-2112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nzila AM, Mberu EK, Sulo J, Dayo H, Winstanley PA, Sibley CH, Watkins WM. Towards an understanding of the mechanism of pyrimethamine-sulfadoxine resistance in Plasmodium falciparum: Genotyping of dihydrofolate reductase and dihydropteroate synthase of Kenyan parasites. Antimicrob. Agents Chemother. 2000a;44:991–996. doi: 10.1128/aac.44.4.991-996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nzila AM, Nduati E, Mberu EK, Sibley CH, Monks SA, Winstanley PA, Watkins WM. Molecular evidence of greater selective pressure for drug resistance exerted by the long-acting antifolate pyrimethamine/sulfadoxine compared with the shorter-acting chlorproguanil/dapsone on Kenyan Plasmodium falciparum. J. Inf. Dis. 2000b;181:2023–2028. doi: 10.1086/315520. [DOI] [PubMed] [Google Scholar]
- Nzila-Mounda A, Mberu EK, Sibley CH, Plowe CV, Winstanley PA, Watkins WM. Kenyan Plasmodium falciparum field isolates: correlation between pyrimethamine and chlorcycloguanil activity in vitro and point mutations in the dihydrofolate reductase domain. Antimicrob. Agents Chemother. 1998;42:164–169. doi: 10.1128/aac.42.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell RA, Preiser PR, Williamson DH, Moore PW, Cowman AF, Crabb BS. An alteration in concatameric structure is associated with efficient segregation of plasmids in transfected Plasmodium falciparum parasites. Nucleic Acids Res. 2001;29:716–724. doi: 10.1093/nar/29.3.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashley TV, Volpe F, Pudney M, Hyde JE, Sims PFG, Delves CJ. Isolation and molecular characterization of the bifunctional hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase gene from Toxoplasma gondii. Mol. Biochem. Parasitol. 1997;86:37–47. [PubMed] [Google Scholar]
- Patel O, Satchell J, Baell J, Fernley R, Coloe P, Macreadie I. Inhibition studies of sulfonamide-containing folate analogs in yeast. Microb. Drug Resist. Mech. Epidemiol. Dis. 2003;9:139–146. doi: 10.1089/107662903765826723. [DOI] [PubMed] [Google Scholar]
- Patel OG, Mberu EK, Nzila AM, Macreadie IG. Sulfa drugs strike more than once. Trends Parasitol. 2004;20:1–3. doi: 10.1016/j.pt.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Peterson DS, Walliker D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc. Natl. Acad. Sci. U.S.A. 1988;85:9114–9118. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowe CV, Cortese JF, Djimde A, Nwanyanwu OC, Watkins WM, Winstanley PA, Estrada-Franco JG, Mollinedo RE, Avila JC, et al. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulfadoxine use and resistance. J. Infect. Dis. 1997;176:1590–1596. doi: 10.1086/514159. [DOI] [PubMed] [Google Scholar]
- Plowe CV, Kublin JG, Doumbo OK. P. falciparum dihydrofolate reductase and dihydropteroate synthase mutations: epidemiology and role in clinical resistance to antifolates. Drug Resist. Update. 1998;1:389–396. doi: 10.1016/s1368-7646(98)80014-9. [DOI] [PubMed] [Google Scholar]
- Price RN, Nosten F. Drug resistant falciparum malaria: clinical consequences and strategies for prevention. Drug Resist. Update. 2001;4:187–196. doi: 10.1054/drup.2001.0195. [DOI] [PubMed] [Google Scholar]
- Rastelli G, Pacchioni S, Sirawaraporn W, Sirawaraporn R, Parenti MD, Ferrari AM. Docking and database screening reveal new classes of Plasmodium falciparum dihydrofolate reductase inhibitors. J. Med. Chem. 2003;46:2834–2845. doi: 10.1021/jm030781p. [DOI] [PubMed] [Google Scholar]
- Ravanel S, Cherest H, Jabrin S, Grunwald D, Surdin-Kerjan Y, Douce R, Rebeille F. Tetrahydrofolate biosynthesis in plants: molecular and functional characterization of dihydrofolate synthetase and three isoforms of folylpolyglutamate synthetase in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 2001;98:15360–15365. doi: 10.1073/pnas.261585098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- Reeder JC, Rieckmann KH, Genton B, Lorry K, Wines B, Cowman AF. Point mutations in the dihydrofolate reductase and dihydropteroate synthetase genes and in vitro susceptibility to pyrimethamine and cycloguanil of Plasmodium falciparum isolates from Papua New Guinea. Am. J. Trop. Med. Hyg. 1996;55:209–213. doi: 10.4269/ajtmh.1996.55.209. [DOI] [PubMed] [Google Scholar]
- Robello C, Navarro P, Castanys S, Gamarro F. A pteridine reductase gene ptr1 contiguous to a p-glycoprotein confers resistance to antifolates in Trypanosoma cruzi. Mol. Biochem. Parasitol. 1997;90:525–535. doi: 10.1016/s0166-6851(97)00207-7. [DOI] [PubMed] [Google Scholar]
- Roberts F, Roberts CW, Johnson JJ, Kyle DE, Krell T, Coggins JR, Coombs GH, Milhous WH, Tzipori S, et al. Evidence for the shikimate pathway in apicomplexan parasites. Nature. 1998;393:801–805. doi: 10.1038/31723. [DOI] [PubMed] [Google Scholar]
- Roland S, Ferone R, Harvey RJ, Styles VL, Morrison RW. The characteristics and significance of sulfonamides as substrates for Escherichia coli dihydropteroate synthase. J. Biol. Chem. 1979;254:10337–10345. [PubMed] [Google Scholar]
- Salcedo E, Cortese JF, Plowe CV, Sims PFG, Hyde JE. A bifunctional dihydrofolate synthetase-folylpolyglutamate synthetase in Plasmodium falciparum identified by functional complementation in yeast and bacteria. Mol. Biochem. Parasitol. 2001;112:241–254. doi: 10.1016/s0166-6851(00)00370-4. [DOI] [PubMed] [Google Scholar]
- Sardarian A, Douglas KT, Read M, Sims PFG, Hyde JE, Chitnumsub P, Sirawaraporn R, Sirawaraporn W. Pyrimethamine analogs as strong inhibitors of double and quadruple mutants of dihydrofolate reductase in human malaria parasites. Org. Biomol. Chem. 2003;1:960–964. doi: 10.1039/b211636g. [DOI] [PubMed] [Google Scholar]
- Schapira A, Bygbjerg IC, Jepsen S, Flachs H, Bentzon MW. The susceptibility of Plasmodium falciparum to sulfadoxine and pyrimethamine—correlation of in vivo and in vitro results. Am. J. Trop. Med. Hyg. 1986;35:239–245. doi: 10.4269/ajtmh.1986.35.239. [DOI] [PubMed] [Google Scholar]
- Selhub J. Determination of tissue folate composition by affinity-chromatography followed by high-pressure ion-pair liquid-chromatography. Anal. Biochem. 1989;182:84–93. doi: 10.1016/0003-2697(89)90722-7. [DOI] [PubMed] [Google Scholar]
- Shallom S, Zhang K, Jiang L, Rathod PK. Essential protein–protein interactions between Plasmodium falciparum thymidylate synthase and dihydrofolate reductase domains. J. Biol. Chem. 1999;274:37781–37786. doi: 10.1074/jbc.274.53.37781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley CH, Hyde JE, Sims PFG, Plowe CV, Kublin JG, Mberu EK, Cowman AF, Winstanley PA, Watkins WM, Nzila AM. Pyrimethamine-sulfadoxine resistance in Plasmodium falciparum: what next? Trends Parasitol. 2001;17:582–588. doi: 10.1016/s1471-4922(01)02085-2. [DOI] [PubMed] [Google Scholar]
- Sims P, Wang P, Hyde JE. Selection and synergy in Plasmodium falciparum. Parasitol. Today. 1999;15:132–134. doi: 10.1016/s0169-4758(99)01420-9. [DOI] [PubMed] [Google Scholar]
- Sirawaraporn W, Sathitkul T, Sirawaraporn R, Yuthavong Y, Santi DV. Antifolate-resistant mutants of Plasmodium falciparum dihydrofolate reductase. Proc. Natl. Acad. Sci. U.S.A. 1997a;94:1124–1129. doi: 10.1073/pnas.94.4.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirawaraporn W, Sirawaraporn R, Cowman AF, Yuthavong Y, Santi DV. Heterologous expression of active thymidylate synthase dihydrofolate-reductase from Plasmodium falciparum. Biochemistry. 1990;29:10779–10785. doi: 10.1021/bi00500a009. [DOI] [PubMed] [Google Scholar]
- Sirawaraporn W, Yongkiettrakul S, Sirawaraporn R, Yuthavong Y, Santi DV. Plasmodium falciparum: asparagine mutant at residue 108 of dihydrofolate reductase is an optimal antifolate-resistant single mutant. Exp. Parasitol. 1997b;87:245–252. doi: 10.1006/expr.1997.4221. [DOI] [PubMed] [Google Scholar]
- Snewin VA, England SM, Sims PFG, Hyde JE. Characterization of the dihydrofolate reductase-thymidylate synthetase gene from human malaria parasites highly resistant to pyrimethamine. Gene. 1989;76:41–52. doi: 10.1016/0378-1119(89)90006-1. [DOI] [PubMed] [Google Scholar]
- Su X, Ferdig M, Huang Y, Huynh CQ, Liu A, You J, Wootton JC, Wellems TE. A genetic map and recombination parameters of the human malaria parasite Plasmodium falciparum. Science. 1999;286:1351–1353. doi: 10.1126/science.286.5443.1351. [DOI] [PubMed] [Google Scholar]
- Sulo J, Chimpeni P, Hatcher J, Kublin JG, Plowe CV, Molyneux ME, Marsh K, Taylor TE, Watkins WM, Winstanley PA. Chlorproguanil-dapsone versus sulfadoxine-pyrimethamine for sequential episodes of uncomplicated falciparum malaria in Kenya and Malawi: a randomised clinical trial. Lancet. 2002;360:1136–1143. doi: 10.1016/s0140-6736(02)11198-6. [DOI] [PubMed] [Google Scholar]
- Triglia T, Cowman AF. Primary structure and expression of the dihydropteroate synthetase gene of Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 1994;91:7149–7153. doi: 10.1073/pnas.91.15.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triglia T, Cowman AF. Plasmodium falciparum: a homo-logue of p-aminobenzoic acid synthetase. Exp. Parasitol. 1999;92:154–158. doi: 10.1006/expr.1999.4400. [DOI] [PubMed] [Google Scholar]
- Triglia T, Healer J, Caruana SR, Hodder AN, Anders RF, Crabb BS, Cowman AF. Apical membrane antigen 1 plays a central role in erythrocyte invasion by Plasmodium species. Mol. Microbiol. 2000;38:706–718. doi: 10.1046/j.1365-2958.2000.02175.x. [DOI] [PubMed] [Google Scholar]
- Triglia T, Menting JGT, Wilson C, Cowman AF. Mutations in dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 1997;94:13944–13949. doi: 10.1073/pnas.94.25.13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller KL, Muhle RA, Ursos LM, Horrocks P, Verdier-Pinard D, Sidhu ABS, Fujioka H, Roepe PD, Fidock DA. Chloroquine resistance modulated in vitro by expression levels of the Plasmodium falciparum chloroquine resistance transporter. J. Biol. Chem. 2003;278:33593–33601. doi: 10.1074/jbc.M302215200. [DOI] [PubMed] [Google Scholar]
- Walliker D, Quakyi IA, Wellems TE, McCutchan TF, Szarfman A, London WT, Corcoran LM, Burkot TR, Carter R. Genetic analysis of the human malaria parasite Plasmodium falciparum. Science. 1987;236:1661–1666. doi: 10.1126/science.3299700. [DOI] [PubMed] [Google Scholar]
- Wang P, Brobey RKB, Horii T, Sims PFG, Hyde JE. Utilization of exogenous folate in the human malaria parasite Plasmodium falciparum and its critical role in antifolate drug synergy. Mol. Microbiol. 1999;32:1254–1262. doi: 10.1046/j.1365-2958.1999.01437.x. [DOI] [PubMed] [Google Scholar]
- Wang P, Lee CS, Bayoumi R, Djimde A, Doumbo O, Swedberg G, Dao LD, Mshinda H, Tanner M, et al. Resistance to antifolates in Plasmodium falciparum monitored by sequence analysis of dihydropteroate synthetase and dihydrofolate reductase alleles in a large number of field samples of diverse origins. Mol. Biochem. Parasitol. 1997a;89:161–177. doi: 10.1016/s0166-6851(97)00114-x. [DOI] [PubMed] [Google Scholar]
- Wang P, Nirmalan N, Wang Q, Sims PFG, Hyde JE. Genetic and metabolic analysis of folate salvage in the human malaria parasite Plasmodium falciparum. Mol. Biochem. Parasitol. 2004a;135:77–87. doi: 10.1016/j.molbiopara.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Wang P, Read M, Sims PFG, Hyde JE. Sulfadoxine resistance in the human malaria parasite Plasmodium falciparum is determined by mutations in dihydropteroate synthetase and an additional factor associated with folate utilization. Mol. Microbiol. 1997b;23:979–986. doi: 10.1046/j.1365-2958.1997.2821646.x. [DOI] [PubMed] [Google Scholar]
- Wang P, Sims PFG, Hyde JE. A modified in vitro sulfadoxine susceptibility assay for Plasmodium falciparum suitable for investigating Fansidar resistance. Parasitology. 1997c;115:223–230. doi: 10.1017/s0031182097001431. [DOI] [PubMed] [Google Scholar]
- Wang P, Wang Q, Aspinall TV, Sims PFG, Hyde JE. Transfection studies to explore essential folate metabolism and antifolate drug synergy in the human malaria parasite Plasmodium falciparum. Mol. Microbiol. 2004b;51:1425–1438. doi: 10.1111/j.1365-2958.2003.03915.x. [DOI] [PubMed] [Google Scholar]
- Wang P, Wang Q, Sims PFG, Hyde JE. Rapid positive selection of stable integrants following transfection of Plasmodium falciparum. Mol. Biochem. Parasitol. 2002;123:1–10. doi: 10.1016/s0166-6851(02)00105-6. [DOI] [PubMed] [Google Scholar]
- Watkins WM, Mberu EK, Winstanley PA, Plowe CV. The efficacy of antifolate antimalarial combinations in Africa: a predictive model based on pharmacodynamic and pharmacokinetic analyses. Parasitol. Today. 1997;13:459–464. doi: 10.1016/s0169-4758(97)01124-1. [DOI] [PubMed] [Google Scholar]
- Wellems TE, Panton LJ, Gluzman IY, do Rosario VE, Gwadz RW, Walker-Jonah A, Krogstad DJ. Chloroquine resistance not linked to mdr-like genes in a Plasmodium falciparum cross. Nature. 1990;345:253–255. doi: 10.1038/345253a0. [DOI] [PubMed] [Google Scholar]
- Winstanley PA, Ward SA, Snow RW. Clinical status and implications of antimalarial drug resistance. Microbes Infect. 2002;4:157–164. doi: 10.1016/s1286-4579(01)01523-4. [DOI] [PubMed] [Google Scholar]
- Wooden JM, Hartwell LH, Vasquez B, Sibley CH. Analysis in yeast of antimalaria drugs that target the dihydrofolate reductase of Plasmodium falciparum. Mol. Biochem. Parasitol. 1997;85:25–40. doi: 10.1016/s0166-6851(96)02808-3. [DOI] [PubMed] [Google Scholar]
- Yuvaniyama J, Chitnumsub P, Kamchonwongpaisan S, Vanichtanankul J, Sirawaraporn W, Taylor P, Walkinshaw MD, Yuthavong Y. Insights into antifolate resistance from malarial DHFR-TS structures. Nat. Struct. Biol. 2003;10:357–365. doi: 10.1038/nsb921. [DOI] [PubMed] [Google Scholar]
- Zhang K, Rathod PK. Divergent regulation of dihydrofolate reductase between malaria parasite and human host. Science. 2002;296:545–547. doi: 10.1126/science.1068274. [DOI] [PMC free article] [PubMed] [Google Scholar]