Abstract
Neural connections, providing the substrate for functional networks, exist whether or not they are functionally active at any given moment. However, it is not known to what extent brain regions are continuously interacting when the brain is “at rest.” In this work, we identify the major explicit activation networks by carrying out an image-based activation network analysis of thousands of separate activation maps derived from the BrainMap database of functional imaging studies, involving nearly 30,000 human subjects. Independently, we extract the major covarying networks in the resting brain, as imaged with functional magnetic resonance imaging in 36 subjects at rest. The sets of major brain networks, and their decompositions into subnetworks, show close correspondence between the independent analyses of resting and activation brain dynamics. We conclude that the full repertoire of functional networks utilized by the brain in action is continuously and dynamically “active” even when at “rest.”
Keywords: brain connectivity, BrainMap, FMRI, functional connectivity, resting-state networks
Spontaneous fluctuations in the brain have been studied with functional magnetic resonance imaging (FMRI) since it was first noted that, even with the subject at rest, the FMRI time series from one part of the motor cortex were temporally correlated with other parts of the same functional network (1). Following this, several other networks of correlated temporal patterns in the “resting brain” have been identified. These distinct patterns can be separated from each other from a single resting FMRI dataset, because, although each has relatively consistent time courses across its set of involved regions, the different networks have different temporal characteristics from each other (2–4). The networks continue to covary even when the subject is asleep (5) and under anesthesia (6). Furthermore, several networks have been found to be spatially consistent across different subjects (7). Although such “resting state networks” (RSNs), and related networks of deactivation under task, have also been investigated in other modalities such as electroencephalography (EEG) (8) and positron emission tomography (PET) (9), the majority of the research to date has used FMRI. In addition to offering information about the structure and function of the healthy brain, the study of RSNs has already been shown to be of great potential clinical value, providing rich and sensitive markers of disease (10, 11). Although there has been concern that some patterns of spatially extended spontaneous signals may be of nonneural physiological origin (12), these concerns are increasingly being addressed (13), and it has been posited that RSNs do reflect functional networks (see an excellent review in ref. 14). However, to date, it has not been shown to what extent the RSN “functional networks” match the full set of functional networks used by the active brain undergoing a comprehensive set of task types. In this study, we compared network analyses from 36 subjects' resting FMRI data against the entirety of a large database of activation studies to test the hypothesis that the set of functional networks seen in resting data closely matches the set derived from thousands of different activation conditions.
To this end, we have used BrainMap (15, 16), currently the largest database of FMRI and PET brain activation studies. At present, >1,600 journal articles are included; at the end of 2007, this represented 19% of all published imaging studies (17). Each study typically includes several different task conditions and contrasts between these; >7,000 functional maps are summarized in terms of the coordinate locations of peaks of activation (or differential activation between conditions). In addition, a large amount of study information is included in the database, including carefully structured, rich descriptive text detailing the experimental paradigm. Each paradigm is also categorized under one or more of 66 behavioral domain classifications; these provide a more simplistic summary of the experimental tasks but are immediately quantitatively useful. Metaanalysis investigations of such databases often begin by reforming “pseudoactivation images” from the list of activation peak locations (18, 19). It is then possible to investigate cooccurrence of different activation sites across the range of experiments represented in the database. A previous analysis of activation images from BrainMap used such an approach to produce an exploratory tool that allows the user to specify a brain location; the tool then generates an image showing which other brain locations tend to coactivate, across all paradigms, with the seed point (20). Here, we take this further by estimating the primary set of independent networks of activation that represent the major modes of coactivation across all activation images. We have done this using independent component analysis (ICA), a powerful data-driven approach for finding independent patterns in multivariate data. This allows us to identify the major functional networks in the brain as estimated from, and hence representative of, a significant proportion of all functional activation studies carried out to date.
Our ICA-based analyses of BrainMap and the resting FMRI data were carried out independently of each other. In each case, we estimated a set of spatial maps and associated time courses in this fully data-driven (unconstrained) analysis of the major modes, or networks, of covariance across the brain. In the case of the resting FMRI data, the time courses correspond to the average spontaneous fluctuation within the corresponding spatial map, and in the case of BrainMap, “time” is the experiment ID, so each time point in one component's time course describes how strongly that component's spatial map relates to that particular activation image in BrainMap. Thus, the original data are decomposed into d networks, in a way that maximizes the homogeneity of function within each network while maximizing the heterogeneity between them. If d is increased, a greater number of networks will be found, accounting for the original data in a more detailed way; in general, these might be expected to constitute subnetworks of the lower-d decomposition (although this is not mathematically guaranteed with such an approach, because the different levels of the “functional hierarchy” are estimated independently of each other). In this article, we report results from analyses at 2 levels: first, a network dimensionality approximately matching many previous RSN studies (d = 20), and, second, using a much greater level of detail (d = 70), low enough to be supported by the resolution of the datasets but giving 3–4 times more subnetworks than the lower-dimensional results. With these analyses, at 2 levels of the functional hierarchy, we investigated the hypothesis that the major functional networks found in the working brain correspond to the networks of correlated spontaneous fluctuations observed when the subject is at rest.
Results
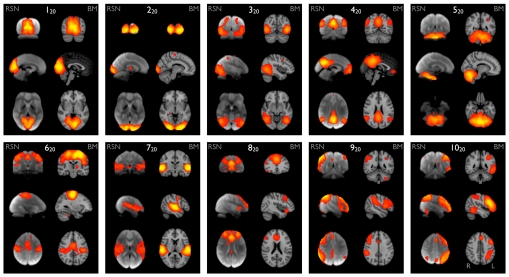
Our primary results stem from ICA decompositions of BrainMap and (independently) of the resting FMRI data, at an ICA dimensionality of 20 components; this matches a common degree of clustering/splitting previously applied via ICA to resting FMRI data (4, 7). We compared components between the resting FMRI and BrainMap datasets through simple spatial cross-correlation of the ICA spatial maps. Of the 20 components generated separately from the 2 datasets, 10 maps from each set were unambiguously paired between datasets, with a minimum correlation r = 0.25 (P < 10−5, corrected for multiple comparisons across all possible pairings and for spatial smoothness); see Fig. 1. The remaining maps were either judged to be artifactual, or of more complex interpretation. See supporting information (SI) for more detailed images of all maps and their classifications.
Fig. 1.
Ten well-matched pairs of networks from the 20-component analysis of the 29,671-subject BrainMap activation database and (a completely separate analysis of) the 36-subject resting FMRI dataset. This figure shows the 3 most informative orthogonal slices for each pair. (Left column of each pair) Resting FMRI data, shown superimposed on the mean FMRI image from all subjects. (Right column of each pair) Corresponding network from BrainMap, shown superimposed on the MNI152 standard space template image. The networks were paired automatically by using spatial cross-correlation, with mean r = 0.53 (0.25:0.79); the weakest of these correlations thus has a significance of P < 10−5 (corrected). All ICA spatial maps were converted to z statistic images via a normalized mixture–model fit, and then thresholded at Z = 3.
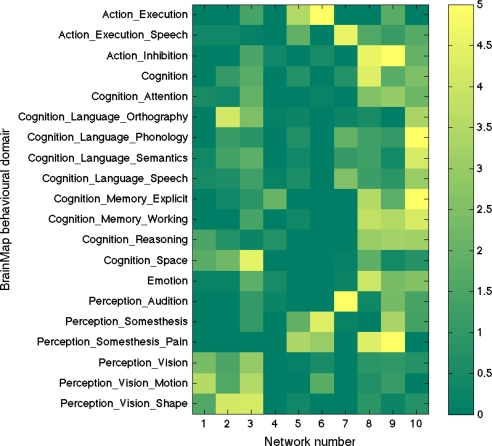
To aid interpretation of the components (and to illustrate the relevance and accuracy of even the simplest form of the experimental descriptions in BrainMap), we extracted the “behavioral domain” categorizations from the BrainMap database for each ICA component shown in Fig. 1. The results (Fig. 2) were found to be in good agreement with known localization of brain function. The 10 primary maps correspond to the 8 RSN maps previously described (4), with the addition of a cerebellar map and with a splitting of one of the previously reported visual maps into 2 distinct maps (220 and 320). The 10 maps correspond to interpretable functional categories and can be considered the “major representative” functional networks as derived independently from both activation metaanalysis and resting data. We describe each of these briefly below. The maps are numbered, with the subscript “20” to distinguish them from the higher-dimensionality “70” decomposition described later. Anatomical and functional descriptions below were derived with reference to the underlying standard-space images in conjunction with several atlases (21–23).
Fig. 2.
A mapping of the 10 primary functional networks shown in Fig. 1 onto the “behavioral domains” (experimental paradigm classifications) in the BrainMap database. Each of the BrainMap-derived ICA spatial maps has an associated experiment-ID “time course” quantifying its relevance to each of the original 7,342 BrainMap activation images. Each one of those activation images is listed, in BrainMap, against 1 or more of 66 possible “behavioral domains.” By multiplying the value at each time point by the corresponding behavioral domain(s) and averaging over all time points (experiments), we can derive a measure of how strongly each network relates to each behavioral domain, subject to interpretational caveats regarding such “reverse inference” (24) (see SI for detail; color scale is arbitrary units). Each row is normalized to have a mean count of 1, to balance for different domains being represented different numbers of times in the database. Of the original 66 behavioral domains, we show here only those that correspond most strongly to the 10 maps, for display brevity.
Maps 120, 220 and 320 (“visual”) correspond to medial, occipital pole, and lateral visual areas. The explicitly visual behavioral domains correspond most strongly to these maps, and paradigms cognition–language–orthography and cognition–space correspond to the occipital pole and lateral visual maps, respectively. We presume that the “orthography” correspondence reflects the visual nature of stimuli used in these studies (e.g., written-word forms).
Map 420 (“default mode network”) includes medial parietal (precuneus and posterior cingulate), bilateral inferior–lateral–parietal and ventromedial frontal cortex. This is often referred to as the default mode network (9), and is possibly the most widely studied RSN in the resting-state FMRI literature. This is also the network that is most commonly seen as deactivating in task-based FMRI experiments; hence, one would not expect this map to correspond strongly to any particular behavioral domain, because more contrasts associated with any given paradigm will, on average, be looking for positive activations rather than deactivations (or negative contrasts). However, there will be some studies that contain a “deactivation” contrast, and hence, we are not surprised that this map is found in our analysis of BrainMap. Indeed, inspection of the full set of experiments reveals that this map does indeed correspond in general to negative contrasts, in particular in cognitive paradigms.
Map 520 (“cerebellum”) covers the cerebellum. Because of limited field of view of the MRI acquisitions in some of the resting FMRI subjects, more inferior parts of the cerebellum are not included in the multisubject RSN analysis. This corresponds most strongly to action–execution and perception–somesthesis–pain domains.
Map 620 (“sensorimotor”) includes supplementary motor area, sensorimotor cortex, and secondary somatosensory cortex. This corresponds closely to the activations seen in bimanual motor tasks and was the first resting state network to be identified in FMRI data (1). This corresponds most strongly to the action–execution and perception–somesthesis paradigms.
Map 720 (“auditory”) includes the superior temporal gyrus, Heschl's gyrus, and posterior insular. It includes primary and association auditory cortices. This corresponds most strongly to action–execution–speech, cognition–language–speech, and perception–audition paradigms.
Map 820 (“executive control”) covers several medial–frontal areas, including anterior cingulate and paracingulate. This corresponds strongly to several cognition paradigms, as well as action–inhibition, emotion, and perception–somesthesis–pain.
Maps 920 and 1020 (“frontoparietal”) cover several frontoparietal areas. These are the only maps to be strongly lateralized, and are largely left–right mirrors of each other. They correspond to several cognition/language paradigms. In addition, map 920 corresponds strongly to perception–somesthesis–pain; this is consistent with the insular areas seen (see SI for more detailed figures showing all maps). Map 1020 corresponds strongly to cognition–language paradigms, which is consistent with the Broca's and Wernicke's areas seen in the map (see SI for slices more clearly showing these areas). Given the known lateralization of language function, it is not surprising that these (mirrored) networks have such different behavioral domain associations.
If the brain is thought of as being organized into functionally distinct (if connected) networks, these can themselves be considered to comprise subnetworks, each with distinct, if related, function. Hence, one would hope to find interpretable network decompositions at a range of levels of detail. Although a thorough investigation into the “full” hierarchy of functional networks and subnetworks is outside the scope of this article, we do include some results from a higher dimensionality than the 20-component results presented above. We generated 70-component ICA decompositions of BrainMap and the resting FMRI data in the hope that we would obtain a richer, more detailed separation of functional subnetworks that could be related to both the 20-component results and, at the 70-component level, between BrainMap and resting FMRI components.
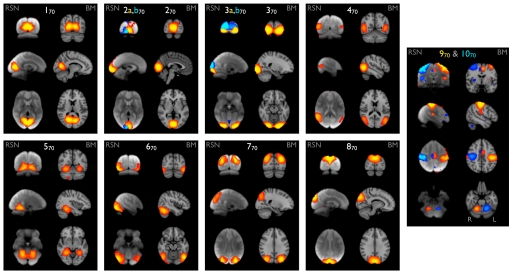
Of the 70-component decompositions, ≈45 of the resting FMRI and 60 of the BrainMap components were nonartifactual. The pairing of components is driven by simple correlation of the spatial maps, aided by association with the original 20-component maps through similarity of component time courses, i.e., by using “functional” similarity. This allowed us to unambiguously identify groups of 70-component RSN–BrainMap pairings with 20-component pairings. In Fig. 3, we show 2 examples: 8 well-matched pairs of networks in the visual cortex, and 2 pairs covering the sensorimotor cortex. There is clear correspondence between the RSNs and BrainMap components and clear functional interpretation of the maps.
Fig. 3.
Eight well-matched pairs of networks in visual areas (1–870), and 2 pairs from the sensorimotor areas (9, 1070), from the 70-component analyses of the BrainMap activation database and the resting FMRI dataset. All Gaussianized ICA maps are thresholded at Z = 4 (higher than for the 20-dimensional results for comparability, because the higher-dimensional analysis, by definition, has reduced ICA residuals).
Maps 1–370 show early visual areas (i.e., covering the same combined area as V1, V2, and V3 combined). In maps 270 and 370 the RSNs are split into 2 components (shown separately in yellow and blue); the combination of these corresponds well with the BrainMap maps. Maps RSN-2a,b70 show a right vs. left visual hemifield distinction, and maps RSN-3a,b70 show an upper vs. lower visual hemifield distinction. At a dimensionality of 100, the BrainMap map BM-370 also splits in a similar way. Map 470 corresponds to area V5 (motion), maps 5,670 cover the ventral/ventrolateral visual stream, and maps 7,870 cover the dorsal visual stream.
Maps 9,1070 cover the sensorimotor cortex (both precentral and postcentral gyri, although overlapping the latter more fully). In the 20-component analyses, this appeared as a single map; at this more detailed decomposition it splits into lateralized maps (one shown in red-yellow, the other in blue). The detail of the splitting is the same in BrainMap and in the resting FMRI data, even including medial cortical regions and cerebellar areas contralateral to the associated cortical areas, consistent with known corticocerebellar connectivity (25).
The hierarchical relationship between the 2 analysis levels is interesting; for example, 120“splits into” (is most strongly related to) 170, 570, and 870; 220 into 270 and 370; 320 into 470, 670 and 770. Hence, in some cases, a lower-dimensional network splits (at higher dimensionality) into left and right subnetworks, but in the majority of cases, left–right symmetry is preserved within the subnetworks, and splitting is into different “subfunctions” rather than into left vs. right.
Discussion
We have shown the major covarying functional networks in the brain, as imaged by FMRI and PET, from thousands of tests of explicit brain activation conditions in >1,600 functional neuroimaging studies. We have found that most of these are very similar to the majority of the networks of spontaneous covariation in the resting brain, as imaged by FMRI. The fact that the set of networks in the 2 domains is highly similar, in at least two-thirds of the (nonartifactual) extracted network components, implies that the resting brain's functional dynamics are fully utilizing the set of functional networks as exhibited by the brain over its range of possible tasks. All regions involved in all functional networks are continuously interacting with each other when the brain is at rest, with the same functional hierarchy that controls all brain action and cognition. The set of covarying networks that can be seen to be paired across the resting and active domains can be (at the simplest level) interpreted in terms of the major functional tasks, for example by mapping the functional networks extracted from BrainMap and resting FMRI data onto the set of BrainMap behavioral domains.
With an ICA dimensionality of 20, the RSN components found are almost identical to those found previously with ICA on different resting FMRI datasets (4, 7). The latter study showed that these primary RSNs are consistent across different individuals, providing convincing evidence that, although the quality of our results (e.g., in terms of spatial detail and signal-to-noise ratio) is aided by having multiple subjects' resting datasets combined, the close matching of the RSNs onto activation networks is not an artifact of combining many subjects together; the set of distinct functional networks is continuously present and identifiable (given sufficient data quality) in all subjects at rest.
When extracting a larger number of components from both BrainMap and resting FMRI data, we probe a different level in the hierarchy of functional networks and their subnetworks. With an ICA dimensionality of 70, we find many more subnetworks than in the 20-component analysis but still find close correspondence between activation networks and resting networks. These “subnetworks” (obviously a relative term) are, in general, subsets of the larger networks found at d = 20, again allowing straightforward interpretation of their functional nature. The primary networks split into subnetworks in both active and resting data in almost identical ways, e.g., into areas of slightly different function or into left vs. right subnetworks. (Note, however, that because the brain is a highly complex set of interconnected functional areas, we do not expect a simple tree-structure hierarchy to be a perfect model of connectivities covering all levels of detail, because that would imply acyclic functional graphs, which is clearly not the case.) There is greater functional (temporal) correlation between subnetworks within a primary network than across primary networks. The mappings found in both domains, and the implied functional hierarchy, will aid in the development of an objective and rich hierarchical functional ontology, one that links functional activity types and spatial localizations in a direct and practically useful way. Achieving this has been one of the primary purposes of BrainMap, and the work presented here complements progress already made by BrainMap towards functional ontologies.
The quality of the correspondence between the BrainMap-derived activation networks and the resting FMRI networks is particularly compelling given the fundamentally different nature of the data feeding into these 2 analyses and potential problems involved in finding a close match. For the resting FMRI analyses, we have data from just 36 subjects (compared with nearly 30,000 subjects in BrainMap), comprising just a few minutes' resting FMRI data from each. For the BrainMap analyses, the pseudoactivation images have to be created purely from the coordinates of the peaks of just a few locations in the brain for each activation condition; the full richness of the original activation maps is not available currently in BrainMap because of necessary practical considerations. The spatial extent of the activations in each pseudoactivation image generated from BrainMap is simply created by adding a Gaussian of fixed size around every activation peak; this is one reason why we see slightly more spatial detail in some of the resting FMRI maps (26). Furthermore, the extent to which different activation paradigms are represented with different frequencies in BrainMap was not adjusted for in our decompositions. This introduces an arbitrary factor into the decompositions, with respect to finding the d “strongest” components in the data (one could attempt to normalize for different numbers of different paradigms in the database, but this would be quite difficult to achieve objectively, and remains to be elucidated in future work). Finally, the BrainMap database comprises results from across a broad range of imaging hardware types, data qualities, analysis software implementations, and task paradigm specifics. Although every effort is made to reduce the final spatial activation information to an objective and comparable representation (the peak locations), the experimental and analytical differences across the >1,600 studies represented in BrainMap lead to a fundamental heterogeneity of information and data quality. The fact that the major functional networks found from the resting and task domains match so closely indicates that BrainMap has been largely successful in its goal of objectively representing the brain's activation dynamics. On the other hand, given the above comments (in particular the lack of normalization of numbers of different paradigms), it is hardly surprising that approximately one-third of the components extracted from the rest and active domains do not achieve a clear 1:1 pairing between the domains. These do cover similar overall spatial extents, i.e., are mostly different linear combinations of each other; further improvement in objective decompositions should be attainable in future, which will provide an optimal integrated decomposition of active and rest data (as opposed to independent analyses carried out to show the resulting similarities).
Note that the ICA decomposition of the BrainMap data in no way used the behavioral paradigm information shown in Fig. 1; the ICA was purely driven by the 7,342 pseudoactivation images, each of which potentially contains a unique spatial activation pattern. In the figure, we have utilized only the crudest level of behavioral information available in BrainMap, but further work can hope to make much richer use of the more detailed paradigm descriptions also encoded in the database. The lack of reliance on behavioral information for our primary analyses is a potential strength when wanting to find the most natural, data-driven, representation of the primary networks. This also helps avoid the problem of “reverse inference” (24) (the likely one-to-many mapping from activation location to function) in the core analysis and may partly explain why our results are so different from those shown in ref. 19, where paradigm information was combined with imaging data before decomposition.
The value of resting FMRI data and the analysis of RSNs has begun to be appreciated. RSNs have already been shown to be of potential clinical value as rich and sensitive markers of disease (10, 11). One obvious strength is that multiple functional networks can be tested in a very short scanning session without having to decide in advance what functional paradigm is most likely to be useful or, indeed, requiring active subject participation. This is particularly important in the clinical setting, where subjects could range from 10-week-old infants (27) to Alzheimer's patients. Our results indicate that the full repertoire of functional dynamics can be investigated in resting FMRI.
The development of an ontology for functional networks (28) and their hierarchy is likely to be of interest to nonclinical neuroscientists to complement other ontologies such as cytoarchitectonic mapping (29) and structural (e.g., lobular) descriptions (30). This work should be able to make contributions in the development of objective, data-driven ontologies, derived directly from functional brain data, from both the full history of activation studies and from the spontaneous dynamics in the resting brain. On a more practical level, although there is continuing debate as to the value and dangers of “functional localizers,” the information that resting data gives us about functional networks and their localizations encourages the use of such data and analyses to aid the spatial correspondence (or labeling) of data, for improved cross-subject analyses in activation studies. Although single-session analyses may not result in a detailed functional decomposition with the same reliability as one can achieve with multisubject datasets, it should still be possible to derive benefit from aligning those networks that are reliably detectable in single-session data with an appropriate network template. We intend to supplement the existing set of complementary brain atlases distributed with FSL (FMRIB Software Library, www.fmrib.ox.ac.uk/fsl), by using the datasets and analyses described in this article to generate a set of functional network maps (such as those shown in Fig. 1) to aid neuroimaging researchers in the interpretation of their functional and structural imaging studies.
Although we may not be surprised to see temporal coherence across functional networks when at rest, we might expect such coherent fluctuations to have minimal amplitude, whereas the amplitudes seen are of the same order as found under explicit activation (7). Although we have shown that activation networks are mirrored in resting data, we must acknowledge that this does not begin to answer the question of why the brain's many regions continue to “function” (with large amplitude fluctuations) when the subject is at rest, and even when the subject is asleep (5) and under anesthesia (6). There are suggestions of rehearsal, learning consolidation, and future preparation as possible explanations (31), but there is not yet conclusive evidence for any of these possibilities. However, the results we have presented here surely provide motivation for further investigations regarding this fundamental question.
Materials and Methods
Image processing and ICA decompositions were carried out with FSL (32, 33).
Resting FMRI.
FMRI time series data from each of 36 healthy adults at rest was acquired over 6 min. Each subject's dataset was transformed into a standard coordinate space, and all subjects' datasets were concatenated temporally. The resulting multisubject dataset was fed into ICA, after an initial PCA-based dimensionality reduction to 20 (and, separately, 70) components, to find the most representative networks of covariation when the brain is at rest (4, 34–38).
BrainMap.
We extracted 7,342 activation-peak images from BrainMap (in standard coordinate space) and applied 12-mm full-width Gaussian blurring to each, producing extended areas of activation (attempting to mimic the nature of the original activation images). We applied ICA to this space × experiment-ID dataset, including an initial PCA-based dimensionality reduction to 20 (and, separately, 70) components to find the most representative functional networks from the entire BrainMap database.
Comparing BrainMap with Resting FMRI.
Spatial maps from BrainMap and resting FMRI were associated with each other primarily via spatial similarity, by using (Pearson) spatial cross-correlation. See SI for more detail on datasets and methodology.
Supplementary Material
Acknowledgments.
We are grateful to Holly Bridge and Paul Matthews for advice. K.L.M. was supported by the Engineering and Physical Sciences Research Council/Royal Academy of Engineering. P.T.F., P.M.F., R.T., and A.R.L. were supported, as part of the Human Brain Project, by National Institute of Mental Health Grant R01-MH074457-01A1 (P.T.F., principal investigator).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905267106/DCSupplemental.
References
- 1.Biswal B, Zerrin Yetkin F, Haughton V, Hyde J. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- 2.Xiong J, Parsons L, Gao J, Fox P. Interregional connectivity to primary motor cortex revealed using MRI resting state images. Hum Brain Mapp. 1999;8:151–156. doi: 10.1002/(SICI)1097-0193(1999)8:2/3<151::AID-HBM13>3.0.CO;2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cordes D, et al. Mapping functionally related regions of brain with functional connectivity MR imaging. Am J Neuroradiol. 2000;21:1636–1644. [PMC free article] [PubMed] [Google Scholar]
- 4.Beckmann CF, De Luca M, Devlin JT, Smith SM. Investigations into resting-state connectivity using independent component analysis. Philos Trans R Soc London. 2005;360(1457):1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukunaga M, et al. Large-amplitude, spatially correlated fluctuations in BOLD fMRI signals during extended rest and early sleep stages. Magn Reson Imaging. 2006;24:979–992. doi: 10.1016/j.mri.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 6.Vincent J, et al. Intrinsic functional architecture in the anaesthetized monkey brain. Nature. 2007;447:83–87. doi: 10.1038/nature05758. [DOI] [PubMed] [Google Scholar]
- 7.Damoiseaux JS, et al. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci USA. 2006;103(37):13848–13853. doi: 10.1073/pnas.0601417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldman R, Stern J, Engel J, Cohen M. Simultaneous EEG and fMRI of the alpha rhythm. NeuroReport. 2002;13(18):2487–2492. doi: 10.1097/01.wnr.0000047685.08940.d0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raichle M, et al. A default mode of brain function. Proc Natl Acad Sci USA. 2001;98(2):676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greicius M, Srivastava G, Reiss A, Menon V. Default-mode network activity distinguishes Alzheimer's disease from healthy aging: Evidence from functional MRI. Proc Natl Acad Sci USA. 2004;101(13):4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filippini N, et al. Distinct patterns of brain activity in young carriers of the APOE-e4 allele. Proc Natl Acad Sci USA. 2009;106:7209–7214. doi: 10.1073/pnas.0811879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birn R, Diamond J, Smith M, Bandettini P. Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in fMRI. NeuroImage. 2006;31:1536–1548. doi: 10.1016/j.neuroimage.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 13.Birn R, Murphy K, Bandettini P. The effect of respiration variations on independent component analysis results of resting state connectivity. Hum Brain Mapp. 2008;29:740–750. doi: 10.1002/hbm.20577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox MD, Raichle ME. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci. 2007;8(9):700–711. doi: 10.1038/nrn2201. [DOI] [PubMed] [Google Scholar]
- 15.Fox PT, Lancaster JL. Mapping context and content: The BrainMap model. Nat Rev Neurosci. 2002;3:319–321. doi: 10.1038/nrn789. [DOI] [PubMed] [Google Scholar]
- 16.Laird A, Lancaster J, Fox P. BrainMap. The social evolution of a human brain mapping database. NeuroInformatics. 2005;3:65–77. doi: 10.1385/ni:3:1:065. [DOI] [PubMed] [Google Scholar]
- 17.Derrfuss J, Mar RA. Lost in localization: The need for a universal coordinate database. NeuroImage. 2009 doi: 10.1016/j.neuroimage.2009.01.053. in press. [DOI] [PubMed] [Google Scholar]
- 18.Turkeltaub P, Eden GF, Jones KM, Zeffiro TA. Meta-analysis of the functional neuroanatomy of single-word reading: Method and validation. NeuroImage. 2002;16(3):765–780. doi: 10.1006/nimg.2002.1131. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen FÅ, Hansen LK, Balslev D. Mining for associations between text and brain activation in a functional neuroimaging database. Neuroinformatics. 2004;2:369–379. doi: 10.1385/NI:2:4:369. [DOI] [PubMed] [Google Scholar]
- 20.Toro R, Fox P, Paus T. Functional coactivation map of the human brain. Cereb Cortex. 2008;18:2553–2559. doi: 10.1093/cercor/bhn014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eickhoff SB, et al. Assignment of functional activations to probabilistic cytoarchitectonic areas revisited. NeuroImage. 2007;36(3):511–521. doi: 10.1016/j.neuroimage.2007.03.060. [DOI] [PubMed] [Google Scholar]
- 22.Talairach J, Tournoux P. Co-Planar Stereotaxic Atlas of the Human Brain. New York: Thieme; 1988. [Google Scholar]
- 23.Worth A, Makris N, Caviness V, Kennedy D. Neuroanatomical segmentation in MRI: Technological objectives. Int J Pattern Recogn Artificial Intell. 1997;11(8):1161–1187. [Google Scholar]
- 24.Poldrack R. Can cognitive processes be inferred from neuroimaging data? Trends Cognit Sci. 2006;10(2):59–63. doi: 10.1016/j.tics.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 25.Mattay VS, et al. Hemispheric control of motor function: A whole brain echo planar fMRI study. Psychiatry Res Neuroimag. 1998;83(1):7–22. doi: 10.1016/s0925-4927(98)00023-7. [DOI] [PubMed] [Google Scholar]
- 26.Salimi-Khorshidi G, Smith SM, Keltner JR, Wager TD, Nichols TE. Meta-analysis of neuroimaging data: A comparison of image-based and coordinate-based pooling of studies. NeuroImage. 2009;45(3):810–823. doi: 10.1016/j.neuroimage.2008.12.039. [DOI] [PubMed] [Google Scholar]
- 27.Fransson P, et al. Resting-state networks in the infant brain. Proc Natl Acad Sci USA. 2007;104(39):15531–15536. doi: 10.1073/pnas.0704380104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poldrack R, Halchenko Y, Hanson SJ. Decoding the large-scale structure of brain function by classifying mental states across individuals. Psychol Sci. 2009 doi: 10.1111/j.1467-9280.2009.02460.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eickhoff S, et al. A new SPM toolbox for combining probabilistic cytoarchitectonic maps and functional imaging data. NeuroImage. 2005;25:1325–1335. doi: 10.1016/j.neuroimage.2004.12.034. [DOI] [PubMed] [Google Scholar]
- 30.Shattuck D, et al. Construction of a 3D probabilistic atlas of human cortical structures. NeuroImage. 2008;39:1064–1080. doi: 10.1016/j.neuroimage.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buckner R, Vincent J. Unrest at rest: Default activity and spontaneous network correlations. NeuroImage. 2007;37:1091–1096. doi: 10.1016/j.neuroimage.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 32.Smith SM, et al. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004;23(S1):208–219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 33.Woolrich MW, et al. Bayesian analysis of neuroimaging data in FSL. NeuroImage. 2009;45:S173–S186. doi: 10.1016/j.neuroimage.2008.10.055. [DOI] [PubMed] [Google Scholar]
- 34.Comon P. Independent component analysis, a new concept? Signal Processing. 1994;36:287–314. [Google Scholar]
- 35.Bell A, Sejnowski T. An information-maximization approach to blind separation and blind deconvolution. Neural Comput. 1995;7:1129–1159. doi: 10.1162/neco.1995.7.6.1129. [DOI] [PubMed] [Google Scholar]
- 36.McKeown M, et al. Analysis of fMRI data by blind separation into independent spatial components. Hum Brain Mapp. 1998;6:160–188. doi: 10.1002/(SICI)1097-0193(1998)6:3<160::AID-HBM5>3.0.CO;2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiviniemi V, Kantola J-H, Jauhiainen J, Hyvarinen A, Tervonen O. Independent component analysis of nondeterministic fMRI signal sources. NeuroImage. 2003;19:253–260. doi: 10.1016/s1053-8119(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 38.Beckmann CF, Smith SM. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans Med Imag. 2004;23(2):137–152. doi: 10.1109/TMI.2003.822821. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.