Abstract
Autophagy adjusts cellular biomass and function in response to diverse stimuli, including infection. Autophagy plays specific roles in shaping immune system development, fueling host innate and adaptive immune responses, and directly controlling intracellular microbes as a cell-autonomous innate defense. As an evolutionary counterpoint, intracellular pathogens have evolved to block autophagic microbicidal defense and subvert host autophagic responses for their survival or growth. The ability of eukaryotic pathogens to deploy their own autophagic machinery may also contribute to microbial pathogenesis. Thus, a complex interplay between autophagy and microbial adaptations against autophagy governs the net outcome of host-microbe encounters.
Introduction
Autophagy is a fundamental biological process that simultaneously touches on multiple aspects of eukaryotic cells and metazoan organisms. In its most generic rendition, autophagy is a process that controls the quality and quantity of intracellular biomass in eukaryotic cells by targeting for autodigestion cytoplasmic components that range in complexity and size from individual proteins to whole organelles. At one end of the spectrum, a delicate process termed chaperone-mediated autophagy directly imports individual cytosolic proteins that contain specific recognition motif sequences into the lysosome. At the other end of the spectrum, macroautophagy acts as a bulk process that captures large portions of the cytosol or sequesters big organelles such as mitochondria and peroxisomes (Figure 1). The general term "autophagy" usually denotes canonical macro-autophagy characterized by its marquee feature, the double-membrane autophagosome. It has also become increasingly evident that autophagy, through its regulators comprised of Atg (Autophagy) and additional factors, interacts in a number of previously unappreciated ways with other pathways and processes in the host cell that do not always easily fit under "autophagy" as defined above. In these secondary roles, Atg proteins interact with other systems in the cell to coordinate various cellular functions (including immunological processes) with classical autophagy functions. Of note, it has also been proposed that Atg factors may have yet a third set of functions completely unrelated to autophagy or coordination between autophagy and other systems, referred to as autophagy-independent functions of Atg genes (Virgin and Levine, 2009).
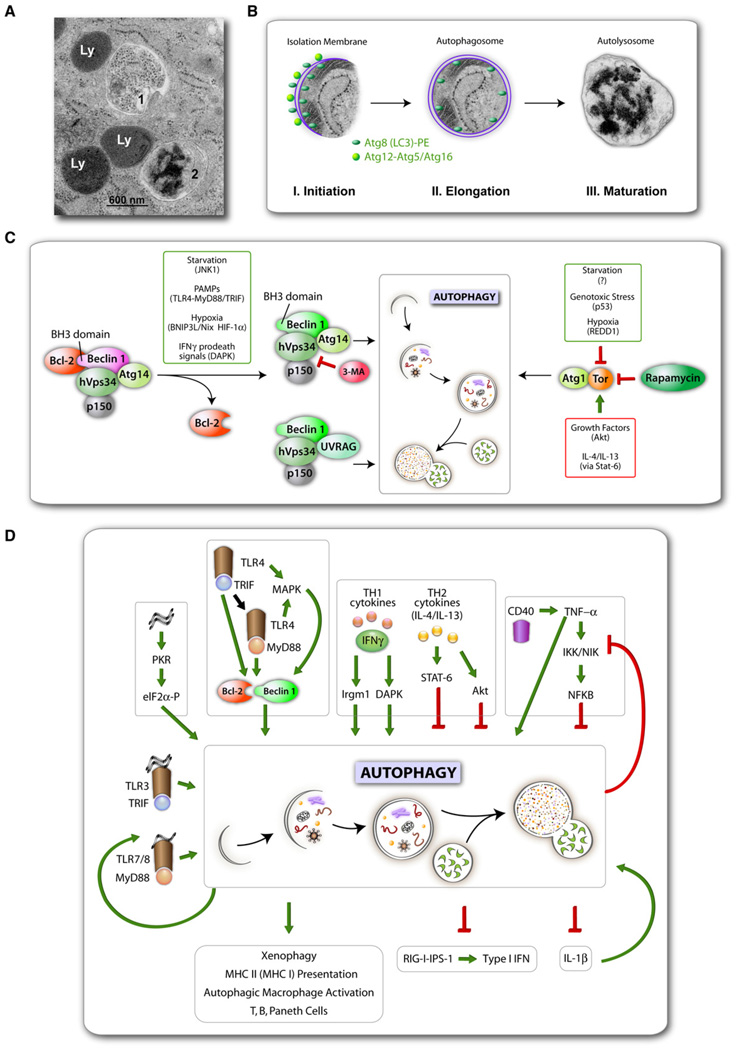
Figure 1. Autophagy Stages, Regulation, and Roles in Immunity.
(A) Electron micrograph comparing the appearance of an autophagosome (1) versus autolysosome (2) with conventional lysosomes (Ly).
(B) A composite of autophagic membrane formation (schematic) with actual autophagosome from an electron micrograph. Three stages can be discerned by ultrastructural morphology: initiation (crescent membrane decorated on both ssides with Atg protein-lipid [Atg8-PE; known as LC3-II] and protein-protein conjugates [Atg12-Atg5, complexed with Atg16]), elongation (growth of isolation membrane) ending in its closure to form an autophagosome, and maturation that involves the formation of degradative autolysosomes through fusion of autophagosomes with lysosomal organelles (electron-dense granular material represents ribosomal degradation intermediates). All micrographs in (A) and (B) are courtesy of Eeva-Lisa Eskalinen (reproduced with permission)
(C) Two signaling systems control induction of autophagy: left, hVps34-Beclin 1; right, Tor-Atg1. Various signals and systems transmitting them that lead to autophagy activation are shown in smaller boxes. Rapamycin induces autophagy by inhibiting Tor, and some immune signals that are transmitted via TLR adapters MyD88 and TRIF or DAPK downstream of IFNγ lead to activation of Beclin 1 (complexed with the phosphatidylinositol 3 kinase hVps34) by its dissociation from Bcl-2. Note that starvation, a classical inducer of autophagy, affects both Bcl-2-Beclin 1 complex (via JNK1) and Tor activity. Th2 cytokines and many growth factors in general inhibit autophagy via Tor (or in the case of immunologically induced autophagy, e.g., by IFNγ, via Stat-6 downstream of IL-4/IL-13).
(D)Immunological inputsand outputs of autophagy. Xenophagy,autophagic Macrophage activation, additional terms, factors, and relationships are described in the text.
A rapidly developing area of autophagy research is the study of immunological functions of autophagy (Levine and Deretic, 2007; Munz, 2009). Since the known immunological functions of chaperone-mediated autophagy (e.g., major histocompatibility complex [MHC] II-restricted endogenous antigen presentation) overlap with a subset of macroautophagy roles, for the purpose of this review, chaperone-mediated autophagy will not be distinguished from macroautophagy. Macroautophagy, which is usually referred to simply as autophagy (with the caveat in the above paragraph), is controlled by several signaling systems relaying nutritional or stress inputs to the core executioner machinery composed of Atg factors, which in turn drive the generation of autophagic organelles to sequester and degrade cytoplasmic targets (Figures 1A and 1B). This homeostatic role of autophagy affects cell survival (Kroemer and Levine, 2008), is reflected in the complexity of its regulation (Figure 1C), and represents the underpinnings of the role of autophagy in health and disease (Levine and Kroemer, 2008; Mizushima et al., 2008), including aging, cancer, neurodegenerative disorders, immunity, infectious diseases, and chronic inflammatory conditions such as Crohn’s disease (CD).
The multilayered intersections (Figure 1D) between immunity and autophagy (the term autophagy being used here in a broad sense, including all functions of Atg factors) span phenomena ranging from cell-autonomous defenses to functions of the entire immune system and can manifest themselves in adaptive and innate immunity, in regulatory and effector immune functions, and in tolerance versus immune activation and inflammation. At the level of the whole immune system, autophagy contributes to positive and negative selection of the CD4 T cell repertoire (Nedjic et al., 2008) and T and B cell homeostasis (Li et al., 2006; Miller et al., 2008; Pua et al., 2007; Pua and He, 2007). Autophagy enables endogenous MHC II antigen presentation (Schmid and Munz, 2007), thus governing thymic selection and central tolerance (Nedjic et al., 2008). This function of autophagy involves the delivery of cytosolic proteins to the lumen of MHC II antigen processing and loading compartments, extends to MHC I presentation (English et al., 2009), affects generation of optimal immune responses to pathogens, and may be of significance for vaccine development (Jagannath et al., 2009; Schmid et al., 2007). Autophagy is furthermore an effector of Th1/Th2 polarization enabling (Andrade et al., 2006; Gutierrez et al., 2004; Ling et al., 2006) or disabling macrophages to utilize autophagy in control of intracellular pathogens (Harris et al., 2007). In innate immunity, autophagy is both a regulator (Jounai et al., 2007; Lee et al., 2007; Tal et al., 2009) and an effector of pattern recognition receptor (PRR) responses to pathogen- (Delgado et al., 2008; Sanjuan et al., 2007; Xu et al., 2007; Yano et al., 2008) and possibly to danger-associated molecular patterns (PAMPs and DAMPs, respectively) (Biswas et al., 2008; Saitoh et al., 2008). Furthermore, autophagy acts as a cell-autonomous defense directly eliminating intracellular microbes or their products, including bacteria, viruses, and protozoa (Levine and Deretic, 2007), in a process termed xenophagy (Levine, 2005).
Clearly, our knowledge of the immunological functions of autophagy continuestorapidly increaseinscope and diversity(Levine and Deretic, 2007). This review summarizes what has been learned regarding the role of autophagy in various branches of immunity and delves into the mechanisms as they pertain to host-pathogen interactions. We explore in depth the status of autophagy as a cell-autonomous innate immunity defense against intracellular pathogens vis-à-vis adaptations that have evolved in pathogens to counter autophagy. These adaptations enable successful intracellular parasites to effectively protect themselves against autophagic inhibition or degradation and even harness autophagy for their own replication. We also highlight another emerging theme: intracellular eukaryotic parasites undergo autophagy themselves, leading to situations where autophagy contributes to both sides of the struggle in host-pathogen interactions.
Autophagy in the Host Cell
The cellular process of autophagy evolved at the beginning of eukaryotic life. The ability of viruses and bacteria to gain access to the interior of the eukaryotic cell generated selective pressures for the development of an effective cellular mechanism to dispose of such microbes. In this context, autophagy may have evolved as a primordial antimicrobial defense mechanism that can degrade intracellular parasites. At the transition to metazoan life, with the evolution of more specialized cells and immune systems, the functions of autophagy broadened so as to maintain its role at the nexus of more complex immune systems. Not only does autophagy function in a cell-autonomous manner to degrade intracellular pathogens, it also helps orchestrate the systemic immune response by functioning as a regulator of innate immunity, adaptive immunity, and inflammation. In this section, we will provide an overview of autophagy in the host cell, with an emphasis on its core molecular mechanisms, its broad roles in immunity, its newly emerging roles in the control of inflammation, and its most primal role as a cell-autonomous mechanism for eliminating intracellular microbes.
Autophagy as a Cell Biological Process
A key morphological manifestation of autophagy is the formation of autophagic organelles in the cytoplasm. They begin as short-lived membrane crescents, termed phagophores or isolation membranes, which give rise to double-membraned autophago-somes (Figure 1B). The autophagosomes undergo maturation into autolysosomes by fusion with lysosomal organelles, followed by loss of the inner of two membranes and degradation of the captured cytoplasmic target(s). While autophagosomes are considered the morphological hallmark of autophagy, their turnover is quite rapid, and often what are actually seen by electron microscopy are autolysosomes. Typically, these structures are delimited by a single membrane and contain lumens packed with remnants of cytoplasmic components, including shredded membranes and electron-dense material from decomposing ribosomes (Figure 1A). The origin of the autophagosomal phag-ophore membrane is not known with certainty, but both older and more recent data suggest involvement of the endoplasmic reticulum (ER) (Axe et al., 2008; Matsunaga et al., 2009).
Autophagosome formation is believed to be driven by two protein-protein and protein-lipid conjugation systems (Figure 1B). The first system yields an Atg5-Atg12 covalent conjugate that associates noncovalently with Atg16L1 (the mammalian equivalent of yeast Atg16) to form a putative E3 enzyme (terminology borrowed from the ubiquitin system) (Hanada et al., 2007), directing the site of the formation of the second protein-lipid conjugate (Fujita et al., 2008). The second system yields LC3-II (Atg8-PE), which has phosphatidylethanolamine at its C terminus that allows it to associate with or assist in autophagic membrane growth. Once formed and membrane-bound, LC3-II becomes an autophagosomal structural protein that executes other functions by interacting with the WXXL motif in the adaptor molecules p62 (Pankiv et al., 2007) and NBR1 (Kirkin et al., 2009), which capture cytoplasmic cargo earmarked for autophagic degradation, such as protein aggregates that are too large for proteasomal degradation. The partnering of LC3 with p62 and NBR1 suggests one potential mechanism by which autophagic targets may be recognized. Although it is not known whether these tag-receptor pairs are ubiquitously used to slate other targets, such as organelles, for autophagy, monoubiqutination has been shown to be sufficient to target an ectopically expressed cytoplasmic protein as well as peroxisomes for autophagic degradation (Kim et al., 2008). To date, no molecular tags of this nature have been reported on a microbe to guide autophagosomes with precision to their microbial targets. However, it is reasonable to speculate that some of the same molecular tags used to deliver cytoplasmic constituents may also be used to deliver microbes to the autophagosome. Yet PAMPs released by microbes stimulate autophagy (Delgado et al., 2009; Yano et al., 2008), and we cannot exclude the possibility that the execution stages of the process may be more stochastic and less perfectly guided toward specific microbial targets.
A multitude of nutritional and stress inputs transduced through protein and lipid kinase signaling cascades that regulate autophagy converge upon two key signaling nodes (Figure 1C): (1) Tor-Atg1 and (2) Beclin 1(Atg6)-hVps34. The Tor-Atg1 system transduces growth, nutritional, and some stress signals to initiate autophagy. The metabolic aspects of autophagy are under negative control by the growth factors, insulin receptor substrate, type I phosphatidylinositol 3 kinase (PI3K), Akt/PKB, and the downstream Tor-Atg1 signaling cascade. How Atg1 sets in motion other Atg factors and downstream morphologically distinguishable events is still not completely clear. It possibly regulates multiprotein complex formation involving a number of other Atg proteins initially recognized only in yeast (Kawamata et al., 2008), but which now appear to have counterparts in mammalian cells (Hosokawa et al., 2009).
The Beclin 1-hVps34 represents another key regulatory node centered on an ancient stress-signaling lipid kinase, known as type III PI3K Vps34. The exact role of the enzymatic product of hVps34 is not fully understood, but it likely plays a pivotal role in early autophagosomal membrane formation (Axe et al., 2008), the targeting of PI3P–binding proteins such as Atg18 to the autophagic membrane (Obara et al., 2008), the localization of LC3 lipidation (Fujita et al., 2008), and autophagosomal maturation into autolysosomes (Liang et al., 2008). Beclin 1 is a key regulator of autophagy, and it exists in functionally distinct hVps34-containing protein complexes (Figure 1C), including several modifier components: (1) Atg14, which plays a role in initiation (Itakura et al., 2008; Matsunaga et al., 2009; Zhong et al., 2009), and (2) UVRAG/VPS38, which plays a role in maturation (Itakura et al., 2008; Liang et al., 2008) and is negatively regulated by Rubicon (Matsunaga et al., 2009; Zhong et al., 2009). Beclin 1 complexes can contain several additional factors such as Ambra1 (Fimia et al., 2007) and Bif-1/Endophilin B1 (Takahashi et al., 2007) that may further modulate its function. Many signals that lead to autophagy activation affect Beclin 1 as the "nerve center" of autophagic control. This includes stress and immunological inputs such as activation by JNK1 kinase (Wei et al., 2008), shown to activate Beclin 1 downstream of starvation (but possibly engaged during innate immune signaling); DAPK (Zalckvar et al., 2009), a kinase activated downstream of IFNγ stimulation; BH3-only proteins such as Bnip3 (in turn regulated by FoxO3 in atrophy or by HIF-1 in hypoxia) (Zhang et al., 2008); and MyD88, reported to occur downstream of PRR stimulation by microbial products (Shi and Kehrl, 2008).
Broad Connections between Autophagy and Immunity
The repertoire of autophagy’s functions in immunity has been expanding at an extraordinary pace. The broad immunological roles of autophagy (Figure 1D), collectively dubbed "immunophagy" (Deretic, 2006), can be categorized as: (1) regulatory versus effector roles, (2) innate versus adaptive immunity functions, (3) Anti-inflammatory versus proinflammatory, and (4) specialized immune-cell-specific versus generic cellular homeostatic roles that are applicable to immune cells.
Among the innate immunity effector functions, the most intrinsic to autophagy (i.e., the engulfment and lysosomal degradation of cytoplasmic components) is its role in direct elimination of intracellular microbes (Figure 1D). This process can manifest itself as xenophagy (Levine, 2005), denoted to describe direct autophagy of intracellular microbes in any cell type. Here, we introduce "autophagic macrophage activation" (APMA) as a general term to denote a collection of autophagy-related processes in cells of the reticulo-endothelial system. APMA includes (1) convergence of phagocytosis and the autophagic machinery (Sanjuan et al., 2007), (2) enhanced microbicidal properties of autolysosomes in comparison to standard phagolysosomes (Alonso et al., 2007), (3) autophagic modulation of PRR signaling (Delgado et al., 2008; Sanjuan et al., 2007; Shelly et al., 2009; Xu et al., 2007), (4) cooperation between immunity related GTPases and autophagy or Atg factors in attacking parasitophorous vacuoles (Gutierrez et al., 2004; Ling et al., 2006; Singh et al., 2006; Zhao et al., 2008), and (5) enhanced antigen presentation (Schmid et al., 2007; English et al., 2009). APMA is thus recognized as a complex outcome of autophagy stimulation in macrophages, representing a unique composite process bringing about a heightened state of activation.
Autophagy in Innate Immunity
The innate immunity functions identified to date encompass both effector outputs (Delgado et al., 2008; Sanjuan et al., 2007; Shelly et al., 2009; Xu et al., 2007) and regulatory roles, some of which are in conjunction with PRRs (Jounai et al., 2007; Lee et al., 2007; Saitoh et al., 2008; Tal et al., 2009). As a regulator of immunity responses, autophagy acts in several ways (Figure 1D). Autophagy is proinflammatory, e.g., when autophagy captures cytosolic viral replication intermediates and delivers them to the lumen of endosomal compartments where they meet their cognate PRRs, as in the case with viral single-stranded RNA and Toll-like receptor (TLR) 7 (Lee et al., 2007). Complementarily, autophagy can also dampen proinflammatory responses, including IL-1β, IL-18 (Saitoh et al., 2008), and type I IFN production (Jounai et al., 2007; Tal et al., 2009) (Figure 1D). As an effector, autophagy is modulated by cytokines (Figure 1D), including IFNγ, TNF-α, IL-4, and IL-13, and acts as an output of both innate and adaptive immunity responses (Deretic, 2009).
The autophagic machinery can be activated upon detection of PAMPs by their cognate PRRs (Delgado et al., 2008; Sanjuan et al., 2007; Shelly et al., 2009; Xu et al., 2007; Yano et al., 2008) or antibody-Fcγ receptor (Huang et al., 2009). The signal transduction pathways between agonist-stimulated receptors and autophagy activation remain to be fully delineated. This is an important frontier, especially since there have been reports of the inability to detect macroautophagy downstream of TLR stimulation (Saitoh et al., 2008) and discrepancies in adaptor usage downstream of certain PRRs in the context of autophagy (Delgado et al., 2008; Sanjuan et al., 2007; Xu et al., 2007). PRR signaling to autophagy may involve the reported complex formed between the TLR adaptors (MyD88 and TRIF) and Beclin 1 and changes in the antiapoptotic protein Bcl-2’s interaction with Beclin 1 upon TLR stimulation (Shi and Kehrl, 2008) (Figure 1D), akin to Bcl-2-Beclin 1 interactions observed during activation of autophagy by nonimmunological signals (Wei et al., 2008) (Figure 1C).
Autophagy can also be activated by reactive oxygen species (ROS) (Scherz-Shouval et al., 2007). Accordingly, ROS produced by NADPH oxidase downstream of TLR or Fcγ receptor stimulation in phagocytes has been shown to activate autophagy (Huang et al., 2009). The report from Sanjuan et al. that LC3-II appears on phagosomes without the appearance of conventional double membranes shortly after particle uptake when particles costimulate TLRs (Sanjuan et al., 2007) called attention to the unconventional roles of Atg proteins. However, the coactivation of NADPH oxidase and ROS production during phagocytosis of opsonized or PAMP-laden particles may in essence mirror the observed induction of autophagy by mitochondrially produced ROS in response to starvation stimuli (Scherz-Shouval et al., 2007) and may explain in part the observations of Sanjuan et al. These events may be best understood within the concept of APMA, as proposed earlier, which is a set of linked events in macrophages and includes connections between ROS production and autophagy.
Importantly, autophagy induction downstream of PRR activation is countered by NF-κB, which is activated concomitantly (Djavaheri-Mergny et al., 2006) (Figure 1D). This may explain why PAMPs do not uncontrollably stimulate autophagy in physiological situations (Delgado et al., 2008) and a report that autophagy could not be detected upon PRR stimulation (Saitoh et al., 2008). Independent of its antimicrobial function, autophagy induced by LPS and TLR4 may act to protect against LPS cytotoxicity, which is of potential relevance for countering myocardial depression in septic shock (Yuan et al., 2009). Other innate immunity mediators such as IL-1β, a proinflammatory cytokine normally generated upon inflammasome activation with PAMPs or the host’s own DAMPs, can also stimulate autophagy (English et al., 2009) (Figure 1D). Consistent with the direct involvement of DAMP pathways, ATP, which is an endogenous activator of the inflammasome, can stimulate autophagy through the P2×7 receptor (Biswas et al., 2008). Thus, both microbial PAMPs and host DAMPs appear to be linked to autophagy.
Autophagy in Adaptive Immunity
We are only just beginning to comprehend what appear to be the critical functions of autophagy in regulating adaptive immune responses, immunological tolerance, and the development and homeostasis of the immune system. At least three distinct processes contribute to these functions (Figure 1D). (1) Autophagy, by the very nature of its ability to capture cytoplasmic proteins, supports MHC II-restricted endogenous antigen presentation of cytosolic self or microbial (e.g., viral) antigens synthesized by host cells (Gannage and Munz, 2009). It may likewise influence MHC I presentation of viral antigens in a process occurring separately from and following the initial canonical ER-dependent cross-presentation pathway (English et al., 2009). (2) Autophagy shapes central tolerance via thymic selection of the T cell repertoire (Nedjic et al., 2008). (3) Autophagy also affects homeostasis of T cells (Pua et al., 2009), B cells (Miller et al., 2008), and specialized granulocytes of the intestinal epithelium known as Paneth cells (Cadwell et al., 2008).
Some additional functions of autophagy may represent more of a "fine tuning" of the immune response. For example, autophagy acts as an effector of immune phenomena such as Th1/Th2 polarization; when induced by Th1 cytokines such as TNF-α (Djavaheri-Mergny et al., 2006) or IFNγ (Harris et al., 2007), macroautophagy (i.e., xenophagy) and APMA kill intracellular microbes (e.g., M. tuberculosis), while Th2 cytokines such as IL-4 and IL-13 inhibit autophagy and protect intracellular pathogens form autophagic elimination (Harris et al., 2007). Additionally, in the context of vaccine development, autophagy has been used to enhance CD4 T cell responses to influenza matrix protein (Schmid et al., 2007) and to enhance BCG vaccine efficacy in animal models of tuberculosis (Jagannath et al., 2009). These studies open a new area of translational research in the application of autophagy in prophylaxis against infectious diseases.
Autophagy in Inflammation
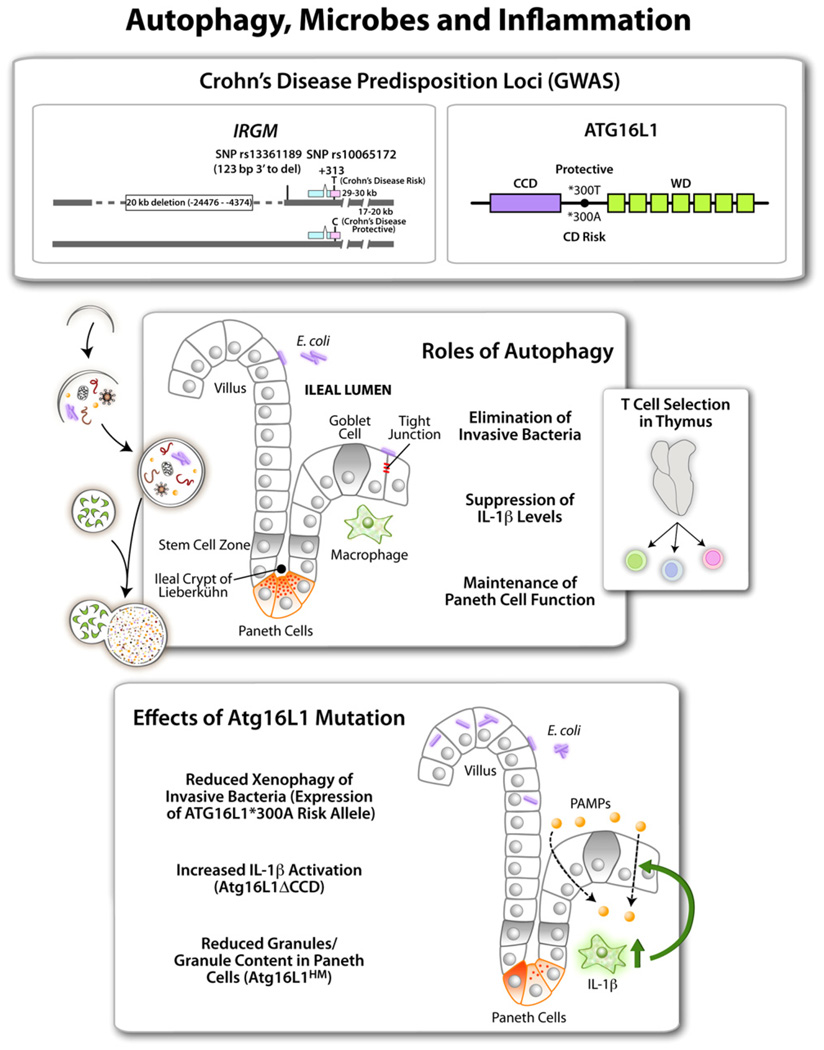
Certain immune responses act as a double-edged sword, either resolving infection or leading to over-exuberant inflammation and tissue damage. Autophagy belongs to this category, in view of the developing connection between inflammatory bowel disease and mutations in autophagy (e.g., ATG16L1) or autophagy-related (e.g., IRGM) genes, as predisposition loci in CD that have been identified in human populations through whole genome association studies (GWAS) (McCarroll et al., 2008; Parkes et al., 2007; Wellcome Trust Case Control Consortium, 2007) (Figure 2). Genetic risks in CD were linked some time ago to innate immunity via Nod2, which like many other PRRs recognizes bacterial products of the enteric flora (Kanneganti et al., 2007). Interestingly, several risk loci identified through GWAS are common to ulcerative colitis and CD, but autophagy genes ATG16L1 and IRGM, along with NOD2, appear to be more specific for CD (Fisher et al., 2008), and IRGM risk alleles may predispose even more specifically to the ileal form of CD (Roberts et al., 2008). ATG16L1 is a part of the basal autophagy apparatus, while IRGM is a member of the vertebrate family of innate immunity effectors called immunity-related GTPases (IRGs) (Bekpen et al., 2009). In the mouse, there are multiple IRG genes, while humans and chimpanzees have only one IRG, IRGM (Bekpen et al., 2009). The human IRGM (Singh et al., 2006) and murine Irgm1 (Gutierrez et al., 2004) and Irga6 (Al-Zeer et al., 2009; Ling et al., 2006; Zhao et al., 2008) have all been shown to play a role in autophagic elimination of intracellular pathogens.
Figure 2. Autophagy in Inflammation.
Top shows two autophagy factors identified as Crohn’s disease (a form of inflammatory bowel disease) susceptibility loci. The IRGM gene has single nucleotide polymorphisms (SNP)and a 20 kb deletion in the promoter region associated with CD. ATG16L1 alleles encode either a protective ATG16L1*300T or a risk form of ATG16L1*300A (CCD, coiled coil domain; green boxes, WD repeats domain [absent in yeast Atg16]). Middle panel shows intestinal epithelium with different cell types along with the proposed functions of autophagy in the ileal epithelium. Side box shows that thymic selection of naive T cell repertoires depends on autophagy, and autophagic anomalies may contribute to inflammation at peripheral sites such as intestinal mucosa. Bottom box lists 3 different effects reported for Atg16L1 (mouse) or ATG16L1 (human epithelial cells) mutations. ATG16L1*300A has been tested in cell lines. Atg16L1HM, hypomorphic Atg16L1 allele, and Atg16LΔCCD construct have been tested in vivo in transgenic mice.
Functional information regarding the role of autophagy in humans in the context of CD is lacking. Nevertheless, relevant information regarding ATG16L1 has been obtained in studies at the cellular level and in vivo in mice, with three published studies identifying different effects that may be additive (Figure 2, middle panel): (1) a diminished capacity of the CD risk ATG16L1*300A to control intracellular enteric pathogens (Kuballa et al., 2008), which fits with the current focus on adherent-invasive E. coli (AIEC) as one of the microbial culprits in CD (Rolhion and Darfeuille-Michaud, 2007); (2) increased susceptibility of Atg16L1-deficient mice to chemically-induced colitis that is linked to elevated IL-1β signaling (Saitoh et al., 2008); and (3) direct or indirect effects in Atg16L1 hypomorphic mice on Paneth cells (Cadwell et al., 2008), the epithelial-type stationary "granulocytes" of the intestinal crypt that guard the overhead stem cell zone from microbial penetration, a long-time suspect in the etiology of CD. The role of IRGM cannot be properly investigated in mice, as the mouse has 19 IRGM-like genes. Human IRGM plays a direct role in antibacterial defenses (Singh et al., 2006), which fits well with a similar role of ATG16L1 in the control of intracellular enteric bacteria (Kuballa et al., 2008). However, additional processes cannot be excluded, given that Irgm1, one of the three putative murine orthologs, shows effects on hematopoietic stem cell proliferation and T cell survival (Feng et al., 2008a, 2008b).
Finally, a comprehensive explanation for the role of autophagy in inflammatory processes, including CD, needs to take into account the endogenous antigen presentation process, whereby thymic epithelial cells present endogenous (self) antigens to developing T cells, influencing their positive and negative selection (Nedjic et al., 2008). Thus, autophagy, through appropriate selection of naive T cells (Figure 2, middle panel) before they exit to the periphery, serves as a guardian of immunological tolerance. This seems to be exceptionally important, as when this process is rendered dissonant between the periphery and the thymus in mice with Atg5−/− thymic implants, the animals develop multiorgan inflammation, including colitis. It remains to be seen whether aberrant autophagy (e.g., risk alleles in CD or yet to be identified potential autophagy defects in other autoimmune or inflammatory diseases) may lead to dissonant thymic selection vis-à-vis endogenous antigen presentation in the periphery.
Autophagy as a Cell-Autonomous Antimicrobial Defense System
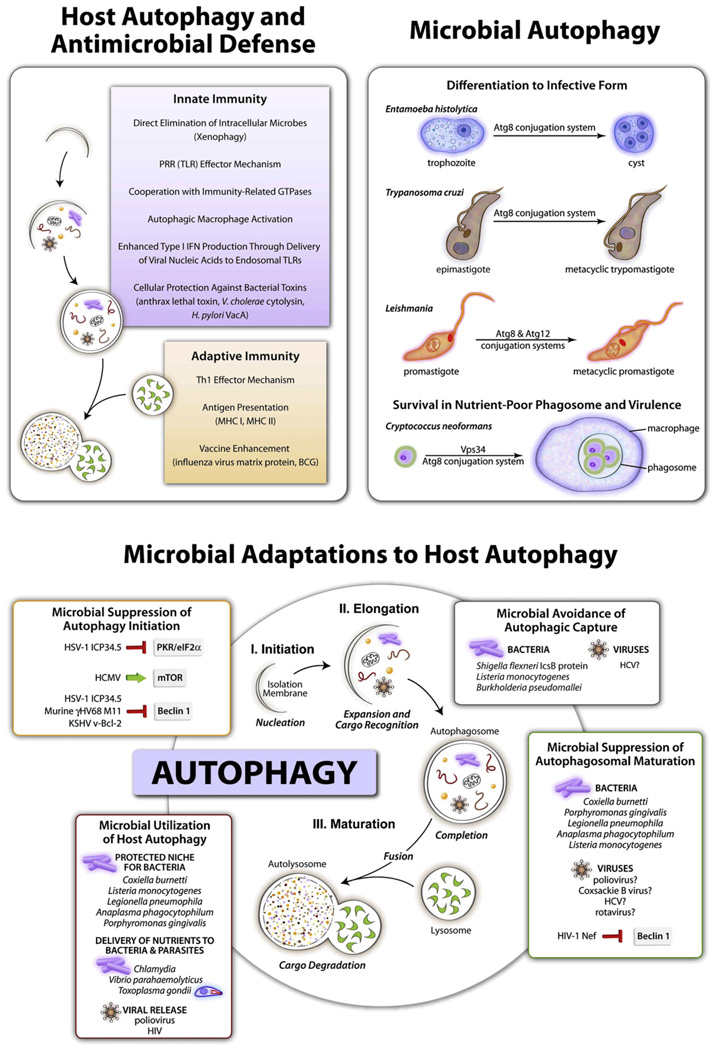
Since autophagosomes can engulf large portions of the cytosol and digest whole organelles such as mitochondria, it is intuitively compelling to think of this process as capable of capturing and eliminating intracellular microbes. Conclusive demonstrations ex vivo and in vivo of this concept, however, have proven less than trivial. A forerunner to the development of this field was a report that ectopic Beclin 1 expression in neurons suppresses viral replication in the brain and reduces morbidity and mortality in experimental animals (Liang et al., 1998). In more recent years, a growing number of microorganisms have been demonstrated to be subject to elimination by autophagy in vitro involving one or more of the processes listed in Figure 3, upper left panel. Only very recently has the autophagic machinery been shown to protect against thus far a small number of infectious diseases in vivo (Table 1). In addition, autophagy may protect host cells against toxic products produced by pathogens, such as Vibrio cholerae cytolysin (Gutierrez et al., 2007), Bacillus anthracis lethal toxin (Tan et al., 2009), and Helicobacter pylori vacuolating toxin (Terebiznik et al., 2009) (Figure 3 and Table 1).
Figure 3. Schematic Depicting Functions of Host Autophagy in Antimicrobial Defense, Functions of Microbial Autophagy, and Microbial Adaptations to Host Autophagy.
See Table 1 and Table 2 for further details of specific host autophagy-pathogen interactions.
Table 1.
Roles of Autophagy in Protection against Microbes
| Microbe | Host organism/ cell type | Autophagy genes or autophagy signals | Effects on host-pathogen interactions | References |
|---|---|---|---|---|
| VIRUSES | ||||
| RNA viruses | ||||
| Alphaviridae | ||||
| Sindbis virus | Mice (neurons) | beclin 1 | Enforced neuronal Beclin 1 expression reduces CNS viral replication, neuronal cell death, and animal mortality | Liang et al., 1998 |
| Rhabdoviridae | ||||
| Vesicular stomatitis virus | Drosophila | Atg8 Atg7 Atg12 Atg18 | Autophagy gene silencing increases viral replication in vivo and decreases fly survival | Shelly et al., 2009 |
| Tobamoviruses | ||||
| Tobacco mosaic virus (TMV) | Plants | BECLIN 1ATG3, ATG7VPS34 | Autophagy gene silencing increases TMV local replication and spread of programmed cell death in vivo | Liu et al., 2005 |
| DNA viruses | ||||
| Herpesviridae | ||||
| Herpes simplex virus 1 (HSV-1) | Mice | beclin 1 | Role for autophagy gene in protection against lethal encephalitis inferred by neuroattenuation of mutant virus that cannot inhibit Beclin 1 autophagy function | Orvedahl et al., 2007 |
| BACTERIA | ||||
| Gram-positive cocci | ||||
| Staphylococcus aureus | Mouse embryonic fibroblasts (MEFs) | Atg5 | Atg5 deletion inhibits bacterial degradation in autolysosomes, leading to delayed bacterial clearance and increased bacterial multiplication | Amano et al., 2006 |
| Group A Streptococcus | MEFs | Atg5 | Atg5 deletion inhibits bacterial degradation in autolysosomes, leading to delayed bacterial clearance and increased bacterial multiplication | Nakagawa et al., 2004 |
| Gram-positive bacilli | ||||
| Listeria monocytogenes | MEFs | Atg5 | Atg5 deletion increases replication of bacterial phospholipase mutants | Py et al., 2007 |
| Mice (macrophages) | Atg5 (macrophages) | Macrophage Atg5 required for in vivo resistance in mice | Zhao et al., 2008 | |
| Drosophila | Atg5, Atg1, peptidoglycan-recognition protein (PGRP) | Autophagy gene silencing results in failure to control L. monocytogenes replication in hemocytes in vitro or in intact fly; PGRP required to signal autophagy-mediated resistance | Yano et al., 2008 | |
| Bacillus anthracis | Mouse macrophage cell line | Autophagy induction may protect cells against anthrax lethal toxin | Tan et al., 2009 | |
| Gram-negative bacilli | ||||
| Burkholderia pseudomallei | Mouse macrophage cell lines and embryonic fibroblasts | Autophagy induction decreases intracellular bacterial survival (but no effect of Atg5 deletion in MEFs on bacterial survival) | Cullinane et al., 2008 | |
| Helicobacter pylori | Human gastric epithelial cells | Autophagy inhibition increases vacuolating toxin stability and toxin-mediated cellular vacuolation | Terebiznik et al., 2009 | |
| Human monocytic cells | Autophagy inhibition enhances and autophagy activation suppresses intracellular bacterial multiplication | Wang et al., 2009b | ||
| Pseudomonas syringae pv. tomato | Arabidopsis thaliana | ATG6 | ATG6 silencing in plants results in increased spreading programmed cell death and bacterial virulence | Patel and Dinesh-Kumar, 2008 |
| Salmonella enterica | MEFs; epithelial cells | Atg5 | Atg5 deletion results in increased intracellular bacterial growth | Birmingham et al., 2006 |
| Shigella flexneri | Dog kidney epithelial cells and MEFs | Atg5 (in MEFs) | IcsB mutant bacteria are targeted by autophagy, leading to decreased intracellular bacterial multiplication | Ogawa et al., 2005 |
| Vibrio cholerae | Human intestinal cell lines; MEFs | Atg5 (in MEFs) | Autophagy protects against toxic effects of bacterial secreted pore-forming toxin, Vibrio cholerae cytolysin | Gutierrez et al., 2007 |
| Mycobacteria | ||||
| Mycobacterium tuberculosis Bacillus Calmette-Guerin | Mouse and human primary macrophages and macrophage cell lines | Atg5 (macrophages Flox/Flox from Atg5 LysM-Cre mice) | Atg5 required for autophagic killing of mycobacteria in macrophages | Zhao et al., 2008 |
| IFNγ and immunity- related GTPases (mouse LRG-47, human IRGM), signaling through P2×7 | Autophagy induction by immune signaling, starvation, or TOR inhibition increases mycobacterial targeting to phagolysosomes and decreases mycobacterial intracellular survival | Alonso et al., 2007; Biswas et al., 2008; Gutierrez et al., 2004; Singh et al., 2006 | ||
| Th1 cytokines activate and Th2 cytokines inhibit autophagy | IL-4 and IL-13 inhibit IFNγ- or starvation-induced autophagic elimination of mycobacteria | Harris et al., 2007 | ||
| PROTOZOA | ||||
| Toxoplasma gondii | Mice (macrophages) | Atg5 | Macrophage Atg5 required for in vivo resistance in mice; mechanism believed to be autophagosome-independent recruitment of Irga6 to parasitophorous vacuole | Ling et al., 2006; Zhao et al., 2008 |
Among viruses, autophagic protection has been shown for vesicular stomatitis virus (VSV) (Shelly et al., 2009), tobacco mosaic virus (TMV) (Liu et al., 2005), herpes simplex virus 1 (HSV-1) (Orvedahl et al., 2007), and human immunodeficiency virus (HIV) (Kyei et al., 2009). HSV-1 and HIV fall prey to autophagy when they are disarmed by inactivation of their specific antiautophagy factors, ICP34.5 and Nef, respectively (Orvedahl et al., 2007; Kyei et al., 2009). Among bacteria, microbes that can invade into host cells (e.g., Gram-positive extracellular pathogens such as Streptococcus pyogenes [Nakagawa et al., 2004] or facultative intracellular pathogens such as M. tuberculosis [Alonso et al., 2007; Biswas et al., 2008; Gutierrez et al., 2004; Singh et al., 2006], Salmonella [Birmingham et al., 2006], and Listeria monocytogenes [Py et al., 2007; Yano et al., 2008; Zhao et al., 2008]) can be eliminated through autophagy. However, as with viruses, the highly evolved intracellular bacterial pathogens possess antiautophagic factors, exemplified by Shigella, where inactivation of the bacterial type III secretion system (T3SS)-dependent effector IcsB is prerequisite to elimination by autophagy (Ogawa et al., 2005). Among protozoans, in vitro and in vivo data exist to support a role for autophagy and/or the autophagic genes in defense against Toxoplasma gondii (Andrade et al., 2006; Ling et al., 2006; Zhao et al., 2008). These examples underscore two concurrent phenomena: (1) autophagy acts as a defense against microbes when they manage to invade the host cell interior, and (2) highly evolved intracellular pathogens have adaptations to protect themselves from autophagic elimination or even harness the host cell autophagic machinery to their own benefit, as will be covered in more detail in subsequent sections.
It is important to keep in mind the above phenomena in interpreting reports using highly adapted intracellular pathogens, as even when the specific antiautophagic adaptations are not yet known, they may exist. Until such factors are identified and inactivated, the true power of autophagic action in eliminating microbes may remain masked. In this context, experiments where pathogens and hosts are slightly "mismatched" are conducive to observing autophagy in action, which may be other wise obscured due to evolutionary adaptations in finely tuned host-pathogen pairs. Examples of this come from using Drosophila to study mammalian pathogens, where infection experiments with L. monocytogenes (Yano et al., 2008) and VSV (Shelly et al., 2009) unambiguously demonstrate that autophagy controls these microbes in vivo. Mouse infection models of human disease have also led to the demonstration of autophagy’s role in vivo, as shown in recent experiments with viruses (Orvedahl et al., 2007) and protozoans (Zhao et al., 2008). The study by Zhao et al. with T. gondii infection using Atg5Flox/Flox LysM-Cre mice (with an Atg5 defect specifically in monocytic cells) has shown that Atg5 function is key in controlling this pathogen in vivo. The details of the study revealed a complex relationship between Atg5 and IRG (in this case Irga6)-dependent processes (Zhao et al., 2008), perhaps not unlike what has been seen with Irgm1 (MacMicking et al., 2003) and autophagy in controlling M. tuberculosis (Gutierrez et al., 2004). The exact details of how IRG and Atg factors work together or whether they work sequentially (e.g., with Atg5 preceding Irga6 recruitment, as implicated in the studies with Irga6 [Al-Zeer et al., 2009; Zhao et al., 2008]) remains to be delineated. Finally, whereas animal experiments remain of high significance to define further the full spectrum of autophagy in antimicrobial defense, GWAS in human populations are of equal interest in exploring whether polymorphisms in autophagy genes predispose to certain infectious diseases, as has been observed with CD.
Microbial Adaptations to Host Autophagy
Microbial pathogens that successfully parasitize eukaryotic cells (i.e., intracellular pathogens) have evolved in the setting of selective pressures imposed by cellular autophagy as a pathway central to innate and adaptive immunity. Consequently, it is not surprising that microbes have developed multipronged strategies to avoid autophagolysosomal degradation and/or to dampen autophagy-dependent activation of host immune responses (Figure 3, bottom panel). Researchers are beginning to delineate such molecular strategies and their potential roles in microbial pathogenesis, although in most cases, our understanding of this area is still quite rudimentary. The types of microbial adaptations identified to date can be broadly categorized as strategies to (1) prevent the induction of autophagy, (2) prevent the maturation of the autophagosome into an autolysosome, (3) avoid pathogen recognition by the autophagic machinery, and (4) utilize functions or components of autophagy to enhance intracellular survival, replication, or extracellular release of intracellular pathogens.
Microbial Suppression of Autophagy Induction
As discussed, the activation of autophagy in an infected cell may represent a fairly ubiquitous response triggered by PRRs that recognize microbe-specific PAMPs. While most studies have been performed using TLR ligands (rather than intact microbes) (reviewed in Delgado et al., 2009), PRR-dependent autophagy induction has recently been shown to protect against L. monocytogenes infection in Drosophila (Yano et al., 2008). It is not yet known whether pathogens possess strategies to block PRR-dependent autophagy induction or how ubiquitously PRRs are used to activate autophagy. What appears more likely, at least based on the limited research to date, is that microbial adaptations to suppress autophagy induction may be focused on targeting some of the more general (i.e., not pathogen-specific) signaling pathways that positively or negatively regulate autophagy. This evidence stems largely from studies done with the three major groups of herpesviruses, the α-, β-, and γ-herpesviruses, although the principles learned from such studies are likely to be extrapolatable to other virus families and perhaps to other types of microbial pathogens.
Herpesviruses block autophagy induction through at least three distinct mechanisms (Table 2 and Figure 3, bottom panel), including blockade of autophagy-stimulatory PKR/eIF2α kinase signaling (Figure 1D) (through HSV-1 ICP34.5-mediated eIF2α dephosphorylation) (Tallóczy et al., 2002), blockade of the autophagy function of Beclin 1 (through HSV-1 ICP34.5, KSHV, or murine γ-HV68 viral Bcl-2 binding of Beclin 1) (Ku et al., 2008a; Orvedahl et al., 2007; Pattingre et al., 2005; Sinha et al., 2008), or activation of autophagy-inhibitory mTOR signaling (by human cytomegalovirus through an as of yet undefined mechanism) (Chaumorcel et al., 2008) (Figure 3, bottom panel). The utilization of different strategies for preventing autophagy induction by a single viral virulence protein (i.e., HSV-1 ICP34.5 blocks both eIF2α phosphorylation and Beclin 1 function), the utilization of different viral structural motifs for targeting Beclin 1 (i.e., HSV-1 ICP34.5 and viral Bcl-2 proteins both bind Beclin 1, but share no structural similarity with each other and bind to nonoverlapping regions of Beclin 1), and the prevention of autophagy induction by all three classes of herpesviruses likely underscore a critically important role for evasion of autophagy in herpesvirus pathogenesis. Indeed, for HSV-1, it has been shown that ICP34.5-mediated blockade of Beclin 1-dependent autophagy is essential for lethal HSV-1 encephalitis (Orvedahl et al., 2007). The role of CMV and γ-herpesvirus evasion of autophagy in viral pathogenesis has not yet been explored, but considering that autophagy is a tumor suppressor pathway (Levine and Kroemer, 2008; Mizushima et al., 2008), it is tempting to speculate that γ-herpesvirus evasion of autophagy might contribute to viral oncogenesis. Another open question is whether other virus families also inhibit autophagy induction; given the numerous viral proteins encoded by the diverse families of viruses that have already been shown to inhibit PKR/eIF2α kinase signaling or to activate mTOR signaling (reviewed in Cooray, 2004 and Sadler and Williams, 2008), it seems likely that suppression of host autophagy induction signaling pathways will be a fairly universal feature of viral infections.
Table 2.
Microbial Adaptations to Evade or Use Autophagy to Promote Survival or Replication
| Microbe | Adaptation | Effects on host-pathogen interactions | Microbial virulence factor/mechanism | References |
|---|---|---|---|---|
| VIRUSES | ||||
| RNA viruses | ||||
| Picornaviridae | ||||
| Poliovirus | Infection induces formation of LC3-positive double membranes | Proposed mechanism for nonlytic virus egress | Poliovirus 2BC and 3 proteins induce GFP-LC3 colocalization with LAMP1 | Jackson et al., 2005; Taylor and Kirkegaard, 2007 |
| Coxsackievirus B3, B4 (CVB3, CVB4) | CVB3 infection induces early but not late stages of autophagy; CVB4 infection induces autophagy in neurons | May increase viral replication or yields | Calpain-dependent (CVB4) | Wong et al., 2008; Yoon et al., 2008 |
| Flaviviridae | ||||
| Dengue virus | Infection induces autophagy | Increases viral yields; colocalization of LC3, viral dsRNA, and endosomal marker may indicate association of viral replication complex with amphisomes | Unknown | Lee et al., 2008; Panyasrivanit et al., 2009 |
| Hepatitis C virus | Infection induces early but not late stages of autophagy in hepatocyte cell lines | Increases HCV replication (without colocalization of viral proteins and autophagosomes) | Activation of unfolded protein response | Ait-Goughoulte et al., 2008; Sir et al., 2008 |
| Pestiviridae | ||||
| Bovine viral diarrheal virus | Incorporation of cellular LC3 into viral genome through RNA recombination | Enhances viral replication and viral cytopathic effects and causes lethal mucosal disease in cattle | Viral LC3 in genome facilitates viral polyprotein processing | Meyers et al., 1998 |
| Orthomyxoviridae | ||||
| Influenza A virus | Infection increases autophagy and autophagic flux | May increase viral replication or yields | Unknown | Zhou et al., 2009 |
| Reoviridae | ||||
| Rotavirus | Viral nonstructural protein NSP4 localizes with LC3 but not LAMP1 | Postulated to play a role in viral morphogenesis, possibly by creating lipid membrane scaffold for formation of viroplasms | Unknown | Berkova et al., 2006 |
| Lentiviridae | ||||
| HIV-1 | Infection inhibits autophagy in primary CD4+ lymphocytes and in macrophage cell lines | Proposed mechanism of viral evasion of innate immunity | Unknown | Zhou and Spector, 2008 |
| Virus may utilize Atg proteins for replication in HeLa cells | Proposed mechanism of viral utilization of autophagic machinery for replication | Unknown | Brass et al., 2008 | |
| Infection induces autophagy gene-dependent cell death in bystander cells | Proposed mechanism of CD4+ T cell depletion | HIV envelope protein triggers autophagy in bystander lymphocytes by binding to CD4 and CXCR4 through receptor signaling-independent mechanisms thought to involve fusogenic activity of gp41 | Denizot et al., 2008;Espert et al., 2006 | |
| Infection induces early stages of autophagy and inhibits autophagosomal maturation in macrophages | Proposed mechanism for increasing HIV yields | HIV Gag interacts with the LC3 autophagy protein to augment Gag processing; HIV Nef binds to Beclin 1 and inhibits autophagosome maturation | Kyei et al., 2009 | |
| DNA viruses | ||||
| Herpesviridae | ||||
| Herpes simplex virus 1 (HSV-1) | Infection inhibits autophagy in neurons | Confers neurovirulence | HSV-1 protein ICP34.5 inhibits PKR signaling and binds to Beclin 1 to block autophagy | Orvedahl et al., 2007; Talloczy et al., 2002 |
| Bovine herpesvirus type 1 (BHV-1) | BHV-1 WT virus induces apoptosis in MDCK cells, whereas BHV-1 bICP0 mutant virus induces nonapoptotic cell death with autophagy | Proposed mechanism of regulating of cell death | BHV-1 bICP0 may inhibit autophagy | Geiser et al., 2008 |
| Human cytomegalovirus (HCMV) | Infection inhibits autophagy in primary fibroblasts | Activates mTOR pathway and rapamycin-insensitive signals | Chaumorcel et al., 2008 | |
| γ-herpesviruses | ||||
| KSHV, murine γ-HV68 | Viral Bcl-2-like proteins inhibit autophagy | Unknown | KSHV vBcl-2 and γ-HV68 M11 inhibit autophagy by binding to Beclin 1 | Ku et al., 2008b; Pattingre et al., 2005; Sinha et al., 2008 |
| Epstein-Barr virus (EBV) | EBV LMP1 protein induces autophagy | Proposed mechanism to decrease LMP1 levels and block its cytostatic effects on B cells | Unknown | Lee and Sugden, 2008 |
| Hepadnaviridae | ||||
| Hepatitis B virus | HBV X protein transfection enhances autophagy in hepatocytes | Unknown | HBV X protein increases Beclin 1 promoter activity | Tang et al., 2009 |
| BACTERIA | ||||
| Intracellular pathogens | ||||
| Gram-positive bacilli | ||||
| Listeria monocytogenes | Prevents or evades autophagic uptake | May enable cytosolic bacteria to escape lysosomal degradation | Bacterial Prf1-regulated factors, including ActA and phospholipases, function in autophagy evasion | Birmingham et al., 2007; Py et al., 2007 |
| Prevents acidification of autophagy-dependent spacious Listeria-containing phagosomes (SLAPs) | May allow persistent intravacuolar infection in host macrophages | Listeriolysin O, a pore-forming toxin and virulence factor, is essential for SLAP formation | Birmingham et al., 2008 | |
| Gram-negative coccobacilli | ||||
| Coxiella burnetii | Coxiella phagosomes (CPh) and Coxiella replicative vacuoles (CRVs) colocalize with LC3 autophagy marker | Interaction between Coxiella and autophagic pathway may delay lysosomal enzyme recruitment to CPh | Bacterial protein synthesis required for CRV formation and LC3 colocalization | Beron et al., 2002; Romano et al., 2009 |
| Francisella tularensis | Francisella enters LC3-positive vacuoles after intracytoplasmic replication in murine macrophages | Postulated to allow cytoplasmic bacteria to regain access to endocytic compartment to promote bacterial egress through exocytosis (also proposed to be source for tularemic MHC II antigen presentation) | Unknown | Checroun et al., 2006; Hrstka et al., 2007 |
| Downregulates expression of autophagy genes (ATG4A571216L2BECN1) during cytoplasmic replication in human monocytes | Postulated to allow cytoplasmic bacteria to escape autophagic capture | Unknown | Butchar et al., 2008 | |
| Gram-negative bacilli | ||||
| Burkholderia pseudomallei | Prevents or evades bacterial colocalization with LC3 autophagosome marker | May increase intracellular bacterial survival | bopA gene required for bacterial evasion of autophagy | Cullinane et al., 2008 |
| Legionella pneumophila | Legionella replication vacuoles initially have the autophagy protein Atg7 and ER markers and later have LC3 and lysosomal markers | Postulated that bacteria slows autophagosome maturation, allowing time for bacteria to differentiate to acid-tolerant form | Soluble bacterial type IV secretion products are sufficient to induce autophagy | Amer and Swanson, 2005 |
| Shigella flexneri | Evades autophagic capture of cytoplasmic bacteria in epithelial cells | Enhances intracellular multiplication in epithelial cells | Bacterial T3SS effector IcsB blocks autophagic targeting of the Shigella protein VirG that is required for actin-based motility, through competitive inhibition of binding to Atg5 autophagy protein | Ogawa et al., 2005 |
| May induce autophagy in macrophages | May prevent pyroptotic death in macrophages | Mechanism of autophagy induction in macrophages unknown and negatively regulated by caspase-1 and Ipaf | Suzuki et al., 2007 | |
| Other | ||||
| Anaplasma phagocytophilum | Bacterial replicative inclusions contain autophagosomal markers (Beclin 1, LC3) but not lysosomal markers | May shield bacteria from endolysosomal pathway and provide niche to enhance replication | Unknown | Niu et al., 2008 |
| Chlamydia trachomatis | Chlamydial inclusion bodies juxtaposed to LC3-positive structures | Postulated that bacteria utilize nutrients derived from autophagy to promote intracellular growth | Unknown | Al-Younes et al., 2004 |
| Autophagy upregulated upon active bacterial growth | Postulated that bacteria have evolved to neither need to inhibit nor to utilize autophagy (since growth is unaffected by augmentation or inhibition of autophagy) | Unknown | Pachikara et al., 2009 | |
| Extracellular pathogens | ||||
| Gram-negative bacilli | ||||
| Vibrio parahaemolyticus | Induces host autophagy | Unknown; proposed that bacteria may utilize nutrients released by autophagically active cells for their proliferation | Bacterial T3SS effectors injected into host cell required for autophagy induction | Burdette et al., 2008 |
| Bacteria that can invade intracellularly | ||||
| Gram-positive cocci | ||||
| Staphylococcus aureus | Resides in autophagosomes after HeLa cell invasion | Postulated to promote intracellular bacterial replication, escape into host cytoplasm, and host cell killing | Bacterial localization with autophagosomes requires agr, a global regulator of S. aureus virulence | Schnaith et al., 2007 |
| Gram-negative bacilli | ||||
| Porphyromonas gingivalis | Traffics through autophagosome-like structures in human coronary artery endothelial cells | May prevent formation of autolysosomes and pathogen destruction | Unknown | Dorn et al., 2001 |
| PROTOZOA | ||||
| Entamoeba histolytica (Eh) Entamoeba invadens (Ei) | Parasite autophagy occurs during proliferation and encystation (Ei) | May facilitate growth of trophozoites and encystation (Ei) | Eh and Ei possess Atg8 but not Atg5-Atg12 autophagy protein conjugation systems | Picazarri et al., 2008 |
| Leishmania amazonensis (La) Leishmania major (Lma) Leishmania mexicana (Lme) | Parasite may exploit IFNg-induced host autophagy response (La) | Increased intracellular La (but not Lma) parasite load in macrophages during starvation or IFNγ-induced autophagy in mouse strain-specific manner | Mechanism of increased parasite load with host autophagy induction unknown | Pinheiro et al., 2008 |
| Parasite autophagy promotes differentiation and survival during starvation (Lma, Lme) | Parasite autophagy important for transformation to mammalian infective form and parasite virulence | Lma Vps and Atg proteins function in endosome sorting (Vps4), autophagy (Vps4, Atg4, Atg8 homologs, Atg12), and differentiation (Vps4, Atg4); Lme lysosomal cysteine peptidases (CPA, CPB) required for autophagy and differentiation | Besteiro et al., 2006; Williams et al., 2006; Williams et al., 2009 | |
| Toxoplasma gondii | Induces autophagy | Autophagy promotes parasite intracellular proliferation in nutrient-limiting conditions in vitro | Calcium-, Atg5-, and Beclin 1-dependent but Tor-independent | Wang et al., 2009b |
| Trypanosoma cruzi | Uses host LC3-positive membranes for cellular entry | Host autophagy enhances parasite invasion | Mechanism of host autophagic membrane recruitment to parasite unknown | Romano et al., 2009 |
| Parasite autophagy promotes differentiation/ development and survival during starvation | Parasite autophagy important for parasite maintenance and survival | Parasite autophagy mediated by conserved Atg proteins (Atg8) but not Atg5-Atg12 protein conjugation system and TOR inhibition | Alvarez et al., 2008 | |
| FUNGI | ||||
| Aspergillus fumigatus (Af) | Fungal autophagy protein functions in metal ion homeostasis | Af Atg1 required for metal ion homeostasis | Richie et al., 2007 | |
| Cryptococcus neoformans (Cn) | Fungal autophagy activated during infection in mammalian Cells | Increases fungal multiplication and lethal infection in mouse model | Cn Class III PI3K/Vps34 and Atg8 required for fungal autophagy and virulence | Hu et al., 2008 |
| Magnaporthe grisea (Mg) | Fungal autophagy activated during infection in plants (rice) | Required for fungal spore (conidium) autophagic cell death, fungal invasion, and pathogenesis in plants | and virulence Mg Atg8 required for conidial cell death; Mg Atg1 required for appressorium invasion in plants | Liu et al., 2007; Veneault-Fourrey et al., 2006 |
It is not yet clear whether other intracellular pathogens besides viruses also actively suppress initiation of the autophagy pathway or rather focus uniquely on blocking pathogen recognition by autophagosomes and/or the maturation of pathogencontaining autophagosomes into acidified autolysosomes. Certain bacterial virulence factors (such as Listeria-encoded Prf1-regulator factors, ActA and phospholipases [Birmingham et al., 2007; Py et al., 2007], Burkholderia pseudomallei BopA [Cullinane et al., 2008], and Shigella-encoded IcsB [Ogawa et al., 2005]) are necessary for bacterial evasion of autophagy, as defined either by increased colocalization with GFP-LC3 or increased growth of replication-deficient bacterial mutants in autophagy-deficient cells. However, to date, there is no published evidence indicating that bacteria, fungi, or parasites block the induction of autophagy in infected cells (although a microarray analysis of Francisella tularensis-infected macrophages revealed downregulation of several autophagy genes [Butchar et al., 2008]). Moreover, in many reports, autophagy induction can be enhanced in bacterially infected cells by starvation or rapamycin, suggesting the absence of microbial mechanisms that can completely block autophagy induction. Nonetheless, it remains an open question whether nonviral pathogens suppress autophagy induction. An interesting possibility is that the known activity of M. tuberculosis in the inhibition of hVps34/PI3-dependent trafficking pathways in macrophages (Vergne et al., 2004) may extend to inhibition of the Beclin 1/hVps34 autophagy complex. At yet another level, highly virulent strains of M. tuberculosis elicit more IL-4 and IL-13 (Manca et al., 2004), the Th2 cytokines known to inhibit autophagy (Petiot et al., 2000), and suppress aspects of APMA (Harris et al., 2007). Other important and outstanding questions are whether bacteria and parasites possess mechanisms to block PRR-dependent autophagy induction and/or IRG autophagy induction.
Microbial Suppression of Autophagosomal Maturation
The greatest threat to an intracellular pathogen imposed by autophagy is not the process of autophagic sequestration per se, but rather the danger imposed by delivery to an autolysosome. Accordingly, it is not surprising that several viruses and intracellular bacteria seem to block fusion of the autophagosome or autophagy protein-dependent fusion of another pathogen containing compartment (i.e., phagosome or pathogen-containing vacuole) with the lysosome (Table 2). For some viruses and intracellular bacteria, it is argued that autophagosome formation may enhance intracellular microbial survival, replication, or extracellular release (see section below); in such cases, enhanced autophagy is usually accompanied by a block in autophagosomal maturation (see Table 2). In this manner, microbes may "avail themselves" of promicrobial functions of autophagy while simultaneously blocking autophagy’s antimicrobial functions (Figure 3, bottom panel).
Perhaps the earliest described example of this concept was with Porphyromonas gingivalis, a bacterial periodontal pathogen that also localizes to atherosclerotic plaques. Based largely on electron and light microscopy studies, this organism is believed to traffic to autophagosomes as a mechanism of evading the conventional endocytic trafficking to lysosomes (Dorn et al., 2001). The bacterial determinants that direct this trafficking and the precise details of the cellular trafficking events in infected cells are not yet defined. In the case of Legionella pneumophila infection, soluble bacterial type IV secretion products are sufficient to induce autophagy, the bacterial replication vacuoles have autophagy markers early after infection, and it is postulated that the bacteria delays autophagosome maturation, allowing time for the bacteria to differentiate into an acid-tolerant form (Amer and Swanson, 2005). A somewhat similar scenario is postulated for Coxiella burnetii replicative vacuoles and Anaplasma phagocytophilum bacterial replicative inclusions; both structures contain autophagosomal but not lysosomal markers (Beron et al., 2002; Niu et al., 2008; Romano et al., 2009), suggesting that the bacteria possess an as of yet unidentified mechanism to block or at least delay autophagosomal fusion with the lysosome. An important area of future research will be to identify specific bacterial factors that interfere with autophagosomal maturation; presumably, pharmacological inhibition of such targets would result in a substantial decrease in the survival of intracellular bacteria that seek refuge in "arrested" autophagosomes. It also will be important to determine whether bacterial evasion of autophagosomal maturation truly contributes to bacterial pathogenesis using in vivo models of infection with bacteria that contain mutations in putative antiautophagosomal maturation bacterial factors. Interestingly, a known Listeria virulence factor, the pore-forming toxin listeriolysin O is required for the formation of structures termed "spacious Listeria-containing phagosomes," which require the autophagy machinery for their formation and fail to acidify, thus potentially allowing persistent intravacuolar infection in host macrophages (Birmingham et al., 2008).
The literature to date suggests that several families of viruses may also block autophagosomal maturation. Poliovirus infection induces the formation of LC3-positive double-membraned structures (initially thought to serve as a scaffold for viral RNA replication and later also postulated to play a role in nonlytic viral egress from the cell) (Jackson et al., 2005; Taylor and Kirkegaard, 2007), but does not induce the formation of autolysosomes. Similarly, coxsackie B virus and hepatitis C virus induce early stages of autophagy, but not late stages of autophagy, in virally infected cells (Ait-Goughoulte et al., 2008; Sir et al., 2008; Wong et al., 2008; Yoon et al., 2008), and the rotavirus nonstructural protein NSP4 colocalizes with LC3 but not lysosomal markers (Berkova et al., 2006). One obvious interpretation of these findings is that these viruses possess specific mechanisms to prevent autophagosomal fusion with the lysosome. However, it is also possible that the lipidation of LC3 (detected by western blot analysis) or membrane localization of lipidated LC3 (detected by light microscopy), which are the usual markers of "early autophagy," do not truly represent the formation of classical autophagosomes that will invariably fuse with late endosomes/lysosomes in the absence of specific autophagosomal maturation-blocking factors. This possibility is supported by evidence that LC3 dots may represent the targeting of LC3 to structures other than autophagosomes, including phagosomes, double-membrane scaffolds for viral RNA replication complexes, or protein aggregates as well as by evidence that "autophagy" proteins may possess or may be co-opted for autophagy-independent functions (Virgin and Levine, 2009).
The identification of specific viral factors that antagonize autophagosomal fusion with the lysosome will be important to distinguish between these possibilities, and as suggested for bacteria, the identification of such factors may represent novel targets for antiviral therapeutics. Along these lines, a recent study reports that HIV-1 Nef, an important pathogenic factor required for HIV disease progression, inhibits autophagic maturation in macrophages through its interaction with the autophagy protein, Beclin 1 (Kyei et al., 2009) (Figure 3, bottom panel). This function of Nef inhibits autophagic degradation of HIV biosynthesis intermediates or virions, thereby enhancing HIV virus yields. These observations provide the first demonstration that a viral virulence factor can target autophagosomal maturation. They also underscore the importance of Beclin 1 as a core component of the autophagic machinery (Figure 1C) that functions both in autophagy initiation (which is antagonized by herpesvirus-encoded proteins) and autophagosomal maturation (which is antagonized by HIV Nef). In future studies, it will be interesting to determine whether Nef has a similar function in virally-infected CD4 T cells and whether this function of Nef contributes to HIV pathogenesis. It will also be interesting to determine whether other viral virulence proteins block the autophagosomal maturation function of Beclin 1 and/or other autophagy proteins.
Microbial Avoidance of Autophagic Capture
Another strategy employed by intracellular bacteria to escape the undesirable fate of lysosomal destruction is to avoid capture by the autophagosome (Figure 3, bottom panel). While bacteria that reside in phagosomes or other vacuolar compartments may seek to avoid lysosomal maturation, the avoidance of autophagic capture may be particularly important for intracellular bacterial pathogens that escape into the cytoplasm. As noted above, evasion of autophagic capture has been described for at least three different intracytoplasmic bacteria, including Shigella flexneri, L. monocytogenes, and Burkholderia pseudomallei (Table 2). The first described, most classic example is that of Shigella escape from autophagy,which seems to utilize a particularly intriguing scheme to escape autophagic envelopment (Ogawa et al., 2005). Shigella possesses a surface protein, VirG, required for actin-based motility that binds to the autophagy protein Atg5 and thereby targets Shigella to the autophagosome. However, the bacterial T3SS effector, IcsB, competitively binds to Atg5, thereby camouflaging its own bacterial target molecule VirG from autophagic capture. It is not yet known whether bacterial T3SS effectors from other organisms exert similar functions or why such a seemingly inefficient mechanism—i.e., the need for one bacterial protein to essentially undo the actions of another protein—evolved. Perhaps we are seeing microbial adaptation to mammalian autophagy in evolutionary progress, and Shigella will ultimately undergo further adaptations in VirG itself to avoid capture by Atg5. As the specific identities of host cell-derived molecular tags for microbial targeting to the autophagosome become known, another interesting question will be whether intracytoplasmic bacteria also possess mechanisms to block such host cell tags, in addition to their own microbial tags.
Less is known about whether viruses possess specific mechanisms to avoid autophagic capture. In the case of hepatitis C virus (HCV) infection, two studies have shown that the virus induces early stages of autophagy at least in part by activating the unfolded protein response (Ait-Goughoulte et al., 2008; Sir et al., 2008). Yet, late stages of autophagy (i.e., the formation of autolysosomes) or the colocalization of HCV with markers of early autophagosomes are not observed. These observations imply that HCV may not only possess mechanisms to block autophagosomal maturation (similar to what presumably happens with poliovirus, coxsackie B, rotavirus, and HIV) but also mechanisms to block autophagic capture of HCV. Particularly for viruses such as HCV that establish persistent infection, it seems likely that future studies will identify specific molecular mechanisms by which they evade autophagic capture.
Microbial Utilization of the Host Autophagic Machinery for Intracellular Survival, Replication, or Cellular Egress
In parallel with strategies to block autophagy induction, autohagolysosomal maturation, or autophagic capture, microbes also have evolved mechanisms to utilize aspects of host autophagy to their own advantage (Figure 3, bottom panel). Postulated benefits of host autophagy for microbes include the promotion of viral replication or morphogenesis via utilization of the autophagic machinery, the shielding of bacteria from the endolysosomal pathway via the utilization of autophagosomes as a protective intracellular niche, and the enhanced survival or growth of bacteria, fungi, or parasites through the provision of autophagy-generated nutrients. Although in vitro data support some of these postulates, whether such mechanisms are important in microbial pathogenesis in vivo remains to be explored, and there is no current evidence that autophagy gene deletion in the host attenuates microbial disease. Thus, unlike the role of host autophagy in protection against infection (in which autophagy gene deletion exacerbates microbial disease [Table 1]) or microbial antagonism of autophagy (in which, for example, HSV-1 evasion of autophagy is essential for lethal encephalitis [Table 2]), the physiological significance of microbial utilization of autophagy for "promicrobial" effects remains to be established.
Several viruses are believed to induce autophagy to foster their own replication, morphogenesis, cellular egress, or pathogenicity (Table 2). One theory, originally developed in the context of poliovirus research, is that double-membrane autophagosome-like structures (that contain the autophagy protein LC3) serve as lipid membrane-scaffolds that enhance viral replication (Jackson et al.,2005). In a some what analogous fashion,the rotavirus NSP4 protein, a protein with pleiotropic functions in viral morphogenesis and pathogenesis, colocalizes with LC3-positive vesicular compartments and is postulated to play a role in the formation of viroplasms and/or the packaging of the rotavirus genome or transcription (Berkova et al., 2006). However, in poliovirus-infected cells, siRNA-mediated knockdown of the autophagy genes, LC3 and Atg12, markedly inhibits the release of infectious virus while only minimally affecting viral replication (Taylor and Kirkegaard, 2007), suggesting that the primary function of poliovirus’s utilization of the autophagic machinery may be for nonlytic viral release. A similar theme is emerging with HIV infection in macrophages, as pharmacological stimulation of autophagy increases extracellular viral yields, whereas pharmacological or genetic inhibition of autophagy decreases extracellular viral yields (Kyei et al., 2009). Interestingly, the HIV Gag precursor protein directly interacts with the LC3 autophagy protein, and this interaction may facilitate Gag processing and HIV biogenesis, suggesting a potential biosynthetic, rather than catabolic, role of the host autophagic machinery. For some other viruses, such as HCV, which induces early stages of autophagy, but which does not colocalize with autophagosomes or autophagosomal markers (Ait-Goughoulte et al., 2008; Sir et al., 2008), it is completely unknown how autophagy induction may function to increase viral RNA levels. Beyond this utilization of autophagic machinery for enhancing viral production, it is also worth noting that, in some contexts, the autophagic machinery may play a role in features associated with viral pathogenesis; for example, at least in vitro, HIV-induced death of bystander lymphocytes requires the autophagy genes Atg7 and beclin 1 (Espert et al., 2006).
As noted above, autophagosomes or cellular compartments that seem to require autophagic machinery for their formation may provide a safe "haven" for several bacteria, including L. monocytogenes, C. burnetii, L. pneumophila, A. phagocytophilum, and P. gingivalis (Table 2). In parallel with suppression of autophagosomal fusion with lysosomes and/or acidification of pathogen-containing compartments, the bacterial autophagosomal-like compartments may enable the bacteria to persist (and potentially multiply intracellularly) in a nonacidic compartment. It will be interesting to further determine the cellular fate, ability to persist intracellularly, and ability to replicate intracellularly of such organisms in target cells that lack critical components of the autophagy machinery. The enhanced pathogenicity of L. monocytogenes-infected mice with macrophage-specific deletion of Atg5 (Zhao et al., 2008) and the wild-type levels of replication observed in L. pneumophila-infected Dictyostelium discoideum lacking Atg1, Atg5, Atg6, Atg7, or Atg8 (Otto et al., 2004) suggest that speculations based upon in vitro studies regarding the "microbe-friendly" role of autophagy in microbial replication may not always correlate with the actual role of autophagy in microbial pathogenesis in vivo. Besides shielding bacteria from the endolysosomal pathway, it has also been proposed that bacterial localization to LC3-positive compartments may allow cytoplasmic bacteria to regain access to the endocytic compartment to promote bacteria egress through exocytosis (i.e., during Francisella infection) (Checroun et al., 2006).
Another theory is that autophagy helps "feed" intracellular pathogens, particularly those that reside in sequestered vacuoles that lack access to cytoplasmic nutrients. This concept emerged decades ago, when it was first reported that rickettsiae induce the formation of autophagosomes in polymorphonuclear cells (Rikihisa, 1984), although a later study suggested that autophagy may be involved in killing rickettsia in endothelial cells (Walker et al., 1997). In recent years, it has been postulated that autophagy may function in infected cells to deliver nutrients to Chlamydia (Al-Younes et al., 2004), Vibrio parahaemolyticus (Burdette et al., 2008), and T. gondii (Wang et al., 2009a), thereby enhancing intracellular pathogen survival and proliferation. As with other beneficial functions of autophagy for microbes, the physiological relevance of these findings is not yet known. Moreover, somewhat akin to the discrepant conclusions between in vitro and in vivo studies with Listeria and Legionella, although T. gondii has impaired growth in Atg5-deficient MEFs, leading to the conclusion that host cell autophagy plays a role in promoting parasite growth through nutrient recovery (Wang et al., 2009a), T. gondii has increased virulence in mice with macrophage-specific deletion of Atg5 (Zhao et al., 2008).
Microbial Autophagy
Pathogens such as protozoans, fungi, and helminths are themselves eukaryotes and hence have their own autophagic machinery (Figure 3, top right panel).An emerging area of research in microbial pathogenesis is the function of autophagy directly within such eukaryotic pathogens. The molecular machinery of autophagy was originally identified in genetic screens performed in the yeast S. cerevisiae, and autophagy is also present in disease-causing fungi, as well as in disease-causing protozoans and helminths. Recent analyses of autophagy in eukaryotic pathogens reveals some interesting common themes, including (1) the identification of potential differences between microbial autophagy and host autophagy in terms of what constitutes the core autophagic machinery and (2) the identification of an essential role for microbial autophagy in stress adaptation and development, both of which may be important for mammalian infectivity and virulence.
The Molecular Machinery of Microbial Autophagy
A critical question is whether the autophagic machinery truly differs in certain pathogenic protozoans from that found in yeasts and mammalian cells, or whether the lack of apparent structural homology in some autophagy genes has resulted in the mere failure of "genome mining" to identify conserved components. In yeast (S. cerevisiae), 17 genes encoding proteins comprising the core machinery of autophagy have been identified (Suzuki and Ohsumi, 2007), most of which are conserved in higher eukaryotes. Interestingly, two protozoan parasite pathogens, Entamoeba and Trypanosoma cruzi, are reported to contain all of the major genes of the Atg8 conjugation system (e.g., Atg3, Atg4, Atg7, Atg8) but to lack the entire Atg5-Atg12 conjugation system (Alvarez et al., 2008; Picazarri et al., 2008). This observation raises the interesting possibility that either the Atg5-Atg12 protein conjugation system is not absolutely essential for autophagy or that autophagy in these organisms is somehow qualitatively different from that in other eukaryotic organisms. However, a recent study found an atypical Atg12-like protein encoded by Leishmania major, which, when truncated before a scissile C-terminal glycine, complements Atg12 deficiency in yeast, as well as more typical Atg5 and Atg10 proteins that restore autophagy in atg5D and atg10D yeast (Williams et al., 2009). These three Leishmania-encoded Atg12, Atg5, and Atg10 proteins have syntenic homologs in Trypanosoma bruci and Trypanosoma cruzi (Williams et al., 2009), suggesting that perhaps the Atg5-Atg12 protein conjugation systems are present but have been "missed" in certain protozoan organisms. Future studies are required to address this possibility and determine whether or not fundamental differences exist between the core autophagic machinery in certain protozoan pathogens and other eukaryotic organisms.
Microbial Autophagy in Microbial Pathogenesis
Two major functions of microbial autophagy may be relevant to microbial pathogenesis: the requirement for fungal and protozoan autophagy for survival during nutrient stress and the requirement for protozoan autophagy in developmental transitions to mammalian infective forms. Of potential medical relevance, autophagy in the dimorphic human fungal pathogen, Cryptococcus neoformans, is important for intracellular survival in macrophages (which is postulated to represent nutrient self-supplying function in the environment of a nutrient-poor phagosome) and for virulence, since deletion of the fungal class III PI3K VPS34 gene or ATG8 knockdown results in decreased replication and lethality in a mouse model of cryptococcosis (Hu et al., 2008) (Figure 3, top right panel). Thus, it will be interesting to see whether it is possible to specifically target the cryptococcal but not mammalian autophagic machinery in the treatment of human cryptococcal disease. For protozoan parasites, the autophagic machinery has been shown as essential for (Figure 3, top right panel): (1) Entamoeba to undergo the developmental transition from the trophozoite to the cyst stage, a process essential for its transmission and reinfection (Picazarri et al., 2008); (2) Leishmania to differentiate into the infective metacyclic promastigote form (Besteiro et al., 2006; Williams et al., 2006, 2009); and (3) Trypanosoma cruzi to differentiate into the infective metacyclic trypomastigote (Alvarez et al., 2008). These highdensity and/or low-nutrient-dependent stress-induced differentiation events allow the parasites to transition from a form adapted to survive in insects to a form capable of infecting mammalian hosts. While it is not yet clear whether autophagy continues to be important for parasite differentiation inside the mammalian host, Leishmania mutants lacking lysosomal cysteine peptidases and defective in autophagy are also defective in differentiation and lack virulence in macrophages and mice. Efforts to target lysosomal cysteine peptidases and/or the Atg4 protease as treatment of T. cruzi caused Chagas disease, and Leishmanias are being considered (Alvarez et al., 2008; Williams et al., 2006).
Conclusion
A number of key recent studies have expanded the scope of autophagy’s role in immunity from its original incarnation as a putative antimicrobial defense mechanism to a full-range immunological process that participates in innate immunity, adaptive immunity, and inflammation. The ability of autophagy to control intracellular microbes, initially documented only in vitro or in ex vivo experiments, has now been confirmed in in vivo models of infection. In support of the importance of autophagy as a critical defense against microbes, it is now evident that highly evolved intracellular pathogens have specialized antiautophagy adaptations that allow intracellular parasites to prevent, block, or elude autophagic elimination. Autophagy is intertwined with innate immunity regulators and can be stimulated by agonists of innate immunity receptors with feedback loops that amplify or inhibit recognition of microbial products or output signals. At an adaptive immunity level, autophagy controls T cell populations by shaping the naive T cell repertoire in the thymus and antigen-specific T cells in the periphery.B cells are likewise affected, while effector cells such as macrophages can be activated through autophagy. When autophagy is aberrant or "out of sync" in central immunological organs versus the periphery, pathological inflammation can ensue. Coming full circle, autophagy as a cell-autonomous defense of the host cell has met its adversarial counterpart in autophagy that occurs within protozoan and fungal pathogens. Apparently, autophagy works tirelessly to protect the master eukaryotic cell, often on both sides of the host-pathogen relationship.
ACKNOWLEDGMENTS
We thank Angela Diehl for artwork, Dr. Eeva-Lisa Eskelinein for electron micrographs, and Dr. Herbert "Skip" Virgin for discussions. V.D. was supported by grants AI069345, AI45148, and AI42999 from the National Institutes of Health; 107160-44-RGRL from amfAR; a Bill and Melinda Gates Grand Challenge Explorations grant (#52068); and a grant from the Crohn’s and Colitis Foundation of America. B.L. was supported by grants AI151267 and CA109618 from the National Institutes of Health and a Senior Scholars Award in Global Infectious Diseases from the Ellison Medical Foundation.
REFERENCES
- Ait-Goughoulte M, Kanda T, Meyer K, Ryerse JS, Ray RB, Ray R. Hepatitis C virus genotype 1a growth and induction of autophagy. J. Virol. 2008;82:2241–2249. doi: 10.1128/JVI.02093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Younes HM, Brinkmann V, Meyer TF. Interaction of Chlamydia trachomatis serovar L2 with the host autophagic pathway. Infect. Immun. 2004;72:4751–4762. doi: 10.1128/IAI.72.8.4751-4762.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Zeer MA, Al-Younes HM, Braun PR, Zerrahn J, Meyer TF. IFN-gamma-inducible Irga6 mediates host resistance against Chlamydia trachomatis via autophagy. PLoS ONE. 2009;4:e4588. doi: 10.1371/journal.pone.0004588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc. Natl. Acad. Sci. USA. 2007;104:6031–6036. doi: 10.1073/pnas.0700036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez VE, Kosec G, Sant’Anna C, Turk V, Cazzulo JJ, Turk B. Autophagy is involved in nutritional stress response and differentiation in Trypanosoma cruzi. J. Biol. Chem. 2008;283:3454–3464. doi: 10.1074/jbc.M708474200. [DOI] [PubMed] [Google Scholar]
- Amano A, Nakagawa I, Yoshimori T. Autophagy in innate immunity against intracellular bacteria. J. Biochem. (Tokyo) 2006;140:161–166. doi: 10.1093/jb/mvj162. [DOI] [PubMed] [Google Scholar]
- Amer AO, Swanson MS. Autophagy is an immediate macrophage response to Legionella pneumophila. Cell. Microbiol. 2005;7:765–778. doi: 10.1111/j.1462-5822.2005.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade RM, Wessendarp M, Gubbels MJ, Striepen B, Subauste CS. CD40 induces macrophage anti-Toxoplasma gondii activity by triggering autophagy-dependent fusion of pathogen-containing vacuoles and lysosomes. J. Clin. Invest. 2006;116:2366–2377. doi: 10.1172/JCI28796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekpen C, Marques-Bonet T, Alkan C, Antonacci F, Leogrande MB, Ventura M, Kidd JM, Siswara P, Howard JC, Eichler EE. Death and resurrection of the human IRGM gene. PLoS Genet. 2009;5:e1000403. doi: 10.1371/journal.pgen.1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkova Z, Crawford SE, Trugnan G, Yoshimori T, Morris AP, Estes MK. Rotavirus NSP4 induces a novel vesicular compartment regulated by calcium and associated with viroplasms. J. Virol. 2006;80:6061–6071. doi: 10.1128/JVI.02167-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beron W, Gutierrez MG, Rabinovitch M, Colombo MI. Coxiella burnetii localizes in a Rab7-labeled compartment with autophagic characteristics. Infect. Immun. 2002;70:5816–5821. doi: 10.1128/IAI.70.10.5816-5821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besteiro S, Williams RA, Morrison LS, Coombs GH, Mottram JC. Endosome sorting and autophagy are essential for differentiation and virulence of Leishmania major. J. Biol. Chem. 2006;281:11384–11396. doi: 10.1074/jbc.M512307200. [DOI] [PubMed] [Google Scholar]
- Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J. Biol. Chem. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- Birmingham CL, Canadien V, Gouin E, Troy EB, Yoshimori T, Cossart P, Higgins DE, Brumell JH. Listeria monocytogenes evades killing by autophagy during colonization of host cells. Autophagy. 2007;3:442–451. doi: 10.4161/auto.4450. [DOI] [PubMed] [Google Scholar]
- Birmingham CL, Canadien V, Kaniuk NA, Steinberg BE, Higgins DE, Brumell JH. Listeriolysin O allows Listeria monocytogenes replication in macrophage vacuoles. Nature. 2008;451:350–354. doi: 10.1038/nature06479. [DOI] [PubMed] [Google Scholar]
- Biswas D, Qureshi OS, Lee WY, Croudace JE, Mura M, Lammas DA. ATP-induced autophagy is associated with rapid killing of intracellular mycobacteria within human monocytes/macrophages. BMC Immunol. 2008;9:35. doi: 10.1186/1471-2172-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Burdette DL, Yarbrough ML, Orvedahl A, Gilpin CJ, Orth K. Vibrio parahaemolyticus orchestrates a multifaceted host cell infection by induction of autophagy, cell rounding, and then cell lysis. Proc. Natl. Acad. Sci. USA. 2008;105:12497–12502. doi: 10.1073/pnas.0802773105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butchar JP, Cremer TJ, Clay CD, Gavrilin MA, Wewers MD, Marsh CB, Schlesinger LS, Tridandapani S. Microarray analysis of human monocytes infected with Francisella tularensis identifies new targets of host response subversion. PLoS ONE. 2008;3:e2924. doi: 10.1371/journal.pone.0002924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumorcel M, Souquere S, Pierron G, Codogno P, Esclatine A. Human cytomegalovirus controls a new autophagy-dependent cellular antiviral defense mechanism. Autophagy. 2008;4:46–53. doi: 10.4161/auto.5184. [DOI] [PubMed] [Google Scholar]
- Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl. Acad. Sci. USA. 2006;103:14578–14583. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooray S. The pivotal role of phosphatidylinositol 3-kinase-Akt signal transduction in virus survival. J. Gen. Virol. 2004;85:1065–1076. doi: 10.1099/vir.0.19771-0. [DOI] [PubMed] [Google Scholar]
- Cullinane M, Gong L, Li X, Lazar-Adler N, Tra T, Wolvetang E, Prescott M, Boyce JD, Devenish RJ, Adler B. Stimulation of autophagy suppresses the intracellular survival of Burkholderia pseudomallei in mammalian cell lines. Autophagy. 2008;4:744–753. doi: 10.4161/auto.6246. [DOI] [PubMed] [Google Scholar]
- Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado M, Singh S, De Haro S, Master S, Ponpuak M, Dinkins C, Ornatowski W, Vergne I, Deretic V. Autophagy and pattern recognition receptors in innate immunity. Immunol. Rev. 2009;227:189–202. doi: 10.1111/j.1600-065X.2008.00725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denizot M, Varbanov M, Espert L, Robert-Hebmann V, Sagnier S, Garcia E, Curriu M, Mamoun R, Blanco J, Biard-Piechaczyk M. HIV-1 gp41 fusogenic function triggers autophagy in uninfected cells. Autophagy. 2008;4:998–1008. doi: 10.4161/auto.6880. [DOI] [PubMed] [Google Scholar]
- Deretic V. Autophagy as an immune defense mechanism. Curr. Opin. Immunol. 2006;18:375–382. doi: 10.1016/j.coi.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Deretic V. Multiple regulatory and effector roles of autophagy in immunity. Curr. Opin. Immunol. 2009;21:53–62. doi: 10.1016/j.coi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djavaheri-Mergny M, Amelotti M, Mathieu J, Besancon F, Bauvy C, Souquere S, Pierron G, Codogno P. NF-kappaB activation represses tumor necrosis factor-alpha-induced autophagy. J. Biol. Chem. 2006;281:30373–30382. doi: 10.1074/jbc.M602097200. [DOI] [PubMed] [Google Scholar]
- Dorn BR, Dunn WA, Jr, Progulske-Fox A. Porphyromonas gingivalis traffics to autophagosomes in human coronary artery endothelial cells. Infect. Immun. 2001;69:5698–5708. doi: 10.1128/IAI.69.9.5698-5708.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, Alexander D, Leib D, Norbury C, Lippe R, et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat. Immunol. 2009;10:480–487. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espert L, Denizot M, Grimaldi M, Robert-Hebmann V, Gay B, Varbanov M, Codogno P, Biard-Piechaczyk M. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J. Clin. Invest. 2006;116:2161–2172. doi: 10.1172/JCI26185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng CG, Weksberg DC, Taylor GA, Sher A, Goodell MA. The p47 GTPase Lrg-47 (Irgm1) links host defense and hematopoietic stem cell proliferation. Cell Stem Cell. 2008a;2:83–89. doi: 10.1016/j.stem.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng CG, Zheng L, Jankovic D, Bafica A, Cannons JL, Watford WT, Chaussabel D, Hieny S, Caspar P, Schwartzberg PL, et al. The immunity-related GTPase Irgm1 promotes the expansion of activated CD4+ T cell populations by preventing interferon-gamma-induced cell death. Nat. Immunol. 2008b;9:1279–1287. doi: 10.1038/ni.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo, S., Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- Fisher SA, Tremelling M, Anderson CA, Gwilliam R, Bumpstead S, Prescott NJ, Nimmo ER, Massey D, Berzuini C, Johnson C, et al. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn’s disease. Nat. Genet. 2008;40:710–712. doi: 10.1038/ng.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannage M, Munz C. Macroautophagy in immunity and tolerance. Traffic. 2009;10:615–620. doi: 10.1111/j.1600-0854.2009.00883.x. [DOI] [PubMed] [Google Scholar]
- Geiser V, Rose S, Jones C. Bovine herpesvirus type 1 induces cell death by a cell-type-dependent fashion. Microb. Pathog. 2008;44:459–466. doi: 10.1016/j.micpath.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Saka HA, Chinen I, Zoppino FC, Yoshimori T, Bocco JL, Colombo MI. Protective role of autophagy against Vibrio cholerae cytolysin, a pore-forming toxin from V. cholerae. Proc. Natl. Acad. Sci. USA. 2007;104:1829–1834. doi: 10.1073/pnas.0601437104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- Harris J, De Haro SA, Master SS, Keane J, Roberts EA, Delgado M, Deretic V. T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity. 2007;27:505–517. doi: 10.1016/j.immuni.2007.07.022. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrstka R, Krocova Z, Cerny J, Vojtesek B, Macela A, Stulik J. Francisella tularensis strain LVS resides in MHC II-positive autophagic vacuoles in macrophages. Folia Microbiol. (Praha) 2007;52:631–636. doi: 10.1007/BF02932193. [DOI] [PubMed] [Google Scholar]
- Hu G, Hacham M, Waterman SR, Panepinto J, Shin S, Liu X, Gibbons J, Valyi-Nagy T, Obara K, Jaffe HA, et al. PI3K signaling of autophagy is required for starvation tolerance and virulenceof Cryptococcus neoformans. J. Clin. Invest. 2008;118:1186–1197. doi: 10.1172/JCI32053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Canadien V, Lam GY, Steinberg BE, Dinauer MC, Magalhaes MA, Glogauer M, Grinstein S, Brumell JH. Activation of antibacterial autophagy by NADPH oxidases. Proc. Natl. Acad. Sci. USA. 2009;106:6226–6231. doi: 10.1073/pnas.0811045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson WT, Giddings TH, Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RL, Jr, Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat. Med. 2009;15:267–276. doi: 10.1038/nm.1928. [DOI] [PubMed] [Google Scholar]
- Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K, et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl. Acad. Sci. USA. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti TD, Lamkanfi M, Nun˜ez G. Intracellular NOD-like receptors in host defense and disease. Immunity. 2007;27:549–559. doi: 10.1016/j.immuni.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophago-some formation. Mol. Biol. Cell. 2008;19:2039–2050. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc. Natl. Acad. Sci. USA. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat. Rev. Mol. Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku B, Woo JS, Liang C, Lee KH, Hong HS, E X, Kim KS, Jung JU, Oh BH. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68. PLoS Pathog. 2008a;4:e25. doi: 10.1371/journal.ppat.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku B, Woo JS, Liang C, Lee KH, Jung JU, Oh BH. An insight into the mechanistic role of Beclin 1 and its inhibition by prosurvival Bcl-2 family proteins. Autophagy. 2008b;4:519–520. doi: 10.4161/auto.5846. [DOI] [PubMed] [Google Scholar]
- Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn’s disease associated ATG16L1 variant. PLoS ONE. 2008;3:e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong C, Wu L, Kominami E, Ueno T, Yamamoto A, et al. Autophagy pathway intersects with HIV-1 biogenesis and regulates viral yields in macrophages. J. Cell Biol. 2009 doi: 10.1083/jcb.200903070. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Sugden B. The latent membrane protein 1 oncogene modifies B-cell physiology by regulating autophagy. Oncogene. 2008;27:2833–2842. doi: 10.1038/sj.onc.1210946. [DOI] [PubMed] [Google Scholar]
- Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- Lee YR, Lei HY, Liu MT, Wang JR, Chen SH, Jiang-Shieh YF, Lin YS, Yeh TM, Liu CC, Liu HS. Autophagic machinery activated by dengue virus enhances virus replication. Virology. 2008;374:240–248. doi: 10.1016/j.virol.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B. Eating oneself and uninvited guests; autophagy-related pathways in cellular defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat. Rev. Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Capan E, Zhao Y, Zhao J, Stolz D, Watkins SC, Jin S, Lu B. Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. J. Immunol. 2006;177:5163–5168. doi: 10.4049/jimmunol.177.8.5163. [DOI] [PubMed] [Google Scholar]
- Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J. Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling YM, Shaw MH, Ayala C, Coppens I, Taylor GA, Ferguson DJ, Yap GS. Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J. Exp. Med. 2006;203:2063–2071. doi: 10.1084/jem.20061318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XH, Lu JP, Zhang L, Dong B, Min H, Lin FC. Involvement of a Magnaporthe grisea serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis. Eukaryot. Cell. 2007;6:997–1005. doi: 10.1128/EC.00011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Czymmek K, Tallo´ czy, Z., Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- MacMicking JD, Taylor GA, McKinney JD. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science. 2003;302:654–659. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- Manca C, Reed MB, Freeman S, Mathema B, Kreiswirth B, Barry CE, 3rd,, Kaplan G. Differential monocyte activation underlies strainspecific Mycobacterium tuberculosis pathogenesis. Infect. Immun. 2004;72:5511–5514. doi: 10.1128/IAI.72.9.5511-5514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat. Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers G, Stoll D, Gunn M. Insertion of a sequence encoding light chain 3 of microtubule-associated proteins 1A and 1B in a pestivirus genome: connection with virus cytopathogenicity and induction of lethal disease in cattle. J. Virol. 1998;72:4139–4148. doi: 10.1128/jvi.72.5.4139-4148.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BC, Zhao Z, Stephenson LM, Cadwell K, Pua HH, Lee HK, Mizushima NN, Iwasaki A, He YW, Swat W, et al. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008;4:309–314. doi: 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munz C. Enhancing immunity through autophagy. Annu. Rev. Immunol. 2009;27:423–449. doi: 10.1146/annurev.immunol.021908.132537. [DOI] [PubMed] [Google Scholar]
- Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, et al. Auto-phagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- Niu H, Yamaguchi M, Rikihisa Y. Subversion of cellular autophagy by Anaplasma phagocytophilum. Cell. Microbiol. 2008;10:593–605. doi: 10.1111/j.1462-5822.2007.01068.x. [DOI] [PubMed] [Google Scholar]
- Obara K, Sekito T, Niimi K, Ohsumi Y. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J. Biol. Chem. 2008;283:23972–23980. doi: 10.1074/jbc.M803180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- Orvedahl A, Alexander D, Tallo´czy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib D, Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Otto GP, Wu MY, Clarke M, Lu H, Anderson OR, Hilbi H, Shuman HA, Kessin RH. Macroautophagy is dispensable for intracellular replication of Legionella pneumophila in Dictyostelium discoideum. Mol. Microbiol. 2004;51:63–72. doi: 10.1046/j.1365-2958.2003.03826.x. [DOI] [PubMed] [Google Scholar]
- Pachikara N, Zhang H, Pan Z, Jin S, Fan H. Productive Chlamydia trachomatis lymphogranuloma venereum 434 infection in cells with augmented or inactivated autophagic activities. FEMS Microbiol. Lett. 2009;292:240–249. doi: 10.1111/j.1574-6968.2009.01494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Panyasrivanit M, Khakpoor A, Wikan N, Smith DR. Co-localization of constituents of the dengue virus translation and replication machinery with amphisomes. J. Gen. Virol. 2009;90:448–456. doi: 10.1099/vir.0.005355-0. [DOI] [PubMed] [Google Scholar]
- Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, Roberts RG, Nimmo ER, Cummings FR, Soars D, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat. Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Dinesh-Kumar SP. Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy. 2008;4:20–27. doi: 10.4161/auto.5056. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- Picazarri K, Nakada-Tsukui K, Nozaki T. Autophagy during proliferation and encystation in the protozoan parasite Entamoeba invadens. Infect. Immun. 2008;76:278–288. doi: 10.1128/IAI.00636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro RO, Nunes MP, Pinheiro CS, D’Avila H, Bozza PT, Takiya CM, Corte-Real S, Freire-de-Lima CG, Dosreis GA. Induction of autophagy correlates with increased parasite load of Leishmania amazonensis in BALB/c but not C57BL/6 macrophages. Microbes Infect. 2008;11:181–190. doi: 10.1016/j.micinf.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Pua HH, He YW. Maintaining T lymphocyte homeostasis: another duty of autophagy. Autophagy. 2007;3:266–267. doi: 10.4161/auto.3908. [DOI] [PubMed] [Google Scholar]
- Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J. Exp. Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J. Immunol. 2009;182:4046–4055. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- Py BF, Lipinski MM, Yuan J. Autophagy limits Listeria monocytogenes intracellular growth in the early phase of primary infection. Autophagy. 2007;3:117–125. doi: 10.4161/auto.3618. [DOI] [PubMed] [Google Scholar]
- Richie DL, Fuller KK, Fortwendel J, Miley MD, McCarthy JW, Feldmesser M, Rhodes JC, Askew DS. Unexpected link between metal ion deficiency and autophagy in Aspergillus fumigatus. Eukaryot. Cell. 2007;6:2437–2447. doi: 10.1128/EC.00224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikihisa Y. Glycogen autophagosomes in polymorphonuclear leukocytes induced by rickettsiae. Anat. Rec. 1984;208:319–327. doi: 10.1002/ar.1092080302. [DOI] [PubMed] [Google Scholar]
- Roberts RL, Hollis-Moffatt JE, Gearry RB, Kennedy MA, Barclay ML, Merriman TR. Confirmation of association of IRGM and NCF4 with ileal Crohn’s disease in a population-based cohort. Genes Immun. 2008;9:561–565. doi: 10.1038/gene.2008.49. [DOI] [PubMed] [Google Scholar]
- Rolhion N, Darfeuille-Michaud A. Adherent-invasive Escherichia coli in inflammatory bowel disease. Inflamm. Bowel Dis. 2007;13:1277–1283. doi: 10.1002/ibd.20176. [DOI] [PubMed] [Google Scholar]
- Romano PS, Arboit MA, Vázquez CL, Colombo MI. The autophagic pathway is a key component in the lysosomal dependent entry of Trypanosoma cruzi into the host cell. Autophagy. 2009;5:6–18. doi: 10.4161/auto.5.1.7160. [DOI] [PubMed] [Google Scholar]
- Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid D, Munz C. Innate and adaptive immunity through autophagy. Immunity. 2007;27:11–21. doi: 10.1016/j.immuni.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaith A, Kashkar H, Leggio SA, Addicks K, Kronke M, Krut O. Staphylococcus aureus subvert autophagy for induction of caspase-independent host cell death. J. Biol. Chem. 2007;282:2695–2706. doi: 10.1074/jbc.M609784200. [DOI] [PubMed] [Google Scholar]
- Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy plays an essential anti-viral role in Drosophila against vesicular stomatitis virus. Immunity. 2009;30:588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J. Biol. Chem. 2008;283:33175–33182. doi: 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- Sinha S, Colbert CL, Becker N, Wei Y, Levine B. Molecular basis of the regulation of Beclin 1-dependent autophagy by the gammaherpesvirus 68 Bcl-2 homolog M11. Autophagy. 2008;4:989–997. doi: 10.4161/auto.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir D, Chen WL, Choi J, Wakita T, Yen TS, Ou JH. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–2161. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Franchi L, Toma C, Ashida H, Ogawa M, Yoshikawa Y, Mimuro H, Inohara N, Sasakawa C, Nun˜ez G. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007;3:e111. doi: 10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mul JJ, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat. Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc. Natl. Acad. Sci. USA. 2009;106:2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallo´czy Z, Jiang W, Virgin HW, 4th, Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc. Natl. Acad. Sci. USA. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YK, Kusuma CM, St John LJ, Vu HA, Alibek K, Wu A. Induction of autophagy by anthrax lethal toxin. Biochem. Biophys. Res. Commun. 2009;379:293–297. doi: 10.1016/j.bbrc.2008.12.048. [DOI] [PubMed] [Google Scholar]
- Tang H, Da L, Mao Y, Li Y, Li D, Xu Z, Li F, Wang Y, Tiollais P, Li T, et al. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology. 2009;49:60–71. doi: 10.1002/hep.22581. [DOI] [PubMed] [Google Scholar]
- Taylor MP, Kirkegaard K. Modification of cellular autophagy protein LC3 by poliovirus. J. Virol. 2007;81:12543–12553. doi: 10.1128/JVI.00755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terebiznik MR, Raju D, Va´zquez CL, Torbricki K, Kulkarni R, Blanke SR, Yoshimori T, Colombo MI, Jones NL. Effect of Heliocobacter pylori’s vacuolating cytotoxin on the autophagy pathway in gastric epithelial cells. Autophagy. 2009;5:370–379. doi: 10.4161/auto.5.3.7663. [DOI] [PubMed] [Google Scholar]
- Veneault-Fourrey C, Barooah M, Egan M, Wakley G, Talbot NJ. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science. 2006;312:580–583. doi: 10.1126/science.1124550. [DOI] [PubMed] [Google Scholar]
- Vergne I, Chua J, Singh SB, Deretic V. Cell biology of mycobacterium tuberculosis phagosome. Annu. Rev. Cell Dev. Biol. 2004;20:367–394. doi: 10.1146/annurev.cellbio.20.010403.114015. [DOI] [PubMed] [Google Scholar]
- Virgin HW, Levine B. Autophagy genes in immunity. Nat. Immunol. 2009;10:461–470. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DH, Popov VL, Crocquet-Valdes PA, Welsh CJ, Feng HM. Cytokine-induced, nitric oxide-dependent, intracellular antirickettsial activity of mouse endothelial cells. Lab Invest. 1997;76:129–138. [PubMed] [Google Scholar]
- Wang Y, Weiss LM, Orlofsky A. Host cell autophagy is induced by Toxoplasma gondii and contributes to parasite growth. J. Biol. Chem. 2009a;284:1694–1701. doi: 10.1074/jbc.M807890200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH, Wu J-J, Lei H-Y. The autophagic induction in Helicobacter pylori-infected macrophage. Exp. Biol. Med. 2009b;234:171–180. doi: 10.3181/0808-RM-252. [DOI] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RA, Tetley L, Mottram JC, Coombs GH. Cysteine peptidases CPA and CPB are vital for autophagy and differentiation in Leishmania mexicana. Mol. Microbiol. 2006;61:655–674. doi: 10.1111/j.1365-2958.2006.05274.x. [DOI] [PubMed] [Google Scholar]
- Williams RA, Woods KL, Juliano L, Mottram JC, Coombs GH. Characterization of unusual families of ATG8-like proteins and ATG12 in the protozoan parasite Leishmania major. Autophagy. 2009;5:159–172. doi: 10.4161/auto.5.2.7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, Zhang J, Si X, Gao G, Mao I, McManus BM, Luo H. Autophagosome Supports Coxsackievirus B3 Replication In Host Cells. J. Virol. 2008;82:43–53. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, Takada H, Goldman WE, Fukase K, Silverman N, et al. Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nat. Immunol. 2008;9:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SY, Ha YE, Choi JE, Ahn J, Lee H, Kweon HS, Lee JY, Kim DH. Coxsackievirus B4 uses autophagy for replication after calpain activation in rat primary neurons. J. Virol. 2008;82:11976–11978. doi: 10.1128/JVI.01028-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Perry CN, Huang C, Iwai-Kanai E, Carreira RS, Glembotski CC, Gottlieb RA. LPS-induced autophagy is mediated by oxidative signaling in cardiomyocytes and is associated with cytoprotection. Am. J. Physiol. Heart Circ. Physiol. 2009;296:H470–H479. doi: 10.1152/ajpheart.01051.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-X(L) and induction of autophagy. EMBO Rep. 2009;10:285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J. Biol. Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, Cadwell K, Delgado MA, Ponpuak M, Green KG, et al. Autophagosomeindependent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4:458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Spector SA. Human immunodeficiency virus type-1 infection inhibits autophagy. AIDS. 2008;22:695–699. doi: 10.1097/QAD.0b013e3282f4a836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Jiang X, Liu D, Fan Z, Hu X, Yan J, Wang M, Gao GF. Autophagy is involved in influenza A virus replication. Autophagy. 2009;5:321–328. doi: 10.4161/auto.5.3.7406. [DOI] [PubMed] [Google Scholar]