Abstract
Respiratory diseases provide an attractive target for gene silencing using small nucleic acids since the respiratory epithelium can be reached by inhalation therapy. Natural surfactant appears to facilitate the uptake and distribution of these types of molecules making aerosolized nucleic acids a possible new class of therapeutics. This article will review the rationale for the use of External Guide Sequence (EGS) in targeting specific mRNA molecules for RNase P-mediated intracellular destruction. Specific destruction of target mRNA results in gene-specific silencing similar to that instigated by siRNA via the RISC complex. The application of EGS molecules specific for influenza genes are discussed as well as the potential for synergy with siRNA. Furthermore, EGS could be adapted to target other respiratory diseases of viral etiology as well as conditions such as asthma.
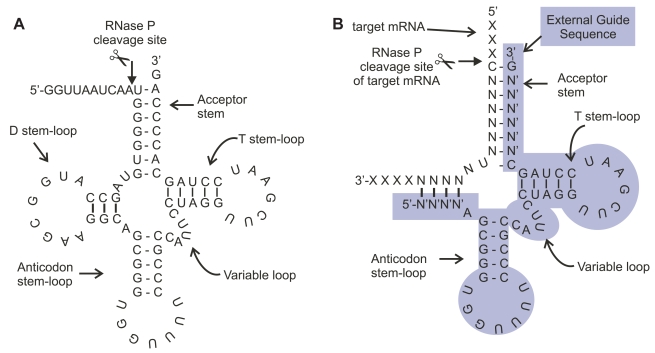
This article will review the use of External Guide Sequence (EGS) directed gene expression silencing which promotes messenger RNA (mRNA) cleavage by RNase P, and the potential applications of this technology to treatment and prophylaxis of influenza and other respiratory diseases such as post viral exacerbations of asthma (Dreyfus et al 2004; Nyce 1997; Nyce and Metzger 1997; Popescu 2005; Trian et al 2006; Ulanova et al 2006). Gene silencing with RNase P is based upon the discovery that the ubiquitous RNA enzyme RNase P, required for processing of precursor transfer RNA (pre-tRNA) to the mature transfer RNA (tRNA) can also be programmed to degrade mRNA (Baer et al 1988; Gopalan et al 2002; Guerrier-Takada et al 1988; Guerrier-Takada et al 1989; Guerrier-Takada and Altman 2000; Raj and Liu 2003). Since RNase P recognizes the structure of pre-tRNA and certain conserved sequence elements of that structure (Figure 1A), a single stranded RNA termed EGS, can be designed to bind non-covalently to target mRNA resulting in a bimolecular structure that resembles pre-tRNA and is recognized and cleaved by RNase P (Figure 1B). EGSs, as a therapeutic agent, fall into the category of oligonucleotides – respiratory diseases as a whole may be particularly amenable to gene silencing using small nucleic acids since the respiratory epithelium is accessible by inhalation delivery and appears to spontaneously take up these types of molecules (Finotto, Buerke et al 2001; Finotto, De Sanctis et al 2001; Massaro et al 2004; Moschos et al 2007; Nyce 1997; Thomas et al 2007).
Figure 1.
(A) The structure of pre-tRNA (Gln): The structure of precursor transfer RNA (pre-tRNA), a typical substrate for RNase P. RNase P is an abundant RNA enzyme that processes a precursor tRNA transcript through cleavage of a 5′ leader sequence from the transcript. Following processing, the tRNA can accept an amino acid and function in protein synthesis. (B) A synthetic external guide sequence (EGS, highlighted) derived from the structure of pre-tRNA (Gln, Figure 1A) bound to target mRNA forms a pre-tRNA (Gln)-like structure resulting in cleavage of a non-natural substrate mRNA (eg, target). RNase P recognizes the structure as precursor RNA and cleaves the mRNA (depicted as scissors). Thus an EGS can be designed that binds to a target mRNA through altered stem sequences maintaining a conserved stem and loop structure resembling a tRNA precursor. The EGS mRNA hybrid is then recognized as a substrate for RNase P.
EGS, a short single-stranded RNA, binds by Watson-Crick base-pairing to target mRNA and directs it to RNase P (Guerrier-Takada and Altman 2000). The bimolecular structure of EGS and mRNA forms a substrate for site specific cleavage of the mRNA moiety and inactivation of the mRNA target (Figure 2). After cleavage of the target, the EGS is released and can bind new target, thereby facilitating additional cycles of target cleavage. The rate of scissile bond cleavage is limited primarily by the concentrations of EGS and mRNA species complementary to the EGS in the nucleus and the binding affinity (Km) of the bimolecular structure to RNase P, which for the native pre-tRNA is a relatively low 9 nM (Ziehler et al 2000) RNase P concentrations are high because it is utilized for generation of tRNA in all cell types with a nucleus and active protein synthesis, thus this molecule is not rate-limiting in the reaction (Doersen et al 1985; Koski et al 1976; Robertson et al 1972). In HeLa cells, it is estimated there are 20,000 copies of RNase P per cell (Bartkiewicz et al 1989).
Figure 2.
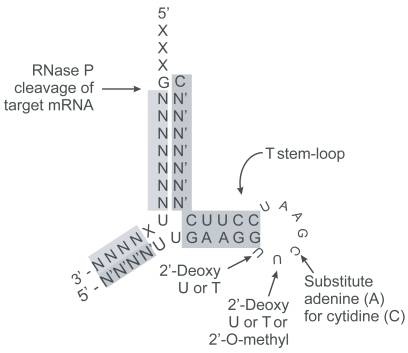
EGS can be minimized to approximately 32 nucleotides of single stranded nuclease resistant RNA by incorporation of modified bases conserving the T stem loop necessary for recognition by RNase P. The yellow highlighted nucleotides correspond to regions in the target sequences homologous to the EGS. The pink highlighted regions are 2′-O-methyl modified nuclease resistant nucleotide moieties. The minimized EGS contains only the T stem-loop structure. Other sequences such as the variable and anti-codon loops of tRNA are not required for cleavage by RNase P but may affect binding affinity.
EGS can be expressed from viral or other expression systems in different cell types and directed at different targets (Dreyfus et al 2004; Kovrigina et al 2005; Zhang and Altman 2004, Kovrigina et al 2003; Guerrier-Takada and Altman 2000; Plehn-Dujowich and Altman 1998; Li et al 2006; Barnor et al 2004; Zhu et al 2004; Kitano et al 2000; Endo et al 2001; Kraus et al 2002; Hnatyszyn et al 2001). Alternatively, EGS can be transiently expressed following high copy number transfection or electroporation of plasmid containing an RNA Pol III promoter driving the EGS sequence (Dreyfus et al 2004; Rangarajan et al 2004). For therapeutic applications in humans such as for asthma or influenza, EGS can be chemically synthesized as nuclease resistant small molecules (Ma et al 1998, 2000; Zhu et al 2004), avoiding the need for vector and promoter sequences (Figure 3). Transient transfection of these nuclease resistant molecules circumvents the risk of somatic gene mutations which could lead to malignant transformation of recipient cells when viruses or virally derived DNA sequences are used.
Figure 3.
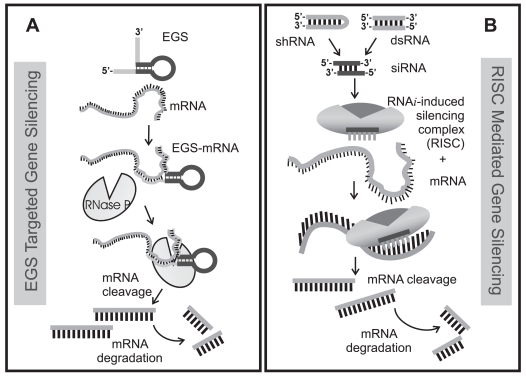
Comparison of (A) EGS directed targeting of mRNA to RNase P with (B) small double-stranded RNA (dsRNA) or regulatory short hairpin RNA (shRNA) directed targeting of mRNA to RISC. Both pathways require a nucleotide guide sequence – for RNase P, EGS is the guide while for RISC, the antisense strand of a double-stranded small interfering RNA (siRNA) is incorporated into the RISC complex. siRNAs can be directly introduced into the cell or are generated by the enzyme, Dicer from short hairpin RNA generated from a DNA template or from dsRNA. In this view, the EGS that binds to target mRNA and forms a substrate recognized by RNase P is directly analogous by function to the antisense strand of the 21–23 nucleotide siRNA that is incorporated into RISC and guides Slicer mediated mRNA cleavage. Both RNase P and the argonaute protein Slicer introduce a single cleavage site in the target – the cleaved mRNA is recognized as abnormal and further degraded by cellular nucleases.
In a research setting, using EGS expressed at high copy number in vivo from vectors is more convenient and cost effective for screening large numbers of constructs than using stably transfected cell lines or transfection of chemically synthesized EGS (Dreyfus et al 2004). We described a relatively simple screening assay in which EGS and reporter genes are transiently electroporated into a well characterized and publicly available B-lymphoblastoid cell line that expresses a functional IL-4 receptor to demonstrate the ability of EGS to silence IL-4 signaling using the human IL-4 promoter fused to the luciferase gene. We have also used a Jurkat T lymphoblastoid cell line expressing both functional IL-4and adenosine receptors important mediators of asthma and other respiratory diseases (Georas et al 1998), to confirm these results. In this type of in vitro cellular assay, EGS potency and specificity are assessed by silencing of a luciferase reporter gene. Moreover, the assay can be scaled up and automated for large numbers of candidate EGS.
The relevance of IL-4/13 receptor as a target for EGS or other gene silencing has been reviewed previously (Dreyfus et al 2004). The effects of active EGS that are RNase P dependent can be distinguished from non-specific effects of the EGS on reporter expression since they are decreased by mutation of the EGS T loop required for cleavage by RNase P. Some interesting conclusions are evident regarding potency and specificity of the anti-IL4 receptor EGS in this type of assay (Dreyfus and Fuleihan, unpublished observations). Two EGS molecules designed to target two distinct regions in the IL-4/13 common receptor mRNA each independently exhibited RNase P dependent inactivation of IL-4 signaling. Of the two EGSs, one which targets a site near the start codon of the IL-4/13 common receptor mRNA is more potent than another EGS targeting a sequence located more distal to the start codon even though both target sequences are similarly accessible based on digestion by nuclease T1 in vitro. Also, an EGS targeting the murine homologue of the human IL-4/13 receptor chain was still able to cause RNase P dependent inactivation of the human target despite the presence of mismatches in 2 of 11 nucleotides. However, these mismatches resulted in a greater than 50% decrease in target site inactivation when tested in tissue culture.
Thus, RNase P dependent EGS targeting is dependent on multiple factors related to target site accessibility and conformation in vivo – factors that are not entirely predictable using sequence based algorithms or in vitro methods. Targets can be cleaved in vivo even if significant mismatches are present between EGS and target although this cleavage is less efficient than correctly paired EGS. Methods such as DNA microarray analysis could be used to look for evidence of off target/non-specific effects of EGS. Once an optimal candidate EGS or multiple EGS are identified using an unbiased automated process capable of screening large numbers of randomized or otherwise modified EGS, these optimized EGS would then be further characterized in vitro and in vivo in human primary cells and animal models prior to human clinical trials. In the absence of a readily available reporter gene for monitoring EGS activity in vivo, a reporter gene could be constructed which expresses the target gene mRNA fused upstream of the luciferase mRNA so that cleavage of the target mRNA would proportionately reduce luciferase expression.
For in vivo applications, EGS delivery methods must be optimized to permit sufficient concentrations of EGS to enter the respiratory tract and gain access to cells of the respiratory epithelium as nuclease resistant small molecules with minimal non-specific or pro-inflammatory properties. To accomplish this, an inhaler could be optimized for delivery of EGS in a transfection reagent composed of lipids. The concentration of EGS in the cell nuclei will be the limiting factor in the rate of EGS-directed target cleavage. Since EGS molecules are RNA oligonucleotides, many of the issues encountered in delivering siRNA are applicable to EGS. In the case of siRNA, published data suggest a therapeutic effect against influenza even with the administration of unmodified RNA duplex by hydrodynamic tail vein injection in mice (Tompkins et al 2004; Larson et al 2007; Herweijer and Wolff 2007; Bradley et al 2005).
RNAi is an evolutionarily conserved expression silencing pathway which also plays a role in antiviral defense, particularly against those viruses with an RNA genome that replicate in the cytoplasm. However, RNA viruses such as influenza and vaccinia have developed means of undermining the RNAi pathway (Li and Ding 2005; Schott et al 2005; Schutz and Sarnow 2006). Viral proteins often target the enzymes in RISC, thereby attenuating the RNAi pathway rather than directly targeting the regulatory RNA mediator molecules. While the role of RNase P in antiviral defense has not specifically been explored, it is interesting to note that a putative RNase P RNA gene has been identified in camel-pox and vaccinia viruses, members of the orthopoxviruses (Yang et al 2005). These RNase P molecules were found to be enzymatically inactive and a role for these RNA enzymes in the viral life cycle has yet to be established.
We suggest that synergy may be possible between gene silencing with EGS/RNase P and gene silencing with siRNA/RNAi. As shown in Figure 3, gene silencing with EGS and RNase P and gene silencing with siRNA and the RNA-induced silencing complex (RISC) share similar characteristics. In both cases a small RNA sequence with complementarity to the target serves as a guide to cleavage of the mRNA by an endogenous RNase. Both schemes result in the cleavage of many targets per guide sequence. In the RNase P mediated gene silencing pathway, the small single stranded RNA is termed an EGS, while with RISC, the RNA is termed siRNA. In contrast, antisense technology utilizes modified DNA oligonucleotides which act pleiotropically at the level of transcription, splicing, protein synthesis and can also activate RNase H resulting in destruction of the complementary mRNA (Monia et al 1996; Monia et al 1993; Lima et al 1992; Chan et al 2006).
What then are the relative advantages and disadvantages of the RNase P pathway compared to RNAi? Advantages of gene silencing with EGS/RNase P may include more rapid onset of action than siRNA/RNAi (Zhang and Altman 2004). Experiments using EGS and shRNA expressed from identical vectors against an identical target suggest that the onset of action of EGS can be detected within 24 hours while under identical conditions, shRNA effects were evident only after 48 hours. If this 24 hour difference in onset of action is a general phenomenon that distinguishes the two, it could be highly significant for therapeutic applications. For example, in the treatment of respiratory viral infections such as influenza, a difference in onset of action of 24 hours could result in significantly different outcomes in viral titer as well as severity of symptoms and complications. In addition, where a gene silencing strategy is used for prophylactic purposes following exposure to virus, a 24 hour difference in onset of action could be highly significant since viral replication peaks within 48 hours of exposure.
Another potential advantage is that EGS is essentially single-stranded. Thus, Toll receptor 3 recognizing short, double-stranded RNA would not be expected to be activated (Kariko, Bhuyan et al 2004; Kariko, Ni et al 2004; Akira 2006; Matsukura et al 2006).
Another important point of comparison between EGS and siRNA is potency and specificity which are related, but not identical issues. A less potent drug can have a similar efficacy to a more potent drug if a higher concentration of the less potent compound is used. However, where there are significant off-target effects, increasing dosage will almost certainly lead to more non-specific side-effects such as inflammatory responses or the triggering of pro-apoptotic proteins such as p21 in the case of siRNA (Bridge et al 2003; Jackson et al 2003; Jackson and Linsley 2004; Scacheri et al 2004). At the current time, very little is known about the specificity of either siRNA or EGS under in vivo conditions and particularly in the inflammatory setting predicted to be elicited by a viral infection such as influenza, or a chronic inflammatory disease, such as asthma. Further experiments using comprehensive methods such as DNA microarrays and proteomics will be required to conclusively resolve issues of specificity of both siRNA and EGS in specific in vivo applications.
Limited in vitro evidence suggests siRNA appear equipotent to EGS if one can design the EGS based on RNase accessibility. However, an EGS targeted to an identical site as siRNA may or may not be as potent. This is due primarily to the accessibility of the EGS to its target site. For RISC, these rules are less well understood because of the involvement of helicases which interact with RISC and functions in the modulation of the secondary structure of the target mRNA and/or RISC loading (Bernstein et al 2001; Robb and Rana 2007). The availability of premade cocktails of siRNA as well as the widespread use of siRNA render this the method of choice for gene expression silencing in life sciences discovery. However, in many applications, it is also preferable to use a second, independent method, to validate results. In this context, EGS may be particularly useful given its specificity and relative ease of design and use. Finally, EGS may be the tool of choice where effective and specific siRNAs cannot be designed.
Once a target is validated, EGS can be optimized through unbiased randomization procedures leading to perhaps 100-fold or more increased potency against a specified mRNA target with only small changes in the EGS sequence (Yang et al 2006; Zhou et al 2002). EGS optimization would not be expected to increase non-specific inflammatory and/or pro-apoptotic responses in vivo however, this remains to be tested. siRNA, in contrast, cannot be optimized beyond the point at which the enzyme Slicer, a component of RISC, becomes rate-limiting (Koller et al 2006). In the context of influenza treatment, inflammation may have a beneficial additive or synergistic effect with siRNA but could also exert detrimental effects leading to the development of acute lung injury (ALI) or adult respiratory distress syndrome (ARDS) in the case of reassorted zoonotic strains. Additionally, post-viral exacerbations such as asthma may also be promoted by siRNA induced inflammatory responses. It would probably be most fair to say that the relative potency and specificity of EGS vs siRNA in vivo for treatment of disease cannot be determined on theoretical grounds because of the many interacting and unknown variables, but will require direct comparisons of the two therapies against a particular target when this is possible. It is certainly plausible that there will be some genes or tissues for which EGS silencing is more potent and specific in vivo, and other genes for which the opposite is found, which would suggest a rationale for using both drugs to target a gene of interest in therapeutic applications.
A clear advantage of siRNA is the ability to inactivate viruses such as the respiratory pathogens RSV (Respiratory Syncytial Virus) and rhinovirus that replicate in the cell cytoplasm (Phipps et al 2004; Strayer et al 2005; Kong et al 2007). Since RNase P is only present in the cell nucleus, pathogens whose transcripts reside exclusively in the cell cytoplasm cannot be targeted directly. However, a number of viruses and intracellular pathogens rely on host proteins for key steps in their life cycle (Elliott et al 2007; Fauci 1996; Fauci and Challberg 2005; Gu et al 2004). Where the identity of such proteins is known, it may be possible to target these using EGS. Furthermore, the pathogenicity of these viruses may occur through induction of cellular inflammatory cytokines such as interferon or IL-13 (Liu et al 2005); inactivation of these virally triggered cytokines by EGS or inhibition of the NF-κB transcription factor which acts as a master switch, might still be an effective immunotherapy for these cytoplasmic pathogens. Using EGS, it may be possible to differentially inactivate specific subunits of NF-κB which would still allow other subunits to engage in transcriptional activation. Alternatively, the receptors for the pathogens could also be silenced with EGS (Barth, Liang et al 2006; Barth, Schnober et al 2006; Julg and Goebel 2005; Weber et al 2006; Lederman et al 2006), preventing infection or reducing spread and virus load in an ongoing infection. Since the pathogenesis of influenza as well as RSV involves modulation of cytokines such as decreased expression of interferon, and increased synthesis of IL-4 and IL-13 that play a role in post viral exacerbations of asthma (Elliott et al 2007; Oh et al 2002; Minor et al 1976; Singh and Busse 2006; Tekkanat et al 2001; Wang et al 2004; Yasuda et al 2005), it is likely that a combination of anti-viral and anti-cytokine therapies may be required for efficacy in both influenza and post viral asthma.
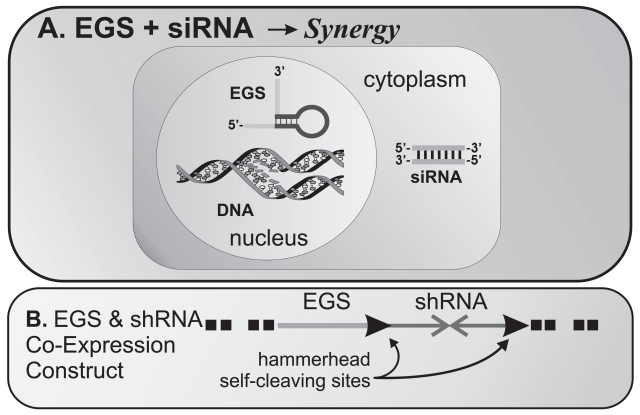
Perhaps the most intriguing possibility is that EGS and siRNA gene silencing may synergize since they use different mechanisms to inactivate target mRNA yet utilize similar or identical methods of delivery. Figure 4 depicts a scheme by which synergy might be attained by the co-expression of EGS and short hairpin RNA if expressed from DNA or EGS and siRNA if administered as RNA oligonucleotides. Advances in delivery of one modality will almost certainly benefit the delivery of the other in a particular disease process. We hypothesize that for pharmacological applications against human respiratory diseases such as influenza and asthma, as well as other diseases which are accessible to either modality, some combination of EGS and siRNA may be optimal in terms of onset of action, specificity, and potency.
Figure 4.
(A) Potential for synergy between EGS and siRNA: Co-expression of EGS in the nucleus and siRNA in the cytoplasm could effectively synergize to promote gene expression silencing since the two use different pathways. (B) Stable transfection of cDNA constructs encoding EGS and shRNA could also achieve the same results – as shown, we propose a self-cleaving transcript using hammerhead ribozymes encoded in the flanking regions of the shRNA sequences.
Acknowledgments and disclosures
The authors thank Dr Sidney Altman of Yale University for his generous assistance and for making available constructs and supplies for the synthesis of EGS directed at the human and murine IL-4 receptor described in this work. Dr David H Dreyfus is currently a member of the Clinical Faculty at the Yale University School of Medicine, Department of Pediatrics, New Haven, CT and is also the founder and Medical Director of Keren Pharmaceutical, Inc. a biotechnology company focused on the use of EGS technology in human therapeutics and research applications. Dr Fuleihan’s present address is the Department of Pediatrics, Children’s Memorial Hospital, Chicago, IL ( r-fuleihan@northwestern.edu). Dr S Mark Tompkins is an Assistant Professor in the Department of Infectious Diseases, at the University of Georgia College of Veterinary Medicine and is a consultant to Keren Pharmaceutical. Dr Lucy Y Ghoda, is an Investigator at the Webb-Waring Institute, and is a faculty member in the Division of Pulmonary Sciences, Department of Medicine, School of Medicine, University of Colorado Health Sciences Center. She is Scientific Director of Keren Pharmaceutical.
References
- Akira S. TLR signaling. Curr Top Microbiol Immunol. 2006;311:1–16. doi: 10.1007/3-540-32636-7_1. [DOI] [PubMed] [Google Scholar]
- Baer MF, Reilly RM, Mccorkle GM, et al. The recognition by RNase P of precursor tRNAs. J Biol Chem. 1988;263:2344–51. [PubMed] [Google Scholar]
- Barnor JS, Endo Y, Habu Y, et al. Effective inhibition of HIV-1 replication in cultured cells by external guide sequences and ribonuclease P. Bioorg Med Chem Lett. 2004;14:4941–4. doi: 10.1016/j.bmcl.2004.07.021. [DOI] [PubMed] [Google Scholar]
- Barth H, Liang TJ, Baumert TF. Hepatitis C virus entry: molecular biology and clinical implications. Hepatology. 2006;44:527–35. doi: 10.1002/hep.21321. [DOI] [PubMed] [Google Scholar]
- Barth H, Schnober EK, Zhang F, et al. Viral and cellular determinants of the hepatitis C virus envelope-heparan sulfate interaction. J Virol. 2006;80:10579–90. doi: 10.1128/JVI.00941-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkiewicz M, Gold H, Altman S. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes Dev. 1989;3:488–99. doi: 10.1101/gad.3.4.488. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, et al. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bradley SP, Rastellini C, Da Costa MA, et al. Gene silencing in the endocrine pancreas mediated by short-interfering RNA. Pancreas. 2005;31:373–9. doi: 10.1097/01.mpa.0000179730.69081.64. [DOI] [PubMed] [Google Scholar]
- Bridge AJ, Pebernard S, Ducraux A, et al. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet. 2003;34:263–4. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- Chan JH, Lim S, Wong WS. Antisense oligonucleotides: from design to therapeutic application. Clin Exp Pharmacol Physiol. 2006;33:533–40. doi: 10.1111/j.1440-1681.2006.04403.x. [DOI] [PubMed] [Google Scholar]
- Doersen CJ, Guerrier-Takada C, Altman S, et al. Characterization of an RNase P activity from HeLa cell mitochondria. Comparison with the cytosol RNase P activity. J Biol Chem. 1985;260:5942–9. [PubMed] [Google Scholar]
- Dreyfus DH, Matczuk A, Fuleihan R. An RNA external guide sequence ribozyme targeting human interleukin-4 receptor alpha mRNA. Int Immunopharmacol. 2004;4:1015–27. doi: 10.1016/j.intimp.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Elliott J, Lynch OT, Suessmuth Y, et al. Respiratory syncytial virus NS1 protein degrades STAT2 by using the Elongin-Cullin E3 ligase. J Virol. 2007;81:3428–36. doi: 10.1128/JVI.02303-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y, Miyano-Kurosaki N, Kitano M, et al. Effective inhibition of HIV-1 replication in cultured cells by external guide sequences and ribonuclease. Nucleic Acids Res Suppl. 2001:213–14. doi: 10.1093/nass/1.1.213. [DOI] [PubMed] [Google Scholar]
- Fauci AS. Host factors and the pathogenesis of HIV-induced disease. Nature. 1996;384:529–34. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- Fauci AS, Challberg MD. Host-based antipoxvirus therapeutic strategies: turning the tables. J Clin Invest. 2005;115:231–3. doi: 10.1172/JCI24270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finotto S, Buerke M, Lingnau K, et al. Local administration of antisense phosphorothioate oligonucleotides to the c-kit ligand, stem cell factor, suppresses airway inflammation and IL-4 production in a murine model of asthma. J Allergy Clin Immunol. 2001;107:279–86. doi: 10.1067/mai.2001.113049. [DOI] [PubMed] [Google Scholar]
- Finotto S, De Sanctis GT, Lehr HA, et al. Treatment of allergic airway inflammation and hyperresponsiveness by antisense-induced local blockade of GATA-3 expression. J Exp Med. 2001;193:1247–60. doi: 10.1084/jem.193.11.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georas SN, Cumberland JE, Burke TF, et al. Stat6 inhibits human interleukin-4 promoter activity in T cells. Blood. 1998;92:4529–38. [PubMed] [Google Scholar]
- Gopalan V, Vioque A, Altman S. RNase P: variations and uses. J Biol Chem. 2002;277:6759–62. doi: 10.1074/jbc.R100067200. [DOI] [PubMed] [Google Scholar]
- Gu W, Ogose A, Kawashima H, et al. High-level expression of the coxsackievirus and adenovirus receptor messenger RNA in osteosarcoma, Ewing’s sarcoma, and benign neurogenic tumors among musculoskeletal tumors. Clin Cancer Res. 2004;10:3831–8. doi: 10.1158/1078-0432.CCR-03-0345. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C, Altman S. Inactivation of gene expression using ribonuclease P and external guide sequences. Methods Enzymol. 2000;313:442–56. doi: 10.1016/s0076-6879(00)13028-9. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C, Lumelsky N, Altman S. Specific interactions in RNA enzyme-substrate complexes. Science. 1989;246:1578–84. doi: 10.1126/science.2480641. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C, Van Belkum A, Pleij CW, et al. Novel reactions of RNAase P with a tRNA-like structure in turnip yellow mosaic virus RNA. Cell. 1988;53:267–72. doi: 10.1016/0092-8674(88)90388-1. [DOI] [PubMed] [Google Scholar]
- Herweijer H, Wolff JA. Gene therapy progress and prospects: hydrodynamic gene delivery. Gene Ther. 2007;14:99–107. doi: 10.1038/sj.gt.3302891. [DOI] [PubMed] [Google Scholar]
- Hnatyszyn H, Spruill G, Young A, et al. Long-term RNase P-mediated inhibition of HIV-1 replication and pathogenesis. Gene Ther. 2001;8:1863–71. doi: 10.1038/sj.gt.3301606. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–7. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Linsley PS. Noise amidst the silence: off-target effects of siRNAs. Trends Genet. 2004;20:521–4. doi: 10.1016/j.tig.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Julg B, Goebel FD. CCR5 antagonists: a new tool in fighting HIV. J HIV Ther. 2005;10:68–71. [PubMed] [Google Scholar]
- Kariko K, Bhuyan P, Capodici J, et al. Exogenous siRNA mediates sequence-independent gene suppression by signaling through toll-like receptor 3. Cells Tissues Organs. 2004;177:132–8. doi: 10.1159/000079987. [DOI] [PubMed] [Google Scholar]
- Kariko K, Ni H, Capodici J, et al. mRNA is an endogenous ligand for Toll-like receptor 3. J Biol Chem. 2004;279:12542–50. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- Kitano M, Miyano-Kurosaki N, Endo Y, et al. Effective suppression of HIV-1 gene expression by a mammalian tRNA 3′ processing endoribonuclease and external guide sequence oligozymes. Nucleic Acids Symp Ser. 2000:207–8. doi: 10.1093/nass/44.1.207. [DOI] [PubMed] [Google Scholar]
- Koller E, Propp S, Murray H, et al. Competition for RISC binding predicts in vitro potency of siRNA. Nucleic Acids Res. 2006;34:4467–76. doi: 10.1093/nar/gkl589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Zhang W, Lockey RF, et al. Respiratory syncytial virus infection in Fischer 344 rats is attenuated by short interfering RNA against the RSV-NS1 gene. Genet Vaccines Ther. 2007;5:4. doi: 10.1186/1479-0556-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski RA, Bothwell AL, Altman S. Identification of a ribonuclease P-like activity from human KB cells. Cell. 1976;9:101–16. doi: 10.1016/0092-8674(76)90056-8. [DOI] [PubMed] [Google Scholar]
- Kovrigina E, Wesolowski D, Altman S. Coordinate inhibition of expression of several genes for protein subunits of human nuclear RNase P. Proc Natl Acad Sci USA. 2003;100:1598–602. doi: 10.1073/pnas.0337661100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovrigina E, Yang L, Pfund E, et al. Regulated expression of functional external guide sequences in mammalian cells using a U6 RNA polymerase III promoter. RNA. 2005;11:1588–95. doi: 10.1261/rna.2140505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus G, Geffin R, Spruill G, et al. Cross-clade inhibition of HIV-1 replication and cytopathology by using RNase P-associated external guide sequences. Proc Natl Acad Sci USA. 2002;99:3406–11. doi: 10.1073/pnas.052651199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson SD, Jackson LN, Chen LA, et al. Effectiveness of siRNA uptake in target tissues by various delivery methods. Surgery. 2007;142:262–9. doi: 10.1016/j.surg.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederman MM, Penn-Nicholson A, Cho M, et al. Biology of CCR5 and its role in HIV infection and treatment. JAMA. 2006;296:815–26. doi: 10.1001/jama.296.7.815. [DOI] [PubMed] [Google Scholar]
- Li H, Trang P, Kim K, et al. Effective inhibition of human cytomegalovirus gene expression and growth by intracellular expression of external guide sequence RNA. RNA. 2006;12:63–72. doi: 10.1261/rna.2184706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HW, Ding SW. Antiviral silencing in animals. FEBS Lett. 2005;579:5965–73. doi: 10.1016/j.febslet.2005.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima WF, Monia BP, Ecker DJ, et al. Implication of RNA structure on antisense oligonucleotide hybridization kinetics. Biochemistry. 1992;31:12055–61. doi: 10.1021/bi00163a013. [DOI] [PubMed] [Google Scholar]
- Liu L, Xu Z, Fuhlbrigge RC, et al. Vaccinia virus induces strong immunoregulatory cytokine production in healthy human epidermal keratinocytes: a novel strategy for immune evasion. J Virol. 2005;79:7363–70. doi: 10.1128/JVI.79.12.7363-7370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Benimetskaya L, Lebedeva I, et al. Intracellular mRNA cleavage induced through activation of RNase P by nuclease-resistant external guide sequences. Nat Biotechnol. 2000;18:58–61. doi: 10.1038/71924. [DOI] [PubMed] [Google Scholar]
- Ma MY, Jacob-Samuel B, Dignam JC, et al. Nuclease-resistant external guide sequence-induced cleavage of target RNA by human ribonuclease P. Antisense Nucleic Acid Drug Dev. 1998;8:415–26. doi: 10.1089/oli.1.1998.8.415. [DOI] [PubMed] [Google Scholar]
- Massaro D, Massaro GD, Clerch LB. Noninvasive delivery of small inhibitory RNA and other reagents to pulmonary alveoli in mice. Am J Physiol Lung Cell Mol Physiol. 2004;287:L1066–70. doi: 10.1152/ajplung.00067.2004. [DOI] [PubMed] [Google Scholar]
- Matsukura S, Kokubu F, Kurokawa M, et al. Synthetic double-stranded RNA induces multiple genes related to inflammation through Toll-like receptor 3 depending on NF-kappaB and/or IRF-3 in airway epithelial cells. Clin Exp Allergy. 2006;36:1049–62. doi: 10.1111/j.1365-2222.2006.02530.x. [DOI] [PubMed] [Google Scholar]
- Minor TE, Dick EC, Baker JW, et al. Rhinovirus and influenza type A infections as precipitants of asthma. Am Rev Respir Dis. 1976;113:149–53. doi: 10.1164/arrd.1976.113.2.149. [DOI] [PubMed] [Google Scholar]
- Monia BP, Johnston JF, Sasmor H, et al. Nuclease resistance and antisense activity of modified oligonucleotides targeted to Ha-ras. J Biol Chem. 1996;271:14533–40. doi: 10.1074/jbc.271.24.14533. [DOI] [PubMed] [Google Scholar]
- Monia BP, Lesnik EA, Gonzalez C, et al. Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J Biol Chem. 1993;268:14514–22. [PubMed] [Google Scholar]
- Moschos SA, Williams AE, Lindsay MA. Cell-penetrating-peptide-mediated siRNA lung delivery. Biochem Soc Trans. 2007;35:807–10. doi: 10.1042/BST0350807. [DOI] [PubMed] [Google Scholar]
- Nyce JW. Respirable antisense oligonucleotides as novel therapeutic agents for asthma and other pulmonary diseases. Expert Opin Investig Drugs. 1997;6:1149–56. doi: 10.1517/13543784.6.9.1149. [DOI] [PubMed] [Google Scholar]
- Nyce JW, Metzger WJ. DNA antisense therapy for asthma in an animal model. Nature. 1997;385:721–5. doi: 10.1038/385721a0. [DOI] [PubMed] [Google Scholar]
- Oh JW, Lee HB, Park IK, et al. Interleukin-6, interleukin-8, interleukin-11, and interferon-gamma levels in nasopharyngeal aspirates from wheezing children with respiratory syncytial virus or influenza A virus infection. Pediatr Allergy Immunol. 2002;13:350–6. doi: 10.1034/j.1399-3038.2002.02018.x. [DOI] [PubMed] [Google Scholar]
- Phipps KM, Martinez A, Lu J, et al. Small interfering RNA molecules as potential anti-human rhinovirus agents: in vitro potency, specificity, and mechanism. Antiviral Res. 2004;61:49–55. doi: 10.1016/j.antiviral.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Plehn-Dujowich D, Altman S. Effective inhibition of influenza virus production in cultured cells by external guide sequences and ribonuclease P. Proc Natl Acad Sci USA. 1998;95:7327–32. doi: 10.1073/pnas.95.13.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu FD. Antisense- and RNA interference-based therapeutic strategies in allergy. J Cell Mol Med. 2005;9:840–53. doi: 10.1111/j.1582-4934.2005.tb00383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj SM, Liu F. Engineering of RNase P ribozyme for gene-targeting applications. Gene. 2003;313:59–69. doi: 10.1016/s0378-1119(03)00677-2. [DOI] [PubMed] [Google Scholar]
- Rangarajan S, Raj ML, Hernandez JM, et al. RNase P as a tool for disruption of gene expression in maize cells. Biochem J. 2004;380:611–6. doi: 10.1042/BJ20040442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb GB, Rana TM. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol Cell. 2007;26:523–37. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Robertson HD, Altman S, Smith JD. Purification and properties of a specific Escherichia coli ribonuclease which cleaves a tyrosine transfer ribonucleic acid presursor. J Biol Chem. 1972;247:5243–51. [PubMed] [Google Scholar]
- Scacheri PC, Rozenblatt-Rosen O, Caplen NJ, et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc Natl Acad Sci USA. 2004;101:1892–7. doi: 10.1073/pnas.0308698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott DH, Cureton DK, Whelan SP, et al. An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2005;102:18420–4. doi: 10.1073/pnas.0507123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz S, Sarnow P. Interaction of viruses with the mammalian RNA interference pathway. Virology. 2006;344:151–7. doi: 10.1016/j.virol.2005.09.034. [DOI] [PubMed] [Google Scholar]
- Singh AM, Busse WW. Asthma exacerbations. 2: aetiology. Thorax. 2006;61:809–16. doi: 10.1136/thx.2005.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer DS, Feitelson M, Sun B, et al. Paradigms for conditional expression of RNA interference molecules for use against viral targets. Methods Enzymol. 2005;392:227–41. doi: 10.1016/S0076-6879(04)92014-9. [DOI] [PubMed] [Google Scholar]
- Tekkanat KK, Maassab HF, Cho DS, et al. IL-13-induced airway hyperreactivity during respiratory syncytial virus infection is STAT6 dependent. J Immunol. 2001;166:3542–8. doi: 10.4049/jimmunol.166.5.3542. [DOI] [PubMed] [Google Scholar]
- Thomas M, Lu JJ, Chen J, et al. Non-viral siRNA delivery to the lung. Adv Drug Deliv Rev. 2007;59:124–33. doi: 10.1016/j.addr.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompkins SM, Lo CY, Tumpey TM, et al. Protection against lethal influenza virus challenge by RNA interference in vivo. Proc Natl Acad Sci USA. 2004;101:8682–6. doi: 10.1073/pnas.0402630101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trian T, Girodet PO, Ousova O, et al. RNA interference decreases PAR-2 expression and function in human airway smooth muscle cells. Am J Respir Cell Mol Biol. 2006;34:49–55. doi: 10.1165/rcmb.2005-0187OC. [DOI] [PubMed] [Google Scholar]
- Ulanova M, Schreiber AD, Befus AD. The future of antisense oligonucleotides in the treatment of respiratory diseases. BioDrugs. 2006;20:1–11. doi: 10.2165/00063030-200620010-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SZ, Bao YX, Rosenberger CL, et al. IL-12p40 and IL-18 modulate inflammatory and immune responses to respiratory syncytial virus infection. J Immunol. 2004;173:4040–9. doi: 10.4049/jimmunol.173.6.4040. [DOI] [PubMed] [Google Scholar]
- Weber J, Piontkivska H, Quinones-Mateu ME. HIV type 1 tropism and inhibitors of viral entry: clinical implications. AIDS Rev. 2006;8:60–77. [PubMed] [Google Scholar]
- Yang L, Wesolowski D, Li Y, et al. Analysis of putative RNase P RNA from orthopoxviruses. J Mol Biol. 2005;354:529–35. doi: 10.1016/j.jmb.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Yang YH, Li H, Zhou T, et al. Engineered external guide sequences are highly effective in inducing RNase P for inhibition of gene expression and replication of human cytomegalovirus. Nucleic Acids Res. 2006;34:575–83. doi: 10.1093/nar/gkj431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda H, Suzuki T, Zayasu K, et al. Inflammatory and bronchospastic factors in asthma exacerbations caused by upper respiratory tract infections. Tohoku J Exp Med. 2005;207:109–18. doi: 10.1620/tjem.207.109. [DOI] [PubMed] [Google Scholar]
- Zhang H, Altman S. Inhibition of the expression of the human RNase P protein subunits Rpp21, Rpp25, Rpp29 by external guide sequences (EGSs) and siRNA. J Mol Biol. 2004;342:1077–83. doi: 10.1016/j.jmb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Zhou T, Kim J, Kilani AF, et al. In vitro selection of external guide sequences for directing RNase P-mediated inhibition of viral gene expression. J Biol Chem. 2002;277:30112–20. doi: 10.1074/jbc.M200183200. [DOI] [PubMed] [Google Scholar]
- Zhu J, Trang P, Kim K, et al. Effective inhibition of Rta expression and lytic replication of Kaposi’s sarcoma-associated herpesvirus by human RNase P. Proc Natl Acad Sci USA. 2004;101:9073–8. doi: 10.1073/pnas.0403164101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziehler WA, Day JJ, Fierke CA, et al. Effects of 5′ leader and 3′ trailer structures on pre-tRNA processing by nuclear RNase P. Biochemistry. 2000;39:9909–16. doi: 10.1021/bi000603n. [DOI] [PubMed] [Google Scholar]