SUMMARY
Purine riboswitches discriminate between guanine and adenine by at least 10,000-fold based on the identity of a single pyrimidine (Y74) that forms a Watson-Crick base pair with the ligand. To understand how this high degree of specificity for closely related compounds is achieved through simple pairing, we investigated their interaction with purine analogs with varying functional groups at the 2- and 6-positions that have the potential to alter interactions with Y74. Using a combination of crystallographic and calorimetric approaches, we find that binding these purines is often facilitated by either small structural changes in the RNA or tautomeric changes in the ligand. This work also reveals that, along with base pairing, conformational restriction of Y74 significantly contributes to nucleobase selectivity. These results reveal that compounds that exploit the inherent local flexibility within riboswitch binding pockets can alter their ligand specificity.
Keywords: riboswitch, RNA, X-ray crystallography, isothermal titration calorimetry, RNA-ligand binding
Riboswitches are untranslated mRNA structures that transduce small molecule binding into a change in gene expression (Montange and Batey, 2008; Winkler and Breaker, 2005). The central feature of all riboswitches is the aptamer domain that binds cellular metabolites with both high affinity and specificity. Their ability to recognize small molecules and their abundance in numerous bacterial species including clinically important pathogens, make them an attractive new target for antibiotic development (Blount and Breaker, 2006). Furthermore, RNA is already a well-proven target as the ribosomal RNA binds a large number of clinically important drugs (Wright, 2007).
Riboswitches are already proposed to be the targets for several known antimicrobial agents (Blount et al., 2007; Sudarsan et al., 2005). For example, the thiamine analog pyrithiamine, originally utilized to study thiamine metabolism, binds thiamine pyrophosphate (TPP) riboswitches, repressing thiamine and TPP biosynthesis (Sudarsan et al., 2005; Winkler et al., 2002). Roseoflavin, an antimicrobial flavin mononucleotide (FMN) analog, binds the FMN riboswitch and mutations that confer resistance map to these RNA elements (Blount and Breaker, 2006; Lee et al., 2009). The lysine-responsive riboswitch binds to the L-aminoethylcysteine, a lysine analog toxic to bacteria, causing downregulation of lysine biosynthesis genes (Blount et al., 2007), although recent evidence indicates its primary target is lysyl-tRNA synthetase (Ataide et al., 2007). Other known riboswitches are attractive antimicrobial targets because they regulate expression of a diverse set of genes required for pathogenicity or survival (Blount and Breaker, 2006). Since the above compounds are close chemical analogs of the riboswitch's natural effector, identifying novel riboswitch-targeting compounds requires, in part, a clear understanding of how these RNAs discriminate between their effector and chemically related molecules found in the cellular environment. An ideal model system to address this issue is the purine riboswitch, which is known to bind a variety of purine derivatives (Mandal et al., 2003; Mandal and Breaker, 2004a) as well as some pyrimidines (Gilbert et al., 2006a).
The guanine and adenine classes of the purine riboswitch family encapsulate the purine nucleobase ligand within the center of a three-way junction that renders it virtually solvent inaccessible (Batey et al., 2004; Serganov et al., 2004). Within this binding pocket, a single pyrimidine residue (Y74) discriminates between guanine/hypoxanthine and adenine by forming a Watson-Crick base pair with the ligand (Figure 1A). Multiple sequence alignment, biochemical studies, and structural analysis have all indicated that ligand specificity for guanine and adenine is dictated by the identity of this single pyrimidine (Gilbert et al., 2006b; Mandal et al., 2003; Mandal and Breaker, 2004b). It is striking that the ~10,000-fold selectivity of the purine riboswitches for guanine/hypoxanthine versus adenine is comparable to that observed for the A and B families of DNA polymerases (McCulloch and Kunkel, 2008).
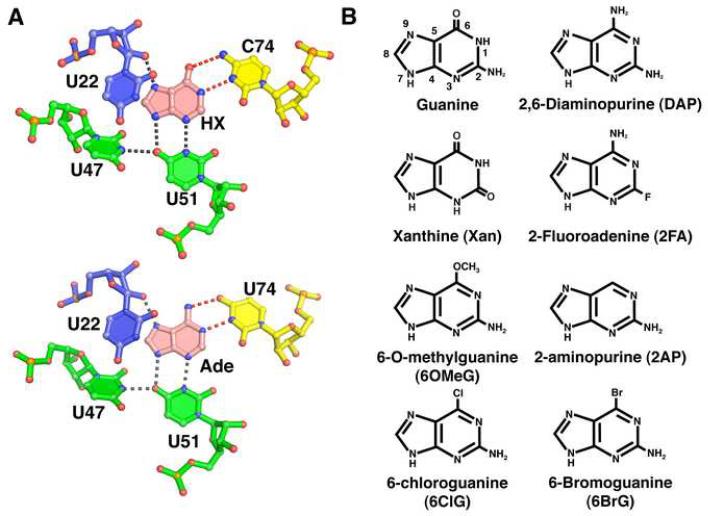
Figure 1. Ligand binding site of the purine riboswitch and chemicals.
(A) Details of hypoxanthine (top, PDB 1U8D) and adenine (bottom, PDB 1Y26) bound to the guanine and adenine riboswitches, respectively. The specificity pyrimidine (C74 or U74, yellow) forms hydrogen bonds (red dashed lines) to the Watson-Crick face of the ligand (pink), while U51, U47, and U22 interact with the other faces of the purine nucleobase. Residues are colored according to their strand position in the junction region. Note that the hydrogen bonding pattern for ligand recognition is identical. (B) Chemical structures of purine derivatives characterized in this study.
However, the observed specificity cannot be solely governed by this single pairing interaction between the ligand and Y74. Almost universally, RNA uses a variety of base mismatches alongside Watson-Crick base pairs to increase its conformational and structural diversity (Westhof and Fritsch, 2000). From an energetic perspective, a G•U pair is as favorable as an A-U pair in an A-form helix and represents ~10% of all base pairs in ribosomal RNA (Gautheret et al., 1995). Wobble G•U and A+•C pairs are easily accommodated within a helix with minimal perturbation to the A-form geometry (Leontis and Westhof, 1998). Thus, the purine riboswitch must contain specific mechanisms to prevent formation of these mismatches in the binding pocket, and the resulting misregulation of gene expression.
To gain insights into how the purine riboswitch achieves its specificity, we have challenged guanine- and adenine-binding variants of this RNA with a series of purine analogs (Figure 1B). Since the interaction of the ligand with Y74 is central to specificity, we focused primarily on 2- and 6-position purine analogs. In this study, a combination of isothermal titration calorimetry (ITC) and X-ray crystallography was used to fully assess the affinity and specificity of each compound for guanine and adenine binding variants of the purine riboswitch as well as reveal the structural basis of the interaction. We found that while the presence of an exocyclic functional group at the 2-position of the ligand is important for enhancing binding affinity, nonideal functional groups at this site are tolerated as long as they do not add steric bulk. Interestingly, purine analogs containing alternative functional groups at the 6-position bind the guanine and adenine riboswitches equally well, in some instances shifting Y74 into the minor groove and altering the specificity of ligand binding. Ligand specificity is further enhanced by a conserved base triple adjacent to the binding pocket that facilitates spatial restriction of Y74. Together, these data reveal that molecular adaptation within the ligand binding pocket of the purine riboswitch can substantially increase the diversity of compounds bound by this RNA.
RESULTS
Functional group substitution at the 2-position affects purine ligand affinity
The observed binding affinity of some 2-substituted purine analogs and available crystal structures cannot be easily reconciled (Batey et al., 2004; Gilbert et al., 2006b; Mandal et al., 2003; Serganov et al., 2004). For example, N2-methylguanine and xanthine appear to bind the guanine riboswitch with 100 μM and 50 nM affinities, respectively, as determined by in-line probing (Mandal et al., 2003; Mandal and Breaker, 2004a). However, examination of the crystal structure suggests that the RNA should not tolerate the increase in steric bulk of the methylation or the electrostatic clash of placing a carbonyl group at the 2-position between two other carbonyls (Figure 1A). It is important to note that hypoxanthine and adenine, both of which lack functionality at the 2-position, bind reasonably tightly to the guanine and adenine riboswitch—each in the 200 nM range—indicating that the 2-position of the ligand is not essential for binding.
The B. subtilis xpt-pbuX guanine riboswitch (GR) was used to re-examine the binding affinity of various 2-position purine derivatives by ITC (Table 1). The 2-position derivatives in this study were chosen to directly characterize the response to nonideal steric or electrostatic interactions between the ligand and the RNA. Substitutions at the 2-position revealed a clear preference of the riboswitch for smaller functional groups at this position. Ligands with large functional groups at the 2-position such, as N2-methylguanine and 2,6-dichloroguanine, do not bind the purine riboswitch (data not shown). Thus, groups that are sterically bulkier than the amino group found in guanine are strongly disruptive to purine riboswitch binding.
Table 1.
Binding affinity of purine derivatives as determined by ITC.
| compound | KD(μM) | |
|---|---|---|
| GR | GRA | |
| guanine1 | 0.004 ± 0.003 | n.d.3 |
| hypoxanthine1 | 0.76 ± 0.07 | n.d. |
| 2,6-diaminopurine2 | 4.1 ± 0.6 | 0.017 ± 0.006 |
| 2-aminopurine2 | 4.4 ± 1.2 | 0.25 ± 0.03 |
| adenine4 | n.d. | 0.47 ± 0.04 |
| xanthine | 39 ± 4.2 | n.d. |
| 2-fluoroadenine | ---4 | 4.0 ± 1.1 |
| 6-chloroguanine | 0.89 ± 0.06 | 0.72 ± 0.04 |
| 6-O-methylguanine | 23 ± 6 | 20 ± 6 |
| 6-bromoguanine | 2.1 ± 0.6 | 2.7 ± 1.2 |
| 6-methylaminopurine | --- | 105 ± 4 |
Values reported for ligand binding to GR have been published previously (Batey et al., 2004).
Values reported for ligand binding to GRA have been published previously (Gilbert et al., 2006b).
n.d.: no detectable binding
---: not determined
On the other hand, derivatives that create electrostatic clashes are still able to productively bind. Xanthine (2-hydroxyguanine) binds weakly to GR RNA (KD = 39 μM), while 2-fluoroadenine (2FA) binds weakly to GRA RNA (4.0 μM). The 2-carbonyl and 2-fluoro moieties of xanthine and 2FA, while presumably creating an electrostatic clash with two adjacent carbonyls in the binding pocket (O2(U51) and O2(C74), Figure 1A), are also the smallest functional groups tested. This indicates that favorable hydrogen bonding and electrostatic interactions at this site in the binding pocket are less important than the steric accommodation of the ligand by the riboswitch.
pH dependence of xanthine binding
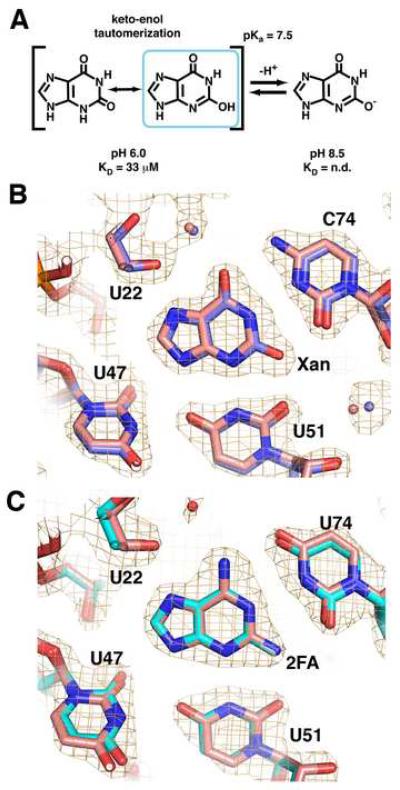
An additional complication to xanthine recognition is that N3 has a pKa of 7.6 (Rogstad et al., 2003). Above this pH, the N3 of xanthine becomes deprotonated, resulting in a negatively charged imino that tautomerizes to an enolate at O2 (Figure 2A). The presence of a formal negative charge leads to a greater electrostatic clash with the RNA as well as eliminating a hydrogen bond between the N3 of xanthine and U51. Consistent with this, at pH 8.5, xanthine binding to the guanine riboswitch could not be detected by ITC (Supplemental Table 1). Xanthine bound to the guanine riboswitch with a slightly stronger affinity at pH 6.0 as compared to pH 7.5 (KD = 32 μM). As a control, we measured the affinity of GR for hypoxanthine between pH 7.5 and pH 6.0 (Supplemental Table 1) and found no change, indicating that the pH dependence of xanthine binding is not due to titratable groups in the RNA.
Figure 2. 2-position purine derivatives bound to the purine riboswitch.
(A) Chemical structure of xanthine, emphasizing the keto-enol tautomerization of the N3/O2 atoms and the deprotonation of the enolic proton. (B) Simulated annealing omit map of the X:GR complex in which xanthine, U51, and C74 are removed and contoured at 1.0σ orange cage); the X:GR model is depicted with blue carbon atoms. Superposition of the xanthine-bound (blue) and hypoxanthine-bound (pink) guanine riboswitches shows that base positioning within the binding pocket is unaltered. (C) Simulated-annealing omit map of the 2FA:GRA complex with 2FA, U51, and C74 omitted and contoured at 1.0σ model of 2FA:GRA complex is pink). Superposition of the 2FA-bound (cyan) and 2,6-diaminpurine-bound (pink) GRA RNAs reveals the binding pocket is also unaltered.
To further examine how xanthine productively interacts with GR RNA, we crystallized the RNA-ligand complex and solved its structure. The resulting structure of the GR:xanthine complex (data and refinement statistics for this and all subsequent structures are summarized in Table 2) reveals that xanthine binds in exactly the same position as guanine and hypoxanthine (Figure 2B). Clearly, the riboswitch RNA does not adjust to compensate for the electrostatic clash between three carbonyl groups; the O2 of xanthine is positioned equidistant between the O2 of C74 and the O2 of U51 (2.7-2.8 Å).
Table 2.
Crystallographic and refinement statistics
| RNA:ligand | GR:Xanthine | GR:2FA | GR:OMG | GR:6ClG | GRA:OMG | GRA:6ClG | GR(A21G/U75C): 6ClG | GR:2AP |
|---|---|---|---|---|---|---|---|---|
| Data collection | ||||||||
| Space group | C2 | C2 | C2 | C2 | C2 | C2 | C2 | C2 |
| Cell dimensions | ||||||||
| a, b, c (Å) | 131.35, 35.10, 42.25 | 132.46, 35.08, 41.89 | 133.18, 35.27, 42,12 | 132.10, 35.12, 41.81 | 130.94, 35.04, 42.19 | 132.60, 35.17, 41.79 | 132.51, 35.12, 41.76 | 127.42, 35.16, 42.24 |
| β(°) | 90.38 | 90.10 | 90.45 | 90.03 | 89.16 | 90.38 | 90.57 | 91.18 |
| Resolution (Å) | 20-1.90 (1.97-1.90)a | 20-1.95 (2.02-1.95) | 20-1.85 (1.92-1.85) | 20-1.56 (1.62-1.56) | 20-2.15 (2.23-2.15) | 20-1.8 (1.86-1.80) | 20-2.1 (2.18-2.10) | 20-2.4 (2.49-2.4) |
| Rmerge | 0.064 (0.197) | 0.059 (0.315) | 0.066 (0.323) | 0.080 (0.377) | 0.096 (0.357) | 0.048 (0.201) | 0.085 (0.352) | 0.080 (0.284) |
| I/σI | 10.2 (3.2) | 10.2 (3.2) | 11.1 (2.0) | 13.1 (2.1) | 11.6 (3.0) | 15.8 (3.8) | 8.5 (3.5) | 8.1 (3.0) |
| Completeness (%) | 95.6 (80.6) | 95.6 (80.6) | 88.1 (53.1) | 99.0 (94.0) | 90.9 (91.4) | 91.8 (69.5) | 98.8 (97.6) | 94.2 (86.0) |
| Redundancy | 3.58 (3.21) | 3.58 (3.21) | 3.22 (2.48) | 6.73 (5.49) | 6.65 (6.67) | 3.56 (3.31) | 4.91 (4.94) | 1.83 (1.12) |
| Refinementb | ||||||||
| Resolution (Å) | 20-1.90 | 20-1.95 | 20-1.90 | 20-1.70 | 20-2.15 | 20-1.90 | 20-2.1 | 20-2.4 |
| Rwork / Rfree | 21.0/24.6 | 21.8/26.8 | 22.6/27.6 | 21.6/23.4 | 23.8/28.3 | 22.7/25.1 | 22.0/25.6 | 22.9/27.8 |
| Average B-factors (Å2) | 26.6 | 39.8 | 34.9 | 28.1 | 40.1 | 28.0 | 40.0 | 40.1 |
| Luzzati error (Å)c | 0.28 | 0.39 | 0.36 | 0.24 | 0.47 | 0.29 | 0.37 | 0.43 |
| rmsd bond lengths (Å) | 0.004 | 0.013 | 0.012 | 0.003 | 0.005 | 0.004 | 0.001 | 0.003 |
| rmsd bond angles (°) | 1.1 | 1.6 | 1.70 | 0.9 | 1.0 | 1.0 | 1.0 | 0.6 |
| PDBID | 3GAO | 3GOT | 3FO6 | 3GER | 3GES | 3FO4 | 3GOG | 3G4M |
Highest resolution shell is shown in parenthesis.
Refinement was against all data within the stated resolution range, with a random 10% omitted for use in calculation of Rfree
Cross-validated Luzzati coordinate error
The simplest explanation for these observations is that a tautomer of xanthine at neutral pH most fully satisfies the hydrogen bonding requirements of the riboswitch. Below pH 8, xanthine has two tautomeric forms at the 2-position: a keto- and an enoltautomer (Figure 2A). The keto-tautomer cannot form hydrogen bonds to O2(C74), O4(U51), and N3(U51). The enol-tautomer, however, partially alleviates this situation by containing a hydroxyl group that can hydrogen bond to either carbonyl on C74 or U51 (Figure 2B) along with re-establishing the hydrogen bond between the N3 of xanthine and N3(U51). Thus, the riboswitch RNA likely binds and stabilizes the enol tautomer of xanthine as it presents a hydrogen bonding donor/acceptor pattern most similar to guanine and hypoxanthine. Stabilization of the enol tautomer form of a nucleobase has been observed in other base-pairing and RNA-ligand interactions. For example, 2-hydroxyadenosine forms a Watson-Crick like base-pair with thymidine in duplex DNA as the rare 2-enol tautomer (Robinson et al., 1998). Similarly, the thymine pyrophosphate (TPP) riboswitch binds the enolic tautomer of oxythiamine pyrophosphate (Thore et al., 2008) and an in vitro selected aptamer induces changes in both the conformation and electronic properties of malachite green (Nguyen et al., 2002).
2-fluoroadenine (2FA) binds an adenine-responsive RNA using weak halogen bonds
By ITC, 2FA binds to GRA (a variant of GR RNA that contains a C74U mutation that makes it specific for adenine (Gilbert et al., 2006b)) with an apparent KD of 4.0 μM (Table 1). The weaker binding of 2FA to GRA compared to 2,6-diaminopurine (Krel = 240) indicates that there is an energetic penalty for accommodating the 2-fluoro moiety in the binding pocket. The 1.95 Å resolution structure of 2FA bound to GRA RNA, again reveals an unaltered arrangement of the ligand and RNA (Figure 2C). The close distance between the fluoro group and the O2 carbonyl oxygens of U51 and U74 (2.9 and 2.8 Å, respectively) indicate that these groups engage in a weak halogen bond, as is observed between halogen atoms and oxygen/nitrogen in a number of other ligand-macromolecular structures (Auffinger et al., 2004). In a separate study, 2-chloroadenine did not bind the riboswitch (Mandal and Breaker, 2004a); the chloro group has a larger atomic radius than either fluorine or oxygen and therefore is likely sterically blocked from proper binding. Thus, the binding of xanthine and 2FA indicate that the RNA can accommodate nonideal electrostatic interactions but not additional steric bulk at the 2-position of purine analogs.
Purine riboswitch specificity at the 6-position of the ligand
Previous studies of analogs binding to purine riboswitches indicate that different modifications at the 6-position of the purine ligand are tolerated (Mandal et al., 2003; Mandal and Breaker, 2004a). To further interrogate the effect of modifications at this position, we tested by ITC the binding of a number of 6-position purine derivatives to GR (Table 1, Figure 1B). The purine riboswitch recognizes these compounds to varying degrees. Single atom substituted compounds like 6-thioguanine, 6-chloroguanine (6ClG), and 6-bromoguanine (6BrG) bind with KDs of 0.073 μM, 0.89 μM, and 2.1 μM, respectively (Table 1). In comparison, 2-aminopurine (2AP) affinity, in which the functional group is absent, binds GR with KD = 4.4 μM, indicating that, despite the fact that these substitutions have slightly more steric bulk relative to a carbonyl group, they contribute favorably to the ligand-RNA interaction. The moderate affinity of 2AP for the guanine riboswitch has been observed in previous studies as well (Lemay and Lafontaine, 2007; Mandal et al., 2003).
Many bulkier additions at this position completely abrogate binding to GR or GRA, as observed with the compounds O-benzylguanine, kinetin, 6-methylaminopurine, and 6-methylmercaptoguanine (data not shown). However, not all bulky additions to the 6-position preclude binding. Adding a methyl group to the O6 of guanine significantly decreases the binding affinity relative to guanine (KD = 23 μM; Krel = 5750) (Table 1); this is much weaker than the KD ~0.5 μM reported previously by in-line probing (Mandal et al., 2003). Interestingly, 2,6,-diaminopurine (DAP), a purine analog with an amine functional group in the place of the O6 on guanine, binds GR with a binding affinity of 4.1 μM, similar to the affinity of 2AP (Table 1). A previous study done by in-line probing also noted the similarity in affinity for DAP and 2AP by the guanine riboswitch (Mandal et al., 2003). These data indicate a more complicated relationship between the 6-position of the ligand and the RNA.
Structural perturbation of the binding pocket
Despite only potentially having one canonical hydrogen bond to C74, it is notable that the above 6-position derivatives bind GR with higher affinity than purine—a compound that interacts with U74 via a single hydrogen bond—binds GRA (>100 μM) (Gilbert et al., 2006b). This suggests that these compounds may have means to compensate for a potentially weak interaction with C74. To explore this issue further, we crystallized GR in complex with 6OMeG, 6ClG, and 2AP (Table 2). These ligands were chosen among the 6-position derivatives because they present two different challenges to the binding pocket: the potential disruption of two hydrogen bonding interactions in the case of 6ClG and 2AP and a bulky methyl group in the case of 6OMeG.
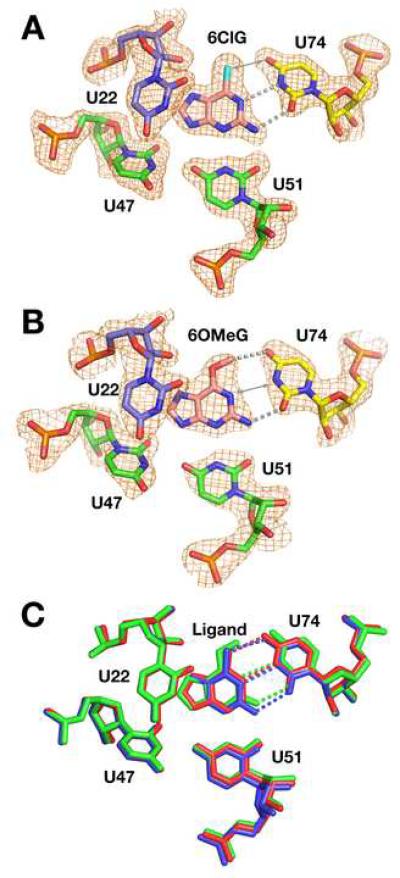
These structures reveal two distinct means of accommodating 6-substituted purines. The crystal structure of the 6ClG:GR complex displays an interaction mode similar to wild type (Figure 3A). C74 forms the expected hydrogen bond with the N2 of the ligand along with a productive interaction between its N4 and the chloro group of 6ClG (3.1 Å). However, this geometry places N1(6ClG) only 3.0 Å from N3(C74), which is expected to be electrostatically unfavorable. This could potentially be alleviated by protonating C74 as observed in other RNAs (Das and Piccirilli, 2005), but we do not have direct evidence for this. Thus, the RNA pays a moderate energetic penalty for nonideal interactions with the ligand as compared with guanine but retains the same binding mode. In contrast, the structure of 6OMeG bound GR structure reveals a clear conformational change in the binding pocket (Figure 3B). While the positions of U22, U51, and the base triples above and below the ligand are identical to previous structures (Figure 3B), C74 shifts towards the minor groove by 1.4 Å (Luzzati cross-validated coordinate error = 0.34 Å). This, along with a small rotation of the ligand in the binding pocket, allows room for the O-methyl group in the binding pocket, creating a new two hydrogen bond donor/acceptor pattern between C74 and the ligand (Figure 3C). This involves N4(C74) hydrogen bonding to N1(6OMeG) instead of O6, and N3(C74) interacting with the exocyclic amine at the 2-position of 6OMeG. The stacking between C74 and the adjacent bases (A73 and U75) also diminishes slightly upon minor groove shifting (Supplemental Figure 1A). An identical shift of C74 towards the minor groove is observed in the crystal structure of 2AP bound to GR (Figure 3D). These structures thus reveal that C74 is afforded a degree of conformational flexibility that allows it to optimize favorable hydrogen bonding interactions with the ligand.
Figure 3. 6-position derivatives binding to a guanine binding RNA (GR).
(A) Binding pocket of the 6ClG:GR structure. The electron density map of the ligand and surrounding nucleotides (orange mesh, contoured at 1σ) is a simulated annealing omit map in which the shown nucleotides have been omitted from the model used to calculate the map. Hydrogen bonding interactions between the ligand and the specificity pyrimidine (C/U74) are depicted as grey dashed lines. Similar maps are shown throughout Figures 3, 5, and 6. (B) Ligand and binding pocket of the 6OMeG:GR structure. (C) Superposition of structures of wild type hypoxanthine:GR (red), 6ClG:GR (blue), and 6OMeG:GR (green). While U22, U47, and U51 in each structure are in nearly identical positions, the ligand and C74 in the 6OMeG:GR have shifted. The largest positional shift is C74, which is moved towards the minor groove, allowing for an altered hydrogen bonding pattern with the ligand. (D) Ligand and binding pocket of the 2AP:GR structure.
Given that guanine versus adenine specificity is determined by ligand pairing with Y74, it is surprising that the RNA allows any degree of conformational flexibility at this position. It is likely that GR also productively binds DAP by minor-groove shifting C74, creating a two hydrogen bond interaction. As some 6-substituted compounds (halogen derivatives) do not promote minor-groove wobbling whereas others do (O-methylation, amino-, or none), what is the magnitude of the energetic barrier for this movement of C74?
Energetic penalty associated with adaptive binding
To address this issue, we calculated an approximate free energy associated with shifting C74 from the free energies associated with different ligands binding to GR and GRA using data obtained from this and previous work (Gilbert et al., 2006b). The simplest means for calculating this value is by directly comparing 2AP binding to GRA, a two hydrogen bond Watson-Crick interaction (Gilbert et al., 2006b; Mandal and Breaker, 2004a), with the two hydrogen bond minor groove wobble pairing interaction with GR (Figure 4A). Assuming that the hydrogen bonds in the two complexes are nearly energetically identical, the free energy difference (ΔΔG) between these two complexes, 1.8 kcal mol-1, corresponds to the energetic penalty for the minor groove shift of C74. A second means of calculating this penalty, using data derived from binding of DAP, yields a difference of 1.7 kcal mol-1 (Figure 4B). Thus, the energetic cost of minor groove wobbling C74 is small and comparable in magnitude to the penalty for elimination of the first of three hydrogen bonds between U74 and the ligand. For ligands that can only form one productive hydrogen bond with the RNA through their Watson-Crick faces, this indicates that minor groove wobbling has a sufficiently low energetic barrier to allow at least partial recovery of binding affinity by re-establishing two hydrogen bonds. In computational searches for novel compounds that may target the purine riboswitch, this effect will clearly have to be accounted for.
Figure 4. Energetic cost of minor groove wobbling.
(A) Calculation of the energetic penalty associated with shifting C74 into the minor groove by directly comparing the same ligand, 2AP, binding to the GRA in the unshifted form and GR in the shifted position. (B) A second independent means of this calculation yields the apparent energetic penalty by a direct measure of the loss of a third hydrogen bond.
Interactions of 6-position derivatives with adenine-responsive GRA
The B. subtilis xpt-pbuX guanine riboswitch carrying a C74U mutation (GRA) becomes highly specific for adenine and adenine derivatives (Gilbert et al., 2006a; Gilbert et al., 2006b). The uridine residue introduces a different pattern of hydrogen-bond donor/acceptor pairs to the ligand that favors ligands with hydrogen bond acceptors at their N1 position like 6ClG, 6BrG, and 6OMeG (Figure 1B). Analogous to the adenine derivatives 2AP and DAP that bind GR, these ligands might productively bind GRA, despite being considered guanine derivatives.
To examine this possibility, we characterized the binding of these compounds to GRA by ITC and X-ray crystallography. In each case, the binding affinity for GRA was similar to that of GR; 6ClG, 6BrG, and 6OMeG interact with both RNAs with nearly the same affinity (Table 1). In contrast to their interactions with GR, where the 6-position halogens increases the affinity of the ligand for the guanine riboswitch compared to 2AP, the 6-chloro and 6-bromo groups reduce ligand affinity for GRA (Table 1). Strikingly, all three modifications at the 6-position abrogate the specificity imparted by Y74; the guanine and adenine classes apparently cannot be distinguished by these ligands. From a drug discovery perspective, these types of compounds might be able to target all purine riboswitches within a cell.
To address how these derivatives are capable of binding both GR and GRA, we crystallized complexes of 6OMeG and 6ClG bound to GRA and determined their structures (Table 2). In both structures the ligand binding pocket is unaltered (Figure 5) as U22, U51 and the base triples above and below the binding pocket superimpose perfectly on the wild type ligand bound complex. In the GRA:6ClG complex, U74 is positioned identically to C74 in the GR:6ClG complex (Figure 5A). However, unlike C74 in the GR:6OMeG ligand bound complex, U74 is not shifted toward the minor groove in the GRA:6OMeG complex (Figure 5B). Instead, the methyl group is placed 3.0 Å away from O4(U74), indicating a productive C-H•••O hydrogen bond as has been observed in other nucleic acids (Auffinger et al., 1996; Wahl and Sundaralingam, 1997).
Figure 5. 6-position derivatives complexed to an adenine binding RNA (GRA).
(A) Structure of 6ClG bound to GRA; double arrow represents a potential halogen bond between the chloro group and U74. (B) Binding pocket of the 6OMeG:GRA complex; arrow denotes a long hydrogen bond (3.5 Å) between N1(6OMeG) and N3(U74). (C) Superposition of structures of the wild type hypoxanthine:GR complex (red), 6ClG:GRA (blue), and 6OMeG:GRA (green). Most atoms in each structure superimpose well, except for a minor repulsion between the ligand and U74 in the 6OMeG:GRA complex to accommodate the methyl group at the Watson-Crick interface.
The preference of 6OMeG to form a Watson-Crick base pair with U74 over C74 has important implications. It is notable that 6-O-methylguanine lesions in DNA are among the most carcinogenic because they base pair with thymidine in a Watson-Crick conformation that is not recognized by DNA polymerases as a damage site (Warren et al., 2006). Alternatively, a 6-O-methylguanine:cytidine base pair shifts the cytidine into the minor-groove. It is then recognized as a misincorporation event and repaired during DNA replication with a high fidelity polymerase (Johnson and Beese, 2004). 6OMeG binding to the adenine riboswitch versus the guanine riboswitch mimics this behavior observed in DNA. Despite the fact that the ligand-Y74 pair is embedded in the complex architecture of the three-way junction, it still behaves in a fashion similar to isolated Watson-Crick pairs in a pure duplex.
A phylogenetically conserved base pair adjacent to the binding site contributes to specificity
While Y74 is the primary means of achieving specificity and affinity, do flanking sequences have a secondary influence on binding? In phylogeny, the flanking A21-U75 pair is >90% conserved and only appears as an U-A transversion in the guanine riboswitch—never in the adenine riboswitch—and never as G-C or C-G base pairs (Gilbert et al., 2007; Mandal et al., 2003; Mandal and Breaker, 2004a). Mutagenesis of this pair to the other Watson-Crick pairs revealed that its identity has a negligible affect on ligand binding affinity (Gilbert et al., 2007; Lemay and Lafontaine, 2007).
To explore this possibility, we first tested the ability of the A21U/U75A and A21G/U75C mutants of GR and GRA to interact with the natural effectors guanine, hypoxanthine, and adenine. Binding of guanine and DAP (previously reported in (Gilbert et al., 2007)) to all 21-75 mutants of GR and GRA, respectively, yielded binding affinities near wild type (Table 3). Conversely, binding of hypoxanthine and adenine to the A21U/U75A and A21G/U75C mutants of GR and GRA, respectively, was sharply reduced compared to wild type (Table 3). Thus, these mutants exhibit decreased affinity for ligands binding through two-hydrogen bonds along the Watson-Crick interface. 6ClG and DAP also bind the guanine riboswitch through two hydrogen bonds and have a reduced affinity for A21G/U75C mutant RNA.
Table 3.
Binding affinities for A21-U75 pair mutants.
| KD(μM) | ||||
|---|---|---|---|---|
| GR(A21U/U75A) | GR(A21G/U75C) | GRA(A21U/U75A) | GRA(A21G/U75C) | |
| Guanine | 0.003±0.0002 | 0.008±0.001 | ---1 | --- |
| Hypoxanthine | 3.3±0.32 | 9.5±0.22 | --- | --- |
| 2,6-Diaminopurine | --- | 13±1.1 | 0.047±0.009 | 0.076±0.015 |
| Adenine | --- | --- | 2.1±0.19 | 6.0±1.1 |
| 6-Chloroguanine | --- | 24±6.0 | --- | --- |
---: not determined
As a basis of comparison to previous work, we crystallized and solved the structure of 6ClG bound to GR(A21G/U75C) RNA as this ligand showed the greatest sensitivity to the identity of the 21-75 pair. In this complex, C74 is observed shifted towards the minor groove (Figure 6A), in contrast to the previous 6ClG containing complexes (superposition shown in Figure 6B, Supporting Figure 1B). The G-C pair at the 21-75 position allows for the repositioning of C74 in the pocket in order to compensate for the weak bonding between the 6-chloro group and the C74(N4) group. Like observed previously, the minor groove wobbling of C74 diminishes the stacking interactions with the adjacent nucleotides, and that this effect may contribute to the ability of the G21-C75 pair to promote the greater flexibility of the specificity pyrimidine. The combination of the binding data and structure suggest that for adenine and hypoxanthine—which interact via two hydrogen bonds with the purine riboswitch— flanking sequences play a role in ensuring proper positioning of Y74, ensuring a Watson-Crick interaction with the ligand, thereby maximizing both affinity and specificity.
Figure 6. Structure of 6ClG bound to GR RNA containing an A21G/U75C mutation.
(A) Binding pocket and mutated pair (cyan). C74 is clearly shifted towards the minor groove (compare with Figure 4A), to allow for an alternative hydrogen bonding pattern to occur. (B) Superposition of the wild type HX:GR (red) and 6ClG:GR(A21G/U75G) (blue) complexes emphasizing the ligand:C74 interaction and the base pair (21-75) below it. Mutation of the 21-75 pair does not cause this pair to adjust its position, but allows C74 to shift towards the minor groove when binding 6ClG.
A comparison of the binding affinities of two ligands (hypoxanthine and DAP) to GR and GR(A21G/U75C) RNAs reveals that C74 likely has a reduced energetic penalty for shifting in the context of different 21-75 base pairs. Hypoxanthine, a ligand that interacts with C74 through a two hydrogen bond Watson-Crick pair, exhibits a Krel = 13 between these two RNAs (Table 3). The difference in DAP (a ligand that binds GR using a two hydrogen bond minor groove wobble pair) affinity for wild type GR and GR(A21G/U75C) is less in magnitude (Krel = 3.2) (Table 3). This indicates that the energetic penalty for shifting C74 into the minor groove is reduced approximately fourfold in the mutant form of the RNA. The increased conformational flexibility of C74 in the context of the A21G/U75C mutation and the ligand affinity of the RNA for various purine analogues suggest that the near universal base-pair conservation at the top of the P1 helix is an important indirect specificity determinant.
DISCUSSION
In the context of cellular metabolism the guanine and adenine riboswitches face the challenge of discriminating between closely related purine compounds with sufficient fidelity to prevent inappropriate regulatory responses. To further understand the basis of their specificity for their natural effectors, we have investigated these RNAs interactions with non-natural purine compounds, revealing several novel aspects of ligand recognition. First, as might be expected, the tautomeric states and pKa(s) of the ligand influence how it will interact with the RNA. In the example of xanthine, the RNA stabilizes the enol tautomer to maximize the number of hydrogen bonds with the ligand. This provides additional support to the previous finding that stability of the nucleobasepurine riboswitch complex is dominated by hydrogen bonding (Gilbert et al., 2006b). Second, we have found that certain ligands can target both the adenine and guanine riboswitches, binding them with near equal affinity. Compounds like 2AP, 6ClG, and 6OMeG, present a challenge to the discriminating power of this RNA, in part through the unexpected ability of C74 to minor groove wobble. Again, this effect appears to be the direct result of maximizing the number of hydrogen bonds between the ligand and RNA to achieve highest possible affinity. These data indicate that if a ligand or RNA can form two productive hydrogen bonds with Y74, then either (or both) can adapt through conformational changes or alterations in the functional groups to form the complex.
The ability of the pyrimidine 74 to minor groove wobble is surprising in light of the fact that it is the primary determinant of specificity for guanine versus adenine. However, riboswitches evolved in the cellular environment where it is only challenged with a few related purine compounds, primarily guanine, hypoxanthine, xanthine, and adenine. The selection pressure imposed upon the RNA would be to prevent major groove wobbling that allows G/HX/X-U pairs, which we have not observed. Indeed, we were not able to detect xanthine binding to GRA RNA, which theoretically could be accommodated via a major groove wobble by U74 to mimic a U•U mismatch that is often found in RNA helices. This suggests that major groove wobbling of Y74 has been selected against to ensure proper specificity. Minor groove wobbling, conversely, would not assist in cross-recognition of cellular purine nucleobases, and was not apparently selected against. The binding pocket of the purine riboswitch may have other yet undiscovered means of conformational flexibility that would allow it to recognize more diverse compounds. This idea is reinforced by the recent discovery of a third class of purine riboswitch that recognizes 2'-deoxyguanosine (Kim et al., 2007), accommodating the additional 2'-deoxyribose moiety in part by an alternative conformation of the J2/3 loop around the binding pocket (Edwards and Batey, 2008). GR RNA is able to bind 2'-deoxyguanosine with an affinity of 12 µM, which is slightly greater than its affinity for 6OMeG (Table 1). Based upon the crystal structure of a mutant form of the GR RNA in complex with 2'-deoxyguanosine, the accommodation of this bulkier ligand is through a displacement of U51 towards the minor groove and possibly stabilization of a tautomeric form of either the ligand or U51.
Another aspect of purine riboswitch specificity is revealed by our investigation of mutations in the base pair of the P1 helix directly adjacent to the ligand. While there is a high degree of conservation at this pair, it is not obvious why this should be the case in light of previous data. Our results indicate ligands that bind using three (G and DAP) or two (HX and A) hydrogen bonds to Y74 behave differently; mutations have a significantly greater affect on HX/A binding than on G/DAP. A structure of 6ClG (two hydrogen bonds) bound to one of the mutant pairs reveals that the presence of alternative base pairs at the 21-75 position relaxes the positioning of Y74 such that it is energetically less costly to minor groove wobble. While we have asserted that this poses little problem for specificity, the increased local flexibility in this region of the RNA may impact the RNA in the free state. It has been shown that the binding pocket is conformationally restricted in the free state and this is important for efficient ligand binding (Ottink et al., 2007; Stoddard et al., 2008). Thus, the strong selective pressure for the A21-U75 pair at this position may be in part for proper positioning of the Y74 base for ligand recognition in the free state.
Analysis of riboswitch aptamer domains bound to chemical analogs of their natural ligands has revealed that their ability to recognize a diverse set of compounds is facilitated by small changes in conformation around the binding pocket. For example, the TPP riboswitch is capable of binding a number of TPP analogs, some of which have antimicrobial activity (Winkler et al., 2002). Crystal structures of these compounds bound to the bacterial (Edwards and Ferre-D'Amare, 2006) or eukaryotic (Thore et al., 2008) TPP riboswitch reveals that small local changes in the binding pocket increases the number of interactions with the ligand. Furthermore, these data suggest a correlation between weaker binding ligands and structural rearrangement of a highly ordered binding pocket (Edwards and Ferre-D'Amare, 2006). Another example is the lysine riboswitch, which binds a number of lysine analogs with reasonably high affinity (Blount et al., 2007; Sudarsan et al., 2003). Like the purine riboswitch, this RNA has a binding pocket that fully encapsulates the ligand in a multi-helix junction that undergoes local limited conformational changes upon ligand binding (Garst et al., 2008; Serganov et al., 2008). Structures of lysine analogs bound to this riboswitch show that small adjustments to the RNA backbone are sufficient to accommodate certain analogs, presumably by optimizing RNA-ligand hydrogen bonding (Serganov et al., 2008).
Together, analyses of these riboswitches suggest molecular adaptability of these binding pockets is a common feature. This inherent plasticity is rooted in the fact that riboswitches, like the majority of biological RNAs, use induced-fit mechanisms in which binding is accompanied by a conformational change in the RNA (Leulliot and Varani, 2001; Williamson, 2000). In the case of riboswitches, ligand-induced conformational changes in the aptamer domain are essential for communication with downstream sequences that interface with the expression machinery (Batey et al., 2004; Stoddard et al., 2008). While riboswitches have evolved to discriminate between a limited set of chemically related metabolites in the cellular environment, this specificity is easily challenged by non-natural analogs that can exploit the inherent local flexibility of the binding pocket. Thus, it is likely that riboswitches may indeed be excellent targets for structure-based drug design efforts that incorporate computational strategies that can account for conformational flexibility (Guilbert and James, 2008; Pinto et al., 2008).
METHODS
RNA preparation
All RNAs used in this study are variants of the B. subtilis xpt-pbuX guanine riboswitch that were synthesized and purified using methods previously described (Batey et al., 2004; Gilbert et al., 2006b). RNA was transcribed using dsDNA templates generated by PCR using T7 RNA polymerase and purified by 12% denaturing PAGE. Gel-extracted RNA was exchanged into 10 mM K+-HEPES, pH 7.5 and maintained at -20 °C until use.
Isothermal Titration Calorimetry
Determination of the apparent equilibrium dissociation constants (KD) was performed using ITC by previously described methods (Gilbert et al., 2007; Gilbert et al., 2006a; Stoddard et al., 2008). Purified RNA was dialyzed against 1 L of buffer containing 50 mM K+-HEPES, pH 7.5, 100 mM KCl, and 10 mM MgCl2 at 4 °C. To ensure buffer matching between the titrant (ligand) and titrate (RNA), solid ligand was dissolved in the same solution the RNA was dialyzed against to yield a final concentration ~8-fold higher than the RNA. ITC measurements were performed at 30 °C using 5 µcal sec-1 reference power. Each titration consisted of either sixty-five 5 µL injections (for tightly binding ligands) or thirty-two 10 µL injections (for weakly binding ligands) with a 0.5 µL sec-1 injection rate with 180 second intervals between injections (MicroCal, 2003). The concentration of RNA and ligand was adjusted for each compound such that the c value for each experiment was between 0.5 and 500, reflecting the experimental concentration range in which useful data can be obtained (Turnbull and Daranas, 2003; Wiseman et al., 1989). Data was fit to a single-site binding model using Origin ITC software (Microcal Software Inc.).
X-ray Crystallography
Diffraction quality crystals were grown by the hanging drop vapor diffusion method by mixing a 1:1 ratio of riboswitch:ligand complex with mother liquor containing 15-20% PEG 3000, 480-720 mM ammonium acetate, 10-12 mM cobalt hexammine, 10 mM K+-HEPES, pH 7.5. Needle-like crystals >200 μm in length grew within one week and were cryoprotected in mother liquor plus 30% 2-methyl-2,4-pentanediol (MPD) for 5-10 minutes and flash frozen in liquid nitrogen. Diffraction data was collected using CuKαX-ray radiation with an R-AXIS IV++ detector and an RU-200/Confocal blue optic source (Rigaku MSC).
Data was indexed, integrated, and scaled using D*TREK (Pflugrath, 1999) in the Crystal Clear package (Rigaku MSC). Initial electron density maps were calculated by molecular replacement in CNS (Brunger et al., 1998) using the previously reported GR:HX complex (PDB 1U8D) as the search model in which the solvent, ligands, and Y74 were removed. In all cases, the initial 2Fo-Fc map showed clear electron density corresponding to the position of the ligand and the specificity pyrimidine (Y74). The ligand, ions, and Y74 were built into the models using PyMOL v0.99 and solvent added using automated water picking; all model refinement was performed using CNS. Final 2Fo-Fc simulated-annealing omit maps with the U51, U47, U22, Y74, and the ligand removed, unless otherwise indicated, were calculated in all cases by slow cooling from 2500 K in 25 K steps in order to definitively show that movements in the binding pocket reflected their actual positions in the molecular structure and were not the subject of model bias. Structure factors and coordinates have been deposited in the RCSB Protein Data Bank.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Colby Stoddard and Quentin Vicens for critical reading of this manuscript and useful discussions. This work was supported by a grant from the National Institutes of Health (GM073850).
ABBREVIATIONS
- 2FA
2-fluoroadenine
- 6ClG
6-chloroguanine
- 6OMeG
6-O-methylguanine
- A
adenine
- DAP
2,6-diaminopurine
- G
guanine
- GR
xpt-pbuX guanine responsive riboswitch from B. subtilis
- GRA
GR riboswitch carrying a C74U mutation
- HX
hypoxanthine
- ITC
isothermal titration calorimetry
- R
purine
- UTR
untranslated region
- X
xanthine
- Y
pyrimidine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ataide SF, Wilson SN, Dang S, Rogers TE, Roy B, Banerjee R, Henkin TM, Ibba M. Mechanisms of resistance to an amino acid antibiotic that targets translation. ACS Chem Biol. 2007;2:819–827. doi: 10.1021/cb7002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffinger P, Hays FA, Westhof E, Ho PS. Halogen bonds in biological molecules. Proc Natl Acad Sci U S A. 2004;101:16789–16794. doi: 10.1073/pnas.0407607101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffinger P, LouiseMay S, Westhof E. Molecular dynamics simulations of the anticodon hairpin of tRNA(Asp): Structuring effects of C-H•••O hydrogen bonds and of long-range hydration forces. J Am Chem Soc. 1996;118:1181–1189. [Google Scholar]
- Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- Blount KF, Breaker RR. Riboswitches as antibacterial drug targets. Nat Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- Blount KF, Wang JX, Lim J, Sudarsan N, Breaker RR. Antibacterial lysine analogs that target lysine riboswitches. Nat Chem Biol. 2007;3:44–49. doi: 10.1038/nchembio842. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Das SR, Piccirilli JA. General acid catalysis by the hepatitis delta virus ribozyme. Nat Chem Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- Edwards AL, Batey RT. A Structural Basis for the Recognition of 2'-Deoxyguanosine by the Purine Riboswitch. J Mol Biol. 2008;385:938–948. doi: 10.1016/j.jmb.2008.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TE, Ferre-D'Amare AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Garst AD, Heroux A, Rambo RP, Batey RT. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem. 2008;283:22347–22351. doi: 10.1074/jbc.C800120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautheret D, Konings D, Gutell RR. G.U base pairing motifs in ribosomal RNA. RNA. 1995;1:807–814. [PMC free article] [PubMed] [Google Scholar]
- Gilbert SD, Love CE, Edwards AL, Batey RT. Mutational analysis of the purine riboswitch aptamer domain. Biochemistry. 2007;46:13297–13309. doi: 10.1021/bi700410g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SD, Mediatore SJ, Batey RT. Modified pyrimidines specifically bind the purine riboswitch. J Am Chem Soc. 2006a;128:14214–14215. doi: 10.1021/ja063645t. [DOI] [PubMed] [Google Scholar]
- Gilbert SD, Stoddard CD, Wise SJ, Batey RT. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J Mol Biol. 2006b;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Guilbert C, James TL. Docking to RNA via root-mean-square-deviation-driven energy minimization with flexible ligands and flexible targets. J Chem Inf Model. 2008;48:1257–1268. doi: 10.1021/ci8000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SJ, Beese LS. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116:803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- Kim JN, Roth A, Breaker RR. Guanine riboswitch variants from Mesoplasma florum selectively recognize 2'-deoxyguanosine. Proc Natl Acad Sci U S A. 2007;104:16092–16097. doi: 10.1073/pnas.0705884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ER, Blount KF, Breaker RR. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol. 2009;6 doi: 10.4161/rna.6.2.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay JF, Lafontaine DA. Core requirements of the adenine riboswitch aptamer for ligand binding. RNA. 2007;13:335–350. doi: 10.1261/rna.142007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Westhof E. Conserved geometrical base-pairing patterns in RNA. Q Rev Biophys. 1998;31:399–455. doi: 10.1017/s0033583599003479. [DOI] [PubMed] [Google Scholar]
- Leulliot N, Varani G. Current topics in RNA-protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol. 2004a;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004b;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MicroCal L. VP-ITC Microcalorimeter user's Manual. Northhampton; MA: 2003. [Google Scholar]
- Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu Rev Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- Nguyen DH, DeFina SC, Fink WH, Dieckmann T. Binding to an RNA aptamer changes the charge distribution and conformation of malachite green. J Am Chem Soc. 2002;124:15081–15084. doi: 10.1021/ja027635d. [DOI] [PubMed] [Google Scholar]
- Ottink OM, Rampersad SM, Tessari M, Zaman GJ, Heus HA, Wijmenga SS. Ligand-induced folding of the guanine-sensing riboswitch is controlled by a combined predetermined induced fit mechanism. RNA. 2007;13:2202–2212. doi: 10.1261/rna.635307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 10):1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- Pinto IG, Guilbert C, Ulyanov NB, Stearns J, James TL. Discovery of Ligands for a Novel Target, the Human Telomerase RNA, Based on Flexible-Target Virtual Screening and NMR. J Med Chem. 2008;51:7205–7215. doi: 10.1021/jm800825n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson H, Gao YG, Bauer C, Roberts C, Switzer C, Wang AH. 2'-Deoxyisoguanosine adopts more than one tautomer to form base pairs with thymidine observed by high-resolution crystal structure analysis. Biochemistry. 1998;37:10897–10905. doi: 10.1021/bi980818l. [DOI] [PubMed] [Google Scholar]
- Rogstad KN, Jang YH, Sowers LC, Goddard WA., 3rd First principles calculations of the pKa values and tautomers of isoguanine and xanthine. Chem Res Toxicol. 2003;16:1455–1462. doi: 10.1021/tx034068e. [DOI] [PubMed] [Google Scholar]
- Serganov A, Huang L, Patel DJ. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature. 2008;455:1263–1267. doi: 10.1038/nature07326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard CD, Gilbert SD, Batey RT. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA. 2008;14:675–684. doi: 10.1261/rna.736908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker RR. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem Biol. 2005;12:1325–1335. doi: 10.1016/j.chembiol.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thore S, Frick C, Ban N. Structural basis of thiamine pyrophosphate analogues binding to the eukaryotic riboswitch. J Am Chem Soc. 2008;130:8116–8117. doi: 10.1021/ja801708e. [DOI] [PubMed] [Google Scholar]
- Turnbull WB, Daranas AH. On the value of c: can low affinity systems be studied by isothermal titration calorimetry? J Am Chem Soc. 2003;125:14859–14866. doi: 10.1021/ja036166s. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Sundaralingam M. C-H•••O hydrogen bonding in biology. Trends in Biochemical Sciences. 1997;22:97–102. doi: 10.1016/s0968-0004(97)01004-9. [DOI] [PubMed] [Google Scholar]
- Warren JJ, Forsberg LJ, Beese LS. The structural basis for the mutagenicity of O(6)-methyl-guanine lesions. Proc Natl Acad Sci U S A. 2006;103:19701–19706. doi: 10.1073/pnas.0609580103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E, Fritsch V. RNA folding: beyond Watson-Crick pairs. Structure. 2000;8:R55–65. doi: 10.1016/s0969-2126(00)00112-x. [DOI] [PubMed] [Google Scholar]
- Williamson JR. Induced fit in RNA-protein recognition. Nat Struct Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- Wright GD. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat Rev Microbiol. 2007;5:175–186. doi: 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.