Summary
Structure-specific endonucleases mediate repair of DNA structures formed from replication fork collapse or during double-strand break (DSB) repair. Here we identify BTBD12 as the human ortholog of the budding yeast DNA repair factor Slx4p and D. melanogaster MUS312. Human SLX4 forms a multiprotein complex with the ERCC4(XPF)-ERCC1, MUS81-EME1, and SLX1 endonucleases, and also associates with MSH2/MSH3 mismatch repair complex, telomere binding complex TERF2(TRF2)-TERF2IP(RAP1), the protein kinase PLK1 and the uncharacterized protein C20orf94. Depletion of SLX4 causes sensitivity to mitomycin C and camptothecin, and reduces the efficiency of DSB repair in vivo. SLX4 complexes cleave 3’-flap, 5’-flap and replication fork structures; yet unlike other endonucleases associated with SLX4, the SLX1-SLX4 module promotes symmetrical cleavage of static and migrating Holliday junctions (HJs), identifying SLX1-SLX4 as a HJ resolvase. Thus, SLX4 assembles a modular tool-kit for repair of specific types of DNA lesions and is critical for cellular responses to replication fork failure.
Introduction
Repair of DNA damage is critical to the survival of all organisms. The fidelity of these repair processes requires the concerted efforts of a highly organized and coordinated group of repair proteins. Critical to the orchestration of these repair events is a signal transduction pathway referred to as the DNA damage response (Harper and Elledge, 2007). At the apex of this network are the protein kinases ATM and ATR that sense different types of DNA damage structures and in turn phosphorylate a host of proteins that elaborate an extensive cellular response to genotoxic stress (Matsuoka et al., 2007).
Many types of DNA repair such as mismatch repair, homologous recombination and DNA crosslink repair require the breakage and rejoining of DNA strands via structure-specific endonucleases. One endonuclease class is typified by dimeric endonucleases ERCC4(XPF)-ERCC1 and MUS81-EME1 that contain structurally related non-catalytic and catalytic subunits (Ciccia et al., 2008). ERCC4-ERCC1 (Rad1p-Rad10p in budding yeast) plays important roles in both nucleotide excision repair and in interstrand crosslink (ICL) repair, and prefers to cleave splayed arm structures (Ciccia et al., 2008). MUS81-EME1 (Mus81p-Mms4p in budding yeast) plays a distinct role in ICL repair, and both yeast and mammalian cells lacking MUS81 are highly sensitive to agents that cause replication fork collapse (Bastin-Shanower et al., 2003; McPherson et al., 2004; Hanada et al., 2006). A role for MUS81-EME1 in incision at an ICL to form double-strand breaks (DSBs) has been demonstrated in mouse embryo fibroblasts lacking MUS81, while cells lacking ERCC1 do not display this defect (Hanada et al., 2006). This is consistent with the preference of MUS81-EME1 for replication fork and 3’ flap substrates (Constantinou et al., 2002; Ciccia et al., 2003). ERCC4-ERCC1 and MUS81-EME1 may also have roles downstream of the incision step, including resolution of Holliday junctions (HJ).
A structurally distinct class of endonucleases typified by Slx1p has been identified in yeast. Slx1p contains an N-terminal domain related to the UvrC-intron-endonuclease (URI) class of bacterial endonucleases (known as a GIY domain) and a C-terminal PHD-related domain required for Slx1p activity (Fricke and Brill, 2003; Coulon et al., 2004). Slx1p binds the Slx4p regulatory subunit, a protein lacking known interaction domains (Fricke and Brill, 2003; Coulon et al., 2004). Slx1p-Slx4p displays a preference for 5’-flap structures in vitro and slx4Δ, but not slx1Δ, mutants are highly sensitive to MMS and camptothecin (Flott et al., 2007; Fricke and Brill, 2003; Deng et al., 2005). Slx4 is phosphorylated by Tel1 and Mec1 in response to DNA damage (Flott et al., 2007). Interestingly, Slx4p also forms an Slx1p–independent complex with Rad1p-Rad10p, and this complex is required for DSB repair during single strand annealing (Flott et al., 2007; Li et al., 2008). Vertebrate orthologs of Slx4p or Slx1p have not been characterized.
A central aspect of meiotic recombination is the generation and resolution of double HJs, which are formed during the process of homologous recombination. HJs are also formed during homology-dependent repair of DSBs in mitotic cells, although other pathways involving helicases provide an alternative route for HJ resolution. HJs physically link two homologous chromosomes and must be cleaved or unwound to allow resolution of these structures into two unconnected dsDNA molecules. As this is a critical transition, the identity of resolvase enzymes has been a central focus of the field. While MUS81-EME1 (and its yeast ortholog Mus81p-Mms4p) and Slx1p-Slx4p display a preference for flap structures and replication forks, they have also been shown to display activity towards migrating or nicked HJs, albeit in a largely non-symmetrical manner (Boddy et al., 2001; Ciccia et al., 2003; Constantinou et al., 2002; Fricke and Brill, 2003; Osman et al., 2003; Chen et al., 2001; Gaillard et al., 2003; Taylor and McGowan, 2008; Coulon et al., 2004) and Mus81 mutants are defective in resolution of cruciform structures formed by palindromic sequences that mimic a HJ in vivo (Cote and Lewis, 2008). However, MUS81-EME1 and Slx1p-Slx4p prepared in E. coli are largely devoid of activity toward static HJs (Ciccia et al., 2003; Fricke and Brill, 2003) leading to the conclusion that these enzymes alone are not efficient HJ resolvases. In contrast, both static and migrating HJs are symmetrically cleaved by XPG-related HJ resolvases: Yen1p from S. cerevisiae and GEN1 from multi-cellular eukaryotes (Ip et al., 2008). Nevertheless, yen1Δ cells display no obvious recombination phenotypes unless combined with an mms4Δ background, and S. pombe lacks Yen1p, suggesting that in S. pombe, other nucleases (most likely Mus81p-Mms4p) must be primarily responsible for endonucleolytic resolution of recombination intermediates involving HJs (Ip et al., 2008). These data raise the question as to whether there might exist additional HJ resolvases in multicellular eukaryotes and, if so, the nature of particular biochemical pathways in which these enzymes participate (Haber and Heyer, 2001; Hollingsworth and Brill, 2004; Ciccia et al., 2008).
Here, we identify a BTB-domain containing protein – BTBD12 – as the apparent vertebrate ortholog of yeast Slx4p and Drosophila MUS312, and demonstrate a central role for BTBD12 (referred to hereafter as SLX4) in the response of cells to interstrand crosslinks, DNA-protein adducts and homology-directed DSB-repair. Human SLX4 assembles into a multiprotein complex containing 3 structure-specific endonucleases -ERCC4-ERCC1, MUS81-EME1, and SLX1 - as well as with the mismatch repair complex MSH2-MSH3, components of the telomere shelterin complex TERF2 (TRF2)-TERF2IP(RAP1), a novel open reading (C20orf94), and polo-like kinase 1 (PLK1). SLX4 complexes exhibit endonucleolytic activity towards both static and mobile HJs, as well as towards replication fork and flap structures. SLX1 in association with its binding domain from SLX4 promotes symmetrical cleavage of static HJs, while asymmetric cleavage of HJs by the SLX1-SLX4 module provides a nicked HJ which can be targeted by MUS81-EME1 to produce gapped duplex products. We therefore propose that the SLX4 complex represents a multi-functional tool-kit for DNA repair, incorporating endonucleases with distinct specificities for the resolution of diverse forms of deleterious DNA structures.
Results
BTBD12 (SLX4) associates with known DNA repair factors
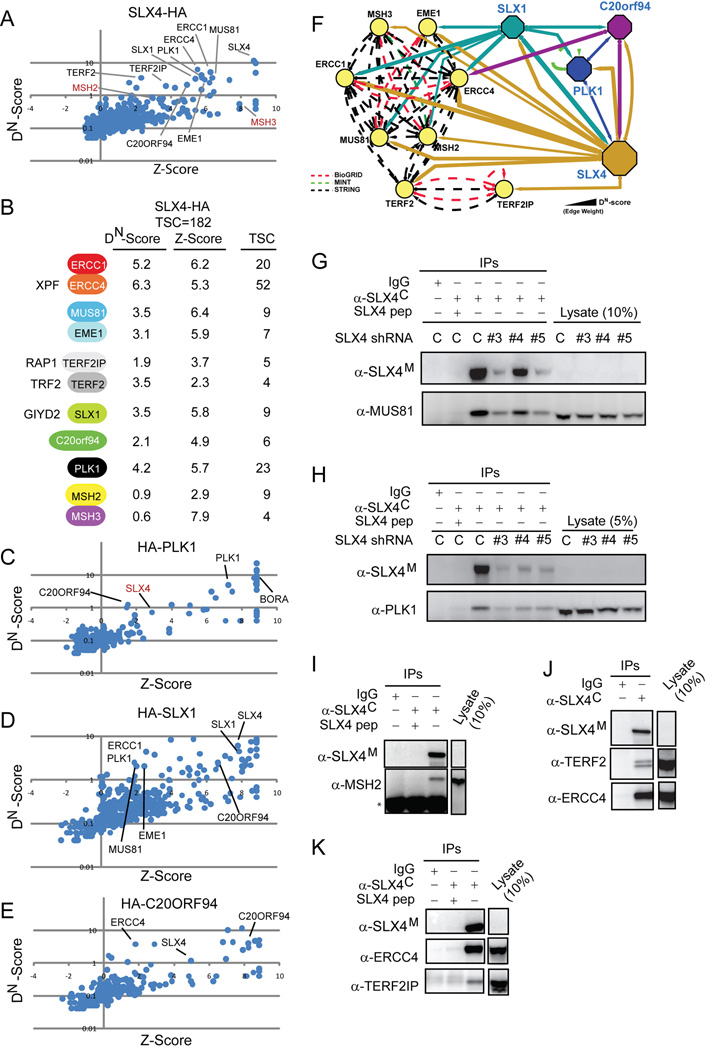
Human BTBD12 (referred to hereafter as SLX4; gene ID 84464) is a largely uncharacterized 200 KDa protein that features a central BTB domain but lacks other previously characterized domains based on analysis via SMART and PFAM at high stringency. Our attention was initially drawn to SLX4 because it was identified as a target of the ATM/ATR protein kinases (Matsuoka et al., 2007; Mu et al., 2007). To gain insight into SLX4 function, we performed a proteomic analysis of SLX4 complexes purified from 293TREX cells harboring an inducible SLX4-HA lentivirus. Proteins associated with SLX4-HA were identified by LC-MS/MS (Table S1). Proteomic data was processed using a newly developed proteomics platform called CompPASS (Comparative Proteomics Analysis Software Suite), which facilitates the identification of high-confidence candidate interacting proteins (HCIPs) in parallel mass spectral studies (see Experimental Procedures) (Sowa et al., 2009). Proteins with DN-Scores of >1 and or Z-scores of >3.5 constitute HCIPs (Sowa et al., 2009).
A number of proteins previously linked with DNA repair were reproducibly identified as HCIPs in the SLX4-HA immune complex (Figure 1A, B, Table S1, Figure S11), including structurally related heterodimeric flap endonucleases – ERCC4(XPF)-ERCC1 and MUS81-EME1 - and the apparent human ortholog of a conserved family of endonucleases typified by yeast Slx1p (Ciccia et al., 2008; Fricke and Brill, 2003; Coulon et al., 2004). In humans, SLX1 is encoded by 2 genes encoding identical proteins (GIYD1/2; Gene IDs 548593 and 79008). SLX1 orthologs (Supplemental Figure S3, Figure S5B) share an N-terminal domain with similarity to the bacterial uvrC nucleotide excision repair protein (referred to as a GIY domain) as well as a functionally important C-terminal PHD-like domain (Fricke and Brill, 2003). SLX4 was also found to associate with the telomere interacting protein TERF2 (TRF2) and its partner protein TERF2IP (RAP1), the PLK1 protein kinase, the MSH2-MSH3 mismatch repair complex, and a previously uncharacterized open reading frame C20orf94 lacking known protein interaction domains (Figure 1A, B). C20orf94 is conserved in vertebrates but apparently absent in non-vertebrates (Figure S4, Figure S5C). These results indicate that SLX4 interacts with a number of proteins implicated in DNA repair and telomere maintenance.
Figure 1. Human SLX4 associates with structure-specific endonucleases, telomere recognition proteins, miss-match repair proteins, and PLK1.
(A) Proteomic analysis of SLX4-HA purified from 293TREX cells was performed as described in Experimental Procedures. High confidence candidate interacting proteins (HCIPs) were determined using CompPASS to derive Z and DN-Scores. Proteins marked by red symbols fall below the stringent threshold set for HCIPs based on a DN-Score = 1.
(B) Summary of DN-Scores, Z-Scores, and total spectral counts (TSCs) for major SLX4 associated proteins. Alternative names are indicated.
(C–E) Plots of Z-Score versus DN-Scores for SLX4 interacting proteins. The indicated proteins were stably expressed in 293T cells, purified and analyzed by LC-MS/MS.
(F) Interaction network for the human SLX4 complex. Solid lines; interactions observed by mass spectrometry. Dotted lines; interactions in BIOGRID (red), STRING (black), or MINT (green) databases.
(G–K) SLX4 was immunoprecipitated (+/− antigenic peptide) from U2OS cells with or without SLX4 depletion and immunoblotted. * indicates heavy chain
Validation of interactions within the human SLX4 complex
Proteomic analyses on HA-PLK1, HA-SLX1 and HA-C20orf94 complexes from 293T using the CompPASS platform revealed reciprocal association with SLX4, as well as other components of the SLX4 complex (Figure 1C–E, Table S1). PLK1 was found to associate with C20orf94, in addition to its known target, BORA; SLX1 also associated with MUS81, EME1, C20orf94, PLK1, and ERCC1; and C20orf94 was found to associate with ERCC4 (Figure 1F).
We examined association of endogenous SLX4 with its interacting proteins in U2OS cells using a rabbit antibody raised against the C-terminus of SLX4 for immunoprecipitation (α-SLX4C) and a mouse antibody generated against an 1151 residue fragment (aa684–1834) for immunoblotting (α-SLX4M) (see Experimental Procedures and Figure S1). Neither of these antibodies was capable of detecting endogenous SLX4 by immunoblotting of crude cell lysates (Figure 1G–K, data not shown) or immunoflourescence (data not shown). SLX4 was found to associate with endogenous ERCC4, MUS81, and PLK1, and these interactions were reduced by either addition of competing peptide or by depletion of SLX4 (Figure 1G, H, J, K). Association with endogenous SLX4 was also observed for TERF2, TERF2IP, and MSH2 (Figure 1I–K). In transfection studies in 293T cells, we found that MYC-SLX4 associated with endogenous ERCC4, MUS81, and MSH2 (Figure 2D–F). Transiently expressed GFP-SLX4 formed nuclear foci that partially co-localized with TERF2IP in HeLa cells, and fully co-localized with ERCC4 and MUS81 in U2OS cells (Figure S7, Figure S8), providing further evidence of a physical interaction. Moreover, HA-FLAG-PLK1 was found to reciprocally associate with ERCC4 and depletion of SLX4 led to a reduction in this association (Figure 2D, E, S6E), suggesting that SLX4 bridges the interaction between PLK1 and ERCC4. Consistent with a functionally relevant interaction between PLK1 and SLX4, MYC-SLX4 immune complexes contain an activity that can phosphorylate SLX4 in situ and this phosphorylation is dramatically reduced upon addition of a small-molecule PLK1 inhibitor (BI2536) (Figure S6). Antibodies against C20orf94 or human SLX1 that are of sufficient affinity to detect endogenous proteins are not yet available, and we have therefore not been able to examine endogenous SLX1 or C20orf94 for their interaction with SLX4 immunologically.
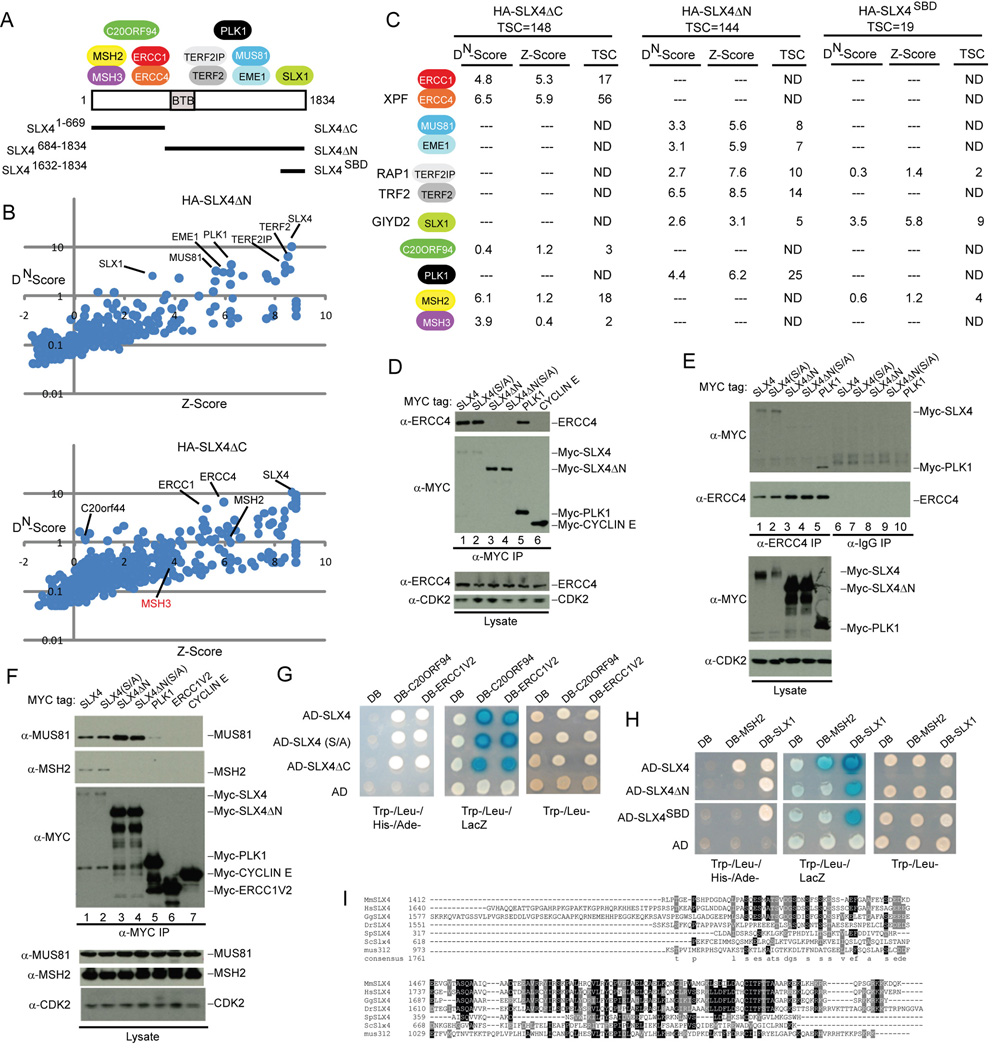
Figure 2. Structural anatomy of the human SLX4 complex.
(A) Summary of the interaction data showing the approximate location of binding between SLX4 and associated proteins.
(B) DN-Score versus Z-Score plots for HA-SLX4ΔN and HA-SLX4ΔC showing the identity of proteins identified by mass spectrometry (see Figure 1 legend for details).
(C) Summary of proteomic data for HA-SLX4ΔN, HA-SLX4ΔC, and HA-SLX4SBD. TSC, total spectral counts. The full list of interacting proteins is provided in Table S1.
(D–F) Domain mapping of SLX4 and its associated proteins. The indicated MYC-tagged proteins were expressed in 293T cells, immunoprecpitated as indicated, and immunoblotted. α-CDK2; loading control.
(G, H) Interaction of SLX4 with target proteins using the yeast two-hybrid system. The indicated proteins were cloned into activation domain (AD) or DNA-binding domain (DB) vectors and two-hybrid analysis performed as described under Experimental Procedures.
(I) Sequence relationship between vertebrate, yeast, Drosophila and C. elegans SLX4 in the C-terminal region involved in binding SLX1.
Previous experiments have demonstrated a functional interaction between the ERCC4-ERCC1 complex and the TERF2-TERF2IP complex (Zhu et al., 2003), and our data now reveal a physical interaction between these complexes and SLX4, SLX1, PLK1, MUS81-EME1, MSH2-MSH3, and C20orf94 (Figure 1F).
Structural anatomy of the SLX4 complex
SLX4/BTBD12 contains a BTB domain (residues 691–794) but otherwise lacks previously annotated domains that would be predictive of function (Figure 2A). While a subset of BTB proteins function as adaptors for Cul3 ubiquitin ligases (Xu et al., 2003), SLX4 does not appear to bind Cul3 (Table S1). The association of SLX1 and ERCC4-ERCC1 with SLX4/BTBD12 suggested the possibility that this BTB protein may be the functional counterpart of budding yeast Slx4p and Drosophila MUS312, neither of which contain recognizable BTB domains based on SMART or PFAM. Budding yeast Slx4p interacts with both Slx1p and with the Rad1p-Rad10p complex, albeit in what appears to be a mutually exclusive manner (Flott et al., 2007). In addition, MUS312 has been shown to interact with the Drosophila ortholog of ERCC4, MEI-9 (Yildiz et al., 2002) Comparison of the vertebrate SLX4 sequence with MUS312 and Slx4p using CLUSTALW revealed that the extreme C-terminus of vertebrate SLX4 orthologs displays weak sequence similarity with MUS312 and Slx4p (Figure 2I and S2, S5A). Residues 1683–1835 of human SLX4 display 17% identity with residues near the C-terminus of MUS312 (residues 920–1011), and 11% identity with residues 620–748 of budding yeast Slx4p.
To examine how SLX4 assembles these DNA repair components and to examine the structural relationship between vertebrate SLX4 and yeast Slx4p, we performed a series of SLX4 domain (Figure 2A, Figure S14A) mapping studies using proteomic (Figure 2B,C), transfection-based interaction (Figure 2D–F, Figure S6C, Figure S14B), and two-hybrid analyses (Figure 2G and H, S6B). These studies revealed that MSH2-MSH3, ERCC4-ERCC1, and C20orf94 associate with the N-terminal region of SLX4 (residues 1–669; SLX4ΔC), while TERF2-TERF2IP, MUS81-EME1, PLK1, and SLX1 bind the C-terminal portion of the protein (residues 684–1834; SLX4ΔN) (Figure 2A). SLX1 associated with the extreme C-terminus of SLX4 (residues 1632–1834) (Figure 2A, C), a region displaying the highest conservation between yeast Slx4p, MUS312, and vertebrate SLX4 (Figure 2I), which we refer to hereafter as the SLX1-binding domain (SBD). In contrast, MUS81 associated with a region encompassing residues 1328–1648 (Figure S14), while PLK1 associated with SLX4 at least in part through a candidate polo-box binding motif (Elia et al., 2003) centered on S1453 (Figure S6B, C). Thus, distinct regions of SLX4 are employed to assemble multiple DNA repair and telomere protection factors.
SLX4 is required for resistance to DNA damaging agents that produce interstrand crosslinks or DNA-protein adducts
Given the known roles of structure-specific nucleases in resistance to DNA damage in yeast and mammals, we examined whether human cells depleted of SLX4 were sensitive to DNA damaging agents. For this purpose, we employed a previously described Multi-color Competition Assay (MCA) which measures the relative resistance of cells to DNA damage by flow cytometry (Smogorzewska et al., 2007) (see Experimental Procedures).
Depletion of SLX4 in U2OS and HeLa cells by RNAi led to reduced resistance to MMC and CPT, with the severity of the phenotype paralleling the extent of SLX4 knockdown (Figure 3A–D, Figure S9, Figure S13). As with SLX4 in yeast, mild sensitivity toward MMS was observed (Figure S13A, E). Little or no sensitivity was observed with UV and IR, in contrast to the results obtained with depletion of ATR (Figure 3A, 3C). Even though most of the SLX4 protein was depleted by RNAi as judged by IP-westerns (Figure 3B, 3D), these experiments likely represent partial loss of function since in other experiments (Figure S13C–E) the MMC sensitivity of SLX4-depleted cells was even more profound. For comparison, depletion of ERCC4 resulted in sensitivity to MMC and UV (again paralleling the extent of knockdown) while the most potent siRNA targeting MUS81 resulted in weak sensitivity to MMC and more substantial sensitivity to camptothecin (Figure 3E, F). We note that substantial depletion of MUS81 (<5% MUS81 remaining) leads to little or no phenotype while further depletion leads to a clear reduction in resistance (Figure 3E, F). Thus, it appears that low levels of MUS81 are capable of supporting its key functions in this context. We also examined depletion of SLX1. Weak sensitivity was observed in the context of MMC, CPT, and MMS, but not UV (Figure 3E, F; Figure S13E), upon treatment with the siRNA that gave the strongest depletion of SLX1 (SLX1#4) (Figure 3F). However, as with MUS81, we cannot exclude the possibility that residual protein is sufficient to provide significant resistance to these damaging agents (Figure 3F). The sensitivity observed with SLX1 depletion paralleled that found with the strongest siRNA targeting GEN1 in the context of MMC (Figure 3E, 3F). Taken together, these data indicate that SLX4 is important for the cells ability to respond to DNA interstrand crosslinks and protein-DNA adducts.
Figure 3. Reduction in components of the SLX4 complex sensitizes cells to DNA damaging agents.
(A–D) The Multi-color Competition Assay (MCA) was used to examine the effect of depletion of SLX4 on the sensitivity of U2OS and HeLa cells to DNA damaging agents using independent shRNAs and siRNAs (panels A and C). Corresponding knockdown is shown (panel B and D). *: p<0.05; **: p<0.005, students T-test. Error bars are STDEV across 3 technical replicates.
(E–F) Effect of siRNA depletion of ERCC4, MUS81, SLX1, and GEN1 on the resistance of HeLa cells to DNA damage (panel E). The indicated HeLa cell extracts were examined for protein expression after siRNA transfection (panel F).
(G) U2OS cells expressing GFP-SLX4 were subjected to laser micro-irradiation and after 60 min, cells were processed for γH2AX staining and/or imaging of GFP-SLX4. Nuclei were visualized with DAPI. Arrowheads indicate position of stripes.
SLX4 is recruited to sites of DNA damage
Several targets of ATM are recruited to sites of DNA damage (Harper and Elledge, 2007). We used laser micro-irradiation to examine whether SLX4 localizes to sites of DNA damage. In order to visualize SLX4, we tagged its N-terminus with GFP and expressed it stably in U2OS or HT1080 cells using a lentivirus (see Supplemental Experimental Procedures). GFP-SLX4 localized to numerous small foci in the nucleus, a subset of which co-localize with TERF2IP and presumably reflect telomeres, as described above (see Figure S7). In cells receiving laser irradiation, GFP-SLX4 was found to accumulate at the site of DNA damage (21% in HT1080 cells and 14% in U2OS cells), as assessed by co-localization with γH2AX, a marker of DSBs (Figure 3G, Figure S17). The absence of complete SLX4/γH2AX co-localization in these experiments may reflect cell cycle dependent responses of SLX4 to the damage signal or low levels of expression that limit detection. Recruitment of GFP-SLX4 to damage stripes was also observed in cells carrying inactivating mutations in FANCA, FANCD2, or ATM (not shown).
SLX4 complexes catalyze cleavage of static HJ structures
ERCC4-ERCC1 preferentially cleaves Y and bubble structures in vitro, while the preferred substrates for MUS81-EME1 complexes are 3’-flaps and replication forks (RF) structures (Ciccia et al., 2008; Ciccia et al., 2003; Constantinou et al., 2002). Preparations of MUS81-EME1 from mammalian cells display weak non-symmetrical activity toward migrating HJs (Constantinou et al., 2002; Chen et al., 2001) and more efficiently cleave nicked HJs (Taylor and McGowan, 2008), but MUS81-EME1 purified from bacteria are devoid of activity toward static HJs (Ciccia et al., 2003). In contrast, Slx1p/Slx4p complexes from bacteria preferentially cleave 5’-flap structures, but display weak activity toward migrating HJs, also in a non-symmetrical manner (Fricke and Brill, 2003). Certain migrating HJs have been shown to display some single-stranded character indicative of base-pair “breathing”, while static junctions do not (West, 1995), suggesting the possibility that any single-stranded nature of migrating junctions could allow for cleavage at flap or bubble-like structures that appear transiently. Thus, cleavage of static HJs, in conjunction with formation of ligatable products with migrating junctions, is considered a more rigorous test of HJ resolvase activity (Ip et al., 2008).
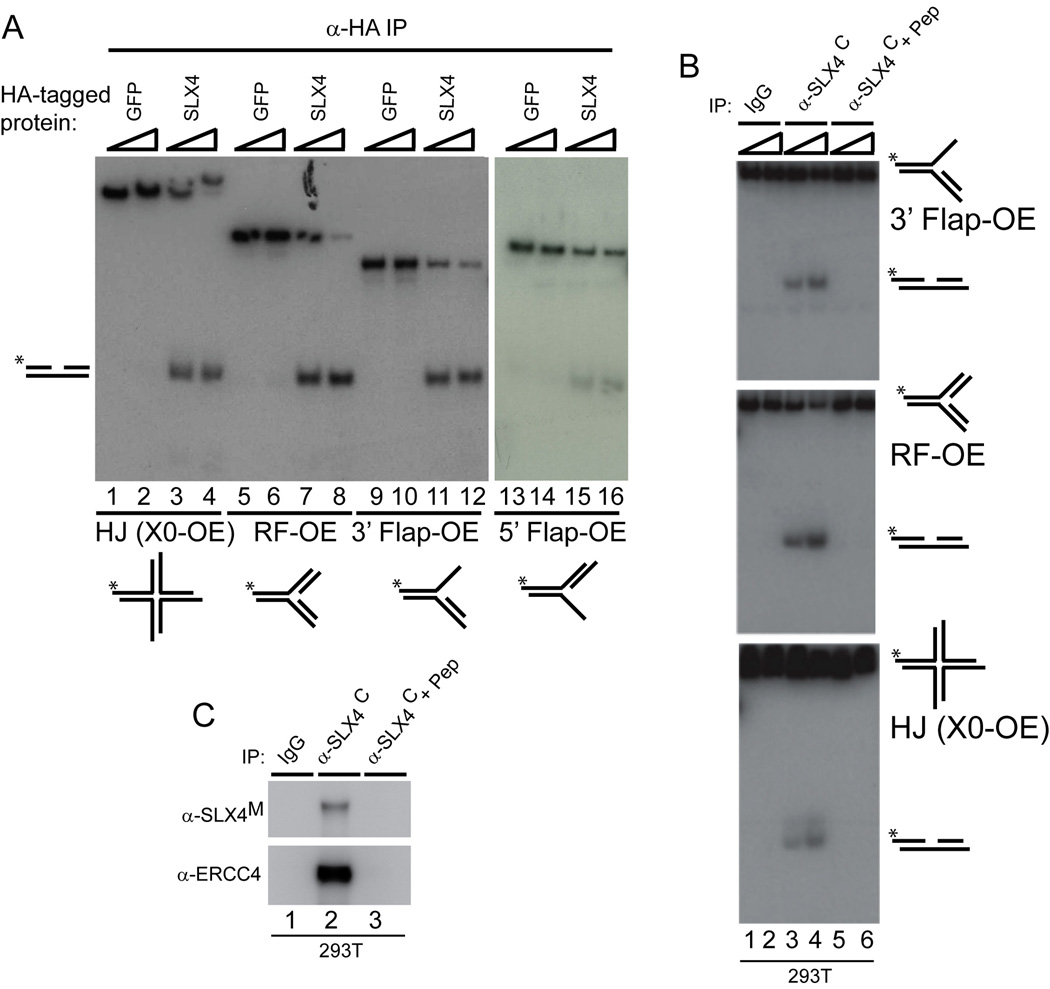
We therefore examined whether SLX4-HA complexes (or GFP-HA as a control) purified from 293T cells displayed endonucleolytic activity toward radiolabeled replication fork (RF), 3’-flap, 5’-flap, and static HJ (X0) substrates using native gel electrophoresis. Substrates containing 1 or 2 nucleotide overhanging 5’ ends are designated “OE” while substrates containing blunt ended duplexes are designated by “BE” (Figure S10). SLX4-HA complexes, but not control GFP-HA complexes, contained robust activity toward replication forks, 3’ and 5’-flaps, and, importantly, static HJs (X0-OE) (Figure 4A).
Figure 4. SLX4 associates with structure-specific endonuclease activity.
(A) SLX4-HA or GFP-HA (0.5mg, 1.5mg) complexes were immunoprecipitated from 293T cells and were incubated with 32P-end-labeled DNA substrates (30 min) prior to electrophoresis on native gels. Labeled strand indicated by *.
(B, C) Endogenous SLX4 complexes were immunoprecipitated from 293T cells (0.5, 1.5mg) and incubated with 32P-end-labeled DNA substrates (30 min) prior to electrophoresis on native gels (panel B) or immunoblotting (panel C).
To determine whether endogenous SLX4 associated with endonuclease activity, 293T cell extracts were subjected to immunoprecipitation with α-SLX4C in the presence or absence of competing antigenic peptide (Figure 4B, C). SLX4 complexes again supported robust activity towards flap, replication fork and static HJ (X0-OE) substrates (Figure 4B, lanes 3 and 4). Importantly, this activity was absent when the immunoprecipitation was performed in the presence of antigenic peptide (Figure 4B, lanes 5 and 6, Figure 4C). Thus, we find that SLX4 can support cleavage of different types of DNA structures using two different sources of SLX4 complexes from human cells. The ability of SLX4 complexes to cleave static HJs is interesting in light of the finding that, thus far, only a single mammalian enzyme, GEN1, has been shown to have robust activity toward static HJs (Ip et al., 2008).
Flap and HJ cleavage activities map to the C-terminus of SLX4
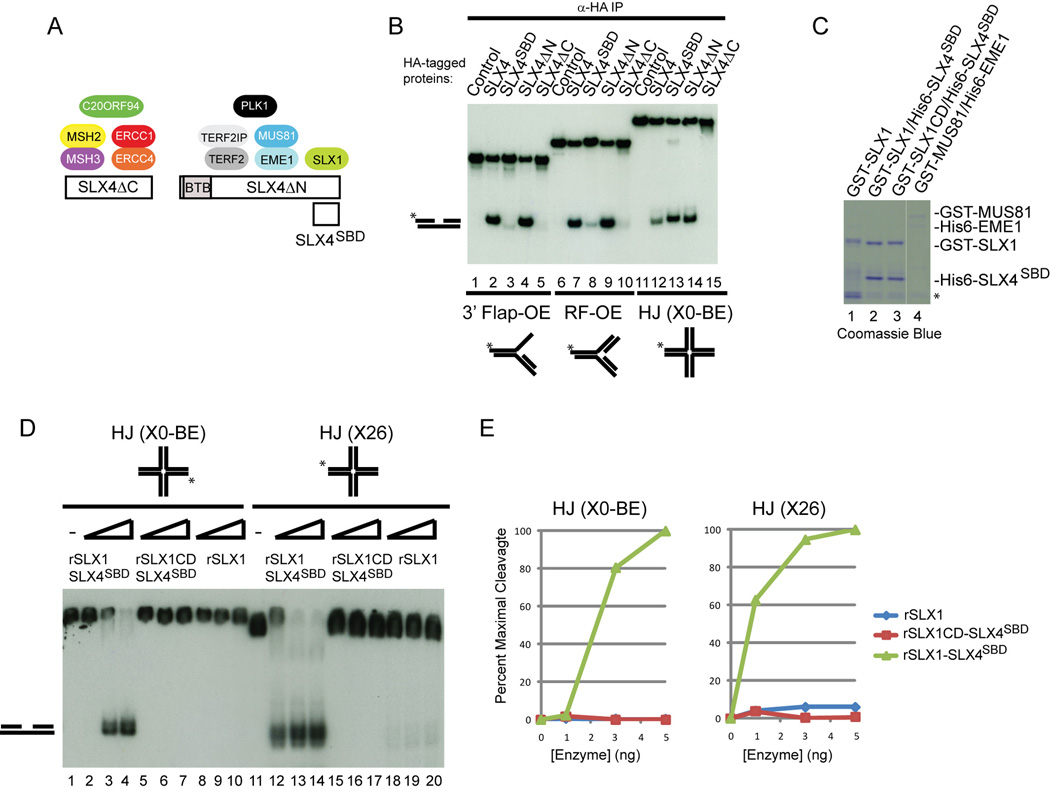
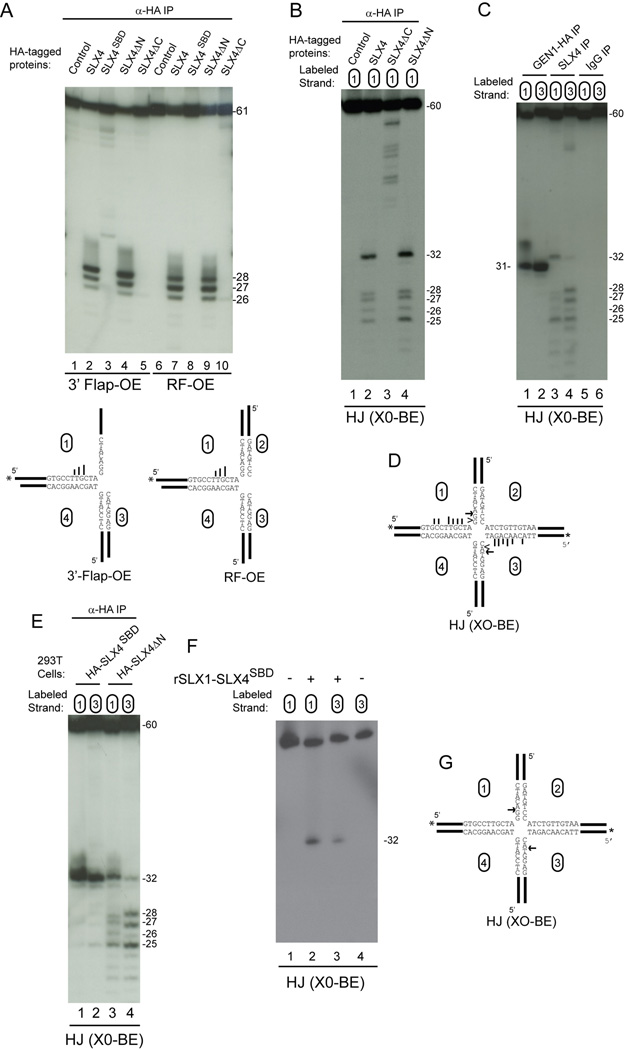
We took advantage of the spatial assembly of ERCC4, MUS81, and SLX1 endonucleases on SLX4 (Figure 5A) to examine where the flap endonuclease and HJ cleavage activity resides. Extracts from 293T cells stably expressing SLX4-HA, HA-SLX4SBD, HA-SLX4ΔC, and HA-SLX4ΔN were immunoprecipitated and incubated with 3’-flap-OE, replication fork (RF-OE), or static HJ (XO-BE) substrates prior to analysis of the reaction products on native gels (Figure 5A, B). Similar levels of endonucleases are present in association with these SLX4 fragments as assessed by mass spectrometry (Figure 1B, Figure 2C). Full length HA-SLX4, as well as HA-SLX4ΔN, displayed robust activity against all three substrates (Figure 5B, lanes 2, 4, 7, 9, 12, and 14). In contrast, HA-SLX4ΔC supported very weak activity toward 3’-flap and replication fork structures, and no activity towards the XO structure (Figure 5B, lanes 5,10, and 15). HA-SLX4SBD, which associates specifically with SLX1, displayed low levels of activity toward 3'-flap and RF structures but displayed robust activity towards the static HJ substrate (Figure 5B, lanes 3, 8, and 13). These data reveal that the N-terminal region of SLX4, which associates with ERCC4-ERCC1, C20orf94, and MSH2-MSH3, is not required for efficient processing of replication fork, HJ and 3’-flap structures in vitro, consistent with the known preference of ERCC4-ERCC1 for bubble structures, and indicate that the C-terminal region of SLX4 encompassing residues 684–1834 contains the major flap and HJ cleavage activity.
Figure 5. SLX4-dependent cleavage of HJs maps to the C-terminal MUS81-EME1 and SLX1 binding domain (SBD).
(A) Schematic of SLX4 deletion fragments and relevant interactions.
(B) The indicated SLX4 complexes (from 1.5mg of cell extract) were incubated with radiolabeled substrates (30 min) labeled on strand 1 prior to native gel electrophoresis and autoradiography.
(C) Purified endonucleases produced in bacteria.
(D, E) SLX4SBD stimulates static and migrating HJ cleavage by SLX1. The indicated recombinant GST-SLX1-His6-SLX4SBD complexes (1, 3, or 5 ng) were incubated with XO-BE or X26 HJs at 37°C for 30 min and reaction products analyzed on native gels (panel D) or by phosphoimaging (panel E). SLX1CD: SLX1E82A.
In order to verify HJ cleavage by the SLX4SBD fragment in association with SLX1 and to examine whether SLX4 regulates the activity of SLX1, we prepared GST-SLX1 in bacteria in the presence or absence of His6-SLX4SBD (rSLX1-SLX4SBD; Figure 5C), and assayed these proteins for cleavage of static (XO-BE) and mobile (X26) HJs. In addition, we prepared a GST-SLX1E82A-His6-SLX4SBD complex in which a previously identified residue important to coordinating catalytic activity (E82) (Coulon et al., 2004) was mutated to alanine (Figure 5C). The rSLX1-SLX4SBD complex displayed robust activity towards both HJ structures (Figure 5D, E). In contrast, neither rSLX1 alone nor the rSLX1 E82A-SLX4SBD complex displayed appreciable activity toward either substrate. Quantitative analysis of the reaction products indicated a >100-fold increase in activity is conferred by binding of SLX4SBD to SLX1 (Figure 5E). As expected based on studies with yeast Slx1p (Fricke and Brill, 2003), rSLX1-SLX4SBD complexes also cleaved a 5’-flap substrate (Figure S15).
Cleavage specificity of the SLX4 complex toward 3’-flap and replication fork substrates
Previous studies have shown that MUS81-dependent cleavage of migrating HJs in vitro occurs at up to 10 distinct positions primarily on the 5’ side of the homologous core, with the precise preference reflecting the source of the MUS81 complex and the substrate employed (Constantinou et al., 2002; Chen et al., 2001; Boddy et al., 2001). Nicked HJs are clipped 2–5 and 3–6 nucleotides 5’ of the branch point by S. pombe and S. cerevisiae Mus81 complexes, respectively (Osman et al., 2003; Gaillard et al., 2003). In contrast, S.cerevisiae Slx1p-Slx4p (purified from bacteria) or S. pombe Slx1-Slx4 purified from endogenous sources cleaves migrating HJs in the 3’ side of the homologous core (Fricke and Brill, 2003; Coulon et al., 2004). We found that flap (3’-flap-OE) and replication fork (RF-OE) substrates had a virtually identical pattern of cleavage by both full length SLX4 and SLX4ΔN, and produced major products of 26, 27, and 28 nucleotides in length, reflecting cleavage 3–5 nucleotides 5’ to the branch site of these substrates (Figure 6A, lanes 2, 4, 7, 9). The products seen with SLX4 and SLX4ΔN complexes are most consistent with the known activity of MUS81-EME1. In contrast, SLX4ΔC complexes produced very weak cleavage 5 nucleotides before the branch site of RF-OE (Figure 6A, lane 10), and SLX4SBD produced low levels of a product of 32 nucleotides with the 3’-flap substrate but no products with RF-OE (Figure 6A, lanes 3 and 8). The simplest interpretation of these data is that MUS81-EME1 in association with SLX4 is responsible for cleavage 5’ to the junction with 3’-flap and RF structures.
Figure 6. Cleavage specificity of SLX4 and SLX1-SLX4SBD complexes.
(A) The indicated SLX4 complexes purified from 293T cells were incubated with radiolabeled substrates (30 min) prior to 12% polyacrylamide-urea gel electrophoresis and autoradiography. In the structural model (bottom) longer bars represent more efficient cleavage.
(B) HJ cleavage activity of SLX4 maps to the C-terminus containing the MUS81-EME1 and SLX1 binding sites. The indicated SLX4 immune complexes derived from 1.5 mg of 293T cell extract were incubated with HJ (X0-BE) labeled on strand 1 (30 min) and the products separated on denaturing gels.
(C) Symmetrical cleavage of HJ (X0-BE) by SLX4 complexes and GEN1-HA. The indicated immune complexes were incubated with HJ (X0-BE) radiolabeled on either strand 1 or strand 3 and products resolved on a denaturing gel prior to autoradiography.
(D) Major sites of cleavage observed with SLX4 (arrow and bars) or GEN1 (arrowhead) complexes.
(E–G) Symmetrical cleavage of HJ (X0-BE) by SLX1-SLX4SBD complexes. The indicated SLX4 complexes purified from 293T cells (panel E) or bacteria (panel F) were incubated with HJ (X0-BE) radiolabeled on strand 1 or strand 3 and products resolved on a denaturing gel prior to autoradiography. In panel G, the major site of HJ (X0-BE) cleavage by SLX1-SLX4SBD is indicated by the arrow.
Symmetrical cleavage of static HJs by SLX1-SLX4
Having localized HJ cleavage activity to the C-terminal region of SLX4, we then used denaturing gels to examine HJ cleavage specificity (Figure 6). SLX4 and SLX4ΔN preferentially cleaved the X0-BE substrate labeled on strand 1 at nucleotide 32, two nucleotides 3’ to the branch site, and also produced nicks 3–5 nucleotides 5’ to the branch site (Figure 6B, lanes 2, and 4). Thus, the cleavage activities of SLX4 and SLX4ΔN are reminiscent of a superposition of the known specificities of MUS81 and Slx1p, although significantly more specific than generally observed in previous studies (Constantinou et al., 2002; Chen et al., 2001; Fricke and Brill, 2003; Coulon et al., 2004).
An advantageous property of a resolvase is the ability to cleave two opposing strands of a HJ in a symmetrical manner such that they can be directly re-ligated to form a contiguous dsDNA strand without further enzymatic editing. Recent studies have shown that GEN1 symmetrically cleaves X0-BE structures primarily at nucleotide 31, a result we have validated here with GEN1-HA purified from 293T cells (Figure 6C, lanes 1 and 2). To examine the symmetry of cleavage of X0-BE by SLX4 complexes, we performed in vitro cleavage assays with X0-BE labeled on either strand 1 or strand 3 (Figure 6C, lanes 3 and 4, and Figure 6D). Cleavage was observed at nucleotide 32 on both strands and in addition, there was largely symmetrical cleavage from nucleotides 25–28 on both strand 1 and strand 3 by both endogenous SLX4 and HA-SLX4ΔN complexes (Figure 6C lanes 3 and 4, and 5E, lanes 3 and 4). The correspondence of products found with SLX4 and SLX4ΔN suggests that ERCC4-ERCC1 does not appreciably contribute to the cleavage of this substrate in vitro.
Given that yeast Slx1p-Slx4p, like GEN1, cleaves 3’ to the junction (Fricke and Brill, 2003; Coulon et al., 2004) and that SLX1-SLX4SBD displays the ability to produce nicked duplexes from static HJs (Figure 5B, lane 13), we hypothesized that the symmetrical cut at nucleotide 32 in X0-BE reflected the activity of SLX1. To address this possibility, we examined the specificity of cleavage by HA-SLX4SBD complexes, which contain SLX1 but not MUS81-EME1 or ERCC4-ERCC1 (Figure 2, Figure 6E). The HA-SLX4SBD complex displayed robust cleavage of X0-BE at nucleotide 32 on both strand 1 and strand 3 (Figure 6E, lanes 1 and 2, and 6G). To demonstrate that this activity was inherent to the SLX1-SLX4SBD complex, we assayed XO-BE substrates with bacterial SLX1-SLX4SBD, and again found symmetrical cleavage at nucleotide 32 (Figure 6F). These data indicate that SLX1-SLX4 displays an activity analogous to GEN1 but cleaves 1 nucleotide 3’ to the site of cleavage by GEN1 (Figure 6C compare lanes 1 and 2 with lanes 3 and 4, and Figure 6D).
The hallmark of symmetrical cleavage of HJs is the production of nicked duplexes that can be ligated without further processing. In order to examine ligation of products produced by SLX4 complexes, we turned to a migrating HJ substrate (X26) previously employed to assess the resolvase activity of GEN1 and Yen1p (Ip et al., 2008). The radiolabeled 5’ strand in this substrate (53-nucleotides) when symmetrically cleaved and ligated yields a 60-nucleotide product. As reported previously (Ip et al., 2008), GEN1 produced two major products, which efficiently formed a 60-nucleotide product upon ligase treatment (Figure 7A, lanes 2 and 8). SLX1-SLX4SBD complexes from both human cells (Figure 7A, lanes 3 and 9, Figure 7B) and bacteria (Figure 7C) produced two major products that were converted to a 60-nucleotide species upon addition of ligase. Quantification of these products indicated 50% conversion to the ligated form. SLX4 and SLX4ΔN complexes (Figure 7B) produced a number of products, potentially reflecting the activity of both MUS81-EME1 and SLX1 on the migrating X26 substrate (Figure 7A, lanes 4 and 5) and ligation products were also detected (lanes 10 and 11). This, together with the largely symmetrical cleavage activity of SLX1-SLX4SBD complexes on static HJs suggests that SLX1 can function to resolve these structures.
Figure 7. SLX1-SLX4 complexes resolve HJs through an ordered pathway in vitro and are required for efficient homology-directed repair of DSBs in vivo.
(A) The indicated SLX4 and GEN1 complexes were incubated with an X26 substrate +/−T4 DNA ligase prior to separation on a denaturing gel and autoradiography.
(B) The indicated SLX4-HA immune complexes were immunoblotted for ERCC4 and MUS81.
(C) Recombinant SLX1-SLX4SBD complexes generate cleaved X26 structures that are ligatable.
(D) Requirement of SLX1 and MUS81 for cleavage of static HJs. 293T/SLX4-HA cells were transfected with control siRNA or siRNAs targeting SLX1 (SLX1#4) or MUS81 (MUS81-17). After 72 h, SLX4-HA complexes were assayed in cleavage assays using HJ X0-BE substrates labeled on either strand 1 or strand 3. Products were resolved on denaturing gels prior to autoradiography.
(E) SLX4-HA immune complexes corresponding to panel D were immunoblotted for ERCC4 and MUS81.
(F) qPCR analysis of mRNA isolated from cells transfected with control siRNA or siRNA targeting SLX1 (#4). mRNA abundance is relative to GAPDH.
(G–I) MUS81-EME1 complexes cleave nicked HJs that mimic those produced by SLX1-SLX4. Bacterial GST-MUS81-His6-EME1 complexes were incubated (37°C, 30 min) with synthetic nicked HJs (XO1 strand nicked at nucleotide 32) labeled on either strand 1 or strand 3 and the products analyzed on either native (panel G) or denaturing (panel H) gels. The positions of cleavage of the nicked XO structure labeled on strand 3 are shown in panel I.
(J) GST-MUS81-His6-EME1 (12.5, 25, 50 ng) was incubated (37°C, 30 min) with XO-BE in the presence or absence of 3 ng of recombinant SLX1-SLX4SBD, and the products analyzed on denaturing gels.
(K) DR-GFP U2OS were infected with viral vectors expressing the indicated shRNAs followed by I-SceI expression 48h later. The extent of homologous recombination was determined by flow cytometry. *, p < 0.05; students T-test. Error bars are STDEV across 3 technical replicates.
(L) Model depicting distinct roles of SLX1 and MUS81 within the SLX4 complex for HJ cleavage (see text for details).
Evidence for ordered processing of HJs by SLX1 and MUS81 endonucleases in vitro
The data presented above indicate that SLX1-SLX4SBD can symmetrically cleave both static and migrating HJs, but in the context of full-length SLX4, a more complex set of products reminiscent of the activities of both SLX1 and MUS81 are evident (Figure 6C). Given that MUS81-EME1 is most active toward nicked HJs (Gaillard et al., 2003; Osman et al., 2003; Taylor and McGowan, 2008) we considered an ordered pathway in which asymmetrical cleavage of static HJs by SLX1-SLX4 complexes produces a nicked substrate that is then processed by MUS81-EME1 as the route to these products. In this regard, the presence of a 5’ end within 3 nucleotides of the branch point stimulates MUS81-dependent cleavage 5’ to the branch point on the opposing strand in the context of flap substrates (Bastin-Shanower et al., 2003). Asymmetric processing at nucleotide 32 in X0-BE by SLX1 would therefore be expected to create a favored substrate for MUS81. To test this idea, we first examined the effect of depletion of SLX1 and MUS81 on the activity of purified SLX4-HA complexes in vitro. Depletion of MUS81 (Figure 7E) reduced the abundance of products that represented nicking 2'5 nucleotides 5’ to the branch point but had little effect on symmetrical cleavage at nucleotide 32 (Figure 7D, compare lanes 3 and 7, 4 and 8). In contrast, depletion of SLX1 (Figure 7F) resulted in a reduction in cleavage efficiency for sites both 3’ and 5’ to the branch site (Figure 7D, compare lanes 3 and 5, 4 and 6), with the reduction in the extent of cleavage at nucleotide 32 being most dramatic, but did not affect MUS81 association with SLX4 (Figure 7E). The simplest interpretation of these data is that cleavage activity of SLX1 3’ to the branch point is a pre-requisite for robust cleavage activity by MUS81 5’ to the branch point.
To test this directly, we examined the ability of recombinant MUS81-EME1 (Figure 5C) to process synthetic nicked XO-BE structures that mimic a single cut by SLX1 at 32 nucleotides on strand 1 using both native and denaturing gels. The expected products reflecting cleavage of strand 3 by MUS81-EME1 were obtained (Figure 7G–I). MUS81-EME1 could also process nicked junctions created in situ by cleavage of XO-BE structures by rSLX1-SLX4SBD (Figure 7J), although in this case the low abundance of MUS81-dependent cleavage products likely reflects a preference for SLX1-SLX4SBD to symmetrically cleave this HJ structure as opposed to making a single asymmetric cut.
These data are consistent with the hypothesis that SLX1 can symmetrically cleave HJ structures and that a nicked intermediate can be acted upon by MUS81 to produce gapped duplexes.
SLX4 is required for efficient repair of DSBs in vivo
The ability of SLX4 to support cleavage of recombination intermediates and flap structures in vitro suggested a possible role in resolution of structures formed during homology-directed repair (HDR) of DSBs. To examine this question, we employed U2OS cells carrying a DR-GFP substrate (Xia et al., 2006). This substrate contains 2 nonfunctional GFP open reading frames, including one GFP coding sequence that is interrupted by a recognition site for the I-SceI endonuclease (SceGFP). Expression of I-SceI leads to formation of a DSB in the SceGFP allele, which is repaired by HDR using a nearby iGFP lacking N- and C-terminal GFP sequences, thereby producing functional GFP (Xia et al., 2006). HDR-reporter cells expressing a control firefly luciferase shRNA displayed robust production of GFP-positive cells after I-SceI expression and this was reduced by ∼25% upon shRNA-mediated depletion of SLX4 (Figure 7K) in a manner that correlated with the extent of depletion seen with these vectors (Figure 3B). Cell cycle analyses demonstrated that small changes in cell cycle distribution did not correlate with the effects on HR (Figure S16). The extent of inhibition of DSB repair upon SLX4 depletion is comparable to that seen with depletion of FANCD2 and FANCI (Smogorzewska et al., 2007).
Discussion
The mammalian SLX4 complex: a tool-kit for DNA repair
Homologous recombination allows the error free repair of a variety of types of DNA damage including DSBs and replication blocks and proceeds via the formation of single or double 4-stranded HJs. The resolution of this topologically constrained structure into 2 separate dsDNA molecules is critical to the successful completion of the repair process, however the identities of the endonucleases (a.k.a. resolvases) responsible for these reactions and an understanding of the repair processes in which they participate remain incompletely understood. Here, we identify a complex of nucleases, brought together through their association with a scaffolding protein originally named BTBD12, which is capable of resolving HJs in vitro and promoting ICL repair and homology-directed DSB repair in vivo. These findings allow us to propose that this complex acts as a HJ resolvase in mammals and for the reasons described below, we have renamed BTBD12 as mammalian SLX4. The accompanying paper (Fekairi et al., 2009) also identifies SLX4 as an endonuclease scaffold required for ICL repair and possessing HJ resolvase activity.
SLX4 is the apparent mammalian ortholog of yeast Slx4p and Drosophila MUS312. Like Slx4p, SLX4 associates with both an SLX1 ortholog and with ERCC4(XPF)-ERCC1, but in contrast to Slx4p, SLX4 also interacts with MUS81-EME1 and with other proteins linked with DNA repair (MSH2-MSH3) and telomere function (TERF2-TERF2IP) independently of DNA damage (Figure S7). The interaction between SLX4 and SLX1 occurs via the extreme C-terminus of SLX4, a region that displays sequence similarity with the C-termini of Slx4p and MUS312. Previously reported BLAST analyses identified Slx4p related proteins only in yeast, yet Slx1p orthologs are broadly conserved in eukaryotes, leading to the conclusion that Slx4p is either unique to budding yeast or that the gene is under little selective pressure (Fricke and Brill, 2003). Our identification of an apparent mammalian Slx4p ortholog suggests the latter interpretation. Outside of the extreme C-terminal region of SLX4, there are only small pockets of sequence similarity between SLX4, Slx4p, and MUS312 (Figure S2). It remains to be determined whether these regions constitute the sites of interaction with common components within orthologous complexes.
In addition to endonucleases, other SLX4-associated proteins may coordinate repair functions. For example, the functions of ERCC4(XPF) in processing of telomere structures via TERF2 (Zhu et al., 2003) may be coordinated by SLX4, which has independently been found at telomeres (Dejardin and Kingston, 2009). Likewise, SLX4 may facilitate MUS81-dependent recombination at telomeres (Zeng et al., 2009) Moreover, our data indicate that PLK1 can phosphorylate SLX4, suggesting a possible role for this kinase in conferring cell cycle control via SLX4 function. An understanding of the roles played by PLK1, as well as the novel SLX4-binding protein C20orf94, will require further study.
Role of SLX4 in resistance to DNA damage
We have found that SLX4 is required for resistance to MMC, CPT, and MMS, but found little evidence of a role in resistance to UV or IR. Similar to SLX4 depletion, reduction of SLX1 to undetectable levels in HeLa cells resulted in weak sensitivity to MMC and CPT but no significant sensitivity to UV, suggesting that both SLX1 and SLX4 are required for full resistance to interstrand and DNA-protein crosslinks. Consistent with a role for SLX4 in homologous recombination, we found that depletion of SLX4 reduces the efficiency of repair of I-SceI-induced DSBs by ∼25% in vivo. Coupled with our biochemical data, these results suggest that one role of the SLX4 complex in homology-directed repair may be through HJ resolution. While non-meiotic mammalian HR is thought to occur primarily by synthesis-dependent strand annealing, which does not appear to involve HJ resolution, our results suggest that the fraction of HR events measured by this assay involving HJ resolution may be higher than previously expected. Alternatively, SLX4 may be involved in cleaving other HR intermediates, e.g., involving flaps, at least in mitotic cells. A full assessment of the extent of DNA-damage sensitivities of cells lacking SLX4 and SLX1, as well as the role of these genes in HR, will require creation of null alleles of these genes. Further studies are also required to determine whether the sensitivities of cells lacking MUS81 or ERCC4 to CPT and MMC reflect a common function with SLX4.
SLX4 complexes display a unique Holliday junction resolvase activity
The assembly of endonucleases on SLX4 endows this complex with multiple types of DNA cleavage activity. While the MUS81-EME1 module promotes flap and RF cleavage, the SLX1 endonuclease promotes symmetrical cleavage of HJs. SLX1 cleaves static HJs with high specificity, 2 nucleotides 3’ to the branch point. This activity is analogous to GEN1, the only vertebrate enzyme known to symmetrically cleave static HJs, which cleaves 1 nucleotide 3’ to the branch point. Yeast Slx1p-Slx4p displays somewhat relaxed specificity relative to the mammalian enzyme reported here, producing multiply nicked products that are largely non-symmetrical (Coulon et al., 2004; Fricke and Brill, 2003). This led to the conclusion that yeast Slx4p-Slx1p is not a HJ resolvase (Coulon et al., 2004; Fricke and Brill, 2003). The biochemical basis for the differences in specificity between yeast and human SLX1 remains to be determined.
SLX4 complexes display not only symmetrical cleavage of static HJs by SLX1 but also display products that represent cleavage 5’ to the junction, reminiscent of those produced by MUS81. Depletion of MUS81 affected cleavage 5’ to the junction while depletion of SLX1 reduced cleavage both 3’ and 5’ to the junction. This, together with the finding that MUS81-EME1 can act on nicked HJs of the type produced by asymmetric SLX1-dependent cleavage, suggests a model in which two types of products can be produced on HJs (Figure 7L). Symmetrical cleavage by SLX1-SLX4 produces a nicked duplex that can be religated. In contrast, asymmetric cleavage by SLX1-SLX4 provides an intermediate that can be acted on by associated MUS81-EME1 to create a gapped duplex, which cannot be ligated directly. While each of these reactions occur in vitro, it is likely that the relative rates of these two reactions in vivo are controlled in order to achieve the desired outcome. For example, the organization of the SLX1-SLX4 complex on HJ structures in vivo may bias the reaction toward synchronous symmetrical cleavage across the junction, thereby rendering the products immune to MUS81 nicking activity. Our results may also help to explain differences in static HJ cleavage activity seen previously with MUS81 (Taylor and McGowan, 2008; Constantinou et al., 2002; Chen et al., 2001; Ciccia et al., 2003). Depletion experiments indicate that a small fraction of cellular MUS81 is associated with SLX4 (Figure S12E). Thus, the bulk of MUS81 complexes from human cells have much lower specific HJ cleavage activity when compared to SLX4 complexes containing an identical quantity of MUS81 (Figure S12A–D). One explanation for this is that SLX1/SLX4 present in a fraction of mammalian MUS81 complexes promotes nicking of static HJs which are then further processed by MUS81. Variable levels of SLX1-SLX4 proteins in MUS81-EME1 preparations from mammalian cells could contribute to differing levels of HJ cleavage activity seen previously (Taylor and McGowan, 2008; Constantinou et al., 2002; Chen et al., 2001; Ciccia et al., 2003). The identification of the SLX4 complex provides a new platform through which to further elucidate the enzymology of DNA repair and should enable a much deeper understanding of many different repair processes as the functions of the other components within the complex are elucidated.
Experimental Procedures
Detailed Experimental Procedures are provided in the Supplemental Material.
Cell Culture, plasmids, antibodies, and RNAi
Cell culture was performed as described (Smogorzewska et al., 2007). All open reading frames (Table S2) were transferred to the indicated expression vectors using λ recombinase. Anti-SLX4C (against residues 1813–1826) was raised in rabbits and affinity purified using immobilized antigenic peptide. All other antibodies were from commercial sources: TERF2IP (Rap1), Bethyl (A300–306A); SLX4M, Novus (H00084464-B01); PLK1, Santa Cruz (F-8); MSH2, Abcam (ab52266); MUS81, Abcam (ab14387); ERCC4 (XPF), Thermo (clone 219); γH2AX, Millipore (JBW301); TRF2, Millipore (4A794). shRNA and siRNA sequences used in this study are provided in Table S2.
Protein purification and mass spectrometry
293 (or 293T) cells stably expressing the indicated proteins (as HA-fusions) using either lenti or retroviral vectors were used for immunoprecipitation and complexes analyzed by mass spectrometry as described (Sowa et al., 2009). Briefly, ∼107 cells were lysed in 4 ml of lysis buffer (50mM Tris-HCl pH 7.5, 150mM NaCl, 0.5% Nonidet P40, 1mM EDTA) + protease inhibitors (ROCHE), and phosphatase inhibitors. Cleared lysates were filtered through 0.45 µM spin filters (Millipore Ultrafree®-CL) and immunoprecipitated with 30 µl of anti-HA resin (Sigma). Complexes were washed with lysis buffer, exchanged into PBS, eluted with HA peptide and precipitated with 10% TCA. Processing of samples for mass spectrometry as well as analysis of proteomic data using CompPASS is described elsewhere (Sowa et al., 2009). Proteomic data is provided in Table S1. Complexes for immunoblotting or for DNA cleavage assays were prepared similarly. For immunoblotting, anti-HA immune complexes were eluted with 2X SDS-sample buffer prior to electrophoresis. For cleavage assays, complexes were washed with cleavage buffer (50mM Tris pH 8.0, 5mM MgCl2 , 40mM NaCl, 1mM DTT, 100µg/ml BSA) prior to substrate addition.
MCA and HR Assays
MCA assays were performed in U2OS or HeLa cells as described previously (Smogorzewska et al., 2007). Briefly, dsRed-U2OS cells were infected with control shRNA vectors and GFP-U2OS cells were infected with the indicated SLX4 shRNA vectors. Seven to ten days later, cells were mixed in equal numbers and incubated in the presence of the indicated DNA damaging agent for 10–12 days. The ratio of GFP-positive to dsRed-positive cells was determined by flow cytometry. HeLa cell MCA assays were performed similarly using the indicated siRNAs. HR assays were performed using I-SceI-induced cleavage of a DR-GFP substrate in U2OS cells as previously described (Xia et al., 2006).
In vitro cleavage assays
In vitro cleavage of DNA substrates (Table S2) was performed using the indicated SLX4 complexes in conjunction with previously described 5’ 32P-labeled DNA substrates (Ciccia et al., 2003; Ip et al., 2008) in cleavage buffer. Enzyme complexes were obtained either by immunoprecipitation from 293T cells or by expression in E. coli. GST-SLX1 and His6-SLX4SBD were co-expressed using pET60 and pColA vectors and purified using sequential GSH-Sepharose/Nickel-NTA resins. GST-MUS81/His6-EME1 (Ciccia et al., 2003) was prepared similarly. Reaction products were subjected to native or denaturing polyacrylamide gel and visualized by autoradiography. Ligation experiments were supplemented with 400 units of T4 DNA ligase (1h) prior to electrophoresis on denaturing gels. Oligonucleotide markers (not shown) were included in each denaturing gel to determine cleavage sites.
Supplementary Material
Acknowledgements
We thank Clare McGowan, Richard Wood, Orlando Scharer, Joyce Reardon, Laura Niedernhofer, Maria Jasin, Steve West and Guang Hu for reagents, John Rouse, Jim Haber, Jeff Sekelsky, Monica Colaiacovo, and Pierre-Henri Gaillard for communicating results prior to publication, and Alberto Ciccia and Maria Jasin for discussions. This work was supported by grants from the National Institutes of Health to S.J.E., S.P.G., and J.W.H., and by a grant from the Stewart Trust to J.W.H. A.S. is supported by T32CA09216 to the Pathology Department at the Massachusetts General Hospital and by an award from the Burroughs Wellcome Fund. B.C.O. is supported by a post-doctoral fellowship from the American Cancer Society. S.J.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bastin-Shanower SA, Fricke WM, Mullen JR, Brill SJ. The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1-Rad10. Mol Cell Biol. 2003;23:3487–3496. doi: 10.1128/MCB.23.10.3487-3496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR, 3rd, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- Chen XB, Melchionna R, Denis CM, Gaillard PH, Blasina A, Van de Weyer I, Boddy MN, Russell P, Vialard J, McGowan CH. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol Cell. 2001;8:1117–1127. doi: 10.1016/s1097-2765(01)00375-6. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Constantinou A, West SC. Identification and characterization of the human mus81-eme1 endonuclease. J Biol Chem. 2003;278:25172–25178. doi: 10.1074/jbc.M302882200. [DOI] [PubMed] [Google Scholar]
- Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu Rev Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- Constantinou A, Chen XB, McGowan CH, West SC. Holliday junction resolution in human cells: two junction endonucleases with distinct substrate specificities. Embo J. 2002;21:5577–5585. doi: 10.1093/emboj/cdf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote AG, Lewis SM. Mus81-dependent double-strand DNA breaks at in vivo-generated cruciform structures in S cerevisiae. Mol. Cell. 2008;31:800–812. doi: 10.1016/j.molcel.2008.08.025. [DOI] [PubMed] [Google Scholar]
- Coulon S, Gaillard PH, Chahwan C, McDonald WH, Yates JR, 3rd, Russell P. Slx1-Slx4 are subunits of a structure-specific endonuclease that maintains ribosomal DNA in fission yeast. Mol Biol Cell. 2004;15:71–80. doi: 10.1091/mbc.E03-08-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009;136:175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Brown JA, You D, Brown JM. Multiple Endonucleases function to repair covalent topoisomerase I complexes in Saccharomyces cerevisiae. Genetics. 2005;170:591–600. doi: 10.1534/genetics.104.028795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia AE, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong M-Q, Ruse C, Yates JR, Russell P, III, Fuchs RP, McGowan CH, Gillard P-HL. Human SLX4 is a Holliday junctions resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009 doi: 10.1016/j.cell.2009.06.029. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flott S, Alabert C, Toh GW, Toth R, Sugawara N, Campbell DG, Haber JE, Pasero P, Rouse J. Phosphorylation of Slx4 by Mec1 and Tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Mol Cell Biol. 2007;27:6433–6445. doi: 10.1128/MCB.00135-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke WM, Brill SJ. Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 2003;17:1768–1778. doi: 10.1101/gad.1105203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard PH, Noguchi E, Shanahan P, Russell P. The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol Cell. 2003;12:747–759. doi: 10.1016/s1097-2765(03)00342-3. [DOI] [PubMed] [Google Scholar]
- Haber JE, Heyer WD. The fuss about Mus81. Cell. 2001;107:551–554. doi: 10.1016/s0092-8674(01)00593-1. [DOI] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, Kanaar R. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. Embo J. 2006;25:4921–4932. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Hollingsworth NM, Brill SJ. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 2004;18:117–125. doi: 10.1101/gad.1165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- Li F, Dong J, Pan X, Oum JH, Boeke JD, Lee SE. Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10-dependent processing of recombination intermediates. Mol Cell. 2008;30:325–335. doi: 10.1016/j.molcel.2008.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- McPherson JP, Lemmers B, Chahwan R, Pamidi A, Migon E, Matysiak-Zablocki E, Moynahan ME, Essers J, Hanada K, Poonepalli A, et al. Involvement of mammalian Mus81 in genome integrity and tumor suppression. Science. 2004;304:1822–1826. doi: 10.1126/science.1094557. [DOI] [PubMed] [Google Scholar]
- Mu JJ, Wang Y, Luo H, Leng M, Zhang J, Yang T, Besusso D, Jung SY, Qin J. A proteomic analysis of ataxia telangiectasia-mutated (ATM)/ATM-Rad3-related (ATR) substrates identifies the ubiquitin-proteasome system as a regulator for DNA damage checkpoints. J Biol Chem. 2007;282:17330–17334. doi: 10.1074/jbc.C700079200. [DOI] [PubMed] [Google Scholar]
- Osman F, Dixon J, Doe CL, Whitby MC. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, Elledge SJ. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett E, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009 doi: 10.1016/j.cell.2009.04.042. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor ER, McGowan CH. Cleavage mechanism of human Mus81-Eme1 acting on Holliday-junction structures. Proc Natl Acad Sci U S A. 2008;105:3757–3762. doi: 10.1073/pnas.0710291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SC. Holliday junctions cleaved by Rad1? Nature. 1995;373:27–28. doi: 10.1038/373027a0. [DOI] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Xu L, Wei Y, Reboul J, Vaglio P, Shin TH, Vidal M, Elledge SJ, Harper JW. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425:316–321. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- Yildiz O, Majumder S, Kramer B, Sekelsky JJ. Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol Cell. 2002;10:1503–1509. doi: 10.1016/s1097-2765(02)00782-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng S, Xiang T, Pandita TK, Gonzalez-Suarez I, Gonzalo S, Harris CC, Yang Q. Telomere recombination requires the MUS81 endonuclease. Nat Cell Biol. 2009;11:616–623. doi: 10.1038/ncb1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T. ERCC1/XPF removes the 3' overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol Cell. 2003;12:1489–1498. doi: 10.1016/s1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.