Abstract
In the clinical management of early-stage cutaneous melanoma, it is critical to determine which patients are cured by surgery alone and which should be treated with adjuvant therapy. To assist in this decision, many groups have made an effort to use molecular information. However, although there are hundreds of studies that have sought to assess the potential prognostic value of molecular markers in predicting the course of cutaneous melanoma, at this time, no molecular method to improve risk stratification is part of recommended clinical practice. To help understand this disconnect, we conducted a systematic review and meta-analysis of the published literature that reported immunohistochemistry-based protein biomarkers of melanoma outcome. Three parallel search strategies were applied to the PubMed database through January 15, 2008, to identify cohort studies that reported associations between immunohistochemical expression and survival outcomes in melanoma that conformed to the REMARK criteria. Of the 102 cohort studies, we identified only 37 manuscripts, collectively describing 87 assays on 62 distinct proteins, which met all inclusion criteria. Promising markers that emerged included melanoma cell adhesion molecule (MCAM)/MUC18 (all-cause mortality [ACM] hazard ratio [HR] = 16.34; 95% confidence interval [CI] = 3.80 to 70.28), matrix metalloproteinase-2 (melanoma-specific mortality [MSM] HR = 2.6; 95% CI = 1.32 to 5.07), Ki-67 (combined ACM HR = 2.66; 95% CI = 1.41 to 5.01), proliferating cell nuclear antigen (ACM HR = 2.27; 95% CI = 1.56 to 3.31), and p16/INK4A (ACM HR = 0.29; 95% CI = 0.10 to 0.83, MSM HR = 0.4; 95% CI = 0.24 to 0.67). We further noted incomplete adherence to the REMARK guidelines: 14 of 27 cohort studies that failed to adequately report their methods and nine studies that failed to either perform multivariable analyses or report their risk estimates were published since 2005.
Cutaneous malignant melanoma (CMM), which accounted for 62 500 new cases of cancer in 2008, is the sixth most common malignancy in men and the seventh most common in women in the United States (1). Although 80% of new lesions are localized to the skin where effective surgical resections result in more than 95% 5-year survival (1), disease can recur in individuals with localized lesions despite appropriate management (2). Because adjuvant therapy is not broadly indicated for localized melanoma due to unfavorable risk–benefit ratios (3), there is a critical need to identify, at the time of diagnosis, the subset of patients most likely to benefit from adjuvant treatment to improve overall survival outcomes. Although, in addition to localization, nine clinicopathologic prognostic markers have been identified for CMM and have been used to establish clinically validated risk stratifications among melanoma patients (4,5), risk models based on these markers do not account for all of the observed variability in melanoma-related survival. Indeed, in melanoma (6–8) as in other cancers (9,10), tumors with identical clinical and histological parameters have markedly different mRNA expression profiles, and tumor subgroups classified by gene expression can be strongly associated with differential survival.
Immunohistochemistry (IHC) is a widely accepted and well-documented method for characterizing patterns of protein expression while preserving tissue and cellular architecture (11). The introduction of tissue microarray (TMA) technology, in which samples from several hundred individual tissue blocks can be spotted on a single glass slide (12), extends the rigor of IHC-based biomarker assays both by facilitating high-throughput analysis of candidate proteins across large patient cohorts and by substantially reducing misclassification of expression across the cohort through the application of consistent staining conditions and reagents (13). However, unlike genomic or proteomic experiments that can be performed in parallel on a massive scale, IHC/TMA experiments must be done serially using a candidate gene approach and data from individual experiments must be combined to establish multimarker prognostic discriminators.
Several recent reviews have been published, each of which surveyed published IHC data on melanoma and focused on prognostic applications (14–16). However, none of these surveys prioritized the available data according to REMARK study design or methodological assessment quality metrics (17). In addition, even among the high-quality studies, the heterogeneity in experimental procedures such as antigen retrieval, choice and final dilution of primary antibody, and antibody validation through appropriate positive and negative controls and interobserver variability in describing the staining patterns, selection of cut points, and assignment of specimens to categories could have influenced the direction, magnitude, or statistical significance of the proposed association (18,19). Furthermore, none of these reviews limited inclusion to proteins evaluated in a multivariable setting adjusted for known clinical prognostic characteristics, a REMARK requirement (17). Because new molecular markers need to enhance the current routine estimators of prognosis to be adopted for use in the clinic, studies that do not extend statistical analyses beyond univariate survival measures are less valuable than studies that do.
In this systematic review and meta-analysis, we sought to determine the candidate biomarkers for which there was sufficient evidence to support prospective validation in a controlled clinical environment and to identify the functional pathways for which the data either suggest a lack of involvement in melanoma prognosis or the need for additional investigation due to insufficient rigor among previously executed studies. We identified the subset of candidate IHC-based protein predictors of melanoma outcome from the published literature that were evaluated according to robust sampling, laboratory, and statistical methods. Then, by applying a systems-based approach to the eligible data, we examined which tumor-sustaining pathways and component proteins are prognostic for melanoma all-cause mortality (ACM), melanoma-specific mortality (MSM), and disease-free survival (DFS).
Materials and Methods
Search Strategy
To identify all primary research articles that evaluated levels of candidate protein expression, as measured by IHC, as a prognostic factor among individuals diagnosed with CMM, we searched the PubMed medical literature database on January 15, 2008, without language restrictions, using the following three independent queries:
{[melanoma] AND ([prognos*] or [surviv*]) AND [cutaneous] AND ([gene] OR [protein])}.
{([melanoma] AND [immunohistoch*] AND ([prognos*] OR [surviv*])) NOT [uvea*]}.
{([melanoma] AND [immunohistoch*] AND [progress*]) NOT [uvea*]}.
One reviewer (B. E. G. Rothberg) inspected the title and abstract of each electronic citation to identify those manuscripts that were likely to report the assay of melanoma samples by IHC and obtained their full texts. Supplemental PubMed searches by names of authors contributing to five or more potentially relevant manuscripts were performed to identify any additional manuscripts not included in the primary queries. In those cases in which several publications derived from the same set of IHC data, only the study presenting the largest dataset was included. Five manuscripts that were not published in English were translated into English for further evaluation.
Methodological and Validity Assessment
We used published guidelines for reporting IHC-based tumor marker studies (17) and quality metrics for evaluating IHC-based studies for inclusion in cancer-related meta-analyses (19) as inclusion criteria for this review. Studies were eligible if they met each of the following six criteria: 1) prospective or retrospective cohort design with a clearly defined source population and justifications for all excluded eligible cases; 2) assay of primary cutaneous tumor specimens; 3) clear descriptions of methods for tissue handling and IHC, including antigen retrieval, selection and preparation of both primary and secondary antibodies, as well as visualization techniques; 4) a clear statement on the choice of positive and negative controls and on the outcome of the assay to ensure that the primary antibody used was a well-validated reagent; 5) statistical analysis using multivariable proportional hazards modeling that adjusted for clinical prognostic factors; and 6) reporting of the resultant adjusted hazard ratios (HRs) and their 95% confidence intervals (CIs). Because acral lentigionous melanomas, mucosal melanomas, and ocular melanomas display different clinical courses and molecular phenotypes from the more common cutaneous superficial spreading and nodular histological subtypes (20–22), studies describing results on non-Caucasian populations as well as those specific for acral lentiginous, mucosal, choroidal, or uveal melanomas were excluded. Studies were also excluded if they did not describe protein expression levels in melanoma cells and limited analysis to the associated stroma or vasculature. Within each study, only assays that evaluated proteins corresponding to mapped genetic loci were included; IHC reagents that targeted nonspecific “activities” or uncharacterized antigens were eliminated from further consideration. When authors described having assessed multivariable proportional hazards but the manuscript did not meet inclusion criteria because details describing the cohort, IHC methods, or the hazard ratio and 95% confidence interval were omitted, the corresponding author was contacted in an attempt to obtain the missing information. Letters were sent to 26 investigators, and responses were received from nine of them. Six responses provided missing IHC methods and/or risk estimate information for seven manuscripts (23–29), one additional response reported an indeterminate risk estimate that could not be used in meta-analysis (30), and one response (31) indicated that the authors no longer had the information that we had requested.
Data Extraction
One investigator (B. E. G. Rothberg) reviewed each eligible manuscript and extracted data on the characteristics of the study, including number and type of melanoma tumors assayed, IHC methodology, and results. The data recorded about each study for metrics included first author’s name, institution, and country of origin; journal and year of publication; sample size; starting material (frozen vs paraffin embedded, whole slides vs TMA); clinical covariates incorporated in the multivariable statistical analysis; outcomes assessed; mention of blinding of those who assessed IHC staining to outcome; and the set of candidate proteins selected for analysis. We also redacted additional data concerning methods within each study, including primary antibody and dilution used, secondary signal amplification and coloration methods, IHC stain scoring scheme, survival analysis cut points and reference group, the computed multivariable hazard ratio and its 95% confidence interval, and the corresponding P value. When results were presented without confidence intervals or SEs, the P value was used to estimate the SE via the z-statistic.
Statistical Data Analysis
All eligible individual protein assays were first sorted according to outcome and then according to the protein's major biological function. Protein function was determined following comprehensive review of the current scientific literature and classified according to the six acquired capabilities of cancer as defined by Hanahan and Weinberg (32): limitless replicative potential, evading apoptosis, insensitivity to antigrowth signals, self-sufficiency in growth signals, tissue invasion and metastasis, and sustained angiogenesis. To accommodate melanoma-associated antigens (eg, gp100, MelanA/MART-1) and immunomodulatory molecules (eg, major histocompatability complex class II), the Hanahan–Weinberg classification system was supplemented by two additional melanoma-specific functional categories: altered immunocompetence and melanocyte differentiation. For the set of proteins evaluated in a single study within one of the study outcomes, the summary hazard ratio (95% confidence interval) represents the value reported in that study. For proteins assayed in multiple studies, fixed effects summary hazard ratio and 95% confidence interval were calculated using the generic inverse variance method (33) and random effects models according to the Der Simonian–Laird method (34). Interstudy heterogeneity was assessed using the I 2 statistic (35). An observed hazard ratio of more than 1 implied a worse outcome for the test group relative to the reference group and would be considered statistically significant if the 95% confidence interval did not overlap with 1 (P < .05). Meta-analyses were conducted using the REVMAN systematic review and meta-analysis software package, version 4.2 (Cochrane Collaboration; www.cochrane.org). To determine whether the proteins demonstrating statistically significant associations with outcome were equally distributed across the Hanahan–Weinberg acquired capabilities of cancer, the proportion of such proteins in each individual functional group was compared with their overall proportion using the one-sample test for a proportion (36).
Results
Excluded Studies
The literature search strategy identified 1797 manuscripts for consideration (Figure 1; Supplementary Table 1, available online). Following the title and abstract search of these, as well as the supplemental-directed author searches, 515 manuscripts were identified that suggested the execution of IHC experiments on cutaneous melanoma samples, and full-text versions of these were retrieved. Of these, 30 studies recruited non-Caucasian patients’ samples, six solely evaluated staining of stromal or vascular elements and 24, after careful review of the study methods, did not perform IHC on human melanoma samples, so all 60 of these studies were excluded from further analysis. The remaining 455 studies, which collectively described IHC results for 387 unique proteins, were first triaged according to study design. Whereas 102 met the criteria for cohort study, 353 manuscripts were excluded for inappropriate study design. Among those excluded, 16 were case–control studies, 284 were cross-sectional analyses limited to determining the association between levels of marker expression with melanocytic lesion progression or with clinicopathologic parameters, and 53 were classified as case series for which the investigators failed to provide details on either the source population of melanocytic tumors or the sampling strategy. This latter group included 14 reports that performed multivariable proportional hazards modeling of survival outcomes (37–50), of which four included sample size greater than 70 (37,41,44,45) and one that met all other inclusion criteria, except for study design (43).
Figure 1.
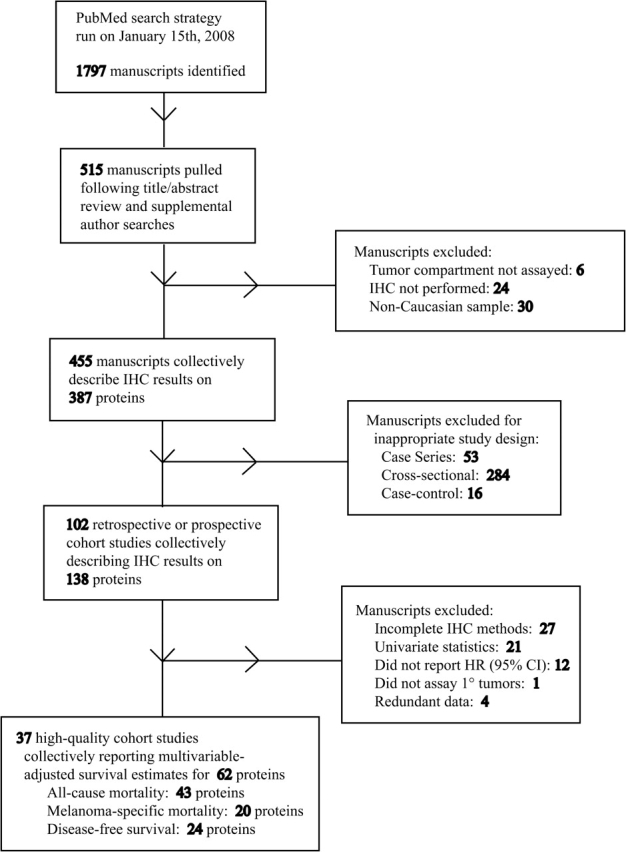
Flow diagram of the literature search and study selection protocol. CI = confidence interval; HR = hazard ratio; IHC = immunohistochemistry.
Among the 102 cohort studies, an additional 65 studies were excluded according to methodological or statistical criteria. Twenty-seven studies failed to completely describe their IHC methods and enumerate their positive and negative controls for antibody specificity validation (5,51–76), and an additional 21 methodologically robust manuscripts limited their analysis to univariate log-rank or proportional hazards computations (77–97). Eleven studies conducted multivariable analyses on methodologically robust data but failed to publish a hazard ratio (95% confidence interval) (31,98–107). One study restricted its analysis to metastatic lesions (108), and four studies had data that were completely redundant with larger included studies (109–112). We also excluded the study by Mihic-Probst et al. (30). In this otherwise eligible study, the choice of a p16/INK4A cut point at 50% of cells stained led to no events among the patients with high expression to yield an indeterminate multivariable hazard ratio that cannot be combined in meta-analysis.
Included Studies
Thirty-seven high-quality cohort studies from 21 independent research groups met the eligibility criteria for this systematic review by presenting multivariable survival estimates for differential levels of candidate protein expression as measured by IHC on primary cutaneous melanoma samples (Table 1). The included studies consist of one prospective cohort study (29) and 36 retrospective cohort studies (23–28,113–142). All 37 studies sampled archival formalin-fixed, paraffin-embedded tissue blocks. Of these, in 23 studies, IHC was performed on individual whole-slide tissue sections, and in 14 studies, TMAs were created using 1.5-mm- (113,114), 1.0-mm- (134,135), or 0.6-mm- (23,27,115–117,128–132) diameter cores from representative tissue regions. Among the TMA subset, the five studies performed at the Restoration of Appearance and Function Trust Institute (Middlesex, UK) included redundant sampling of individual tissue blocks (128–132). Third studies (23,115,117) used immunofluorescence-based staining, with the remaining 34 studies reporting data obtained from chromogenic stains. Twenty-three studies (23,29,114–118,122–125,129–132,134–136,138,139,141,142), including four that documented automated image capture and staining analysis (23,115,117,125), indicated that staining assessment was blinded to outcome status, but blinding status was unknown for the remaining 14 studies. Effective sample size of included melanoma patients ranged from 37 to 1270, with six studies including 75 or fewer individuals (26,28,114,133,136,141), 16 including 76–150 individuals (24,25,27,113,118,122,125–132,138,139), and 12 including between 151 and 300 individuals (23,115–117,119–121,123,124,137,140,142). Three studies (29,134,135), among them the prospective cohort that enrolled 1270 individuals (29), included more than 300 individuals.
Table 1.
Main sample, study, and methodological characteristics of the high-quality cohort studies eligible for this systematic review*
| First author, year (reference) | Sample characteristics |
|||||||
| Preservation | Sectioning | Sample size | Blinding | Adjustment | Proteins assayed | Primary antibody | Endpoints | |
| Alonso, 2007 (113) | FFPE | TMA | 127 | N/A | Breslow (mm) | Glypican-3 | Santa Cruz polyclonal (1:25) | DFS |
| N-cadherin | Zymed monoclonal 3B9 (1:10) | |||||||
| Osteonectin | Novocastra monoclonal 15G12 (1:25) | |||||||
| Osteopontin | Abcam polyclonal (1:1500) | |||||||
| Protein kinase C-α | Santa Cruz monoclonal H7 (1:25) | |||||||
| Alonso, 2004 (114) | FFPE | TMA | 60 | Blinded | Breslow (mm) | Bcl-2 | Dako monoclonal 124 (1:25) | ACM |
| Bcl-6 | Dako monoclonal PG-B6p (1:10) | |||||||
| Bcl-xL | Zymed monoclonal 2H12 (1:10) | |||||||
| BMI-1 | Santa Cruz polyclonal (1:100) | |||||||
| Caveolin | Transduction Labs monoclonal 2297 (1:25) | |||||||
| c-Kit | Dako polyclonal (1:50) | |||||||
| Cyclin A | Novocastra monoclonal 6E6 (1:100) | |||||||
| Cyclin B1 | Novocastra monoclonal 7A9 (1:25) | |||||||
| Cyclin D1 | Dako monoclonal DCS-6 (1:100) | |||||||
| Cyclin D3 | Novocastra monoclonal DCS-22 (1:10) | |||||||
| Cyclin E | Novocastra monoclonal 13A3 (1:10) | |||||||
| cdk-1 | Transduction Labs monoclonal 1 (1:1500) | |||||||
| cdk-2 | NeoMarkers monoclonal 8D4 (1:500) | |||||||
| cdk-6 | Chemicon monoclonal K6.83 (1:10) | |||||||
| Double minute-2 | Oncogene monoclonal IF2 (1:10) | |||||||
| gp100/pmel17 | Biogenex monoclonal HMB45 (1:10) | |||||||
| Ki-67 | Dako monoclonal MIB-1 (1:100) | |||||||
| Mel-18 | Santa Cruz polyclonal (1:100) | |||||||
| MelanA/MART-1 | Dako monoclonal A103 (1:10) | |||||||
| MHC class II | CNIO monoclonal J576 (1:150) | |||||||
| MLH-1 | Pharmingen monoclonal G168-15 (1:100) | |||||||
| Mum-1/IRF4 | Santa Cruz polyclonal (1:200) | |||||||
| p16/INK4a | Santa Cruz monoclonal F12 (1:50) | |||||||
| p21/WAF1/CIP1 | Oncogene monoclonal EA10 (1:50) | |||||||
| p27/KIP1 | Transduction Labs monoclonal 57 (1:1000) | |||||||
| p53 | Novocastra monoclonal DO-7 (1:50) | |||||||
| PH1 | MA Vidal (Madrid, Spain) monoclonal (1:2) | |||||||
| Protein kinase C-β | Serotec monoclonal 28 (1:500) | |||||||
| pRb | Transduction Labs monoclonal G3-245 | |||||||
| RING1B | MA Vidal (Madrid, Spain) monoclonal (1:1) | |||||||
| RYBP | MA Vidal (Madrid, Spain) monoclonal (1:25) | |||||||
| Skp2 | Zymed monoclonal 1G12E9 (1:10) | |||||||
| STAT-1 | Santa Cruz monoclonal C-136 (1:50) | |||||||
| Survivin | R&D Systems polyclonal (1:1500) | |||||||
| Topoisomerase II | Dako monoclonal Ki-S1 (1:400) | |||||||
| Berger, 2005 (115) | FFPE | TMA | 214 | Blinded | Breslow (mm) | AP-2α | Santa Cruz polyclonal C18 (1:1600) | MSM |
| Clark level | ||||||||
| Ulceration | ||||||||
| Microsatellitosis | ||||||||
| TILs | ||||||||
| Berger, 2004 (23) | FFPE | TMA | 203 | Blinded | Breslow (mm) | HDM-2 | Novocastra monoclonal 1B10 (1:100) | MSM |
| Age at diagnosis | ||||||||
| Ulceration | ||||||||
| TILs | ||||||||
| Anatomic site | ||||||||
| Microsatellitosis | ||||||||
| Berger, 2003 (116) | FFPE | TMA | 269 | Blinded | Breslow (mm) | ATF-2 | Santa Cruz polyclonal (1:50) | MSM |
| Clark level | ||||||||
| Ulceration | ||||||||
| Microsatellitosis | ||||||||
| TILs | ||||||||
| Divito, 2004 (117) | FFPE | TMA | 159 | Blinded | Breslow (mm) | Bcl-2 | Dako monoclonal 124 (1:30) | MSM |
| Clark level | ||||||||
| Ulceration | ||||||||
| Gender | ||||||||
| Age at diagnosis | ||||||||
| Ekmekcioglu, 2006 (118) | FFPE | Whole sections | 132 stage III | Blinded | Age at diagnosis | iNOS | Transduction Labs monoclonal (dilution N/A) | MSM |
| Gender | ||||||||
| Stage at Dx | ||||||||
| Ferrier, 2000 (119) | FFPE | Whole sections | 214 T3–T4, N0 | N/A | Breslow (mm) | tPA | American Diagnostica polyclonal (dilution N/A) | ACM |
| Ulceration | DFS | |||||||
| Gender | ||||||||
| Treatment | ||||||||
| Anatomic site | ||||||||
| Florenes, 2001 (120) | FFPE | Whole sections | 172 | N/A | Breslow (mm) | Cyclin A | Novocastra monoclonal 6E6 (1:50) | ACM |
| Age at diagnosis | Ki-67 | Dako polyclonal A047 (1:200) | DFS | |||||
| Gender | ||||||||
| Anatomic site | ||||||||
| Histological type | ||||||||
| Florenes, 2000 (121) | FFPE | Whole sections | 172 | N/A | Breslow (mm) | Cyclin D1 | Oncogene Research Products DCS-6 (1:200) | ACM |
| Histological type | Cyclin D3 | Dako monoclonal DCS-22 (1:25) | DFS | |||||
| Hofbauer, 2004 (24) | FFPE | Whole sections | 91 | N/A | Breslow (mm) | MHC class I | Monoclonal vf135-1D6 (S. Ferrone; dilution N/A) | MSM |
| MAGE-3 | Monoclonal 57B (G. Spagnoli; dilution N/A) | |||||||
| MelanA/MART-1 | Novocastra monoclonal A103 (dilution N/A) | |||||||
| gp100/Pmel17 | Enzo Diagnostics monoclonal HMB45 (dilution N/A) | |||||||
| Tyrosinase | Novocastra monoclonal T311 (dilution N/A) | |||||||
| Ilmonen, 2004 (122) | FFPE | Whole sections | 98 | Blinded | Breslow (mm) | Tenascin-C | Biohit monoclonal 143DB7 | MSM |
| Clark level | DFS | |||||||
| Ulceration | ||||||||
| Karjalainen, 2000 (123) | FFPE | Whole sections | 282 stage I | Blinded | Breslow (mm) | CD44 | R&D Systems monoclonal 2C5 (1:2000) | DFS |
| Ulceration | ||||||||
| Karjalainen, 1998 (124) | FFPE | Whole sections | 273 | Blinded | Breslow (mm) | AP-2α | Santa Cruz polyclonal C18 (dilution N/A) | DFS |
| Clark level | ||||||||
| Ulceration | ||||||||
| Gender | ||||||||
| Korabiowska, 2002 (125) | FFPE | Whole sections | 76 | Blinded | Breslow (mm) | Ku70 | Santa Cruz polyclonal M19 (1:50) | ACM |
| Ku80 | Santa Cruz polyclonal M20 (1:50) | |||||||
| Li, 2004 (25) | FFPE | Whole sections | 104 | N/A | Breslow (mm) | Skp2 | Zymed monoclonal (1:50) | ACM |
| Clark level | ||||||||
| Ulceration | ||||||||
| Mitotic index | ||||||||
| TILs | ||||||||
| Regression | ||||||||
| Vascular invasion | ||||||||
| McDermott, 2000 (126) | FFPE | Whole sections | 145 | N/A | Breslow (mm) | nm23 | Novocastra antibody (1:50) | ACM |
| Stage at Dx | ||||||||
| Regression | ||||||||
| Niezabitowski, 1999 (127) | FFPE | Whole sections | 93 | N/A | Breslow (mm) | gp100/pmel17 | Biogenex monoclonal HMB45 (1:80) | ACM |
| Stage at Dx | Ki-67 | Immunotech monoclonal MIB-1 (Neat) | DFS | |||||
| PCNA | Dako monoclonal PC10 (1:200) | |||||||
| Pacifico, 2006 (128) | FFPE | TMA | 84 | Blinded | Breslow (mm) | CD44v3 | Novocastra monoclonal VFF-327v3 (1:25) | ACM |
| Clark level | ||||||||
| Age at diagnosis | ||||||||
| Gender | ||||||||
| Ulceration | ||||||||
| Anatomic site | ||||||||
| Pacifico, 2005 (129) | FFPE | TMA | 76 | Blinded | Breslow (mm) | nm23 | Novocastra monoclonal 37.6 (1:50) | ACM |
| Clark level | ||||||||
| Age at diagnosis | ||||||||
| Gender | ||||||||
| Ulceration | ||||||||
| Anatomic site | ||||||||
| Pacifico, 2005 (130) | FFPE | TMA | 76 | Blinded | Breslow (mm) | P-cadherin | Novocastra monoclonal 56C1 (1:50) | ACM |
| Clark level | ||||||||
| Age at diagnosis | ||||||||
| Gender | ||||||||
| Ulceration | ||||||||
| Anatomic site | ||||||||
| Pacifico, 2005 (131) | FFPE | TMA | 76 | Blinded | Breslow (mm) | MCAM | Novocastra monoclonal N1238 (1:50) | ACM |
| Clark level | ||||||||
| Age at diagnosis | ||||||||
| Gender | ||||||||
| Ulceration | ||||||||
| Anatomic site | ||||||||
| Pearl, 2008 (132) | FFPE | TMA | 76 | Blinded | Breslow (mm) | MCAM | Novocastra monoclonal N1238 (1:50) | DFS |
| Age at diagnosis | ||||||||
| Gender | ||||||||
| Ulceration | ||||||||
| Piras, 2007 (133) | FFPE | Whole sections | 50 | N/A | Breslow (mm) | Survivin | Novus rabbit polyclonal (1:200) | DFS |
| Rangel, 2008 (134) | FFPE | TMA | 345 | Blinded | Breslow (mm) | Osteopontin | Abcam polyclonal ab8448 (1:200) | MSM |
| Clark level | ||||||||
| Ulceration | ||||||||
| Age at diagnosis | ||||||||
| Gender | ||||||||
| Anatomic site | ||||||||
| Rangel, 2006 (135) | FFPE | TMA | 343 | Blinded | Breslow (mm) | NCOA3/AIB-1 | Abcam monoclonal ab14139 (1:10) | MSM |
| Clark level | DFS | |||||||
| Ulceration | ||||||||
| Age at diagnosis | ||||||||
| Gender | ||||||||
| Anatomic site | ||||||||
| Scala, 2005 (136) | FFPE | Whole sections | 71 | Blinded | Breslow (mm) | CXCR4 | R&D Systems monoclonal 44716 (dilution N/A) | ACM |
| Age at diagnosis | DFS | |||||||
| Gender | ||||||||
| Ulceration | ||||||||
| SLN status | ||||||||
| Soltani, 2005 (26) | FFPE | Whole sections | 37 | N/A | Breslow (mm) | MAP-2 | Zymed monoclonal (dilution N/A) | DFS |
| Clark level | ||||||||
| Age at diagnosis | ||||||||
| Gender | ||||||||
| Straume, 2005 (27) | FFPE | TMA | 119 | N/A | Breslow (mm) | Id1 | Santa Cruz polyclonal SC-488 (1:100) | MSM |
| Clark level | DFS | |||||||
| Anatomic site | ||||||||
| Ulceration | ||||||||
| Straume, 2000 (137) | FFPE | Whole sections | 187 | N/A | Clark level | Ki-67 | Dako polyclonal A047 (1:50) | MSM |
| Anatomic site | p16 | Santa Cruz polyclonal SC-468 (1:500) | ||||||
| Vascular invasion | p53 | Dako monoclonal DO-7 (1:100) | ||||||
| Thies, 2002 (138) | FFPE | Whole sections | 100 | Blinded | Stage at Dx | L1-CAM | U. Schumacher lab polyclonal (1:100) | DFS |
| Ulceration | ||||||||
| Mitotic index | ||||||||
| Thies, 2002 (139) | FFPE | Whole sections | 100 | Blinded | Breslow (mm) | CEACAM | U. Schumacher lab monoclonal 4D1/C2 (8 μg/mL) | DFS |
| Ulceration | ||||||||
| Mitotic index | ||||||||
| Tran, 1998 (28) | FFPE | Whole sections | 66 | N/A | Breslow (mm) | Cyclin A | Novocastra monoclonal 6E6 (1:20) | ACM |
| Clark level | ||||||||
| Mitotic index | ||||||||
| Vaisanen, 2008 (140) | FFPE | Whole sections | 157 | Blinded | Breslow (mm) | MMP-2 | Diabor monoclonal CA-4001 (5 μg/mL) | MSM |
| Age at diagnosis | MMP-9 | Diabor monoclonal GE-231 (10 μg/mL) | ||||||
| Gender | ||||||||
| Vaisanen, 1998 (141) | FFPE | Whole sections | 50 | N/A | Breslow (mm) | MMP-2 | T. Turpeenniemi-Hujanen polyclonal (dilution N/A) | ACM |
| Clark level | ||||||||
| Age at diagnosis | ||||||||
| Gender | ||||||||
| Weinlich, 2007 (142) | FFPE | Whole sections | 158 | Blinded | Breslow (mm) | Metallothionein | Dako monoclonal E9 (dilution N/A) | MSM |
| Age at diagnosis | DFS | |||||||
| Gender | ||||||||
| Ulceration | ||||||||
| SLN status | ||||||||
| Weinlich, 2006 (29) | FFPE | Whole sections | 1270 | Blinded | Breslow (mm) | Metallothionein | Dako monoclonal E9 (dilution N/A) | MSM |
| Clark level | DFS | |||||||
| Age at diagnosis | ||||||||
| Gender | ||||||||
| Anatomic site | ||||||||
FFPE = formalin fixed, paraffin embedded; TMA = tissue microarray; N/A = not available; DFS = disease-free survival; ACM = all-cause mortality; MSM = melanoma-specific mortality; TILs = tumor-infiltrating lymphocytes; Dx = diagnosis; MHC = major histocompatability complex.
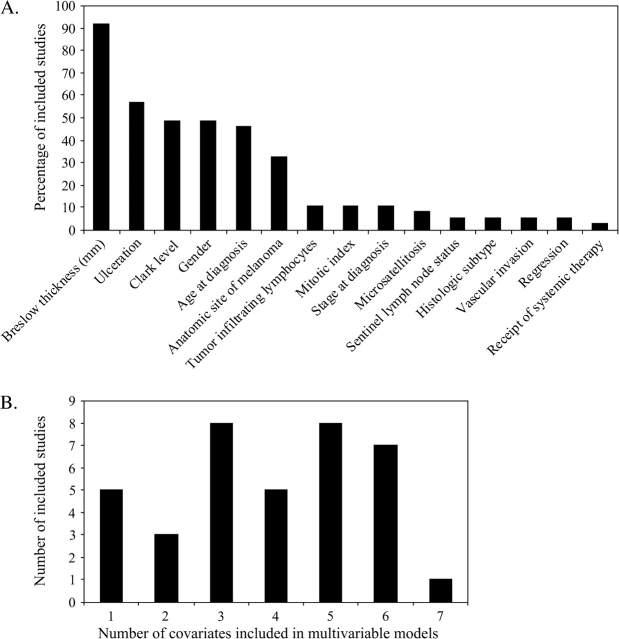
Fifteen unique clinicopathologic factors were incorporated in one or more of the eligible multivariable analyses (Figure 2, A). Breslow thickness as measured in millimeters (143), the strongest and most reproducible clinical prognostic factor (4,144), was the most commonly occurring clinical covariate, with inclusion in 34 analyses. All five studies that included a single clinical covariate adjusted for Breslow thickness (24,113,114,125,133). Clark level of dermal invasion (145), which overlaps with (146) and, in smaller populations (n ∼ 1000), can be collinear with Breslow thickness (147), was considered in 18 studies, of which 17 simultaneously adjusted for Breslow thickness, and one study (137) used Clark level of invasion as the exclusive measure of tumor thickness. Two studies (118,138) did not include any measure for tumor thickness. Ulceration, a validated prognostic factor that prompts tumor upstaging when present (4,5,148), was adjusted for in 21 of 37 studies. Other common adjustment parameters included gender (18 of 37 studies), age at diagnosis (17 of 37 studies), and anatomic location of the melanoma (12 of 37 studies). Twenty-one studies included three to five clinical parameters in their multivariable proportional hazards models; eight studies included less than three parameters, and another eight included more than five covariates (Figure 2, B).
Figure 2.
Characteristics of studies included in this meta-analysis. A) Frequencies with which adjustments were made for various clinicopathologic parameters. B) Distributions of the total number of clinicopathologic covariates that were adjusted for across the 37 eligible cohort studies included.
Collectively, these 37 studies present data on 62 unique proteins. The majority of eligible manuscripts (n = 28) restricted their analysis to a single candidate protein marker and eight additional studies considered between two to five proteins. Only Alonso et al. (114) reported multivariable hazard ratios on a large series of proteins, with data available for 35 evaluated markers. Twenty-two of the 62 candidate biomarkers were evaluated for two outcomes, and two candidate biomarkers, Ki-67 and gp100, were evaluated across all three outcomes. Stratified by outcome, data were available on 43 proteins for ACM, 20 proteins for MSM, and 24 proteins for DFS. For 79 of the 87 unique marker–outcome combinations, a multivariable hazard ratio and associated 95% confidence interval were available only from a single study, with that value extracted as the corresponding summary estimate. For the remaining eight marker–outcome combinations, data were available from two or more studies and were combined using both fixed effects general inverse variance and Der Simonian–Laird random effects modeling to obtain a single summary hazard ratio and 95% confidence interval (Figure 3). For four of these associations (cyclin D1–ACM, Skp2–ACM, nm23–ACM, and metallothionein-1–DFS), which each combined two individual studies to create summary estimates, the fixed effects summary point estimate and 95% confidence interval were identical to the random effects summary statistic. For the remaining four studies, the random effects analysis yielded a more conservative result than the fixed effects estimate.
Figure 3.
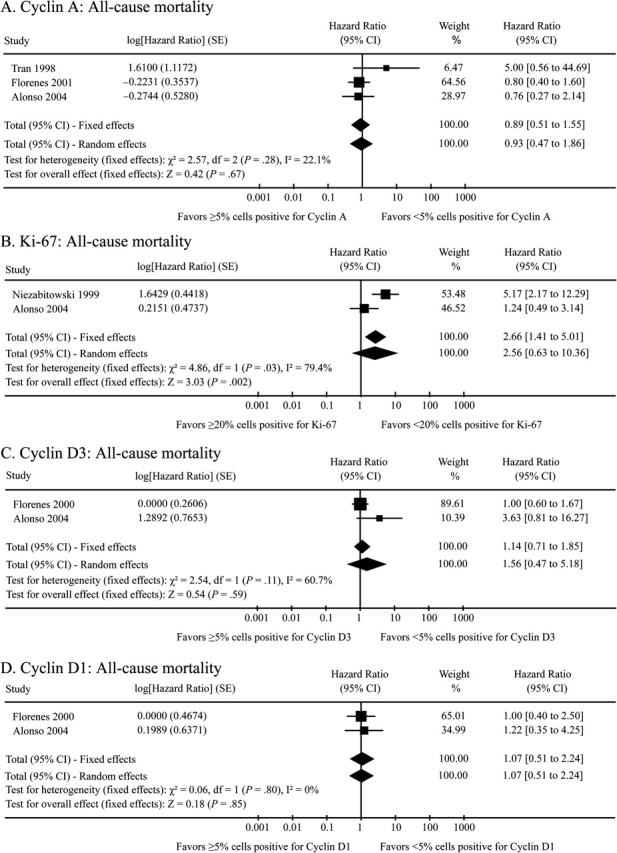
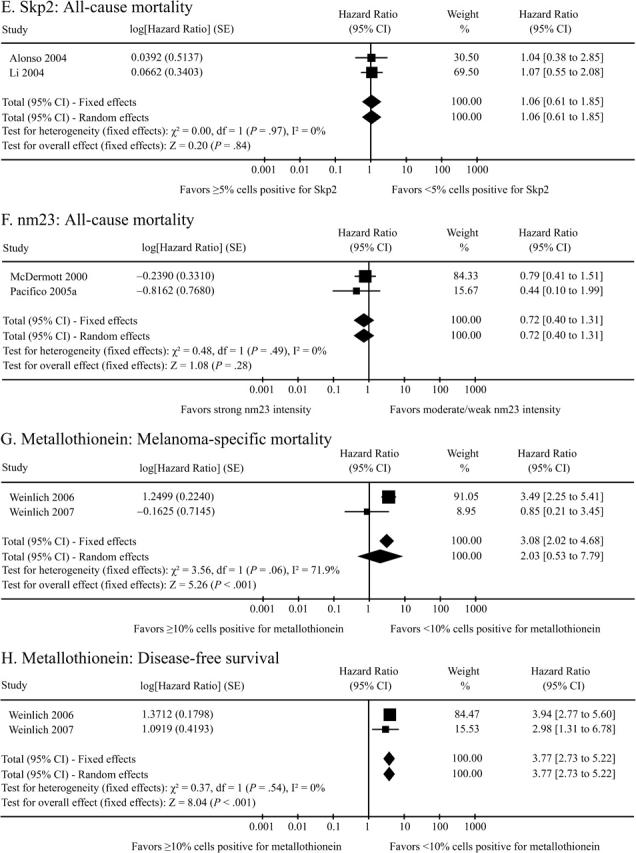
Forest plots of the data from the contributing studies for each of the eight biomarker–outcome comparisons for which eligible data were presented in two or more studies. For each study, hazard ratio, 95% confidence interval (CI), and relative weight are shown. SEs are shown for log of the hazard ratio. Combined fixed effects hazard ratios and tests for heterogeneity (I 2) were based on the generic inverse variance method. Combined random effects hazard ratios were calculated according to the Der Simonian–Laird method.
All-Cause Mortality
The 43 proteins evaluated for ACM were sorted among seven of the eight modified Hanahan–Weinberg functional capabilities (Table 2); no eligible assays were available for “sustained angiogenesis.” Thirteen (30.2%) of the 43 candidates had a statistically significant association with ACM at P < .05. Of the eight cell cycle proteins evaluated, only cyclin E (P = .03) had a statistically significant association with ACM, but two of four cell cycle regulators (p16/INK4A [P = .02] and p27/KIP1 [P = .02]) showed statistically significant associations. Four of eight DNA-damage checkpoint and repair proteins (Ki-67 [P = .002], PCNA [P = .03], Ku70 [P < .001], and Ku80 [P < .001]) also showed statistically significant multivariable associations. Among the regulators of tissue invasion and metastasis, chemokine receptor CXCR4 (P = .02), matrix metalloproteinase (MMP)-2 (P = .006), MCAM/MUC18 (P < .001), and tissue plasminogen activator (tPA; P = .04) were statistically significant. None of the four polycomb transcriptional repressor complex proteins assayed demonstrated any statistically significant associations. Overall, the proportions of biomarker candidates with statistically significant associations to ACM observed among evading apoptosis, insensitivity to antigrowth signals, limitless replicative potential, and tissue invasion and metastasis did not differ more than would be expected by chance (P > .05), with only the functional category of tissue invasion and metastasis (four proteins that had statistically significant associations among seven assays; 57.14%) approaching statistical significance (z-score = 1.55; P = .12).
Table 2.
Summary multivariable hazard ratios and 95% confidence intervals for eligible proteins, organized according to Hanahan–Weinberg functional capabilities: all-cause mortality*
| Protein | Total n | Reference group | HR (95% CI) | P value† | References |
| Altered immunocompetence | |||||
| MHC class II (HLA-DR, -DP, -DQ) | 60 | No nuclear stain | 1.47 (0.45 to 4.79) | .53 | (114) |
| STAT-1 | 60 | No nuclear stain | 0.64 (0.24 to 1.70) | .37 | (114) |
| Evading apoptosis | |||||
| Bcl-2 | 57 | <10% cells + | 3.42 (0.45 to 25.96) | .24 | (114) |
| Bcl-xL† | 60 | <10% cells + | 8.07 (1.77 to 36.89) | .007 | (114) |
| Survivin | 59 | No nuclear stain | 1.62 (0.45 to 5.79) | .46 | (114) |
| Insensitivity to antigrowth signals | |||||
| Bcl-6† | 59 | No stain | 3.98 (1.37 to 11.60) | .01 | (114) |
| BMI-1 | 57 | <10% cells + | 0.92 (0.33 to 2.59) | .88 | (114) |
| Mel-18 | 59 | <50% cells + | 1.31 (0.16 to 10.97) | .81 | (114) |
| p16/INK4a† | 60 | <50% cells + | 0.29 (0.10 to 0.83) | .02 | (114) |
| p21/WAF1 | 60 | <10% cells + | 1.98 (0.77 to 5.06) | .15 | (114) |
| p27/KIP1† | 60 | <10% cells + | 3.08 (1.20 to 7.91) | .02 | (114) |
| PH1 | 60 | No nuclear stain | 1.50 (0.50 to 4.54) | .47 | (114) |
| pRb | 60 | <10% cells + | 3.40 (0.42 to 27.79) | .25 | (114) |
| RING1B | 59 | No nuclear stain | 2.89 (0.66 to 12.63) | .16 | (114) |
| Limitless replicative potential | |||||
| Cyclin A | 298 | <5% cells + | 0.89 (0.51 to 1.55) | .67 | (28,114,120) |
| Cyclin B1 | 57 | <5% cells + | 0.73 (0.21 to 2.55) | .62 | (114) |
| Cyclin D1 | 231 | <5% cells + | 1.07 (0.51 to 2.24) | .85 | (114,121) |
| Cyclin D3 | 232 | <5% cells + | 1.14 (0.71 to 1.85) | .59 | (114,121) |
| Cyclin E† | 60 | <10% cells + | 2.89 (1.09 to 7.71) | .03 | (114) |
| Cyclin-dependent kinase-1 | 60 | <5% cells + | 0.83 (0.32 to 2.11) | .69 | (114) |
| Cyclin-dependent kinase-2 | 60 | <5% cells + | 0.38 (0.12 to 1.18) | .10 | (114) |
| Cyclin-dependent kinase-6 | 60 | <5% cells + | 1.86 (0.72 to 4.78) | .20 | (114) |
| Double minute-2 | 60 | No nuclear stain | 2.49 (0.71 to 8.70) | .15 | (114) |
| Ki-67† | 153 | <20% cells + | 2.66 (1.41 to 5.01) | .002 | (114,127) |
| Ku70† | 76 | No stain | 0.87 (0.82 to 0.92) | <.001 | (125) |
| Ku80† | 76 | No stain | 0.85 (0.79 to 0.92) | <.001 | (125) |
| nm23 | 221 | Weak/moderate stain | 0.72 (0.40 to 1.31) | .28 | (126,129) |
| p53 | 60 | <10% cells + | 2.19 (0.50 to 9.51) | .30 | (114) |
| PCNA† | 93 | ≤35% cells + | 2.27 (1.56 to 3.31) | .03 | (127) |
| Skp2 | 163 | <5% cells + | 1.06 (0.61 to 1.85) | .84 | (25,114) |
| Topoisomerase II | 60 | <10% cells + | 0.78 (0.26 to 2.40) | .67 | (114) |
| Melanocyte differentiation | |||||
| gp100 | 60 | <50% cells + | 2.29 (0.30 to 17.67) | .43 | (114) |
| MelanA/MART-1 | 60 | <50% cells + | 1.57 (0.35 to 7.01) | .56 | (114) |
| Self-sufficiency in growth signals | |||||
| c-Kit | 60 | <50% cells + | 0.65 (0.22 to 1.98) | .45 | (114) |
| Mum-1/IRF4 | 60 | <30% cells + | 1.64 (0.65 to 4.11) | .29 | (114) |
| Protein kinase C-β | 60 | <10% cells + | 0.70 (0.28 to 1.73) | .44 | (114) |
| Tissue invasion and metastasis | |||||
| Caveolin | 60 | <50% cells + | 0.73 (0.24 to 2.23) | .58 | (114) |
| CD44 (variant 3) | 84 | No stain | 0.53 (0.19 to 1.47) | .22 | (128) |
| CXCR4† | 71 | No stain | 2.07 (1.15 to 3.71) | .02 | (136) |
| Matrix metalloproteinase-2† | 50 | <34% cells + | 4.5 (1.5 to 13.0) | .006 | (141) |
| MCAM/MUC18† | 76 | No stain | 16.34 (3.80 to 70.28) | <.001 | (131) |
| P-cadherin | 78 | No stain | 0.44 (0.17 to 1.13) | .09 | (130) |
| Tissue plasminogen activator† | 214 | <5% cells | 1.90 (1.00 to 3.76), 6%–50%; 0.4 (0.01 to 1.61), >50% | .04 | (119) |
HR = hazard ratio; CI = confidence interval; MHC = major histocompatability complex.
For associations representing data from a single study, P values were determined by multivariable Cox proportional hazards modeling. For associations representing data from multiple studies, combined summary HRs are those calculated for the fixed effects general inverse variance method. Proteins with statistically significant values (P < .05) are marked.
Melanoma-Specific Mortality
Twelve of the 20 candidate biomarkers with eligible data for MSM demonstrated a statistically significant association with this outcome (Table 3). All eight modified Hanahan–Weinberg functional capabilities were represented, and all but altered immunocompetence possessed at least one candidate biomarker statistically significantly associated with MSM. Two functional categories, limitless replicative potential and self-sufficiency in growth signals, included statistically significant associations for more than 50% of assayed candidates with 3/4 and 3/3 proteins, respectively, showing statistically significant associations with MSM. The melanocyte differentiation category was unique in returning 33% or fewer statistically significant candidates, with only gp100 (P = .045) yielding a marginally statistically significant result. The small number of protein candidates within each functional category precluded analysis of proportions.
Table 3.
Summary multivariable hazard ratios and 95% confidence intervals for eligible proteins, organized according to Hanahan–Weinberg functional capabilities: melanoma-specific mortality*
| Protein | Total n | Reference group | HR (95% CI) | P value† | References |
| Altered immunocompetence | |||||
| MHC class I (HLA-A, -B, -C) | 91 | ≤50% cells + | 1.13 (0.82 to 1.56) | .45 | (24) |
| MAGE-3 | 91 | ≤50% cells + | 0.86 (0.63 to 1.18) | .36 | (24) |
| Evading apoptosis | |||||
| Bcl-2† | 159 | ≤50 percentile AQUA score | 0.64 (0.48 to 0.86) | .03 | (117) |
| Insensitivity to antigrowth signals | |||||
| Id1 | 119 | <10% cells + | 1.31 (0.68 to 2.52) | .42 | (27) |
| p16/INK4a† | 187 | Weak stain | 0.4 (0.24 to 0.67) | .007 | (137) |
| Limitless replicative potential | |||||
| Double minute-2 | 203 | ≤25 percentile AQUA score | 0.99 (0.48 to 2.06) | .99 | (23) |
| Ki-67† | 187 | <16% cells + | 3.7 (1.6 to 8.9) | .003 | (137) |
| Metallothionein† | 1428 | <10% cells + | 3.08 (2.02 to 4.68) | <.001 | (29,142) |
| p53† | 187 | No stain | 8.9 (2.7 to 29.0) | <.001 | (137) |
| Melanocyte differentiation | |||||
| gp100† | 91 | ≤50% cells + | 0.63 (0.40 to 0.99) | .045 | (24) |
| MelanA/MART-1 | 91 | ≤50% cells + | 0.95 (0.82 to 1.09) | .79 | (24) |
| Tyrosinase | 91 | ≤50% cells + | 1.47 (0.83 to 2.58) | .19 | (24) |
| Self-sufficiency in growth signals | |||||
| AP-2α† | 214 | ≤25%-ile cytoplasmic/ nuclear AQUA score ratio | 2.14 (1.22 to 3.76) | .008 | (115) |
| ATF-2† | 269 | High nuclear combined with low cytoplasmic AQUA scores | 0.55 (0.42 to 0.75) | <.001 | (116) |
| NCOA3/AIB-1† | 343 | Negative/weak stain | 1.91 (1.44 to 2.53) | .021 | (135) |
| Sustained angiogenesis | |||||
| iNOS† | 132 | <5% cells + | 4.63 (2.60 to 8.25) | <.001 | (118) |
| Tissue invasion and metastasis | |||||
| Matrix metalloproteinase-2† | 157 | <20% cells + | 2.6 (1.32 to 5.07) | .006 | (140) |
| Matrix metalloproteinase-9 | 157 | <20% cells + | 0.8 (0.34 to 1.56) | .46 | (140) |
| Osteopontin† | 345 | Weak stain | 1.55 (1.24 to 1.94) | .049 | (134) |
| Tenascin-C | 98 | No stain | 1.12 (1.00 to 1.25) | .12 | (122) |
HR = hazard ratio; CI = confidence interval; MHC = major histocompatability complex; AQUA = automated quantitative analysis
P values were determined by multivariable Cox proportional hazards modeling. Proteins with statistically significant values (P < .05) are marked.
Eight MSM candidates were also evaluated for ACM. Among these, five proteins showed concordant associations between the two outcomes. Elevated p16/INK4A was protective for both ACM and MSM, whereas elevated MMP-2 or Ki-67 increased risk of both ACM and MSM. Changes in MelanA/MART-1 or double minute-2 (HDM-2) were not associated with either ACM or MSM. Discordant results were observed for three candidates: gp100, p53, and Bcl-2. In 2004, Alonso et al. (114) reported null associations for these with ACM, but other groups presented statistically significant associations with MSM in separate reports. To determine whether publication bias could have contributed to this discrepancy, results from the 14 methodologically robust studies that omitted the hazard ratio were reviewed. Two studies were identified that evaluated the association of p53 with MSM (100,106) and one with ACM (127). All three studies indicated that following adjustment for Breslow thickness, p53 was not statistically significantly associated with outcome. Whereas Niezabitowski et al. (n = 93) and Karjalainen et al. (n = 283) only indicated that P > .05, and Talve et al. (n = 80) specified P = .96. Without the actual point estimates and precise P values, however, we cannot determine whether the missing data would be sufficient to cancel the strong association (HR = 8.9; 95% CI = 2.7 to 29.0) observed by Straume et al. (137).
Disease-Free Survival
Twenty-four proteins representing six of eight functional capabilities were assayed for DFS, and 15 (62.5%) statistically significant associations were found (Table 4). Three categories, limitless replicative potential, self-sufficiency in growth signals, and tissue invasion and metastasis, each had four or more proteins assayed for DFS, and the number of statistically significant candidates was equally distributed among these categories (50%–75%; P = .70) and not different from the overall proportion of statistically significant markers (P > .40). Seventeen DFS candidates were also evaluated for either or both of the survival outcomes, with nine yielding concordant results for disease-free and overall survival. Among these, Id1, cyclin D1, and cyclin D3 were not associated with either outcome, whereas differential levels of PNCA, NCOA3/AIB-1, AP-2α, CXCR4, MCAM/MUC18, and metallothionein were predictive of both overall and DFS with similar directionality and magnitude of each association. The remaining eight proteins yielded discordant results, and statistically significant results were only observed for a subset of the evaluated outcomes. Survivin (P = .017), cyclin A (P < .001), and tenascin-C (sp) (P = .04) were statistically significant for DFS but not for mortality outcomes. Conversely, osteopontin (P = .10) and tPA (P = .15) were not associated with DFS but achieved statistical significance for mortality. Ki-67, which was independently statistically significant for both mortality outcomes (ACM, P = .002; MSM, P < .001), did not achieve statistical significance for DFS (P = .26) among eligible studies, and gp100 was statistically significantly associated with both MSM (P = .045) and DFS (P = .01) but not with ACM (P = .43) despite similar selection of cut points. Qualitative discordance, in which a protein would be protective for DFS but promoting of either mortality endpoint, was not observed.
Table 4.
Summary multivariable hazard ratios and 95% confidence intervals for eligible proteins, organized according to Hanahan–Weinberg functional capabilities: disease-free survival*
| Protein | Total n | Reference group | HR (95% CI) | P value | References |
| Evading apoptosis | |||||
| Survivin† | 50 | No nuclear stain | 7.32 (1.43 to 37.38) | .017 | (133) |
| Insensitivity to antigrowth signals | |||||
| Id1 | 119 | <10% cells + | 1.39 (0.74 to 2.60) | .31 | (27) |
| Limitless replicative potential | |||||
| Cyclin A† | 172 | <5% cells + | 3.7 (3.4 to 4.1) | .001 | (120) |
| Cyclin D1 | 172 | <5% cells + | 0.9 (0.3 to 2.2) | .74 | (121) |
| Cyclin D3 | 172 | <5% cells + | 1.5 (0.9 to 2.6) | .13 | (121) |
| Ki-67 | 172 | <5% cells + | 0.7 (0.4 to 1.0) | .26 | (120) |
| Microtubule-associated protein-2† | 37 | <70% cells + | 0.18 (0.06 to 0.56) | .003 | (26) |
| Metallothionein† | 1428 | <10% cells + | 3.77 (2.73 to 5.22) | <.001 | (29,142) |
| PCNA† | 93 | ≤15% cells + | 4.00 (2.05 to 7.81) | .039 | (127) |
| Melanocyte differentiation | |||||
| gp100† | 93 | ≤90% cells + | 1.86 (1.36 to 2.54) | .01 | (127) |
| Self-sufficiency in growth signals | |||||
| AP-2α† | 273 | Strong stain | 3.12 (1.42 to 6.82), moderate; 2.52 (1.23 to 5.20), weak | .01 | (124) |
| Glypican-3 | 127 | <10% cells + | 1.19 (0.62 to 2.26) | .60 | (113) |
| NCOA3/AIB-1† | 343 | Negative/weak stain | 1.69 (1.38 to 2.07) | .001 | (135) |
| Protein kinase C-α† | 127 | <50% cells + | 2.03 (1.19 to 3.44) | .009 | (113) |
| Tissue invasion and metastasis | |||||
| CD44 (all variants)† | 282 | ≤90% cells + | 0.57 (0.35 to 0.95) | .03 | (123) |
| CEACAM-1† | 100 | <20% cells + | 7.17 (3.22 to 15.95) | <.001 | (139) |
| CXCR4† | 71 | No stain | 1.65 (1.10 to 2.46) | .01 | (136) |
| L1-CAM† | 100 | <20% cells + | 4.38 (2.08 to 9.23) | <.001 | (138) |
| MCAM/MUC18† | 76 | No stain | 14.83 (5.20 to 42.24) | .01 | (132) |
| N-cadherin | 127 | <5% cells + | 1.49 (0.86 to 2.58) | .16 | (113) |
| Osteonectin/SPARC | 127 | <10% cells + | 1.48 (0.89 to 2.46) | .13 | (113) |
| Osteopontin | 127 | <10% cells + | 1.72 (0.91 to 3.23) | .10 | (113) |
| Tenascin-C† | 98 | No stain | 1.20 (1.11 to 1.43) | .04 | (122) |
| Tissue plasminogen activator | 214 | <5% cells + | 1.5 (0.82 to 2.63), 6%–50%; 0.6 (0.23 to 1.43), >50% | .15 | (119) |
HR = hazard ratio; CI = confidence interval.
For associations representing data from a single study, P values were determined by multivariable Cox proportional hazards modeling. For associations representing data from multiple studies, combined summary HRs are those calculated for the fixed effects general inverse variance method. Proteins with statistically significant values (P < .05) are marked.
Discussion
In response to the need for independently prognostic molecular markers for CMM that are readily assayable on routinely acquired clinical specimens, we conducted a systematic review and meta-analysis of the published melanoma IHC literature to identify the subset of proteins for which the data support validation as prognostic biomarkers of melanoma outcomes. Using stringent inclusion and exclusion criteria that examined patient selection, as well as laboratory and statistical methods (17,19), we identified 37 high-quality cohort studies that published multivariable survival point estimates and SEs for 62 unique proteins. Individual biomarker assay data were organized according to outcome (ACM, MSM, or DFS) and, within each outcome, according to functional groupings that reflected the acquired capabilities of cancer as defined by Hanahan and Weinberg (32).
In terms of functional capabilities, proteins that facilitate tissue invasion and metastasis were most likely associated with melanoma prognosis as numerous subclasses displayed statistically significant results with one or more outcome. Increased expression of three cellular adhesion molecules, melanocyte-specific MCAM/MUC18, neuron-specific L1-CAM, and glandular tissue–associated CEACAM-1, was statistically significantly associated with worse DFS. MCAM/MUC18 was also evaluated for mortality and yielded concordant results. Overexpression of CEACAM-1 and L1-CAM typically occurs at the leading edge of tumors (139,149), and both CEACAM-1 and MCAM interact with β3-integrin (150,151). Thus, both findings support the involvement of these molecules with abnormal tumor–stroma interactions. We also found statistically significant results for multiple members of the matricellular protein family, which consists of secreted molecules that interface between the extracellular matrix and the cell surface receptors (152). Increased levels of osteopontin expression were statistically significantly associated with worse MSM and trended toward statistical significance for DFS. Increased levels of tenascin-C were associated with worse DFS and trended toward worse MSM. Elevated osteonectin expression trended toward, but did not achieve, statistical significance for worsened DFS. Among the proteases, increased levels of tPA (P = .04) and MMP-2 (72 kDa type IV collagenase; P = .006), but not of MMP-9 (92 kDa type IV collagenase; P = .46), were statistically significant for mortality outcomes. Although only two MMPs were addressed in studies eligible for this analysis, the observed differences in their prognostic value suggest that only a subset of melanoma-expressed MMPs affect outcome. Evaluation of additional MMPs will be necessary to test this hypothesis. Available data as well as data from a recent, otherwise eligible study published after the cutoff date for inclusion do not support independent prognostic roles for cadherins or catenins (113,130,153).
Among proteins that contributed to limitless replicative potential, effectors of DNA replication and repair (eg, Ki-67, PCNA, metallothionein, Ku70, Ku80, and microtubule-associated protein-2) were most consistently associated with disease-free and overall survival. In contrast, cyclins and cyclin-dependent kinases were not associated with melanoma prognosis. Cyclin E was the only cyclin among five cyclins examined to achieve statistical significance, but because these data were from a single small cohort study (114), validation in an independent, larger cohort is needed. The statistically significant association between cyclin A and DFS was mitigated by three separate studies showing no association between cyclin A and mortality. Cyclin-dependent kinase inhibitors were statistically significantly associated with mortality. Elevated levels of p16/INK4A demonstrated protective effects for both ACM (P = .02) and MSM (P = .007), consistent with the established role for p16/INK4A in regulating aberrant cell proliferation in cells of melanocytic origin (154). The paradoxically increased mortality observed with elevated p27/KIP1 levels (P = .02) supports recent observations that p27/KIP1 dysregulation occurs through its cytoplasmic sequestration rather than through protein degradation; cytoplasmic accumulation of p27/KIP has been associated with increased metastatic potential (155).
Although strengths of our study include a broad, unbiased survey of the available literature and the application of standard systematic review and meta-analysis methods to objectively identify the subset of studies with robust data for summarization, there were several limitations inherent to our approach. We did not extend our search criteria to meeting abstracts or other sources of unpublished data that may contain increased proportions of null results. Although limiting our search to published manuscripts risks publication bias for studies with statistically significant associations, these alternate sources likely contain inadequate methodological descriptions to satisfy our inclusion criteria. We also elected to divide the standard oncological endpoint of overall survival into ACM and MSM. Whereas ACM is considered more robust because it avoids nondifferential outcome misclassification due to cause-of-death misadjudication (156), it also requires adjustment for age where MSM does not (157). We separated ACM and MSM outcomes in anticipation of different adjustment covariates and potential sources of outcome measurement error. In doing so, we were not able to combine biomarker data across the two mortality outcomes, which compromised our ability to calculate robust summary estimates of individual biomarkers through meta-analysis.
This study is also limited because, for 38 of the included proteins, summary data across all outcomes were derived from association data presented in a single study, which, in 29 cases, included fewer than 100 samples. False-positive as well as false-negative results, the latter due to insufficient statistical power, cannot be ruled out. Validation of these results in additional, independent studies is warranted. For the subset of proteins that were evaluated in two or more studies, the cross-study heterogeneity in the execution of IHC experiments as well as the categorization and statistical adjustments for the clinicopathologic criteria may also contribute to measurement error of biomarker to outcome associations. Although the authors of the majority of these manuscripts adjusted for Breslow thickness, their approaches to parameterization varied, ranging from continuous assessment to binary categorizations. Both positive and negative confounding of risk estimates could arise from inconsistent adjustment for other accepted clinicopathologic prognostic factors such as ulceration, gender, age, and stage at diagnosis.
Variability in assessment of protein expression and subsequent cut-point selection across studies must also be considered as a potential source of bias. First, although four studies reported automated image capture and digitized assessment of candidate biomarker expression, in the remaining studies, one or more of the investigators visually determined levels of protein expression, which could contribute to misclassification, especially among the 14 studies for which blinding status of these pathologists was not known. Next, for the majority of markers, selection of cut points to define categories of protein expression was arbitrary and could vary from study to study. For Ki-67, two studies (114,127) selected a cut point at 20% cells staining positively, one study selected 16% (137), and one study selected 5% (120). Similar variability was also observed for p16/INK4A, gp100, MMP-2, and osteopontin. Validation and adoption of consensus cut points across the melanoma community could facilitate replication of results. Finally, as automated image capture platforms that calculate expression as a continuous parameter gain popularity, the challenge of combining results reflecting dose–response relationships must be addressed. Of the eligible data in this review, six of 87 associations relied on quantiles of expression (23,117), reported a dose–response (119,124), or defined expression based on ratios of subcellular localizations (115,116). Reporting of such results requires extra rigor because meta-analysis of these data is hampered if the reporting is not done correctly or consistently. Although the most simple and straightforward method to report such data for combination in a meta-analysis is categorical parameterization and estimation of hazard ratio for all categories relative to a baseline category, this approach consumes more df than estimation of dose–response from categorized data. If dose–response is estimated from categorized data, the data must be accompanied by the exposure value assigned to each category so that the hazard ratio per unit increase can be extracted for meta-analysis (158).
The execution of this systematic review and meta-analysis has illuminated gaps in IHC-based melanoma prognostic biomarker research. Most notable is the limited and highly selective number of proteins with eligible data. Several factors may have contributed to the paucity of rigorously studied candidate proteins. First, unlike genome-wide massively parallel genomics or proteomics platforms, IHC analyses must begin with candidate nominations that are based on a priori biological rationales, followed by their prioritization for execution in serial assays. The strong influence of research trends leads to significant selection bias in candidate prioritization. The rather comprehensive evaluation of cell cycle proteins and their regulators originates from the well-characterized increased risk of familial melanoma in individuals with heritable mutations in the gene encoding the p16/INK4A cyclin-dependent kinase inhibitor (159). Examination of the proliferation markers Ki-67 and PCNA as well as the DNA-damage regulator p53 was supported by their long-standing roles in regulating the progression of many cancers (160–162). Conversely, proteins that have not been often linked to direct involvement in melanoma are less likely to have been rigorously examined as potential prognostic biomarkers. For example, selective expression of chemokines and their cognate receptors on tumor cells contributes to metastasis during both initial invasion and selective homing to distinct target organ sites (163,164). Although basic research has associated expression of chemokine receptors CXCR1, CXCR2, CXCR3, CXCR4, CCR5, CCR7, and CCR10 with metastatic behavior of melanomas (165–171), rigorous prognostic data are only available for CXCR4, for which increased expression is associated with poorer outcome (136). Although CXCR1, CXCR2, CXCR3, CCR9, CCR10, and CCXR1 have been evaluated, these analyses were either limited to correlations with progression (172–174) or did not meet the criteria defining a cohort (175).
Another factor driving candidate selection is reagent availability. Even if mRNA expression profiling were used to generate an unbiased list of candidate genes for subsequent independent validation by IHC, such an approach would be thwarted if no validated antibodies against selected candidates were to exist. Many transcripts that are highlighted in microarray experiments lack functional characterization or are only annotated according to their clone identifier from high-throughput transcript sequencing projects (eg, KIAA, German Cancer Research Center [DKFZ]), and the corresponding proteins, if they exist, are least likely to have commercially available antibodies. For example, a comparative analysis of DNA microarray data suggested that CITED, an X-linked gene that regulates the transcription of tyrosinase and dopachrome tautomerase, was one of few genes consistently associated with melanoma progression across multiple studies (176). Despite these results, IHC correlates are limited to a single descriptive cross-sectional study that considered a small sample of eight nevi and 14 primary melanomas using a proprietary rabbit polyclonal antibody obtained from a collaborating laboratory (177).
The attrition of IHC-based studies lacking one or more inclusion criteria most severely limited the number of analyzable proteins. Our three parallel keyword-based PubMed searches identified 455 manuscripts describing IHC staining patterns for 387 distinct proteins. Yet, only 37 studies that collectively published 87 assays on 62 unique proteins met all eligibility criteria for inclusion in this systematic review. For 173 proteins, best analysis was restricted to cross-sectional correlations with melanocytic lesion progression or clinicopathologic criteria, and many proteins showed statistically significant associations with these endpoints. Although statistical significance in such cases does not guarantee prognostic relevance, none of these candidates were pursued in prognostic experiments. For example, among growth signaling proteins, statistically significant associations were most frequent among either transcription factors (ATF-2, AP-2α) or transcriptional coactivators (NCOA3/AIB-1), suggesting altered transcriptional regulation as a pivotal step in regulation of melanoma-specific survival. Because only five additional high-quality assays across all three outcomes reported associations for growth factor receptors and intermediate signal transduction molecules, we cannot rule out the possibility that upstream signaling components share a similar role. Eleven studies reported data on c-Kit (31,109,114,178–185), with only one eligible for inclusion in this review (114). Additional signal transduction components with melanoma IHC data that did not meet eligibility criteria included c-Met (79,186,187), epidermal growth factor receptor (188–190), fibroblast growth factor receptor-1 (95,179,191), trk-C (192,193), akt (54,93,194), PTEN (93,195,196), p42/22 extracellular signal–related kinases (85,97), p38 mitogen–activated protein kinase (84), jun amino-terminal kinase (84), and c-myc (69,197–201). Another functional capability lacking eligible data despite numerous published experiments is sustained angiogenesis. Although only a single protein from this group, iNOS (118), was available for inclusion in our study, all 18 reports regarding VEGF (31,94,95,98,110,182,202–213) as well as those that evaluated VEGF receptors (94,95,208), ephrins and their receptors (95,214,215), or hypoxia-inducible transcription factors (98) as melanoma biomarkers did not meet inclusion criteria.
Of greatest concern is the subset of 125 proteins for which best evidence came from a study that described a prognostic endpoint but was dropped from this analysis due to methodological inadequacies; this included 13 proteins from case–control studies, 39 from case series and 73 from cohort studies that did not meet all the prespecified inclusion criteria. Despite the fact that the REMARK guidelines outlining minimum reporting criteria for molecular prognostic studies had been published in seven cancer-based peer-reviewed journals from 2005 to 2006, 14 of the 27 cohort studies that failed to adequately describe their IHC methods were published since 2005. An additional 96 potential biomarkers from 35 otherwise robust cohort studies (nine published since 2005) were excluded because either only univariate survival data were published (n = 44) or a multivariable analysis was executed, but the actual hazard ratio and 95% confidence interval were omitted (n = 52). REMARK guidelines state that the investigators must execute a multivariable analysis that includes the marker with all standard prognostic variables and must report this hazard ratio and associated confidence intervals regardless of statistical significance (17). Because of the volume of pre-REMARK manuscripts, we sent letters to 26 investigators requesting methodological details that had been omitted, or, for those who had reported multivariable statistical analysis, the missing point estimate and confidence intervals. Responses were received from nine groups; three groups indicated that they no longer had our requested information, and the remaining 17 queries went unanswered. Taken together, these findings suggest slow uptake and implementation of the REMARK guidelines, at least in the melanoma research community.
Exclusion of the 52 otherwise eligible biomarker assays in which estimated effects and confidence intervals were omitted, of which all but two predated the REMARK guidelines, constitutes an important source of publication bias because 44 described results that were not statistically significant, three indicated indeterminate results, and only five reported statistically significant associations. Omission of these data may contribute to overestimation of the prognostic utility for these markers and for their assigned functional pathways. Four excluded assays described associations between mortality and Ki-67, with three (total n = 206) yielding results that were not statistically significant (28,104,107) and one (105) demonstrating a statistically significant relationship. Although summary estimates among the eligible data were statistically significant, substantial interstudy heterogeneity was also observed, which suggests that these omitted studies will likely influence the true relationship between Ki-67 expression and ASM or MSM.
Finally, IHC-based prognostic marker studies, by serially investigating individual candidates and estimating their independent effects, evaluate only the marginal effects of individual proteins on prognosis and overlook the complex interplay between molecular pathways and their constituent proteins to support tumor progression. Modeling joint effects for complimentary proteins requires evaluation on the same cohort, entry into a single statistical model, and analysis for effect modification. Third or higher order interactions typically require sophisticated statistical models such as regression tree (CART) analysis for survival outcomes (216).
This systematic review of published IHC-based CMM molecular prognostic marker research supports involvement of cyclin-dependent kinase inhibitors, effectors of DNA replication and cell proliferation, growth-promoting transcription factors, and multiple regulators of tissue invasion and metastasis (the latter including cell adhesion molecules, matricellular proteins, and selected matrix metalloproteinases) in modulating melanoma outcomes. These results, however, need to be validated in adequately powered prospective studies designed to test both joint and marginal effects. At the same time, this study revealed substantial limitations in areas ranging from the choice of assayed proteins to the consistency and quality of published studies that strongly impacted the set of candidates available for consideration. The persistence of incomplete adoption of the 2005 REMARK guidelines should be addressed by the collective melanoma research community. This list of shortcomings may explain why molecular prognostic markers have largely failed to be incorporated into guidelines, staging systems, or the standard of care for melanoma patients.
Funding
National Institutes of Health (CA R01 CA 114277 to D.L.R., P50 CA121974 to Ruth Halaban).
Supplementary Material
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Gimotty PA, Guerry D, Ming ME, et al. Thin primary cutaneous malignant melanoma: a prognostic tree for 10-year metastasis is more accurate than American Joint Committee on Cancer staging [published online ahead of print August 9, 2004] J Clin Oncol. 2004;22(18):3668–3676. doi: 10.1200/JCO.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 3.Kirkwood JM, Moschos S, Wang W. Strategies for the development of more effective adjuvant therapy of melanoma: current and future explorations of antibodies, cytokines, vaccines, and combinations. Clin Cancer Res. 2006;12(7 pt 2):2331s–2336s. doi: 10.1158/1078-0432.CCR-05-2538. [DOI] [PubMed] [Google Scholar]
- 4.Balch CM, Soong SJ, Gershenwald JE, et al. Prognostic factors analysis of 17,600 melanoma patients: validation of the American Joint Committee on Cancer melanoma staging system. J Clin Oncol. 2001;19(16):3622–3634. doi: 10.1200/JCO.2001.19.16.3622. [DOI] [PubMed] [Google Scholar]
- 5.Gimotty PA, Elder DE, Fraker DL, et al. Identification of high-risk patients among those diagnosed with thin cutaneous melanomas. J Clin Oncol. 2007;25(9):1129–1134. doi: 10.1200/JCO.2006.08.1463. [DOI] [PubMed] [Google Scholar]
- 6.Bittner M, Meltzer P, Chen Y, et al. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406(6795):536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 7.Onken MD, Ehlers JP, Worley LA, Makita J, Yokota Y, Harbour JW. Functional gene expression analysis uncovers phenotypic switch in aggressive uveal melanomas. Cancer Res. 2006;66(9):4602–4609. doi: 10.1158/0008-5472.CAN-05-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winnepenninckx V, Lazar V, Michiels S, et al. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst. 2006;98(7):472–482. doi: 10.1093/jnci/djj103. [DOI] [PubMed] [Google Scholar]
- 9.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 10.van ‘t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 11.Taylor C. Standardization in immunohistochemistry: the role of antigen retrieval in molecular morphology. Biotech Histochem. 2006;81(1):3–12. doi: 10.1080/10520290600667866. [DOI] [PubMed] [Google Scholar]
- 12.Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4(7):844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 13.Rimm DL, Camp RL, Charette LA, Costa J, Olsen DA, Reiss M. Tissue microarray: a new technology for amplification of tissue resources. Cancer J. 2001;7(1):24–31. [PubMed] [Google Scholar]
- 14.Bosserhoff AK. Novel biomarkers in malignant melanoma [published online ahead of print February 9, 2006] Clin Chim Acta. 2006;367(1–2):28–35. doi: 10.1016/j.cca.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 15.Carlson JA, Ross J, Murphy M. Markers of high-risk cutaneous melanoma: is there a winning combination for individualized prognosis? J Cutan Pathol. 2005;32(10):700–703. doi: 10.1111/j.0303-6987.2005.00389.x. [DOI] [PubMed] [Google Scholar]
- 16.de Wit NJ, van Muijen GN, Ruiter DJ. Immunohistochemistry in melanocytic proliferative lesions. Histopathology. 2004;44(6):517–541. doi: 10.1111/j.1365-2559.2004.01860.x. [DOI] [PubMed] [Google Scholar]
- 17.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97(16):1180–1184. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 18.Gould Rothberg BE, Bracken MB. E-cadherin immunohistochemical expression as a prognostic factor in infiltrating ductal carcinoma of the breast: a systematic review and meta-analysis [published online ahead of print June 22, 2006] Breast Cancer Res Treat. 2006;100(2):139–148. doi: 10.1007/s10549-006-9248-2. [DOI] [PubMed] [Google Scholar]
- 19.Steels E, Paesmans M, Berghmans T, et al. Role of p53 as a prognostic factor for survival in lung cancer: a systematic review of the literature with a meta-analysis. Eur Respir J. 2001;18(4):705–719. doi: 10.1183/09031936.01.00062201. [DOI] [PubMed] [Google Scholar]
- 20.Antonescu CR, Busam KJ, Francone TD, et al. L576P KIT mutation in anal melanomas correlates with KIT protein expression and is sensitive to specific kinase inhibition. Int J Cancer. 2007;121(2):257–264. doi: 10.1002/ijc.22681. [DOI] [PubMed] [Google Scholar]
- 21.Byrd-Miles K, Toombs EL, Peck GL. Skin cancer in individuals of African, Asian, Latin-American, and American-Indian descent: differences in incidence, clinical presentation, and survival compared to Caucasians. J Drugs Dermatol. 2007;6(1):10–16. [PubMed] [Google Scholar]
- 22.Phan A, Touzet S, Dalle S, Ronger-Savle S, Balme B, Thomas L. Acral lentiginous melanoma: a clinicoprognostic study of 126 cases. Br J Dermatol. 2006;155(3):561–569. doi: 10.1111/j.1365-2133.2006.07368.x. [DOI] [PubMed] [Google Scholar]
- 23.Berger AJ, Camp RL, Divito KA, Kluger HM, Halaban R, Rimm DL. Automated quantitative analysis of HDM2 expression in malignant melanoma shows association with early-stage disease and improved outcome. Cancer Res. 2004;64(23):8767–8772. doi: 10.1158/0008-5472.CAN-04-1384. [DOI] [PubMed] [Google Scholar]
- 24.Hofbauer GF, Burkhart A, Schuler G, Dummer R, Burg G, Nestle FO. High frequency of melanoma-associated antigen or HLA class I loss does not correlate with survival in primary melanoma. J Immunother. 2004;27(1):73–78. doi: 10.1097/00002371-200401000-00007. [DOI] [PubMed] [Google Scholar]
- 25.Li Q, Murphy M, Ross J, Sheehan C, Carlson JA. Skp2 and p27kip1 expression in melanocytic nevi and melanoma: an inverse relationship. J Cutan Pathol. 2004;31(10):633–642. doi: 10.1111/j.0303-6987.2004.00243.x. [DOI] [PubMed] [Google Scholar]
- 26.Soltani MH, Pichardo R, Song Z, et al. Microtubule-associated protein 2, a marker of neuronal differentiation, induces mitotic defects, inhibits growth of melanoma cells, and predicts metastatic potential of cutaneous melanoma. Am J Pathol. 2005;166(6):1841–1850. doi: 10.1016/S0002-9440(10)62493-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Straume O, Akslen LA. Strong expression of ID1 protein is associated with decreased survival, increased expression of ephrin-A1/EPHA2, and reduced thrombospondin-1 in malignant melanoma. Br J Cancer. 2005;93(8):933–938. doi: 10.1038/sj.bjc.6602792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran TA, Ross JS, Carlson JA, Mihm MC., Jr Mitotic cyclins and cyclin-dependent kinases in melanocytic lesions. Hum Pathol. 1998;29(10):1085–1090. doi: 10.1016/s0046-8177(98)90418-x. [DOI] [PubMed] [Google Scholar]
- 29.Weinlich G, Eisendle K, Hassler E, Baltaci M, Fritsch PO, Zelger B. Metallothionein—overexpression as a highly significant prognostic factor in melanoma: a prospective study on 1270 patients. Br J Cancer. 2006;94(6):835–841. doi: 10.1038/sj.bjc.6603028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mihic-Probst D, Mnich CD, Oberholzer PA, et al. p16 expression in primary malignant melanoma is associated with prognosis and lymph node status. Int J Cancer. 2006;118(9):2262–2268. doi: 10.1002/ijc.21608. [DOI] [PubMed] [Google Scholar]
- 31.Potti A, Moazzam N, Langness E, et al. Immunohistochemical determination of HER-2/neu, c-Kit (CD117), and vascular endothelial growth factor (VEGF) overexpression in malignant melanoma [published online ahead of print November 21, 2003] J Cancer Res Clin Oncol. 2004;130(2):80–86. doi: 10.1007/s00432-003-0509-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 33.Deeks JJ, Altman DG, Bradburn MJ. Chapter 15: statistical methods for examining heterogeneity and combining results from several studies in meta-analysis. In: Egger M, Smith GD, Altman DG, editors. Systematic Reviews in Health Care: Meta-analysis in Context. 2nd ed. Cornwall, UK: BMJ Press; 2001. pp. 285–312. [Google Scholar]
- 34.Der Simonian R, Laird N. Meta-analysis in clinical trials. Controlled Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 35.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gauvreau K. Hypothesis testing: proportions. Circulation. 2006;114(14):1545–1548. doi: 10.1161/CIRCULATIONAHA.105.586487. [DOI] [PubMed] [Google Scholar]
- 37.Berset M, Cerottini JP, Guggisberg D, et al. Expression of Melan-A/MART-1 antigen as a prognostic factor in primary cutaneous melanoma. Int J Cancer. 2001;95(1):73–77. doi: 10.1002/1097-0215(20010120)95:1<73::aid-ijc1013>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 38.Bjornhagen V, Lindholm J, Auer G. Analysis of nuclear DNA and morphometry, and proliferating cell nuclear antigen in primary and metastatic malignant melanoma. Scand J Plast Reconstr Surg Hand Surg. 1997;31(2):109–118. doi: 10.3109/02844319709085477. [DOI] [PubMed] [Google Scholar]
- 39.Brinck U, Ruschenburg I, Di Como CJ, et al. Comparative study of p63 and p53 expression in tissue microarrays of malignant melanomas. Int J Mol Med. 2002;10(6):707–711. [PubMed] [Google Scholar]
- 40.Bron LP, Scolyer RA, Thompson JF, Hersey P. Histological expression of tumour necrosis factor-related apoptosis-inducing ligand (TRAIL) in human primary melanoma. Pathology. 2004;36(6):561–565. doi: 10.1080/00313020400011268. [DOI] [PubMed] [Google Scholar]
- 41.Chakravarti N, Lotan R, Diwan AH, Warneke CL, Johnson MM, Prieto VG. Decreased expression of retinoid receptors in melanoma: entailment in tumorigenesis and prognosis. Clin Cancer Res. 2007;13(16):4817–4824. doi: 10.1158/1078-0432.CCR-06-3026. [DOI] [PubMed] [Google Scholar]
- 42.Eliopoulos P, Mohammed MQ, Henry K, Retsas S. Overexpression of HER-2 in thick melanoma. Melanoma Res. 2002;12(2):139–145. doi: 10.1097/00008390-200204000-00006. [DOI] [PubMed] [Google Scholar]
- 43.Essner R, Kuo CT, Wang H, et al. Prognostic implications of p53 overexpression in cutaneous melanoma from sun-exposed and nonexposed sites. Cancer. 1998;82(2):309–316. doi: 10.1002/(sici)1097-0142(19980115)82:2<317::aid-cncr10>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 44.Hieken TJ, Farolan M, Ronan SG, Shilkaitis A, Wild L, Das Gupta TK. Beta3 integrin expression in melanoma predicts subsequent metastasis. J Surg Res. 1996;63(1):169–173. doi: 10.1006/jsre.1996.0242. [DOI] [PubMed] [Google Scholar]
- 45.Korabiowska M, Brinck U, Middel P, et al. Proliferative activity in the progression of pigmented skin lesions, diagnostic and prognostic significance. Anticancer Res. 2000;20(3A):1781–1785. [PubMed] [Google Scholar]
- 46.Salti GI, Manougian T, Farolan M, Shilkaitis A, Majumdar D, Das Gupta TK. Micropthalmia transcription factor: a new prognostic marker in intermediate-thickness cutaneous malignant melanoma. Cancer Res. 2000;60(18):5012–5016. [PubMed] [Google Scholar]
- 47.Vielkind JR, Huhn K, Tron VA. The Xiphophorus-derived antibody, XMEL, shows sensitivity and specificity for human cutaneous melanoma but is not a prognostic marker. J Cutan Pathol. 1997;24(10):620–627. doi: 10.1111/j.1600-0560.1997.tb01093.x. [DOI] [PubMed] [Google Scholar]
- 48.Vihinen P, Nikkola J, Vlaykova T, et al. Prognostic value of beta1 integrin expression in metastatic melanoma. Melanoma Res. 2000;10(3):243–251. [PubMed] [Google Scholar]
- 49.Vlaykova T, Talve L, Hahka-Kemppinen M, et al. MIB-1 immunoreactivity correlates with blood vessel density and survival in disseminated malignant melanoma. Oncology. 1999;57(3):242–252. doi: 10.1159/000012038. [DOI] [PubMed] [Google Scholar]
- 50.Zhuang L, Lee CS, Scolyer RA, et al. Mcl-1, Bcl-XL and Stat3 expression are associated with progression of melanoma whereas Bcl-2, AP-2 and MITF levels decrease during progression of melanoma. Mod Pathol. 2007;20(4):416–426. doi: 10.1038/modpathol.3800750. [DOI] [PubMed] [Google Scholar]
- 51.Bachmann IM, Halvorsen OJ, Collett K, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast [published online ahead of print December 5, 2005] J Clin Oncol. 2006;24(2):268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 52.Bachmann IM, Straume O, Puntervoll HE, Kalvenes MB, Akslen LA. Importance of P-cadherin, beta-catenin, and Wnt5a/frizzled for progression of melanocytic tumors and prognosis in cutaneous melanoma. Clin Cancer Res. 2005;11(24 pt 1):8606–8614. doi: 10.1158/1078-0432.CCR-05-0011. [DOI] [PubMed] [Google Scholar]
- 53.Dai DL, Makretsov N, Campos EI, et al. Increased expression of integrin-linked kinase is correlated with melanoma progression and poor patient survival. Clin Cancer Res. 2003;9(12):4409–4414. [PubMed] [Google Scholar]
- 54.Dai DL, Martinka M, Li G. Prognostic significance of activated Akt expression in melanoma: a clinicopathologic study of 292 cases. J Clin Oncol. 2005;23(7):1473–1482. doi: 10.1200/JCO.2005.07.168. [DOI] [PubMed] [Google Scholar]
- 55.Dai DL, Wang Y, Liu M, Martinka M, Li G. Bim expression is reduced in human cutaneous melanomas [published online ahead of print July 19, 2007] J Invest Dermatol. 2008;128(2):403–407. doi: 10.1038/sj.jid.5700989. [DOI] [PubMed] [Google Scholar]
- 56.Gutman M, Even-Sapir E, Merimsky O, Trejo L, Klausner JM, Lev-Chelouche D. The role of interleukin-8 in the initiation and progression of human cutaneous melanoma. Anticancer Res. 2002;22(6A):3395–3398. [PubMed] [Google Scholar]
- 57.Hazan C, Melzer K, Panageas KS, et al. Evaluation of the proliferation marker MIB-1 in the prognosis of cutaneous malignant melanoma. Cancer. 2002;95(3):634–640. doi: 10.1002/cncr.10685. [DOI] [PubMed] [Google Scholar]
- 58.Ilmonen S, Hernberg M, Pyrhonen S, Tarkkanen J, Asko-Seljavaara S. Ki-67, Bcl-2 and p53 expression in primary and metastatic melanoma. Melanoma Res. 2005;15(5):375–381. doi: 10.1097/00008390-200510000-00005. [DOI] [PubMed] [Google Scholar]
- 59.Ilmonen S, Vaheri A, Asko-Seljavaara S, Carpen O. Ezrin in primary cutaneous melanoma. Mod Pathol. 2005;18(4):503–510. doi: 10.1038/modpathol.3800300. [DOI] [PubMed] [Google Scholar]
- 60.Karst AM, Dai DL, Martinka M, Li G. PUMA expression is significantly reduced in human cutaneous melanomas. Oncogene. 2005;24(6):1111–1116. doi: 10.1038/sj.onc.1208374. [DOI] [PubMed] [Google Scholar]
- 61.Kielhorn E, Provost E, Olsen D, et al. Tissue microarray-based analysis shows phospho-beta-catenin expression in malignant melanoma is associated with poor outcome. Int J Cancer. 2003;103(5):652–656. doi: 10.1002/ijc.10893. [DOI] [PubMed] [Google Scholar]
- 62.Korabiowska M, Brinck U, Dengler H, Stachura J, Schauer A, Droese M. Analysis of the DNA mismatch repair proteins expression in malignant melanomas. Anticancer Res. 2000;20(6B):4499–4505. [PubMed] [Google Scholar]
- 63.Korabiowska M, Fischer G, Steinacker A, Stachura J, Cordon-Cardo C, Brinck U. Cytokeratin positivity in paraffin-embedded malignant melanomas: comparative study of KL1, A4 and Lu5 antibodies. Anticancer Res. 2004;24(5B):3203–3207. [PubMed] [Google Scholar]
- 64.Korabiowska M, Ruschenburg I, Betke H, et al. Downregulation of the retinoblastoma gene expression in the progression of malignant melanoma. Pathobiology. 2001;69(5):274–280. doi: 10.1159/000064338. [DOI] [PubMed] [Google Scholar]
- 65.Moretti S, Spallanzani A, Chiarugi A, Fabiani M, Pinzi C. Correlation of Ki-67 expression in cutaneous primary melanoma with prognosis in a prospective study: different correlation according to thickness. J Am Acad Dermatol. 2001;44(2):188–192. doi: 10.1067/mjd.2001.110067. [DOI] [PubMed] [Google Scholar]
- 66.Nikkola J, Vihinen P, Vlaykova T, Hahka-Kemppinen M, Heino J, Pyrhonen S. Integrin chains beta1 and alphav as prognostic factors in human metastatic melanoma. Melanoma Res. 2004;14(1):29–37. doi: 10.1097/00008390-200402000-00005. [DOI] [PubMed] [Google Scholar]
- 67.Ostmeier H, Fuchs B, Otto F, et al. Prognostic immunohistochemical markers of primary human melanomas. Br J Dermatol. 2001;145(2):203–209. doi: 10.1046/j.1365-2133.2001.04335.x. [DOI] [PubMed] [Google Scholar]
- 68.Polsky D, Melzer K, Hazan C, et al. HDM2 protein overexpression and prognosis in primary malignant melanoma. J Natl Cancer Inst. 2002;94(23):1803–1806. doi: 10.1093/jnci/94.23.1803. [DOI] [PubMed] [Google Scholar]
- 69.Ricaniadis N, Kataki A, Agnantis N, Androulakis G, Karakousis CP. Long-term prognostic significance of HSP-70, c-myc and HLA-DR expression in patients with malignant melanoma. Eur J Surg Oncol. 2001;27(1):88–93. doi: 10.1053/ejso.1999.1018. [DOI] [PubMed] [Google Scholar]
- 70.Sauroja I, Smeds J, Vlaykova T, et al. Analysis of G(1)/S checkpoint regulators in metastatic melanoma. Genes Chromosomes Cancer. 2000;28(4):404–414. doi: 10.1002/1098-2264(200008)28:4<404::aid-gcc6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 71.Staibano S, Pepe S, Lo Muzio L, et al. Poly(adenosine diphosphate-ribose) polymerase 1 expression in malignant melanomas from photoexposed areas of the head and neck region. Hum Pathol. 2005;36(7):724–731. doi: 10.1016/j.humpath.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 72.Streit S, Mestel DS, Schmidt M, Ullrich A, Berking C. FGFR4 Arg388 allele correlates with tumour thickness and FGFR4 protein expression with survival of melanoma patients [published online ahead of print May 23, 2006] Br J Cancer. 2006;94(12):1879–1886. doi: 10.1038/sj.bjc.6603181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tucci MG, Lucarini G, Brancorsini D, et al. Involvement of E-cadherin, beta-catenin, Cdc42 and CXCR4 in the progression and prognosis of cutaneous melanoma. Br J Dermatol. 2007;157(6):1212–1216. doi: 10.1111/j.1365-2133.2007.08246.x. [DOI] [PubMed] [Google Scholar]
- 74.van Duinen SG, Ruiter DJ, Broecker EB, et al. Level of HLA antigens in locoregional metastases and clinical course of the disease in patients with melanoma. Cancer Res. 1988;48(4):1019–1025. [PubMed] [Google Scholar]
- 75.Wang Y, Dai DL, Martinka M, Li G. Prognostic significance of nuclear ING3 expression in human cutaneous melanoma. Clin Cancer Res. 2007;13(14):4111–4116. doi: 10.1158/1078-0432.CCR-07-0408. [DOI] [PubMed] [Google Scholar]
- 76.Zhou Y, Dai DL, Martinka M, et al. Osteopontin expression correlates with melanoma invasion. J Invest Dermatol. 2005;124(5):1044–1052. doi: 10.1111/j.0022-202X.2005.23680.x. [DOI] [PubMed] [Google Scholar]
- 77.Andersen K, Nesland JM, Holm R, Florenes VA, Fodstad O, Maelandsmo GM. Expression of S100A4 combined with reduced E-cadherin expression predicts patient outcome in malignant melanoma. Mod Pathol. 2004;17(8):990–997. doi: 10.1038/modpathol.3800151. [DOI] [PubMed] [Google Scholar]
- 78.Bachmann IM, Straume O, Akslen LA. Altered expression of cell cycle regulators cyclin D1, p14, p16, CDK4 and Rb in nodular melanomas. Int J Oncol. 2004;25(6):1559–1565. [PubMed] [Google Scholar]
- 79.Cruz J, Reis-Filho JS, Silva P, Lopes JM. Expression of c-met tyrosine kinase receptor is biologically and prognostically relevant for primary cutaneous malignant melanomas. Oncology. 2003;65(1):72–82. doi: 10.1159/000071207. [DOI] [PubMed] [Google Scholar]
- 80.Dai DL, Martinka M, Bush JA, Li G. Reduced Apaf-1 expression in human cutaneous melanomas. Br J Cancer. 2004;91(6):1089–1095. doi: 10.1038/sj.bjc.6602092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ekmekcioglu S, Ellerhorst J, Smid CM, et al. Inducible nitric oxide synthase and nitrotyrosine in human metastatic melanoma tumors correlate with poor survival. Clin Cancer Res. 2000;6(12):4768–4775. [PubMed] [Google Scholar]
- 82.Florenes VA, Maelandsmo GM, Holm R, Reich R, Lazarovici P, Davidson B. Expression of activated TrkA protein in melanocytic tumors: relationship to cell proliferation and clinical outcome. Am J Clin Pathol. 2004;122(3):412–420. doi: 10.1309/CHFH-EYAT-44WW-P7J3. [DOI] [PubMed] [Google Scholar]
- 83.Henrique R, Azevedo R, Bento MJ, Domingues JC, Silva C, Jeronimo C. Prognostic value of Ki-67 expression in localized cutaneous malignant melanoma. J Am Acad Dermatol. 2000;43(6):991–1000. doi: 10.1067/mjd.2000.109282. [DOI] [PubMed] [Google Scholar]
- 84.Jorgensen K, Davidson B, Florenes VA. Activation of c-jun N-terminal kinase is associated with cell proliferation and shorter relapse-free period in superficial spreading malignant melanoma. Mod Pathol. 2006;19(11):1446–1455. doi: 10.1038/modpathol.3800662. [DOI] [PubMed] [Google Scholar]
- 85.Jorgensen K, Holm R, Maelandsmo GM, Florenes VA. Expression of activated extracellular signal-regulated kinases 1/2 in malignant melanomas: relationship with clinical outcome. Clin Cancer Res. 2003;9(14):5325–5331. [PubMed] [Google Scholar]
- 86.Korabiowska M, Cordon-Cardo C, Betke H, et al. GADD153 is an independent prognostic factor in melanoma: immunohistochemical and molecular genetic analysis. Histol Histopathol. 2002;17(3):805–811. doi: 10.14670/HH-17.805. [DOI] [PubMed] [Google Scholar]
- 87.Korabiowska M, Schlott T, Siems N, et al. Analysis of adenomatous polyposis coli gene expression, APC locus-microsatellite instability and APC promoter methylation in the progression of melanocytic tumours. Mod Pathol. 2004;17(12):1539–1544. doi: 10.1038/modpathol.3800238. [DOI] [PubMed] [Google Scholar]
- 88.Lu F, Dai DL, Martinka M, Ho V, Li G. Nuclear ING2 expression is reduced in human cutaneous melanomas [published online ahead of print June 6, 2006] Br J Cancer. 2006;95(1):80–86. doi: 10.1038/sj.bjc.6603205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McCarthy MM, DiVito KA, Sznol M, et al. Expression of tumor necrosis factor—related apoptosis-inducing ligand receptors 1 and 2 in melanoma. Clin Cancer Res. 2006;12(12):3856–3863. doi: 10.1158/1078-0432.CCR-06-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McCarthy MM, Pick E, Kluger Y, et al. HSP90 as a marker of progression in melanoma [published online ahead of print November 23, 2007] Ann Oncol. 2008;19(3):590–594. doi: 10.1093/annonc/mdm545. [DOI] [PubMed] [Google Scholar]
- 91.Nikkola J, Vihinen P, Vlaykova T, Hahka-Kemppinen M, Kahari VM, Pyrhonen S. High expression levels of collagenase-1 and stromelysin-1 correlate with shorter disease-free survival in human metastatic melanoma. Int J Cancer. 2002;97(4):432–438. doi: 10.1002/ijc.1636. [DOI] [PubMed] [Google Scholar]
- 92.Sirigu P, Piras F, Minerba L, et al. Prognostic prediction of the immunohistochemical expression of p16 and p53 in cutaneous melanoma: a comparison of two populations from different geographical regions. Eur J Histochem. 2006;50(3):191–198. [PubMed] [Google Scholar]
- 93.Slipicevic A, Holm R, Nguyen MT, Bohler PJ, Davidson B, Florenes VA. Expression of activated Akt and PTEN in malignant melanomas: relationship with clinical outcome. Am J Clin Pathol. 2005;124(4):528–536. doi: 10.1309/YT58WWMTA6YR1PRV. [DOI] [PubMed] [Google Scholar]
- 94.Straume O, Akslen LA. Expresson of vascular endothelial growth factor, its receptors (FLT-1, KDR) and TSP-1 related to microvessel density and patient outcome in vertical growth phase melanomas. Am J Pathol. 2001;159(1):223–235. doi: 10.1016/S0002-9440(10)61688-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Straume O, Akslen LA. Importance of vascular phenotype by basic fibroblast growth factor, and influence of the angiogenic factors basic fibroblast growth factor/fibroblast growth factor receptor-1 and ephrin-A1/EphA2 on melanoma progression. Am J Pathol. 2002;160(3):1009–1019. doi: 10.1016/S0002-9440(10)64922-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Torlakovic EE, Bilalovic N, Nesland JM, Torlakovic G, Florenes VA. Ets-1 transcription factor is widely expressed in benign and malignant melanocytes and its expression has no significant association with prognosis. Mod Pathol. 2004;17(11):1400–1406. doi: 10.1038/modpathol.3800206. [DOI] [PubMed] [Google Scholar]
- 97.Zhuang L, Lee CS, Scolyer RA, et al. Activation of the extracellular signal regulated kinase (ERK) pathway in human melanoma. J Clin Pathol. 2005;58(11):1163–1169. doi: 10.1136/jcp.2005.025957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Giatromanolaki A, Sivridis E, Kouskoukis C, Gatter KC, Harris AL, Koukourakis MI. Hypoxia-inducible factors 1alpha and 2alpha are related to vascular endothelial growth factor expression and a poorer prognosis in nodular malignant melanomas of the skin. Melanoma Res. 2003;13(5):493–501. doi: 10.1097/00008390-200310000-00008. [DOI] [PubMed] [Google Scholar]
- 99.Hieken TJ, Ronan SG, Farolan M, Shilkaitis AL, Das Gupta TK. Molecular prognostic markers in intermediate-thickness cutaneous malignant melanoma. Cancer. 1999;85(2):375–382. doi: 10.1002/(sici)1097-0142(19990115)85:2<375::aid-cncr15>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 100.Karjalainen JM, Eskelinen MJ, Kellokoski JK, Reinikainen M, Alhava EM, Kosma VM. p21(WAF1/CIP1) expression in stage I cutaneous malignant melanoma: its relationship with p53, cell proliferation and survival. Br J Cancer. 1999;79(5–6):895–902. doi: 10.1038/sj.bjc.6690143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Maelandsmo GM, Holm R, Nesland JM, Fodstad O, Florenes VA. Reduced beta-catenin expression in the cytoplasm of advanced-stage superficial spreading malignant melanoma. Clin Cancer Res. 2003;9(9):3383–3388. [PubMed] [Google Scholar]
- 102.Mu XC, Tran TA, Ross JS, Carlson JA. Topoisomerase II-alpha expression in melanocytic nevi and malignant melanoma. J Cutan Pathol. 2000;27(5):242–248. doi: 10.1034/j.1600-0560.2000.027005242.x. [DOI] [PubMed] [Google Scholar]
- 103.Ostmeier H, Fuchs B, Otto F, et al. Can immunohistochemical markers and mitotic rate improve prognostic precision in patients with primary melanoma? Cancer. 1999;85(11):2391–2399. doi: 10.1002/(sici)1097-0142(19990601)85:11<2391::aid-cncr14>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 104.Pearl RA, Pacifico MD, Richman PI, Stott DJ, Wilson GD, Grobbelaar AO. Ki-67 expression in melanoma. A potential method of risk assessment for the patient with a positive sentinel node. J Exp Clin Cancer Res. 2007;26(1):109–115. [PubMed] [Google Scholar]
- 105.Ramsay JA, From L, Iscoe NA, Kahn HJ. MIB-1 proliferative activity is a significant prognostic factor in primary thick cutaneous melanomas. J Invest Dermatol. 1995;105(1):22–26. doi: 10.1111/1523-1747.ep12312431. [DOI] [PubMed] [Google Scholar]
- 106.Talve L, Kainu J, Collan Y, Ekfors T. Immunohistochemical expression of p53 protein, mitotic index and nuclear morphometry in primary malignant melanoma of the skin. Pathol Res Pract. 1996;192(8):825–833. doi: 10.1016/S0344-0338(96)80056-2. [DOI] [PubMed] [Google Scholar]
- 107.Talve LA, Collan YU, Ekfors TO. Nuclear morphometry, immunohistochemical staining with Ki-67 antibody and mitotic index in the assessment of proliferative activity and prognosis of primary malignant melanomas of the skin. J Cutan Pathol. 1996;23(4):335–343. doi: 10.1111/j.1600-0560.1996.tb01307.x. [DOI] [PubMed] [Google Scholar]
- 108.Vlaykova T, Talve L, Hahka-Kemppinen M, et al. Immunohistochemically detectable bcl-2 expression in metastatic melanoma: association with survival and treatment response. Oncology. 2002;62(3):259–268. doi: 10.1159/000059574. [DOI] [PubMed] [Google Scholar]
- 109.Potti A, Hille RC, Koch M. Immunohistochemical determination of HER-2/neu overexpression in malignant melanoma reveals no prognostic value, while c-Kit (CD117) overexpression exhibits potential therapeutic implications. J Carcinog. 2003;2(1):8. doi: 10.1186/1477-3163-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Potti A, Moazzam N, Tendulkar K, Javed NA, Koch M, Kargas S. Immunohistochemical determination of vascular endothelial growth factor (VEGF) overexpression in malignant melanoma. Anticancer Res. 2003;23(5A):4023–4026. [PubMed] [Google Scholar]
- 111.Straume O, Akslen LA. Alterations and prognostic significance of p16 and p53 protein expression in subgroups of cutaneous melanoma. Int J Cancer. 1997;74(5):535–539. doi: 10.1002/(sici)1097-0215(19971021)74:5<535::aid-ijc10>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 112.Weinlich G, Bitterlich W, Mayr V, Fritsch PO, Zelger B. Metallothionein-overexpression as a prognostic factor for progression and survival in melanoma. A prospective study on 520 patients. Br J Dermatol. 2003;149(3):535–541. doi: 10.1046/j.1365-2133.2003.05472.x. [DOI] [PubMed] [Google Scholar]
- 113.Alonso SR, Tracey L, Ortiz P, et al. A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Res. 2007;67(7):3450–3460. doi: 10.1158/0008-5472.CAN-06-3481. [DOI] [PubMed] [Google Scholar]
- 114.Alonso SR, Ortiz P, Pollan M, et al. Progression in cutaneous malignant melanoma is associated with distinct expression profiles: a tissue microarray-based study. Am J Pathol. 2004;164(1):193–203. doi: 10.1016/s0002-9440(10)63110-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Berger AJ, Davis DW, Tellez C, et al. Automated quantitative analysis of activator protein-2alpha subcellular expression in melanoma tissue microarrays correlates with survival prediction. Cancer Res. 2005;65(23):11185–11192. doi: 10.1158/0008-5472.CAN-05-2300. [DOI] [PubMed] [Google Scholar]
- 116.Berger AJ, Kluger HM, Li N, et al. Subcellular localization of activating transcription factor 2 in melanoma specimens predicts patient survival. Cancer Res. 2003;63(23):8103–8107. [PubMed] [Google Scholar]
- 117.Divito KA, Berger AJ, Camp RL, Dolled-Filhart M, Rimm DL, Kluger HM. Automated quantitative analysis of tissue microarrays reveals an association between high Bcl-2 expression and improved outcome in melanoma. Cancer Res. 2004;64(23):8773–8777. doi: 10.1158/0008-5472.CAN-04-1387. [DOI] [PubMed] [Google Scholar]
- 118.Ekmekcioglu S, Ellerhorst JA, Prieto VG, Johnson MM, Broemeling LD, Grimm EA. Tumor iNOS predicts poor survival for stage III melanoma patients. Int J Cancer. 2006;119(4):861–866. doi: 10.1002/ijc.21767. [DOI] [PubMed] [Google Scholar]
- 119.Ferrier CM, Suciu S, van Geloof WL, et al. High tPA-expression in primary melanoma of the limb correlates with good prognosis. Br J Cancer. 2000;83(10):1351–1359. doi: 10.1054/bjoc.2000.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Florenes VA, Maelandsmo GM, Faye R, Nesland JM, Holm R. Cyclin A expression in superficial spreading malignant melanomas correlates with clinical outcome. J Pathol. 2001;195(5):530–536. doi: 10.1002/path.1007. [DOI] [PubMed] [Google Scholar]
- 121.Florenes VA, Faye RS, Maelandsmo GM, Nesland JM, Holm R. Levels of cyclin D1 and D3 in malignant melanoma: deregulated cyclin D3 expression is associated with poor clinical outcome in superficial melanoma. Clin Cancer Res. 2000;6(9):3614–3620. [PubMed] [Google Scholar]
- 122.Ilmonen S, Jahkola T, Turunen JP, Muhonen T, Asko-Seljavaara S. Tenascin-C in primary malignant melanoma of the skin. Histopathology. 2004;45(4):405–411. doi: 10.1111/j.1365-2559.2004.01976.x. [DOI] [PubMed] [Google Scholar]
- 123.Karjalainen JM, Tammi RH, Tammi MI, et al. Reduced level of CD44 and hyaluronan associated with unfavorable prognosis in clinical stage I cutaneous melanoma. Am J Pathol. 2000;157(3):957–965. doi: 10.1016/S0002-9440(10)64608-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Karjalainen JM, Kellokoski JK, Eskelinen MJ, Alhava EM, Kosma VM. Downregulation of transcription factor AP-2 predicts poor survival in stage I cutaneous malignant melanoma. J Clin Oncol. 1998;16(11):3584–3591. doi: 10.1200/JCO.1998.16.11.3584. [DOI] [PubMed] [Google Scholar]
- 125.Korabiowska M, Tscherny M, Stachura J, Berger H, Cordon-Cardo C, Brinck U. Differential expression of DNA nonhomologous end-joining proteins Ku70 and Ku80 in melanoma progression. Mod Pathol. 2002;15(4):426–433. doi: 10.1038/modpathol.3880542. [DOI] [PubMed] [Google Scholar]
- 126.McDermott NC, Milburn C, Curran B, Kay EW, Barry Walsh C, Leader MB. Immunohistochemical expression of nm23 in primary invasive malignant melanoma is predictive of survival outcome. J Pathol. 2000;190(2):157–162. doi: 10.1002/(SICI)1096-9896(200002)190:2<157::AID-PATH512>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 127.Niezabitowski A, Czajecki K, Rys J, et al. Prognostic evaluation of cutaneous malignant melanoma: a clinicopathologic and immunohistochemical study. J Surg Oncol. 1999;70(3):150–160. doi: 10.1002/(sici)1096-9098(199903)70:3<150::aid-jso2>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 128.Pacifico MD, Grover R, Richman PI, Daley FM, Buffa F, Wilson GD. CD44v3 levels in primary cutaneous melanoma are predictive of prognosis: assessment by the use of tissue microarray. Int J Cancer. 2006;118(6):1460–1464. doi: 10.1002/ijc.21504. [DOI] [PubMed] [Google Scholar]
- 129.Pacifico MD, Grover R, Richman PI, Buffa F, Daley FM, Wilson GD. nm23 as a prognostic marker in primary cutaneous melanoma: evaluation using tissue microarray in a patient group with long-term follow-up. Melanoma Res. 2005;15(5):435–440. doi: 10.1097/00008390-200510000-00012. [DOI] [PubMed] [Google Scholar]
- 130.Pacifico MD, Grover R, Richman PI, Buffa F, Daley FM, Wilson GD. Identification of P-cadherin in primary melanoma using a tissue microarrayer: prognostic implications in a patient cohort with long-term follow up. Ann Plast Surg. 2005;55(3):316–320. doi: 10.1097/01.sap.0000171429.19320.ce. [DOI] [PubMed] [Google Scholar]
- 131.Pacifico MD, Grover R, Richman PI, Daley FM, Buffa F, Wilson GD. Development of a tissue array for primary melanoma with long-term follow-up: discovering melanoma cell adhesion molecule as an important prognostic marker. Plast Reconstr Surg. 2005;115(2):367–375. doi: 10.1097/01.prs.0000148417.86768.c9. [DOI] [PubMed] [Google Scholar]
- 132.Pearl RA, Pacifico MD, Richman PI, Wilson GD, Grover R. Stratification of patients by melanoma cell adhesion molecule (MCAM) expression on the basis of risk: implications for sentinel lymph node biopsy. J Plast Reconstr Aesthet Surg. 2008;61:265–271. doi: 10.1016/j.bjps.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 133.Piras F, Murtas D, Minerba L, et al. Nuclear survivin is associated with disease recurrence and poor survival in patients with cutaneous malignant melanoma. Histopathology. 2007;50(7):835–842. doi: 10.1111/j.1365-2559.2007.02695.x. [DOI] [PubMed] [Google Scholar]
- 134.Rangel J, Nosrati M, Torabian S, et al. Osteopontin as a molecular prognostic marker for melanoma. Cancer. 2008;112(1):144–150. doi: 10.1002/cncr.23147. [DOI] [PubMed] [Google Scholar]
- 135.Rangel J, Torabian S, Shaikh L, et al. Prognostic significance of nuclear receptor coactivator-3 overexpression in primary cutaneous melanoma. J Clin Oncol. 2006;24(28):4565–4569. doi: 10.1200/JCO.2006.07.3833. [DOI] [PubMed] [Google Scholar]
- 136.Scala S, Ottaiano A, Ascierto PA, et al. Expression of CXCR4 predicts poor prognosis in patients with malignant melanoma. Clin Cancer Res. 2005;11(5):1835–1841. doi: 10.1158/1078-0432.CCR-04-1887. [DOI] [PubMed] [Google Scholar]
- 137.Straume O, Sviland L, Akslen LA. Loss of nuclear p16 protein expression correlates with increased tumor cell proliferation (Ki-67) and poor prognosis in patients with vertical growth phase melanoma. Clin Cancer Res. 2000;6(5):1845–1853. [PubMed] [Google Scholar]
- 138.Thies A, Schachner M, Moll I, et al. Overexpression of the cell adhesion molecule L1 is associated with metastasis in cutaneous malignant melanoma. Eur J Cancer. 2002;38(13):1708–1716. doi: 10.1016/s0959-8049(02)00105-3. [DOI] [PubMed] [Google Scholar]
- 139.Thies A, Moll I, Berger J, et al. CEACAM1 expression in cutaneous malignant melanoma predicts the development of metastatic disease. J Clin Oncol. 2002;20(10):2530–2536. doi: 10.1200/JCO.2002.05.033. [DOI] [PubMed] [Google Scholar]
- 140.Vaisanen AH, Kallioinen M, Turpeenniemi-Hujanen T. Comparison of the prognostic value of matrix metalloproteinases 2 and 9 in cutaneous melanoma. Hum Pathol. 2008;39:377–385. doi: 10.1016/j.humpath.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 141.Vaisanen A, Kallioinen M, Taskinen PJ, Turpeenniemi-Hujanen T. Prognostic value of MMP-2 immunoreactive protein (72 kD type IV collagenase) in primary skin melanoma. J Pathol. 1998;186(1):51–58. doi: 10.1002/(SICI)1096-9896(199809)186:1<51::AID-PATH131>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 142.Weinlich G, Topar G, Eisendle K, Fritsch PO, Zelger B. Comparison of metallothionein-overexpression with sentinel lymph node biopsy as prognostic factors in melanoma. J Eur Acad Dermatol Venereol. 2007;21(5):669–677. doi: 10.1111/j.1468-3083.2006.02051.x. [DOI] [PubMed] [Google Scholar]
- 143.Breslow A. Thickness, cross-sectional areas and depth of invasion in the prognosis of cutaneous melanoma. Ann Surg. 1970;172(5):902–908. doi: 10.1097/00000658-197011000-00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Scoggins CR, Ross MI, Reintgen DS, et al. Gender-related differences in outcome for melanoma patients. Ann Surg. 2006;243(5):693–698. doi: 10.1097/01.sla.0000216771.81362.6b. discussion 698–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Clark WH, Jr, From L, Bernardino EA, Mihm MC. The histogenesis and biologic behavior of primary human malignant melanomas of the skin. Cancer Res. 1969;29(3):705–727. [PubMed] [Google Scholar]
- 146.Chin L, Merlino G, DePinho RA. Malignant melanoma: modern black plague and genetic black box. Genes Dev. 1998;12(22):3467–3481. doi: 10.1101/gad.12.22.3467. [DOI] [PubMed] [Google Scholar]
- 147.Crocetti E, Mangone L, Lo Scocco G, Carli P. Prognostic variables and prognostic groups for malignant melanoma. The information from Cox and Classification and Regression Trees analysis: an Italian population-based study. Melanoma Res. 2006;16(5):429–433. doi: 10.1097/01.cmr.0000222602.80803.e1. [DOI] [PubMed] [Google Scholar]
- 148.Eigentler TK, Buettner PG, Leiter U, Garbe C. Impact of ulceration in stages I to III cutaneous melanoma as staged by the American Joint Committee on Cancer Staging System: an analysis of the German Central Malignant Melanoma Registry. J Clin Oncol. 2004;22(21):4376–4383. doi: 10.1200/JCO.2004.03.075. [DOI] [PubMed] [Google Scholar]
- 149.Gavert N, Sheffer M, Raveh S, et al. Expression of L1-CAM and ADAM10 in human colon cancer cells induces metastasis. Cancer Res. 2007;67(16):7703–7712. doi: 10.1158/0008-5472.CAN-07-0991. [DOI] [PubMed] [Google Scholar]
- 150.Briese J, Schulte HM, Bamberger CM, Loning T, Bamberger AM. Expression pattern of osteopontin in endometrial carcinoma: correlation with expression of the adhesion molecule CEACAM1. Int J Gynecol Pathol. 2006;25(2):161–169. doi: 10.1097/01.pgp.0000189243.49522.ae. [DOI] [PubMed] [Google Scholar]
- 151.Watson-Hurst K, Becker D. The role of N-cadherin, MCAM and beta3 integrin in melanoma progression, proliferation, migration and invasion. Cancer Biol Ther. 2006;5(10):1375–1382. doi: 10.4161/cbt.5.10.3241. [DOI] [PubMed] [Google Scholar]
- 152.Bornstein P, Sage EH. Matricellular proteins: extracellular modulators of cell function. Curr Opin Cell Biol. 2002;14(5):608–616. doi: 10.1016/s0955-0674(02)00361-7. [DOI] [PubMed] [Google Scholar]
- 153.Kreizenbeck GM, Berger AJ, Subtil A, Rimm DL, Gould Rothberg BE. Prognostic significance of cadherin-based adhesion molecules in cutaneous malignant melanoma. Cancer Epidemiol Biomarkers Prev. 2008;17(4):949–958. doi: 10.1158/1055-9965.EPI-07-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sharpless NE, Chin L. The INK4a/ARF locus and melanoma. Oncogene. 2003;22(20):3092–3098. doi: 10.1038/sj.onc.1206461. [DOI] [PubMed] [Google Scholar]
- 155.Denicourt C, Saenz CC, Datnow B, Cui XS, Dowdy SF. Relocalized p27Kip1 tumor suppressor functions as a cytoplasmic metastatic oncogene in melanoma. Cancer Res. 2007;67(19):9238–9243. doi: 10.1158/0008-5472.CAN-07-1375. [DOI] [PubMed] [Google Scholar]
- 156.Black WC, Haggstrom DA, Welch HG. All-cause mortality in randomized trials of cancer screening. J Natl Cancer Inst. 2002;94(3):167–173. doi: 10.1093/jnci/94.3.167. [DOI] [PubMed] [Google Scholar]
- 157.Yood MU, Owusu C, Buist DS, et al. Mortality impact of less-than-standard therapy in older breast cancer patients. J Am Coll Surg. 2008;206(1):66–75. doi: 10.1016/j.jamcollsurg.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 158.Bekkering GE, Harris RJ, Thomas S, et al. How much of the data published in observational studies of the association between diet and prostate or bladder cancer is usable for meta-analysis? Am J Epidemiol. 2008;167(9):1017–1026. doi: 10.1093/aje/kwn005. [DOI] [PubMed] [Google Scholar]
- 159.Bishop JN, Harland M, Randerson-Moor J, Bishop DT. Management of familial melanoma. Lancet Oncol. 2007;8(1):46–54. doi: 10.1016/S1470-2045(06)71010-5. [DOI] [PubMed] [Google Scholar]
- 160.Bartek J, Bartkova J, Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007;26(56):7773–7779. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- 161.Brown DC, Gatter KC. Ki67 protein: the immaculate deception? Histopathology. 2002;40(1):2–11. doi: 10.1046/j.1365-2559.2002.01343.x. [DOI] [PubMed] [Google Scholar]
- 162.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129(4):665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 163.Karnoub AE, Dash AB, Vo AP, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449(7162):557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 164.Singh S, Sadanandam A, Singh RK. Chemokines in tumor angiogenesis and metastasis. Cancer Metastasis Rev. 2007;26(3–4):453–467. doi: 10.1007/s10555-007-9068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ben-Baruch A. Organ selectivity in metastasis: regulation by chemokines and their receptors. Clin Exp Metastasis. 2008;25(4):345–356. doi: 10.1007/s10585-007-9097-3. [DOI] [PubMed] [Google Scholar]
- 166.Horton LW, Yu Y, Zaja-Milatovic S, Strieter RM, Richmond A. Opposing roles of murine duffy antigen receptor for chemokine and murine CXC chemokine receptor-2 receptors in murine melanoma tumor growth. Cancer Res. 2007;67(20):9791–9799. doi: 10.1158/0008-5472.CAN-07-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kawada K, Sonoshita M, Sakashita H, et al. Pivotal role of CXCR3 in melanoma cell metastasis to lymph nodes. Cancer Res. 2004;64(11):4010–4017. doi: 10.1158/0008-5472.CAN-03-1757. [DOI] [PubMed] [Google Scholar]
- 168.Kim SY, Lee CH, Midura BV, et al. Inhibition of the CXCR4/CXCL12 chemokine pathway reduces the development of murine pulmonary metastases. Clin Exp Metastasis. 2008;25(3):201–211. doi: 10.1007/s10585-007-9133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Murakami T, Cardones AR, Hwang ST. Chemokine receptors and melanoma metastasis. J Dermatol Sci. 2004;36(2):71–78. doi: 10.1016/j.jdermsci.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 170.van Deventer HW, O’Connor W, Jr, Brickey WJ, Aris RM, Ting JP, Serody JS. C-C chemokine receptor 5 on stromal cells promotes pulmonary metastasis. Cancer Res. 2005;65(8):3374–3379. doi: 10.1158/0008-5472.CAN-04-2616. [DOI] [PubMed] [Google Scholar]
- 171.Varney ML, Li A, Dave BJ, Bucana CD, Johansson SL, Singh RK. Expression of CXCR1 and CXCR2 receptors in malignant melanoma with different metastatic potential and their role in interleukin-8 (CXCL-8)-mediated modulation of metastatic phenotype. Clin Exp Metastasis. 2003;20(8):723–731. doi: 10.1023/b:clin.0000006814.48627.bd. [DOI] [PubMed] [Google Scholar]
- 172.Seidl H, Richtig E, Tilz H, et al. Profiles of chemokine receptors in melanocytic lesions: de novo expression of CXCR6 in melanoma. Hum Pathol. 2007;38(5):768–780. doi: 10.1016/j.humpath.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 173.Varney ML, Johansson SL, Singh RK. Distinct expression of CXCL8 and its receptors CXCR1 and CXCR2 and their association with vessel density and aggressiveness in malignant melanoma. Am J Clin Pathol. 2006;125(2):209–216. doi: 10.1309/VPL5-R3JR-7F1D-6V03. [DOI] [PubMed] [Google Scholar]
- 174.Longo-Imedio MI, Longo N, Trevino I, Lazaro P, Sanchez-Mateos P. Clinical significance of CXCR3 and CXCR4 expression in primary melanoma. Int J Cancer. 2005;117(5):861–865. doi: 10.1002/ijc.21269. [DOI] [PubMed] [Google Scholar]
- 175.Monteagudo C, Martin JM, Jorda E, Llombart-Bosch A. CXCR3 chemokine receptor immunoreactivity in primary cutaneous malignant melanoma: correlation with clinicopathologic prognostic factors. J Clin Pathol. 2007;60(6):596–599. doi: 10.1136/jcp.2005.032144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hoek KS. DNA microarray analyses of melanoma gene expression: a decade in the mines. Pigment Cell Res. 2007;20(6):466–484. doi: 10.1111/j.1600-0749.2007.00412.x. [DOI] [PubMed] [Google Scholar]
- 177.Sedghizadeh PP, Williams JD, Allen CM, Prasad ML. MSG-1 expression in benign and malignant melanocytic lesions of cutaneous and mucosal epithelium. Med Sci Monit. 2005;11(7):BR189–BR194. [PubMed] [Google Scholar]
- 178.Shen SS, Zhang PS, Eton O, Prieto VG. Analysis of protein tyrosine kinase expression in melanocytic lesions by tissue array. J Cutan Pathol. 2003;30(9):539–547. doi: 10.1034/j.1600-0560.2003.00090.x. [DOI] [PubMed] [Google Scholar]
- 179.Giehl KA, Nagele U, Volkenandt M, Berking C. Protein expression of melanocyte growth factors (bFGF, SCF) and their receptors (FGFR-1, c-kit) in nevi and melanoma. J Cutan Pathol. 2007;34(1):7–14. doi: 10.1111/j.1600-0560.2006.00569.x. [DOI] [PubMed] [Google Scholar]
- 180.Ivan D, Niveiro M, Diwan AH, et al. Analysis of protein tyrosine kinases expression in the melanoma metastases of patients treated with Imatinib Mesylate (STI571, Gleevec) J Cutan Pathol. 2006;33(4):280–285. doi: 10.1111/j.0303-6987.2006.00432.x. [DOI] [PubMed] [Google Scholar]
- 181.Woenckhaus C, Giebel J, Failing K, Fenic I, Dittberner T, Poetsch M. Expression of AP-2alpha, c-kit, and cleaved caspase-6 and -3 in naevi and malignant melanomas of the skin. A possible role for caspases in melanoma progression? J Pathol. 2003;201(2):278–287. doi: 10.1002/path.1424. [DOI] [PubMed] [Google Scholar]
- 182.Stefanou D, Batistatou A, Zioga A, Arkoumani E, Papachristou DJ, Agnantis NJ. Immunohistochemical expression of vascular endothelial growth factor (VEGF) and C-KIT in cutaneous melanocytic lesions. Int J Surg Pathol. 2004;12(2):133–138. doi: 10.1177/106689690401200206. [DOI] [PubMed] [Google Scholar]
- 183.Baldi A, Santini D, Battista T, et al. Expression of AP-2 transcription factor and of its downstream target genes c-kit, E-cadherin and p21 in human cutaneous melanoma. J Cell Biochem. 2001;83(3):364–372. doi: 10.1002/jcb.1235. [DOI] [PubMed] [Google Scholar]
- 184.Moretti S, Pinzi C, Spallanzani A, et al. Immunohistochemical evidence of cytokine networks during progression of human melanocytic lesions. Int J Cancer. 1999;84(2):160–168. doi: 10.1002/(sici)1097-0215(19990420)84:2<160::aid-ijc12>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 185.Montone KT, van Belle P, Elenitsas R, Elder DE. Proto-oncogene c-kit expression in malignant melanoma: protein loss with tumor progression. Mod Pathol. 1997;10(9):939–944. [PubMed] [Google Scholar]
- 186.Natali PG, Nicotra MR, Di Renzo MF, et al. Expression of the c-Met/HGF receptor in human melanocytic neoplasms: demonstration of the relationship to malignant melanoma tumour progression. Br J Cancer. 1993;68(4):746–750. doi: 10.1038/bjc.1993.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Puri N, Ahmed S, Janamanchi V, et al. c-Met is a potentially new therapeutic target for treatment of human melanoma. Clin Cancer Res. 2007;13(7):2246–2253. doi: 10.1158/1078-0432.CCR-06-0776. [DOI] [PubMed] [Google Scholar]
- 188.Sparrow LE, Heenan PJ. Differential expression of epidermal growth factor receptor in melanocytic tumours demonstrated by immunohistochemistry and mRNA in situ hybridization. Australas J Dermatol. 1999;40(1):19–24. doi: 10.1046/j.1440-0960.1999.00310.x. [DOI] [PubMed] [Google Scholar]
- 189.Marincola FM, Hijazi YM, Fetsch P, et al. Analysis of expression of the melanoma-associated antigens MART-1 and gp100 in metastatic melanoma cell lines and in in situ lesions. J Immunother Emphasis Tumor Immunol. 1996;19(3):192–205. doi: 10.1097/00002371-199605000-00004. [DOI] [PubMed] [Google Scholar]
- 190.Rakosy Z, Vizkeleti L, Ecsedi S, et al. EGFR gene copy number alterations in primary cutaneous malignant melanomas are associated with poor prognosis. Int J Cancer. 2007;121(8):1729–1737. doi: 10.1002/ijc.22928. [DOI] [PubMed] [Google Scholar]
- 191.Xerri L, Battyani Z, Grob JJ, et al. Expression of FGF1 and FGFR1 in human melanoma tissues. Melanoma Res. 1996;6(3):223–230. doi: 10.1097/00008390-199606000-00005. [DOI] [PubMed] [Google Scholar]
- 192.Xu X, Tahan SR, Pasha TL, Zhang PJ. Expression of neurotrophin receptor Trk-C in nevi and melanomas. J Cutan Pathol. 2003;30(5):318–322. doi: 10.1034/j.1600-0560.2003.00068.x. [DOI] [PubMed] [Google Scholar]
- 193.Innominato PF, Libbrecht L, van den Oord JJ. Expression of neurotrophins and their receptors in pigment cell lesions of the skin. J Pathol. 2001;194(1):95–100. doi: 10.1002/path.861. [DOI] [PubMed] [Google Scholar]
- 194.Dhawan P, Singh AB, Ellis DL, Richmond A. Constitutive activation of Akt/protein kinase B in melanoma leads to up-regulation of nuclear factor-kappaB and tumor progression. Cancer Res. 2002;62(24):7335–7342. [PubMed] [Google Scholar]
- 195.Singh RS, Diwan AH, Zhang PS, Prieto VG. Phosphoinositide 3-kinase is not overexpressed in melanocytic lesions. J Cutan Pathol. 2007;34(3):220–225. doi: 10.1111/j.1600-0560.2006.00592.x. [DOI] [PubMed] [Google Scholar]
- 196.Packer L, Pavey S, Parker A, et al. Osteopontin is a downstream effector of the PI3-kinase pathway in melanomas that is inversely correlated with functional PTEN. Carcinogenesis. 2006;27(9):1778–1786. doi: 10.1093/carcin/bgl016. [DOI] [PubMed] [Google Scholar]
- 197.Korabiowska M, Brinck U, Mirecka J, Kellner S, Marx D, Schauer A. Antigen Ki-67 and c-myc oncogene as related to histoclinical parameters in pigmented skin lesions. In Vivo. 1995;9(5):433–438. [PubMed] [Google Scholar]
- 198.Boni R, Bantschapp O, Muller B, Burg G. c-myc is not useful as prognostic immunohistochemical marker in cutaneous melanoma. Dermatology. 1998;196(3):288–291. doi: 10.1159/000017922. [DOI] [PubMed] [Google Scholar]
- 199.Konstadoulakis MM, Vezeridis M, Hatziyianni E, et al. Molecular oncogene markers and their significance in cutaneous malignant melanoma. Ann Surg Oncol. 1998;5(3):253–260. doi: 10.1007/BF02303782. [DOI] [PubMed] [Google Scholar]
- 200.Lazaris AC, Theodoropoulos GE, Aroni K, Saetta A, Davaris PS. Immunohistochemical expression of C-myc oncogene, heat shock protein 70 and HLA-DR molecules in malignant cutaneous melanoma. Virchows Arch. 1995;426(5):461–467. doi: 10.1007/BF00193169. [DOI] [PubMed] [Google Scholar]
- 201.Kalogeraki A, Garbagnati F, Darivianaki K, et al. HSP-70, C-myc and HLA-DR expression in patients with cutaneous malignant melanoma metastatic in lymph nodes. Anticancer Res. 2006;26(5A):3551–3554. [PubMed] [Google Scholar]
- 202.Redondo P, Sanchez-Carpintero I, Bauza A, Idoate M, Solano T, Mihm MC., Jr Immunologic escape and angiogenesis in human malignant melanoma. J Am Acad Dermatol. 2003;49(2):255–263. doi: 10.1067/s0190-9622(03)00921-6. [DOI] [PubMed] [Google Scholar]
- 203.Bayer-Garner IB, Hough AJ, Jr, Smoller BR. Vascular endothelial growth factor expression in malignant melanoma: prognostic versus diagnostic usefulness. Mod Pathol. 1999;12(8):770–774. [PubMed] [Google Scholar]
- 204.Vlaykova T, Laurila P, Muhonen T, et al. Prognostic value of tumour vascularity in metastatic melanoma and association of blood vessel density with vascular endothelial growth factor expression. Melanoma Res. 1999;9(1):59–68. doi: 10.1097/00008390-199902000-00008. [DOI] [PubMed] [Google Scholar]
- 205.Simonetti O, Lucarini G, Brancorsini D, et al. Immunohistochemical expression of vascular endothelial growth factor, matrix metalloproteinase 2, and matrix metalloproteinase 9 in cutaneous melanocytic lesions. Cancer. 2002;95(9):1963–1970. doi: 10.1002/cncr.10888. [DOI] [PubMed] [Google Scholar]
- 206.Pritchard-Jones RO, Dunn DB, Qiu Y, et al. Expression of VEGFb, the inhibitory isoforms of VEGF, in malignant melanoma. Br J Cancer. 2007;97(2):223–230. doi: 10.1038/sj.bjc.6603839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Demirkesen C, Buyukpinarbasili N, Ramazanoglu R, Oguz O, Mandel NM, Kaner G. The correlation of angiogenesis with metastasis in primary cutaneous melanoma: a comparative analysis of microvessel density, expression of vascular endothelial growth factor and basic fibroblastic growth factor. Pathology. 2006;38(2):132–137. doi: 10.1080/00313020600557565. [DOI] [PubMed] [Google Scholar]
- 208.Pisacane AM, Risio M. VEGF and VEGFR-2 immunohistochemistry in human melanocytic naevi and cutaneous melanomas. Melanoma Res. 2005;15(1):39–43. doi: 10.1097/00008390-200502000-00007. [DOI] [PubMed] [Google Scholar]
- 209.Marcoval J, Moreno A, Graells J, et al. Angiogenesis and malignant melanoma. Angiogenesis is related to the development of vertical (tumorigenic) growth phase. J Cutan Pathol. 1997;24(4):212–218. doi: 10.1111/j.1600-0560.1997.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 210.Birck A, Kirkin AF, Zeuthen J, Hou-Jensen K. Expression of basic fibroblast growth factor and vascular endothelial growth factor in primary and metastatic melanoma from the same patients. Melanoma Res. 1999;9(4):375–381. doi: 10.1097/00008390-199908000-00006. [DOI] [PubMed] [Google Scholar]
- 211.Salven P, Heikkila P, Joensuu H. Enhanced expression of vascular endothelial growth factor in metastatic melanoma. Br J Cancer. 1997;76(7):930–934. doi: 10.1038/bjc.1997.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Erhard H, Rietveld FJ, van Altena MC, Brocker EB, Ruiter DJ, de Waal RM. Transition of horizontal to vertical growth phase melanoma is accompanied by induction of vascular endothelial growth factor expression and angiogenesis. Melanoma Res. 1997;7(2 suppl 2):S19–S26. [PubMed] [Google Scholar]
- 213.Einspahr JG, Thomas TL, Saboda K, et al. Expression of vascular endothelial growth factor in early cutaneous melanocytic lesion progression. Cancer. 2007;110(11):2519–2527. doi: 10.1002/cncr.23076. [DOI] [PubMed] [Google Scholar]
- 214.Hafner C, Bataille F, Meyer S, et al. Loss of EphB6 expression in metastatic melanoma. Int J Oncol. 2003;23(6):1553–1559. [PubMed] [Google Scholar]
- 215.Easty DJ, Hill SP, Hsu MY, et al. Up-regulation of ephrin-A1 during melanoma progression. Int J Cancer. 1999;84(5):494–501. doi: 10.1002/(sici)1097-0215(19991022)84:5<494::aid-ijc8>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 216.Molinaro AM, Dudoit S, van der Laan MJ. Tree-based multivariate regression and density estimation with right-censored data. J Multivar Anal. 2004;90:154–177. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.