Abstract
Restless legs syndrome (RLS) is a common neurological disorder characterized by an irresistible urge to move the legs at night, which is often accompanied by unpleasant sensations. A recent genomewide association study identified an association between RLS and intronic markers from the MEIS1 gene. Comparative genomic analysis indicates that MEIS1 is the only gene encompassed in this evolutionarily conserved chromosomal segment, i.e. a conservation synteny block, from mammals to fish. We carried out a series of experiments to delineate the role of MEIS1 in RLS pathogenesis and the underlying genetic mechanism. We sequenced all 13 MEIS1 exons and their splice junctions in 285 RLS probands with confirmed clinical diagnosis and did not identify any causative coding or exon–intron junction mutations. We found no evidence of structural variation or disease-associated haplotype differential splicing. However, sequencing of conserved regions of MEIS1 introns 8 and 9 identified a novel single nucleotide polymorphism (C13B_2) significantly associated with RLS (allelic association, P = 1.81E−07). We detected a significant decrease in MEIS1 mRNA expression by quantitative real-time polymerase chain reaction in lymphoblastoid cell lines (LCLs) and brain tissues from RLS patients homozygous for the intronic RLS risk haplotype, compared with those homozygous for the non-risk haplotype. Finally, we found significantly decreased MEIS1 protein levels in the same batch of LCLs and brain tissues from the homozygous carriers of the risk haplotype, compared with the homozygous non-carriers. Therefore, these data suggest that reduced expression of the MEIS1 gene, possibly through intronic cis-regulatory element(s), predisposes to RLS.
INTRODUCTION
Restless legs syndrome (RLS) (MIM 102300) is a common sensorimotor disorder, characterized by an imperative urge to move the legs during night, which is often associated with paresthesias (tingling, pricking or ‘pins and needles’ sensations) or dysesthesias (unpleasant abnormal sensations) of the lower limbs (1). The symptom is induced by rest or inactivity, can be temporarily relieved by leg movements and follows a circadian pattern. Approximately 80% of RLS patients also experience periodic limb movements in sleep (PLMS) (2), which are rhythmic extensions of the big toe and dorsiflexion of the ankle, with occasional flexion at the knee and hip and can be objectively measured by polysomonography in a sleep laboratory (3) or by an ambulatory actigraphy device (4). The pathogenic pathways of RLS are largely unknown. The effective relief of RLS symptoms by dopaminergic treatments indicates dopaminergic dysregulation in the central nervous system of RLS patients (5). Although most cases are of unknown cause; anemia, end-stage renal disease and multiple pregnancies have been shown to increase the risk for RLS, all three involving a possible iron deficiency status, strongly suggesting an innate iron dysregulation as a major cause for RLS (5,6). Though RLS is a chronic disorder that often starts in childhood or adolescence with mild symptoms and slowly progresses with age (1), it is mostly seen in older patients with other age-related comorbidities. RLS can cause chronic sleep deprivation (1), lead to increased risk for cardiovascular disease (7,8) and significantly compromise quality of life for those afflicted (9).
Family and twin studies have indicated a substantial genetic contribution to the pathogenesis of RLS (1,2,10,11). The initial attempts to identify genes responsible for RLS had focused on linkage analyses of large multiplex families. However, although several loci have been mapped, no causative mutations have been identified in the linked families, possibly due to high prevalence, clinical uncertainty and genetic heterogeneity (12). Recently, Winkelmann et al. (13) identified three gene regions, i.e. MEIS1, BTBD9 and MAP2K5/LBXCOR1, associated with clinically diagnosed RLS through a genomewide association study (GWAS) using Affymetrix 500K Arrays and genotyping 401 subjects with familial RLS and 1644 population controls from Germany. The association signals have been confirmed in two independent replication samples, including 255 French-Canadian (FC) cases and 287 controls (13), and in another independent study of 244 patients with RLS and 497 controls (14). Among these three regions, rs2300478 within the MEIS1 intronic region had the most significant P-value in each replication stage and for all three stages combined [P = 8.08E−23, allelic odds ratio (OR): 1.74, 95% CI: 1.57–1.92] (13). It is considered as the leading common genetic risk factor so far identified for RLS (15). The validated replications have positioned MEIS1 as one of the most robust association signals among all first-wave GWASs (16,17), especially in the context of common neuropsychiatric disorders, where some of them probably do not fit with the common disease/common variant hypothesis. It is worth noting, however, the deCODE group, using a different ascertainment strategy, phenotypic definition and different panel of markers, found significant association of RLS and PLMS with single nucleotide polymorphisms (SNPs) within the BTBD9 gene (P = 3.0E−14, allelic OR: 1.7, 95% CI: 1.5–2.0) and reported stronger association with the PLMS measurement than with the RLS phenotype. However, the Icelandic study did not report a genomewide-corrected significant association in the MEIS1 region using the Illumina Hap300 Arrays (18). None of these associated candidate genes lies within a previously reported RLS linkage region; nor are any of these genes directly implicated in iron and/or dopamine pathways (5), indicating upstream or novel pathogenic pathways of RLS. To further define the role of the MEIS1 locus in RLS, we carried out a series of experiments to delineate the sequence variant(s) potentially contributing to RLS phenotype.
RESULTS
We first genotyped the two MEIS1 SNPs previously reported to be significantly associated with RLS (rs12469063 and rs2300478, Fig. 1A) (13) in an autopsy brain sample of 28 cases and 140 controls by TaqMan Assay. We found both SNPs to be significantly associated with the RLS phenotype (Table 1) and to be in strong linkage disequilibrium (LD) (r2 = 0.933 in patients and r2 = 0.876 in controls). The haplotype defined by both risk alleles (GG) was also found to be significantly associated with RLS. The analysis of this small number of brain samples therefore added further evidence to the association of MEIS1 with the RLS phenotype.
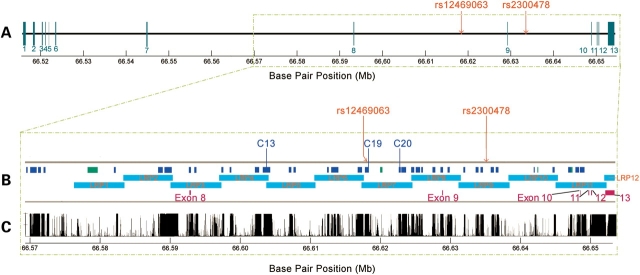
Figure 1.
Physical map, PCR amplification and conservation of MEIS1 gene region. (A) Structure of the MEIS1 gene: blue vertical bars indicate the locations of the 13 exons; red vertical lines indicate the positions of the two associated SNPs. (B) Enlarged region from boxed section (A): blue bars C01–C37 indicate the amplified and sequenced fragments of conserved regions; green bars indicate the amplified and sequenced fragments of predicted gene regions; the long aqua band indicates the region with overlapping long-range PCR fragments as LRP01–LRP12; exons 8–13 are depicted by purple vertical bars. (C) MEIS1 conservation tracks from the UCSC Genome Browser website [Vertebrate Multiz Alignment & PhastCons Conservation (28 Species)] (http://genome.ucsc.edu/).
Table 1.
Genetic association of MEIS1 SNPs rs12469063 and rs2300478 in 28 RLS and 140 control brain samples
| SNP | Risk allele | Non-risk allele | Cases (%) | Controls (%) | OR (95% CI) | P-value |
| rs12469063 | G | A | 43 | 24 | 2.3 (1.3–4.1) | 0.006 |
| rs2300478 | G | T | 44 | 27 | 2.1 (1.2–3.7) | 0.014 |
| SNP | Risk genotype | Non-risk genotype | Cases (%) | Controls (%) | OR | P-value |
| rs12469063 | G/G | A/A | 25 | 2 | 15.9 (3.6–70.8) | 0.00046 |
| rs2300478 | G/G | T/T | 30 | 3 | 12.2 (3.1–47.4) | 0.00017 |
| SNP | Risk haplotype | Non-risk haplotype | Cases (%) | Controls (%) | OR | P-value |
| rs12469063–rs2300478 | GG/GG | AA/TT | 43 | 25 | 2.3 (1.3–4.0) | 0.0095 |
To identify sequence variant(s) within the MEIS1 genomic region that could potentially contribute to the RLS phenotype, we first searched for common structural variation(s) that might disrupt normal MEIS1 function. To date, the public databases (http://projects.tcag.ca/variation; http://genome.ucsc.edu) (19–27) do not report the existence of any copy number or other structural variations larger than 1 kb, or in/del of ≥100 bp, within or encompassing the MEIS1 region. In order to further exclude smaller in/del variants (<1 kb), we designed 12 pairs of long-range polymerase chain reaction (PCR) primers (Supplementary Material, Table S1) to amplify overlapped fragments of 3.0–7.0 kb from the candidate MEIS1 genomic region (Fig. 1B). We did not detect any size variants, estimated to be ≥250 bp based on the resolution of a 0.9% agarose gel electrophoresis, in representative DNA samples from three RLS patients homozygous for the disease-associated haplotype (GG/GG) and two homozygous wild-type controls (AA/TT) (Supplementary Material, Fig. S1). In addition, using sequencing, SNP (see below) and microsatellite (data not shown) genotyping methods, we were able to amplify all exons and selected conserved regions within introns 8 and 9 of MEIS1 in 570 RLS and control samples, without detecting any PCR products of unexpected size, unexplained PCR failures or non-Mendelian inheritance patterns in the family samples genotyped (data not shown). No loss of heterozygosity was observed in RLS patients carrying the risk haplotype, suggesting no large deletion of the MEIS1 gene. Therefore, it is unlikely that a common structural variant within or encompassing the MEIS1 RLS candidate region would be the underlying causative genetic variant.
To search for potential coding variant(s) within the MEIS1 gene, we sequenced all 13 exons (Primers see Supplementary Material, Table S1), including UTRs and exon–intron junctions (NM_002398.2) (Fig. 1A), in 285 patients with a clinically confirmed RLS diagnosis. All variants detected were also sequenced in 285 controls. In all these individuals, we only detected one synonymous coding variant in MEIS1; and this variant along with all the UTR and exon–intron junction variants detected (n = 19) did not show any association with RLS phenotype (Supplementary Material, Table S2). This is in accordance with the high conservation of the coding sequences of MEIS1 across different species (Fig. 1C) and the absence of MEIS1 common missense variants in the public SNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/).
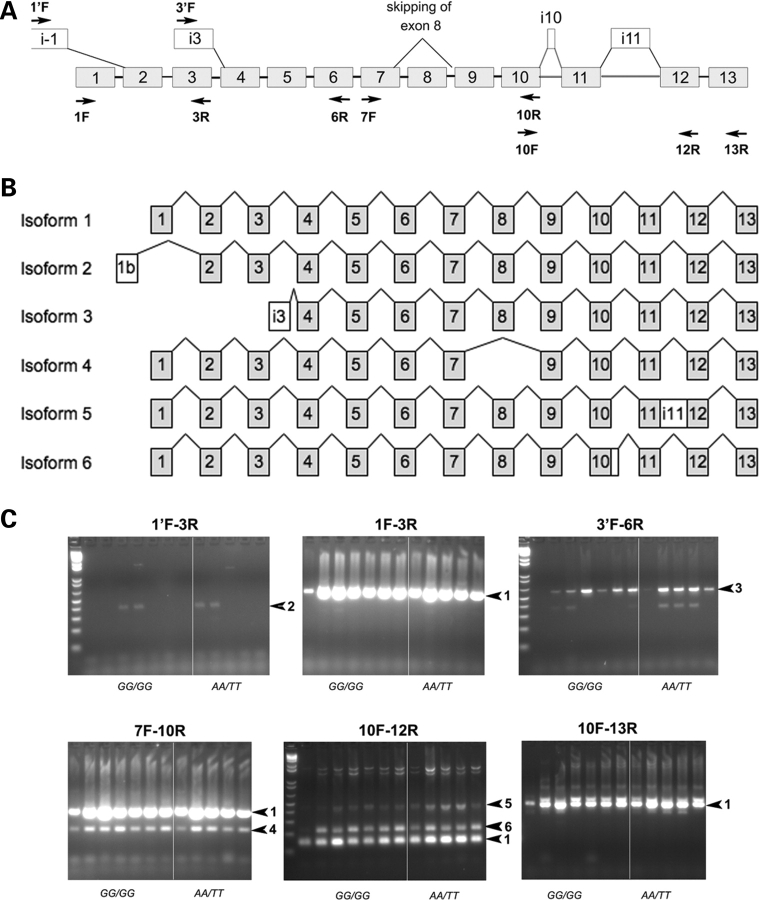
To determine if the risk haplotype is associated with alternative splicing of the MEIS1 gene, different ESTs (UCSC Genome Browser site: http://genome.ucsc.edu/) were used to examine all possible expressed isoforms of MEIS1. Primers were subsequently designed across the entire MEIS1 transcribed sequence in order to identify different isoforms (Fig. 2A) (Supplementary Material, Table S1). Reverse transcriptase PCRs were used to amplify several fragments covering the full span of the MEIS1 cDNA from brain tissue and lymphoblastoid cell lines (LCLs) of RLS patients and controls homozygous for the risk (GG/GG) or non-risk (AA/TT) haplotypes. In addition to the previously described full-length MEIS1 mRNA isoform (NM_002398.2) encoding a 390 amino acid protein, five different transcriptional isoforms were identified (Fig. 2B). However, each of these alternative isoforms was identical in pons (Fig. 2C), thalamus (data not shown) and LCLs (data not shown) of samples with the risk and non-risk haplotypes, suggesting that differential splicing of MEIS1 isoforms is not responsible for RLS in these patients.
Figure 2.
Alternative splicing isoforms of MEIS1. (A) Schematic representation of exons (grey boxes, numbered as in NM_002398.2) and alternate exons (white boxes) predicted on the basis of available EST sequences from GenBank or identified in our experiments. Primers used for PCR amplification of cDNA fragments are shown as arrows. (B) Schematic representation of the different isoforms of MEIS1 identified in our experiments. Isoform 1 is the principal form of MEIS1 (NM_002398). Isoforms 2 (DC424361) and 3 use alternate promoters (AK098174), and produce N-terminal truncated proteins of 377 and 246 amino acids, respectively. Isoform 4 skips exon 8 and prematurely truncates the protein to 252 amino acid by using a STOP codon located in exon 9 (DA057712), and isoform 5 includes the entire unspliced sequence from intron 11, and prematurely truncates the protein to 378 amino acid (AL832770 and BC036511). Isoform 6 includes an additional 81 bp from the 5′ end of intron 10, and is predicted to add a 27 amino acid in-frame segment near the C-terminus of the protein, with a putative PKG phosphorylation site. (C) Detection of alternatively spliced isoforms of MEIS1 in pons brain tissues isolated from RLS patients carrying the homozygous GG/GG genotype (lanes 1–7 in each panel) or AA/TT genotype (lanes 8–12 in each panel). RT-PCR products shown in each panel were obtained using the primer pairs indicated above each panel. The corresponding expected MEIS1-specific bands are indicated by arrows labeled with the number of the isoform.
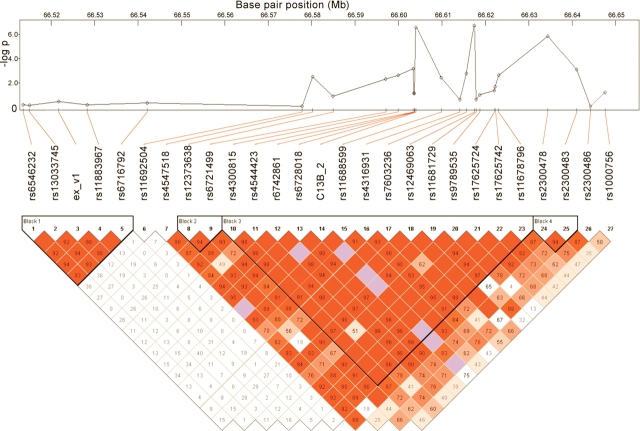
Having ruled out the presence of MEIS1 structural, coding or splicing changes in our RLS samples, we then focused on the non-coding regions of introns 8 and 9, within a single associated LD block previously defined in the German control population (13), as well as in the HapMap Caucasian samples (http://www.hapmap.org). We selected 37 highly conserved fragments (lod > 100) (Fig. 1B) (Primers see Supplementary Material, Table S1) from the UCSC conservation track (Fig. 1C), which completely covered the predicted enhancer elements in this region (http://enhancer.lbl.gov/). We sequenced these fragments in 24 individuals (12 patients and 12 non-RLS controls) carrying the disease-associated haplotype. Forty-three intronic variants were identified; most of these variants were either rare or showed a similar frequency in patients and controls (data not shown). We thus selected four variants with distinctive allele frequencies in cases and controls and genotyped in 285 RLS patients and 285 controls by sequencing. To further define the candidate interval using association in FC samples, we also genotyped 20 additional tagging-SNPs within the MEIS1 region, in addition to two more common SNPs detected in the above-mentioned deep-sequencing performed in 285 RLS patient and 285 control samples (Fig. 3). Genetic association tests indicated that three reported SNPs (rs4544423, rs12469063 and rs2300478) and a novel SNP (C13B_2) showed significant association with the RLS phenotype, both allele-wise and genotype-wise, after correction for multiple testing (Supplementary Material, Table S3). SNP rs4544423 and C13B_2 are located within the same LD block as rs12469063 and rs2300478 (Fig. 3). Therefore, the sequence variant(s) responsible for the association lie in a large genomic region (∼55 kb) encompassing introns 8 and 9, and flanking exon 9. In addition, rs4544423, C13B_2 and rs12469063 are in segments that show significant evolutionary conservation between human and zebrafish (Supplementary Material, Fig. S2), which is highly suggestive of a gene regulatory function (28,29). Given the distances between the associated SNPs and the neighboring genes (1.1 Mb to the upstream gene FLJ16124 and 843 kb to the downstream ETAA1 gene), we considered that the detected SNPs are more likely to act as cis-element(s) for the MEIS1 gene itself, rather than other genes in the vicinity. Furthermore, analysis of the previously characterized syntenic blocks (30), i.e. evolutionarily conserved chromosomal segments that allow the identification of cis-regulatory boundaries, indicated that MEIS1 is the only gene encompassed in this block conserved from mammals to fish; thus suggesting that gene regulatory sequences within the RLS associated region indeed have a high likelihood to regulate in cis the MEIS1 gene.
Figure 3.
Genetic association between the MEIS1 gene and RLS. P-values from association tests of 26 SNPs from the MEIS1 gene region in 285 RLS cases and 285 controls in French-Canadian population and linkage disequilibrium of structure of the MEIS1 gene.
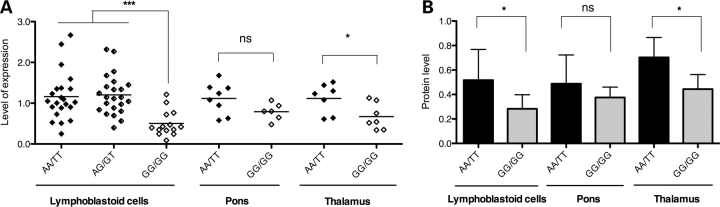
We therefore went on to examine the possibility of a MEIS1 RLS risk-haplotype effect on mRNA and protein expression levels. The expression data from the San Antonio Family Heart Study indicates that MEIS1 expression has a heritability of 0.28 ± 0.056 (P = 1.00E−10) (31), which could be consistent with the existence of cis-regulatory sequence variations. To assess the potential effect of the RLS-associated SNPs on MEIS1 gene expression, we first explored the data sets from the Sanger Centre containing the MEIS1 gene expression data in LCLs and genotyping results of 461 SNPs from the MEIS1 gene and flanking regions in the HapMap Caucasian samples (60 parents) (ftp://ftp.sanger.ac.uk/pub/genevar) (32). Analysis of the two RLS-associated SNPs, rs12469063 and rs2300478, indicated reduced gene expression in LCLs of individuals carrying the GG/GG genotype comparing to heterozygous and homozygous individuals of the AA/TT genotype (data not shown). A similar correlation of genotypes with gene expression levels was observed in microarray data from 40 brain tissues by Turecki's group at McGill University (33) (data not shown). Based on these preliminary findings, we designed additional experiments to validate these results in our RLS patient and control samples. We selected LCLs and brain samples (pons and thalamus) from RLS patients and controls carrying different MEIS1 haplotypes, i.e. GG/GG and AA/TT, and measured the expression of MEIS1 mRNA by quantitative real-time PCR (qRT-PCR) using the TaqMan method (Applied Biosystems). Using one of the MEIS1 TaqMan gene expression assays (Hs 00180020_m1, Applied Biosystems), which covers the exon 6/7 boundary and detects all potential isoforms, we found a significant decrease in MEIS1 mRNA expression in LCLs carrying the GG/GG genotype, compared with the AA/TT genotype (P < 0.001) (Fig. 4A). Similar significant results were obtained by qRT-PCR from thalamus (P = 0.0325) of RLS patients. In RLS pons samples, we detected a similar but not significant trend (P = 0.086).
Figure 4.
MEIS1 mRNA and protein expression levels of different haplotypes. (A) Comparison of MEIS1 mRNA expression between AA/TT and GG/GG genotypes. LCLs from RLS patients and controls (22 AA/TT patients and controls versus 14 GG/GG patients; mean ± SD: 1.16 ± 0.59 versus 0.49 ± 0.28; P < 0.0001). RLS patient and control samples carrying the AA/TT haplotype showed no expression difference in MEIS1 expression. We could not identify control cell lines carrying the GG/GG genotype because of its low frequency in the general population and its strong association with the RLS phenotype. Brain tissues from RLS patients: pons [eight AA/TT (seven females/one male) versus six GG/GG (four females/two males), mean ± SD: 1.12 ± 0.38 versus 0.79 ± 0.21; P = 0.086] and thalamus [seven AA/TT (six females/one male,) versus seven GG/GG (four females/three males), mean ± SD: 1.12 ± 0.36 versus 0.67 ± 0.32; P = 0.0325]. There were no age and post-mortem interval differences between the AA/TT and GG/GG genotype groups (data not shown). (B) Graphical summary of the western blotting experiments illustrating a significant reduction in expression levels of MEIS1 in lymphoblastoid and thalamus cell samples carrying the GG/GG genotype (for lymphocytes: mean ± SD, 0.52 ± 0.25 versus 0.28 ± 0.12, Student's t-test: P < 0.05, for pons: mean ± SD, 0.49 ± 0.24 versus 0.38 ± 0.09, P = 0.39, for thalamus: mean ± SD, 0.70 ± 0.16 versus 0.44 ± 0.12, P < 0.05).
To assess whether the down-regulation of MEIS1 mRNA translated into a decrease in protein levels, we performed western blot analysis in extracts from the same batch of LCLs and RLS brain samples (pons and thalamus) using an antibody against the C-terminus of MEIS1 (1:500; H00004211-A01, Abcam) and an antibody against α-tubulin (1:120 000; ab15246, Abnova) as controls. We detected that MEIS1 protein levels were significantly decreased in LCLs of GG/GG carriers, compared with AA/TT carriers (P = 0.03) (Fig. 4B and Supplementary Material, Fig. S3). This was confirmed in thalamus samples of RLS patients (P = 0.01) and a similar but not significant trend was observed in pons (P = 0.39). Hence, the GG/GG haplotype associated with an increased risk for RLS significantly decreased MEIS1 expression at both the mRNA and protein levels.
DISCUSSION
MEIS1 encodes a homeobox protein belonging to the three amino acid loop extension family of homeodomain-containing proteins. MEIS1 forms a heterodimeric or heterotrimeric complex with PBX and HOX proteins to augment the affinity and specificity of DNA binding by HOX proteins (34), and is part of a HOX transcriptional regulatory network that specifies spinal motor neuron pool identity and connectivity (35). Previous studies in model organisms show that MEIS1 has an important role in limb development (36,37), establishment of the intrasegmental spinal motor neuron identity and connectivity (35), and patterning of sensory organs in the embryonic peripheral nervous system (38). With regard to Meis1 function in post-embryonic neuronal tissues, not much is known other than that it is expressed in the adult mouse brain in cerebellar granule cells, the forebrain and, notably, in dopaminergic neurons of the substantia nigra shown in the Allen Brain Atlas of the mouse brain (http://mouse.brain-map.org) (39). Paradoxically, overexpression of MEIS1 plays a role in leukemia (40,41) and neuroblastoma (42). Recent data in Caenorhabditis elegans showed that unc-62, the MEIS1 ortholog, was among the 64 genes that were found to extend the worm's lifespan significantly when inactivated post-developmentally by RNA interference (RNAi), a similar situation as the reduced expression detected in adult human brain tissues and cell lines from RLS patients in this report, indicating its possible role in aging as well (43). Combined, these data point to a potential functional link between MEIS1 and pathogenic pathways of RLS.
MEIS1, like many other homeobox genes, contains extensive highly conserved segments in its non-coding 5′ and 3′ intergenic and intragenic regions (34,35,44), one of which (intergenic) was already shown to function as an enhancer (http://enhancer.lbl.gov/) and others were predicted to control temporal and spatial gene expression through complex but well-orchestrated interactions with other cis- and/or trans-factors, during development.
Non-coding sequences that make up 98% of our genome contain regions that have been conserved throughout evolution. One vital function that is clearly embedded in these regions is gene regulation, i.e. instructing genes when and where to turn on or off. There are examples in the literature supporting non-coding sequences that function as gene regulatory elements to cause human disease (45–48). In addition, several recent systematic genetic studies in humans have demonstrated the potential causal impact of differential gene expression on complex disease risk (31,49–52). Currently, the majority of GWASs are finding significant disease risk associations with non-coding variants (16,17). However, the pathogenic mechanism and pathways leading to their contribution or causality to disease phenotypes are largely unknown. The MEIS1 RLS-associated non-coding region will be one of the examples to uncover the functional effect of these non-coding variants can have on common human diseases.
LCLs may not reflect in vivo biology of the individuals from whom they were derived; and their gene expression levels are known to be variable with genetic and non-genetic factors. This could either reduce the power to detect the real effect or creates spurious association between different lines and genotypes. Nevertheless, the decreased expression levels of MEIS1 in LCLs in our study were compared with the well-controlled samples; and have been replicated in independent experiments. The results have been further supported by data from RLS patient brain tissues.
The GG susceptibility haplotype appears to have a recessive effect on gene expression at the cellular level, which is different from a dosage effect on the clinical phenotype as defined in the GWAS by Winkelmann et al. (13). The different effect of the same risk haplotype on different phenotypic levels could be due to time-, cell- or tissue-specific gene expression regulation. Furthermore, the decrease in MEIS1 expression detected in adult human samples carrying the risk genotype/haplotype is probably due to yet defined cis-variant(s) interacting with other trans-elements in a time- or tissue-specific fashion. Further refining the minimum haplotype carrying all the RLS-associated sequence variants by deep-sequencing and genotyping in further enlarged human samples from different populations, as well as characterization of the enhancer sequences and activity in vivo systematically are needed to delineate the complex control of the MEIS1 gene expression and its potential direct implication in the RLS phenotype or subphenotype. Iron, as an essential cofactor for many proteins involved in the normal function of neuronal tissue, an important regulator of gene expression and an indispensable nutrition element from diet could act as a transfactor; and dopaminergic dysregulation might manifest as an endophenotype of RLS in the MEIS1 pathways leading to RLS.
MATERIALS AND METHODS
DNA samples
Brain tissue specimens (thalamus and upper pons) were obtained from autopsy brain tissues of 28 individuals with an RLS diagnosis from the Harvard Brain Tissue Resource Center. Final diagnosis was made by an RLS expert (R.A.) based on available questionnaires and medical records, but blinded to the genotype information (detailed clinical description of the samples: Supplementary Material, Table S4). Control brain specimens were also obtained from autopsy brain tissues of 140 individuals from the Douglas Hospital Research Centre Brain Bank. These individuals died either from suicide, accident or other non-neurological diseases. All brain samples were from Caucasians of European ancestry.
The FC samples used in this study included 285 unrelated RLS cases with clinically validated diagnosis, and 285 unspecified population controls, part of which was previously used in the stage 2 of the German GWAS Study (13) (211 cases and 241 controls) (Supplementary Material, Table S5). The FC cases (probands) were ascertained and sampled through the Centre d'étude du sommeil et des rythmes biologiques, a sleep clinic at the University of Montreal, Canada. Ascertainment strategy, clinical assessment procedures and detailed diagnostic criteria of the RLS/PLMS cases were previously described (2,53). The PLMS measurements were performed by one-night polysomnography prior to treatment or with reduced dopaminergic medications. Secondary RLS cases due to known uremia, dialysis, neuropathy and iron deficiency were excluded. All Montreal case and control subjects are Caucasian, and 90% are of confirmed FC ancestry as defined by having four grandparents of FC origin.
Genomic DNA was isolated from autopsy brain tissues and peripheral lymphocytes using standard kits according to manufacturer's protocols (Gentra), and LCLs were established in selected RLS patients and controls from Montreal according to standard procedures. The project has been approved by all involved Institutional Ethical Committees. All participants provided signed informed consent forms.
LCL culture
Selected LCLs from RLS patients and controls previously established by transformation with EB virus using standard protocols were grown at 37°C and 5% CO2 in RPMI 1640 medium (Invitrogen) supplemented with 15% (v/v) heat-inactivated fetal calf serum (Sigma-Aldrich), 0.29 mg/ml of l-glutamine, 100 U/ml of penicillin and 100 µg/ml of streptomycin (Invitrogen). Healthy cells were harvested at an approximate density of 1 × 106 cells/ml.
SNP genotyping
SNP genotyping was carried out by TaqMan Assay using the Applied Biosystems 7900 Fast Real-Time PCR System and the SDS software (versus 2.2.2) for allele calling, in accordance with the manufacturers’ protocols. Five percentage of total genotypes were replicated for selected markers across different DNA plates (384-well format) yielding a concordance rate of 99.98%. Samples (11 RLS patients and 8 controls) with <95% call rate for total SNPs genotyped were eliminated from the analyses. Missed call rates were not significantly different between cases and controls. All markers were in Hardy–Weinberg disequilibrium for the whole sample and each subgroup of cases and controls using χ2 tests (P > 0.05).
Long-range PCR
We selected and quantified three patient DNA samples carrying the GG haplotype and two control DNA samples carrying the AT haplotype (DNA concentration adjusted to 50 ng/µl by picogreen DNA quantification method). PCR amplifications were performed using Long PCR Enzyme Mix according to manufacturer's protocols (Fermentas). The PCR products were electrophoresed on a 0.9% agarose gel (UltraPure, Invitrogen) for 4 h. Lambda DNA/HindIII fragment (Invitrogen) was used as the molecular weight standard.
Bioinformatic annotation of sequence conservation
All sequence positions are based on NCBI Build 36.1. A comparative genomic method was used to identify potential regulatory regions by using the PhastCons program (54) (http://genome.ucsc.edu), which scores conserved elements by pairwise alignment of genomic sequences of several species. The identified conserved regions were further correlated with the predicted enhancer elements (http://enhancer.lbl.gov/). We selected all regions that presented a lod > 100. Using this threshold, we selected 37 different regions for sequencing (Fig. 1. fragments C01–C37 in numerical order, primer information Supplementary Material, Table S1).
Sequencing
Primers (Supplementary Material, Table S1) were designed using the Exon Primer program from the UCSC genome browser (http://genome.ucsc.edu/) and the primer3 program (http://frodo.wi.mit.edu/) so that they would have a uniform Tm of 60°C. PCR products were sequenced by the BigDye terminator chemistry 3.1 (ABI) on an ABI3730 sequencer at the McGill University and Genome Quebec Innovation Centre. Sequence results were first aligned and analyzed using Polyphred (version 3.5) and further validated manually using Mutation Surveyor (version 3.10).
RNA isolation and reverse transcriptase PCR
Total RNA was extracted from 0.2 g of frozen brain tissue using the RNAeasy® Lipid Tissue kit (Qiagen) or from 5 million lymphoblastoid cells using the RNAeasy® kit (Qiagen). Single-stranded cDNA synthesis was performed from 1 µg of total RNA using a mix of oligo-dT and random primers and the Quantitect® Reverse Transcription kit (Qiagen). RNA concentration and purity was determined by UV spectroscopy; and RNA integrity was assessed by visualization of 28S and 18S rRNA on standard denaturing agarose gels (an aliquot of 200 ng RNA/sample loaded on the gel). Only RNAs with clear 28S and 18S rRNA bands and with an approximate 2:1 ratio of 28S and 18S rRNA, as well as the A260/A280 absorbance ratio of 1.8–1.9 were considered intact and pure, and used for further analyses.
Detection of different MEIS1 splice variants
Using the UCSC Genome Browser site (http://genome.ucsc.edu/), different ESTs were used to examine all possible expressed isoforms of MEIS1. Primers were subsequently designed across the entire MEIS1 transcribed sequence in order to identify different isoforms (Fig. 2A and Supplementary Material, Table S1). 0.5 µl of the cDNAs obtained by reverse transcriptase PCR of RNA extracted from RLS and control LCLs and brain tissues was used to amplify MEIS1-specific fragments using the BD Advantage® 2 Taq polymerase (Clontech), according to the manufacturer's protocol. All amplified PCR products were gel-purified and sequenced to validate the sequence content.
Quantitative real-time PCR (qRT-PCR)
qRT-PCR was performed in LCLs and brain tissues from RLS patients, using the TaqMan method (Applied Biosystems) with probes and primers designed to recognize all potential isoforms of MEIS1 (Hs00180020_m1). PCR conditions were as follows: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles at 95°C for 15 s (denaturation) and 60°C for 1 min (annealing and extension). Fluorescent signals were captured using the ABI PRISM® 7900HT Sequence Detection System (Applied Biosystems). The level of expression was determined by converting the threshold cycle (Ct) values using the 2−ΔΔCt method. Expression levels were normalized using the human 18S ribosomal RNA (rRNA) gene (Hs99999901_m1) with commercial primer–probe mix (Applied Biosystems). All experiments were run in triplicate. Independent cDNA synthesis was carried out twice.
Western blot
Protein samples from the same batch of human thalamus, pons and lymphoblastoid cells, used in qRT-PCR, were prepared (Supplementary Material, Method section). Samples were incubated overnight with an antibody against the C-terminus of Meis1 (1:500; H00004211-A01, Abcam). After Meis1 detection, membranes were incubated with an antibody against α-tubulin (1:120 000; ab15246, Abnova). Species-specific HRP-conjugated secondary antibodies (1:10 000) were used after each primary antibody detection to reveal the protein signals (ECL, Perkin Elmer). After Meis1 and α-tubulin detections, the intensities of signals were analyzed by quantitative densitometry (Image J Software; http://rsbweb.nih.gov/ij/).
Statistical analyses
Case–control association analyses were conducted with UNPAHSED (version 3.0.9), based on a likelihood ratio test (55). Allelic, genotypic and haplotypic ORs were estimated by the same program. Statistical analyses of gene expression data on mRNA were performed using Prism 5 (GraphPad Software). Comparisons of different groups were carried out by Krustal–Wallis tests. The density ratios of Meis1 protein over α-tubulin were calculated and an unpaired Student's t-test (two-tailed) was used to compare the samples carrying the AA/TT and the GG/GG haplotypes.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This project was supported by a Canadian Institutes of Health Research grant to G.A.R. and J.M. (MOP 82900) and a grant from the Restless Legs Syndrome Foundation to G.A.R., J.M. and L.X. C.E. and R.A.'s clinical work on RLS was supported by a NIH grant (PO1-AG2119). The RLS brain tissues were provided by the Harvard Brain Tissue Resource Center, which is supported in part by a Public Health Service Grant (R24 MH068855), with permission from the RLS Brain Bank Tissue Review Committee through the RLS Foundation.
ACKNOWLEDGEMENTS
We would like to thank all the RLS patients and families who participated in this study. We thank Annie Levert, Carol Cheung and Francois Gougeon for technical assistance; and Yan Yang, Dan Spiegelman, Sandra Laurent and Laurie Destroismaisons for bioinformatics and high-throughput genotyping and sequencing technical support. We thank Dr Nadav Ahituv's (from UCSF) advice on comparative genomic analyses of the conserved regions. The control brain tissues were obtained from the Douglas Hospital Research Center Brain Bank, Montreal, Canada.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Allen R.P., Picchietti D., Hening W.A., Trenkwalder C., Walters A.S., Montplaisi J. Restless legs syndrome: diagnostic criteria, special considerations, and epidemiology. A report from the restless legs syndrome diagnosis and epidemiology workshop at the National Institutes of Health. Sleep Med. 2003;4:101–119. doi: 10.1016/s1389-9457(03)00010-8. [DOI] [PubMed] [Google Scholar]
- 2.Montplaisir J., Boucher S., Poirier G., Lavigne G., Lapierre O., Lesperance P. Clinical, polysomnographic, and genetic characteristics of restless legs syndrome: a study of 133 patients diagnosed with new standard criteria. Mov. Disord. 1997;12:61–65. doi: 10.1002/mds.870120111. [DOI] [PubMed] [Google Scholar]
- 3.Lugaresi E., Coccagna G., Mantovani M., Lebrun R. Some periodic phenomena arising during drowsiness and sleep in man. Electroencephalogr. Clin. Neurophysiol. 1972;32:701–705. doi: 10.1016/0013-4694(72)90106-x. [DOI] [PubMed] [Google Scholar]
- 4.Sforza E., Johannes M., Claudio B. The PAM-RL ambulatory device for detection of periodic leg movements: a validation study. Sleep Med. 2005;6:407–413. doi: 10.1016/j.sleep.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Earley C.J., Allen R.P., Beard J.L., Connor J.R. Insight into the pathophysiology of restless legs syndrome. J. Neurosci. Res. 2000;62:623–628. doi: 10.1002/1097-4547(20001201)62:5<623::AID-JNR1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 6.Allen R.P., Earley C.J. The role of iron in restless legs syndrome. Mov. Disord. 2007;22(Suppl. 18):S440–S448. doi: 10.1002/mds.21607. [DOI] [PubMed] [Google Scholar]
- 7.Pennestri M.H., Montplaisir J., Colombo R., Lavigne G., Lanfranchi P.A. Nocturnal blood pressure changes in patients with restless legs syndrome. Neurology. 2007;68:1213–1218. doi: 10.1212/01.wnl.0000259036.89411.52. [DOI] [PubMed] [Google Scholar]
- 8.Winkelman J.W., Shahar E., Sharief I., Gottlieb D.J. Association of restless legs syndrome and cardiovascular disease in the Sleep Heart Health Study. Neurology. 2008;70:35–42. doi: 10.1212/01.wnl.0000287072.93277.c9. [DOI] [PubMed] [Google Scholar]
- 9.Allen R.P., Walters A.S., Montplaisir J., Hening W., Myers A., Bell T.J., Ferini-Strambi L. Restless legs syndrome prevalence and impact: REST general population study. Arch. Intern. Med. 2005;165:1286–1292. doi: 10.1001/archinte.165.11.1286. [DOI] [PubMed] [Google Scholar]
- 10.Winkelmann J., Wetter T.C., Collado-Seidel V., Gasser T., Dichgans M., Yassouridis A., Trenkwalder C. Clinical characteristics and frequency of the hereditary restless legs syndrome in a population of 300 patients. Sleep. 2000;23:597–602. [PubMed] [Google Scholar]
- 11.Xiong L., Jang K., Montplaisir J., Levchenko A., Thibodeau P., Gaspar C., Turecki G., Rouleau G.A. Canadian restless legs syndrome twin study. Neurology. 2007;68:1631–1633. doi: 10.1212/01.wnl.0000261016.90374.fd. [DOI] [PubMed] [Google Scholar]
- 12.Xiong L., Turecki G., Levchenko A., Gaspar C., Montplaisir J., Rouleau G.A. Genetics of restless legs syndrome. In: Hening W., Allen R., Chokroverty S., editors. Restless Legs Syndrome. Philadelphia, PA: Elsevier; 2009. pp. 31–49. [Google Scholar]
- 13.Winkelmann J., Schormair B., Lichtner P., Ripke S., Xiong L., Jalilzadeh S., Fulda S., Putz B., Eckstein G., Hauk S., et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat. Genet. 2007;39:1000–1006. doi: 10.1038/ng2099. [DOI] [PubMed] [Google Scholar]
- 14.Vilarino-Guell C., Farrer M.J., Lin S.C. A genetic risk factor for periodic limb movements in sleep. N Engl J Med. 2008;358:425–427. doi: 10.1056/NEJMc072518. [DOI] [PubMed] [Google Scholar]
- 15.Mignot E. A step forward for restless legs syndrome. Nat. Genet. 2007;39:938–939. doi: 10.1038/ng0807-938. [DOI] [PubMed] [Google Scholar]
- 16.Altshuler D., Daly M. Guilt beyond a reasonable doubt. Nat. Genet. 2007;39:813–815. doi: 10.1038/ng0707-813. [DOI] [PubMed] [Google Scholar]
- 17.Manolio T.A., Brooks L.D., Collins F.S. A HapMap harvest of insights into the genetics of common disease. J. Clin. Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stefansson H., Rye D.B., Hicks A., Petursson H., Ingason A., Thorgeirsson T.E., Palsson S., Sigmundsson T., Sigurdsson A.P., Eiriksdottir I., et al. A genetic risk factor for periodic limb movements in sleep. N. Engl. J. Med. 2007;357:639–647. doi: 10.1056/NEJMoa072743. [DOI] [PubMed] [Google Scholar]
- 19.Conrad D.F., Andrews T.D., Carter N.P., Hurles M.E., Pritchard J.K. A high-resolution survey of deletion polymorphism in the human genome. Nat. Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 20.Hinds D.A., Kloek A.P., Jen M., Chen X., Frazer K.A. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat. Genet. 2006;38:82–85. doi: 10.1038/ng1695. [DOI] [PubMed] [Google Scholar]
- 21.Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 22.Locke D.P., Sharp A.J., McCarroll S.A., McGrath S.D., Newman T.L., Cheng Z., Schwartz S., Albertson D.G., Pinkel D., Altshuler D.M., et al. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am. J. Hum. Genet. 2006;79:275–290. doi: 10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCarroll S.A., Hadnott T.N., Perry G.H., Sabeti P.C., Zody M.C., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., et al. Common deletion polymorphisms in the human genome. Nat. Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 24.Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 26.Sharp A.J., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tuzun E., Sharp A.J., Bailey J.A., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., et al. Fine-scale structural variation of the human genome. Nat. Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 28.Prabhakar S., Poulin F., Shoukry M., Afzal V., Rubin E.M., Couronne O., Pennacchio L.A. Close sequence comparisons are sufficient to identify human cis-regulatory elements. Genome Res. 2006;16:855–863. doi: 10.1101/gr.4717506. (Epub June 12, 2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahituv N., Zhu Y., Visel A., Holt A., Afzal V., Pennacchio L.A., Rubin E.M. Deletion of ultraconserved elements yields viable mice. PLoS Biol. 2007;5:e234. doi: 10.1371/journal.pbio.0050234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahituv N., Prabhakar S., Poulin F., Rubin E.M., Couronne O. Mapping cis-regulatory domains in the human genome using multi-species conservation of synteny. Hum. Mol. Genet. 2005;14:3057–3063. doi: 10.1093/hmg/ddi338. (Epub September 9, 2005) [DOI] [PubMed] [Google Scholar]
- 31.Goring H.H., Curran J.E., Johnson M.P., Dyer T.D., Charlesworth J., Cole S.A., Jowett J.B., Abraham L.J., Rainwater D.L., Comuzzie A.G., et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 32.Stranger B.E., Forrest M.S., Dunning M., Ingle C.E., Beazley C., Thorne N., Redon R., Bird C.P., de Grassi A., Lee C., et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sequeira A., Klempan T., Canetti L., ffrench-Mullen J., Benkelfat C., Rouleau G.A., Turecki G. Patterns of gene expression in the limbic system of suicides with and without major depression. Mol. Psychiatry. 2007;12:640–655. doi: 10.1038/sj.mp.4001969. [DOI] [PubMed] [Google Scholar]
- 34.Moens C.B., Selleri L. Hox cofactors in vertebrate development. Dev. Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 35.Dasen J.S., Tice B.C., Brenner-Morton S., Jessell T.M. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell. 2005;123:477–491. doi: 10.1016/j.cell.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 36.Azpiazu N., Morata G. Distinct functions of homothorax in leg development in Drosophila. Mech. Dev. 2002;119:55–67. doi: 10.1016/s0925-4773(02)00295-2. [DOI] [PubMed] [Google Scholar]
- 37.Mercader N., Leonardo E., Azpiazu N., Serrano A., Morata G., Martinez C., Torres M. Conserved regulation of proximodistal limb axis development by Meis1/Hth. Nature. 1999;402:425–429. doi: 10.1038/46580. [DOI] [PubMed] [Google Scholar]
- 38.Kurant E., Pai C.Y., Sharf R., Halachmi N., Sun Y.H., Salzberg A. Dorsotonals/homothorax, the Drosophila homologue of meis1, interacts with extradenticle in patterning of the embryonic PNS. Development. 1998;125:1037–1048. doi: 10.1242/dev.125.6.1037. [DOI] [PubMed] [Google Scholar]
- 39.Lein E.S., Hawrylycz M.J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A.F., Boguski M.S., Brockway K.S., Byrnes E.J., et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. (Epub December 6, 2006) [DOI] [PubMed] [Google Scholar]
- 40.Moskow J.J., Bullrich F., Huebner K., Daar I.O., Buchberg A.M. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol. Cell Biol. 1995;15:5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thorsteinsdottir U., Kroon E., Jerome L., Blasi F., Sauvageau G. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol. Cell Biol. 2001;21:224–234. doi: 10.1128/MCB.21.1.224-234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spieker N., van Sluis P., Beitsma M., Boon K., van Schaik B.D., van Kampen A.H., Caron H., Versteeg R. The MEIS1 oncogene is highly expressed in neuroblastoma and amplified in cell line IMR32. Genomics. 2001;71:214–221. doi: 10.1006/geno.2000.6408. [DOI] [PubMed] [Google Scholar]
- 43.Curran S.P., Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conte I., Bovolenta P. Comprehensive characterization of the cis-regulatory code responsible for the spatio-temporal expression of olSix3.2 in the developing medaka forebrain. Genome Biol. 2007;8:R137. doi: 10.1186/gb-2007-8-7-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pennacchio L.A., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 46.Lettice L.A., Heaney S.J., Purdie L.A., Li L., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 47.Emison E.S., McCallion A.S., Kashuk C.S., Bush R.T., Grice E., Lin S., Portnoy M.E., Cutler D.J., Green E.D., Chakravarti A. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature. 2005;434:857–863. doi: 10.1038/nature03467. [DOI] [PubMed] [Google Scholar]
- 48.Loots G.G., Kneissel M., Keller H., Baptist M., Chang J., Collette N.M., Ovcharenko D., Plajzer-Frick I., Rubin E.M. Genomic deletion of a long-range bone enhancer misregulates sclerostin in Van Buchem disease. Genome Res. 2005;15:928–935. doi: 10.1101/gr.3437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Belbin O., Dunn J.L., Ling Y., Morgan L., Chappell S., Beaumont H., Warden D., Smith D.A., Kalsheker N., Morgan K. Regulatory region single nucleotide polymorphisms of the apolipoprotein E gene and the rate of cognitive decline in Alzheimer's disease. Hum. Mol. Genet. 2007;16:2199–2208. doi: 10.1093/hmg/ddm171. [DOI] [PubMed] [Google Scholar]
- 50.Moffatt M.F., Kabesch M., Liang L., Dixon A.L., Strachan D., Heath S., Depner M., von Berg A., Bufe A., Rietschel E., et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. (Epub July 4, 2007) [DOI] [PubMed] [Google Scholar]
- 51.Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C., Taylor J., Burnett E., Gut I., Farrall M., et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 52.Emilsson V., Thorleifsson G., Zhang B., Leonardson A.S., Zink F., Zhu J., Carlson S., Helgason A., Walters G.B., Gunnarsdottir S., et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 53.Pennestri M.H., Whittom S., Adam B., Petit D., Carrier J., Montplaisir J. PLMS and PLMW in healthy subjects as a function of age: prevalence and interval distribution. Sleep. 2006;29:1183–1187. doi: 10.1093/sleep/29.9.1183. [DOI] [PubMed] [Google Scholar]
- 54.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet. Epidemiol. 2003;25:115–121. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.