Abstract
Protection of chromosome ends from DNA repair and degradation activities is mediated by specialized protein complexes bound to telomere repeats. Recently, it has become apparent that epigenetic regulation of the telomric chromatin template critically impacts on telomere function and telomere-length homeostasis from yeast to man. Across all species, telomeric repeats as well as the adjacent subtelomeric regions carry features of repressive chromatin. Disruption of this silent chromatin environment results in loss of telomere-length control and increased telomere recombination. In turn, progressive telomere loss reduces chromatin compaction at telomeric and subtelomeric domains. The recent discoveries of telomere chromatin regulation during early mammalian development, as well as during nuclear reprogramming, further highlights a central role of telomere chromatin changes in ontogenesis. In addition, telomeres were recently shown to generate long, non-coding RNAs that remain associated to telomeric chromatin and will provide new insights into the regulation of telomere length and telomere chromatin. In this review, we will discuss the epigenetic regulation of telomeres across species, with special emphasis on mammalian telomeres. We will also discuss the links between epigenetic alterations at mammalian telomeres and telomere-associated diseases.
Keywords: epigenetics, silencing, telomeres, telomeric chromatin
Introduction
Telomeres are nucleoprotein structures that protect the ends of linear chromosomes from degradation and from being detected as double-strand DNA breaks (Chan and Blackburn, 2004; Palm and de Lange, 2008). A tri-partite organization of telomeres is a canonical feature of chromosome termini in eukaryotes. Telomeres consist of (i) a capping structure, which protects the end of chromosomes from degradation and from eliciting a DNA damage response (DDR), and also controls the extension of telomeric repeats; (ii) a stretch of double-stranded repetitive and transcribed DNA elements; and (iii) repetitive telomere-associated sequences (TAS) also referred to as subtelomeres (Riethman et al, 2005; Blasco, 2007; Anderson et al, 2008). Whereas yeast, vertebrate, and plant telomeres consist of short-tandem repeats, Drosophila melanogaster chromosomes terminate in arrays of telomere-specific non-long terminal-repeat (LTR) retrotransposons (Pardue and DeBaryshe, 2003; Chan and Blackburn, 2004; Zellinger and Riha, 2007). Telomere function depends on a minimal length of telomeric repeats and the functionality of the associated protein complexes. In addition, higher-order DNA conformations, such as the T-loop, are thought to contribute to telomere function (Griffith et al, 1999). In most species, telomeres are maintained by telomerase, a reverse transcriptase that adds telomeric repeats de novo after every cell division, thereby counteracting incomplete DNA replication of telomeres due to the so-called end-replication problem (Collins and Mitchell, 2002; Chan and Blackburn, 2004). Drosophila melanogaster compensates the lack of telomerase by transposing telomere-specific LTR retrotransposons to chromosome ends (Pardue and Debaryshe, 2008). Alternative pathways involving telomere recombination (ALT, alternative lengthening of telomeres) have been also described in mammals (Collins and Mitchell, 2002; Pardue and DeBaryshe, 2003; Muntoni and Reddel, 2005).
In adult mammalian tissues and adult stem cells, telomerase activity is not sufficient to maintain telomeres during cell division and tissue renewal (Collins and Mitchell, 2002; Flores et al, 2005; Sarin et al, 2005). Progressive telomere shortening leads to telomere dysfunction and elicitation of a DDR, which result in cell cycle arrest/senescence or apoptosis (Harley et al, 1990; d'Adda di Fagagna et al, 2003). In vivo, critically short telomeres result in stem cell dysfunction, premature loss of tissue regeneration, and reduced life span, as shown in the context of telomerase-deficient mice (Blasco et al, 1997; Herrera et al, 1999; Rudolph et al, 1999; Gonzalez-Suarez et al, 2000; Collins and Mitchell, 2002; Blasco, 2005; Garcia-Cao et al, 2006). In contrast, over-expression of telomerase is sufficient to immortalize most human cell types in vitro and leads to a significant extension of the median life span of Tert transgenic mice with increased cancer resistance (Bodnar et al, 1998; Gonzalez-Suarez et al, 2001; Artandi et al, 2002; Canela et al, 2004; Tomas-Loba et al, 2008).
Pioneer studies in yeast indicated the involvement of chromatin modifications in the control of telomere function and telomere length. In particular, reporter genes introduced in proximity to telomeres were found to be silenced, suggesting a repressive chromatin environment at yeast telomeres, which was later also reported for D. melanogaster and mammals (Palladino et al, 1993; Cooper et al, 1997; Baur et al, 2001; Koering et al, 2002; Biessmann et al, 2005; Mason et al, 2008). Whereas telomeric repeats are devoid of histones in yeast, the accumulation of repressive histone modifications at mammalian telomeric and subtelomeric repeats, as well as the hypermethylation of subtelomeric DNA, has been recently shown to have a central function in mammalian telomere-length homeostasis (Blasco, 2007).
Recent discoveries of transcripts derived from yeast and vertebrate telomeres, as well as rasiRNAs derived from Drosophila melanogaster telomeric retrotransposons, suggests the involvement of non-coding RNAs in telomere structure and telomere regulation across species (Savitsky et al, 2006; Azzalin et al, 2007; Schoeftner and Blasco, 2008). Mammalian and yeast telomeric RNAs have been proposed to control telomere structure as well as telomere elongation by telomerase (Azzalin et al, 2007; Luke et al, 2008; Schoeftner and Blasco, 2008).
In this review, we provide an overview on the epigenetic regulation of yeast, D. melanogaster, and vertebrate telomeres, with a special emphasis on the regulation of mammalian telomeric chromatin during development and in the context of telomere-associated diseases.
The telomere-binding proteins
From yeast to man, telomeres are bound by specialized protein complexes that regulate telomere length and telomere capping. In Saccharomyces cerevisiae, Cdc13 binds to the G-strand overhang and controls telomere elongation by telomerase, whereas Rap1 (repressor–activator protein 1) recruits the silent information regulator proteins Sir2, Sir3, Sir4 and the telomere-length regulators Rif1 and Rif2 to telomeres, forming the so-called ‘telosome' (Wright et al, 1992; Tham and Zakian, 2002). Rap1–Rif1 complexes act as a counting mechanism to negatively regulate telomere length (Kyrion et al, 1992; Krauskopf and Blackburn, 1996; Marcand et al, 1999; Levy and Blackburn, 2004). Homologues of S. cerevisiae Rap1 and Rif1 have also been described in Schyzosaccharomyces pombe. In S. pombe, Rap1 and Rif1 are recruited to double-stranded telomeric repeats through association with the telomere repeat-binding protein Taz1, thus regulating telomere length and telomeric silencing (Kanoh and Ishikawa, 2001). The S. pombe G-strand overhang is protected by Pot1. Pot1 associates with Tpz1, Ccq1 and Poz1 and contacts the Taz1–Rap1 complex located at double-stranded telomeric repeats (Miyoshi et al, 2008). Telomere-binding proteins in S. pombe telomeres are highly related to components of the mammalian shelterin complex. In functional analogy to Taz1, the mammalian shelterin components TRF1 and TRF2 bind to double-stranded telomeric repeats and recruit TPP1 (orthologue of S. pombe Tpz1), RAP1, TIN2, and the poly(ADP)-ribosylases TANK1 and TANK2 to telomeres (Palm and de Lange, 2008). The single-stranded 3′overhang is bound by POT1, which contacts with TRF1 and TRF2 at double-stranded telomere regions through TPP1.
D. melanogaster lacks telomerase activity and maintains arrays of telomere-specific LTR retrotransposons by retrotransposition or gene conversion (Biessmann and Mason, 2003). In contrast to yeast and vertebrate telomeres, chromosome capping in D. melanogaster is mediated by an alternative mechanism, which is dependent on the ‘terminin' protein complex containing the heterochromatin protein 1 (HP1), HOAP (HP1/ORC-associated protein), and the modigliani (moi) gene product (Cenci et al, 2005). The dependence of chromosome capping on HP1, a major component of heterochromatin, shows the strong bias of Drosophila melanogaster telomere regulation towards the use of general chromatin regulators.
Epigenetic regulation of yeast telomeres
S. cerevisiae telomeres consist of 350±75 bp of C1–2A/TG1–3 histone-free DNA repeats that terminate in a single 3′ overhang (Wright et al, 1992). Adjacent subtelomeric Y′ and X repeats are assembled into nucleosomes and extend several kilobases towards centromeres (Louis, 1995). The silencing of reporter genes introduced into S. cerevisiae subtelomeric regions, a phenomenon also referred to as ‘telomere position effect' (TPE), provided early evidence for a repressive chromatin environment at telomeres (Gottschling et al, 1990; Tham and Zakian, 2002). As discussed above, histone-free telomeric repeats are bound by Rap1, which recruits the silent information regulator Sir4. Sir4 further attracts Sir2 and Sir3 to telomeres. The NAD-dependent deacetylase activity of Sir2 is essential for telomere repression and the spreading of silencing, whereas Sir3 and Sir4 act as structural components. Sir2 de-acetylates the tails of histones H3 and H4 with preference for acetylated lysine 16 on histone H4 (H4K16Ac), thereby creating a high-affinity-binding site for Sir3 and Sir4 (Hecht et al, 1995; Tanny et al, 1999; Imai et al, 2000; Carmen et al, 2002). Mutations in residues K16–K20 of histone H4, as well as loss of Sir2, result in loss of telomeric repression (Johnson et al, 1990; Aparicio et al, 1991; Tanny et al, 1999). Binding of Sir3 and Sir4 is further enhanced by the production of 2′-O-acetyl-ADP-ribose (O-AADPR), a side product of the NAD+ hydrolysis by Sir2 (Liou et al, 2005; Martino et al, 2009). Thus, a positive-feedback loop based on cycles of histone H3 and H4 de-acetylation, Sir protein recruitment and O-AADPR-mediated stabilization allows the Sir complex to spread along subtelomeric nucleosomes and silence promoters kilobases away from Rap1-determined silencing nucleation. Silencing is further enhanced by the formation of a telomeric fold-back structure and the association of telomeres with the Sir-rich nuclear periphery (Maillet et al, 1996; Strahl-Bolsinger et al, 1997; de Bruin et al, 2000). Spreading of telomeric silencing is antagonized by Sas2, a specific MYST-type family acetylase of the SAS complex that competes with Sir2 in controlling the acetylation status of H4K16 (Osada et al, 2001; Kimura et al, 2002; Suka et al, 2002; Shia et al, 2005). H4K16 acetylation by Sas2 is important for the subsequent incorporation of H2A.Z that forms a chromatin boundary preventing the propagation of silencing (Meneghini et al, 2003; Shia et al, 2006). Hyperacetylated H4K16 also drives Sir3 displacement and allows binding of the histone methyltransferase Dot1 that methylates the histone H3 lysine 79 residue, further antagonizing the spreading of Sir complexes (Park et al, 2002; van Leeuwen and Gottschling, 2002; van Leeuwen et al, 2002; Ng et al, 2002a; Altaf et al, 2007; Fingerman et al, 2007). In addition, the ubiquitination of lysine 123 of H2B by the ubiquitin-ligating enzyme Rad6 is required for efficient H3K79 methylation and the methylation of histone H3K4 by Set1, another marker of telomeric chromatin (Briggs et al, 2002; Dover et al, 2002; Ng et al, 2002b; Sun and Allis, 2002; Shahbazian et al, 2005). Together, this indicates the existence of a network of trans-histone pathways to tune repression at telomeres and subtelomeres.
The role of these epigenetic modifications in the regulation of yeast telomere length is well documented. Several mutations that disrupt telomeric silencing also decrease the length of telomeres (Palladino et al, 1993; Greenwell et al, 1995; Porter et al, 1996; Nislow et al, 1997). In addition, the Rap1 counting pathway seems to be indirectly regulated by the Sir proteins (Marcand et al, 1997). Furthermore, anchoring of telomeres to the nuclear periphery seems to regulate telomere length in cells that are compromised for the Rap1 counting pathway (Gartenberg et al, 2004; Berthiau et al, 2006; Hediger et al, 2006). Notably, deletion of Rif2 can also lead to recombination-dependent telomere elongation (Teng et al, 2000), suggesting a link between telomeric chromatin and recombination. Recently, S. cerevisiae and vertebrate telomeres were shown to be transcribed by RNA Polymerase II, giving rise to single-stranded telomeric repeat-containing RNAs (TERRA/TelRNAs). Yeast TERRA was reported to form RNA/DNA hybrids negatively regulating telomerase-dependent telomere elongation; however, the possible role of TelRNA/TERRA in defining telomeric silencing has not yet been addressed (Azzalin et al, 2007; Luke et al, 2008). Studying the involvement of TERRA in the regulation of yeast telomeric chromatin will reveal novel pathways of telomere control.
Epigenetic regulation of S. pombe telomeres
Telomeres in fission yeast S. pombe share features with S. cerevisiae and mammalian telomeres. Similar to budding yeast, S. pombe telomeric repeats are devoid of nucleosomes; however, telomere-binding proteins and the telomeric chromatin structure are highly related to that of mammals. Mutations in telomere-binding proteins and telomere heterochromatin regulators, such as Taz1, Rap1, Swi6, and Clr1-4, are known to affect telomeric silencing (Thon and Klar, 1992; Allshire et al, 1995; Cooper et al, 1997; Nimmo et al, 1998; Chikashige and Hiraoka, 2001; Kanoh and Ishikawa, 2001; Sugiyama et al, 2007). In addition, disruption of telomeric heterochromatin results in increased subtelomeric recombination, which, similar to mammals, can impact on telomere-length homeostasis (Kanoh et al, 2003; Bisht et al, 2008). Fission yeast telomeric heterochromatin is enriched for Swi6, the orthologue of D. melanogaster HP1. HP1 recruitment to telomeres is dependent on H3K9 methylation by the SET domain-containing histone methyltransferase Clr4 (orthologue of mammalian Suv39h HMTases) that methylates the histone H3 lysine 9 residues at telomeres (Bannister et al, 2001; Nakayama et al, 2001). The chromatin structure of S. pombe telomeres is similar to that found at centromeric regions and the mating-type locus where H3K9 methylation by Clr4 is dependent on the generation of small RNAs derived from heterochromatic regions by Dcr1 (the homologue of mammalian Dicer 1) (Ekwall et al, 1995, 1996; Nakayama et al, 2000, 2001; Reinhart and Bartel, 2002; Motamedi et al, 2004; Noma et al, 2004; Verdel et al, 2004; Kato et al, 2005). However, only the combined ablation of the telomeric repeat-binding protein Taz1 and proteins involved RNAi-mediated heterochromatin formation releases Swi6 from telomeres, suggesting that telomeric heterochromatin is recruited by Taz1 and components of the RNAi machinery (Kanoh et al, 2005). Recently, the multi-enzyme complex SHREC, which mediates heterochromatic transcriptional gene silencing in S. pombe, was shown to be recruited to telomeres by redundant pathways involving Taz1 and Ccq1, as well as the RNAi machinery (Sugiyama et al, 2007). SHREC contains the histone deacetylase Clr3 and the chromatin remodelling factor Mit1 and both activities are required to silence reporter genes at subtelomeres (Sugiyama et al, 2007). Interestingly, in addition to recruiting SHREC, Ccq1, which is functionally linked to the telomeric single-stranded-binding protein Pot1, also recruits telomerase and prevents telomeric recombination (Miyoshi et al, 2008; Tomita and Cooper, 2008). Finally, absence of SpSet1p, a histone H3 lysine 4 methyltransferase associated with transcriptional activation, also results in impaired telomeric silencing and telomere elongation (Kanoh et al, 2003). In summary, the regulation of telomeric heterochromatin in S. pombe illustrates an interplay between the telomere-binding proteins and general chromatin regulators. Given the high similarity between S. pombe and mammalian telomeres, a role for shelterin in telomere chromatin regulation can be anticipated. In this respect, altered nucleosome spacing in cells over-expressing TRF2 provides evidence for such a connection (Benetti et al, 2008b).
The heterochromatin structure of Drosophila telomeres
In contrast to short telomeric repeats in yeast and mammals, D. melanogaster chromosome termini consist of up to 12 kb of tandem arrays of telomere-specific HeT-A, TART and TAHR LTR retrotransposons (Mason and Biessmann, 1995; Mason et al, 2008). These arrays of HeT-A, TART, and TAHR (HTT) retroelements are preferentially maintained by target-primed reverse transcription-based retrotransposition to chromosome ends, or alternatively, by gene conversion. Transposition is dependent on HTT retroelements-encoded reverse transcriptases and occurs to any chromosome end, creating a high heterogeneity in array length (Biessmann et al, 1993; Levis et al, 1993; Walter et al, 1995; Biessmann and Mason, 2003; Abad et al, 2004; Pardue et al, 2005). Telomere capping is mediated by the ‘terminin' complex comprising HP1, the telomere-specific HOAP (HP1/ORC-asociated protein), and the modigliani (moi) gene product (Silva et al, 2004; Bi et al, 2005; Ciapponi et al, 2006; Oikemus et al, 2006; Raffa et al, 2009). Interestingly, HP1, encoded by Su(var)205, is recruited to chromosome ends independently of the sequence content or presence of H3K9me3 and spreads at lower density into adjacent HTT arrays where HP1 uses its chromodomain to bind H3K9me3 (Fanti et al, 1998; Andreyeva et al, 2005; Frydrychova et al, 2008). Su(var)205 mutants display telomere fusions, increased HeT-A transcript levels, and increased retroelement addition leading to telomere elongation (Savitsky et al, 2002). Thus, Drosophila telomere length is controlled by an interaction of H3K9me3 and HP1 in silencing HTT arrays, whereas chromosome capping by HP1 controls the addition of retroelements to chromosome ends (Perrini et al, 2004). In addition to siRNAs and miRNAs, a third RNA silencing system based on the Piwi subfamily of Argonaut proteins has evolved that prevents the spreading of selfish DNA elements such as telomeric retro-transposons in the germline (Hartig et al, 2007). In the first step of the repeat-associated short-interfering (rasi)RNA pathway, rasiRNAs are generated from damaged inactive copies of transposable elements. These antisense rasiRNAs then target transcripts of functional transposons in a process that dependents on the action of the Piwi proteins (Saito et al, 2006; Brennecke et al, 2007; Gunawardane et al, 2007). Complementary relationships of sense and antisense RNA populations indicate the existence of a positive-feedback loop, also described as ‘the ping-pong model' that ensures efficient elimination of transcripts derived from active transposons (Brennecke et al, 2007). Consistent with this model, transcript levels from functional telomere-specific retrotransposons are significantly increased in germline mutants for components of the rasiRNA pathway and the RNA helicase gene spn-E (Savitsky et al, 2006; Klenov et al, 2007; Shpiz et al, 2007). Furthermore, decreased rasiRNA production is accompanied by reduced H3K9me3 and HP1 levels at HTT arrays and by an abundant retrotransposition of HeT-A elements (Savitsky et al, 2006; Klenov et al, 2007). In line with this, Piwi is reported to localize to chromatin in a complex with HP1a, providing further evidence for a role of the rasiRNA pathway in telomere regulation (Brower-Toland et al, 2007; Klenov et al, 2007).
Telomere-associated sequences (TAS) located adjacent to HTT arrays sequences have been reported to have a role in silencing (Mason et al, 2008). TAS are enriched for the K3K27me3 mark and bound by Polycomb proteins, which in turn impact on TPE (Boivin et al, 2003; Mason et al, 2004; Andreyeva et al, 2005; Shanower et al, 2005; Doheny et al, 2008). Interestingly, TAS are also subjected to regulation by the rasiRNA pathway. However, in contrast to HTT repeats where mutations of the rasiRNA pathway result in loss of telomeric heterochromatin, reduced TAS-originated rasiRNAs are associated with a loss of euchromatic marks (Yin and Lin, 2007). This discrepancy in chromatin regulation indicates that repetitive elements in HTT arrays and TAS sequences underlie distinct mechanisms of epigenetic regulation. A functional conservation of the rasiRNA pathway in telomere regulation in the mammalian germline is not known to date.
Vertebrate telomeric heterochromatin
Similar to D. melanogaster and S. pombe, vertebrate telomeres are enriched for the H3K9m3 mark, imposed by the Suv39h1 and Suv39h2 HMTases, the mammalian homologues of S. pombe Clr4 (Peters et al, 2001, 2003; Garcia-Cao et al, 2004). H3K9me3 provides a high-affinity-binding site for HP1 and promotes the imposition of the H4K20me3 mark by the Suv4-20h1 and Suv4-20h2 HMTases (Bannister et al, 2001; Lachner et al, 2001; Nakayama et al, 2001; Schotta et al, 2004, 2008; Benetti et al, 2007b) (Figure 1). In addition to these heterochromatic histone marks, telomeric repeats also contain di-methyalted H3K79, which is mediated by the Dot1L HMTase (San-Segundo and Roeder, 2000; Shanower et al, 2005). Dot1L activity is also required for efficient imposition of the H4K20me3 mark at telomeres, suggesting that both Suv39h HMTases and Dot1L are acting upstream of the Suv4-20h HMTases (Jones et al, 2008) (Figure 1) Interestingly, although telomeres display normal H3K9me3 levels, the abundance of H3K9me2 is markedly reduced at telomeric repeats in cells lacking Dot1L. This suggests that additional H3K9-specific HMTases, such as G9a of ESET, could be involved mediating H3K9me2 at telomeres (Jones et al, 2008). In addition to repressive histone marks, telomeric H3 and H4 histones are under-acetylated (Benetti et al, 2007a). In this regard, lack of the histone deacetylase SIRT6 results in elevated H3K9-acetylation levels at human telomeres and can lead to telomere dysfunction (Michishita et al, 2008).
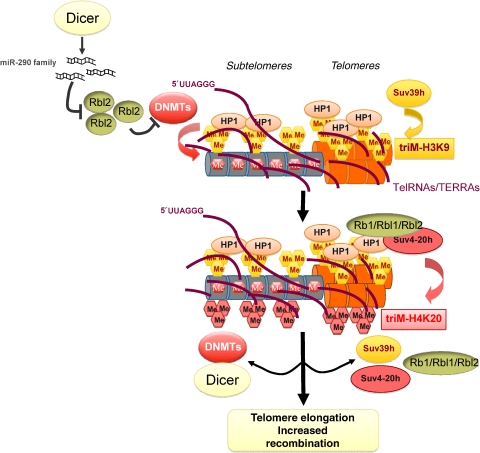
Figure 1.
Assembly of mammalian telomeric and subtelomeric heterochromatin. Scheme showing a model for the assembly of telomeric and subtelomeric heterochromatin. Suv39 h1 and h2 HMTases tri-methylate H3K9, which in turn generates a high-affinity site for HP1. HP1 can recruit Suv4-20 h1 and h2 HMTases to telomeres and subtelomeres, thereby tri-methylating H4K20 at these regions. The Rb family proteins (Rb1, Rbl1, and Rbl2) can directly interact with Suv420 HMTases and with HP1, thus influencing the levels of H4K20m3. Dicer is essential for the maturation of miRNAs including the miR290 cluster. miR290 cluster expression in ES cells results in post-transcriptional repression of Rbl2 (p130), a transcriptional repressor of mammalian DNA methyltransferases (DNMTs). Low Rbl2 levels ensure the establishment of global and subtelomeric DNA methylation patterns in ES cells. A lack of mature miRNA290 cluster results in repression of DNMTs by uncontrolled expression of Rbl2. Consequently, a global decrease in DNA methylation unleashes recombination leading to telomere elongation and increased chromatin compaction at telomeric and subtelomeric repeats mediated by Suv39h and Suv4-20h HMTases. Loss of heterochromatin in cells lacking Dicer, DNMTs, Suv39h, or Suv4-20h HMTases results in increased telomeric recombination and telomere elongation.
Importantly, repressive chromatin marks are also present at subtelomeric repeats. In particular, subtelomeres are enriched for H3K9me3, HP1, H4K20me3, and contain under-acetylated histone H3 and H4 (Benetti et al, 2007a, 2007b) (Figure 1). To this end, however, it is not clear whether subtelomeric heterochromatin is a consequence of spreading of a heterochromatic ‘island' at telomeres or recruited in cis because due to the presence of repetitive elements at subtelomeres.
DNA methylation at subtelomeric repeats
DNA methylation is known to regulate mammalian development and to specify silent chromatin regions in both eu- and heterochromatin (Chen and Li, 2006). In contrast to S. cerevisiae and D. melanogaster, which lack or display low levels of DNA methylation, mammalian subtelomeric regions are heavily methylated (Tommerup et al, 1994; van Overveld et al, 2003; Steinert et al, 2004; Gonzalo et al, 2006) (Figure 1). Importantly, TTAGGG repeats remain unmethylated because of the lack of methylate-able cytosine. It has been proposed that DNA methylation at subtelomeric repeats acts as an additional mechanism in mammals that enforces TPE (van Overveld et al, 2003; Pedram et al, 2006). DNA methylation patterns in mammalian cells are established by three main DNA metyltransferases (DNMTs). De novo methylation patterns are established by DNMT3a and DNMT3b and maintained by DNMT1, which copies parental-strand methylation onto the de novo synthesized daughter strand after DNA replication (Okano et al, 1998). DNA methylation is enriched at repetitive elements such as the pericentric regions and is regarded to prevent frequent recombination events (Bender, 1998; Maloisel and Rossignol, 1998; Dominguez-Bendala and McWhir, 2004; Gonzalo et al, 2006; Jaco et al, 2008). Consistent with this, deficiency of DNMT1 or DNMT3ab causes a dramatic elongation of telomeres, which is driven by increased homologous recombination events between telomeric sister chromatids (Gonzalo et al, 2006). The mechanism of DNMT recruitment to subtelomeres remains however unclear. Whereas DNA methylation at pericentric repeats is reduced in the absence of Suv39h HMTases and an interaction between HP1 and Suv39h1 had been reported, loss of Suv39h HMTases does not affect subtelomeric DNA methylation (Fuks et al, 2003; Lehnertz et al, 2003; Benetti et al, 2007b). This suggests the existence of an alternative pathway of DNMT recruitment to subtelomeres.
Rb family proteins regulate telomeric and subtelomeric chromatin status
A major tumour suppressor pathway in mammals is centred on the family of retinoblastoma (RB) proteins, consisting of RB1, RBL1 and RBL2 (Weinberg, 1995; Lipinski and Jacks, 1999; Classon and Harlow, 2002). RB proteins are transcriptional repressors that control cell cycle genes through interaction with E2F family of transcription factors, as well as by direct recruitment of chromatin regulators to promoters (Harbour and Dean, 2000a, 2000b). In addition to their role at specific promoters, RB family proteins also influence global H4K20me3 and DNA methylation levels, impacting on the epigenetic regulation of telomeres and centromeres (Gonzalo et al, 2005). In particular, RB proteins promote the recruitment of Suv4-20h HMTase and HP1 to telomeres, thereby negatively regulating telomere length and telomere recombination (Gonzalo and Blasco, 2005). In addition, mouse Rbl2 acts as a transcriptional repressor of DNMTs, thereby influencing telomere length and telomere recombination (Kimura et al, 2003; Gonzalo and Blasco, 2005; McCabe et al, 2005; Benetti et al, 2008a) (Figure 1). In particular, the lack of a functional miR290 cluster targeting Rbl2 in embryonic stem (ES) cells deficient for Dicer1 results in elevated levels of Rbl2 (Sinkkonen et al, 2008; Benetti et al, 2008a). In turn, increased Rbl2 levels repress DNMT expression and result in loss of global as well as subtelomeric DNA methylation, which drives increased telomeric recombination and aberrant telomere elongation (Benetti et al, 2008a). Indeed, Dicer1-null ES cells phenocopy telomere defects of DNMT-deficient cells, suggesting that Rbl2 and the miR290 cluster are major determinants controlling DNA methylation in ES cells (Gonzalo et al, 2006; Benetti et al, 2008a) (Figure 1). Remarkably, Dicer deficiency does not result in a loss of heterochromatic histone marks at telomeres, excluding a direct involvement of Dicer1-dependent small RNAs in the assembly of telomeric heterochromatin (Benetti et al, 2008a). The antagonistic role of Rbl2 on DNA methylation is at first glance in contradiction to the reduced DNA methylation levels observed in primary mouse embryonic fibroblasts (MEFs) lacking Rb, Rbl1 and Rbl2 proteins; however, this discrepancy can be explained by the fact that Rbl2 is not expressed in MEFs (Gonzalo and Blasco, 2005). In summary, loss of RB proteins results in improved telomere maintenance due to a more relaxed telomeric chromatin structure. Given the central role of RB proteins as tumour suppressors, it will be very interesting to investigate the contribution of improved telomere maintenance to proliferative capacity of tumour cells lacking RB proteins.
Telomere repeat-associated transcripts (TERRA/TelRNAs)
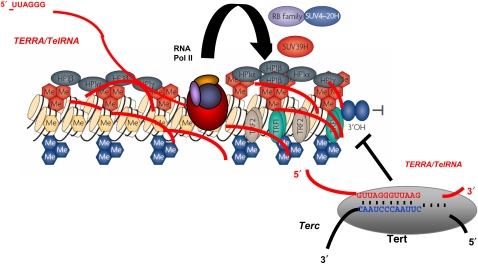
On account of their compact heterochromatic structure, telomeres were not regarded to be permissive for transcription. However, other heterochromatic domains in the genome, such as mouse major satellite or human heterochromatic satellite III repeats, were already shown to be efficiently transcribed by RNA polymerase II, giving rise to non-coding RNAs (Lehnertz et al, 2003; Jolly et al, 2004; Rizzi et al, 2004). Recently, two independent reports showed that the telomeric C-rich strand is frequently transcribed by RNA polymerase II, giving rise to UUAGGG-repeat containing non-coding RNAs (TERRA or TelRNA) (Azzalin et al, 2007; Schoeftner and Blasco, 2008). Although formal evidence is still missing, the detection of subtelomeric sequences in TelRNA/TERRA molecules strongly suggests the existence of transcriptional control elements at subtelomeres (Azzalin et al, 2007). Up to date, transcripts containing telomeric repeats have been described in Mus musculus, Homo sapiens, S. cerevisiae and Danio rerio (Azzalin et al, 2007; Luke et al, 2008; Schoeftner and Blasco, 2008). The fact that retrotransposition events at HTT arrays of D. melanogaster also depend on transcription suggests that transcription is a universal process occurring at the ends of linear, eukaryotic chromosomes. Importantly, telomeric RNAs can be detected at telomeres by RNA-FISH techniques, suggesting that TERRA/TelRNAs can associate with telomeric chromatin in cis, a feature reported earlier for the non-coding XIST RNA that controls mammalian dosage compensation (Azzalin et al, 2007; Payer and Lee, 2008; Schoeftner and Blasco, 2008). Interestingly, in a panel of female mouse cell lines, TERRA/TelRNA form accumulations (Tacs) in the immediate vicinity of the territory of inactive X chromosome (Xi), suggesting an involvement of TERRA/TelRNA in the biology of X inactivation (Schoeftner and Blasco, 2008). TERRA/TelRNA molecules range between ca 100 bp and >9 kb in length and were reported to form intermolecular G-quadruplex structure with single-stranded telomeric DNA, but can also fold into a compact repeated structure containing G-quartets (Azzalin et al, 2007; Schoeftner and Blasco, 2008; Xu et al, 2008; Martadinata and Phan, 2009; Randall and Griffith, 2009). Several lines of evidence exist implicating TelRNA/TERRA in the negative control of telomere length (Schoeftner and Blasco, 2008). Increased TelRNA/TERRA levels by interfering with TelRNA/TERRA decay, such as the impairment of non-sense-mediated RNA decay in human cells or by deletion of the 5′–3′exonuclease Rat1p in S. cerevisiae, are associated with a loss of telomere reserve (Azzalin et al, 2007; Luke et al, 2008). Current models propose a role for TelRNA/TERRA in controlling telomerase activity. In yeast, the formation of a DNA/RNA hybrid between TelRNA/TERRA and telomeres is thought to inhibit elongation by telomerase, whereas in mammals, TelRNA/TERRA was shown to efficiently inhibit telomerase activity in vitro, presumably by base pairing with the template region of the RNA component of telomerase (TERC) (Luke et al, 2008; Schoeftner and Blasco, 2008) (Figure 2). These working models are supported by expression data showing low TelRNA/TERRA levels during mouse embryogenesis and in cancer cells—two biological conditions that are characterized by rapid cell proliferation and dependence on high telomerase activity (Schoeftner and Blasco, 2008). On the other hand, accumulation of TelRNA/TERRA in adult tissues could be coupled to telomerase inhibition and ageing (Schoeftner and Blasco, 2008). Importantly, in immortal cell lines, as well as during nuclear reprogramming, TelRNA/TERRA levels correlate with the average telomere reserve (Schoeftner and Blasco, 2008; Marion et al, 2009). Together with the fact that TelRNA/TERRA can be localized to telomeric DNA repeats this suggests that TelRNA/TERRA could locally control telomerase activity in cis, a mechanism that could explain the preferential elongation of the shortest telomere in yeast and mammals on the molecular level (Marcand et al, 1999; Hemann et al, 2001; Samper et al, 2001; Teixeira et al, 2004; Schoeftner and Blasco, 2008). In addition, this mechanism would also preclude excessive telomere elongation by telomerase (i.e. telomere elongation during nuclear reprogramming, Marion et al, 2009), a condition that was found to be associated with impaired female fertility and fecundity in D. melanogaster (Walter et al, 2007). However, until formal evidence for a direct role of TERRA in telomerase inhibition has been presented, a speculative role of telomerase recruitment by TelRNA/TERRA should be considered (Figure 2).
Figure 2.
TERRA/TelRNAs associate to telomeric chromatin and may be involved in regulation of telomere length. Model for a role of telomeric RNAs in the regulation of telomere length. TERRA/TelRNA acts as a potent inhibitor of telomerase activity in vitro, possibly by formation of RNA:RNA hybrids with the template region of the telomerase RNA component.
Interestingly, long non-coding RNAs transcribed by RNA Pol II have been shown earlier to be involved in the epigenetic regulation of the genome (Bernstein and Allis, 2005). In particular, XIST and rox RNAs are chromatin-associated non-coding RNAs that regulate mammalian and Drosophila melanogaster dosage compensation, respectively (Deng and Meller, 2006; Payer and Lee, 2008). In addition, other non-coding RNAs such as the Air or Kcnq1ot1 RNAs are involved in genomic imprinting (Pauler et al, 2007; Pandey et al, 2008). Functional evidence is still missing, but it is expected that non-coding TelRNA/TERRA may also influence the chromatin status at subtelomeres and telomeres. Although small Dicer1-dependent double-stranded small RNAs are not involved in the generation of telomeric heterochromatin (Benetti et al, 2008a), a possible contribution of small single-stranded TelRNA/TERRA molecules, processed from a larger RNA precursor, has to be considered. In this respect, it will be particularly interesting to explore a possible connection between TelRNA/TERRA and the mammalian Piwi proteins, which generate small single-stranded RNAs from transcripts derived from repetitive elements (Aravin et al, 2007; Carmell et al, 2007; Kuramochi-Miyagawa et al, 2008).
Epigenetic regulation of telomere length and telomere recombination
Heterochromatic marks at telomeres have been proposed to act as negative regulators of telomere elongation (Blasco, 2007). This is exemplified by a substantial elongation of telomeres upon the loss of H3K9me3, HP1, and H4K20me3 marks in cells deficient for the Suv39h or Suv4-20h HMTases (Garcia-Cao et al, 2004; Benetti et al, 2007b) (Figure 1). In both settings, subtelomeric DNA methylation remains unaffected, suggesting that DNMTs can be recruited to subtelomeric regions independently of the Su(var) HMTases. Loss of subtelomeric DNA methylation in DNMT1, DNMT3ab or Dicer deficient ES cells also results in a dramatic telomere elongation, which is accompanied by increased abundance of histone heterochromatic marks at telomeric repeats (Gonzalo et al, 2006; Benetti et al, 2008a) (Figure 1). In both instances, telomere recombination frequencies are increased, suggesting that repressive marks at telomeric and subtelomeric chromatin are essential to repress recombination events (Figure 1) (Gonzalo et al, 2006; Benetti et al, 2007a, 2007b). Consistent with this notion, increased numbers of APB bodies (ALT-associated PML bodies) have been detected in all models for impaired telomeric chromatin (Garcia-Cao et al, 2004; Gonzalo et al, 2006; Benetti et al, 2007a, 2007b, 2008a). Of interest, loss of heterochromatic marks at telomeres does not seem to affect TRF1 and TRF2 binding, indicating that shelterin recruitment is uncoupled from telomeric chromatin regulation (Garcia-Cao et al, 2004; Gonzalo et al, 2006; Benetti et al, 2007a, 2007b, 2008a). However, up to date it cannot be excluded that an altered function of shelterin components contributes to ALT.
TPE experiments suggest a model in which increasing telomere-length augments silencing and thus chromatin compaction (Baur et al, 2001; Koering et al, 2002; Tham and Zakian, 2002). In agreement with this model, progressive telomere shortening in telomerase-deficient MEFs was associated with a continuous loss of H3K9me3, H4K20me3, and HP1 heterochromatic marks at telomeres and subtelomeres, which was accompanied by increased histone H3 and H4 acetylation at these regions (Benetti et al, 2007a). Moreover, subtelomeric DNA methylation was significantly reduced upon telomere shortening (Benetti et al, 2007a). Similarly, telomere shortening in mice over-expressing negative regulators of telomere length, such as TRF2 transgenic mice, also results in the loss of telomeric and subtelomeric heterochromatic features and altered nucleosome spacing (Benetti et al, 2008b). On the other hand, aberrant telomere elongation in the context of DNMT or Dicer1 deficiencies leads to increased density of heterochromatic marks at telomeres (Gonzalo et al, 2006; Benetti et al, 2007a, 2008a). These findings support a model in which the number of TTAGGG repeats at telomeres directs the epigenetic status of heterochromatin in cis and exerts a trans-acting effect on chromatin structure at subtelomeric regions. Telomere shortening, as observed during organismal ageing, causes a switch from a repressive to a more open telomeric chromatin status and favour telomere elongation by telomerase or by unleashing telomere recombination (Benetti et al, 2007a) (Figure 1). In this regard, recombination-based ALT pathways are activated in telomerase-deficient mice (Blasco et al, 1997; Hande et al, 1999; Rudolph et al, 1999; Herrera et al, 2000; Niida et al, 2000; Chang et al, 2003). More recently, an impact of telomere chromatin on telomerase-dependent telomere elongation has been also shown in the context of nuclear reprogramming (see below).
Telomeric chromatin during differentiation and reprogramming
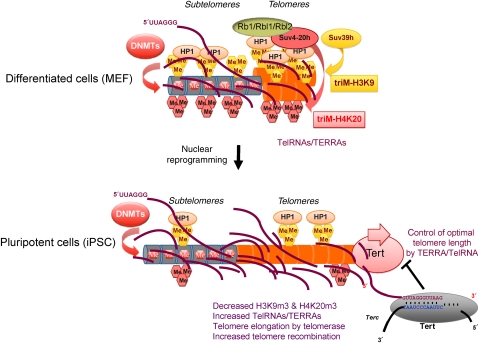
Telomere length is a major regulator of telomeric chromatin status in a given cell type and is assumed to change over the lifetime of organisms due to progressive loss of telomere reserve (Benetti et al, 2007a). In mouse embryos, telomere length is reset to a maximum length until the blastocyst stage in a telomerase-independent manner (Liu et al, 2007). In particular, increased recombination events at telomeres of mouse zygotes and two-cell embryos suggest that ALT is the driving force for the resetting of telomere length at early cleavage embryos (Schaetzlein et al, 2004; Liu et al, 2007). These data suggest that (sub-)telomeres are organized into a relatively open chromatin structure that favours telomeric recombination until the blastocyst stage. Resetting of telomere length can be recapitulated by nuclear cloning using terminally differentiated cells. Animals derived from differentiated cells with short telomeres were shown to display normal telomere length even after several cycles of nuclear transfer (Lanza et al, 2000; Wakayama et al, 2000). More recently, nuclear reprogramming has been achieved in vitro. Retroviral transduction of pluripotency factors into primary MEF, gives rise to induced pluripotent stem cells (iPSC), which are functional equivalents of mouse ES cells (Takahashi and Yamanaka, 2006; Maherali et al, 2007; Takahashi et al, 2007; Nakagawa et al, 2008; Stadtfeld et al, 2008; Wernig et al, 2008). This reprogramming event is accompanied by a dramatic telomerase-dependent telomere elongation that continues post-reprogramming until reaching the length of ES cell telomeres (Marion et al, 2009). During this process, high densities of H3K9m3 and H4K20me3 at telomeres of primary MEF are converted into a more open—ES cell-like—chromatin structure at iPSC telomeres (Marion et al, 2009). In parallel with telomere elongation, TERRA levels are efficiently upregulated in iPSC compared with MEF, a phenomenon that may serve to negatively regulate telomerase activity once iPSC reach the ES cell-like telomere length (Marion et al, 2009) (Figure 3). The reprogramming of telomeres during iPSC generation provides formal evidence that telomeric chromatin structure is defined by cell-type-specific epigenetic programmes that can be reversed by reprogramming. In line with the need for sufficient telomere reserve for stem cell functionality, reprogramming efficacy of telomerase-deficient MEF is dramatically reduced due to the appearance increased chromosome end-to-end fusions (Allsopp et al, 2003; Flores et al, 2005, 2008; Marion et al, 2009). Together, this indicates a complex regulation of telomeric heterochromatin during development and cellular differentiation, which is expected to impact on human disease.
Figure 3.
Reprogramming of telomeres upon induction of plutipotency in differentiated cells. Telomeres in primary MEFs are shorter than in ES cells and are organized into a highly compact chromatin structure with low TelRNA/TERRA expression. Induction of pluripotency by retroviral transduction of Oct4, Sox2, Kfl4, (c-myc), results in nuclear reprogramming and the generation of pluripotent iPS cells, which are functionally equivalent to ES cells. Reprogramming results in a dramatic upregulation of telomerase activity concomitant with a reduction of H3K9me3, H4K20me3, HP1, and DNA methylation at telomeres and subtelomeres as well as an increase in TelRNA/TERRA expression. Telomerase efficiently elongates telomeres until the natural limit of telomere length of pluripotent mouse ES cells has been reached.
Implications of telomere chromatin regulation for human disease
Telomere maintenance is essential for tumour cells to escape cell arrest/senescence and apoptosis. Tumour formation often occurs in the context of altered DNA methylation, loss of H4K20me3, and altered expression of Suv4-20h and Suv39h HMTases (Fraga et al, 2005; Gonzalo and Blasco, 2005; Pogribny et al, 2006; Ting et al, 2006; Tryndyak et al, 2006). Furthermore, loss of H3K9me2 and H3K9me3 in Suv39h HMTase double null-mice results in an increased incidence of B cell lymphomas (Peters et al, 2001). Along this line, it has been recently shown that the methylation status of subtelomeric DNA repeats negatively correlates with telomere length and telomere recombination in a large panel of human cancer cell lines (Vera et al, 2008). This suggests that telomeres suffer epigenetic alterations during tumourigenesis, which in turn are important drivers of telomere length changes in cancer cells. These epigenetic alterations are also expected to impact on the telomeric chromatin structure, improving telomere maintenance by ALT or providing improved access for telomerase to the G-strand overhang (Blasco, 2007). It is not known, however, whether increasing telomere compaction can affect the proliferative potential of cancer cells and impact on telomere homeostasis during organismal ageing.
Some severe premature ageing syndromes are caused by mutations in telomerase components giving rise to human syndromes such as Aplastic anaemia (TERC, TERT) (Yamaguchi et al, 2005), Dyskeratosis congenita (DKC1, TERC) and idiopatic pulmonary fibrosis (Tsakiri et al, 2007), or by mutations in various DNA repair genes such as Ataxia telangiectasia (ATM), Werner (WRN) and Bloom syndromes (BLM), Fanconi anaemia (Fanc genes), and Nijmegen breakage syndrome (NBN) (reviewed in Blasco, 2005). These patients display a substantially increased risk of developing disease states characterized by a premature loss of tissue renewal; however, the possible contribution of epigenetic defects at telomeres is still unclear (Mason et al, 2005). Similarly, accelerated telomere shortening can also occur due to environmental influences. In this regard, human population studies recently linked environmental influences (smoking, obesity, or stress) to an accelerated rate of telomere shortening (Cawthon et al, 2003; Epel et al, 2004; Valdes et al, 2005). Given the important role of epigenetic regulators during organismal ageing, it is tempting to speculate that these factors could also impact on chromatin structure leading to telomere-length abnormalities and disease (Oberdoerffer et al, 2008; Dang et al, 2009).
The recent discovery of TelRNA/TERRA allows making a new link between disease and telomeres. Increased TelRNA/TERRA transcription is linked to telomere shortening in humans and yeast. The fact that TelRNA/TERRA can antagonize telomere maintenance by telomerase, and the presence of decreased TERRA levels in human cancer samples, could point towards a relevant role of TelRNA/TERRA in limiting telomerase-dependent telomere elongation in cancer cells (Schoeftner and Blasco, 2008). This pinpoints TelRNA/TERRA as a candidate for cancer therapies based on the inhibition of telomerase (Harley, 2008). Another interesting line of evidence for a role of TelRNA/TERRA in disease comes from patients suffering from autosomal-recessive ICF (immunodeficiency, centromeric region instability, facial anomalies) syndrome. These patients display subtelomeric DNA methylation defects and abnormally short and or undetectable telomeres on some chromosome arms. Increased TelRNA/TERRA transcription in these patients points towards a role of telomeric transcripts in ICF (Yehezkel et al, 2008).
We are just beginning to understand the complex regulation of telomeric chromatin and the regulation of telomeric transcripts. The detailed investigation of function of RNAs derived from telomeres and the epigenetic control of telomeres will foster our understanding of general telomere regulation. This line of research is also expected to provide important insight into the roles of telomeres during development, ageing, and a panel of important telomere associated human diseases.
References
- Abad JP, De Pablos B, Osoegawa K, De Jong PJ, Martin-Gallardo A, Villasante A (2004) Genomic analysis of Drosophila melanogaster telomeres: full-length copies of HeT-A and TART elements at telomeres. Mol Biol Evol 21: 1613–1619 [DOI] [PubMed] [Google Scholar]
- Altaf M, Utley RT, Lacoste N, Tan S, Briggs SD, Cote J (2007) Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol Cell 28: 1002–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G (1995) Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev 9: 218–233 [DOI] [PubMed] [Google Scholar]
- Allsopp RC, Morin GB, DePinho R, Harley CB, Weissman IL (2003) Telomerase is required to slow telomere shortening and extend replicative lifespan of HSCs during serial transplantation. Blood 102: 517–520 [DOI] [PubMed] [Google Scholar]
- Anderson JA, Song YS, Langley CH (2008) Molecular population genetics of Drosophila subtelomeric DNA. Genetics 178: 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreyeva EN, Belyaeva ES, Semeshin VF, Pokholkova GV, Zhimulev IF (2005) Three distinct chromatin domains in telomere ends of polytene chromosomes in Drosophila melanogaster Tel mutants. J Cell Sci 118: 5465–5477 [DOI] [PubMed] [Google Scholar]
- Aparicio OM, Billington BL, Gottschling DE (1991) Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66: 1279–1287 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ (2007) Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747 [DOI] [PubMed] [Google Scholar]
- Artandi SE, Alson S, Tietze MK, Sharpless NE, Ye S, Greenberg RA, Castrillon DH, Horner JW, Weiler SR, Carrasco RD, DePinho RA (2002) Constitutive telomerase expression promotes mammary carcinomas in aging mice. Proc Natl Acad Sci USA 99: 8191–8196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J (2007) Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 [DOI] [PubMed] [Google Scholar]
- Baur JA, Zou Y, Shay JW, Wright WE (2001) Telomere position effect in human cells. Science 292: 2075–2077 [DOI] [PubMed] [Google Scholar]
- Bender J (1998) Cytosine methylation of repeated sequences in eukaryotes: the role of DNA pairing. Trends Biochem Sci 23: 252–256 [DOI] [PubMed] [Google Scholar]
- Benetti R, Garcia-Cao M, Blasco MA (2007a) Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat Genet 39: 243–250 [DOI] [PubMed] [Google Scholar]
- Benetti R, Gonzalo S, Jaco I, Munoz P, Gonzalez S, Schoeftner S, Murchison E, Andl T, Chen T, Klatt P, Li E, Serrano M, Millar S, Hannon G, Blasco MA (2008a) A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol 15: 268–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetti R, Gonzalo S, Jaco I, Schotta G, Klatt P, Jenuwein T, Blasco MA (2007b) Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J Cell Biol 178: 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetti R, Schoeftner S, Munoz P, Blasco MA (2008b) Role of TRF2 in the assembly of telomeric chromatin. Cell Cycle 7: 3461–3468 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Allis CD (2005) RNA meets chromatin. Genes Dev 19: 1635–1655 [DOI] [PubMed] [Google Scholar]
- Berthiau AS, Yankulov K, Bah A, Revardel E, Luciano P, Wellinger RJ, Geli V, Gilson E (2006) Subtelomeric proteins negatively regulate telomere elongation in budding yeast. EMBO J 25: 846–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Srikanta D, Fanti L, Pimpinelli S, Badugu R, Kellum R, Rong YS (2005) Drosophila ATM and ATR checkpoint kinases control partially redundant pathways for telomere maintenance. Proc Natl Acad Sci USA 102: 15167–15172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann H, Kasravi B, Jakes K, Bui T, Ikenaga K, Mason JM (1993) The genomic organization of HeT-A retroposons in Drosophila melanogaster. Chromosoma 102: 297–305 [DOI] [PubMed] [Google Scholar]
- Biessmann H, Mason JM (2003) Telomerase-independent mechanisms of telomere elongation. Cell Mol Life Sci 60: 2325–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann H, Prasad S, Walter MF, Mason JM (2005) Euchromatic and heterochromatic domains at Drosophila telomeres. Biochem Cell Biol 83: 477–485 [DOI] [PubMed] [Google Scholar]
- Bisht KK, Arora S, Ahmed S, Singh J (2008) Role of heterochromatin in suppressing subtelomeric recombination in fission yeast. Yeast 25: 537–548 [DOI] [PubMed] [Google Scholar]
- Blasco MA (2005) Mice with bad ends: mouse models for the study of telomeres and telomerase in cancer and aging. EMBO J 24: 1095–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA (2007) The epigenetic regulation of mammalian telomeres. Nat Rev Genet 8: 299–309 [DOI] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW (1997) Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91: 25–34 [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE (1998) Extension of life-span by introduction of telomerase into normal human cells. Science 279: 349–352 [DOI] [PubMed] [Google Scholar]
- Boivin A, Gally C, Netter S, Anxolabehere D, Ronsseray S (2003) Telomeric associated sequences of Drosophila recruit polycomb-group proteins in vivo and can induce pairing-sensitive repression. Genetics 164: 195–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ (2007) Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD (2002) Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418: 498. [DOI] [PubMed] [Google Scholar]
- Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H (2007) Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev 21: 2300–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A, Martin-Caballero J, Flores JM, Blasco MA (2004) Constitutive expression of tert in thymocytes leads to increased incidence and dissemination of T-cell lymphoma in Lck-Tert mice. Mol Cell Biol 24: 4275–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ (2007) MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514 [DOI] [PubMed] [Google Scholar]
- Carmen AA, Milne L, Grunstein M (2002) Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J Biol Chem 277: 4778–4781 [DOI] [PubMed] [Google Scholar]
- Cawthon RM, Smith KR, O'Brien E, Sivatchenko A, Kerber RA (2003) Association between telomere length in blood and mortality in people aged 60 years or older. Lancet 361: 393–395 [DOI] [PubMed] [Google Scholar]
- Cenci G, Ciapponi L, Gatti M (2005) The mechanism of telomere protection: a comparison between Drosophila and humans. Chromosoma 114: 135–145 [DOI] [PubMed] [Google Scholar]
- Ciapponi L, Cenci G, Gatti M (2006) The Drosophila Nbs protein functions in multiple pathways for the maintenance of genome stability. Genetics 173: 1447–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classon M, Harlow E (2002) The retinoblastoma tumour suppressor in development and cancer. Nat Rev Cancer 2: 910–917 [DOI] [PubMed] [Google Scholar]
- Collins K, Mitchell JR (2002) Telomerase in the human organism. Oncogene 21: 564–579 [DOI] [PubMed] [Google Scholar]
- Cooper JP, Nimmo ER, Allshire RC, Cech TR (1997) Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385: 744–747 [DOI] [PubMed] [Google Scholar]
- Chan SR, Blackburn EH (2004) Telomeres and telomerase. Philos Trans R Soc Lond B Biol Sci 359: 109–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Khoo CM, Naylor ML, Maser RS, DePinho RA (2003) Telomere-based crisis: functional differences between telomerase activation and ALT in tumor progression. Genes Dev 17: 88–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Li E (2006) Establishment and maintenance of DNA methylation patterns in mammals. Curr Top Microbiol Immunol 301: 179–201 [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Hiraoka Y (2001) Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr Biol 11: 1618–1623 [DOI] [PubMed] [Google Scholar]
- d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP (2003) A DNA damage checkpoint response in telomere-initiated senescence. Nature 426: 194–198 [DOI] [PubMed] [Google Scholar]
- Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL (2009) Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 459: 802–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin D, Kantrow SM, Liberatore RA, Zakian VA (2000) Telomere folding is required for the stable maintenance of telomere position effects in yeast. Mol Cell Biol 20: 7991–8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Meller VH (2006) Non-coding RNA in fly dosage compensation. Trends Biochem Sci 31: 526–532 [DOI] [PubMed] [Google Scholar]
- Doheny JG, Mottus R, Grigliatti TA (2008) Telomeric position effect—a third silencing mechanism in eukaryotes. PLoS One 3: e3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bendala J, McWhir J (2004) Enhanced gene targeting frequency in ES cells with low genomic methylation levels. Transgenic Res 13: 69–74 [DOI] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A (2002) Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem 277: 28368–28371 [DOI] [PubMed] [Google Scholar]
- Ekwall K, Javerzat JP, Lorentz A, Schmidt H, Cranston G, Allshire R (1995) The chromodomain protein Swi6: a key component at fission yeast centromeres. Science 269: 1429–1431 [DOI] [PubMed] [Google Scholar]
- Ekwall K, Nimmo ER, Javerzat JP, Borgstrom B, Egel R, Cranston G, Allshire R (1996) Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J Cell Sci 109(Part 11): 2637–2648 [DOI] [PubMed] [Google Scholar]
- Epel ES, Blackburn EH, Lin J, Dhabhar FS, Adler NE, Morrow JD, Cawthon RM (2004) Accelerated telomere shortening in response to life stress. Proc Natl Acad Sci USA 101: 17312–17315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanti L, Giovinazzo G, Berloco M, Pimpinelli S (1998) The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell 2: 527–538 [DOI] [PubMed] [Google Scholar]
- Fingerman IM, Li HC, Briggs SD (2007) A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: identification of a new trans-histone pathway. Genes Dev 21: 2018–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores I, Canela A, Vera E, Tejera A, Cotsarelis G, Blasco MA (2008) The longest telomeres: a general signature of adult stem cell compartments. Genes Dev 22: 654–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores I, Cayuela ML, Blasco MA (2005) Effects of telomerase and telomere length on epidermal stem cell behavior. Science 309: 1253–1256 [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, Iyer NG, Perez-Rosado A, Calvo E, Lopez JA, Cano A, Calasanz MJ, Colomer D, Piris MA, Ahn N, Imhof A et al. (2005) Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 37: 391–400 [DOI] [PubMed] [Google Scholar]
- Frydrychova RC, Mason JM, Archer TK (2008) HP1 is distributed within distinct chromatin domains at Drosophila telomeres. Genetics 180: 121–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Deplus R, Kouzarides T (2003) The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res 31: 2305–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cao I, Garcia-Cao M, Tomas-Loba A, Martin-Caballero J, Flores JM, Klatt P, Blasco MA, Serrano M (2006) Increased p53 activity does not accelerate telomere-driven ageing. EMBO Rep 7: 546–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cao M, O'Sullivan R, Peters AH, Jenuwein T, Blasco MA (2004) Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet 36: 94–99 [DOI] [PubMed] [Google Scholar]
- Gartenberg MR, Neumann FR, Laroche T, Blaszczyk M, Gasser SM (2004) Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell 119: 955–967 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Suarez E, Samper E, Flores JM, Blasco MA (2000) Telomerase-deficient mice with short telomeres are resistant to skin tumorigenesis. Nat Genet 26: 114–117 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Suarez E, Samper E, Ramirez A, Flores JM, Martin-Caballero J, Jorcano JL, Blasco MA (2001) Increased epidermal tumors and increased skin wound healing in transgenic mice overexpressing the catalytic subunit of telomerase, mTERT, in basal keratinocytes. EMBO J 20: 2619–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S, Blasco MA (2005) Role of Rb family in the epigenetic definition of chromatin. Cell Cycle 4: 752–755 [DOI] [PubMed] [Google Scholar]
- Gonzalo S, Garcia-Cao M, Fraga MF, Schotta G, Peters AH, Cotter SE, Eguia R, Dean DC, Esteller M, Jenuwein T, Blasco MA (2005) Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol 7: 420–428 [DOI] [PubMed] [Google Scholar]
- Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA (2006) DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol 8: 416–424 [DOI] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA (1990) Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762 [DOI] [PubMed] [Google Scholar]
- Greenwell PW, Kronmal SL, Porter SE, Gassenhuber J, Obermaier B, Petes TD (1995) TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell 82: 823–829 [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T (1999) Mammalian telomeres end in a large duplex loop. Cell 97: 503–514 [DOI] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC (2007) A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Hande MP, Samper E, Lansdorp P, Blasco MA (1999) Telomere length dynamics and chromosomal instability in cells derived from telomerase null mice. J Cell Biol 144: 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour JW, Dean DC (2000a) Chromatin remodeling and Rb activity. Curr Opin Cell Biol 12: 685–689 [DOI] [PubMed] [Google Scholar]
- Harbour JW, Dean DC (2000b) The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev 14: 2393–2409 [DOI] [PubMed] [Google Scholar]
- Harley CB (2008) Telomerase and cancer therapeutics. Nat Rev Cancer 8: 167–179 [DOI] [PubMed] [Google Scholar]
- Harley CB, Futcher AB, Greider CW (1990) Telomeres shorten during ageing of human fibroblasts. Nature 345: 458–460 [DOI] [PubMed] [Google Scholar]
- Hartig JV, Tomari Y, Forstemann K (2007) piRNAs—the ancient hunters of genome invaders. Genes Dev 21: 1707–1713 [DOI] [PubMed] [Google Scholar]
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M (1995) Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80: 583–592 [DOI] [PubMed] [Google Scholar]
- Hediger F, Berthiau AS, van Houwe G, Gilson E, Gasser SM (2006) Subtelomeric factors antagonize telomere anchoring and Tel1-independent telomere length regulation. EMBO J 25: 857–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Strong MA, Hao LY, Greider CW (2001) The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107: 67–77 [DOI] [PubMed] [Google Scholar]
- Herrera E, Martinez AC, Blasco MA (2000) Impaired germinal center reaction in mice with short telomeres. EMBO J 19: 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera E, Samper E, Martin-Caballero J, Flores JM, Lee HW, Blasco MA (1999) Disease states associated with telomerase deficiency appear earlier in mice with short telomeres. EMBO J 18: 2950–2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800 [DOI] [PubMed] [Google Scholar]
- Jaco I, Canela A, Vera E, Blasco MA (2008) Centromere mitotic recombination in mammalian cells. J Cell Biol 181: 885–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Kayne PS, Kahn ES, Grunstein M (1990) Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 87: 6286–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Metz A, Govin J, Vigneron M, Turner BM, Khochbin S, Vourc'h C (2004) Stress-induced transcription of satellite III repeats. J Cell Biol 164: 25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B, Su H, Bhat A, Lei H, Bajko J, Hevi S, Baltus GA, Kadam S, Zhai H, Valdez R, Gonzalo S, Zhang Y, Li E, Chen T (2008) The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet 4: e1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Francesconi S, Collura A, Schramke V, Ishikawa F, Baldacci G, Geli V (2003) The fission yeast spSet1p is a histone H3-K4 methyltransferase that functions in telomere maintenance and DNA repair in an ATM kinase Rad3-dependent pathway. J Mol Biol 326: 1081–1094 [DOI] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F (2001) spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol 11: 1624–1630 [DOI] [PubMed] [Google Scholar]
- Kanoh J, Sadaie M, Urano T, Ishikawa F (2005) Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr Biol 15: 1808–1819 [DOI] [PubMed] [Google Scholar]
- Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y (2005) RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science 309: 467–469 [DOI] [PubMed] [Google Scholar]
- Kimura A, Umehara T, Horikoshi M (2002) Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat Genet 32: 370–377 [DOI] [PubMed] [Google Scholar]
- Kimura H, Nakamura T, Ogawa T, Tanaka S, Shiota K (2003) Transcription of mouse DNA methyltransferase 1 (Dnmt1) is regulated by both E2F-Rb-HDAC-dependent and -independent pathways. Nucleic Acids Res 31: 3101–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA (2007) Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res 35: 5430–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koering CE, Pollice A, Zibella MP, Bauwens S, Puisieux A, Brunori M, Brun C, Martins L, Sabatier L, Pulitzer JF, Gilson E (2002) Human telomeric position effect is determined by chromosomal context and telomeric chromatin integrity. EMBO Rep 3: 1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauskopf A, Blackburn EH (1996) Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature 383: 354–357 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, Li E, Matsuda Y, Kimura T, Okabe M, Sakaki Y, Sasaki H, Nakano T (2008) DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22: 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrion G, Boakye KA, Lustig AJ (1992) C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol Cell Biol 12: 5159–5173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120 [DOI] [PubMed] [Google Scholar]
- Lanza RP, Cibelli JB, Blackwell C, Cristofalo VJ, Francis MK, Baerlocher GM, Mak J, Schertzer M, Chavez EA, Sawyer N, Lansdorp PM, West MD (2000) Extension of cell life-span and telomere length in animals cloned from senescent somatic cells. Science 288: 665–669 [DOI] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH (2003) Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 13: 1192–1200 [DOI] [PubMed] [Google Scholar]
- Levis RW, Ganesan R, Houtchens K, Tolar LA, Sheen FM (1993) Transposons in place of telomeric repeats at a Drosophila telomere. Cell 75: 1083–1093 [DOI] [PubMed] [Google Scholar]
- Levy DL, Blackburn EH (2004) Counting of Rif1p and Rif2p on Saccharomyces cerevisiae telomeres regulates telomere length. Mol Cell Biol 24: 10857–10867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D (2005) Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell 121: 515–527 [DOI] [PubMed] [Google Scholar]
- Lipinski MM, Jacks T (1999) The retinoblastoma gene family in differentiation and development. Oncogene 18: 7873–7882 [DOI] [PubMed] [Google Scholar]
- Liu L, Bailey SM, Okuka M, Munoz P, Li C, Zhou L, Wu C, Czerwiec E, Sandler L, Seyfang A, Blasco MA, Keefe DL (2007) Telomere lengthening early in development. Nat Cell Biol 9: 1436–1441 [DOI] [PubMed] [Google Scholar]
- Louis EJ (1995) The chromosome ends of Saccharomyces cerevisiae. Yeast 11: 1553–1573 [DOI] [PubMed] [Google Scholar]
- Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J (2008) The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell 32: 465–477 [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, Plath K, Hochedlinger K (2007) Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 1: 55–70 [DOI] [PubMed] [Google Scholar]
- Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser SM (1996) Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev 10: 1796–1811 [DOI] [PubMed] [Google Scholar]
- Maloisel L, Rossignol JL (1998) Suppression of crossing-over by DNA methylation in Ascobolus. Genes Dev 12: 1381–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S, Brevet V, Gilson E (1999) Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J 18: 3509–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S, Gilson E, Shore D (1997) A protein-counting mechanism for telomere length regulation in yeast. Science 275: 986–990 [DOI] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Tejera A, Schoeftner S, Ortega S, Serrano M, Blasco MA (2009) Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell 4: 141–154 [DOI] [PubMed] [Google Scholar]
- Martadinata H, Phan AT (2009) Structure of propeller-type parallel-stranded RNA G-quadruplexes, formed by human telomeric RNA sequences in K+ solution. J Am Chem Soc 131: 2570–2578 [DOI] [PubMed] [Google Scholar]
- Martino F, Kueng S, Robinson P, Tsai-Pflugfelder M, van Leeuwen F, Ziegler M, Cubizolles F, Cockell MM, Rhodes D, Gasser SM (2009) Reconstitution of yeast silent chromatin: multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol Cell 33: 323–334 [DOI] [PubMed] [Google Scholar]
- Mason JM, Biessmann H (1995) The unusual telomeres of Drosophila. Trends Genet 11: 58–62 [DOI] [PubMed] [Google Scholar]
- Mason JM, Frydrychova RC, Biessmann H (2008) Drosophila telomeres: an exception providing new insights. Bioessays 30: 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JM, Ransom J, Konev AY (2004) A deficiency screen for dominant suppressors of telomeric silencing in Drosophila. Genetics 168: 1353–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PJ, Wilson DB, Bessler M (2005) Dyskeratosis congenita—a disease of dysfunctional telomere maintenance. Curr Mol Med 5: 159–170 [DOI] [PubMed] [Google Scholar]
- McCabe MT, Davis JN, Day ML (2005) Regulation of DNA methyltransferase 1 by the pRb/E2F1 pathway. Cancer Res 65: 3624–3632 [DOI] [PubMed] [Google Scholar]
- Meneghini MD, Wu M, Madhani HD (2003) Conserved histone variant H2A. Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112: 725–736 [DOI] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF (2008) SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452: 492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Kanoh J, Saito M, Ishikawa F (2008) Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science 320: 1341–1344 [DOI] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D (2004) Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119: 789–802 [DOI] [PubMed] [Google Scholar]
- Muntoni A, Reddel RR (2005) The first molecular details of ALT in human tumor cells. Hum Mol Genet 14 (Spec No. 2): R191–R196 [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S (2008) Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol 26: 101–106 [DOI] [PubMed] [Google Scholar]
- Nakayama J, Klar AJ, Grewal SI (2000) A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell 101: 307–317 [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113 [DOI] [PubMed] [Google Scholar]
- Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K (2002a) Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev 16: 1518–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Xu RM, Zhang Y, Struhl K (2002b) Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J Biol Chem 277: 34655–34657 [DOI] [PubMed] [Google Scholar]
- Niida H, Shinkai Y, Hande MP, Matsumoto T, Takehara S, Tachibana M, Oshimura M, Lansdorp PM, Furuichi Y (2000) Telomere maintenance in telomerase-deficient mouse embryonic stem cells: characterization of an amplified telomeric DNA. Mol Cell Biol 20: 4115–4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo ER, Pidoux AL, Perry PE, Allshire RC (1998) Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature 392: 825–828 [DOI] [PubMed] [Google Scholar]
- Nislow C, Ray E, Pillus L (1997) SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol Biol Cell 8: 2421–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal SI (2004) RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet 36: 1174–1180 [DOI] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, Wright SM, Mills KD, Bonni A, Yankner BA, Scully R, Prolla TA, Alt FW, Sinclair DA (2008) SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell 135: 907–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikemus SR, Queiroz-Machado J, Lai K, McGinnis N, Sunkel C, Brodsky MH (2006) Epigenetic telomere protection by Drosophila DNA damage response pathways. PLoS Genet 2: e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E (1998) Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet 19: 219–220 [DOI] [PubMed] [Google Scholar]
- Osada S, Sutton A, Muster N, Brown CE, Yates JR III, Sternglanz R, Workman JL (2001) The yeast SAS (something about silencing) protein complex contains a MYST-type putative acetyltransferase and functions with chromatin assembly factor ASF1. Genes Dev 15: 3155–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T (2008) How shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334 [DOI] [PubMed] [Google Scholar]
- Palladino F, Laroche T, Gilson E, Axelrod A, Pillus L, Gasser SM (1993) SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell 75: 543–555 [DOI] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C (2008) Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 32: 232–246 [DOI] [PubMed] [Google Scholar]
- Pardue ML, DeBaryshe PG (2003) Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres. Annu Rev Genet 37: 485–511 [DOI] [PubMed] [Google Scholar]
- Pardue ML, Debaryshe PG (2008) Drosophila telomeres: a variation on the telomerase theme. Fly (Austin) 2: 101–110 [DOI] [PubMed] [Google Scholar]
- Pardue ML, Rashkova S, Casacuberta E, DeBaryshe PG, George JA, Traverse KL (2005) Two retrotransposons maintain telomeres in Drosophila. Chromosome Res 13: 443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Cosgrove MS, Youngman E, Wolberger C, Boeke JD (2002) A core nucleosome surface crucial for transcriptional silencing. Nat Genet 32: 273–279 [DOI] [PubMed] [Google Scholar]
- Pauler FM, Koerner MV, Barlow DP (2007) Silencing by imprinted noncoding RNAs: is transcription the answer? Trends Genet 23: 284–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Lee JT (2008) X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet 42: 733–772 [DOI] [PubMed] [Google Scholar]
- Pedram M, Sprung CN, Gao Q, Lo AW, Reynolds GE, Murnane JP (2006) Telomere position effect and silencing of transgenes near telomeres in the mouse. Mol Cell Biol 26: 1865–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrini B, Piacentini L, Fanti L, Altieri F, Chichiarelli S, Berloco M, Turano C, Ferraro A, Pimpinelli S (2004) HP1 controls telomere capping, telomere elongation, and telomere silencing by two different mechanisms in Drosophila. Mol Cell 15: 467–476 [DOI] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, Martens JH, Jenuwein T (2003) Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12: 1577–1589 [DOI] [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107: 323–337 [DOI] [PubMed] [Google Scholar]
- Pogribny IP, Ross SA, Tryndyak VP, Pogribna M, Poirier LA, Karpinets TV (2006) Histone H3 lysine 9 and H4 lysine 20 trimethylation and the expression of Suv4-20h2 and Suv-39h1 histone methyltransferases in hepatocarcinogenesis induced by methyl deficiency in rats. Carcinogenesis 27: 1180–1186 [DOI] [PubMed] [Google Scholar]
- Porter SE, Greenwell PW, Ritchie KB, Petes TD (1996) The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res 24: 582–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffa GD, Siriaco G, Cugusi S, Ciapponi L, Cenci G, Wojcik E, Gatti M (2009) The Drosophila modigliani (moi) gene encodes a HOAP-interacting protein required for telomere protection. Proc Natl Acad Sci USA 106: 2271–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall A, Griffith JD (2009) Structure of long telomeric RNA transcripts: the G-rich RNA forms a compact repeating structure containing G-quartets. J Biol Chem 284: 13980–13986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Bartel DP (2002) Small RNAs correspond to centromere heterochromatic repeats. Science 297: 1831. [DOI] [PubMed] [Google Scholar]
- Riethman H, Ambrosini A, Paul S (2005) Human subtelomere structure and variation. Chromosome Res 13: 505–515 [DOI] [PubMed] [Google Scholar]
- Rizzi N, Denegri M, Chiodi I, Corioni M, Valgardsdottir R, Cobianchi F, Riva S, Biamonti G (2004) Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol Biol Cell 15: 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph KL, Chang S, Lee HW, Blasco M, Gottlieb GJ, Greider C, DePinho RA (1999) Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell 96: 701–712 [DOI] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC (2006) Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 20: 2214–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samper E, Flores JM, Blasco MA (2001) Restoration of telomerase activity rescues chromosomal instability and premature aging in Terc−/− mice with short telomeres. EMBO Rep 2: 800–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Segundo PA, Roeder GS (2000) Role for the silencing protein Dot1 in meiotic checkpoint control. Mol Biol Cell 11: 3601–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin KY, Cheung P, Gilison D, Lee E, Tennen RI, Wang E, Artandi MK, Oro AE, Artandi SE (2005) Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature 436: 1048–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky M, Kravchuk O, Melnikova L, Georgiev P (2002) Heterochromatin protein 1 is involved in control of telomere elongation in Drosophila melanogaster. Mol Cell Biol 22: 3204–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V (2006) Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev 20: 345–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaetzlein S, Lucas-Hahn A, Lemme E, Kues WA, Dorsch M, Manns MP, Niemann H, Rudolph KL (2004) Telomere length is reset during early mammalian embryogenesis. Proc Natl Acad Sci USA 101: 8034–8038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA (2008) Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol 10: 228–236 [DOI] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T (2004) A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 18: 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, Callen E, Celeste A, Pagani M, Opravil S, De La Rosa-Velazquez IA, Espejo A, Bedford MT, Nussenzweig A, Busslinger M, Jenuwein T (2008) A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev 22: 2048–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian MD, Zhang K, Grunstein M (2005) Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol Cell 19: 271–277 [DOI] [PubMed] [Google Scholar]
- Shanower GA, Muller M, Blanton JL, Honti V, Gyurkovics H, Schedl P (2005) Characterization of the grappa gene, the Drosophila histone H3 lysine 79 methyltransferase. Genetics 169: 173–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shia WJ, Li B, Workman JL (2006) SAS-mediated acetylation of histone H4 Lys 16 is required for H2A. Z incorporation at subtelomeric regions in Saccharomyces cerevisiae. Genes Dev 20: 2507–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shia WJ, Osada S, Florens L, Swanson SK, Washburn MP, Workman JL (2005) Characterization of the yeast trimeric-SAS acetyltransferase complex. J Biol Chem 280: 11987–11994 [DOI] [PubMed] [Google Scholar]
- Shpiz S, Kwon D, Uneva A, Kim M, Klenov M, Rozovsky Y, Georgiev P, Savitsky M, Kalmykova A (2007) Characterization of Drosophila telomeric retroelement TAHRE: transcription, transpositions, and RNAi-based regulation of expression. Mol Biol Evol 24: 2535–2545 [DOI] [PubMed] [Google Scholar]
- Silva E, Tiong S, Pedersen M, Homola E, Royou A, Fasulo B, Siriaco G, Campbell SD (2004) ATM is required for telomere maintenance and chromosome stability during Drosophila development. Curr Biol 14: 1341–1347 [DOI] [PubMed] [Google Scholar]
- Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W (2008) MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol 15: 259–267 [DOI] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K (2008) Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell 2: 230–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert S, Shay JW, Wright WE (2004) Modification of subtelomeric DNA. Mol Cell Biol 24: 4571–4580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M (1997) SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev 11: 83–93 [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI (2007) SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128: 491–504 [DOI] [PubMed] [Google Scholar]
- Suka N, Luo K, Grunstein M (2002) Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet 32: 378–383 [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD (2002) Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104–108 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D (1999) An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99: 735–745 [DOI] [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J (2004) Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117: 323–335 [DOI] [PubMed] [Google Scholar]
- Teng SC, Chang J, McCowan B, Zakian VA (2000) Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol Cell 6: 947–952 [DOI] [PubMed] [Google Scholar]
- Tham WH, Zakian VA (2002) Transcriptional silencing at Saccharomyces telomeres: implications for other organisms. Oncogene 21: 512–521 [DOI] [PubMed] [Google Scholar]
- Thon G, Klar AJ (1992) The clr1 locus regulates the expression of the cryptic mating-type loci of fission yeast. Genetics 131: 287–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting AH, McGarvey KM, Baylin SB (2006) The cancer epigenome—components and functional correlates. Genes Dev 20: 3215–3231 [DOI] [PubMed] [Google Scholar]
- Tomas-Loba A, Flores I, Fernandez-Marcos PJ, Cayuela ML, Maraver A, Tejera A, Borras C, Matheu A, Klatt P, Flores JM, Vina J, Serrano M, Blasco MA (2008) Telomerase reverse transcriptase delays aging in cancer-resistant mice. Cell 135: 609–622 [DOI] [PubMed] [Google Scholar]
- Tomita K, Cooper JP (2008) Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev 22: 3461–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommerup H, Dousmanis A, de Lange T (1994) Unusual chromatin in human telomeres. Mol Cell Biol 14: 5777–5785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryndyak VP, Kovalchuk O, Pogribny IP (2006) Loss of DNA methylation and histone H4 lysine 20 trimethylation in human breast cancer cells is associated with aberrant expression of DNA methyltransferase 1, Suv4-20h2 histone methyltransferase and methyl-binding proteins. Cancer Biol Ther 5: 65–70 [DOI] [PubMed] [Google Scholar]
- Tsakiri KD, Cronkhite JT, Kuan PJ, Xing C, Raghu G, Weissler JC, Rosenblatt RL, Shay JW, Garcia CK (2007) Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc Natl Acad Sci USA 104: 7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes AM, Andrew T, Gardner JP, Kimura M, Oelsner E, Cherkas LF, Aviv A, Spector TD (2005) Obesity, cigarette smoking, and telomere length in women. Lancet 366: 662–664 [DOI] [PubMed] [Google Scholar]
- van Leeuwen F, Gafken PR, Gottschling DE (2002) Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109: 745–756 [DOI] [PubMed] [Google Scholar]
- van Leeuwen F, Gottschling DE (2002) Genome-wide histone modifications: gaining specificity by preventing promiscuity. Curr Opin Cell Biol 14: 756–762 [DOI] [PubMed] [Google Scholar]
- van Overveld PG, Lemmers RJ, Sandkuijl LA, Enthoven L, Winokur ST, Bakels F, Padberg GW, van Ommen GJ, Frants RR, van der Maarel SM (2003) Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet 35: 315–317 [DOI] [PubMed] [Google Scholar]
- Vera E, Canela A, Fraga MF, Esteller M, Blasco MA (2008) Epigenetic regulation of telomeres in human cancer. Oncogene 27: 6817–6833 [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D (2004) RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama T, Shinkai Y, Tamashiro KL, Niida H, Blanchard DC, Blanchard RJ, Ogura A, Tanemura K, Tachibana M, Perry AC, Colgan DF, Mombaerts P, Yanagimachi R (2000) Cloning of mice to six generations. Nature 407: 318–319 [DOI] [PubMed] [Google Scholar]
- Walter MF, Biessmann MR, Benitez C, Torok T, Mason JM, Biessmann H (2007) Effects of telomere length in Drosophila melanogaster on life span, fecundity, and fertility. Chromosoma 116: 41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MF, Jang C, Kasravi B, Donath J, Mechler BM, Mason JM, Biessmann H (1995) DNA organization and polymorphism of a wild-type Drosophila telomere region. Chromosoma 104: 229–241 [DOI] [PubMed] [Google Scholar]
- Weinberg RA (1995) The retinoblastoma protein and cell cycle control. Cell 81: 323–330 [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Cassady JP, Jaenisch R (2008) c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell 2: 10–12 [DOI] [PubMed] [Google Scholar]
- Wright JH, Gottschling DE, Zakian VA (1992) Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes Dev 6: 197–210 [DOI] [PubMed] [Google Scholar]
- Xu Y, Kimura T, Komiyama M (2008) Human telomere RNA and DNA form an intermolecular G-quadruplex. Nucleic Acids Symp Ser (Oxf) 52: 169–170 [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Calado RT, Ly H, Kajigaya S, Baerlocher GM, Chanock SJ, Lansdorp PM, Young NS (2005) Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N Engl J Med 352: 1413–1424 [DOI] [PubMed] [Google Scholar]
- Yehezkel S, Segev Y, Viegas-Pequignot E, Skorecki K, Selig S (2008) Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum Mol Genet 17: 2776–2789 [DOI] [PubMed] [Google Scholar]
- Yin H, Lin H (2007) An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature 450: 304–308 [DOI] [PubMed] [Google Scholar]
- Zellinger B, Riha K (2007) Composition of plant telomeres. Biochim Biophys Acta 1769: 399–409 [DOI] [PubMed] [Google Scholar]