Abstract
Prokaryotic protein–protein interactions are underrepresented in currently available databases. Here, we describe a ‘gold standard’ dataset (MPI-LIT) focusing on microbial binary protein–protein interactions and associated experimental evidence that we have manually curated from 813 abstracts and full texts that were selected from an initial set of 36 852 abstracts. The MPI-LIT dataset comprises 1237 experimental descriptions that describe a non-redundant set of 746 interactions of which 659 (88%) are not reported in public databases. To estimate the curation quality, we compared our dataset with a union of microbial interaction data from IntAct, DIP, BIND and MINT. Among common abstracts, we achieve a sensitivity of up to 66% for interactions and 75% for experimental methods. Compared with these other datasets, MPI-LIT has the lowest fraction of interaction experiments per abstract (0.9) and the highest coverage of strains (92) and scientific articles (813). We compared methods that evaluate functional interactions among proteins (such as genomic context or co-expression) which are implemented in the STRING database. Most of these methods discriminate well between functionally relevant protein interactions (MPI-LIT) and high-throughput data.
Availability: http://www.jcvi.org/mpidb/interaction.php?dbsource=MPI-LIT.
Contact: raja@jcvi.org
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Microbes represent the vast majority of completely sequenced genomes (Peterson et al., 2001). Recent metagenomics projects have fortified this dominance even more with bacteria representing about 90% of all sequences in the global ocean sampling dataset (Yooseph et al., 2007). Clearly, an understanding of protein function and physiology requires a detailed understanding of their interactions with both other proteins and small molecules. Surprisingly, compared with eukaryotes the protein interactions of microbial species are largely unexplored: while the microbial interaction database (Goll et al., 2008) reports 22 000 microbial interactions, general interaction databases such as IntAct report on the order of 100 000 eukaryotic interactions. Although most interactions from high-throughput studies are reported in public databases, the majority of interactions from small-scale studies remain hidden in the primary scientific literature. Due to the ambiguity of free text, especially of protein names, species/strains and experimental methods, natural language processing algorithms are unable to reliably extract most interacting protein pairs and associated data automatically (Leitner and Valencia, 2008). While manual curation remains key, it poses a number of problems, including curation inconsistency, heterogenous levels of annotation depth and a large volume of text to be analyzed. Such manual curation of protein–protein and genetic interactions has been carried out for Saccharomyces cerevisiae (Reguly et al., 2006) and human (Peri et al., 2003). Here, we report a manually curated dataset (MPI-LIT) that we have extracted from 813 publications focusing on microbial species. This dataset comprises 1237 experimental descriptions (PubMed IDs and experimental methods) that link 940 proteins by 746 binary protein–protein interactions (Table 1). While our dataset does not appear to be large, it is the largest manually curated dataset published so far for microbial protein interactions and thus will serve as a ‘gold standard’ dataset. It can be used to evaluate interaction confidence assessment methods, to estimate the confidence of high-throughput microbial interaction datasets [e.g. generated by yeast-two-hybrid (Y2H) or complex purification studies] and to train automatic literature mining algorithms.
Table 1.
Literature-curation strategy
| Phase | Target | Publications | Experiments | Interactions | Proteins |
|---|---|---|---|---|---|
| I. Literature search | PubMed | 36 852 | – | – | – |
| II. Text analysis | Protein names, | 1732 | 2303 | 2289 | 4046 |
| species, methods | |||||
| III. Protein ID and | Mapped onto | ||||
| PSI-MI mapping | UniProt ID's and PSI-MI | 813 | 1237 | 746 | 940 |
| Microbial binary protein–protein interaction datasets | |||||
| Dataset | Species | Experimentsa | Interactions | Abstracts | Interactions/abstract |
|---|---|---|---|---|---|
| MPI-LIT | 92 | 1237 | 746 | 813 | 0.9 |
| MINT | 63 | 234 | 170 | 136 | 1.25 |
| DIP | 32 | 1404 | 1403 | 109 | 12.8 |
| BIND | 58 | 1576 | 1564 | 102 | 15.3 |
| IntAct | 73 | 13 887 | 13 242 | 196 | 67.5 |
| MPI-UNION | 142 | 15 848 | 15 077 | 501 | 30 |
aExperiment is a unique combination of an interaction, an experimental method and PubMed ID describing a protein–protein interaction.
2 METHODS
2.1 Literature curation strategy
Phase I: PubMed search. We started our curation effort by searching the primary literature via PubMed, similar to previous efforts for yeast (Reguly et al., 2006) and human (Peri et al., 2003). Using somewhat arbitrarily chosen keywords, our PubMed search for ‘ “bacteria” OR “Escherichia coli” OR “Salmonella” OR “Bacillus subtilis” OR “Pseudomonas” AND (interaction OR interact OR interacts OR bind OR binds)’, yielded 36 852 articles as of August 14, 2006 that potentially contain microbial protein interaction data.
Phase II: Text analysis. From these abstracts, we manually extracted the interacting protein pairs, the respective microbial species and the experimental method. During this phase, we were able to identify 4046 protein names and 2303 experimental descriptions (Table 1).
Phase III: Protein ID and controlled vocabulary mapping. As proteins were usually represented by their common names in the publications, we set up an automated protein identification pipeline. We systematically screened microbial versions of the UniProtKB/Swiss-Prot (UniProt-Cons, 2008) and Biothesaurus (Liu et al., 2006) databases to match proteins to the latest stable UniProt accessions based on their common names and species. Using this pipeline, we were able to uniquely identify 1254 proteins out of 4046 proteins (31%). For the remaining 2796 proteins (69%), we could not identify a unique UniProt ID automatically because the strain could not be uniquely identified. For example, a UniProt search for E. coli and RecA results in 11 different UniProt entries from 10 different strains. We addressed such cases by manually mapping the proteins to primary UniProt accessions. Out of the 2796 unassigned proteins, we were able to uniquely identify UniProt accessions for 1790 (44%) proteins. 1006 (25%) proteins could not be matched at all and these proteins were removed from the final curated dataset. Such deleted entries include non-protein entities that were initially identified as proteins such as small-molecules or protein complexes (‘RNA polymerase’), non-microbial proteins and misspelled common names. Independently, we manually mapped the free-text curated experimental methods onto experimental methods defined by the PSI-MI (Proteomics Standards Initiative–Molecular Interactions) controlled vocabulary (Kerrien et al., 2007a).
Interaction versus Experiment: Interactions are defined as unique pairs of UniProt accessions. An interaction experiment is defined by an interaction, an experimental method (PSI-MI) and a publication (PubMed ID). An interaction can be described by more than one experiment whenever such an interaction is reported by a different method and/or different study.
2.2 Datasets
The MPI-UNION dataset has been downloaded from the MPIDB database (Goll et al., 2008) and is the microbial subset of IntAct, DIP, BIND and MINT (as of December 4, 2007) (Alfarano et al., 2005;; Chatr-aryamontri et al., 2007; Kerrien et al., 2007b; Salwinski et al., 2004) filtered for binary interactions i.e. the direct physical associations between two proteins were experimentally characterized. The E. coli K12 STRING scores were downloaded from the STRING database (version 7.1) (von Mering et al., 2007). Escherichia coli K12 gene ontology (GO) annotations were collected from the Gene Ontology Annotation (GOA) Database (as of April 1, 2008).
2.3 GO term enrichments
We used the topGO R package (Alexa et al., 2006) to detect significantly enriched GO terms in the E. coli K12 MPI-LIT subset when compared with all E. coli K12 genes. The classic algorithm based on gene count using the elim method was applied to minimize the false-positive rate (Alexa et al., 2006). The degree of over-representation is assessed with a statistical score. Here, the score is the P-value returned by Fisher's exact test. A GO term was marked as significant when its P-value was smaller than 0.01.
2.4 Interologs
We used the PORC (Putative ORthologous Clusters) database to identify pairs of interacting orthologs (interologs). Compared with other pre-computed clusters such as COGs (Cluster of Orthologous Groups), PORCs only contain one sequence per species.
Data was downloaded from ftp://ftp.ebi.ac.uk/pub/databases/integr8/porc (as of June 22, 2008).
3 RESULTS
3.1 Literature curation quality assessment
3.1.1 Abstract overlap
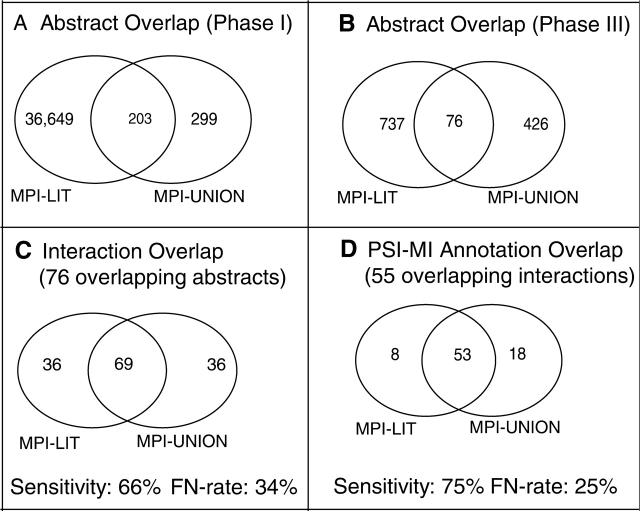
Our PubMed search retrieved 36 852 abstracts of which only 203 (=0.6%) are reported in the MPI-UNION dataset (see Section 2.2). Our PubMed search missed 299 abstracts that are reported to contain microbial interactions in MPI-UNION. This indicates that our query missed some relevant abstracts, probably because of the choice of search terms (see Section 2.1) (Fig. 1A). This might be due to the fact that the search was limited to abstracts. Articles which describe interactions in the full text or even supplementary information, which is true for most of the medium and high-throughput protein-protein interaction (PPI) studies, would have been missed. During Phases II and III of the curation process, we were able to remove non-relevant abstracts and those for which we could not uniquely identify the interacting proteins (Table 1). After Phase III, 813 abstracts (Fig. 1B) describing microbial protein–protein interactions remained. To assess the curation quality, we compared the 76 MPI-LIT articles overlapping with MPI-UNION (Fig. 1B).
Fig. 1.
(A) Overlap between abstracts retrieved from our initial PubMed search and abstracts indexed in ‘MPI-UNION’ (i.e. the union of binary interactions from IntAct, DIP, BIND and MINT). (B) The overlap between abstracts curated for MPI-LIT and MPI-UNION describing protein interactions. (C) Assessment of the sensitivity and false-negative rate of interaction curation. Here, we compared the curation fidelity of overlapping abstracts in MPI-LIT and MPI-UNION: from the 76 overlapping abstracts (Fig. 1B) 69 Interactions were curated both by MPI-LIT and MPI-UNION and 36 unique interactions were curated only by MPI-LIT and MPI-UNION, respectively. (D) Assessment of the sensitivity and false-negative rate of mapping PSI-MI methods to each interaction. We could map experimental methods to 55 out of 69 overlapping interactions (Fig. 1C). Fifty-three of these methods were identical in MPI-LIT and MPI-UNION.
3.1.2 Interaction curation
We estimated the sensitivity of interaction curation by comparing the number of interactions overlapping between MPI-LIT and MPI-UNION. The MPI-LIT curation efforts achieve a sensitivity of 66%. That is, we identified 66% of the reference MPI-UNION interactions (note that strains were merged into species and common protein names were used for the comparison). Vice versa, the MPI-UNION obtained the same sensitivity when using MPI-LIT as a reference. This indicates that independent literature curation efforts, MPI-LIT and MPI-UNION, miss an estimated 34% of interactions (false negatives) (Fig. 1C). A possible reason for false negatives in MPI-LIT is that the Phase II curation was limited to abstracts. If interactions are described in the full text and not mentioned in the abstracts, curators failed to report the interactions. However, we curated full text for all the online available articles in Phase III. Interestingly, both MPI-LIT and MPI-UNION curated 36 interactions each from the common set of 76 articles that were unique to one of the two datasets (Fig. 1C). When we re-examined these interactions in the primary articles, four out of the 36 unique MPI-LIT interactions (which are not reported in MPI-UNION) turned out to be false-positives. The estimated false-positive rate for MPI-LIT is thus 4% (based on four false-positives out of 105 interactions, Fig. 1C). A similar rate (4%) of manual curation errors was also reported in a previous study (Reguly et al., 2006). Vice versa, out of the 36 unique MPI-UNION interactions, one interaction turned out to be false. The estimated false-positive rate for the MPI-UNION dataset is thus 1% (i.e. one false-positive out of 105 interactions). False positives in the MPI-LIT dataset can usually be explained by either curator typos (2%) or wrong protein ID mapping (2%). These wrong entries were removed from the final dataset.
3.1.3 Method curation
Based on 76 articles that are common to both MPI-LIT and MPI-UNION, 69 common interactions have been curated from these articles (Fig. 1C), ignoring strain variations. Of these, 55 interactions were identical when strains were considered too. Each protein interaction can have more than one experimental method if the same interaction is reported from more than one study or from a different experiment. We estimated the sensitivity of experimental method annotation by comparing the PSI-MI terms of these 55 interactions (Fig. 1D). In total, 79 experimental methods were curated for the 55 interactions of the MPI-LIT and MPI-UNION datasets. We mapped all methods onto the first level of the hierarchically organized PSI-MI controlled vocabulary, i.e. biophysical, protein complementation assay, genetic interference, post-transcriptional interference, biochemical, and imaging techniques. For such a merged set, we estimate the average sensitivity to be on the order of 75% for MPI-LIT using MPI-UNION as a reference set. Although, we assume that we have missed a small fraction of experimental descriptions, all the curated descriptions are true positives (Supplementary Table S1).
3.2 The MPI-LIT dataset
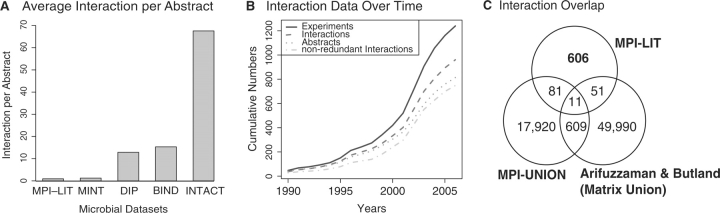
The MPI-LIT dataset covers 1237 experimental descriptions comprising 746 non-redundant bacterial PPIs involving 940 full-length proteins of 92 species/strains extracted from 813 articles (Supplementary Table S2). The coverage of abstracts and species/strains is significantly higher than those compiled by curation efforts represented in the MPI-UNION dataset (Table 1). Notably, the 746 PPIs in MPI-LIT are supported by 1237 experiments. This indicates that on average an interaction is either confirmed by more than one experimental method and/or by multiple publications (Table 1). Most of the interactions in MPI-LIT are reported for E. coli (54%, all strains), B. subtilis (11%) and Salmonella typhimurium (3%) (Supplementary Table S3a). On average, we identified 0.9 non-redundand interactions per article, reflecting the small-scale nature of the source articles. Within MPI-UNION, MINT curated an average of 1.25 non-redundand interactions per article, whereas 13, 15 and 67 were reported on average by DIP, BIND and IntAct, respectively. This indicates an increasing focus on interactions derived from high-throughput experiments (Fig. 2A).
Fig. 2.
(A) Number of interactions per publication in MPI-LIT, MINT, DIP, BIND and IntAct datasets. (B) Graph showing the accumulation of curated microbial protein interaction data over time. (C) Overlapping interactions between different datasets. The MPI-UNION dataset is a microbial subset of IntAct, DIP, BIND and MINT, filtered for binary interactions. Matrix Union is the binary protein interaction dataset predicted for E. coli complex purification data from Arifuzzaman et al. and Butland et al. (Arifuzzaman et al., 2006; Butland et al., 2005) using the matrix model of protein interactions, i.e. assuming that every protein interacts with every other protein in a complex.
We wondered whether certain molecular functions, cellular components and biological processes are enriched in the literature curated dataset. To investigate this, we looked for significantly enriched GO terms in each of the three subontologies. We did this by comparing the frequency of GO terms of genes that are present in the MPI-LIT subset of E. coli K12 interactions with those present in the whole E. coli K12 genome (see Section 2). Table 2 lists the top ten enriched GO terms for the Biological Process subontology. A broad range of processes have been enriched, including ‘protein folding’ and ‘DNA replication’. Processes related to ‘intracellular protein transport across a membrane’ were enriched as well. The presence of proteins involved in most of the cellular processes indicates that there is no strong bias towards certain functional groups in the curated dataset. As expected, in the GO molecular function and cellular component categories ‘protein binding’ and ‘protein complex’ were found to be highly enriched, reflecting the protein interaction nature of the dataset. A list of all enriched terms and highlighted GO graphs can be found in Supplementary Table S4 (Supplementary Figure 1).
Table 2.
Top ten enriched GO terms for the Biological Process subontology
| GO ID | Term | Genome | MPI-LIT | MPI-LIT | P<0.01 |
|---|---|---|---|---|---|
| observed | Expected | ||||
| 0009432 | SOS response | 17 | 14 | 1.99 | 3.50E-11 |
| 0006935 | chemotaxis | 22 | 15 | 2.58 | 6.40E-10 |
| 0065002 | intracellular | 11 | 10 | 1.29 | 4.30E-09 |
| protein transport | |||||
| 0007049 | cell cycle | 57 | 24 | 6.68 | 2.00E-07 |
| 0051301 | cell division | 52 | 24 | 6.09 | 3.50E-07 |
| 0006457 | protein folding | 28 | 14 | 3.28 | 6.10E-07 |
| 00069501 | response to stress | 153 | 53 | 17.93 | 2.00E-06 |
| 0006281 | DNA repair | 68 | 28 | 7.97 | 3.70E-06 |
| 0006260 | DNA replication | 69 | 33 | 8.09 | 8.30E-06 |
| 0006352 | transcription | 7 | 7 | 0.82 | 1.60E-05 |
| initiation |
All numbers refer to proteins interacting in E. coli K12. Genome gives the total number of genes in this category.
We wondered how many of MPI-LIT interactions are recovered in high-throughput bacterial interactome studies. In fact, there is only little overlap with existing high-throughput datasets. For example, of the 355 E. coli K12 interactions in MPI-LIT, only 62 interactions have been found by E. coli complex purification studies (using predicted binary interactions based on the matrix model; Figure 2C). Next, we used Orthology as defined by the PORC database to predict homologous interactions (http://www.ebi.ac.uk/clustr/). Surprisingly, only 2 out of 45 predicted interactions from MPI-LIT were found in a high-throughput study of Treponema pallidum (Titz et al., 2008), 3 out of 58 in Campylobacter jejuni (Parrish et al., 2007), 3 out of 56 in Helicobacter pylori (Rain et al., 2001) and 2 out of 52 in Synechocystis sp. PCC6803 (Sato et al., 2007) (Table 3). There are two possible explanations for this observation: first, these large-scale datasets recovered only a small fraction of all interactions in these species with a large false-negative rate. In fact, we have shown that systematic Y2H screens using full-length proteins probably recover no more than 20–30% of all interactions (Rajagopala et al., 2007). Second, many interactions in bacteria may actually not be conserved between distantly related species such as T. pallidum and E. coli. Most likely, both factors play an important role and each contribution remains to be determined. However, the MPI-LIT dataset covers a relatively small number of interactions and yet a large diversity of species, thus more data and/or curation are needed to substantiate our findings.
Table 3.
MPI-LIT interologs
3.3 Benchmarking PPI confidence estimation methods
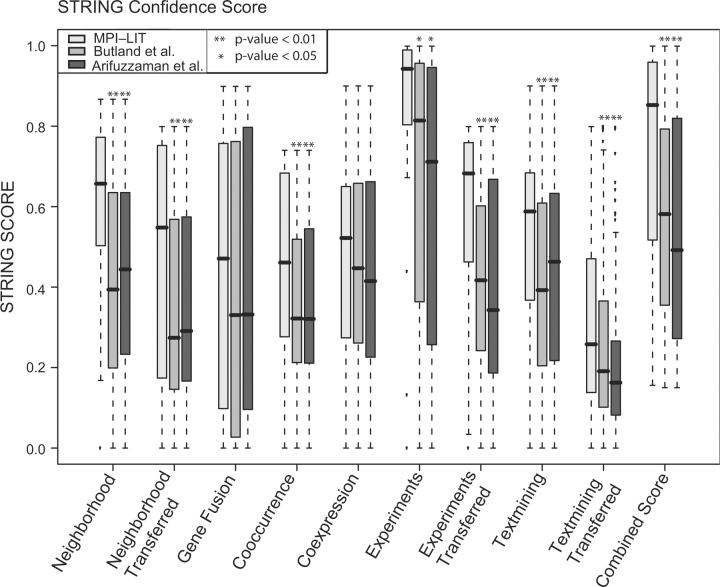
The advance of high-throughput experimental techniques such as Y2H assays (Fields and Song, 1989) and co-immunoprecipitation (Arifuzzaman et al., 2006; Butland et al., 2005) screens has led to the elucidation of large-scale protein interaction networks in different bacterial species (Parrish et al., 2007; Rain et al., 2001; Titz et al., 2008). Unfortunately, in high-throughput screens false positives and false negatives are nearly inevitable. Many computational methods have been proposed to estimate the biological relevance or confidence of high-throughput protein interaction data (Suthram et al., 2006; von Mering et al., 2007). Among them are methods that measure the degree of co-occurrence, gene-neighborhood, gene-fusion, co-expression, interologs, co-pathway membership, co-citation and co-annotation of interacting proteins. Our literature curated list of protein–protein interactions represents biologically relevant interactions as shown by most of the publications we curated. We wondered whether we could use the MPI-LIT dataset to validate confidence estimation methods such as those used by the STRING database of protein–protein associations (von Mering et al., 2007). Most confidence scores have been pre-computed for a variety of microbial genomes in STRING. For each of these methods we obtained values for an E. coli K12 subset of MPI-LIT (355 interactions, 47.6% of all current MPI-LIT interactions) and compared them with those obtained for binary interactions derived from two E. coli high-throughput pull-down experiments (Arifuzzaman et al., 2006; Butland et al., 2005) using the SPOKE model (boxplots for all scores are shown in Fig. 3, statistics can be found in Supplementary Table S5). Known protein interactions in the STRING database have primarily been imported from other interaction databases. Note that 88% of the MPI-LIT interactions are not reported in these databases, so the STRING database does not know that 88% of the MPI-LIT interactions are biologically relevant interactions. Thus, we expected that STRING yields a higher confidence score for MPI-LIT compared with high-throughput interaction data (Arifuzzaman et al., 2006; Butland et al., 2005). One-sided two sample Mann–Whitney U-tests revealed that STRING's gene neighbourhood, co-occurrence, experiments and text mining methods scored MPI-LIT interactions significantly better than either of the high-throughput pull-down datasets (Fig. 3, Mann–Whitney U-test, P<0.05), indicating that the STRING methods are well suited for data quality estimation. In contrast, the gene fusion and co-expression were not able to clearly separate the MPI-LIT and high-throughput datasets. Overall, STRING's probabilistic combined score discriminates very well between MPI-LIT and inferred interactions from high-throughput pull-down experiments (P<0.01 for either pull-down datasets) underlining STRING's usefulness for interaction confidence estimation.
Fig. 3.
Assessment of criteria implemented in the STRING database to score functional associations of proteins. Here, we used the MPI-LIT dataset and two high-throughput complex purification datasets of E. coli (Arifuzzaman et al., 2006; Butland et al., 2005) to asses the protein–protein associations by STRING database. For all methods, except for gene fusion, and co-expression, the MPI-LIT dataset scored significantly better than either of the pull-down datasets (Mann–Whitney U-test, P<0.05). The line in the box indicates the median value (statistics can be found in Supplementary Table S5).
4 DISCUSSION AND CONCLUSIONS
While our dataset is relatively small, it is the largest manually curated functionally validated protein interaction dataset for microbial proteins and thus can serve as a gold standard dataset of true positive microbial interactions. Our curation effort attempted to collect biologically relevant interactions as is shown by most of the publications we curated. However, there are other ways of obtaining gold standard datasets for protein–protein interactions. Edwards et al. used structural data as gold standards (Edwards et al., 2002). Unfortunately, there are not that many crystal structures available of microbial protein complexes (439 unique 3D complexes based on the UniProt Knowledgebase Release 13.1). Hence, structural biology is still of limited use, although this may change with the shift of structural genomics towards complexes. Another source of gold standard interactions are protein pairs that are supported by multiple experiments (Han et al., 2004). We have integrated our literature data with additional evidences such as 3D structures, interaction conservation, co-purification and predicted interacting domains. The integrated MPI-LIT dataset can be filtered for such supporting evidences and queried at the Microbial Protein Interaction Database at www.jcvi.org/mpidb (Goll et al., 2008).
Considering the fact that small-scale protein interaction studies are usually believed to be of higher quality than high-throughput data, the MPI-LIT dataset can be used as training set for PPI literature mining algorithms, as a ‘gold standard’ dataset for PPI confidence estimations (as shown in Section 3.3), to predict interologs for other species and for integrative bioinformatics analysis. Given our focus on microbial interactions, we plan to continue our curation efforts and will initially focus on the Journal of Bacteriology and Molecular Microbiology to increase the coverage of this reference dataset. We also started to coordinate our literature curation activity as an observer member of the IMEx consortium (http://imex.sourceforge.net/index.html).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Harihara Dixit, Srikanta Dixit, Lakshmi C. Madhusudana and Guruprasad B. Rajegowda from Indgen Life Technologies, Mysore, for their support with literature curation. This project was supported by The J. Craig Venter Institute, Indgen Life Technologies, and NIH grant RO1GM79710.
Conflict of Interest: none declared.
REFERENCES
- Alexa A, et al. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- Alfarano C, et al. The biomolecular interaction network database and related tools 2005 update. Nucleic Acids Res. 2005;33(Database issue):D418–D424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arifuzzaman M, et al. Large-scale identification of protein--protein interaction ofEscherichia coliK-12. Genome Res. 2006;16:686–691. doi: 10.1101/gr.4527806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butland G, et al. Interaction network containing conserved and essential protein complexes inEscherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- Chatr-aryamontri A, et al. MINT: the molecular interaction database. Nucleic Acids Res. 2007;35(Database issue):D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AM, et al. Bridging structural biology and genomics: assessing protein interaction data with known complexes. Trends Genet. 2002;18:529–536. doi: 10.1016/s0168-9525(02)02763-4. [DOI] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Goll J, et al. MPIDB: the microbial protein interaction database. Bioinformatics. 2008;24:1743–1744. doi: 10.1093/bioinformatics/btn285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J-DJ, et al. Evidence for dynamically organized modularity in the yeast protein–protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- Kerrien S, et al. Broadening the horizon--level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol. 2007a;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrien S, et al. IntAct--open source resource for molecular interaction data. Nucleic Acids Res. 2007b;35(Database issue):D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner F, Valencia A. A text-mining perspective on the requirements for electronically annotated abstracts. FEBS Lett. 2008;582:1178–1181. doi: 10.1016/j.febslet.2008.02.072. [DOI] [PubMed] [Google Scholar]
- Liu H, et al. Biothesaurus: a web-based thesaurus of protein and gene names. Bioinformatics. 2006;22:103–105. doi: 10.1093/bioinformatics/bti749. [DOI] [PubMed] [Google Scholar]
- Parrish JR, et al. A proteome-wide protein interaction map forCampylobacter jejuni. Genome Biol. 2007;8:R130. doi: 10.1186/gb-2007-8-7-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peri S, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson JD, et al. The comprehensive microbial resource. Nucleic Acids Res. 2001;29:123–125. doi: 10.1093/nar/29.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rain JC, et al. The protein–protein interaction map ofHelicobacter pylori. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- Rajagopala SV, et al. The protein network of bacterial motility. Mol. Syst. Biol. 2007;3:128. doi: 10.1038/msb4100166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguly T, et al. Comprehensive curation and analysis of global interaction networks inSaccharomyces cerevisiae. J. Biol. 2006;5:11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salwinski L, et al. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32(Database issue):D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, et al. A large-scale protein protein interaction analysis inSynechocystis sp. PCC6803. DNA Res. 2007;14:207–216. doi: 10.1093/dnares/dsm021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthram S, et al. A direct comparison of protein interaction confidence assignment schemes. BMC Bioinform. 2006;7:360. doi: 10.1186/1471-2105-7-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titz B, et al. The binary protein interactome ofTreponema pallidum–the syphilis spirochete. PLoS ONE. 2008;3:e2292. doi: 10.1371/journal.pone.0002292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt-Consortium. The universal protein resource (uniprot) Nucleic Acids Res. 2008;36(Database issue):D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering, et al. STRING 7–recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35(Database issue):D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yooseph S, et al. The sorcerer II global ocean sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.