Abstract
Crustaceans possess remarkably diverse appendages, both between segments of a single individual as well as between species. Previous studies in a wide range of crustaceans have demonstrated a correlation between the anterior expression boundary of the homeotic (Hox) gene Ultrabithorax (Ubx) and the location and number of specialized thoracic feeding appendages, called maxillipeds. Given that Hox genes regulate regional identity in organisms as diverse as mice and flies, these observations in crustaceans led to the hypothesis that Ubx expression regulates the number of maxillipeds and that evolutionary changes in Ubx expression have generated various aspects of crustacean appendage diversity. Specifically, evolutionary changes in the expression boundary of Ubx have resulted in crustacean species with either 0, 1, 2, or 3 pairs of thoracic maxillipeds. Here we test this hypothesis by altering the expression of Ubx in Parhyale hawaiensis, a crustacean that normally possesses a single pair of maxillipeds. By reducing Ubx expression, we can generate Parhyale with additional maxillipeds in a pattern reminiscent of that seen in other crustacean species, and these morphological alterations are maintained as the animals molt and mature. These results provide critical evidence supporting the proposition that changes in Ubx expression have played a role in generating crustacean appendage diversity and lend general insights into the mechanisms of morphological evolution.
Keywords: appendages, arthropods, development, Hox
The morphology and structure of crustacean appendages are as diverse as their assorted functions, and these appendages not only vary between species, but between different segments of the same individual as well. Appendages of the posterior head segments are part of the jaw apparatus that crushes food and moves it to the mouth during feeding. The more posterior appendages of the crustacean trunk serve numerous roles including mating, defense, and locomotion. The pattern of these segmental specializations varies between species, and is often used as a criterion for subdividing crustaceans into various groups. For example, brine shrimp possess appendages throughout the entire trunk that are used in locomotion. These appendages are similar to one another, yet they are quite distinct from the head appendages used in feeding. Other crustaceans, however, possess a variety of specialized appendages within the trunk. Lobsters, for example, have certain anterior, thoracic appendages that are morphologically similar to the mouthparts and serve as additional feeding appendages. These modified thoracic appendages are called maxillipeds (“jaw-feet”), and crustaceans may possess up to 3 pairs of these specialized appendages.
A striking correlation has been found between the anterior expression boundary of the Hox gene Ultrabithorax (Ubx) and the position and number of maxillipeds that develop in crustaceans (1–4). Hox genes are members of a highly conserved family of transcription factors that specify regional identity in diverse animal body plans (5). Experimentally altering boundaries of Hox gene expression has produced dramatic phenotypes in organisms such as flies and mice. Therefore, it is possible that shifting Hox expression may have similar morphological consequences in crustaceans and could provide one potential mechanism contributing to the evolution of crustacean appendage diversity.
To test these hypotheses regarding the role of Ubx in crustacean appendage specification and evolution, we characterized Ubx in the malacostracan amphipod crustacean Parhyale hawaiensis, an emerging model system. We examined both mRNA and protein expression, and found Ubx expression throughout the walking and grasping appendages of the second through eighth thoracic appendages, but no expression in the maxilliped appendage of the first thoracic segment. We then developed an siRNA-based approach to knock down gene function in Parhyale, and used this technique to functionally test the developmental role of PhUbx directly in this crustacean. By reducing Ubx expression in Parhyale, we were able to transform multiple walking legs to a maxilliped-like identity. The extent of transformation varied among thoracic segments in a pattern that replicates the morphological variation naturally occurring among wild crustacean species that possess multiple pairs of maxillipeds.
Results
Ubx Expression Correlates with Appendage Identity in Parhyale.
Molecular characterization of the Parhyale ortholog of Ultrabithorax (PhUbx) revealed that alternative splicing generates 2 different mRNAs (PhUbx-I and PhUbx-II) that vary only in the first few amino acids they encode (see Fig. S1). We initially analyzed the pattern of PhUbx expression by in situ hybridization with a probe that recognizes both mRNAs and immunostaining with polyclonal antisera that recognizes both protein products (Fig. 1). At stage 23–25 (staging convention of ref. 6), a time point when the maxillipeds on the first thoracic segment (T1) can be easily distinguished from the remaining thoracic appendages (on T2–T8), the PhUbx mRNA, and protein patterns are identical (Fig. 1 A and C). Consistent with previous studies, expression in Parhyale correlates with appendage identity—PhUbx expression is absent from the jaw-like maxilliped appendages of T1, but present in the rest of the thoracic appendages: the grasping gnathopods of T2 and T3, and the walking legs of T4–T8 (Fig. 1 A and B). We also consistently observe that expression in T2 and T3 is lower than expression in T4–T8 (Fig. 1 A and C), and at late stages of development, T2 shows slightly lower expression than T3, especially in the most distal parts of the limb (Fig. 1A).
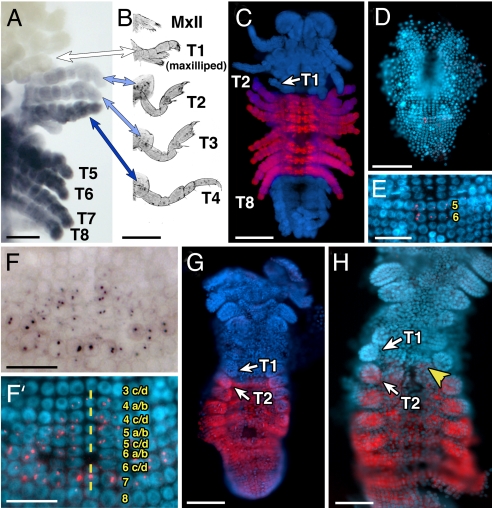
Fig. 1.
Parhyale Ubx expression during embryonic development. (A) and (F) are brightfield images of PhUbx expression (purple) and (C–E and F′–H) are DAPI images (highlighting nuclei) with PhUbx expression overlaid in red. All images display a ventral view of the embryo with anterior to the top except for (B), which shows ventral views of hatchling appendages. (A) PhUbx mRNA in a stage 23 Parhyale embryo is expressed at lower levels in the second and third thoracic segments (T2 and T3) and appendages, at higher levels throughout the remaining thoracic segments and appendages (T4–T8), and is absent from the T1 segment and maxillipeds. (B) Hatchling thoracic (T1–T4) and head (MxII) appendages corresponding to the embryonic appendages shown in (A). Colors of each arrow reflect the level of PhUbx expression in that appendage. (C) Ventral view of a stage 24 Parhyale embryo stained with an antibody recognizing PhUbx protein. The PhUbx protein expression boundaries and levels are consistent with the mRNA expression in (A). (D) PhUbx mRNA expression is initially detected around stage 12 in 2 parasegment precursor rows (PSPR 5 & 6) that will give rise to parts of the second through fourth thoracic segments. (E) Higher magnification of embryo in (D). As development proceeds, PhUbx is expressed in the more posterior parasegment precursor row cells (rows 7 & 8 in F′) as well as some of the more anterior row cells that will contribute to the posterior neuroectoderm of the first thoracic segment (F and F′). (F′) Embryonic ventral midline is marked by yellow dashes, numbers indicate PSPRs and their subsequent daughter cells. By stage 18, PhUbx mRNA (H) and protein (G) are expressed in the appendages of T2–T8. In addition, low levels of PhUbx transcript are detected in the abdomen and expression is maintained in neurons of the T1 neuromere (yellow arrowhead). [Scale bars, (E, F, and F′) 25 μm, (A and H) 50 μm, (C, D, and G) 100 μm, (B) 200 μm.]
We then looked at earlier stages to understand how this expression pattern forms. In Parhyale, each parasegment arises from individual rows of cells (called parasegment precursor rows, or PSPRs) that become organized during early germband stages (6). We found that PhUbx transcripts are first detected at stage 12 in the most medial cells of PSPR 5 and 6 (Fig. 1 D and E)—the progeny of these PSPRs will produce ectodermal derivatives stretching from posterior T2 to anterior T4. At stage 14 (Fig. 1F and F′), expression expands laterally in PSPR 5 and 6, anteriorly into progeny of PSPR 4, and posteriorly into PSPRs that form the remaining thoracic segments. PhUbx mRNA expression in PSPR 4 is detected only after this row has divided into a/b and c/d progeny. At stages 15–16, PhUbx expression in the 4 a/b row (and subsequently formed 4a and 4b rows) is confined to the medial (ventral) region and does not extend laterally to the cells that contribute to the posterior portion of the developing T1 appendage. This leads to an expression pattern at stage 18 (Fig. 1H) where Ubx mRNA expression has a parasegmental anterior boundary in the medially located neuroectoderm, but a segmental boundary more laterally so that expression is seen throughout the T2 limb primordia, but not in the T1 limb primordia. This same pattern can also be seen for PhUbx protein at stage 18 (Fig. 1G). At these stages, expression is also seen throughout the abdomen. By stage 25, however, abdominal expression is restricted to a small number of segmentally repeated cells. It should be noted that there is a significant delay between the appearance of PhUbx mRNA and PhUbx protein. PhUbx protein is not detected before stage 16–17. Even for mRNA, PhUbx transcripts are restricted to the nucleus until stage 15. Probes specific to each splice variant revealed an identical expression pattern with respect to segmental boundaries, although expression of PhUbx-I appears weaker than PhUbx-II, but this difference in signal intensity may be due to the necessarily short length of the PhUbx-I specific probe.
Knockdown of PhUbx in Parhyale.
To test the hypothesis that Ubx defines the distinction between the maxillipeds and the remaining walking and grasping legs of the thorax in crustaceans we developed a strategy to knock down gene function during Parhayle development using injection of siRNAs, a technique effective in several diverse animal taxa. To demonstrate the efficacy of this technique in Parhyale, we chose the Dll gene as a positive control. Dll and its vertebrate homologs, the Dlx genes, function in appendage development in invertebrates and vertebrates (reviewed in ref. 7). Reduction in Dll activity results in the truncation of appendages in a wide variety of arthropods including Drosophila (8), Tribolium (9) and the spider Cupiennius salei (10). We isolated 3 Dll genes from Parhyale (Fig. S2). Of these, PhDll-early (PhDll-e) shows a similar expression pattern (Fig. 2A) to Distal-less in other arthropods and to Dll protein expression in Parhyale as detected with a broadly cross-reactive antibody (6). PhDll-late1 and PhDll-late2 (PhDll-l1 and PhDll-l2), are expressed much later in development (stage 23) in a subset of the patterns seen for PhDll-e, and thus we focused on PhDll-e for functional analysis. We designed a chemically modified siRNA oligo (Stealth siRNAs, Invitrogen) targeting a nonconserved region of PhDll-e. Injection of that siRNA at the 1-cell stage produced truncated appendages in hatchlings (11 of 80 animals; Fig. 2C, E, and F) consistent with the Dll knockdown phenotype observed in diverse arthropods (8–10). Control injections of siRNAs designed to target DsRed gave no morphological abnormalities (n = 64). These results demonstrate that siRNA injection knocks down gene function in Parhyale.
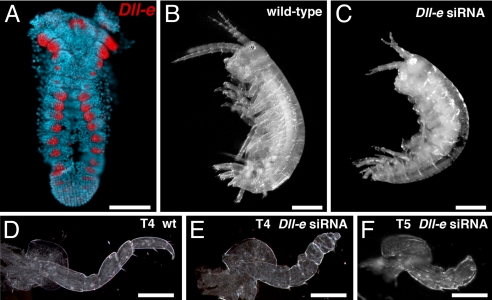
Fig. 2.
Parhyale Dll expression and phenotype. (A) Parhyale Dll-e mRNA is expressed in the developing limb primordia in a pattern that matches what has been previously described for Dll protein using a cross-reactive antibody (6). (B) Wild-type Parhyale displaying a normal complement of limbs. (C) Injection of Dll-e siRNA results in hatchlings with truncated appendages. (D--F) Higher magnification views of dissected limbs from wild-type (D) and siRNA knockdown animals (E and F). Knockdown of Dll-e causes truncation or elimination of the more distal limb segments (E and F). [Scale bars, (A–C) 200 μm, (D–F) 100 μm.]
We then used the same approach to examine PhUbx function. Three nonoverlapping siRNAs were designed to nonconserved regions of PhUbx (yellow highlighted regions in Fig. S1A) and injected as a pool or individually (Table S1). The PhUbx siRNA injections resulted in the transformation of T2 and T3 toward a T1-like maxilliped (Table S1 and Figs. 3 and 4) based on several morphological criteria, two of which were frequently and easily observed. First, the only thoracic appendages in Parhyale that contain branches are those found on the first thoracic segment (T1). These branches develop on 2 of the proximal limb segments, the basis and the ischium (see Fig. S3 for details), and they give T1 appendages their ability to serve as jaws and process food. The remaining thoracic limb appendages do not possess these branches in wild-type Parhyale, but PhUbx siRNA-treated animals develop T2 and T3 appendages with branches on the basis and ischium (arrow in Fig. 3D and asterisks in Fig. 4F, G, I, and J). In addition, the branches on transformed appendages have shapes and bristle patterns similar to those of T1 appendages. Second, T1 appendages have much shorter basis segments than T2–T8 appendages. PhUbx siRNA-injected animals also possess shortened T2 and T3 basis segments (Fig. 4F, G, I, and J). Less frequently, we observed additional morphological changes that confirm a transformation of T2 and T3 appendages to a T1 maxilliped-like identity. These included missing coxal plates from T2 and T3 appendages, missing gills from T3 appendages, altered comb-like bristles on the carpus of T2, more rounded and shortened T2 propodi, altered joints between the carpus and propodus in T2, and T2 dactyls that extended from a medial location. Also, the T2 limbs themselves were often positioned more ventrally and held against the mouth like T1 maxillipeds. In addition, some PhUbx siRNA-injected animals displayed defects in gut development. The fact that none of the appendage or gut phenotypes were seen with control siRNA injections and that 3 unique PhUbx siRNAs individually produce the same phenotypes suggests our transformations result from specifically knocking down PhUbx function. We also immunostained PhUbx siRNA-injected embryos at stage 23 and found that PhUbx protein levels were clearly reduced, although not eliminated (Fig. S4). Most PhUbx-knockdown animals did not survive long after hatching (possibly due to gut defects), but some survived for up to 3 months (becoming adults). In the surviving individuals none of the morphological changes described above underwent reversion, even after many molts.
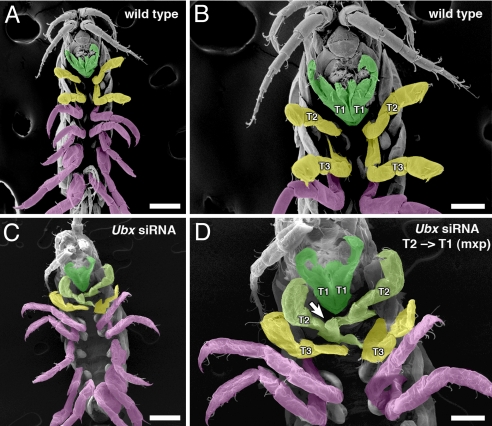
Fig. 3.
Transformation of thoracic appendage identity by PhUbx knockdown. Ventral view scanning electron micrograph (SEM) of a wild-type Parhyale hatchling (A and B) and a hatchling of an embryo injected with PhUbx siRNAs (C and D). Anterior is toward the top. Higher magnifications of (A and C) are shown in (B and D) respectively. Appendage identity is indicated by color: green for maxillipeds (T1), yellow for gnathopods (T2 and T3), and magenta for walking legs (T4–T8). (A and B) In wild-type hatchlings, the first thoracic (T1) segment bears 1 pair of branched appendages, called maxillipeds (green), which function in feeding and are held against the other mouthparts of the head. The remaining thoracic appendages (T2–T8) lack these branches. (C and D) The T1 appendages (green) in PhUbx siRNA-injected animals appear unaffected (A and B). However, the second thoracic appendages of the siRNA-injected hatchlings (light green shading in C and D) possess additional branches (arrow) on the same limb segments (basis and ischium) as the maxillipeds, indicating transformation of appendage identity. The more posterior thoracic appendages in this PhUbx siRNA-injected animal retain their wild-type identity, but in more severely affected animals the third thoracic appendages are partially transformed to a more maxilliped-like identity (Fig. 4). [Scale bars, (A and C) 100 μm, (B and D) 50 μm.]
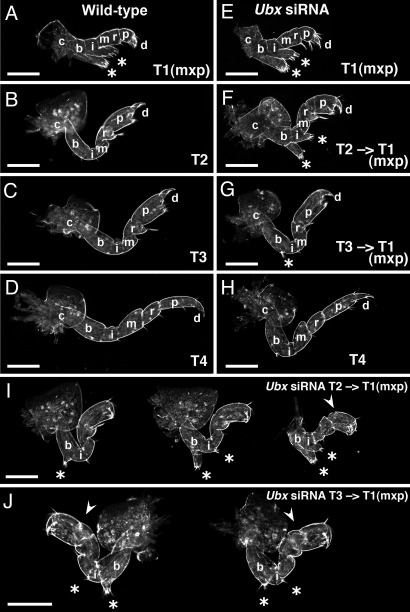
Fig. 4.
Characterization of transformations in PhUbx knockdown hatchling appendages. Darkfield view of a complement of appendages from the left side of a single representative wild-type (A–D) and a single representative PhUbx siRNA-injected (E–H) Parhyale hatchling. The first thoracic (T1) appendages, or maxillipeds, display no morphological differences between uninjected (A) and injected animals (E). The second thoracic (T2) appendages of the siRNA-injected hatchling (F) display a dramatic transformation from walking-leg to maxilliped-like identity in which the basis (b) is shortened, and the basis and ischium (i) develop branches (*) similar to those on wild-type maxillipeds of T1 (A). (G) The T3 appendage appears slightly shorter in the injected animal and a small branch (*) indicates weak transformation. The T4 appendages appear very similar in both (D and H). (I) Three additional examples of T2 to maxilliped transformations. The severity of transformation of T2 appendages toward maxilliped identity varied among PhUbx siRNA hatchlings from single small branches on the basis (*, appendage on left) to more dramatic phenotypes (appendage on right) with reduced basis length, pronounced branches on the basis and ischium (*), absence of the comb-like bristle, absence of the coxal plate, and a short/round propodus (arrowhead). (J) Pair of T3 appendages from the same individual showing T3 to maxilliped transformations. Some PhUbx siRNA hatchlings also displayed multiple maxilliped-like (T1-like) characters in their T3 appendages including shortened basis, loss or reduction of the comb-like bristle on the carpus, addition of bristles or a branch with distal bristles on the basis and ischium (*), and an altered joint on the propodus (arrowhead). (Scale bars, 100 μm in all panels.) Additional limb segment labels described in Fig. S1.
Discussion
The expression pattern of Parhyale Ubx is consistent with that seen in other crustaceans: Ubx expression is absent from all maxillipeds, but present in the remaining thoracic appendages (and the segments bearing these appendages). Thus, our expression data supports the hypothesis that in crustaceans Ubx plays a role in distinguishing between segments bearing jaw-like feeding appendages and the remaining thoracic segments, which possess primarily locomotory appendages. However, by developing methods for knocking down Ubx function in Parhyale we are able to go beyond correlation to functionally test, and strongly support, this hypothesis. While wild-type Parhyale have just a single pair of maxillipeds positioned on the first thoracic segment, Parhyale with reduced Ubx expression develop additional maxillipeds through the homeotic transformation of more posterior thoracic appendages.
Remarkably, the phenotypes we obtain by experimentally knocking down PhUbx closely resemble the patterns and morphologies seen during crustacean evolution. First, our manipulations transform only the T2 and T3 appendages toward a maxilliped identity while leaving the more posterior thoracic appendages unaffected. Likewise, evolution has produced malacostracan crustacean species possessing jaw-like appendages on T1-T3, but no species develop appendages with this morphology posterior to T3. This may reflect the lack of adaptive value for jaw-like appendages posterior to T3, but it might also reflect some of the underlying mechanisms that control appendage diversity in these species. In Parhyale, T4–T8 express higher levels of PhUbx than T2–T3, and the lack of T4–T8 transformations in our experiments may be due to the fact that we reduced, but did not eliminate, Ubx protein expression. This highlights the potentially important role that variation in levels of expression plays, both in generating differences between segments within an individual during development and in the evolutionary changes that have occurred during crustacean diversification. Finally, there may be additional regulatory genes, including the more posterior Hox gene abdominal-A (abdA), expressed in T4-T8 that maintain the identity of these appendages irrespective of the state of PhUbx expression.
Second, siRNA knockdowns generate a gradation of transformation in T2 and T3, with T2 more strongly affected than T3 (Fig. 4). This morphologically graded series of maxillipeds is reminiscent of the wild-type pattern observed in other crustaceans. For example, crayfish possess 3 pairs of morphologically distinct maxillipeds on their T1-T3 segments. In these crustaceans, the T1 appendage most resembles the jaw appendages, T2 is larger and less jaw-like, and T3 is the least jaw-like with only some of the branches and bristles characteristic of maxillipeds. Additional Hox genes, such as Sex combs reduced (Scr) and Antp, may play a role in specifying this morphological variation. Previous studies in crayfish have shown that Ubx is initially expressed in T2–T8, but then expression retracts from T2 and T3 early in development (3). Scr is expressed in T1, and it retains this boundary even as Ubx expression retracts in crayfish (3). Thus, both T1 and T2 appendages in crayfish eventually lack Ubx expression, but differ in Scr expression. Likewise, we have not observed an expansion of Scr expression in PhUbx knockdown Parhyale embryos, and this may, in part, account for differences between wild-type maxillipeds on T1 and the T2–T3 appendages transformed toward maxilliped identity in Parhyale in which Ubx has been knocked down.
In summary, our results, plus those described in the accompanying paper by Pavlopoulos et al. (11), clearly indicate that Parhyale Ubx plays a role in distinguishing the T1 maxillipeds from the more posterior thoracic appendages and provide critical functional support for the hypothesis that changes in Ubx expression have played an important role in the evolution of crustacean diversity. Our data also suggest that variation in maxilliped number and morphology in crustaceans could have initially arisen from relatively subtle and gradual changes in the global levels of Ubx expression, which were then subsequently refined by alterations in cis or trans regulators resulting in the segment specific patterns of Ubx expression seen in extant crustaceans. Such a scenario would also allow for gradual changes in morphology without the need to invoke the appearance of “hopeful monsters.” Continued comparative, molecular, and genetic analysis of Ubx expression and function should yield further insights into the mechanisms that control morphological evolution.
Materials and Methods
Cloning Parhyale Ubx, and Dll.
Parhyale embryos of mixed ages were collected to make cDNA as previously described (12). The degenerate PCR primers for Ubx were described previously (3) and for Dll were GGNAARAARATGMGNAARCC; CCADATYTTNACYTGNGTYTG, and GCNGCNARYTCNGCNCKYTCNGG. The initial Parhyale sequences were extended by 5′ and 3′ RACE and Genome Walker (Clontech). Full-length sequences were amplified by PCR from cDNA using gene specific primers (SuperScript, Gibco BRL). Sequence accession numbers: PhDll-e, FJ617569; PhUbx-I, FJ628448; and PhUbx-II, FJ628449.
In Situ Hybridization and Antibody Staining.
Embryo dissection, fixation, in situ hybridization, and antibody staining followed previously published protocols (13). For the production of a polyclonal antibody to PhUbx, we amplified the entire PhUbx-I ORF using the oligos TATAGTCGACTCAGCTCATCCTCGACGATGGA and ATATGCGGCCGCTTAGTTTTGTCCGGGGTTTTG, digested the product with SalI-NotI, and subcloned in frame into the pGEX-4T-1 vector. The resulting construct was used to produce and purify the bacterial GST-fusion protein as described in the manufacturer's protocol (Amersham Biosciences). Polyclonal antibodies against the purified fusion protein were raised in 3 rats at Covance Research Products. All 3 antisera give identical staining patterns and recognize both PhUbx-I and PhUbx-II protein. Monoclonal FP6.87 (14) recognizes Ubx and abdA proteins in a range of crustaceans and was a gift from Rob White.
RNAi in Parhyale.
Morpholino mediated knockdown has recently been used to address vasa function in early Parhyale embryos (15), and our own pilot experiments with morpholino and dsRNA injections have shown that these techniques work, but lead to relatively transient knockdown in Parhyale with poor penetrance at late germband stages. Stealth siRNA duplexes (Invitrogen) were designed against PhDll and PhUbx using the BLOCK-IT RNAi Designer tool (Invitrogen). For PhUbx and DsRed, these siRNAs target 3 nonoverlapping 25 nucleotide regions in nonconserved coding regions. The siRNA sequences (forward strand) are as follows:
PhDll-e AGAUGCAGAACAAUCUUGCGCUGAU;
PhUbx-A GCCTGCACCACGACAAGATGCTTAT;
PhUbx-B CCAAGACCACATGACTCCAAATCCT;
PhUbx-C CAGAATGGATACGGTGCCAAAGATA;
DsRed-A AGUACGGCUCCAAGGUGUACGUGAA;
DsRed-B AGGUGAAGUUCAUCGGCGUGAACUU;
DsRed-C CCGUAAUGCAGAAGAAGACUAUGGG;
siRNAs were resuspended in water and mixed with a small amount of Phenol Red. Embryo injection was carried out as described previously (13). Approximately 44 picoliters of 67–200 μM PhUbx siRNAs or 16 picoliters of 400 μM PhDll siRNAs were injected into 1-cell embryos or both cells of 2-cell embryos.
Microscopy.
Images were captured with a Spot Camera and figures assembled with Photoshop (Adobe). To create overlapping images for histochemical plus DAPI images, the histochemical signal was overlaid on the DAPI images by inverting the brightfield image, eliminating the blue and green channels, and applying the screen command to allow the DAPI signal to show through. Parhyale hatchling appendages were removed individually, placed on microscope slides, arranged in 70% glycerol, coverslipped, and viewed with darkfield optics on a Zeiss Axiophot microscope. For scanning electron microscopy (SEM), Parhyale hatchlings were fixed in 3.7% formaldehyde in filtered seawater.
Supplementary Material
Acknowledgments.
We thank Paul Liu, Ron Parchem, and Crystal Chaw for valuable comments and suggestions; Pat Kysar, Grete Adamson, Guangwei Min, and Reena Zalpuri for help with SEM. N.H.P. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903105106/DCSupplemental.
References
- 1.Averof M, Patel NH. Crustacean appendage evolution associated with changes in Hox gene expression. Nature. 1997;388:682–686. doi: 10.1038/41786. [DOI] [PubMed] [Google Scholar]
- 2.Abzhanov A, Kaufman TC. Novel regulation of the homeotic gene Scr associated with a crustacean leg-to-maxilliped appendage transformation. Development. 1999;126:1121–1128. doi: 10.1242/dev.126.6.1121. [DOI] [PubMed] [Google Scholar]
- 3.Abzhanov A, Kaufman TC. Embryonic expression patterns of the Hox genes of the crayfish Procambarus clarkii (Crustacea, Decapoda) Evol Dev. 2000;2:271–283. doi: 10.1046/j.1525-142x.2000.00066.x. [DOI] [PubMed] [Google Scholar]
- 4.Shiga Y, Yasumoto R, Yamagata H, Hayashi S. Evolving role of Antennapedia protein in arthropod limb patterning. Development. 2002;129:3555–3561. doi: 10.1242/dev.129.15.3555. [DOI] [PubMed] [Google Scholar]
- 5.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 6.Browne WE, Price AL, Gerberding M, Patel NH. Stages of embryonic development in the amphipod crustacean, Parhyale hawaiensis. Genesis. 2005;42:124–149. doi: 10.1002/gene.20145. [DOI] [PubMed] [Google Scholar]
- 7.Panganiban G, Rubenstein JLR. Developmental functions of the Distal-less/Dlx homeobox genes. Development. 2002;129:4371–4386. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- 8.Cohen SM, Bronner G, Kuttner F, Jurgens G, Jackle H. Distal-less encodes a homeodomain protein required for limb development. Nature. 1989;338:432–434. doi: 10.1038/338432a0. [DOI] [PubMed] [Google Scholar]
- 9.Beermann A, et al. The Short antennae gene of Tribolium is required for limb development and encodes the orthologue of the Drosophila Disal-less protein. Development. 2001;128:287–297. doi: 10.1242/dev.128.2.287. [DOI] [PubMed] [Google Scholar]
- 10.Schoppmeier M, Damen WG. Double-stranded RNA interference in the spider Cupiennius salei: The role of Distal-less is evolutionarily conserved in arthropod appendage formation. Dev Genes Evol. 2001;211:76–82. doi: 10.1007/s004270000121. [DOI] [PubMed] [Google Scholar]
- 11.Pavlopoulos A, et al. Probing the evolution of appendage specialization by Hox gene misexpression in an emerging model crustacean. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0902804106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price AL, Patel NH. Investigating divergent mechanisms of mesoderm development in arthropods: The expression of Ph-twist and Ph-mef 2 in Parhyale hawaiensis. J Exp Zool (Mol Dev Evol) 2008;310B:24–40. doi: 10.1002/jez.b.21135. [DOI] [PubMed] [Google Scholar]
- 13.Rehm EJ, Hannibal RL, Chaw RC, Vargas-Vila MA, Patel NH. Emerging Model Organisms: A Laboratory Manual. Volume 1. New York: Cold Spring Harbor Press; 2009. pp. 373–404. [Google Scholar]
- 14.Kelsh R, Weinzierl RO, White RA, Akam M. Homeotic gene expression in the locust Schistocerca: An antibody that detects conserved epitopes in Ultrabithorax and abdominal-A proteins. Dev Genet. 1994;15:19–31. doi: 10.1002/dvg.1020150104. [DOI] [PubMed] [Google Scholar]
- 15.Kizil GO, Havemann J, Gerberding M. Germ cells in the crustacean Parhyale hawaiensis depend on Vasa protein for their maintenance but not for their formation. Dev Biol. 2009;327:230–239. doi: 10.1016/j.ydbio.2008.10.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.