Abstract
This review summarizes the essential characteristics of matrix-assisted laser desorption/ionization (MALDI) time-of-flight mass spectrometry (TOF MS), especially as they relate to its applications in quantitative analysis. Approaches to quantification by MALDI-TOF MS are presented and published applications are critically reviewed.
Keywords: quantification, quantitative analysis, MALDI, mass spectrometry, biomarkers
OVERVIEW
Since its inception in 1987 [1], matrix-assisted laser desorption/ionization (MALDI) time-of-flight mass spectrometry (TOF MS) has been applied for the analysis of a wide range of biomolecules. Initial applications were almost exclusively for the qualitative analysis of biopolymers because MALDI-TOF MS provided a fast and accurate approach to molecular mass and purity information: Is my material the right mass and is it free from contaminants? Because of the speed of analysis, ease of use, relative low equipment cost, ease of data interpretation and limited potential for cross-contamination between samples/users, MALDI-TOF MS systems were considered walk-up instruments where investigators run their own samples and determine, within minutes, whether they were on the right track with their work.
There were, however, two specific areas of application where MALDI-TOF MS was initially viewed as impractical: i.e. the analysis of low mass analytes and quantitative applications. Low mass analysis is complicated by the vast molar excess of the matrix that can swamp any analyte-specific signal in the low m/z region of the spectrum. Quantitative analysis was viewed as implausible because crystallization does not yield a uniform distribution of the analyte and matrix across the target surface and this gives rise to regions where the analyte signal is especially intense relative to other locations on the target surface. MALDI MS was therefore viewed as inherently irreproducible because, for a given amount of analyte loaded onto the target, the measured ion intensity varies.
Subsequently, it has been shown that both these perceived limitations can be overcome and quantification can be routine, both for low and high mass analytes. Now, an extensive list of published applications includes quantification of amino acids, lipids, natural products, drugs, polymers, herbicides, metabolites, toxins, oligonucleotides, carbohydrates, peptides and proteins. (Selected applications from these areas are discussed at the end of this review.) Here, we focus on the quantitative applications of MALDI and illustrate applications relating to both low and high-molecular mass analytes. We also discuss the strategies for applying MALDI to relative and absolute quantification. While MALDI is most commonly combined with TOF analysers, other analyser options are possible and applications employing these are also presented. Our aim is not to provide an exhaustive catalogue of published applications, but to discuss the general principles and to illustrate the central considerations in quantitative MALDI by way of examples. As with any approach, MALDI offers some advantages and some limitations relative to the available alternatives. Performance characteristics of the system and the assay such as throughput, precision, accuracy, cost, sensitivity, selectivity, linear dynamic range, and, of course, practicality, are discussed for some of these applications.
A REVIEW OF THE ESSENTIAL FEATURES OF MALDI AS THEY RELATE TO QUANTIFICATION
For a typical MALDI analysis, solutions of a sample (pure or a mixture; ∼10 µM) and matrix (∼10 mM) are pre-mixed and a small volume (∼1 µl) is applied directly to the sample plate. (The matrix is selected such that it has a large extinction coefficient at the emission wavelength of the laser.) The solvent is generally allowed to evaporate at room temperature and pressure to yield a heterogeneous crystalline surface. Inside the mass spectrometer the laser is pulsed onto the crystalline surface and the matrix immediately sublimes, concomitantly desorbing and ionizing the analyte molecules. The m/z values for the analyte ions are then measured. The MALDI mass spectrum is a plot of intensities on the ordinate axis and the corresponding mass/charge values on the x-axis. The intensity scale is the relative abundance of ions, but generally the region below about m/z 500 is not shown because it is saturated with signals from matrix-derived ions. MALDI mass spectra of pure compounds are normally dominated by a single ion corresponding to the protonated molecule; multiply charged ions are rare, except for very large molecules and even then they are usually of low relative abundance. MALDI spectra of mixtures typically show multiple ions corresponding to the protonated forms of some if not all of the molecular components present in the sample. It is noteworthy that peak heights for equimolar loadings of different analytes may vary significantly. The proclivity of MALDI to yield singly charged ions is in contrast to electrospray and offers significant advantages in quantification, especially for mixtures.
Characteristically, there is substantial variability in the noise level, baseline and peak intensities in a collection of MALDI spectra generated from the same sample. Variations in ion current are observed with consecutive laser shots fired at the same position on the target surface (shot-to-shot reproducibility), across different locations on the target surface (region-to-region reproducibility) and between identical loadings of the same sample onto different targets (sample-to-sample reproducibility). To minimize this variability, multiple spectra are usually acquired from different locations across the target surface and these are averaged (or added) to yield a more representative spectrum.
Fluctuations in laser power and changes in detector response alter signal intensity, but the primary contributor to variability is heterogeneous incorporation of the analyte into the co-crystallized matrix–analyte complex. This results in ‘hot-spots’ on the target surface where the ratio of analyte to matrix is optimal and where the analyte signal is high relative to other locations. This variability can be circumvented, at least in part, by pre-mixing the sample and matrix solutions and by facilitating faster crystallization times. Faster crystallization generates smaller crystals and a more homogeneous incorporation of the analytes into the crystalline lattice. For example, when 2,5-dihydroxybenzoic acid (DHB) is allowed to crystallize slowly (i.e. from predominantly aqueous solutions at room temp and pressure) large needle-like crystals form and sample incorporation is highly variable. When crystallization is rapid (i.e. when the matrix is prepared in a volatile solvent and/or the target is dried at elevated temperature and/or reduced pressure) the resulting crystals are small and crystallization of the analyte–matrix is much more uniform [2].
Competitive ionization/ion suppression is an additional factor that can obliterate any attempt to quantify by MALDI, especially in complex samples. In a mixture, some analytes will have higher affinities for charge than others (e.g. R-terminated peptides > K-terminated peptides) and will more successfully compete for the available protons [3]. Therefore, when quantifying across a series of samples it is essential to keep the sample composition (or sample matrix) constant. Sample preparation methods should aim to reduce the complexity of the sample, thereby minimizing the background and eliminating potentially interfering peaks, but at the same time, these methods must be highly reproducible and minimize the potential to introduce contaminants that result in higher variability and adversely influence signal intensity (i.e. ion suppression). Although ion suppression is not unique to MALDI, in electrospray LC-MS applications online separation helps minimize the potential for other sample components to influence the ion current of the target analyte. In contrast, few quantitative MALDI applications employ ‘online’ separation of the analytes; rather, multiple analytes are present on the target at the same time and actively compete for the available charge for each desorption/ionization event (i.e. laser shot).
There are, however, several very positive attributes of MALDI, not the least of which is its sensitivity. There have been several estimates of the amount of material required to generate a spectrum, but it is clear that attomoles or less are sufficient to generate a good quality spectrum [4]. A quick survey of the literature also illustrates the remarkable versatality of this ionization method: i.e. thermally labile, low mass analytes; involatile, high mass biopolymers; even alkanes and polyethylenes [5] can all be ionized by MALDI. Additional advantages of the approach include the ability to analyse complex mixtures, the potential for high-throughput, the speed of the analysis, ease of automation and the low cost per analysis.
In summary, MALDI offers some important attributes, but its application to quantitative analysis requires careful optimization of the experimental parameters and a good understanding of how confounding factors could compromise a study. In particular, sample preparation (e.g. choice of matrix compound, concentration, solvents and crystallization conditions) is critical and must be optimized in order to reduce the variability introduced during this step. It is also important to acquire and average many single-shot spectra from several positions within a given sample spot to gain representative sample data. Ideally, the laser power should be automatically adjusted to limit the acceptable analyte and internal standard signal intensity to below saturation, but above background noise. Consequently, criteria for spectral acceptance based on minimum and maximum peak height and the signal-to-noise ratio should be established and adhered to before the spectra are averaged.
QUANTITATIVE ASSAYS IN BIOLOGICAL INVESTIGATIONS: GENERAL CONSIDERATIONS
Why quantify?
Qualitative analyses of complex mixtures are often made, but without a quantitative dimension these determinations are of limited value and can be misleading. For example, by generating a list of the components in a sample the analyst is indicating the presence of specific species and implying the absence of others. But it is important to appreciate that the exclusion of an analyte from the list may simply mean that it was not recovered, was not characterized, or that it was present below its limit of detection. Lists must therefore be interpreted with caution and this is perhaps no more true than in proteomics: a field rapidly transitioning from a qualitative to quantitative science. When analysing complex biological samples it is important to have reliable measures of absolute analyte levels, or at least precise determinations of the differences in analyte levels between two or more samples.
Quantitative relationships
Quantification requires that the magnitude of the measured property—in this case the ion current—is proportional to the concentration or amount of the analyte in the sample matrix. Ideally, from sample to sample, the process should be reproducible and the response identical for equal amounts of analyte. However, as already discussed, there is significant variability in the MALDI signal even when the same sample is analysed repeatedly. The process of applying the sample to the target is irreproducible, the target surface is heterogeneous and changes in laser intensity along with fluctuations in detector response all contribute to signal variability. There is therefore a poor correlation between uncorrected signal intensity and analyte amount. But, there are ways to optimize the experimental conditions and design experiments that allow for meaningful quantitative comparisons by MALDI. These approaches are common to other areas of quantitative analysis and their essential features are reviewed below.
Relative quantification without an internal standard (or profile analysis)
For a given sample run repeatedly, the relative signal intensities within the mass spectrum are surprisingly constant. Because of this, comparisons can be made between similar samples (e.g. a series of plasma samples) and the changes in relative signal intensities against an essentially ‘constant’ background can provide useful information. No meaning is ascribed to absolute peak intensities and typically hundreds of unidentified features per profile are recorded without the incorporation of any internal standards. The approach utilizes classification algorithms to examine spectra in a reproducible manner in order to determine the differences between populations. The most common iteration of this approach is MALDI protein profiling [6–13].
MALDI profiling offers exceptionally high-throughput and has therefore become an attractive tool for the discovery and validation of biomarkers. Throughput is critical because for any approach to be of value in clinical science it must be feasible to analyse hundreds (or even thousands) of samples so that sufficient representation of the natural population variation in human-derived samples can be determined. In addition, the method must be sufficiently precise to observe relative differences between distinct populations.
Profiling has now been applied in thousands of studies, on a broad range of sample types, and across a wide range of disease conditions. (Some specific examples using this approach are discussed later in this review.) Early studies based on this approach showed excellent predictive value, but they were soon criticized, in large part because of problems in the study design and implementation [14–16]. However, profiling can be used to make meaningful comparisons if rigid protocols for both experimental design and data analysis are followed. Specifically, pre-analytic variables must be controlled and the available samples split into a training set (i.e. to develop a classification algorithm) and a blinded test set(s) (i.e. to verify the classification is sensitive and specific).
Albrethsen [17] has reviewed the critical issue of reproducibility in protein profiling and has discussed approaches to improve the analytical performance of MALDI protein profiling including automated sample processing, pre-fractionation, immunocapture, pre-structured target surfaces, standardized matrix (co)crystallization, internal standard peptides, quality-control samples, replicate measurements, and improved algorithms for normalization and peak detection. Protein profiling can be combined with techniques like LC-MALDI and other fractionation approaches to help accommodate the large dynamic range of protein abundances in biological fluids such as serum, plasma and urine. This can enhance coverage, but the reproducibility and throughput of these additional steps must be carefully evaluated. Several strategies have been reported [18–20], but these are yet to be comprehensively evaluated and other reproducible, high-throughput sample fractionation methods are certainly needed.
Relative quantification with an internal standard
With this approach a fixed amount of an exogenous component (i.e. an internal standard) is added to all samples and peak heights or areas of individual endogeneous analytes are measured relative to it. Signals (discrete m/z values) that increase relative to that of the internal standard when two or more samples are compared can be considered to reflect increases in the amounts of the analyte; decreases correspond to reduced levels. Incorporation of the standard adjusts for some of the variability inherent in the process of sample preparation, ionization and ion detection. A single set of samples run under identical conditions, at the same time, and all containing the same amount of internal standard, is considered a single experiment. In biological studies, the objective is frequently to establish statistically significant differences between populations: i.e. to establish that analyte levels in one group are different to another. This exercise can be undertaken without knowing the absolute amounts and by simply comparing relative amounts between two or more samples/groups (e.g. different by environment, drug, age or disease). Because absolute quantities are not determined by this approach, comparisons between different experiments must be made with extreme caution. There is no common point of reference between the two different studies, the studies may have been performed at different times, with different instrument conditions/settings or even on different instruments by different operators.
Although the absolute amount of the internal standard need not be known, it must remain constant, must generate a signal that can be measured precisely (i.e. above the limit of quantification, but not saturated) and must not significantly suppress the signals for the analytes in the sample. In addition, it is important that the mass of the standard be close to that of the analyte(s) of interest, but be distinct from that of any sample components. Multiple analytes can be compared against a single internal standard by this approach, but there will be some compromise in quantitative precision for some.
Absolute quantification with an internal standard
Relative changes can be transformed to absolute amounts through a modification of the above procedure. In addition to the samples under evaluation, a calibration curve is prepared that also contains the same (fixed) amount of the internal standard and varying amounts of a single specific analyte of interest. In this way, the constant of proportionality for one analyte can be established and ion abundance (or intensity) ratios can be converted to absolute amounts. If the objective is to determine absolute amounts for several different species, separate calibration curves should be constructed for each analyte because the constant of proportionality will be different in each instance.
The internal standard serves as a mimic of the analyte and therefore its chemical and physical properties should match those of the analyte as closely as possible. In this way, the internal standard will behave in a manner close to or identical to the analyte at all stages of the assay process (e.g. extraction, crystallization, ionization/ion suppression). Although the conversion from relative to absolute data requires substantial additional effort, absolute quantification allows results to be reliably compared across experiments or between laboratories.
Absolute quantification by standard addition
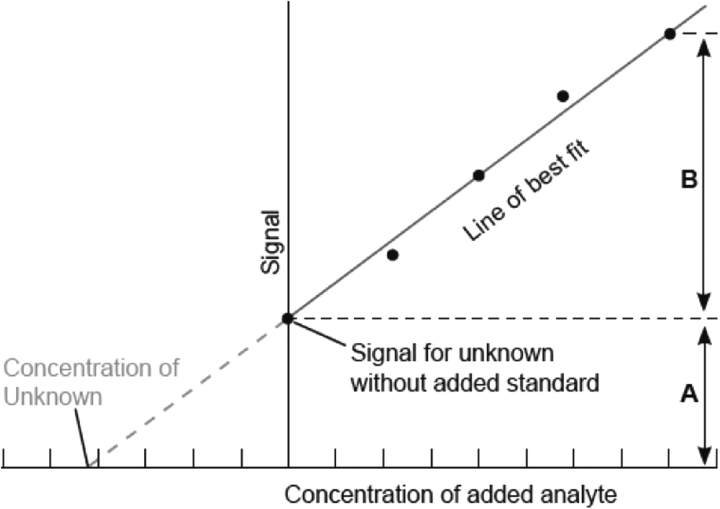
When the matrix is complex or highly variable between samples, as discussed previously, other components can influence instrument response. In these circumstances, standard addition analysis is the preferred approach. Here, each unknown sample is divided into two or more portions and known amount(s) of the analyte are added to these portions (i.e. a spike). The original sample and the spiked samples are then analysed. The samples incorporating the spike(s) will show greater analytical responses than the unadulterated sample due to the additional amounts of analyte added to them. These response differences provide a calibration point (or points) to determine the analyte concentration in the original sample. (The process of calculating the amount in the original sample is shown in Figure 1.)
Figure 1:
This figure illustrates the process of converting analyte signal response to amount by the standard addition method.
FITNESS-FOR-PURPOSE
In biology, the most common objective is to compare analyte values between distinct populations. In this setting, the individual data points are comprised of the analyte values, the associated errors in the analytical method (i.e. instrument errors + sample preparation errors) and the inherent biological variation in the samples. For any analytical method, the ‘purpose’ should be precisely defined and all available resources optimally utilized to address that purpose—nothing more or less. The focus should be on the efficiency to deliver what is required for a specific purpose. Resources should not be squandered on attempting to produce a higher quality product than is necessary, nor should they be under-utilized to yield a method that is inadequate for the intended purpose. One interpretation of this principle is that, given the variations inherent in biological samples, it is unnecessary to measure any analyte with a precision greater than the variability seen in the population. The utility of MALDI as a quantitative tool should be considered in this context; every task is different and what the method can offer—its strengths and weaknesses—must relate to the problem at hand. There are unquestionably instances where MALDI provides extremely valuable data that are well suited to the intended purpose and in subsequent sections we discuss the ways in which quantitative MALDI has been applied to the study of biological samples and comment on their fitness-for-purpose.
THE INTERNAL STANDARD
Internal standards are incorporated into quantitative mass spectrometric analyses to compensate for systematic and random errors during analysis, but their inclusion in MALDI assays, and certainly those aimed at absolute quantification, is arguably more important than in most other applications.
The internal standard is selected based on its ability to mimic the chemical and physical properties of the analyte of interest throughout all stages of the analytical process. For quantification of low molecular mass analytes, either by GC-MS or LC-MS, the optimal internal standard is a stable isotope-labelled form (isotopomer) of the analyte. The same principle holds true for MALDI applications: stable isotope-labelled standards are optimal, at least in the low mass region. Such a standard guarantees identical extraction, crystallization and gas-phase behaviour. But, isotopomers become increasingly impractical as the molecular mass of the analyte increases. For example, to generate a stable isotope labelled form of a protein with a molecular mass of 50 000 and with an isotopic distribution that is distinct from that of its native counterpart (i.e. non-overlapping) is complex and costly. In fact, it is by no means certain that this additional cost and effort is warranted and that it will result in a significant improvement in precision relative to that offered by the careful selection of a more practical alternative. For example, a structural analogue internal standard that maintains the critical solution phase properties (e.g. acid–base properties and solubility) and that is close in mass to the analyte, but sufficiently distinct to allow for appropriate resolution, is certainly more practical to design and prepare, and may not offer any significant compromise in either precision or accuracy. A structural analogue of a peptide can be prepared where amino acids with similar chemical and physical properties are interchanged (e.g. substitution of aspartic acid for glutamic acid or alanine for glycine) and these have been shown to compensate for crystallization irregularities and subsequent desorption and gas phase effects. The importance of a vicinal mass for the internal standard has also been reported based on experiments with a series of acylcarnitines having a fixed charge site and growing alkyl chain length [21]. In the final analysis, however, the acceptable degree of difference between the internal standard and the analyte is often only determined by experimentation and must be viewed in the context of the analytical objective.
In multi-component assays, a single compound sometimes serve as the internal standard for several analytes, but because of their chemical and physical diversity, this can lead to differing responses to variations in experimental conditions. For example, different analytes will give different recoveries during extraction and different responses during crystallization and ionization. One standard is unlikely to optimally correct for all these factors, but the hope is that important biological variations will not be obscured by the sources of variability introduced in the experimental design.
DATA ANALYSIS: SPECIAL CONSIDERATIONS IN PROFILING
Although profiling might not be considered an orthodox example of quantification, it is, nonetheless, a quantitative exercise. Profile analysis is one of the major applications of MALDI-TOF MS and therefore we include discussion of the critical role of the software in the process; to our minds it has rarely received due attention.
It is important that the necessary spectral pre-processing methods are performed in such a way that they do not introduce bias into the resulting data sets. Different mass spectra can show changes in intensity and shifts in the m/z axis, often in a non-linear manner. It is therefore important that mass spectra are standardized before the peak picking process begins, but many commercial algorithms begin the analysis with peak lists generated prior to standardization. Further, in complex mass spectra, peak widths can be influenced by the presence of proximate peaks and the peak picking and the peak intensity measurement process must take these local effects into account. With rigorous spectral pre-processing consisting of regional (in terms of m/z) background determinations and noise estimation, alignment using common peaks in training sample sets and normalization methods using partial ion current techniques, MALDI mass spectra are highly reproducible across laboratories and even across slightly different hardware [22].
Coefficients of variation (CVs) for each m/z value within technical replicates of biological samples such as crude serum vary markedly and typically range from a few percent to unusable. The role of normalization in this process cannot be overestimated. The standard approach to normalization is based on the total ion current (TIC). Here, each spectrum is divided by the sum (or more accurately the square root of the sum of the squared intensities), but this approach is not optimal for several reasons. First, large peaks are the most important contributors in the TIC normalization of spectra, but often they contain little relevant quantitative information, they are saturated, and/or their intensities fluctuate dramatically from sample to sample. When employing TIC normalization, these fluctuations are telegraphed via the normalization process to other peaks, thereby rendering the whole analysis less sensitive and artificially increasing CVs. Even worse, TIC normalization can lead to an over-training of classification when large peaks present in one clinical group are less prevalent in another. (As an example, lysates prepared from tumor, rather than normal tissue, may contain higher levels of haemoglobin and other blood products.) In these circumstances, TIC normalization inappropriately suppresses all peaks in the group. An alternative normalization approach is based on partial ion current (PIC) and uses only a suitably selected subset of peaks (i.e. those whose CVs are below a threshold) for normalization. In its most trivial implementation, highly fluctuating regions (sample to sample) are excluded from the normalization process.
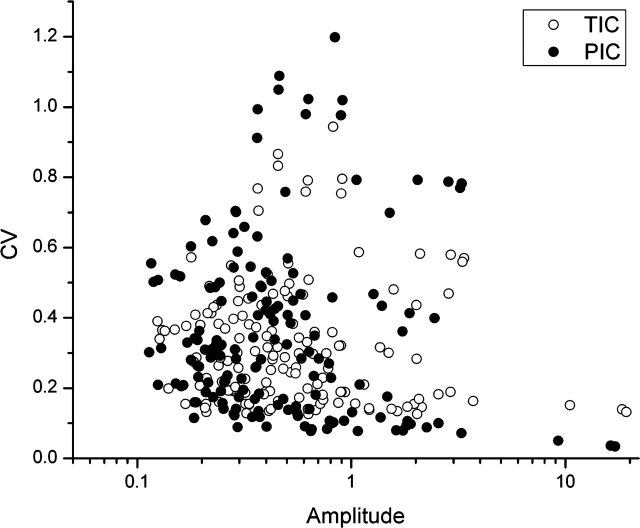
More sophisticated selection tools have been developed based on an iterative algorithm using CVs as selection criterion [23]. As an example, Figure 2 compares PIC to TIC using CVs from replicate analyses of MALDI-TOF mass spectra of urine samples, i.e. the same sample was run 19 times and the peak intensities of these 19 replicate runs were compared. Based on PIC, the CVs are about half what they would be if TIC normalization had been adopted. This improvement in precision increases the sensitivity of any biomarker analysis accordingly. This section falls short of addressing other issues. There are numerous other experimental and computational considerations that determine the success (or otherwise) of profiling approaches, but they are beyond the scope of this review.
Figure 2:
A comparison of CVs obtained using TIC (open circles) and PIC (solid circles).
MODIFICATIONS TO THE GENERAL MALDI APPROACH
Variants of the basic MALDI approach have been reported, most involving modifications relating to the matrix and its application, and some of these offer potential benefits for quantification. For example, desorption/ionization on silicon (DIOS) allows for the analysis of proteins and related small molecules in the absence of matrix. In common with MALDI, DIOS yields little or no fragmentation and is relatively tolerant of moderate amounts of contaminants commonly found in biological samples. The absence of a matrix offers special benefits in the low mass region [23].
Gross and colleagues have reported the use of ionic matrices in quantitative applications of oligodeoxynucleotides, peptides and small proteins [24, 25]. They report that good calibrations, linearity and reproducibility were achieved over a broad concentration range for all the tested ionic liquid matrices (ILMs) in spite of their different physical states. However, the SD is higher for ILMs than for solids with visible crystals. The experimental results indicate various ILMs yield different sensitivities owing to changes in their cation components and the slopes of the calibration curves correlate with the inverse of the peptide molecular weights.
Caprioli and colleagues reported on an acoustic reagent multi-spotter that provides improved reproducibility for depositing matrix onto sample surfaces. Although primarily developed for imaging of tissue sections, this approach may also offer benefits in quantification. Their acoustic droplet ejector provides better control of conditions affecting protein extraction and matrix crystallization, and offers the ability to deposit matrix onto small surface features accurately. Mass spectral quality and reproducibility was found to be better than that obtained with manual pipette spotting [26].
Gross and colleagues also developed an alternative small droplet deposition method using an induction-based fluidics (IBF) technique. This allowed the investigators to dispense nanolitre drops and they report that this improves the signal intensity and sensitivity achieved in MALDI-MS [27].
Solvent-free or dry prepared sample methods have also been developed and show some advantages over traditional wet sample preparation methods [28]. For example, solvent-free sample preparation methods enable analysis of insoluble materials and reportedly provide higher quality mass spectra. The surfaces of these samples have been examined and the authors report remarkably smooth and homogeneous sample morphologies [26].
There are numerous other refinements to the general MALDI process that have the potential to improve the precision and accuracy of quantification, but they are yet to be assessed rigorously in a quantitative setting.
EXAMPLES OF QUANTITATIVE MALDI APPLICATIONS FROM THE LITERATURE
Quantitative MALDI has been employed to analyse a wide range of low and high mass molecules from many different sample types. The published applications cover the spectrum of quantitative analyses from relative to absolute, but few of the assays have been applied in a practical setting and even fewer are used on a routine basis. In the text that follows, we present some examples of where MALDI has been employed as the ionization method and the application is quantitative in nature. In almost all instances, these reports amount to a proof-of-principle, rather than an approach that is likely to be applied in a routine setting.
Low molecular mass analytes
These studies mirror more conventional quantitative applications of mass spectrometry (i.e. GC-MS and LC-MS) and typically incorporate stable isotope-labelled internal standards. In some of the first reports in this area, we set out to demonstrate that MALDI can be applied to the analysis of low molecular mass analytes and yield reliable quantitative data [29]. We reported three examples: a standard curve for 3,4-dihydroxyphenylalanine (DOPA) was prepared by using an isotopomer (i.e. [13C6]DOPA) as the internal standard; [2H16]-acetylcholine was employed as an internal standard for the quantification of acetylcholine; and in the final example, the peptide H-Ser-Ala-Leu-Arg-His-Tyr-NH2 was quantified by employing a structural analog as the internal standard (i.e. Ac-Ser-Ile-Arg-His-Tyr-NH2). In each instance Pearson correlations for calibration curves (r2 > 0.95) demonstrated that MALDI can be a viable approach for the quantitative analysis of low molecular mass analytes. This early work helped to dispel the notion that MALDI could not deliver quantitative results, but we did not undertake any practical sample analysis at this stage.
MALDI-TOF MS has also been applied to the quantification of lysine, alanine and glucose [30]. The method is based on using stable isotopes as internal standards and the authors report fast, sensitive and reproducible quantification of these compounds. The quantitative method was demonstrated for aqueous standard solutions with concentrations of the analytes between 10 μM and 100 mM. The mean SDs from five replicate measures of each analyte were 4.3% (lysine), 3.7% (alanine) and 3.2% (glucose). This level of precision is impressive and comparable to that achievable with the best of quantitative methods. A noteworthy additional feature of this study is that the authors also demonstrated excellent agreement between these data and those obtained by more conventional quantitative techniques.
In 2002, we reported a more comprehensive and practical study in which automated MALDI-TOF MS was used to quantify a range of components in biological extracts and fluids [31]. Growth hormone was measured in rat pituitary tissue; insulin in human pancreatic tissue; homovanillic acid in human urine; and LVV-hemorphin-7, epinephrine and norepinephrine in human adrenal and pheochromocytoma tissues. Internal standards included compounds of similar molecular mass, structural analogues and isotopomers. These were incorporated into each assay and the practical limitations of quantitative MALDI-TOF MS are discussed. Although we had no need to routinely apply any of these assays, they were cost-effective, fast and offered excellent performance characteristics. Based on this study, these or similar methods could be applied in a routine setting.
Sleno and Volmer [32] have also addressed some of the practical and theoretical aspects of quantification of low mass analytes by MALDI-TOF MS. They quantified quinidine, danofloxacin, ramipril and nadolol by MALDI and carefully selected the internal standards to match the structure and fragmentation pathways of the analytes. The impact of different matrices was also examined as well as the role of laser energy and pulse rate. Results for the quantification of small molecules with molecular masses <500 Da by MALDI were generally good. In a companion study, the same authors focused their attention on the selection of internal standards for low mass quantification by MALDI and discussed the importance of a vicinal mass for the internal standard [21]. Both these studies were performed on a prototype MALDI-triple quadrupole instrument equipped with a high repetition rate laser.
Several more recent reports also describe key issues for the successful application of MALDI-TOF MS to quantifying drugs. van Kampen and colleagues [33] examined such issues as the matrix (i.e. nature and preparation), the inclusion of Li+ to facilitate formation of cationized drug species, automation and data analysis procedures. Of special note, they utilized a high molecular mass matrix, meso-tetrakis (pentafluorophenyl) porphyrin because it eliminated chemical noise in the low m/z range. Their method was applied to the quantification of clinically relevant concentrations of lopinavir, an HIV protease inhibitor, in extracts of peripheral blood mononuclear cells.
Notari and colleagues [34] recently applied MALDI-TOF/TOF MS to the quantification of abacavir, amprenavir, didanosine, efavirenz, nevirapine and stavudine in the plasma of HIV-infected patients. They used standard additional analysis and therefore it was not necessary to incorporate stable isotope-labelled analogues to achieve precise and accurate quantification. Standard addition is a powerful approach, but it requires that each sample be analysed multiple times, and consequently is slow and labour-intensive relative to approaches based on the use of an internal standard and calibration curve. The authors point out, however, that the high-throughput and full automation achievable with MALDI makes it feasible to adopt this approach in a routine setting, especially when the matrix is complex, or highly variable between samples.
Kovaric and colleagues [35] also investigated aspects of the quantitative analysis of pharmaceutical compounds in human plasma based on MALD-TOF MS operated in the multiple reaction monitoring mode. Talinolol was selected as a model analyte in these studies and liquid–liquid extraction and protein precipitation were evaluated, both with and without chromatographic separation. They report acceptable precision and accuracy when liquid–liquid extraction was employed without any LC separation and note that a full calibration curve and its quality control samples (20 samples) can be analysed within a minute. Combining LC with MALDI analysis improves the linearity down to 50 pg/ml, but reduces the throughput by (at least) 2-fold. These authors argue that matrix effects are still a significant issue with MALDI but can be monitored in a similar way to that used for LC-MS analysis based on electrospray.
Wagner and colleagues [36] have also highlighted the potential for high-throughput in their studies. These authors quantified saquinavir in human plasma samples without prior chromatographic separation. They report that the method was precise and accurate, but that the spotting and crystallization processes need to be automated and are one of the main sources of analytical variation. They too point out that compared to approaches requiring online LC separation or infusion, quantitative analysis by MALDI combined with selected reaction monitoring (SRM) can be undertaken within a few seconds, and consequently, potentially thousands of measurements can be undertaken per day.
Peptides and proteins
Analysis of one or a few proteins
Peptides and proteins can be measured directly as intact molecules, or for proteins, it is also possible to base quantification on one or more chemical or enzymatic cleavage products. Examples of both approaches have been reported. Kiernan and colleagues [37] developed and applied a sensitive mass spectrometric immunoassay to the quantitative analysis of C-reactive protein (CRP) in human plasma. In this study, multiplexed antibody-based retrieval and MALDI-TOF MS were used to target retinol-binding protein, C-reactive protein, serum amyloid P component and an added exogenous internal reference standard (i.e. staphylococcal enterotoxin B). The three analytes of interest and the exogenous internal standard were isolated from 35 plasma samples using a high-throughput robotic system and eluted directly onto the MALDI target. Their approach allowed semi-quantitative analysis of both retinol-binding protein and serum amyloid P component, and absolute quantification of C-reactive protein. The CRP values measured by this approach were compared to CRP values determined by a high-sensitivity latex immunoturbidometric assay. While the values between assays showed the same general trend, the mass spectrometric assay consistently returned lower numbers. The authors speculate that the different values between assays result from the different reagents (antibodies and standards) used in the two approaches.
There are several notable features to this study. First, the automated approach—both sample preparation and mass spectrometry were automated—allows for high-throughput analysis and provides the highest level of reproducibility (coefficient of variation for CRP <15%); this is a critical consideration if these types of assays are to enter the clinical diagnostic arena. Second, the authors collected and averaged 500 spectra from different regions within each sample spot, thereby minimizing the shot-to-shot and region-to-region variability. Finally, the mass spectrometric immunoassay has the capacity to independently quantify variant forms of the proteins of interest. This is generally not feasible with immunoaffinity-based assays that use either spectroscopic or turbidometric detection.
Our group has also quantified protein isoforms, in both relative and absolute terms, based on MALDI-TOF MS [38]. The utility of the approach was demonstrated by quantifying the α and β protein isoforms of myosin heavy chain (MyHC) in human atrial tissue. Isoform-specific tryptic peptides of α-MyHC (726–741) and β-MyHC (724–739) were identified and calibration curves were constructed by plotting ion current ratios against molar ratios for the two synthetic peptides. For atrial samples, MyHC was digested by trypsin, the ion current ratio was determined for the two tryptic peptides, and this was converted to the peptide ratio, then the isoform ratio. The molar ratio was converted to absolute values by employing a third peptide as an internal standard (i.e. an analogue of the two peptides being measured). A known quantity of the IS was added to the MyHC digests and the measured ion current ratios were converted to the actual quantities of the isoform-specific peptides and then the quantity of each protein isoform. The accuracy of the method was confirmed by comparing these results to those determined by an established method for the MyHC isoform ratio determination. This approach is generally applicable to the analysis of protein isoforms that have differences in primary sequence or chemical modification. This is often difficult to achieve with traditional antibody-based assays.
Recently, Bizzarri and colleagues [39] undertook absolute quantification of troponin T in mouse cardiac tissue by MALDI-TOF MS. After protein extracted from heart tissue, cardiac troponin T was isolated by high-performance liquid chromatography and digested with trypsin. Quantification was based on measurement of a cardiac troponin T-specific tryptic peptide (YEINVLR). Calibration curves were generated in the sample matrix by adding varying amounts of the cardiac troponin T-specific synthetic peptide and a fixed amount of an internal standard, which was a structural analogue of the native peptide (YEIQVLR). The authors investigated critical aspects of assay performance including enzymatic digestion efficiency, matrix effects, recovery, linearity, precision, limit of detection and limit of quantification. Although one limitation of the assay is the requirement for extensive sample clean-up, this study represents an excellent example of assay development and validation. Nevertheless, this work too is more of a proof-of-principle, rather than a practical routine application.
Comprehensive peptide and/or protein applications (peptidomics and proteomics)
A strategy based on isotope labelling of peptides and liquid chromatography combined with MALDI has been employed to accurately quantify and identify differentially expressed proteins between an E-cadherin-deficient human carcinoma cell line (SCC9) and its transfectants expressing E-cadherin (SCC9-E) [40]. Proteins extracted from each cell line were digested with trypsin and the resulting peptides were labelled individually with either [2H0]- or [2H2]-formaldehyde. The labelled peptides were combined and the peptide mixture was separated and fractionated by a strong cation exchange (SCX) column. Peptides from each SCX fraction were further separated on a reversed phase column. The effluents were spotted onto a MALDI target with a heated droplet LC-MALDI interface and after mixing with a MALDI matrix, individual sample spots were analysed by MALDI on a quadrupole TOF MS. An initial MS scan was used to quantify the dimethyl-labelled peptide pairs and MS/MS analysis was performed on the peptide pairs demonstrating a relative peak intensity change of greater than 2-fold. The MS/MS spectra were subjected to database searching for protein identification. The strategy was employed to detect and compare relative peak intensity changes for 5480 peptide pairs and amongst these, 320 peptide pairs showed changes of >2-fold. MS/MS analysis led to the identification of 49 differentially expressed proteins between the parental SCC9 cells and SCC9-E transfectants. The accuracy of the MS quantification and subcellular localization for six of the differentially expressed proteins were validated by immunoblotting and immunofluorescence assays.
Gutierrez et al. [41] have also quantified peptides by MALDI-TOF MS. They examined the influence of factors such as crystal heterogeneity, peptide ionization efficiency and data handling. In their studies, they report an overall coefficient of variance of 4.4% and a quantitative working range of 0.58–37.5 ng for bovine insulin.
Off-line capillary liquid chromatography was combined with MALDI-TOF MS and TOF/TOF-MS for the identification and quantification of neuropeptides in microwave-fixed rat brain tissue [42]. The effluent was mixed with matrix solution and transferred to a MALDI target plate by pulsed electric field deposition. Consistent with other reports, their detection limits were in the low amol range and they detected over 400 distinct peaks. TOF/TOF-MS allowed identification of 10 peptides, including three novel peptides. Quantification was evaluated using substance P as the analyte and [15N3]-labelled substance P as an internal standard. Measured levels for substance P were about 7-fold higher than previously reported in the rat striatum and the authors attributed this to the unique properties of microwave fixation. They conclude that their approach could serve as a versatile tool for neuropeptide analysis in brain tissue.
A variant of stable isotope labelling combined with mass spectrometry, termed isotope-coded protein label (ICPL), has been reported to yield high accuracy, reproducibility and good sequence coverage. The utility of the approach was demonstrated by comparative analysis of two differentially spiked proteomes [43].
Pan et al. [44] employed a targeted quantitative proteomics approach based on MALDI TOF/TOF MS to search for, identify and quantify selected peptides in human cerebrospinal spinal fluid (CSF). The approach involved isotopically labelled synthetic peptides as references for targeted identification, quantification and mass spectrometric analysis, and LC MALDI-TOF/TOF was used to selectively identify and quantify the targeted peptides in the proteome of CSF without prior depletion of abundant proteins. Candidate markers were quantified in CSF samples of patients with Parkinson's disease (PD) and Alzheimer's disease (AD) along with normal controls.
Griffin et al. [45] described an approach to the quantitative analysis of complex protein mixtures by MALDI on a quadrupole TOF (QqTOF) mass spectrometer [46]. Proteins were labelled on cysteinyl residues with an isotope-coded affinity tag reagent, enzymatically digested, the labelled peptides purified by a multi-dimensional procedure, and then eluted directly onto a MALDI sample target. After the addition of matrix, the sample spots were analysed by MALDI-QqTOF MS. The effectiveness of the approach was demonstrated by the quantification and identification of protein expression in Saccharomyces cerevisiae grown on two different carbon sources.
Liao and Turko [47] recently reported another approach to proteome analysis based on ammonium sulphate depletion of serum, labelling of unfolded proteins with native acrylamide and deuterated [2H3]-acrylamide, separation of the unfolded acrylamide-modified proteins, trypsinolysis and relative quantification by MALDI MS. There is minimal sample handling before isotopic labelling and extensive separation of unfolded proteins after isotopic labelling. The authors report the quantification of a large number of serum proteins, including those with an abundance of 10–5 less than albumin, and suggest that this is a robust and inexpensive workflow suitable for the quantitative profiling of protein changes in the serum proteome.
Protein profiling studies
There are hundreds of examples in the literature of where protein profiling has been used to uncover different protein signatures in two or more populations. Studies have utilized many sample types including serum, plasma and urine. As discussed in previous sections, many of the early studies were not appropriately validated or replicated. We recently reported a study that establishes the reproducibility of the profiling approach both within and between laboratories. We developed and tested the ability of a predictive algorithm based on MALDI-TOF MS analysis of pre-treatment serum to identify patients who are likely to benefit from treatment with epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) [22]. Some but not all patients with non-small cell lung cancer (NSCLC) respond to treatment with EGFR TKIs. Sera collected from NSCLC patients before treatment with gefitinib or erlotinib were analysed by MALDI independently at two institutions. An algorithm to predict outcomes after treatment with EGFR TKIs was developed from a training set of 139 patients derived from three cohorts. The algorithm was then tested in two independent validation cohorts of 67 and 96 patients who were treated with gefitinib and erlotinib, respectively, and in three control cohorts of patients who were not treated with EGFR TKIs. The clinical outcomes of survival and time to progression were analysed. An algorithm based on eight distinct m/z features was developed based on outcomes after EGFR TKI therapy in the training set patients. Classifications based on spectra acquired at the two institutions had a concordance of 97.1%, showing that high reproducibility can be achieved using MALDI-TOF MS. For both validation cohorts, the classifier identified patients who showed improved outcomes after EGFR TKI treatment. In one cohort, median survival of patients in the predicted ‘good’ and ‘poor’ groups was 207 and 92 days, respectively [hazard ratio (HR) of death in the good versus poor groups = 0.50, 95% confidence interval (CI) = 0.24–0.78]; in the other cohort, median survivals were 306 versus 107 days (HR = 0.41, 95% CI = 0.17–0.63). The classifier did not predict outcomes in patients who did not receive EGFR TKI treatment. This MALDI algorithm was not merely prognostic but could classify NSCLC patients for good or poor outcomes after treatment with EGFR TKIs and it may assist in the pre-treatment selection of appropriate subgroups of NSCLC patients for treatment with EGFR TKIs. This is only one example of a rapidly expanding literature on profile analysis. Early studies were marred by design problems and lacked thorough validation but increasingly the potential of the approach is being realized in newly published work.
Phosphorylation analysis
There are few mass spectrometric approaches to quantify the extent of phosphorylation of a substrate, in large part because this is a difficult problem confounded by the physiochemical differences between unphosphorylated and phosphorylated peptides. In one of the first practical applications, Matsumoto and colleagues [48] used MALDI-TOF MS to quantify the phosphopeptide produced by calcium/calmodulin-dependent protein kinase II (CaMK II). They measured both the substrate peptide and the resulting product directly by MALDI to improve accuracy, and their approach eliminated the need for radiolabelled materials. They found that the phosphorylation ratio obtained from MALDI-TOF was consistently smaller than that obtained from HPLC and they attributed this to non-linearity of the detector under the analysis conditions.
Huang and colleagues [49] coupled stable isotope dimethyl labelling with immobilized metal affinity chromatography (IMAC) enrichment to quantify protein phosphorylation at MS-determined phosphorylation sites. They studied two model phosphoproteins and also applied their analysis to pregnant rat uteri, both with and without treatment with 8-bromo-cGMP. They acknowledge some limitations with their approach, notably low-throughput, but suggest that they have a working approach to this challenging task.
Parker and colleagues have generated calibration curves from a set of synthetic peptides of known input ratios. Towards developing a universally applicable approach to phosphorylation analysis they used these to determine subtleties in sequence-dependent differences for relative desorption/ionization efficiencies and to predict relative signal strengths for other peptide sequences [50].
Each of these applications provides useful data in a given setting, but no universally applicable, high-throughput and accurate approach to quantitative phosphorylation analysis has yet been developed.
Oligosaccharide analysis
MALDI has been applied to both the qualitative and quantitative analysis of oligosaccharides of lichenase-hydrolysed water-soluble β-glucan from barley [51]. For this application, MALDI proved to be a rapid, accurate and sensitive approach that could also offer primary structural features on water-soluble β-glucan from different barley varieties.
Inulin, a class of fructo-oligosaccharide derived from plants, is an additive in baked goods, dairy products, infant formula and dietary supplements. In order to gain a better understanding of the role of inulin, MALDI combined with Fourier transform ion cyclotron resonance MS was used for qualitative and quantitative analysis of bacterial growth. The method employed an internal standard and reference to a calibration curve to quantify the consumption of fructo-oligosaccharide by Bifidobacterium longum bv. infantis. The method described was designed to be more rapid, precise and robust than other existing methods [52]. In a similar application, MALDI-MS has also been applied to the analysis of oligosaccharides in aqueous food extracts. Fructo-oligosaccharides were measured by standard addition and the assay was applied to samples including red onions, shallots and elephant garlic [53].
Bile acids
Six cholic acid derivatives: taurocholic acid (TCA), taurochenodeoxycholic acid (TCDCA), taurolithocholic acid (TLCA), glycocholic acid (GCA), glycochenodeoxycholic acid (GCDCA) and glycolithocholic acid (GCDCA) have been quantified by MALDI-TOF MS. Urine samples were pre-concentrated and purified by solid phase extraction (SPE). The method was optimized to eliminate ion suppression and proved to be reproducible from day to day [54]. The same group has applied essentially the same procedure to the quantification of bile acids in plasma [55].
Lipids
Fujiwaki et al. [56] employed MALDI-TOF MS to quantify sphingolipids in cardiac valve tissue from a patient with Fabry disease. Crude lipids were extracted from the tissue with chloroform/methanol and then saponified with base. Six different species of ceramide trihexose were detected in the tissue extracts. The peak heights for these species were compared to a fixed amount of the exogenous internal standard sphingosylphosphorylcholine. A linear response was reported for one of the species from 0 to 50 ng with a good Pearson correlation (r > 0.95). The authors showed that ceramide trihexose species were markedly increased in tissue obtained from a patient with Fabry disease, relative to a healthy control, and they plan to apply the method to body fluids and other tissues in the hopes of developing a diagnostic test.
The phospholipid composition of serum lipoproteins has been quantified by MALDI-TOF MS [57]. Lipoproteins were isolated from serum and the bound lipids were extracted and analysed by MALDI. A variety of lipids were detected including phospholipids (PLs), lysophospholipids (lysoPLs), sphingolipids (SLs), triglycerides (TGs), cholesteryl esters (CEs) and free cholesterol. MALDI analysis also allowed for the characterization of individual fatty acid chains; however, as expected, the correlation between ion peak intensity and lipid concentration was poor due to the diverse nature of the lipid mixture (i.e. the wide variation in polarity). The authors quantified lipids with similar chemical characteristics but with different fatty acid chains by using 4-cholesten-3-one as an internal standard. They also investigated the influence of sample solvent, laser shot frequency and laser intensity on assay performance.
MALDI-TOF MS was recently used to determine the SLs in liver and spleen specimens from patients with Niemann-Pick disease types A and C, and Gaucher disease. Crude lipids were extracted from tissue with chloroform and methanol, mild alkaline treatment of the crude lipids was performed and a SL fraction was prepared and analysed. Sphingosylphosphorylcholine was used as the internal standard for quantification of sphingomyelin and ceramide monohexoside (CMH). The authors report that accumulated sphingomyelin and CMH in small amounts of tissues from sphingolipidosis patients can be quantified for diagnostic purposes and also for biochemical pathophysiology evaluation [58].
Quantification of other compounds by MALDI
There are numerous other examples of quantification by MALDI. For example, quantification of DNA oligonucleotides by MALDI-TOF MS has been demonstrated [59]. The authors used the sequence of interest and a synthetic oligonucleotide internal standard with a single base mutation. The natural and mutant sequences were co-amplified by PCR and a third primer was designed to anneal to the region immediately upstream of the mutation site. Depending on the specific mutation introduced and the ddNTP/dNTP mixtures used, either one or two bases were added to the third primer to produce two extension products from the natural and mutant templates. Finally, the two extension products were detected and quantified by MALDI-TOF MS. This method is generally applicable to the quantification of DNA oligonucleotides.
MALDI-TOF MS has been used to identify and quantify coccidiostats in poultry feeds. DHB was found to be the best matrix and the coccidiostats were shown to form predominantly [M + Na]+ ions, with some [M + K]+ and [M − H + 2Na]+ ions. A Sep-pak C18 cartridge purification procedure was developed for sample work-up and limits of detection for lasalocid, monensin, salinomycin and narasin standards were reportedly 251, 22, 24 and 24 fmol, respectively [60].
Wang and Sporns [61] used a MALDI-TOF MS method to quantify four flavonol glycosides in almond seedcoats (i.e. isorhamnetin rutinoside, isorhamnetin glucoside, kaempferol rutinoside and kaempferol glucoside) by incorporating rutin (quercetin-3-rutinoside) as the internal standard; and a method has been reported to detect and quantify curcumin and two curcuminoid metabolites in mouse serum and mouse lung cell cultures. Standard curves were prepared in serum and showed correlation coefficients of 0.94–0.99. Alcoholic extraction, concentration and addition of dilute hydrochloric acid to stabilize the curcumin were found to be essential to the reproducibility of the protocol. Untreated and curcumin-treated mouse lung fibrotic and non-fibrotic cell cultures were analysed by MALDI and curcumin uptake was calculated. Curcumin was not detected in untreated cells [62].
A combinatorial extraction method and an automated MALDI-TOF MS procedure were used to improve the clinical analysis of the immunosuppressant drug cyclosporin A. Cyclosporin extracts from whole blood were analysed by MALDI and electrospray mass spectrometry, allowing for their identification and quantification. The combinatorial approach was devised to optimize the extraction by generating an array of solvent systems to be used for extraction from blood, and an automated analysis was performed by MALDI-TOF MS to identify successful extractions [63].
Key Point.
Quantitative applications of MALDI present some additional challenges over and above those associated with alternative ionization methods. Despite this, following quickly after the development of technique, there were reports of quantitative applications that spanned a wide range of compounds. Most applications, at least in the early phase, were demonstrations of the potential of MALDI to provide precise quantitative data in the low atto- to femtomole range; few of these reports detail what might eventually become a practical assay. Subsequently, a second generation of studies demonstrated the practical benefits of MALDI ionization, not the least of which are its high-throughput, versatility, low cost, reliability and ease of use. These attributes make MALDI attractive as a tool, especially for profile analysis and targeted quantification by standard addition. It is fair to say that each of these manuscripts covers some of the important issues in the application of MALDI as a quantitative tool, but most fall short of providing rugged assays that have been thoroughly tested in a routine setting. There is little doubt that practical applications are feasible, even optimal for some tasks, but we are yet to see wide adoption of MALDI-TOF MS in a routine analytical setting. This is in part because commercially available hardware and software have been developed with qualitative applications in mind, and when these are applied to quantification, they deliver suboptimal performance. In future, we look forward to seeing applications of MALDI to quantification that are fit for their intended purpose and that take advantage of the unique feature of MALDI ionization, especially its exceptional throughput when it is combined with TOF analysis. These developments will best be realized through symbiotic collaboration between biological scientists and instrument developers.
FUNDING
National Institutes of Health (to M.W.D. and S.W.H.); Cystic Fibrosis Foundation.
Biographies
Mark W. Duncan is a professor in the School of Medicine, University of Colorado, Denver. He also holds an appointment as professor within the Obesity Research Center, King Saud University, Riyadh, Saudi Arabia.
Heinrich Roder is chief technology officer, Biodesix Inc., Broomfield, CO, USA.
Stephen W. Hunsucker was previously an assistant professor at the University of Colorado, Denver, but he has recently relocated to Biodesix Inc, CO, USA.
References
- Karas M, Bachmann D, Bahr U, et al. Matrix-assisted ultraviolet laser desorption of non-volatile compounds. Int J Mass Spectrom Ion Process. 1987;78:53. [Google Scholar]
- Nicola AJ, Gusev AI, Proctor A, et al. Application of the fast-evaporation sample preparation method for improving quantification of angiotensin II by matrix-assisted laser desorption/ionization. Rapid Commun Mass Spectrom. 1995;9:1164–71. doi: 10.1002/rcm.1290091216. [DOI] [PubMed] [Google Scholar]
- Krause E, Wenschuh H, Jungblut PR. The dominance of arginine-containing peptides in MALDI-derived tryptic mass fingerprints of proteins. Anal Chem. 1999;71:4160–5. doi: 10.1021/ac990298f. [DOI] [PubMed] [Google Scholar]
- Cerpa-Poljak A, Jenkins A, Duncan MW. Recovery of peptides and proteins following matrix-assisted laser desorption ionization mass spectrometry. Rapid Commun Mass Spectrom. 1995:233–9. [Google Scholar]
- Wallace WE. Reactive MALDI mass spectrometry: application to high mass alkanes and polyethylene. Chem Commun. 2007:4525–7. doi: 10.1039/b711932a. [DOI] [PubMed] [Google Scholar]
- Rubin RB, Merchant M. A rapid protein profiling system that speeds study of cancer and other diseases. Am Clin Lab. 2000;19:28–9. [PubMed] [Google Scholar]
- Zhou H, Roy S, Schulman H, et al. Solution and chip arrays in protein profiling. Trends Biotechnol. 2001;19:S34–9. doi: 10.1016/S0167-7799(01)01798-X. [DOI] [PubMed] [Google Scholar]
- Petricoin EF, Ardekani AM, Hitt BA, et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572–7. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- Petricoin EF, Liotta LA. Proteomic analysis at the bedside: early detection of cancer. Trends Biotechnol. 2002;20:S30–4. doi: 10.1016/s1471-1931(02)00204-5. [DOI] [PubMed] [Google Scholar]
- Petricoin EF, Ornstein DK, Paweletz CP, et al. Serum proteomic patterns for detection of prostate cancer. J Natl Cancer Inst. 2002;94:1576–8. doi: 10.1093/jnci/94.20.1576. [DOI] [PubMed] [Google Scholar]
- Conrads TP, Zhou M, Petricoin EF, et al. Cancer diagnosis using proteomic patterns. Expert Rev Mol Diagn. 2003;3:411–20. doi: 10.1586/14737159.3.4.411. [DOI] [PubMed] [Google Scholar]
- Grizzle WE, Adam BL, Bigbee WL, et al. Serum protein expression profiling for cancer detection: validation of a SELDI-based approach for prostate cancer. Dis Markers. 2003;19:185–95. doi: 10.1155/2004/546293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidransky D, Irizarry R, Califano JA, et al. Serum protein MALDI profiling to distinguish upper aerodigestive tract cancer patients from control subjects. J Natl Cancer Inst. 2003;95:1711–7. doi: 10.1093/jnci/djg099. [DOI] [PubMed] [Google Scholar]
- Baggerly KA, Morris JS, Coombes KR. Reproducibility of SELDI-TOF protein patterns in serum: comparing datasets from different experiments. Bioinformatics. 2004;20:777–85. doi: 10.1093/bioinformatics/btg484. [DOI] [PubMed] [Google Scholar]
- Baggerly KA, Morris JS, Edmonson SR, et al. Signal in noise: evaluating reported reproducibility of serum proteomic tests for ovarian cancer. J Natl Cancer Inst. 2005;97:307–9. doi: 10.1093/jnci/dji008. [DOI] [PubMed] [Google Scholar]
- Diamandis EP. Serum proteomic profiling by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry for cancer diagnosis: next steps. Cancer Res. 2006;66:5540–1. doi: 10.1158/0008-5472.CAN-05-4503. [DOI] [PubMed] [Google Scholar]
- Albrethsen J. Reproducibility in protein profiling by MALDI-TOF mass spectrometry. Clin Chem. 2007;53:852–8. doi: 10.1373/clinchem.2006.082644. [DOI] [PubMed] [Google Scholar]
- Villanueva J, Philip J, Entenberg D, et al. Serum peptide profiling by magnetic particle-assisted, automated sample processing and MALDI-TOF mass spectrometry. Anal Chem. 2004;76:1560–70. doi: 10.1021/ac0352171. [DOI] [PubMed] [Google Scholar]
- Harkins JBt, Katz BB, Pastor SJ, et al. Parallel electrophoretic depletion, fractionation, concentration, and desalting of 96 complex biological samples for mass spectrometry. Anal Chem. 2008;80:2734–43. doi: 10.1021/ac702214n. [DOI] [PubMed] [Google Scholar]
- Righetti PG, Boschetti E, Lomas L, et al. Protein equalizer technology: the quest for a ‘democratic proteome’. Proteomics. 2006;6:3980–92. doi: 10.1002/pmic.200500904. [DOI] [PubMed] [Google Scholar]
- Sleno L, Volmer DA. Assessing the properties of internal standards for quantitative matrix-assisted laser desorption/ionization mass spectrometry of small molecules. Rapid Commun Mass Spectrom. 2006;20:1517–24. doi: 10.1002/rcm.2498. [DOI] [PubMed] [Google Scholar]
- Taguchi F, Solomon B, Gregorc V, et al. Mass spectrometry to classify non-small-cell lung cancer patients for clinical outcome after treatment with epidermal growth factor receptor tyrosine kinase inhibitors: a multicohort cross-institutional study. J Natl Cancer Inst. 2007;99:838–46. doi: 10.1093/jnci/djk195. [DOI] [PubMed] [Google Scholar]
- Thomas JJ, Shen Z, Crowell JE, et al. Desorption/ionization on silicon (DIOS): a diverse mass spectrometry platform for protein characterization. Proc Natl Acad Sci USA. 2001;98:4932–7. doi: 10.1073/pnas.081069298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong DW, Zhang LK, He L, et al. Ionic liquids as matrixes for matrix-assisted laser desorption/ionization mass spectrometry. Anal Chem. 2001;73:3679–86. doi: 10.1021/ac010259f. [DOI] [PubMed] [Google Scholar]
- Li YL, Gross ML. Ionic-liquid matrices for quantitative analysis by MALDI-TOF mass spectrometry. J Am Soc Mass Spectrom. 2004;15:1833–7. doi: 10.1016/j.jasms.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Hanton SD, McEvoy TM, Stets JR. Imaging the morphology of solvent-free prepared MALDI samples. J Am Soc Mass Spectrom. 2008;19:874–81. doi: 10.1016/j.jasms.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Tu T, Sauter AD, Jr, Sauter AD, III, et al. Improving the signal intensity and sensitivity of MALDI mass spectrometry by using nanoliter spots deposited by induction-based fluidics. J Am Soc Mass Spectrom. 2008;00 doi: 10.1016/j.jasms.2008.03.017. [DOI] [PubMed] [Google Scholar]
- Hanton SD, Parees DM. Extending the solvent-free MALDI sample preparation method. J Am Soc Mass Spectrom. 2005;16:90–3. doi: 10.1016/j.jasms.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Duncan MW, Matanovic G, Cerpa-Poljak A. Quantitative analysis of low molecular weight compounds of biological interest by matrix-assisted laser desorption ionization. Rapid Commun Mass Spectrom. 1993;7:1090–4. doi: 10.1002/rcm.1290071207. [DOI] [PubMed] [Google Scholar]
- Wittmann C, Heinzle E. MALDI-TOF MS for quantification of substrates and products in cultivations of Corynebacterium glutamicum. Biotechnol Bioeng. 2001;72:642–7. [PubMed] [Google Scholar]
- Bucknall M, Fung KY, Duncan MW. Practical quantitative biomedical applications of MALDI-TOF mass spectrometry. J Am Soc Mass Spectrom. 2002;13:1015–27. doi: 10.1016/S1044-0305(02)00426-9. [DOI] [PubMed] [Google Scholar]
- Sleno L, Volmer DA. Some fundamental and technical aspects of the quantitative analysis of pharmaceutical drugs by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun Mass Spectrom. 2005;19:1928–36. doi: 10.1002/rcm.2006. [DOI] [PubMed] [Google Scholar]
- van Kampen JJ, Burgers PC, de Groot R, et al. Qualitative and quantitative analysis of pharmaceutical compounds by MALDI-TOF mass spectrometry. Anal Chem. 2006;78:5403–11. doi: 10.1021/ac060436i. [DOI] [PubMed] [Google Scholar]
- Notari S, Mancone C, Alonzi T, et al. Determination of abacavir, amprenavir, didanosine, efavirenz, nevirapine, and stavudine concentration in human plasma by MALDI-TOF/TOF. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;863:249–57. doi: 10.1016/j.jchromb.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Kovarik P, Grivet C, Bourgogne E, et al. Method development aspects for the quantitation of pharmaceutical compounds in human plasma with a matrix-assisted laser desorption/ionization source in the multiple reaction monitoring mode. Rapid Commun Mass Spectrom. 2007;21:911–9. doi: 10.1002/rcm.2912. [DOI] [PubMed] [Google Scholar]
- Wagner M, Varesio E, Hopfgartner G. Ultra-fast quantitation of saquinavir in human plasma by matrix-assisted laser desorption/ionization and selected reaction monitoring mode detection. J Chromatogr B Analyt Technol Biomed Life Sci. 2008 doi: 10.1016/j.jchromb.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Kiernan UA, Addobbati R, Nedelkov D, et al. Quantitative multiplexed C-reactive protein mass spectrometric immunoassay. J Proteome Res. 2006;5:1682–7. doi: 10.1021/pr0601133. [DOI] [PubMed] [Google Scholar]
- Helmke SM, Yen CY, Cios KJ, et al. Simultaneous quantification of human cardiac alpha- and beta-myosin heavy chain proteins by MALDI-TOF mass spectrometry. Anal Chem. 2004;76:1683–9. doi: 10.1021/ac035144l. [DOI] [PubMed] [Google Scholar]
- Bizzarri M, Cavaliere C, Foglia P, et al. A label-free method based on MALDI-TOF mass spectrometry for the absolute quantitation of troponin T in mouse cardiac tissue. Anal Bioanal Chem. 2008 doi: 10.1007/s00216-008-2113-x. [DOI] [PubMed] [Google Scholar]
- Ji C, Li L, Gebre M, et al. Identification and quantification of differentially expressed proteins in E-cadherin deficient SCC9 cells and SCC9 transfectants expressing E-cadherin by dimethyl isotope labeling, LC-MALDI MS and MS/MS. J Proteome Res. 2005;4:1419–26. doi: 10.1021/pr050094h. [DOI] [PubMed] [Google Scholar]
- Gutierrez JA, Dorocke JA, Knierman MD, et al. Quantitative determination of peptides using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Biotechniques. 2005;Suppl:13–17. doi: 10.2144/05386su02. [DOI] [PubMed] [Google Scholar]
- Wei H, Nolkrantz K, Parkin MC, et al. Identification and quantification of neuropeptides in brain tissue by capillary liquid chromatography coupled off-line to MALDI-TOF and MALDI-TOF/TOF-MS. Anal Chem. 2006;78:4342–51. doi: 10.1021/ac052196x. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Kellermann J, Lottspeich F. A novel strategy for quantitative proteomics using isotope-coded protein labels. Proteomics. 2005;5:4–15. doi: 10.1002/pmic.200400873. [DOI] [PubMed] [Google Scholar]
- Pan S, Rush J, Peskind ER, et al. Application of targeted quantitative proteomics analysis in human cerebrospinal fluid using a liquid chromatography matrix-assisted laser desorption/ionization time-of-flight tandem mass spectrometer (LC MALDI TOF/TOF) platform. J Proteome Res. 2008;7:720–30. doi: 10.1021/pr700630x. [DOI] [PubMed] [Google Scholar]
- Griffin TJ, Gygi SP, Rist B, et al. Quantitative proteomic analysis using a MALDI quadrupole time-of-flight mass spectrometer. Anal Chem. 2001;73:978–86. doi: 10.1021/ac001169y. [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rist B, Gerber SA, et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–9. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- Liao WL, Turko IV. Strategy combining separation of isotope-labeled unfolded proteins and matrix-assisted laser desorption/ionization mass spectrometry analysis enables quantification of a wide range of serum proteins. Anal Biochem. 2008;377:55–61. doi: 10.1016/j.ab.2008.03.016. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Kahn ES, Komori N. Nonradioactive phosphopeptide assay by matrix-assisted laser desorption ionization time-of-flight mass spectrometry: application to calcium/calmodulin-dependent protein kinase II. Anal Biochem. 1998;260:188–94. doi: 10.1006/abio.1998.2691. [DOI] [PubMed] [Google Scholar]
- Huang SY, Tsai ML, Wu CJ, et al. Quantitation of protein phosphorylation in pregnant rat uteri using stable isotope dimethyl labeling coupled with IMAC. Proteomics. 2006;6:1722–34. doi: 10.1002/pmic.200500507. [DOI] [PubMed] [Google Scholar]
- Parker L, Engel-Hall A, Drew K, et al. Investigating quantitation of phosphorylation using MALDI-TOF mass spectrometry. J Mass Spectrom. 2008;43:518–27. doi: 10.1002/jms.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G, Vasanthan T. MALDI-MS and HPLC quantification of oligosaccharides of lichenase-hydrolyzed water-soluble beta-glucan from ten barley varieties. J Agric Food Chem. 2000;48:3305–10. doi: 10.1021/jf0001278. [DOI] [PubMed] [Google Scholar]
- Seipert RR, Barboza M, Ninonuevo MR, et al. Analysis and quantitation of fructooligosaccharides using matrix-assisted laser desorption/ionization Fourier transform ion cyclotron resonance mass spectrometry. Anal Chem. 2008;80:159–65. doi: 10.1021/ac7017298. [DOI] [PubMed] [Google Scholar]
- Wang J, Sporns P, Low NH. Analysis of food oligosaccharides using MALDI-MS: quantification of fructooligosaccharides. J Agric Food Chem. 1999;47:1549–57. doi: 10.1021/jf9809380. [DOI] [PubMed] [Google Scholar]
- Mims D, Hercules D. Quantification of bile acids directly from urine by MALDI-TOF-MS. Anal Bioanal Chem. 2003;375:609–16. doi: 10.1007/s00216-003-1771-y. [DOI] [PubMed] [Google Scholar]
- Mims D, Hercules D. Quantification of bile acids directly from plasma by MALDI-TOF-MS. Anal Bioanal Chem. 2004;378:1322–6. doi: 10.1007/s00216-003-2475-z. [DOI] [PubMed] [Google Scholar]
- Fujiwaki T, Tasaka M, Takahashi N, et al. Quantitative evaluation of sphingolipids using delayed extraction matrix-assisted laser desorption ionization time-of-flight mass spectrometry with sphingosylphosphorylcholine as an internal standard. Practical application to cardiac valves from a patient with Fabry disease. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;832:97–102. doi: 10.1016/j.jchromb.2005.12.039. [DOI] [PubMed] [Google Scholar]
- Hidaka H, Hanyu N, Sugano M, et al. Analysis of human serum lipoprotein lipid composition using MALDI-TOF mass spectrometry. Ann Clin Lab Sci. 2007;37:213–21. [PubMed] [Google Scholar]
- Fujiwaki T, Tasaka M, Yamaguchi S. Quantitative evaluation of sphingomyelin and glucosylceramide using matrix-assisted laser desorption ionization time-of-flight mass spectrometry with sphingosylphosphorylcholine as an internal standard Practical application to tissues from patients with Niemann-Pick disease types A and C, and Gaucher disease. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;870:170–6. doi: 10.1016/j.jchromb.2008.05.013. [DOI] [PubMed] [Google Scholar]
- Ding C. Qualitative and quantitative DNA and RNA analysis by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Methods Mol Biol. 2006;336:59–71. doi: 10.1385/1-59745-074-X:59. [DOI] [PubMed] [Google Scholar]
- Wang J, Sporns P. MALDI-TOF MS quantification of coccidiostats in poultry feeds. J Agric Food Chem. 2000;48:2807–11. doi: 10.1021/jf000193+. [DOI] [PubMed] [Google Scholar]
- Frison-Norrie S, Sporns P. Identification and quantification of flavonol glycosides in almond seedcoats using MALDI-TOF MS. J Agric Food Chem. 2002;50:2782–7. doi: 10.1021/jf0115894. [DOI] [PubMed] [Google Scholar]
- May LA, Tourkina E, Hoffman SR, et al. Detection and quantitation of curcumin in mouse lung cell cultures by matrix-assisted laser desorption ionization time of flight mass spectrometry. Anal Biochem. 2005;337:62–9. doi: 10.1016/j.ab.2004.10.037. [DOI] [PubMed] [Google Scholar]
- Wu J, Chatman K, Harris K, et al. An automated MALDI mass spectrometry approach for optimizing cyclosporin extraction and quantitation. Anal Chem. 1997;69:3767–71. doi: 10.1021/ac970276y. [DOI] [PubMed] [Google Scholar]