Abstract
Background
In recent years there has been an increasing problem with Staphylococcus aureus strains that are resistant to treatment with existing antibiotics. An important starting point for the development of new antimicrobial drugs is the identification of "essential" genes that are important for bacterial survival and growth.
Results
We have developed a robust microarray and PCR-based method, Transposon-Mediated Differential Hybridisation (TMDH), that uses novel bioinformatics to identify transposon inserts in genome-wide libraries. Following a microarray-based screen, genes lacking transposon inserts are re-tested using a PCR and sequencing-based approach. We carried out a TMDH analysis of the S. aureus genome using a large random mariner transposon library of around a million mutants, and identified a total of 351 S. aureus genes important for survival and growth in culture. A comparison with the essential gene list experimentally derived for Bacillus subtilis highlighted interesting differences in both pathways and individual genes.
Conclusion
We have determined the first comprehensive list of S. aureus essential genes. This should act as a useful starting point for the identification of potential targets for novel antimicrobial compounds. The TMDH methodology we have developed is generic and could be applied to identify essential genes in other bacterial pathogens.
Background
There is an urgent need for the development of novel antimicrobial agents to counter the increasing problem of multiply resistant strains of Staphylococcus aureus [1]. A first step in the development of new classes of antibiotic is the identification of potential targets within the pathogen genome. Priority targets are genes and gene products that are important for bacterial survival and growth [2]. Several methods of identifying such "essential" genes are described in a recent volume [3]. The most rigorous method is the systematic construction of defined knockout mutants across the whole genome. This has been applied to Bacillus subtilis [4], with essential genes defined as those for which a mutant could not be obtained, in many cases being verified by conditional-lethal constructs. However this process is time-consuming and expensive.
An alternative is the use of transposon mutagenesis to generate a library of random mutants. In general, no transposons will be present within essential genes, since the presence of an intragenic transposon will disrupt gene function. The transposon insertion sites can be determined through the use of a transposon-specific primer to amplify the DNA flanking the transposon by PCR. If the library is saturated with a large number of transposon insertions, then essential genes can be identified by genetic footprinting [5]. Alternatively, the primer can be used to sequence the transposon junction directly [6]. However these approaches require a separate PCR and sequencing reaction for every mutated gene; consequently a whole genome screen is again time-consuming and expensive. A higher throughput can be obtained using microarrays to identify simultaneously the location of many inserts [7]; we term these "tag-array" approaches. The regions flanking transposons can be amplified using PCR and hybridised to an amplicon microarray [8], however this is labour intensive and in some cases may be non-reproducible [9]. An alternative is to use a custom transposon with one or two outward-facing promoters, from which labelled RNA run-offs are produced [9]. The use of high-density tiling microarrays can improve resolution [10], but this is still inferior to sequencing-based methods. Small genes (less than ~300 bp) are likely to be problematic since they are only covered by a small number of probes on the array. Large transposon libraries may also be problematic since the signal from a particular probe may be influenced by RNA derived from multiple transposons inserted within the same region.
In order to overcome the limitations of current tag-array approaches we have developed a simple method, Transposon-Mediated Differential Hybridisation (TMDH) [11,12], that combines the advantages of both sequencing- and array-based approaches to determine the repertoire of genes required for the survival and growth of the target organism. Application of TMDH to an analysis of the S. aureus genome identified 351 essential genes. Many of these may represent potential targets for the development of new therapeutic approaches to fight this important pathogen. A comparison with the essential gene list of B. subtilis provides insight into the changes in core metabolism that have occurred since the divergence of the two organisms.
Results
Development of the TMDH procedure
The TMDH procedure is outlined in Figure 1. Genomic DNA from the TMDH mutant library is digested using an appropriate restriction enzyme and amplified using linker PCR. Transcription is induced from the transposon T7 promoters in the presence of fluorescently-labelled dNTPs, and DNaseI is used to remove DNA, leaving labelled RNA run-offs. The labelled RNA is hybridised to a microarray, and the retrieved data used to screen for the presence of transposons as detailed below. Following the screening process, candidate essential genes are further interrogated for the presence of transposon inserts using PCR and DNA sequencing.
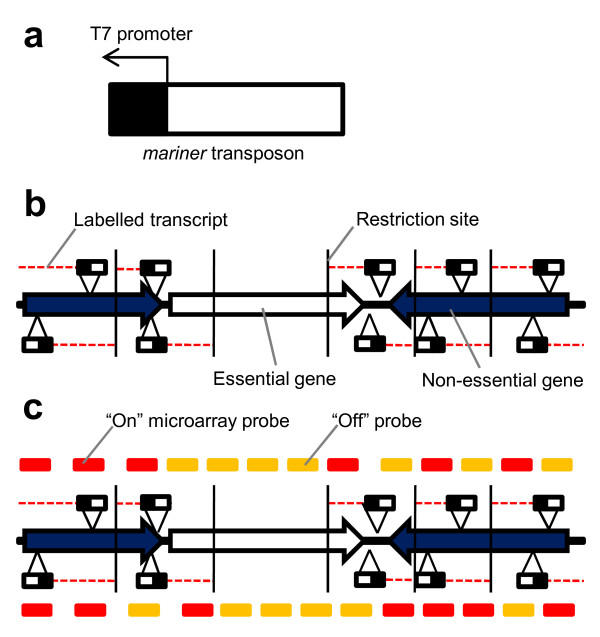
Figure 1.
Diagrammatic representation of the TMDH process. a) Illustration of the mariner transposon b) A saturated genome-wide transposon library is produced. Cells with a transposon disrupting an essential gene will not survive. Genomic DNA from the library is restriction digested, and Cy5-labelled RNA run-offs are produced from the T7 promoters by in vitro transcription. c) The labelled RNA is hybridised to a tiling oligonucleotide microarray. Probes that are downstream of a transposon give an "on" signal, other probes give an "off" signal. Note that some probes within an essential gene can give "on" signals, if they can be influenced by transposons outside the gene. Such probes are "non-informative" (see Results and Figure 4). Similarly, probes within a non-essential gene may give an "off" signal if they are upstream of the first transposon.
The restriction digest is critical for the TMDH procedure as it provides boundaries to limit the extension of the RNA run-offs, preventing transcripts produced from transposons within non-essential regions from extending into adjacent essential genes. However, for any particular restriction enzyme the distribution of restriction sites may be sub-optimal for some genes, limiting the number of "informative probes" that can be used to assess gene essentiality (see below). To minimise this problem two TMDH experiments were performed, one using the restriction enzyme AluI (AG^CT), the other using RsaI (GT^AC). Even using two enzymes, some genes were still potentially problematic, but sufficiently few that an additional enzyme was not deemed necessary; the affected genes were investigated using PCR (see below).
For each TMDH experiment, chromosomal DNA from the library was digested using the appropriate restriction enzyme, and oligonucleotide linkers were ligated to the restriction fragments. Linker PCR was performed using a linker-specific primer and a transposon-specific primer. In vitro transcription was performed from the T7 promoters, with direct incorporation of Cy5-UTP. The reaction was treated with DNaseI and the resultant labelled RNA was purified and used for hybridisation to the microarray.
Construction of a TMDH transposon library in S. aureus
Full details are presented in the Methods section. Briefly, a TMDH transposon, with an outward facing T7 promoter, was initially developed for use in E. coli based on the transposon Tn5 (EZ:Tn R6k ori Kan transposon, Epicentre) [12]. The construct was adapted for use in S. aureus by the addition of mariner mosaic ends (ME) and an erythromycin resistance gene. It was incorporated into a temperature-sensitive (ts) plasmid that contains a chloramphenicol resistance gene and is stable for replication in S. aureus at 30°C and below.
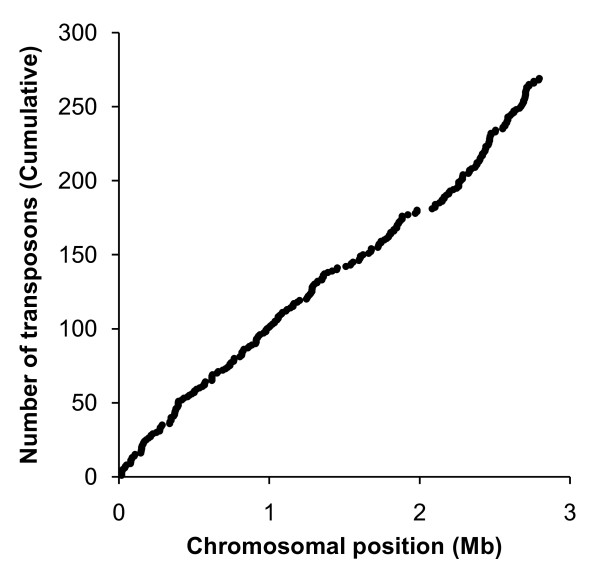
A large transposon library of around a million mutants was generated in S. aureus SH1000, essentially using the procedure described by Bae et al. [13]. A serial dilution of the resulting culture was used to determine library size (~106 mutants) and DNA sequencing and a preliminary TMDH analysis were used to demonstrate that the transposon had integrated throughout the S. aureus chromosome (see Figure 2).
Figure 2.
Cumulative distribution of transposons identified in preliminary experiment. Transposons were located using either the PCR/sequencing method or an initial microarray-based screen. From these experiments it was concluded that the mariner transposon inserted throughout the S. aureus chromosome and hence was suitable for use in TMDH.
Strain selection
The TMDH method is designed to work for strains for which sequence information can be obtained. This includes non-sequenced strains that are related to strains with a genome sequence. Our experiments were carried out in the S. aureus strain SH1000. This was derived from the genome sequenced strain NCTC 8325 (GenBank accession: CP000253) through phage curing followed by reconstruction of the rsbU gene [14-16]. The microarray probes that were relevant to TMDH in strain SH1000, and their positions relative to the NCTC 8325 genome sequence, were determined using BLAST (see Methods).
Microarray procedure
A set of 60-mer oligonucleotide probes was designed based upon the S. aureus MW2 genome sequence (GenBank accession: BA000033). The probes were spaced approximately every 100 bases on both strands across the whole genome, with the exception of repetitive regions. In-house inkjet printers capable of generating around 22,000 features per slide were used to produce microarrays [17]. As the total number of probes to span the genome exceeded this capacity, each experiment was performed using three separate slides. Following hybridisation of the labelled RNA and washing, the arrays were scanned using an Agilent G2500A scanner, and the images analyzed using the Agilent Feature Extractor software. Full details of the microarray procedure are available in the Methods section.
Development of software to determine location of transposon insertions
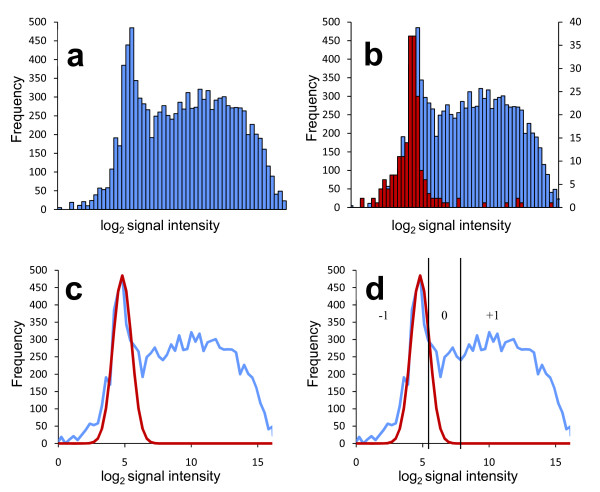
For TMDH the primary interest is in a discrete binary property, i.e. the presence or absence of transposons. Microarray data are continuously variable, so it is necessary to adopt a strategy for scoring transposons as present or absent. The log2 of the spot intensities measured from each microarray show a mixed distribution (see Figure 3a). In the absence of any transposons, low signal intensities with an approximately normal distribution would be expected ("off" signals). In contrast, "on" signals would be expected to have higher intensities but follow an irregular distribution, reflecting the multiple factors that influence the signal if transposons are present. These include the number of transposons present, their distance from probes, and local sequence elements that may affect transcription from the promoter located in the transposon.
Figure 3.
Illustration of the TMDH microarray scoring system (see text). a) Histogram of log2 of the raw microarray signals. b) As a), but overlaid with histogram of log2 of the microarray signals for the probes which do not hybridise to the S. aureus NCTC 8325 genome sequence, and hence indicate the background signal level in the absence of labelled transcript. c) A normal curve is fitted empirically to the data to model the distribution of background signals. d) Cut-offs are defined to classify the probe signals as reflecting the presence (+1) or absence (-1) of transposons. Probes with an intermediate signal are scored as 0, and effectively omitted from the analysis.
To test our hypothesis that the low intensity signals represent the "off" distribution, we exploited the differences between the genomes of the S. aureus strains SH1000, in which the library was constructed, and MW2, the genome sequenced strain used to design the microarray probes. Probes designed to hybridise to MW2-specific genomic islands will not hybridise to any target from the SH1000 genome, so act as negative controls. Plotting the data from these probes demonstrates that the lower region of the full distribution corresponds to the signal produced in the absence of any specific hybridisation (Figure 3b). The presence of a small number of higher signals within the negative control dataset suggests some non-specific hybridisation. This issue is addressed in the microarray analysis method detailed below.
For the analysis of TMDH data we developed a method for determining cut-off values to distinguish the "on" and "off" signals. The method is analogous to one commonly used in the analysis of microarray data derived from comparative genome hybridisation [CGH, also referred to as genomotyping; [18]]. The "off" distribution is modelled using a normal distribution fit empirically to the microarray data. A low cut-off point is defined at the point where the number of probes predicted to show that particular signal by the fitted distribution drops below the observed number. A high cut-off is defined at the point where the fitted distribution explains close to 0% of the observed data at that intensity. Probes that gave a signal above the high cut-off were assigned a score of +1, since they were likely to be influenced by the presence of transposons. Probes with a signal below the lower cut-off were given a score of -1, as it is likely that their signal represents the background level without any influence from the RNA produced from the transposon promoters. Probes with an intermediate score were assigned a score of 0, since it was not possible to infer unambiguously the presence or absence of transposons. This method accounts for between-array signal variation, since the curve-fitting procedure is carried out independently for each array. It should also be noted that no account is taken of within-array variation. Standard normalisation approaches cannot be applied due to the high proportion of "on" signals within the dataset.
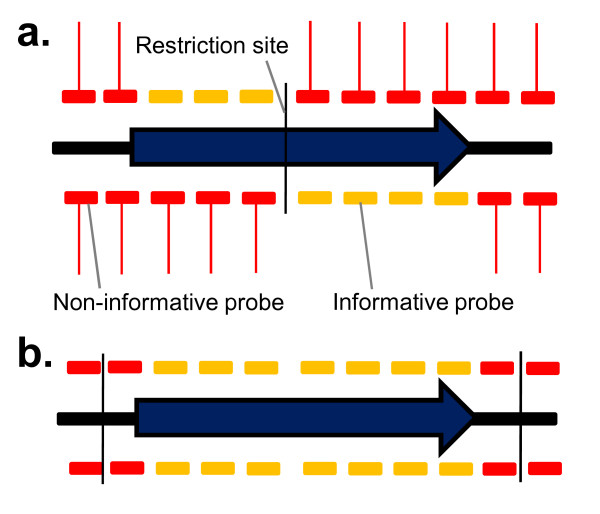
In TMDH, a probe that hybridises within an essential gene may still give an "on" signal if it is downstream of a transposon that has integrated outside the gene, since RNA run-offs are defined by restriction enzyme sites, and not gene boundaries. Therefore the position of the restriction sites is critical. A method was developed to score only "informative" probes for each restriction fragment (see Figure 4). Probes were only considered informative if they could not be influenced by a transposon outside of the gene. These included probes downstream of an intragenic restriction site (shown in Figure 4 as vertical black lines). If no transposon was evident anywhere within a particular restriction fragment, then all probes from within that fragment were considered informative, since there could be no interference from outside the gene.
Figure 4.
Procedure for identifying "informative probes" for the automated scoring system. a) Probes are only informative for a gene if they are downstream of an intragenic restriction site. Other probes may be influenced by transposons located outside the gene, and give an "on" microarray signal (red lines) even if the gene is essential. b) The exception to this rule is if there are no "on" signals anywhere within a restriction fragment. In that case all the probes within the fragment that overlap the gene are informative, since there are no interfering signals.
To determine a list of candidate essential genes in an automated manner, we examined the score of informative probes overlapping each gene, across all arrays using both restriction enzymes. A simple sum of the scores was found to be the most robust indicator of the presence/absence of transposons. This exploits the presence of multiple probes per gene to minimize the impact of any aberrant signals due to non-specific hybridisation or the lack of normalisation. Genes with a total score of -4 or lower were automatically classified as essential. This cut-off was chosen empirically to minimize the number of false positive genes that were considered unlikely to be essential based on their annotation or prior experimental evidence, whilst retaining most known essential genes. However, the cut-off also results in the omission of genes that had fewer than 4 informative probes, due to their short length or the distribution of restriction sites. The R scripts used to implement the TMDH scoring system are available upon request.
PCR-based identification of essential genes
Since the automated analysis of microarray data was not expected to be comprehensive, we devised a complementary PCR-based footprinting approach to generate a robust list of essential genes (see Figure 5). A PCR primer was designed between 50 and 300 bases upstream of the target gene. An outward-facing primer was also designed based on the mariner transposon sequence. These primers can be used to amplify a range of products, each corresponding to one of the transposon inserts in the library. The size of the products can be determined on an agarose gel, and from each of these the location of the insert can be determined. If a gene is essential, no transposons should be found within the boundaries of the gene, so no PCR products should be seen within the corresponding size range. Most of these genes were investigated further by DNA sequencing, and the gene was considered non-essential if any of the PCR products was confirmed to be derived from an intragenic transposon. An exception to this rule was made if a single PCR product was identified in the region corresponding to the C-terminal portion of the protein where a transposon insertion was considered unlikely to disrupt functionality. If a PCR product could not be obtained, the ability of the gene-specific primer to generate a PCR product was assessed by linker PCR (see Methods).
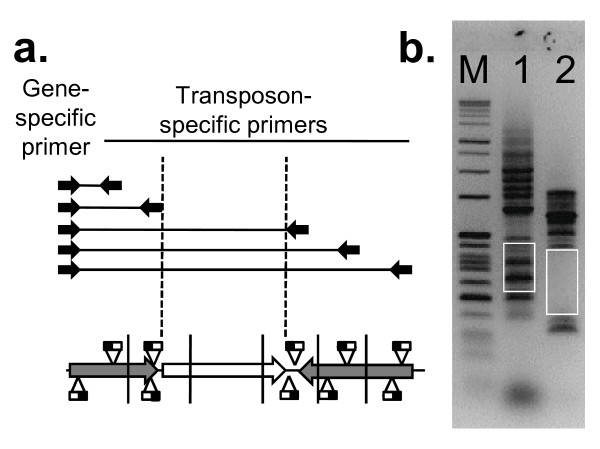
Figure 5.
PCR footprinting strategy used to confirm or reject putative essential genes. a) Diagrammatic representation of the strategy. PCR is performed using a gene specific primer 50–300 bases upstream of the start codon, and a primer corresponding to the transposon sequence. b) Agarose gel showing sample results. M – size markers, lane 1 – SAOUHSC_00862 (non-essential), lane 2 – SAOUHSC_01672 (essential). Each band represents a PCR product of a different size, corresponding to a transposon insertion in a different position. The white boxes represent the product size range expected for transposon insertions within the gene. For essential genes this region does not contain bands, since cells with insertions in this region are not viable.
Manual inspection of the raw microarray data using an online genome browser (http://www-tmdh.vet.cam.ac.uk) allowed us to identify a number of candidate essential genes (mostly small essential genes that are not detectable by the automated analysis). These genes were investigated using the PCR method. To validate the microarray analysis we also performed the PCR confirmation step on all genes identified as essential by the automated method, and also on all genes not identified as essential in S. aureus, but which had an essential orthologue in B. subtilis [4].
S. aureus Essential Gene List
Following the preliminary microarray screen and automated analysis, 274 candidate essential S. aureus genes were identified. These, together with a further 235 candidates chosen as potentially essential by manual inspection of the microarray data, or because of their presence in the B. subtilis essential gene list [4], were further investigated by PCR and sequencing. Following this process, 351 S. aureus genes were identified that were not disrupted by transposons and constitute our putative S. aureus essential gene list. The genes were classified into the same functional categories as used in the B. subtilis study [4], and a summary of the findings is presented in Table 1. Additional file 1 contains a full list of the 351 essential genes in S. aureus, together with a comparison with the essential gene list from B. subtilis [4] and a number of previously published S. aureus "essential gene" studies [13,19-21]. The results are also presented in a more compact form in Additional file 2. The full results of the PCR/sequencing analysis are available in Additional file 3. Unless otherwise stated, gene names below are those from the B. subtilis GenBank entry, with the S. aureus gene name in parentheses if different. Genes that are unnamed and do not have a B. subtilis orthologue are referred to by the S. aureus NCTC 8325 systematic nomenclature, with the prefix "SAOUHSC_".
Table 1.
Tabulation of S. aureus essential genes by category.
| Category | Number of essential genes in S. aureus | Number of S. aureus essential genes with an essential orthologue in B. subtilis | Number of B. subtilis essential genes with no essential orthologue in S. aureus |
| DNA metabolism | 27 | 22 | 3 |
| DNA replication | 16 | 16 | 0 |
| DNA packaging and segregation | 11 | 6 | 3 |
| RNA metabolism | 25 | 14 | 2 |
| Basic transcription machinery | 4 | 4 | 0 |
| RNA modification | 10 | 7 | 2 |
| RNA regulation | 11 | 3 | 0 |
| Protein synthesis | 102 | 86 | 7 |
| Ribosomal proteins | 49 | 47 | 4 |
| tRNA synthetase | 22 | 20 | 1 |
| tRNA modification | 2 | 2 | 0 |
| Translation factors | 13 | 10 | 1 |
| Protein folding | 5 | 2 | 0 |
| Protein modification | 3 | 0 | 0 |
| Protein translocation | 8 | 5 | 1 |
| Cell envelope | 66 | 43 | 22 |
| Lipids | 15 | 11 | 0 |
| Phospholipids | 4 | 4 | 1 |
| Cell wall/amino sugar | 3 | 3 | 3 |
| Diaminopimelate biosynthesis | 0 | 0 | 4 |
| Peptidoglycan biosynthesis | 17 | 10 | 2 |
| Teichoic acid biosynthesis | 12 | 8 | 0 |
| Cell division | 12 | 7 | 0 |
| Cell shape | 0 | 0 | 3 |
| Na/H transporter | 0 | 0 | 9 |
| Cell envelope – other | 3 | 0 | 0 |
| Carbon metabolism | 20 | 9 | 0 |
| Glycolysis | 11 | 9 | 0 |
| Pentose phosphate | 4 | 0 | 0 |
| Intermediary metabolism | 3 | 0 | 0 |
| Regulation | 2 | 0 | 0 |
| Respiratory pathways | 11 | 4 | 9 |
| Isoprenoid/Mevalonate biosynthesis | 8 | 1 | 2 |
| Menaquinone biosynthesis | 1 | 1 | 7 |
| Thioredoxin | 2 | 2 | 0 |
| Nucleotides | 12 | 8 | 2 |
| Purine biosynthesis | 3 | 2 | 2 |
| Purine metabolism | 1 | 0 | 0 |
| Purine/Pyrimidine biosynthesis | 3 | 3 | 0 |
| Pyrimidine biosynthesis | 5 | 3 | 0 |
| Cofactors | 24 | 13 | 2 |
| Acetyl CoA/CoA | 5 | 1 | 0 |
| Folate | 6 | 1 | 2 |
| NAD biosynthesis | 4 | 4 | 0 |
| SAM | 1 | 1 | 0 |
| Fe-sulphate cluster | 7 | 6 | 0 |
| Riboflavin biosynthesis | 1 | 0 | 0 |
| Other/unknown | 64 | 14 | 2 |
| Amino acid transporter | 1 | 0 | 0 |
| GTP binding | 6 | 6 | 0 |
| Other | 4 | 3 | 1 |
| Unknown | 53 | 5 | 1 |
Influence of experimental conditions on the essential gene list
Several studies have been carried out to determine the minimal set of genes that is essential for growth and replication of a bacterial cell [22-24]. However, any attempt to determine this experimentally will inevitably be influenced by the conditions under which the experiment is performed. A gene may be scored as essential in a particular assay because it is required for survival following exposure to a particular stress inherent in the method, or because it is involved in the uptake or metabolism of the particular nutrients provided in the growth media. An example of this in our method is the requirement for extended incubation of S. aureus at 44°C to remove temperature-sensitive replicons. Consequently, genes required for high temperature survival will be scored as putatively essential. To test this, we examined defined mutations in mrpF, dlt, tagO, fur and sarA (classified as essential from the TMDH score) and all were shown to confer heat sensitivity in S. aureus (data not shown). Furthermore, mutations in clpP and dnaK can cause a growth defect at high temperatures [25,26].
Transposon insertions may not be tolerated within a particular gene even if it is not truly "essential", since they may have a polar effect on the function of a downstream essential gene within the same operon. Examples of potential polarity issues include rimM and recU (possibly polar on trmD and pbp2, respectively). Under our experimental conditions, false positives may also be identified if transposon mutagenesis results in a reduced growth rate, since such mutants may be out-competed within the pool. Conversely, false negatives are possible since it may be possible to insert a transposon close to the 3' end of some essential genes without impairing gene function. Insertions close to the 5' end may also be possible if a suitable alternative initiation codon is available. This can also be considered an advantage of the technique, since the transposon screen effectively acts as a truncation assay that gives an indication of the minimal portion of the gene sequence required for function.
Evaluation of the microarray method as a screen
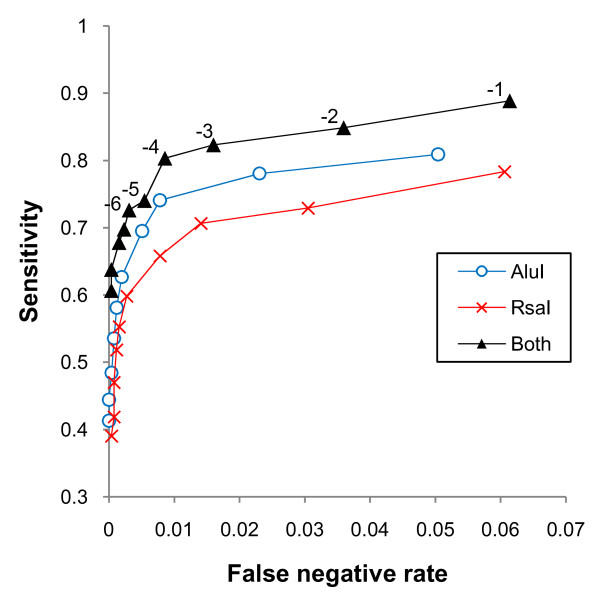
The microarray procedure alone was not expected to comprehensively identify the complete S. aureus essential gene list, but was applied as a screen to reduce the workload for the more robust but laborious PCR-based analysis. However, for some applications an exhaustive essential gene list may not be necessary, so it is useful to evaluate the efficacy of the microarray screen. The robust S. aureus essential gene list obtained using PCR footprinting allows the sensitivity (percentage of essential genes identified as such in the screen) and specificity (percentage of non-essential genes correctly identified by the screen) of the microarray procedure to be evaluated. Figure 6 shows a ROC curve of sensitivity against false positive rate (1-specificity) for cut-off values from -1 to -10. The curve suggests that a cut-off value of -4 would give an optimal balance between false positives and false negatives in future studies. This gives a specificity of 80.3%, with a sensitivity of 99.1%.
Figure 6.
ROC curves showing sensitivity against false positive rate (1-specificity) for the TMDH microarray screens using individual restriction enzymes, and the combined data from both, for cut-off values from -1 to -10.
The use of multiple restriction enzymes for TMDH has theoretical advantages, but increases the expense of the method since it involves an additional set of microarrays. Knowledge of the robust essential gene list allows us to evaluate our use of two restriction enzymes, by investigating the data from each microarray screen separately. The analysis of the individual microarray screens is also shown in Figure 6. In these analyses, a cut-off value of -3 is optimal. The AluI experiment performed better than the experiment using RsaI, with specificities of 74.1% and 70.1%, respectively, but both are inferior to the combined dataset. The choice of whether to use a single or multiple enzymes for future studies will depend on the application and available resources.
Discussion
DNA metabolism
The core set of 16 genes involved in DNA replication from the B. subtilis essential gene list are all scored as essential in S. aureus using TMDH. Of the 9 essential B. subtilis genes involved in DNA packaging and segregation, 6 were essential in S. aureus. The exceptions were smc, ypuG (scpA) and ypuH (scpB). The products of these three genes form a complex that is involved in chromosome condensation and segregation [27]. The reason for their apparent dispensability in S. aureus is unclear.
The essential B. subtilis DNA methylation genes ydiO and ydiP do not have homologues in S. aureus. Five additional genes in this category that were not essential in B. subtilis were identified as putatively essential in S. aureus: recU, ruvA, ruvB, yrrK and yvcI. These are all associated with recombination and repair, and could be essential to survive the experimental procedure, and thus may be an artefact of the particular methodology that has been used to generate the transposon library.
RNA metabolism
The four components of the basic transcription machinery (rpoA, rpoB, rpoC and sigA) are all essential in both B. subtilis and S. aureus. There are eight essential genes involved in RNA modification in B. subtilis: cca, cspR, rnpA, rnc, trmU, trmD, ykqC and yqjK. All of these are also essential in S. aureus, with the exception of rnc, which encodes a ribonuclease, and cspR, which encodes a putative RNA methylase. The trmU gene is split into two parts in the NCTC 8325 genome sequence, but this is due to a frameshift that occurs in a run of five consecutive adenine residues and most likely represents a sequencing error, or a consequence of slipped strand mis-pairing; the gene is intact in all other available S. aureus genome sequences. Three additional essential RNA modification genes were identified in S. aureus, rimM, ymfA and thdF. Insertions in rimM may not be tolerated due to polar effects on the essential trmD gene. ymfA is a paralogue of ykqC that is non-essential in B. subtilis, but is evidently more important in S. aureus. The ThdF protein (also known as TrmE) is a tRNA modification GTPase that is conserved across bacteria and eukaryotes, and is essential in some Escherichia coli genetic backgrounds [28]. The three essential B. subtilis RNA regulation genes, yycF (vicR), yycG (vicK) and nusA that are conserved in S. aureus are all essential, together with eight additional putative regulators: glnR, sarA, rnr, yvhJ (msrR), lexA, greA, yjbD and fur.
Protein synthesis
There is no direct experimental evidence that the B. subtilis complement of 52 ribosomal proteins is essential, but they are considered as such since the ribosome itself is essential [4]. Most of the ribosomal genes appear essential in S. aureus. However, the rpmG genes, of which there are two copies in both genomes, can each be mutated and are therefore not individually essential in S. aureus. The two S. aureus copies are more similar to each other than the two B. subtilis genes, both showing more similarity to B. subtilis rpmGA than to rpmGB. rpmE is non-essential in B. subtilis, due to the presence of two paralogous copies [29], but essential in S. aureus, which possesses only a single copy. The rplI and rpsT genes are essential in B. subtilis but not in S. aureus. The reason for this is unclear. rplK is not essential in B. subtilis but was identified as essential by TMDH, possibly due to its involvement in the heat shock response [30]. rpsJ is not listed as essential as it is not annotated in the NCTC 8325 genome, but an ORF is present that is predicted to encode a protein identical to RpsJ from other S. aureus strains. It is likely that this is an oversight in annotation and rpsJ is identified by TMDH as essential in S. aureus NCTC 8325.
Of 24 tRNA synthetases listed as essential in B. subtilis, 20 are similarly essential in S. aureus. Two others, glyS and glyQ, which are adjacent in the B. subtilis genome and encode the α- and β-subunits of a glycyl-tRNA synthetase, are not conserved at the sequence level between B. subtilis and S. aureus. However, at the equivalent position within a syntenic region, S. aureus has an alternative glyS gene that is essential. This encodes a class-II glycyl-tRNA synthetase, similar to that encoded by Bacillus cereus [31]. The thrS and thrZ genes of B. subtilis can compensate for the absence of each other [4], but only one copy (thrS) is present in S. aureus, and as may be predicted this is essential. The asnS gene is essential in B. subtilis but not in S. aureus. yacA, a gene originally classified as essential but of unknown function in B. subtilis [4], is involved in the modification of tRNA-Ile [32] and is also essential in S. aureus
The ten genes identified as essential translation accessory factors in B. subtilis are all essential in S. aureus. The S. aureus list includes three additional genes, homologues of efp, yrvI and smpB. The SmpB protein is associated with tmRNA-mediated rescue of stalled ribosomes, and is important for B. subtilis growth at high temperatures [33]. B. subtilis efp mutants are viable but defective in sporulation [34], however this gene is essential for protein synthesis in E. coli [35]. The yrvI (dtd) gene in B. subtilis lacks a start codon, and cell extracts have not shown any D-Tyr-tRNATyr deacylase activity [36], suggesting that the gene is non-functional in B. subtilis. The gene is intact in other Bacillus genomes, so the inactivation may have occurred during the adoption of B. subtilis as a laboratory strain. The gene is conserved across most bacteria and eukaryotes, and has been suggested to counteract the toxicity of D-tyrosine [37].
The two genes essential for protein folding in B. subtilis, groES and groEL, are also essential in S. aureus. Three other protein folding genes, dnaJ, dnaK and grpE, are scored as essential in S. aureus, however dnaK mutants are temperature sensitive [26] so may be viable but unable to survive the procedure for generating the transposon library. A homologue of the essential B. subtilis post-translational modification gene map is also essential in S. aureus, however, it shows more similarity to the non-essential B. subtilis gene yflG. Deformylation is essential in B. subtilis, but the genome contains two genes associated with this process (def and ykrB), and either can be inactivated singly [4]. In S. aureus only one deformylation gene, ykrB (pdf1) is present, and it is essential. Five of the six B. subtilis genes essential for protein translocation are also essential in S. aureus. The exception is prsA, which encodes a protein involved in extracellular folding of secreted proteins. Three additional protein translocation genes, sipT (spsB), secDF and yqjG, are essential in S. aureus. In B. subtilis, yqjG has a paralogue, spoIIIJ, that is absent from S. aureus; in B. subtilis the double knockout is lethal [38]. sipT encodes one of four closely related type I signal peptidases in B. subtilis, whereas only two are present in S. aureus (spsA and spsB). spsB, which is most closely related to B. subtilis sipT, is essential [39], and has been targeted for the development of novel antimicrobial agents [40]. SpsA, however, lacks residues essential for catalytic activity and is not essential [39].
Cell Envelope/Cell Wall and Associated Proteins
Sixteen genes involved in the production of the cell membrane are essential in B. subtilis [4]. Of these, only gpsA, which encodes an NAD-dependent glycerol-3-phosphate dehydrogenase, is not essential in S. aureus. Mutants in this gene in other bacteria are auxotrophic for glycerol [41]. Four additional cell envelope genes are essential in S. aureus: fabH, fabI, ywpB and yuxO. The first three of these are non-essential in B. subtilis due to the presence of a second gene with an overlapping function, fabHB, fabL and ycsD, respectively [42,43,4]. The reason for the essential nature of yuxO, a possible thioesterase [44], is unclear. Three additional genes that are thought to encode membrane-associated proteins, pbpX (fmtA), yfiX (fmtC) and ypbE (ebpS), are also essential in S. aureus but not B. subtilis. pbpX may be involved in resistance to high temperatures. EbpS in S. aureus is an elastin binding protein [45] and although a mutant can be isolated it shows a growth defect [46]. The multi-subunit sodium-hydrogen antiporter encoded by the mrp (mnh) operon is essential in B. subtilis but not in S. aureus, most likely due to the presence of a paralogous system in the S. aureus genome. A transposon-insertion mutant of mnhD in S. aureus retains halotolerance and loses transmembrane potential during postexponential growth [47], leading to the suggestion that the mnh operon is involved in electron transport. It is possible that the two systems have diverged following a duplication event, but retain the ability to functionally compensate for one another.
The essential B. subtilis genes involved in amino sugar biosynthesis prior to peptidoglycan polymerization are all also essential in S. aureus, with the exception of yvyH, which is present as three copies (capG, capP and mnaA) in the S. aureus genome. Diaminopimelate biosynthesis is not important in S. aureus as it uses lysine in its peptidoglycan. Of the six B. subtilis essential genes involved in the process, only four (asd, dapA, dapB and ykuQ) are retained in S. aureus. These are required for lysine biosynthesis [48] but are not essential. Of the two racemases that convert L-glutamate and L-alanine into the corresponding D-isomers, racE is essential in S. aureus, but alr is present as two copies neither of which is individually essential. The other genes essential for the synthesis of peptidoglycan precursors in B. subtilis (ddl, murAA, murB, murC, murD, murE, murF, murG and mraY) are all similarly essential in S. aureus. Three S. aureus genes absent from B. subtilis, femA, femB and fmtB (femX), are essential in the formation of the pentaglycine interpeptide bridge, which is characteristic of S. aureus peptidoglycan [49]. Also essential in S. aureus but not B. subtilis are ponA (pbp2), yrvJ (lytH) and SAOUHSC_02107. The essential genes involved in teichoic acid biosynthesis in B. subtilis are all essential in S. aureus. The genes dltA, B, C and D are also scored as essential, but this may be because they are part of the sigma X regulon which is associated with survival at high temperatures in B. subtilis [50].
Perhaps unsurprisingly, given the morphological differences between the two species, the orthologues of the essential B. subtilis cell shape determining genes, mreC and rodA, are not essential in S. aureus, and there is no orthologue of the mreB gene. The essential B. subtilis gene ylaN is involved in the determination of cell shape, with a similar phenotype to rodA [51], and is included in this category. Like rodA it is not essential in S. aureus. The seven essential cell division-related genes described for B. subtilis are all essential in S. aureus, together with five others: ylmF, ezrA, yyaA, gidA and ypsB. YlmF can complement the activity of the FtsZ-binding protein FtsA in B. subtilis, to the extent that overexpression of YlmF complemented the otherwise lethal ftsA null mutant [52]. These overlapping roles seem to have diverged in S. aureus, since both genes are essential. ezrA encodes a negative regulator of FtsZ ring formation, which can be knocked out in B. subtilis resulting in multiple FtsZ rings [53]. The nucleoid occlusion protein YyaA (also known as Noc) is essential for cell division in the absence of the Min system in B. subtilis; the genes encoding the Min system are not present in the S. aureus genome. gidA mutants affect cell division in E. coli cells grown on glucose [54], and GidA may be involved in tRNA modification [55]. It is also essential in Helicobacter [56]. YpsB localizes to the cell division site in B. subtilis, but its role is unclear [57].
Carbon Metabolism and Respiration
The core essential genes involved in glycolysis in B. subtilis are also essential in S. aureus. Three additional components of the glycolysis pathway are essential in S. aureus: pgi, gap, and pykA. The essentiality of these three genes means that all components of the pathway from glucose to pyruvate are essential in S. aureus, with the exception of the initial conversion of glucose to glucose-6-phosphate. The pentose phosphate pathway also appears to be more important in S. aureus than B. subtilis, with the component genes rpe (cfxE), zwf, yqiI (gnd) and SAOUHSC_02612 all scored as essential. A number of genes that play an intermediary or regulatory role in metabolism are also essential. These are: sucC, which encodes the beta subunit of succinyl-CoA synthetase, glnA, which encodes glutamine synthetase, yvcK, which is of unknown function but is required for growth on Krebs cycle intermediates and carbon sources metabolized by the pentose phosphate pathway [58], the HPr (Ser) kinase/phosphatase gene hprK, and the phosphotransferase system component ptsH. None of these is essential in B. subtilis, although glnA is essential in Salmonella enterica serovar Typhimurium, Mycobacterium tuberculosis and Haemophilus influenzae [59]. Interestingly, glnR, which encodes a repressor of glnA, is also scored as essential. The genes are within an operon, so the essentiality of glnR could be due to polar effects on glnA.
Perhaps the most striking differences between the essential gene complements of S. aureus and B. subtilis are in the respiratory pathways. The menaquinone biosynthesis pathway, which is essential in B. subtilis, seems to be dispensable in S. aureus, with the exception of the final step catalyzed by the menA gene product. The isoprenoid pathway is essential in B. subtilis but in S. aureus it is replaced by the mevalonate pathway, where it is itself essential, as it is in other Gram positive cocci [60].
Nucleotides and Cofactors
There are 10 essential genes involved in nucleotide biosynthesis in the B. subtilis list. Of these 8 are essential in S. aureus. guaB was described as essential in the B. subtilis study, but this was a likely consequence of a lack of guanine in the growth medium [4]: it is not required in S. aureus, but the downstream gene that encodes GMP synthase, guaA, is essential. hprT, which was a surprising inclusion in the B. subtilis list, does not play an essential role in S. aureus. Three other genes, relA, which encodes GTP pyrophosphokinase, pyrH (smbA), which encodes uridylate kinase, and thyA, which encodes thymidilate synthase, were all essential in S. aureus. relA, which mediates the stringent response, has previously been reported as being essential for in vitro growth of S. aureus [61].
Of the genes involved in cofactor biosynthesis, those required for the production of NAD and SAM are essential in both S. aureus and B. subtilis. The genes required for the iron-sulphate cluster in B. subtilis are also essential in S. aureus, with an additional requirement for ferredoxin encoded by fer, which is not essential in B. subtilis. The pathways to produce CoA and folate are more divergent. In B. subtilis, only the last gene in the CoA biosynthesis pathway was essential, suggesting that the immediate precursor (dephospho-CoA) could be scavenged from the medium [4], but that process does not seem to occur in S. aureus under our experimental conditions, since three other genes involved in CoA biosynthesis were essential, ylbI, yloI and SAOUHSC_02371. These allow the synthesis of CoA from the precursor pantothenate, which is presumably obtained from the growth medium. Similarly, folate is not obtained from the growth medium, so all the genes required for its synthesis (yciA, folB, folK, folP, folC and dfrA) are essential. There are two non-homologous genes in B. subtilis, yciA and mtrA, that encode a type I GTP cyclohydrolase (FolE) that catalyses the initial stages of the pathway from GTP to folate [62]. The S. aureus genome contains an orthologue of only one of these, yciA, which is essential. The riboflavin biosynthesis gene ribC was not identified as essential in B. subtilis in Kobayashi et al. [4], but had been in an earlier study [63], and is essential in S. aureus.
Genes of unknown function
Sixty-six putatively essential genes without a clear indication of their role have been identified in S. aureus. Of these, 14 are also essential in B. subtilis. Six members of the family of low molecular mass GTP binding proteins, proposed to have ribosome-associated functions [64], are indispensible in B. subtilis and we have found them also to be essential in S. aureus. Two of the genes, yneS and yfnI have recently been functionally characterised. YneS (subsequently renamed to PlsY) is a glycerol-3-phosphate acyl transferase required for fatty acid and phospholipid biosynthesis and is essential in B. subtilis [65]. YfnI (renamed to LtaS) is essential for lipoteichoic acid biosynthesis in S. aureus but not in B. subtilis due to the presence of numerous paralogues [66]. Elucidation of the function of further unknowns will shed light on important facets of cellular physiology and form the basis for potential new antibiotic targets.
Conclusion
To aid in the selection of targets for drug discovery, we have developed TMDH, a method for the genome-wide identification of genes essential for survival and growth in vitro. An advantage of our system over other tag-array methods is the software developed for analysis of the microarray data. Use of this automated scoring system allows unbiased prediction of essential genes from even large mutant libraries (in this case a million mutants were used). This method could be applied in isolation to identify potential targets for drug discovery or, as is the case here, used as a preliminary screen to reduce dramatically the workload required for a PCR footprinting and sequencing follow-on strategy to locate unambiguously transposon inserts, and hence infer the essential genes. Our approach incorporates the advantages of tag-array methods, genome-wide coverage and efficiency, and the high resolution associated with PCR/sequencing data.
Application of TMDH to the genome of S. aureus allowed the construction of a list of 351 genes that were not disrupted by transposon insertions. It is expected that most of these are essential, although some could be included on the list for other reasons, such as the presence of "cold spot" regions that are refractory to transposon insertion, the possibility of transposon inserts exhibiting polar effects on downstream genes or an inability of some transposon mutants to survive the experimental process. Nevertheless, the list of genes makes biological sense, corresponds well with the previously published non-comprehensive S. aureus essential gene lists [13,19-21] and the rigorously determined B. subtilis list [4], and should act as a useful starting point for the identification of potential targets for novel antimicrobial compounds. The comparison with the B. subtilis essential gene list sheds new light on the biology of both organisms. The major differences are in the respiratory pathways and genes associated with cell morphology, both of which are likely to reflect evolution in different environmental niches. In a number of cases genes that are essential in one organism are non-essential in the other due to the presence of multiple paralogous copies that can presumably compensate for each other. This comparative approach can be used to identify essential processes that are not dependent on a single gene product, but may still represent potential targets for drug development.
TMDH is a powerful technology that can be used for the determination of the essential gene repertoire of a target organism. Our analysis of the S. aureus genome is the first such comprehensive study in this organism and highlights the potential of the approach. The application of TMDH to a range of pathogens may highlight important targets for the development of novel antimicrobial therapies. TMDH also has additional potential applications, notably for the identification of conditionally essential genes. One such application has been the use of TMDH to identify Salmonella enterica serovar Typhimurium genes required for infection of BALB/c mice [67].
Methods
Construction of transposons for TMDH in Gram positive and Gram negative bacteria
A TMDH-compatible transposon was constructed for use in E. coli based on Tn5. The EZ:Tn R6k ori Kanr transposon (Epicentre) was used as a PCR template with oligonucleotides: 5'-CAGCTGTCTCTTATACACATCTCCCTATAGTGAGTCGTATTACCCATAATACCCATAATAGCTGTTTGCCAgtcgactctagagg-3' and 5'-CAGCTGTCTCTTATACACATCTCTTCTATAGTGTCACCTAAATAGGGATAACAGGGTAATGaattcgttaatacagatgt-3'. These PCR primers introduce a T7 RNA polymerase site with the homing endonuclease site PI-PspI, and an SP6 RNA polymerase site with the homing endonuclease site for I-SceI, respectively. PCR was carried out using a Roche Expand High Fidelity Kit: 96°C for 3 min, then 5 cycles of 96°C for 30 s, 25°C for 90 s and 72°C for 2 min 30 s, then a further 25 cycles at 96°C for 30 s, 50°C for 90 s and 72°C for 2 min 30 s, with a final extension step of 4 minutes followed by cooling to 4°C. The PCR product was separated by gel electrophoresis, and the band corresponding to the expected size of 2044 bp cut out and purified using a gel extraction kit (Qiagen). In order to evaluate the construct, purified transposon DNA was electroporated into 40 μl TransforMax EC100D pir+ electrocompetent E. coli (Epicentre) using 0.1 cm cuvettes at 200Ω, 25 μF and 20 kV/cm. Following electroporation, cells were grown in 1 ml SOC medium (Gibco Life Technologies) for 1 h at 37°C. Cells were then plated out on L-Agar (Sigma) containing kanamycin at 30 μg/ml (LB-kan). DNA was purified from a panel of colonies and their DNA sequence determined on a Beckman CEQ DNA sequencer (using the manufacturer's recommended conditions) with kan-2-FP-1 (Epicentre) as a DNA sequencing primer. One of the plasmids (pETBlueHGK5) was selected for further study.
Construction of a mariner transposon for TMDH analysis of S. aureus
The mariner mutagenesis system uses two temperature-sensitive plasmids to introduce transposons into a target strain [13], one bearing the mariner transposon (TS1), the other the mariner transposase gene (pFA544). The mariner plasmids were the kind gift of Dr Fredrik Aslund, Karolinska Institute, Stockholm, Sweden.
In order to construct a mariner transposon designed to function in S. aureus the following oligonucleotides:
5'-TAACAGGTTGGCTGATAAGTCCCCGGTCTCCCTATAGTGAG-3' and 5'-TAACAGGTTGGCTGATAAGTCCCCGGTCTCTTCTATAGTGTC-3' were used to introduce mariner ME ends to the Tn5 template described above. The resulting fragment was gel purified (Qiagen), ligated into the EcoRV restriction site of pETBlue-1 (Novagen), transformed into E. coli and its DNA sequence determined. Clone 5 pETBlueHGK5 was found to contain the expected DNA sequence and was used in future experiments. An erythromycin (erm) resistance determinant was amplified from pIL253 [68], using the primers: 5'-GATATCGAAGCAAACTTAAGAGTGT-3' and 5'-GATATCTACAAAAGCGACTCATAGA-3' using standard conditions and cloned into pCR2.1 (Invitrogen). The erm cassette was excised from this vector following digestion with EcoRV (NEB) purified and ligated into HincII (NEB) linearised pETBlueHGK5. Following transformation and DNA preparation (Qiagen) the DNA sequence of the resulting construct was determined on a Beckman CEQ DNA sequencer, (using the manufacturer's recommended conditions) and one of the clones (pHGK5erm3) was found to contain the correct sequence. In order to generate a construct that would function in S. aureus, plasmid TS1 was digested with the restriction endonuclease BamHI (NEB) and subsequently made blunt-ended by treatment with Klenow fragment (Gibco). The blunt-end fragment was purified and ligated with a BglII-SmaI fragment from pHGK5erm3 comprising the mariner TMDH transposon cassette that was also treated with Klenow (as above). The ligation mixture was then transformed into E. coli PIR1 cells (Epicentre). Following transformation, cells were plated on LB-Kan (30 μg/ml) and incubated overnight at 37°C. DNA-mini-preps (Qiagen) were sequenced on a Beckman CEQ DNA sequencer using the manufacturer's recommended conditions, with the sequencing primers: 5'-AAGATACTGCACTATCAACACACTC-3' and 5'-CTACCCTGTGGAACACCTACATCT-3'. One of the plasmids (pMARGK2b) was found to contain the correct DNA sequence and was used for further studies. This plasmid comprises the TMDH transposon cassette, a temperature sensitive origin of replication for growth in S. aureus (stable replication at temperatures of 30°C or below), and a chloramphenicol resistance gene for selection in S. aureus at 5 μg/ml.
Construction of a S. aureus TMDH library
The plasmid pMARGK2b was introduced into S. aureus RN4220 by electroporation (0.5 μg pMARGK2b: 2.3 kV (0.1 cm cuvettes), 25 μF, 100Ω) using a Gene Pulser (Biorad). Following electroporation, cells were plated out onto Brain Heart Infusion (BHI) agar (Oxoid) containing chloramphenicol and erythromycin at 5 μg/ml. Following successful growth in RN4220, pMARGK2b was transferred to S. aureus SH1000 with the transducing bacteriophage phi-11 using standard methods [15]. Plasmid pFA544 carrying the mariner transposase gene was subsequently introduced into S. aureus SH1000 carrying pMARGK2b by phi-11 transduction. Transductants were propagated on BHI agar containing chloramphenicol, erythromycin and tetracycline (all at 5 μg/ml) at 32°C for 48 h. The resulting colonies were harvested and inoculated into 600 ml of BHI broth containing chloramphenicol, erythromycin and tetracycline (all at 5 μg/ml) and incubated overnight at 30°C. Following growth, bacteria (100 ml) were recovered by centrifugation at 4000 g for 5 min and resuspended in 5 ml BHI broth containing 50% (v/v) glycerol and stored in 0.5 ml aliquots at -80°C.
In order to generate a S. aureus SH1000 TMDH mutant library, 0.5 ml of the glycerol stock was inoculated into 100 ml of BHI broth (at room temperature) containing chloramphenicol, erythromycin and tetracycline (all at 5 μg/ml) and incubated at 37°C until the culture reached an A600 of 0.4. Bacteria from a 30 ml sample of this culture were recovered by centrifugation (4000 g for 5 min) and the pellet was resuspended in 600 ml BHI broth containing 5 μg/ml erythromycin (pre-warmed to 44°C) and grown until the culture reached an A600 of 0.4. Bacteria from 30 ml of this culture were recovered by centrifugation at 4000 g for 5 min and the pellet resuspended in 600 ml BHI broth containing 5 μg/ml erythromycin at 44°C overnight. 10-fold dilutions were made of the resulting primary library (between 10-3 and 10-6) and 100 μl aliquots were plated onto BHI plates containing chloramphenicol, tetracycline or erythromycin (all at 5 μg/ml) and grown at 37°C overnight. The resulting growth pattern demonstrated that the bacteria were sensitive to tetracycline and chloramphenicol whilst maintaining resistance to erythromycin. This suggested that the mariner TMDH cassette had successfully transposed into the S. aureus SH1000 chromosome and the two temperature-sensitive replicons had been lost. This method generated a TMDH library comprising around 106 mutants. In order to demonstrate that transposon insertion had occurred throughout the S. aureus genome, chromosomal DNA was prepared from a number of colonies and the DNA sequence flanking the transposon insertion site determined using the primer 5'-TAGCCAGTTTCGTCGTTAAATGCCC-3' that binds 310 bases 5' from the end of the transposon. The site of transposon integration was also analyzed for a panel of mutants using the TMDH protocol (see below). DNA sequencing and a preliminary TMDH analysis demonstrated that the transposon had integrated throughout the S. aureus chromosome (see Figure 2).
In vitro transcription (IVT) reactions
In order to evaluate the suitability of the novel mariner transposon for the TMDH protocol, preliminary in vitro transcription (IVT) reactions using both SP6 (SP6 MEGAscript kit; Ambion) and T7 (T7 MEGAshortscript kit, Ambion) were carried out following the manufacturer's recommended conditions. 10 μg of genomic DNA from a TMDH library was digested overnight with a target enzyme and purified on a QIAquick column (Qiagen) using the gel extraction protocol, eluted in 200 μl of water, and analyzed by agarose gel electrophoresis to assess the completeness of digestion. For each reaction, 20 μl digested DNA (~1 μg), was reduced in volume by vacuum drying and incorporated into either SP6 or T7 IVT reactions (Ambion) overnight. RNA from IVT reactions was treated with DNaseI, purified on a MinElute column (Qiagen), eluted with 14 μl of water and quantified by measurement at OD260. Transcription was observed from both the SP6 and T7 IVT reactions only when the specific RNA polymerase was included in reactions bearing the cognate RNA polymerase binding site.
IVT reactions for mariner SH1000 TMDH experiments were produced from DNA fragments flanking the sites of transposon insertion by a Linker PCR (LPCR) strategy. Template for the LPCR reaction was generated by ligating annealed oligonucleotide linkers to restriction digested genomic DNA. LPCR, using a primer pair corresponding to the transposon and to the linker, permits the specific amplification of DNA flanking the site of transposon insertion including the SP6 or T7 promoter sites. Purified genomic DNA (10 μg) from the TMDH library was digested separately with either RsaI or AluI (NEB), in 400 μl, overnight at 37°C. Digested DNA was then purified using a Gel Extraction Kit (Qiagen) and eluted in 50 μl of water. Restriction digests were analyzed by agarose gel (0.8%, w/v, in TBE) electrophoresis to check for completeness. 100 μM each of complementary oligonucleotide pairs (5'-cgactggacctgga-3' and 5'-gataagcagggatcggaacctccaggtccagtcg-3') were annealed by heating to 95°C for 3 min followed by cooling to room temperature over a 1 hr period. The RsaI or AluI linkers were ligated with restriction digested DNA using standard conditions overnight and were purified using a Gel Extraction Kit (Qiagen). We found that during the linker ligation step linkers may self-ligate, giving rise to background signal in subsequent PCR steps. To reduce this possibility, the DNA samples were re-digested with the appropriate restriction enzyme (i.e. linker-ligated RsaI was re-digested with RsaI while linker-ligated AluI was re-digested with AluI). The DNA was then re-purified using a Gel Extraction Kit column (Qiagen). LPCR amplification of the DNA flanking the transposon insertion site was carried out with HotStarTaq, (Qiagen) using the primer pairs: 5'-ctaccctgtggaacacctacatct-3' and 5'-gataagcagggatcggaacc-3' (for the SP6 end) and 5'-cttcgatgactggcaaacagc-3' and 5'-gataagcagggatcggaacc-3' (for the T7 end). Control PCR reactions were performed using PCR primers alone, as well as reactions omitting template. LPCR conditions were as follows; 95°C for 15 min; 30 cycles of 94°C for 30 s, 55°C for 1 min and 72°C for 2 min; 72°C for 5 min, pause at 4°C. Products from each template (AluI, RsaI, T7 and SP6 ends) were purified using the Qiaquick Gel Extraction Kit (Qiagen). Purified DNA was analyzed on a 0.8% (w/v) agarose gel run in TBE.
Labelling IVT reactions
IVT reactions were carried out using MEGAscript or MEGAshortscript (Ambion) reagents with direct incorporation of either Cy5-UTP or Cy3-UTP (Perkin Elmer). For the T7 promoter, 3.75 μl 10 mM Cy3-UTP was used with 2 μl buffer (10× Ambion stock); 2 μl each of 75 mM ATP, CTP and GTP, and 1.5 μl of UTP; 1 μl (up to 4.75 μl) of template (10 ug of digested gDNA) and 2 μl of enzyme. For the SP6 promoter, 2.5 μl 10 mM Cy5-UTP was used with 2 μl buffer (10× Ambion stock); 2 μl each of 50 mM ATP, CTP and GTP; 1.5 μl 50 mM UTP; 1 μl (up to 6 μl) template and 2 μl enzyme. The reactions were performed overnight at 37°C in the dark in 20 μl reaction volumes.
Labelled RNA was subsequently treated with DNaseI (according to the Ambion protocol) and purified using RNeasy reagents (Qiagen). The concentration of LPCR-derived labelled RNA was determined using a spectrophotometer (1 AU A260 = 40 μg/ml RNA).
Array hybridisations, washing and scanning
Preliminary whole-genome TMDH experiments showed there was no increase in signal resolution by using both T7 and SP6 IVT reactions (labelled with Cy3 and Cy5 respectively), consequently only T7 (Cy3) labelling was used. However, in preliminary experiments to map the position of a small number of transposons, arrays were hybridised with both Cy3 (T7) and Cy5 (SP6) labelled RNA. 100 μl of labelled RNA was added to the hybridisation solution. This consisted of 46.6 μl 12 × MES, 112.0 μl 5 M NaCl, 62.0 μl formamide, 22.4 μl 0.5 M EDTA (pH 8.0), 56.0 μl 10% Triton-X-100 and 161.0 μl water, giving a total volume of 560 μl.
A stock solution of 1 l of MES buffer (2-(N-morpholino) ethanesulphonic acid) was made from 70.4 g 2N-morpholenoethene sulphonic acid (MES free acid) and 193.3 g of MES sodium salt (pH 6.5 – 6.7) and filter sterilized through a 0.2 μM) filter. RNA for each array-hybridisation was made up to 50 μl with water, 50 μl of formamide added and the mix denatured by incubation at 55°C for 5 min. The resulting 100 μl volume was then added to the remaining components (460 μl) of hybridisation solution. Hybridisation solutions were added to each microarray in a Hybridisation Chamber (Agilent) and incubated at 50°C for 18 hr. Post-hybridisation washes of the arrays were carried out at room temperature using two 40 ml SSPE (Sigma)-based solutions of 6 × SSPE, 0.005% N-lauryl sarcosine for 5 min, followed by 0.06 × SSPE, 0.18% PEG-200 for 5 min. Arrays were then blown-dry using dry-nitrogen and scanned on an Agilent G2500A Scanner. The raw microarray data have been deposited in ArrayExpress (accession number E-MEXP-1588).
PCR on genomic DNA from the transposon library
Individual PCR experiments were carried out with a gene-specific primer corresponding to a region of genomic DNA 50–300 bp 5' to the ATG start site (see Additional file 3) in conjunction with a transposon-specific primer. PCRs were carried out using HotStarTaq polymerase (Qiagen) using the manufacturer's recommended conditions for the initial set-up (95°C for 15 min) followed by 30 cycles of 95°C for 45 sec, 50°C for 45 sec and 72°C for 2 min, with a final extension step of 72°C for 10 min. PCR was carried out in a 50 μl reaction mix with a final primer concentration of 0.2 μM. Following PCR, fragments were cloned using TOPO cloning reagents (Invitrogen), and the resulting colonies picked. Colony PCRs were performed on the colonies and PCR products of the appropriate sizes were sequenced directly. DNA sequencing was carried out on an ABI 3100 Automated Capillary DNA Sequencer using the manufacturer's recommended conditions either using plasmid DNA or on colony-PCR products using BigDyeTerminator Cycle Sequencing reagents (Applied Biosystems). Full results are shown in Additional file 3.
Mapping microarray probes to S. aureus NCTC 8325 genome
To characterize the differences from the MW2 genome used to design the microarray, the probes were used as query sequences in a blastn search of the NCTC 8325 genome sequence (accession CP000253). A probe was considered to match uniquely the NCTC 8325 genome if the top hit showed >= 80% identity over >= 50% of the length of the probe, and the second hit did not fulfil both of those criteria. Uniquely matching probes were used in the TMDH analysis, with their hybridisation position on the NCTC 8325 genome determined from the BLAST result. Differences between the experimental strain SH1000 and the genome sequenced strain NCTC 8325 were determined by hybridising genomic DNA from SH1000 against the MW2 TMDH microarray. The probes were scored using the trinary method from GACK [18], and only probes that were scored as present were included in the TMDH analysis.
Linker PCR
For 87 genes the standard protocol failed to generate PCR product. Typically, this may be due to either problems with the gene-specific primer (e.g. mis-synthesis or sequence error), or the lack of transposons within a specific genomic region suitable for generating a PCR product (e.g. the transposon-specific primer is too far away from the gene-specific primer to generate PCR product under the conditions used). In order to determine whether the gene-specific primer was able to generate PCR product, linker-PCR experiments were carried out using a linker that effectively introduced a PCR primer binding site proximal to the gene-specific primer. Linkers (5'-gataagcagggatcggaacctccaggtccagtcg-3' and 5'-cgactggacctgga-3') were annealed as described above. Genomic DNA from the TMDH library was digested with a 4-base pair blunt-end restriction endonuclease (either AluI (AG↓CT) or RsaI (GT↓AC)). The restriction enzyme selection was based on a bioinformatic analysis of the DNA sequence of the target gene and chosen to be around 500–1000 bp away from the gene-specific primer-binding site. Blunt-end ligation of the linker onto either AluI or RsaI fragments resulted in the production of short fragments of DNA that could only be amplified optimally with PCR using a gene-specific primer in combination with the linker-specific primer (5'gataagcagggatcggaacc-3') using HotStarTaq polymerase (Qiagen) conditions described above. As the distance of the gene-specific primer from the selected restriction site was known, the size of the resulting PCR product could be used to determine whether or not the gene-specific primer was functional. Linker-PCR results are included in Additional file 3.
Comparison with other published essential gene lists
Previous studies have identified many S. aureus genes as putatively essential [13,19-21]. As these were identified in a variety of strains, the equivalent proteins encoded by strain NCTC 8325 were identified by a blastp search, with an E-value cut-off of 0.001. BLAST results and xBASE genome alignments [69] were inspected manually to determine the most likely orthologue if multiple hits were obtained. The NCTC 8325 homologues of genes from the B. subtilis essential gene list [4] were determined using the same method. In the case of the S. aureus list published by Ji et al. [20], the published format used an obsolete nomenclature, and only 138 of their 168 putative essential genes could be identified. Forsyth et al. [19] did not publish their essential list in full, only those genes with homologues in Mycoplasma genitalium were named and it is this subset of their data that is included here.
Authors' contributions
IGC & DJM invented the TMDH approach [11]. IGC, DJM, AGA & PJO developed the transposons [12]. IGC, DJM, SJP, SEP, ML, AGA & PJO designed the experiments. AGA, PJO, GS, ML, SJP, JG-L, SJF & SEP performed the experiments. MH and TAB designed the arrays, MH, TAB and RRC designed the PCR primers and KS synthesized arrays. RRC designed the software for the TMDH analysis and RRC, AGA, PJO, GS, MH, TAB, JG-L, SJF, DJM & IGC analyzed the data. RRC, AGA, JG-L, SJF, DJM & IGC wrote the paper. All authors read and approved the final manuscript. In the opinion of the authors RRC and AGA made an equivalent contribution and should be regarded as joint first authors. Similarly, DJM and IGC should be regarded as joint senior authors.
Supplementary Material
Essential S. aureus genes, and comparison with the essential gene complement of B. subtilis and previous S. aureus essential gene studies. A comprehensive list of S. aureus essential genes identified in this study. The names Bae, Ko, Forsyth and Ji refer to the primary authors of previously published S. aureus essential gene studies[13,19-21]. The B. subtilis essential gene data is derived from the study of Kobayashi et al. [4].
Essential S. aureus genes, and comparison with the essential gene complement of B. subtilis. A compact version of the data presented in Additional file 1, suitable for printing.
Results of PCR/sequencing analysis and linker PCR. Full results from the PCR/sequencing analysis used to validate the microarray screen. Column C indicates the position of the PCR primer relative to the start codon of the target gene. Columns F and G indicate the size range of the PCR products that correspond to an intragenic transposon. Column K indicates the location of the insertion as determined from the sequence data (where available). For intragenic inserts the first number is the position of the insert relative to the start of the gene. LPCR indicates that no PCR products could be obtained, but the gene-specific primer was verified by linker PCR.
Acknowledgments
Acknowledgements
We acknowledge Ann Chubb, Kerstin Williams, Simon Orange, David Jackson, Peter Corish and Tim Fell (Arrow Therapeutics Ltd.) for help with the initial development of TMDH and Mark Pallen (University of Birmingham) for help with S. aureus MW2 genome annotation. This work was supported by the BBSRC (grant references APG19115 and BBD0179471).
Contributor Information
Roy R Chaudhuri, Email: roy.chaudhuri@gmail.com.
Andrew G Allen, Email: andy.allen@pfizer.com.
Paul J Owen, Email: powen@cancertechnology.com.
Gil Shalom, Email: gilshalom@hotmail.com.
Karl Stone, Email: Karl.Stone@ogt.co.uk.
Marcus Harrison, Email: marcus.harrison@ogt.co.uk.
Timothy A Burgis, Email: t.burgis@imperial.ac.uk.
Michael Lockyer, Email: MLockyer@arrowt.co.uk.
Jorge Garcia-Lara, Email: J.Garcia-Lara@sheffield.ac.uk.
Simon J Foster, Email: s.foster@sheffield.ac.uk.
Stephen J Pleasance, Email: sp476@cam.ac.uk.
Sarah E Peters, Email: sep34@cam.ac.uk.
Duncan J Maskell, Email: djm47@cam.ac.uk.
Ian G Charles, Email: I.Charles@sheffield.ac.uk.
References
- Livermore DM. The need for new antibiotics. Clin Microbiol Infect. 2004;10:1–9. doi: 10.1111/j.1465-0691.2004.1004.x. [DOI] [PubMed] [Google Scholar]
- Haselbeck R, Wall D, Jiang B, Ketela T, Zyskind J, Bussey H, Foulkes JG, Roemer T. Comprehensive essential gene identification as a platform for novel anti-infective drug discovery. Current pharmaceutical design. 2002;8:1155–1172. doi: 10.2174/1381612023394818. [DOI] [PubMed] [Google Scholar]
- Osterman AL, Gerdes SY, eds . Microbial Gene Essentiality: Protocols and Bioinformatics. Totowa, N.J.: Humana Press; 2007. [Google Scholar]
- Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, et al. Essential Bacillus subtilis genes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerley BJ, Rubin EJ, Camilli A, Lampe DJ, Robertson HM, Mekalanos JJ. Systematic identification of essential genes by in vitro mariner mutagenesis. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:8927–8932. doi: 10.1073/pnas.95.15.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune W, Menard C, Hobom U, Odenbreit S, Messerle M, Koszinowski UH. Rapid identification of essential and nonessential herpesvirus genes by direct transposon mutagenesis. Nature Biotechnology. 1999;17:360–364. doi: 10.1038/7914. [DOI] [PubMed] [Google Scholar]
- Baldwin DN, Salama NR. Using genomic microarrays to study insertional/transposon mutant libraries. Methods in enzymology. 2007;421:90–110. doi: 10.1016/S0076-6879(06)21010-3. [DOI] [PubMed] [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Molecular microbiology. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- Bijlsma JJ, Burghout P, Kloosterman TG, Bootsma HJ, de Jong A, Hermans PW, Kuipers OP. Development of genomic array footprinting for identification of conditionally essential genes in Streptococcus pneumoniae. Applied and environmental microbiology. 2007;73:1514–1524. doi: 10.1128/AEM.01900-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterberg KM, Luecke J, Bruegl AS, Reznikoff WS. Phenotypic screening of Escherichia coli K-12 Tn5 insertion libraries, using whole-genome oligonucleotide microarrays. Applied and environmental microbiology. 2005;71:451–459. doi: 10.1128/AEM.71.1.451-459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles IG, Maskell DJ. Transposon Mediated Differential Hybridisation. International Patent Number WO2001/007651. 2001.
- Maskell DJ, Charles IG, Allen A, Owen P. Transposon. International Patent Number WO2003/074700. 2003.
- Bae T, Banger AK, Wallace A, Glass EM, Aslund F, Schneewind O, Missiakas DM. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12312–12317. doi: 10.1073/pnas.0404728101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iandolo JJ, Worrell V, Groicher KH, Qian Y, Tian R, Kenton S, Dorman A, Ji H, Lin S, Loh P, et al. Comparative analysis of the genomes of the temperate bacteriophages phi 11, phi 12 and phi 13 of Staphylococcus aureus 8325. Gene. 2002;289:109–118. doi: 10.1016/s0378-1119(02)00481-x. [DOI] [PubMed] [Google Scholar]
- Novick RP. Genetic systems in staphylococci. Methods in enzymology. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, Foster SJ. sigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. Journal of bacteriology. 2002;184:5457–5467. doi: 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, Lefkowitz SM, Ziman M, Schelter JM, Meyer MR, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nature Biotechnology. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- Kim CC, Joyce EA, Chan K, Falkow S. Improved analytical methods for microarray-based genome-composition analysis. Genome biology. 2002;3:RESEARCH0065. doi: 10.1186/gb-2002-3-11-research0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsyth RA, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, Wall D, Wang L, Brown-Driver V, Froelich JM. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Molecular microbiology. 2002;43:1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
- Ji Y, Zhang B, Van SF, Horn , Warren P, Woodnutt G, Burnham MK, Rosenberg M. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science (New York, NY) 2001;293:2266–2269. doi: 10.1126/science.1063566. [DOI] [PubMed] [Google Scholar]
- Ko KS, Lee JY, Song J-H, Baek JY, Oh WS, Chun J, Yoon HS. Screening of Essential Genes in Staphylococcus aureus N315 Using Comparative Genomics and Allelic Replacement Mutagenesis. Journal of Microbiology and Biotechnology. 2006;16:623–632. [Google Scholar]
- Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, Bult CJ, Kerlavage AR, Sutton G, Kelley JM, et al. The minimal gene complement of Mycoplasma genitalium. Science (New York, NY) 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, Hutchison CA, 3rd, Smith HO, Venter JC. Essential genes of a minimal bacterium. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:425–430. doi: 10.1073/pnas.0510013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CA, Peterson SN, Gill SR, Cline RT, White O, Fraser CM, Smith HO, Venter JC. Global transposon mutagenesis and a minimal Mycoplasma genome. Science (New York, NY) 1999;286:2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- Frees D, Qazi SN, Hill PJ, Ingmer H. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Molecular microbiology. 2003;48:1565–1578. doi: 10.1046/j.1365-2958.2003.03524.x. [DOI] [PubMed] [Google Scholar]
- Singh VK, Utaida S, Jackson LS, Jayaswal RK, Wilkinson BJ, Chamberlain NR. Role for dnaK locus in tolerance of multiple stresses in Staphylococcus aureus. Microbiology (Reading, England) 2007;153:3162–3173. doi: 10.1099/mic.0.2007/009506-0. [DOI] [PubMed] [Google Scholar]
- Soppa J, Kobayashi K, Noirot-Gros MF, Oesterhelt D, Ehrlich SD, Dervyn E, Ogasawara N, Moriya S. Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members ScpA and ScpB. Molecular microbiology. 2002;45:59–71. doi: 10.1046/j.1365-2958.2002.03012.x. [DOI] [PubMed] [Google Scholar]
- Cabedo H, Macian F, Villarroya M, Escudero JC, Martinez-Vicente M, Knecht E, Armengod ME. The Escherichia coli trmE (mnmE) gene, involved in tRNA modification, codes for an evolutionarily conserved GTPase with unusual biochemical properties. The EMBO journal. 1999;18:7063–7076. doi: 10.1093/emboj/18.24.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanamiya H, Akanuma G, Natori Y, Murayama R, Kosono S, Kudo T, Kobayashi K, Ogasawara N, Park SM, Ochi K, et al. Zinc is a key factor in controlling alternation of two types of L31 protein in the Bacillus subtilis ribosome. Molecular microbiology. 2004;52:273–283. doi: 10.1111/j.1365-2958.2003.03972.x. [DOI] [PubMed] [Google Scholar]
- Zhang S, Scott JM, Haldenwang WG. Loss of ribosomal protein L11 blocks stress activation of the Bacillus subtilis transcription factor sigma(B) Journal of bacteriology. 2001;183:2316–2321. doi: 10.1128/JB.183.7.2316-2321.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okstad OA, Hegna I, Lindback T, Rishovd AL, Kolsto AB. Genome organization is not conserved between Bacillus cereus and Bacillus subtilis. Microbiology (Reading, England) 1999;145:621–631. doi: 10.1099/13500872-145-3-621. [DOI] [PubMed] [Google Scholar]
- Soma A, Ikeuchi Y, Kanemasa S, Kobayashi K, Ogasawara N, Ote T, Kato J, Watanabe K, Sekine Y, Suzuki T. An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Molecular cell. 2003;12:689–698. doi: 10.1016/s1097-2765(03)00346-0. [DOI] [PubMed] [Google Scholar]
- Shin JH, Price CW. The SsrA-SmpB ribosome rescue system is important for growth of Bacillus subtilis at low and high temperatures. Journal of bacteriology. 2007;189:3729–3737. doi: 10.1128/JB.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y, Inaoka T, Kasai K, Ito Y, Okamoto S, Satsu H, Tozawa Y, Kawamura F, Ochi K. Expression profiling of translation-associated genes in sporulating Bacillus subtilis and consequence of sporulation by gene inactivation. Biosci Biotechnol Biochem. 2003;67:2245–2253. doi: 10.1271/bbb.67.2245. [DOI] [PubMed] [Google Scholar]
- Aoki H, Dekany K, Adams SL, Ganoza MC. The gene encoding the elongation factor P protein is essential for viability and is required for protein synthesis. The Journal of biological chemistry. 1997;272:32254–32259. doi: 10.1074/jbc.272.51.32254. [DOI] [PubMed] [Google Scholar]
- Calendar R, Berg P. D-Tyrosyl RNA: formation, hydrolysis and utilization for protein synthesis. Journal of molecular biology. 1967;26:39–54. doi: 10.1016/0022-2836(67)90259-8. [DOI] [PubMed] [Google Scholar]
- Soutourina J, Plateau P, Delort F, Peirotes A, Blanquet S. Functional characterization of the D-Tyr-tRNATyr deacylase from Escherichia coli. The Journal of biological chemistry. 1999;274:19109–19114. doi: 10.1074/jbc.274.27.19109. [DOI] [PubMed] [Google Scholar]
- Murakami T, Haga K, Takeuchi M, Sato T. Analysis of the Bacillus subtilis spoIIIJ gene and its Paralogue gene, yqjG. Journal of bacteriology. 2002;184:1998–2004. doi: 10.1128/JB.184.7.1998-2004.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregg KM, Wilding I, Black MT. Molecular cloning and expression of the spsB gene encoding an essential type I signal peptidase from Staphylococcus aureus. Journal of bacteriology. 1996;178:5712–5718. doi: 10.1128/jb.178.19.5712-5718.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruton G, Huxley A, O'Hanlon P, Orlek B, Eggleston D, Humphries J, Readshaw S, West A, Ashman S, Brown M, et al. Lipopeptide substrates for SpsB, the Staphylococcus aureus type I signal peptidase: design, conformation and conversion to alpha-ketoamide inhibitors. European journal of medicinal chemistry. 2003;38:351–356. doi: 10.1016/s0223-5234(03)00040-0. [DOI] [PubMed] [Google Scholar]
- Mindich L. Membrane biogenesis: mitochondria, chloroplasts and bacteria. Plenum Publishing Corp, New York; 1975. Studies on bacterial membrane biogenesis using glycerol auxotrophs; pp. 429–452. [Google Scholar]
- He X, Reynolds KA. Purification, characterization, and identification of novel inhibitors of the beta-ketoacyl-acyl carrier protein synthase III (FabH) from Staphylococcus aureus. Antimicrobial agents and chemotherapy. 2002;46:1310–1318. doi: 10.1128/AAC.46.5.1310-1318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath RJ, Su N, Murphy CK, Rock CO. The enoyl-[acyl-carrier-protein] reductases FabI and FabL from Bacillus subtilis. The Journal of biological chemistry. 2000;275:40128–40133. doi: 10.1074/jbc.M005611200. [DOI] [PubMed] [Google Scholar]
- Dillon SC, Bateman A. The Hotdog fold: wrapping up a superfamily of thioesterases and dehydratases. BMC bioinformatics. 2004;5:109. doi: 10.1186/1471-2105-5-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PW, Rosenbloom J, Abrams WR, Rosenbloom J, Mecham RP. Molecular cloning and expression of the gene for elastin-binding protein (ebpS) in Staphylococcus aureus. The Journal of biological chemistry. 1996;271:15803–15809. doi: 10.1074/jbc.271.26.15803. [DOI] [PubMed] [Google Scholar]
- Downer R, Roche F, Park PW, Mecham RP, Foster TJ. The elastin-binding protein of Staphylococcus aureus (EbpS) is expressed at the cell surface as an integral membrane protein and not as a cell wall-associated protein. The Journal of biological chemistry. 2002;277:243–250. doi: 10.1074/jbc.M107621200. [DOI] [PubMed] [Google Scholar]
- Bayer AS, McNamara P, Yeaman MR, Lucindo N, Jones T, Cheung AL, Sahl HG, Proctor RA. Transposon disruption of the complex I NADH oxidoreductase gene (snoD) in Staphylococcus aureus is associated with reduced susceptibility to the microbicidal activity of thrombin-induced platelet microbicidal protein 1. Journal of bacteriology. 2006;188:211–222. doi: 10.1128/JB.188.1.211-222.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltshire MD, Foster SJ. Identification and analysis of Staphylococcus aureus components expressed by a model system of growth in serum. Infection and immunity. 2001;69:5198–5202. doi: 10.1128/IAI.69.8.5198-5202.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer S, Ehlert K, Tschierske M, Labischinski H, Berger-Bachi B. The essential Staphylococcus aureus gene fmhB is involved in the first step of peptidoglycan pentaglycine interpeptide formation. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9351–9356. doi: 10.1073/pnas.96.16.9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Decatur A, Sorokin A, Helmann JD. The Bacillus subtilis sigma(X) protein is an extracytoplasmic function sigma factor contributing to survival at high temperature. Journal of bacteriology. 1997;179:2915–2921. doi: 10.1128/jb.179.9.2915-2921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt A, Rawlins JP, Thomaides HB, Errington J. Functional analysis of 11 putative essential genes in Bacillus subtilis. Microbiology (Reading, England) 2006;152:2895–2907. doi: 10.1099/mic.0.29152-0. [DOI] [PubMed] [Google Scholar]
- Ishikawa S, Kawai Y, Hiramatsu K, Kuwano M, Ogasawara N. A new FtsZ-interacting protein, YlmF, complements the activity of FtsA during progression of cell division in Bacillus subtilis. Molecular microbiology. 2006;60:1364–1380. doi: 10.1111/j.1365-2958.2006.05184.x. [DOI] [PubMed] [Google Scholar]
- Levin PA, Kurtser IG, Grossman AD. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9642–9647. doi: 10.1073/pnas.96.17.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Meyenburg K, Jorgensen BB, Nielsen J, Hansen FG. Promoters of the atp operon coding for the membrane-bound ATP synthase of Escherichia coli mapped by Tn10 insertion mutations. Molecular and General Genetics. 1982;188:240–248. doi: 10.1007/BF00332682. [DOI] [PubMed] [Google Scholar]
- Yim L, Moukadiri I, Bjork GR, Armengod ME. Further insights into the tRNA modification process controlled by proteins MnmE and GidA of Escherichia coli. Nucleic acids research. 2006;34:5892–5905. doi: 10.1093/nar/gkl752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karita M, Etterbeek ML, Forsyth MH, Tummuru MK, Blaser MJ. Characterization of Helicobacter pylori dapE and construction of a conditionally lethal dapE mutant. Infection and immunity. 1997;65:4158–4164. doi: 10.1128/iai.65.10.4158-4164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry SE, Edwards DH. The Bacillus subtilis DivIVA protein has a sporulation-specific proximity to Spo0J. Journal of bacteriology. 2006;188:6039–6043. doi: 10.1128/JB.01750-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorke B, Foulquier E, Galinier A. YvcK of Bacillus subtilis is required for a normal cell shape and for growth on Krebs cycle intermediates and substrates of the pentose phosphate pathway. Microbiology (Reading, England) 2005;151:3777–3791. doi: 10.1099/mic.0.28172-0. [DOI] [PubMed] [Google Scholar]
- Zhang R, Ou HY, Zhang CT. DEG: a database of essential genes. Nucleic acids research. 2004:D271–272. doi: 10.1093/nar/gkh024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding EI, Brown JR, Bryant AP, Chalker AF, Holmes DJ, Ingraham KA, Iordanescu S, So CY, Rosenberg M, Gwynn MN. Identification, evolution, and essentiality of the mevalonate pathway for isopentenyl diphosphate biosynthesis in gram-positive cocci. Journal of bacteriology. 2000;182:4319–4327. doi: 10.1128/jb.182.15.4319-4327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry D, Li T, Rosenberg M, McDevitt D. The rel gene is essential for in vitro growth of Staphylococcus aureus. Journal of bacteriology. 2000;182:4995–4997. doi: 10.1128/jb.182.17.4995-4997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yacoubi B, Bonnett S, Anderson JN, Swairjo MA, Iwata-Reuyl D, de Crecy-Lagard V. Discovery of a new prokaryotic type I GTP cyclohydrolase family. The Journal of biological chemistry. 2006;281:37586–37593. doi: 10.1074/jbc.M607114200. [DOI] [PubMed] [Google Scholar]
- Mack M, van Loon AP, Hohmann HP. Regulation of riboflavin biosynthesis in Bacillus subtilis is affected by the activity of the flavokinase/flavin adenine dinucleotide synthetase encoded by ribC. Journal of bacteriology. 1998;180:950–955. doi: 10.1128/jb.180.4.950-955.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh PC, Morimoto T, Matsuo Y, Oshima T, Ogasawara N. The GTP-binding protein YqeH participates in biogenesis of the 30S ribosome subunit in Bacillus subtilis. Genes & genetic systems. 2007;82:281–289. doi: 10.1266/ggs.82.281. [DOI] [PubMed] [Google Scholar]
- Paoletti L, Lu YJ, Schujman GE, de Mendoza D, Rock CO. Coupling of fatty acid and phospholipid synthesis in Bacillus subtilis. Journal of bacteriology. 2007;189:5816–5824. doi: 10.1128/JB.00602-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundling A, Schneewind O. Synthesis of glycerol phosphate lipoteichoic acid in Staphylococcus aureus. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8478–8483. doi: 10.1073/pnas.0701821104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri RR, Peters SE, Pleasance SJ, Northern H, Willers C, Paterson GK, Cone DB, Allen AG, Owen PJ, Shalom G, et al. Comprehensive Identification of Salmonella enterica serovar Typhimurium Genes Required for Infection of BALB/c Mice. PLoS Pathogens. [DOI] [PMC free article] [PubMed]
- Simon D, Chopin A. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie. 1988;70:559–566. doi: 10.1016/0300-9084(88)90093-4. [DOI] [PubMed] [Google Scholar]
- Chaudhuri RR, Loman NJ, Snyder LA, Bailey CM, Stekel DJ, Pallen MJ. xBASE2: a comprehensive resource for comparative bacterial genomics. Nucleic acids research. 2008:D543–546. doi: 10.1093/nar/gkm928. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Essential S. aureus genes, and comparison with the essential gene complement of B. subtilis and previous S. aureus essential gene studies. A comprehensive list of S. aureus essential genes identified in this study. The names Bae, Ko, Forsyth and Ji refer to the primary authors of previously published S. aureus essential gene studies[13,19-21]. The B. subtilis essential gene data is derived from the study of Kobayashi et al. [4].
Essential S. aureus genes, and comparison with the essential gene complement of B. subtilis. A compact version of the data presented in Additional file 1, suitable for printing.
Results of PCR/sequencing analysis and linker PCR. Full results from the PCR/sequencing analysis used to validate the microarray screen. Column C indicates the position of the PCR primer relative to the start codon of the target gene. Columns F and G indicate the size range of the PCR products that correspond to an intragenic transposon. Column K indicates the location of the insertion as determined from the sequence data (where available). For intragenic inserts the first number is the position of the insert relative to the start of the gene. LPCR indicates that no PCR products could be obtained, but the gene-specific primer was verified by linker PCR.