Abstract
A population genetic analysis of the long-wavelength opsin (OPN1LW, “red”) color vision gene in a global sample of 236 human nucleotide sequences had previously discovered nine amino acid replacement single nucleotide polymorphisms, which were found at high frequencies in both African and non-African populations and associated with an unusual haplotype diversity. Although this pattern of nucleotide diversity is consistent with balancing selection, it has been argued that a recombination “hot spot” or gene conversion within and between X-linked color vision genes alone may explain these patterns. The current analysis investigates a closely related primate with trichromatism to determine whether color vision gene amino acid polymorphism and signatures of adaptive evolution are characteristic of humans alone. Our population sample of 56 chimpanzee (Pan troglodytes) OPN1LW sequences shows three singleton amino acid polymorphisms and no unusual recombination or linkage disequilibrium patterns across the ∼5.5-kb region analyzed. Our comparative population genetic approach shows that the patterns of OPN1LW variation in humans and chimpanzees are consistent with positive and purifying selection within the two lineages, respectively. Although the complex role of color vision has been greatly documented in primate evolution in general, it is surprising that trichromatism has followed very different selective trajectories even between humans and our closest relatives.
Keywords: opsin, long-wavelength, primates
Introduction
Through large-scale comparisons of the human and chimpanzee genomes, a number of studies have attempted to identify the genes underlying the evolutionary change between us and our closest primate relatives (e.g., Clark et al. 2003; Bustamante et al. 2005; Chimpanzee Sequencing and Analysis Consortium 2005). Of the genes most evolutionarily diverged, those with sensory perception functions were overrepresented, such as olfactory (Niimura and Nei 2003; Gilad et al. 2003; Nozawa et al. 2007) and bitter-taste receptor genes (Wang et al. 2004; Fischer et al. 2005; Go et al. 2005). Although these analyses capture “ancient” events, comparative population genetic approaches examining sensory perception would reveal how chimpanzee and human lineages have recently diverged (Stone and Verrelli 2006). For example, haplotype analyses of the bitter-taste receptor gene TAS2R38 in humans are consistent with balancing selection (Wooding et al. 2004), whereas chimpanzee polymorphism shows that they have independently evolved a similar bitter-taste sensitivity (Wooding et al. 2006).
One of the most widely studied examples of adaptive diversification of sensory perception in primates is that of X-linked color vision variation. Trichromatic color vision in catarrhines (Old World monkeys and apes, including humans) is due to an autosomal-linked S-opsin (OPN1SW, “blue”) gene and an X-linked gene duplication ∼30–40 Ma that resulted in L- (OPN1LW, “red”) and M-opsin (OPN1MW, “green”) genes (Nathans et al. 1986; Jacobs et al. 1996). The OPN1LW and OPN1MW genes each code for a 364 amino acid protein (>98% similar). Many have estimated ranges of color vision perception from these proteins’ amino acid sequences given the strong correlation between opsin wavelength absorption maxima (λmax) and single amino acid variants. In fact, only ∼5 amino acid residues may account for the difference in λmax between the L- and M-opsins (Merbs and Nathans 1992; Asenjo et al. 1994; Sharpe et al. 1998; Yokoyama and Radlwimmer 2001). The OPN1LW and OPN1MW genes are separated by ∼24 kb, and recombination and gene conversion in this array still frequently occurs, resulting in deletions and duplications that are responsible for “red/green” color blindness in ∼8% of the male human population (Hayashi et al. 1999; Sharpe et al. 1999). Interestingly, several variants are found within the human L-opsin protein that also have functional, and even behavioral, implications (Merbs and Nathans 1992; Winderickx et al. 1992, 1993; Asenjo et al. 1994; Sharpe et al. 1998; Jameson et al. 2001).
Several studies have proposed that this L-opsin amino acid variation persists because purifying selection maintains only a few specific amino acid residues to distinguish M- and L-opsins (Merbs and Nathans 1992; Winderickx et al. 1992, 1993), and thus, these L-opsin amino acid variants are simply “neutral.” However, in the first DNA sequence analysis of OPN1LW in a global population sample of 236 human “color normals,” Verrelli and Tishkoff (2004) found an unusual haplotype structure associated only with high-frequency amino acid polymorphisms and observed no fixed amino acid differences since our divergence from chimpanzees, which argued that positive selection has shaped human color vision variation within populations.
Opsin protein variation is found in several primates and is believed to confer color vision perception that is adaptively tuned for foraging efficiency, as in the detection of ripe fruits or immature leaves (e.g., Mollon 1989; Dominy and Lucas 2001; Martin and Ross 2005). Although studies have examined OPN1LW-coding regions among primates (e.g., Deeb et al. 1994; Tan and Li 1999), there are very few comparative population genetic analyses of exons and introns with tests of selection at primate color vision genes. In fact, we lack these data for even another great ape to compare with the high human OPN1LW variation. In continuing with our comparative genetic analysis of primate color vision systems (Verrelli and Tishkoff 2004; Perry et al. 2007), we conducted the first DNA sequence analysis of exons and introns at OPN1LW in a chimpanzee population to determine the selective forces shaping color vision in our closest primate relative.
Materials and Methods
Primate Samples
The major question of interest here involves how color vision gene variation differs between humans and chimpanzees. Thus, although several chimpanzee subspecies are recognized, Pan troglodytes verus appears to have nuclear genetic diversity comparable to that of humans (Chimpanzee Sequencing and Analysis Consortium 2005). We have collected a sample of population genetic data from this subspecies and, additionally, a few individuals from other chimpanzee subspecies for comparison. Nucleotide sequences were collected from DNA samples of 45 unrelated chimpanzees: 40 from the western African P. t. verus, 3 from the central African Pan troglodytes troglodytes, 1 from the eastern African Pan troglodytes schweinfurthii, and 1 Nigerian Pan troglodytes vellerosus chimpanzee purported to be an additional subspecies (Gonder et al. 1997). Samples were imported in accordance with the Convention on International Trade in Endangered Species of Wild Fauna and Flora under permit 99US013176/9. One gorilla (Gorilla gorilla gorilla) was also sampled for our analysis. In addition to these new data, we add 5 P. t. verus, 1 P. t. troglodytes, and 236 human OPN1LW nucleotide sequences from Verrelli and Tishkoff (2004).
The human data set includes 163 males from 11 subpopulations within sub-Saharan Africa and 73 males from 8 groups outside of this geographic region (referred to as “African” and “non-African,” respectively). Because OPN1LW is X linked, sampling males enables the unambiguous determination of polymorphisms (i.e., no heterozygous sites) and haplotype phase. All Pan troglodytes were males except for 5 P. t. verus. As is the case with our other analyses of these 5 females (e.g., Verrelli et al. 2006), each of these individuals had two different haplotypes that could be resolved empirically (see below). Thus, our chimpanzee analyses reflect a total of 4 P. t. troglodytes, 1 P. t. schweinfurthii, 1 P. t. vellerosus, and 50 P. t. verus OPN1LW sequences.
OPN1LW Fragment Amplification and Sequencing
As in humans, the OPN1LW and OPN1MW genes are situated in the same orientation and location on the X chromosome and separated by ∼24 kb in chimpanzees. Exons 1 and 6 are identical in nucleotide sequence for OPN1LW and OPN1MW human genes, but exons 2–5 code for amino acid residues that greatly influence λmax (Merbs and Nathans 1992; Asenjo et al. 1994; Sharpe et al. 1998). As in Verrelli and Tishkoff (2004) and Winderickx et al. (1992), primers in exons 2 and 5 were used to amplify single OPN1LW copies by polymerase chain reaction (PCR). This ∼5,464-bp product (a few indels vary the size), which includes all of intron 2 through a partial exon 5, was prepared for DNA sequencing using shrimp alkaline phosphatase and exonuclease I (US Biochemicals, Cleveland, OH). All nucleotide sequence data were obtained with an ABI 3730 automated sequencer (Applied Biosystems, Foster City, CA), and sequence files were aligned using the Sequencher v. 4.5 program (Gene Codes, Ann Arbor, MI). Exon 5 codes for two major amino acid differences that account for the largest differences between the L- and M-cone opsin λmax and are used to predict functional absorbance from inferred amino acid sequence: the Tyr277Phe (∼7 nm) and Thr285Ala (∼14 nm) variants (Neitz et al. 1991; Yokoyama and Radlwimmer 2001). Expressed L-cone opsins have the Tyr277 and Thr285 residues (Deeb et al. 1992); and thus, all PCR fragments were first analyzed for their exon 5 nucleotide sequence to verify that they were only L-cone opsins. In the case of the aforementioned 5 P. t. verus females, the ∼5.5-kb product was cloned using the Invitrogen TOPO-XL kit and a clone was sequenced along with the initial PCR product. All single nucleotide polymorphisms (SNPs) identified in the cloned sequence that did not appear in the initial PCR sequence were considered cloned artifacts and ignored.
Data Analysis and Statistical Tests
Because our data collection reflects a random sample of chimpanzees with respect to OPN1LW genetic variation (i.e., we have no a priori phenotypic evidence for color vision variation), all statistics and neutrality tests should reflect the frequencies of variants found in the natural population. There has been little genetic evidence to date to suggest any significant underlying demographic history within chimpanzee subspecies groups, such as population expansion or substructuring. However, human population samples from sub-Saharan Africa and outside this geographic region show markedly different genetic patterns with higher nucleotide diversity, less linkage disequilibrium (LD), and a greater effective population size (Ne) for the former compared with the latter (e.g., Tishkoff and Verrelli 2003a). Therefore, we present all estimates of nucleotide diversity from our African and non-African humans separately to appropriately compare with our chimpanzee sample. We also present population genetic parameter estimates for P. troglodytes overall as well as comparisons among the subspecies.
We have used the Rozas et al. (2003) DnaSP v. 4.1 program for our population and comparative species genetic analyses. “Silent” SNPs (which refer to introns and synonymous sites in exons, throughout the study) do not alter the amino acid sequence and, here, are used to best reflect neutral evolution in all tests. Locus-specific estimates of the population parameter θ = 3Neμ (for X-linked genes) were calculated using the number of SNPs as θW of Watterson (1975), which is corrected by sample size through a coalescent estimation. These measures were compared with the estimate θπ, which is based on the average number of pairwise differences among all sequences, and thus, sample size is also taken into consideration. These two estimates, θW and θπ, are expected to be equal under neutrality. This can be assessed using Tajima's (1989) D test, which examines the SNP frequency spectrum and evaluates whether the number of SNPs are higher or lower than expected, given their frequencies and the number of sequences sampled. Positive D values are consistent with balancing selection, whereas negative D values imply directional selection. Changes in demographic history, such as population structure or expansion, may also be consistent with positive or negative D values, respectively; thus, the statistical significance of all measures of diversity and neutrality tests were computed using coalescent simulations and permutations in DnaSP. For example, we simulated 1,000 genealogies with observed values of θW to estimate how often we would find the observed values of θπ and Tajima's D by chance alone (i.e., neutrality), given the sample sizes of humans and chimpanzees in our analysis. In using coalescent simulations to determine statistical significance, we effectively account for all differences in sample sizes across humans and chimpanzees that may otherwise effect statistical tests of polymorphism data.
To examine the haplotype structure at OPN1LW, we calculated LD and estimated the recombination parameter ρ = 3Nec (where c is the recombination rate per base pair) across the ∼5.5-kb region in chimpanzees for comparison with humans. We used the LDhat program of McVean et al. (2002), which applies the approximate likelihood method of Hudson (2001), to estimate ρ. LDhat conducts a permutation analysis to determine whether pairwise comparisons among SNPs exhibit significantly more or less LD given the locus-specific estimate of ρ and SNP frequencies. As determined by LDhat, uninformative SNPs (i.e., rare) were omitted for these analyses.
Finally, to examine older lineage-specific changes in selective pressures, we compared chimpanzee OPN1LW polymorphism at silent and replacement sites with fixation between chimpanzee and human OPN1LW at silent and replacement sites with the McDonald and Kreitman (1991) test of neutrality. If amino acid replacement sites are evolving according to neutrality, we may expect that the ratio of replacement polymorphism to fixation will approximate this same ratio at silent sites. The OPN1LW sequence from the single gorilla individual was used to facilitate the polarization of fixed differences within human and chimpanzee lineages.
Results
Chimpanzee OPN1LW Diversity
In humans, single X chromosomes can have more than one OPN1LW copy (e.g., Sjoberg et al. 1998). If there are SNPs among paralogous OPN1LW copies, they would be easily detectable from our nucleotide sequences given that the large majority of our sampled individuals are males. For example, the presence of multiple OPN1LW gene copies with different nucleotide sequences would be apparent from “heterozygous” sites (paralogous sequence SNPs). Such duplications were previously seen in ∼5% of human PCR amplifications, and these individuals were omitted from all analyses (Verrelli and Tishkoff 2004). However, in the current study, we did not observe a single instance of heterozygous sites within chimpanzee individuals.
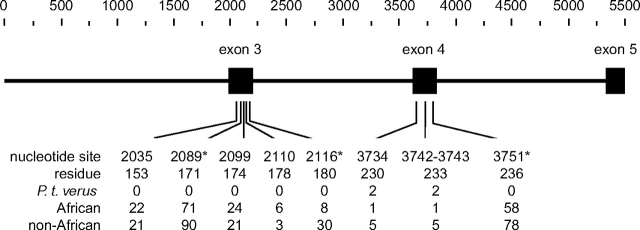
Figure 1 shows a schematic of the OPN1LW gene and the region sequenced for our chimpanzee and human population samples. Our analysis involves 456 bp in exons 3–5 and 5,008 bp in introns 2–4 that have a total of 5,118 effectively neutral silent sites. In our sample of 56 chimpanzee chromosomes, we find 42 variants, which include an 11-bp deletion in intron 2, a 2-bp deletion, and a 1-bp insertion in intron 3 (where insertion/deletion status was deduced from comparison with human sequence). Of the 39 SNPs, there are one silent and three replacement SNPs in exon 4, which are found as “singletons” in our population sample, and involve two amino acid changes in a single individual (Ile230Thr, site 3734 and Ala233Ser, sites 3742–3743; figs. 1 and 2). Otherwise, the remaining 35 SNPs are distributed over introns.
FIG. 1.—
Diagram of the ∼5.5 kb of the OPN1LW gene region analyzed in humans and chimpanzees. Exon 3 (169 bp), exon 4 (166 bp), and partial exon 5 (121 bp) are shown as black boxes with introns 2 (1,987 bp), 3 (1,467 bp), and 4 (1,554 bp) separating them. Starting at the beginning of intron 2, the nucleotide sites of the nine human amino acid replacement SNPs are shown below the gene with their respective amino acid residue positions and their frequencies (%) in population samples of Pan troglodytes verus chimpanzees (n = 50), African (163), and non-African (73) humans. Asterisks denote significant differences between the latter two samples using a chi-square statistical test (P < 0.01).
FIG. 2.—
Haplotypes for the 56 chimpanzee sequences and their frequencies. Samples (Ptt, Pan troglodytes troglodytes; Pts, Pan troglodytes schweinfurthii; Ptvel, Pan troglodytes vellerosus; Ptv, Pan troglodytes verus) are shown with their inferred derived state for each variable site (nucleotide position numbers start with intron 2 as in Verrelli and Tishkoff 2004), which were estimated with comparisons to human and gorilla sequences. Coding region SNPs are labeled as “R” and “S” for replacement and silent sites, respectively, and insertions and deletions are denoted by “+” and “−”, respectively.
For silent site diversity at OPN1LW, chimpanzees are far less variable than humans. Humans show silent site diversity (π = 0.31%, after multiplying by 4/3 for it being X linked) that is over three times that found in the nuclear genome on average, and given the relatively few replacement sites in exons (∼300), the nine replacement SNPs are unusually high. In addition, all replacement SNPs are found across African and non-African populations and several show significant frequency differences between the two (table 1 and fig. 1). After correcting for being X linked, chimpanzee silent site diversity (π = 0.08%) is average compared with the genome (Deinard and Kidd 1999; Kaessmann et al. 1999; Stone et al. 2002; Gilad et al. 2003; Yu et al. 2003; Fischer et al. 2004; Wooding et al. 2005; Verrelli et al. 2006), which demonstrates that the silent site discrepancy between the two species is likely due to an excess of human variation.
Table 1.
Population Diversity Estimates and Tests of Neutrality at OPN1LW
| Silent a |
Replacement |
MKb | ||||||
| Sample | nc | Sd | πe | TDf | S | π | TD | P value |
| Pan troglodytes | 56 | 36 | 0.065 | −1.86* | 3 | 0.032 | −1.68* | 0.096 |
| Pan troglodytes verus | 50 | 15 | 0.045 | −0.94 | 3 | 0.036 | −1.70* | 0.021* |
| Pan troglodytes troglodytes | 4 | 19 | 0.205 | 0.14 | 0 | 0 | NA | NA |
| Homo sapiens | 236 | 74 | 0.193 | −0.58 | 9 | 0.605 | 0.90* | 0.026* |
| African | 163 | 67 | 0.213 | −0.15 | 9 | 0.606 | 0.71* | 0.014* |
| Non-African | 73 | 36 | 0.123 | −0.48 | 9 | 0.553 | 0.07 | 0.002* |
NOTE.—Samples are described in the Materials and Methods. NA, not applicable.
Includes coding and noncoding regions (see the Results).
McDonald–Kreitman test (see the Results).
Number of chromosomes.
Number of SNPs.
Average pairwise sequence differences (%).
Tajima's D test (*P < 0.05, see the Results).
Although our sample of P. t. troglodytes is not considered a population sample per se, this subspecies appears to have greater nucleotide diversity than humans have, which is true for nuclear genes in general. This subspecies of chimpanzee also shows silent diversity similar to that of African humans, whereas P. t. verus has considerably less and similar to that found in non-African humans. Interestingly, silent site diversity among P. t. troglodytes haplotypes is even greater than that found among chimpanzee subspecies (table 2). On the other hand, consistent with other nuclear loci (Yu et al. 2003; Fischer et al. 2004; Wooding et al. 2005; Verrelli et al. 2006), nucleotide diversity among P. t. verus haplotypes is about half that found among subspecies (tables 1 and 2).
Table 2.
Divergence at OPN1LW Silent Sites between Chimpanzees and Humans
| Sample | Pan troglodytes troglodytes | Pan troglodytes schweinfurthii | Pan troglodytes verus | Pan troglodytes vellerosus |
| P. t. schweinfurthii | 0.191 | |||
| P. t. verus | 0.172 | 0.093 | ||
| P. t. vellerosus | 0.151 | 0.117 | 0.070 | |
| Human | 1.221 | 1.182 | 1.163 | 1.182 |
NOTE.—Average pairwise SNP differences (%) among groups (see the Results section).
Silent site diversity in chimpanzees appears to be significantly skewed toward an excess of rare SNPs, as indicated by the negative Tajima's D values (table 1). However, as has been documented before for these samples (Verrelli et al. 2006), SNPs are likely common within subspecies, but when small samples of each (e.g., one for each of P. t. schweinfurthii and P. t. vellerosus) are combined into one sample, it appears that these SNPs are singletons at low frequencies. In fact, when the P. t. verus sample as well as the smaller sample of P. t. troglodytes is analyzed alone, no excess of rare alleles is seen (table 1). When analyzed separately, African and non-African human samples also show no significant skew toward rare frequency silent SNPs. Altogether, it appears that silent site diversity in humans and chimpanzees is consistent with neutrality.
In contrast to silent site diversity, humans and chimpanzees show very different patterns of variation for amino acid replacement sites. Although both species possess replacement polymorphisms, these SNPs in P. t. verus are very rare in frequency (all three found as singletons in one individual, fig. 2), which results in a significantly high negative Tajima's D value. In contrast, human replacement SNPs are not only high in number but several are high in frequency (table 1 and fig. 1), which results in a significantly high positive Tajima's D under a standard neutral model of human population expansion (Verrelli and Tishkoff 2004).
Recombination and LD Analyses
Analyses of LD and recombination are only truly valid when conducted for within-population samples; and thus, we included only our samples of P. t. verus and humans for these comparisons. In addition, the amount of variation found within and between the chimpanzee subspecies at OPN1LW also warrants that these groups be analyzed separately, as is the case for African and non-African human samples. For chimpanzee OPN1LW, only 8 pairwise comparisons in the P. t. verus sample of 16 SNPs were significantly correlated given the overall maximum likelihood (ML) estimate of recombination at OPN1LW, which finds very little evidence of historical crossing-over across the ∼5.5-kb region (ρ ≈ 0). These few correlations were not clustered in any specific gene region and include SNPs across introns 2–4. Thus, no unusual pattern of LD or recombination was apparent in the chimpanzee sample.
This pattern in chimpanzees is in sharp contrast to the estimates of LD and recombination found in humans. As previously noted, overall, there is far more diversity in the human sample, and this is associated with ML estimates of recombination that are also very high in both Africans and non-Africans (ρ > 100). Given these estimates, it is not surprising that only 56 of the >700 African pairwise comparisons and 25 of the >300 non-African pairwise comparisons show significant LD, all of which are spread across introns 2–4. Surprisingly, given the overall high levels of gene-specific recombination, significantly high levels of localized recombination were still detected in both human population samples as a hot spot in the 169 bp of exon 3. In fact, 125 of the African and 59 of the non-African correlations had significantly less LD given the gene-specific estimate of ρ, yet 86 and 49 of these correlations, respectively, are clustered in only an 86-bp region centered on exon 3. The HOTSPOTTER program designed by Li and Stephens (2003) was used to show that this clustering was significant; the haplotype diversity in this region appears to have recombination rates that are 20 and 5 times greater in Africans and non-Africans, respectively, compared with the rest of the ∼5.5 kb (Verrelli and Tishkoff 2004).
OPN1LW Interspecific Analyses
Silent site fixation at OPN1LW (45 differences, 0.9%) is similar to other nuclear gene comparisons between humans and chimpanzees (1.1%; Fischer et al. 2004; Wooding et al. 2005; Chimpanzee Sequencing and Analysis Consortium 2005; Verrelli et al. 2006), yet there is no amino acid fixation. In spite of this, both chimpanzees and humans show high levels of amino acid polymorphism, and analyses of P. t. verus as well as African and non-African human samples show significant McDonald–Kreitman tests of neutrality (table 1). However, measures of diversity (π) and SNP frequency distributions (Tajima's D) of replacement SNPs in the two species indicate that, although unexpectedly high in number, these SNPs are demonstrably rare in frequency for chimpanzees but very common for human populations (table 1).
Discussion
Amino acid replacement polymorphisms are not common for the human genome in general, and yet when present, are often associated with low frequencies or even singletons that imply at least weak purifying selection (i.e., Ohta 1992) acting across populations (e.g., Bustamante et al. 2005; Boyko et al. 2008; Lohmueller et al. 2008). Focus remains on those genes that show different patterns of amino acid polymorphism across populations and species and thus point to phenotypes under positive selection that is unique to the human or chimpanzee lineages. One example is of the human G6PD locus that shows several amino acid polymorphisms that have recently risen to high frequencies with strong LD in geographic regions of malarial endemism (Tishkoff et al. 2001; Verrelli et al. 2002; Saunders et al. 2005). Yet, G6PD in chimpanzees and all other great apes shows strong purifying selection and no significant LD and unusual haplotype structure, thus implying that positive selection for malarial resistance has targeted this locus recently only in humans (Verrelli et al. 2006).
In this current study, we have examined another phenotype, trichromatism, which shows very different patterns of genetic diversity between humans and chimpanzees. Some have suggested that the abundance of amino acid polymorphism at human OPN1LW is the result of gene conversion (Winderickx et al. 1993) and not necessarily adaptive evolution. Although Verrelli and Tishkoff (2004) used human polymorphism data to address this question, the current comparative study tests a second hypothesis. If this pattern in humans is simply due to structural gene organization (i.e., gene conversion) and weak purifying selection, given that chimpanzees may also experience gene conversion in this tandem gene array (e.g., Zhou and Li 1996), we may expect to see patterns of amino acid variation in chimpanzees similar to those in humans.
First, if gene conversion is the sole explanation for OPN1LW diversity, then we may expect that both silent and amino acid polymorphism, and fixation, show similar patterns. In spite of typical silent diversity and fixation, chimpanzee replacement SNPs are singletons found on only one haplotype (fig. 2). These two amino acid polymorphisms, at residues 230 and 233, are similarly found only together in 1% and 5% of haplotypes in African and non-African humans, respectively (fig. 1), and are identical to that found at OPN1MW. That is, this identical “block” of replacement SNPs found rarely in both humans and chimpanzees is clearly the result of single–gene conversion events that transport this variation from OPN1MW to OPN1LW. As is true in humans, we find that chimpanzee introns at OPN1LW are more similar to OPN1MW than they are to their OPN1LW homologs in other primates, which is the most obvious impact of long-term gene conversion. However, in contrast to chimpanzees, humans have nine replacement SNPs whose derived allele frequencies range from 1% to 90% (fig. 1), showing a discrepancy in how gene conversion alters amino acid variation in the two species. It is possible that gene conversion is simply higher in humans, yet we may not expect OPN1LW silent divergence to be higher in chimpanzees than in humans (31 vs. 14, with gorilla as outgroup). In addition, several high-frequency human OPN1LW replacement SNPs are not found in OPN1MW (Winderickx et al. 1993), which is also unexpected given that gene conversion should be unbiased if it is simply a neutral mechanism that drives variation.
Another signature of gene conversion is the unusual pattern of haplotype diversity that extends across exon 3 in humans. Recombination hot spots within and between human and chimpanzee genomes have recently been identified (Wall et al. 2003; Crawford et al. 2004; Ptak et al. 2004, 2005; Myers et al. 2005; Winckler et al. 2005; Coop et al. 2008). It is possible that these hot spots over small areas (<1 kb) are the result of gene conversion that shuffles small tracks preferentially (Ardlie et al. 2001; Frisse et al. 2001; Przeworski and Wall 2001; Reich et al. 2002; Wall 2004). At OPN1LW, the entire ∼5.5-kb region shows no dramatic pattern of LD and no recombination rate variation in chimpanzees. Furthermore, the fact that recombination rates in the Xq28 region are generally low (e.g., Ardlie et al. 2001; Frisse et al. 2001; Matise et al. 2007) indicates that the observed pattern in humans, and not chimpanzees, is unusual.
It is difficult to reconcile how a neutral substitution model could explain the lack of amino acid fixation between the two species in the last 5–7 My since their divergence, the observation of only three replacement singletons in chimpanzees, as well as explain several high-frequency replacement SNPs in human populations. It is also difficult to employ a neutral model of gene conversion or a recombination hot spot to explain the haplotype structure associated with human replacement SNPs, given only one replacement haplotype from gene conversion and the lack of unusual LD in chimpanzees. Although it is apparent that gene conversion occurs in both species in introns and exons, replacement SNPs are found at different frequencies in human and chimpanzee OPN1LW, many of which have measured functional affects (Merbs and Nathans 1992; Asenjo et al. 1994; Sharpe et al. 1998). Based on our data, it appears that certain residues that are implicated in changes to opsin λmax and spectral tuning, especially 277 and 285 (>7–14 nm) and to a lesser degree 230 and 233 (<5 nm), are under purifying selection in both species. This is unlike other amino acid sites, such as the 180 variant in humans, which although may alter the λmax < 5 nm, shows very different patterns of frequency and haplotype variation in the two species. Thus, several pieces of information support the conclusion that the different patterns of amino acid variation in humans and chimpanzees are the result of selective and not neutral processes alone. In fact, as our comparative population genetic studies of human and chimpanzees mount, like G6PD and OPN1LW, it will be interesting to see how often unusual haplotype diversity in the genome is more often explained by different selective pressures and not by varying recombination rates across species (Tishkoff and Verrelli 2003b; Wall and Pritchard 2003; Reed and Tishkoff 2006; Verrelli et al. 2006; Clark et al. 2007).
Data collected for OPN1MW show very little amino acid polymorphism in humans and no amino acid fixation between humans and chimpanzees (Winderickx et al. 1993; Deeb et al. 1994; Verrelli BC, unpublished data), demonstrating the effects of strong purifying selection. However, OPN1MW copy number among humans is highly variable, with individuals having 1–5 copies (Sjoberg et al. 1998; Sharpe et al. 1999; Carroll et al. 2002). Thus, population genetic parameters (as estimated in the current study) would be inappropriate because assumptions of homology for OPN1MW gene copies among individuals would be invalidated. As previously discussed, humans rarely show evidence for OPN1LW duplications (Sjoberg et al. 1998; Verrelli and Tishkoff 2004); yet, there has been no published study of OPN1LW copy number variation in other primates. In general, studies of copy number variation have only recently shown the magnitude of this diversity within and between human and chimpanzee genomes (Cheng et al. 2005; Perry et al. 2006; Redon et al. 2006). In an additional study, we have examined copy number variation at the X-linked opsin region in 30 human and 30 chimpanzee males with a high-resolution microarray–based comparative genomic hybridization platform comprised of ∼385,000 X-chromosome oligonucleotide probes (median spacing = 347 bp; Perry GH, unpublished data). These data suggest that single X chromosomes in humans are variable not only for the number of OPN1MW gene copies but are also consistent with some individuals having more than one OPN1LW copy. In contrast, there was not a single chimpanzee X chromosome with copy number variation of either opsin gene, which confirms our PCR results in showing no OPN1LW heterozygous sites within chimpanzees. Interestingly, this contrasting copy number variation between humans and chimpanzees may indicate yet another level upon which selection differentially acts within species at X-linked opsin genes.
The global human analysis by Verrelli and Tishkoff (2004) first proposed that human OPN1LW replacement SNPs may be explained by historical balancing selection for spectral tuning. For example, the amino acid variant at residue 180 in humans that reaches frequencies ∼30% in Africans (fig. 1) but which is not found in chimpanzees has been hypothesized to shift L-opsin λmax further into the “red/orange” portion of visible light (Asenjo et al. 1994; Sharpe et al. 1998; Carroll et al. 2002). As OPN1LW is X linked, balancing selection for heterozygotes would have only occurred in females and lead to “tetrachromacy” (blue, green, red, and red/orange opsins) as some have speculated (e.g., Jameson et al. 2001). In New World monkeys, where only one X-linked opsin gene exists and males are dichromatic, balancing selection favors heterozygous females that carry two different functional alleles because it effectively makes them trichromatic. Therefore, although it is possible that further variation may be under balancing selection in female humans, it is interesting to question why this variation has not been adaptive in chimpanzees. It is possible that this color vision variation only recently became adaptive for sex-specific behavior in human hunter–gatherer societies, especially for the need to distinguish among colored fruits and other food items on shaded backgrounds (Mollon et al. 1984; Dominy and Lucas 2001; Regan et al. 2001; Lucas et al. 2003; Surridge et al. 2003).
Finally, it has been proposed that sexual selection and mate recognition may have further shaped red color vision intraspecific variation (Fernandez and Morris 2007). Thus, specifically L-opsin spectral tuning may be responsible for adaptive changes in behavior among populations. This hypothesis is strengthened by the functional opsin variation between African and non-African populations as evidenced by the significant differences in L-opsin amino acid haplotype frequencies (Verrelli and Tishkoff 2004) and L- to M-opsin expression ratios (McMahon et al. 2008) found between these two groups. As our molecular population genetic analyses of other closely and distantly related primates continue (e.g., Perry et al. 2007), we can reveal how color vision became phenotypically unique during human and primate evolution in general.
Acknowledgments
Chimpanzee samples were provided by H. Vasken Aposhian, the Detroit Zoological Institute (Detroit, MI), the Lincoln Park Zoo (Chicago, IL), the Primate Foundation of Arizona (Mesa, AZ), the Riverside Zoo (Scottsbluff, NE), the Southwest Foundation for Biomedical Research (San Antonio, TX), the Sunset Zoo (Manhattan, KS), the Welsh Mountain Zoo (North Wales, UK), the Yerkes National Primate Research Center (Atlanta, GA), and the New Iberia Research Center (Lafayette, LA), where samples were supported by a National Institutes of Health–National Center for Research Resources grant U42-RR015087 to the University of Louisiana at Lafayette. This work was supported by the Center for Evolutionary Functional Genomics in The Biodesign Institute and the School of Life Sciences at Arizona State University and National Science Foundation grant BCS-0715972 (to B.C.V. and A.C.S.).
References
- Ardlie K, Liu-Cordero SN, Eberle MA, Daly M, Barrett J, Winchester E, Lander ES, Kruglyak L. Lower-than-expected linkage disequilibrium between tightly linked markers in humans suggests a role for gene conversion. Am J Hum Genet. 2001;69:582–589. doi: 10.1086/323251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asenjo AB, Rim J, Oprian DD. Molecular determinants of human red/green color discrimination. Neuron. 1994;12:1131–1138. doi: 10.1016/0896-6273(94)90320-4. [DOI] [PubMed] [Google Scholar]
- Boyko AR, Williamson SH, Indap AR, et al. (14 co-authors) Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 2008;4(5):e1000083. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, Fledel-Alon A, Williamson S, et al. (14 co-authors) Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- Carroll J, Neitz J, Neitz M. Estimates of L:M cone ratio from ERG flicker photometry and genetics. J Vis. 2002;2:531–542. doi: 10.1167/2.8.1. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Ventura M, She X, et al. (12 co-authors) A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Clark AG, Glanowski S, Nielsen R, et al. (17 co-authors) Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- Clark VJ, Ptak SE, Tiemann I, Qian Y, Coop G, Stone AC, Przeworski M, Arnheim N, Di Rienzo A. Combining sperm typing and linkage disequilibrium analyses reveals differences in selective pressures or recombination rates across human populations. Genetics. 2007;175:795–804. doi: 10.1534/genetics.106.064964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford DC, Bhangale T, Li N, Hellenthal G, Rieder MJ, Nickerson DA, Stephens M. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat Genet. 2004;36:700–706. doi: 10.1038/ng1376. [DOI] [PubMed] [Google Scholar]
- Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science. 2008;319:1395–1398. doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- Deeb SS, Jorgensen AL, Battisti L, Iwasaki L, Motulsky AG. Sequence divergence of the red and green visual pigments in great apes and humans. Proc Natl Acad Sci USA. 1994;91:7262–7266. doi: 10.1073/pnas.91.15.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeb SS, Lindsey DT, Hibiya Y, Sanocki E, Winderickx J, Teller DY, Motulsky AG. Genotype-phenotype relationships in human red/green color-vision defects: molecular and psychophysical studies. Am J Hum Genet. 1992;51:687–700. [PMC free article] [PubMed] [Google Scholar]
- Deinard A, Kidd K. Evolution of a HOXB6 intergenic region within the great apes and humans. J Hum Evol. 1999;36:687–703. doi: 10.1006/jhev.1999.0298. [DOI] [PubMed] [Google Scholar]
- Dominy NJ, Lucas PW. Ecological importance of trichromatic vision to primates. Nature. 2001;410:363–366. doi: 10.1038/35066567. [DOI] [PubMed] [Google Scholar]
- Fernandez AA, Morris MR. Sexual selection and trichromatic color vision in primates: statistical support for the preexisting-bias hypothesis. Am Nat. 2007;170:10–20. doi: 10.1086/518566. [DOI] [PubMed] [Google Scholar]
- Fischer A, Gilad Y, Man O, Pääbo S. Evolution of bitter taste receptors in humans and apes. Mol Biol Evol. 2005;22:432–436. doi: 10.1093/molbev/msi027. [DOI] [PubMed] [Google Scholar]
- Fischer A, Wiebe V, Paabo S, Przeworski M. Evidence for a complex demographic history of chimpanzees. Mol Biol Evol. 2004;21:799–808. doi: 10.1093/molbev/msh083. [DOI] [PubMed] [Google Scholar]
- Frisse L, Hudson RR, Bartoszewicz A, Wall JD, Donfack J, Di Rienzo A. Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am J Hum Genet. 2001;69:831–843. doi: 10.1086/323612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Bustamante CD, Lancet D, Paabo S. Natural selection on the olfactory receptor gene family in humans and chimpanzees. Am J Hum Genet. 2003;73:489–501. doi: 10.1086/378132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go Y, Satta Y, Takenaka O, Takahata N. Lineage-specific loss of function of bitter taste receptor genes in humans and nonhuman primates. Genetics. 2005;170:313–326. doi: 10.1534/genetics.104.037523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonder MK, Oates JF, Disotell TR, Forstner MRJ, Morales JC, Melnick DJ. A new west African chimpanzee subspecies? Nature. 1997;388:337. doi: 10.1038/41005. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Motulsky AG, Deeb SS. Position of a ‘green-red’ hybrid gene in the visual pigment array determines colour-vision phenotype. Nat Genet. 1999;22:90–93. doi: 10.1038/8798. [DOI] [PubMed] [Google Scholar]
- Hudson RR. Two-locus sampling distributions and their application. Genetics. 2001;159:1805–1817. doi: 10.1093/genetics/159.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs GH, Neitz M, Deegan JF, Neitz J. Trichromatic colour vision in New World monkeys. Nature. 1996;382:156–158. doi: 10.1038/382156a0. [DOI] [PubMed] [Google Scholar]
- Jameson KA, Highnote SM, Wasserman LM. Richer color experience in observers with multiple photopigment opsin genes. Psychon Bull Rev. 2001;8:244–261. doi: 10.3758/bf03196159. [DOI] [PubMed] [Google Scholar]
- Kaessmann H, Wiebe V, Paabo S. Extensive nuclear DNA sequence diversity among chimpanzees. Science. 1999;286:1159–1162. doi: 10.1126/science.286.5442.1159. [DOI] [PubMed] [Google Scholar]
- Li N, Stephens M. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics. 2003;165:2213–2233. doi: 10.1093/genetics/165.4.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller KE, Indap AR, Schmidt S, et al. (12 co-authors) Proportionally more deleterious genetic variation in European than in African populations. Nature. 2008;451:994–997. doi: 10.1038/nature06611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas PW, Dominy NJ, Riba-Hernandez P, et al. (12 co-authors) Evolution and function of routine trichromatic vision in primates. Evolution. 2003;57:2636–2643. doi: 10.1111/j.0014-3820.2003.tb01506.x. [DOI] [PubMed] [Google Scholar]
- Martin RD, Ross CF. The evolutionary and ecological context of primate vision. In: Kremers J, Silveira L, Martin P, editors. Structure, function, and evolution of the primate visual system. New York: John Wiley; 2005. pp. 1–36. [Google Scholar]
- Matise TC, Chen F, Chen W, et al. (13 co-authors) A second-generation combined linkage physical map of the human genome. Genome Res. 2007;17:1783–1786. doi: 10.1101/gr.7156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merbs SL, Nathans J. Absorption spectra of human cone pigments. Nature. 1992;356:433–435. doi: 10.1038/356433a0. [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- McMahon C, Carroll J, Awua S, Neitz J, Neitz M. The L:M cone ratio in males of African descent with normal color vision. J Vis. 2008;8:1–9. doi: 10.1167/8.2.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G, Awadalla P, Fearnhead P. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics. 2002;160:1231–1241. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollon JD. “Tho' she kneel'd in that place where they grew…”: The uses and origins of primate colour vision. J Exp Biol. 1989;146:21–38. doi: 10.1242/jeb.146.1.21. [DOI] [PubMed] [Google Scholar]
- Mollon JD, Bowmaker JK, Jacobs GH. Variations of color-vision in a New World primate can be explained by polymorphism of retinal photopigments. Proc R Soc Lond B Biol Sci. 1984;222:373–399. doi: 10.1098/rspb.1984.0071. [DOI] [PubMed] [Google Scholar]
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- Nathans J, Thomas D, Hogness DS. Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science. 1986;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- Neitz M, Neitz J, Jacobs GH. Spectral tuning of pigments underlying red-green color vision. Science. 1991;252:971–974. doi: 10.1126/science.1903559. [DOI] [PubMed] [Google Scholar]
- Niimura Y, Nei M. Evolution of olfactory receptor genes in the human genome. Proc Natl Acad Sci USA. 2003;100:12235–12240. doi: 10.1073/pnas.1635157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M, Kawahara Y, Nei M. Genomic drift and copy number variation of sensory receptor genes in humans. Proc Natl Acad Sci USA. 2007;104:20421–20426. doi: 10.1073/pnas.0709956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. The nearly neutral theory of molecular evolution. Annu Rev Ecol Syst. 1992;23:263–286. [Google Scholar]
- Perry GH, Martin RD, Verrelli BC. Signatures of functional constraint at aye-aye opsin genes: the potential of adaptive color vision in a nocturnal primate. Mol Biol Evol. 2007;24:1963–1970. doi: 10.1093/molbev/msm124. [DOI] [PubMed] [Google Scholar]
- Perry GH, Tchinda J, McGrath SD, et al. (12 co-authors) Hotspots for copy number variation in chimpanzees and humans. Proc Natl Acad Sci USA. 2006;103:8006–8011. doi: 10.1073/pnas.0602318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M, Wall JD. Why is there so little intragenic linkage disequilibrium in humans? Genet Res. 2001;77:143–151. doi: 10.1017/s0016672301004967. [DOI] [PubMed] [Google Scholar]
- Ptak SE, Hinds DA, Koehler K, Nickel B, Patil N, Ballinger DG, Przeworski M, Frazer KA, Pääbo S. Fine scale recombination patterns differ between chimpanzees and humans. Nat Genet. 2005;37:429–434. doi: 10.1038/ng1529. [DOI] [PubMed] [Google Scholar]
- Ptak SE, Roeder AD, Stephens M, Gilad Y, Pääbo S, Przeworski M. Absence of the TAP2 human recombination hotspot in chimpanzees. PLoS Biol. 2004;2:e155. doi: 10.1371/journal.pbio.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, et al. (43 co-authors) Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed FA, Tishkoff SA. Positive selection can create false hotspots of recombination. Genetics. 2006;172:2011–2014. doi: 10.1534/genetics.105.052183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan BC, Julliot C, Simmen B, Vienot F, Charles-Dominique P, Mollon JD. Fruits, foliage and the evolution of primate colour vision. Philos Trans R Soc Lond B Biol Sci. 2001;356:229–283. doi: 10.1098/rstb.2000.0773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DE, Schaffner SF, Daly MJ, McVean G, Mullikin JC, Higgins JM, Richter DJ, Lander ES, Altshuler D. Human genome sequence variation and the influence of gene history, mutation and recombination. Nat Genet. 2002;32:135–142. doi: 10.1038/ng947. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Saunders MA, Slatkin M, Garner C, Hammer MF, Nachman MW. The extent of linkage disequilibrium caused by selection on G6PD in humans. Genetics. 2005;171:1219–1229. doi: 10.1534/genetics.105.048140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe LT, Stockman A, Jagle H, Knau H, Klausen G, Reitner A, Nathans J. Red, green, and red-green hybrid pigments in the human retina: correlations between deduced protein sequences and psychophysically measured spectral sensitivities. J Neurosci. 1998;18:10053–10069. doi: 10.1523/JNEUROSCI.18-23-10053.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe LT, Stockman A, Jagle H, Nathans J. Opsin genes, cone photopigments, color vision, and color blindness. In: Gegenfurtner KR, Sharpe LT, editors. Color vision: from genes to perception. Cambridge: 1999. pp. 3–51. [Google Scholar]
- Sjoberg SA, Neitz M, Balding SD, Neitz J. L-cone pigment genes expressed in normal colour vision. Vision Res. 1998;38:3213–3219. doi: 10.1016/s0042-6989(97)00367-2. [DOI] [PubMed] [Google Scholar]
- Stone AC, Griffiths RC, Zegura SL, Hammer MF. High levels of Y-chromosome nucleotide diversity in the genus Pan. Proc Natl Acad Sci USA. 2002;99:43–48. doi: 10.1073/pnas.012364999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone AC, Verrelli BC. Focusing on comparative ape population genetics in the post-genomic age. Curr Opin Genet Dev. 2006;16:586–591. doi: 10.1016/j.gde.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Surridge AK, Osorio D, Mundy NI. Evolution and selection of trichromatic vision in primates. Trends Ecol Evol. 2003;18:198–205. [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Li W-H. Trichromatic vision in prosimians. Nature. 1999;402:36. doi: 10.1038/46947. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Varkonyi R, Cahinhinan N, et al. (17 co-authors) Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science. 2001;293:455–462. doi: 10.1126/science.1061573. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Verrelli BC. Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu Rev Genomics Hum Genet. 2003a;4:293–340. doi: 10.1146/annurev.genom.4.070802.110226. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Verrelli BC. Role of evolutionary history on haplotype block structure in the human genome: implications for disease mapping. Curr Opin Genet Dev. 2003b;13:569–575. doi: 10.1016/j.gde.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Verrelli BC, McDonald JH, Argyropoulos G, Destro-Bisol G, Froment A, Drousiotou A, Lefranc G, Helal AN, Loiselet J, Tishkoff SA. Evidence for balancing selection from nucleotide sequence analyses of human G6PD. Am J Hum Genet. 2002;71:1112–1128. doi: 10.1086/344345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrelli BC, Tishkoff SA. Signatures of selection and gene conversion associated with human color vision variation. Am J Hum Genet. 2004;75:363–375. doi: 10.1086/423287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrelli BC, Tishkoff SA, Stone AC, Touchman JW. Contrasting histories of G6PD molecular evolution and malarial resistance in humans and chimpanzees. Mol Biol Evol. 2006;23:1592–1601. doi: 10.1093/molbev/msl024. [DOI] [PubMed] [Google Scholar]
- Wall JD. Close look at gene conversion hot spots. Nat Genet. 2004;36:114–115. doi: 10.1038/ng0204-114. [DOI] [PubMed] [Google Scholar]
- Wall JD, Frisse LA, Hudson RR, Di Rienzo A. Comparative linkage-disequilibrium analysis of the beta-globin hotspot in primates. Am J Hum Genet. 2003;73:1330–1340. doi: 10.1086/380311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Pritchard JK. Haplotype blocks and linkage disequilibrium in the human genome. Nat Rev Genet. 2003;4:587–597. doi: 10.1038/nrg1123. [DOI] [PubMed] [Google Scholar]
- Wang X, Thomas SD, Zhang J. Relaxation of selective constraint and loss of function in the evolution of human bitter taste receptor genes. Hum Mol Genet. 2004;13:2671–2678. doi: 10.1093/hmg/ddh289. [DOI] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetic models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Winckler W, Myers SR, Richter DJ, et al. (11 co-authors) Comparison of fine-scale recombination rates in humans and chimpanzees. Science. 2005;308:107–111. doi: 10.1126/science.1105322. [DOI] [PubMed] [Google Scholar]
- Winderickx J, Battisti L, Hibiya Y, Motulsky AG, Deeb SS. Haplotype diversity in the human red and green opsin genes: evidence for frequent sequence exchange in exon 3. Hum Mol Genet. 1993;2:1413–1421. doi: 10.1093/hmg/2.9.1413. [DOI] [PubMed] [Google Scholar]
- Winderickx J, Lindsey DT, Sanocki E, Teller DY, Motulsky AG, Deeb SS. Polymorphism in red photopigment underlies variation in colour matching. Nature. 1992;356:431–433. doi: 10.1038/356431a0. [DOI] [PubMed] [Google Scholar]
- Wooding S, Bufe B, Grassi C, Howard MT, Stone AC, Vazquez M, Dunn DM, Meyerhof W, Weiss RB, Bamshad MJ. Independent evolution of bitter-taste sensitivity in humans and chimpanzees. Nature. 2006;440:930–934. doi: 10.1038/nature04655. [DOI] [PubMed] [Google Scholar]
- Wooding S, Kim UK, Bamshad MJ, Larsen J, Jorde LB, Drayna D. Natural selection and molecular evolution in PTC, a bitter-taste receptor gene. Am J Hum Genet. 2004;74:637–646. doi: 10.1086/383092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding S, Stone AC, Dunn DM, Mummidi S, Jorde LB, Weiss RK, Ahuja S, Bamshad MJ. Contrasting effects of natural selection on human and chimpanzee CC chemokine receptor 5. Am J Hum Genet. 2005;76:291–301. doi: 10.1086/427927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Radlwimmer FB. The molecular genetics and evolution of red and green color vision in vertebrates. Genetics. 2001;158:1697–1710. doi: 10.1093/genetics/158.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Jensen-Seaman MI, Chemnick L, Kidd JR, Deinard AS, Ryder O, Kidd KK, Li WH. Low nucleotide diversity in chimpanzees and bonobos. Genetics. 2003;164:1511–1518. doi: 10.1093/genetics/164.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YH, Li WH. Gene conversion and natural selection in the evolution of X-linked color vision genes in higher primates. Mol Biol Evol. 1996;13:780–783. doi: 10.1093/oxfordjournals.molbev.a025638. [DOI] [PubMed] [Google Scholar]