Abstract
Strains of Xanthomonas campestris pv. vesicatoria (Xcv) carrying avrBs2 are specifically recognized by Bs2 pepper plants, resulting in localized cell death and plant resistance. Agrobacterium-mediated transient expression of the Xcv avrBs2 gene in plant cells results in Bs2-dependent cell death, indicating that the AvrBs2 protein alone is sufficient for the activation of disease resistance-mediated cell death in planta. We now provide evidence that AvrBs2 is secreted from Xcv and that secretion is type III (hrp) dependent. N- and C-terminal deletion analysis of AvrBs2 has identified the effector domain of AvrBs2 recognized by Bs2 pepper plants. By using a truncated Pseudomonas syringae AvrRpt2 effector reporter devoid of type III signal sequences, we have localized the minimal region of AvrBs2 required for type III secretion in Xcv. Furthermore, we have identified the region of AvrBs2 required for both type III secretion and translocation to host plants. The mapping of AvrBs2 sequences sufficient for type III delivery also revealed the presence of a potential mRNA secretion signal.
Bacterial spot disease of tomato and pepper is caused by Xanthomonas campestris pathovar vesicatoria (Xcv). Most Xc pathovars infecting a wide host range contain avrBs2 in the chromosome (1). Such prevalence suggests that the conservation of avrBs2 in Xc may be crucial for the maintenance of pathogen fitness. In fact, Xcv strains possessing natural or introduced mutations at the avrBs2 locus are less virulent on susceptible hosts (1–3). Susceptibility to Xcv often leads to devastating losses in commercial production of crop plants in regions with high humidity and heavy rainfall. Fortunately, resistance to strains of Xcv carrying avrBs2 has been identified in a wild species of pepper (Capsicum chacoense) and introduced into cultivated pepper (Capsicum annuum) by traditional breeding.
Molecular studies show that the Bs2 gene from C. chacoense specifically recognizes and confers resistance to strains of Xcv that contain avrBs2 (4, 5). The Bs2 gene (4) is a member of the nucleotide-binding site-leucine-rich repeat class of plant disease resistance genes (6, 7) and is predicted to reside in the plant cytoplasm. The successful engineering of Bs2 resistance in transgenic tomatoes provides a source of protection to Xcv strains containing avrBs2 (4). However, durability of Bs2 resistance in the field is challenged by the emergence of Xcv strains that are able to cause disease in previously resistant pepper plants (8, 9). The characterization of natural field isolates overcoming Bs2 resistance has revealed that Xcv is evolving under selection pressure at the avrBs2 locus to evade recognition and to maintain avrBs2-dependent virulence in its host (2). Although the virulence function of avrBs2 is not known, transient expression of avrBs2 in plant cells has confirmed that AvrBs2 protein is recognized in Bs2 plants and that this effector is sufficient for the activation of Bs2-dependent disease resistance in planta (4). The predicted AvrBs2 protein shares homology with agrocinopine synthase of Agrobacterium tumefaciens and the glycerophosphoryl diester phosphodiesterase UgpQ of Escherichia coli (3), suggesting that AvrBs2 may function in plant cells as an enzyme to synthesize or hydrolyze phosphodiester linkages.
The hrp locus encoding the type III pathway is essential for Xcv to induce avrBs2-dependent localized cell death in resistant Bs2 pepper plants and to cause disease in susceptible bs2 pepper plants (10). This suggested to us that Xcv delivers AvrBs2 to the plant cytoplasm via the type III secretion pathway. Type III secretion systems function to target virulence proteins to host cells (11, 12). The conservation of the type III pathway in distantly related Gram-negative pathogenic bacteria (12, 13) has revealed a shared mechanism used by pathogens for the delivery of specialized virulence factors. Type III effector proteins are structurally unrelated and do not share a conserved signal peptide for export. However, extensive analysis of type III effector proteins, Yops, from Yersinia, has identified two modular domains at the N terminus of Yops sufficient for secretion and translocation (14). Secretion signals in some Yops are confined to the N-terminal 15–17 codons (15, 16), whereas translocation signals are located in the first 50–100 codons (15, 17). A specific chaperone binds within the translocation domain and facilitates the secretion and translocation of the respective Yop (18). Frameshift mutagenesis within some Yop secretion domains can be tolerated, suggesting that the secretion signal is the mRNA (16).
Significantly less is known about type III signals in effectors from phytopathogenic bacteria. Only recently have methods been established to study in vitro secretion from Xcv (19) and Pseudomonas syringae (20, 21). The translocation of a plant pathogen effector into a host cell has not yet been formally demonstrated. Technical limitations stem from the inability to target some reporters through the type III apparatus (20) and to identify reporters sensitive enough for detection in planta. However, indirect evidence does exist for the delivery of effectors to plant cells (11). What is known about plant pathogen type III signals is that the signals are similarly localized to N termini (20, 22), secretion signals can be encoded in the mRNA (22), and chaperones may exist (23). Moreover, the type III secretion systems of Xcv (19) and Erwinia chrysanthemi (20, 22, 24) secrete proteins from both plant and animal pathogens, implying that the recognition of type III secretion signals may be functionally conserved among type III pathogens.
Herein we have explored the mechanism for Xcv delivery of AvrBs2 to pepper plants. We have taken advantage of Xcv for studying type III trafficking because in vitro secretion of protein is robust (19). Moreover, the promiscuous secretion of proteins through the type III apparatus in Xcv (19) suggested to us that AvrRpt2, a P. syringae type III effector, would be a novel, sensitive reporter for detecting type III delivery of AvrBs2 to plants. Some P. syringae type III effector proteins are secreted by the type III pathways of unrelated animal and plant bacterial pathogens, including Xcv (19, 20, 22, 24). In some cases, the P. syringae effectors can induce localized cell death in resistant plants, inferring that the effectors are being translocated to the host cell by the heterologous type III apparatus (20, 24). This indicated to us that effectors devoid of type III targeting signals would be sensitive protein reporters to elucidate not only type III secretion signals but also type III translocation signals. We explicitly chose the P. syringae AvrRpt2 protein as a reporter because Xcv cannot secrete or translocate this effector. Furthermore, the C terminus of AvrRpt2, devoid of type III signals, is specifically recognized by RPS2 and is sufficient to induce cell death in planta (20). Thus, we predicted that the N terminus of AvrBs2 fused to the C-terminal domain of AvrRpt2 would be sufficient to secrete and translocate the fusion protein, providing us with a tool to further map AvrBs2 type III signals.
We now report that Xcv secretes AvrBs2 by the type III pathway. N-terminal and C-terminal deletion analysis of AvrBs2 has identified the effector domain of AvrBs2 specifically recognized by Bs2 in pepper plants. By using chimeric AvrBs2-AvrRpt2 fusion proteins, we have localized the minimal region of AvrBs2 required for in vitro type III secretion in Xcv. Furthermore, we have identified the region of AvrBs2 required for both type III secretion and translocation to plants. We also show that an mRNA signal within the first 18 codons of AvrBs2 is recognized by Xcv and required for the induction of Bs2 resistance in pepper.
Materials and Methods
Strains and Growth.
Strains used in this study were E. coli DH5α, Xcv strain 85–10 hrpG* (85*) and 85–10 hrpG* (85*) ΔhrcV (19, 25), Xcv strain GM98–38 mutant at the avrBs2 locus (2), Xc pv. campestris (Xcc) strain 8004 (M. Daniels, Sainsbury Laboratory, Norwich, U.K.), and A. tumefaciens C58C1 (pCH32) (4). E. coli and A. tumefaciens strains were grown on Luria agar medium (26) at 37°C and 28°C, respectively. Xcv and Xcc strains were grown on NYGA (27) at 28°C. Vectors were mobilized from E. coli into Xcv, Xcc, and A. tumefaciens by triparental matings by using standard methods.
Plasmid Construction.
PCR was used to construct gene fusions, deletions, and frameshifts. Numbering herein refers to the codon of the gene described. For AvrBs2, codon 1 represents the second AUG in the predicted ORF1 (3). PCR-generated DNA fragments were cloned into pCR4Blunt-TOPO (Invitrogen). Primers and conditions used for PCR will be available on request. The sequence of DNA constructs was verified by cycle sequencing.
For expression in Xc, constructs were cloned into pVSP61 (DNA Plant Technology, Oakland, CA) or a derivative, pDD62. For pDD62, the HindIII-EcoRI fragment of pVSP61 was replaced with a linker containing a BamHI site, a XhoI site, and stop codons in all three reading frames downstream of the lac promoter. HindIII and EcoRI sites were eliminated in this step. To construct pDD62 (avrBs2), the BamHI fragment from p81533b (B. Kearney and B.J.S., unpublished) containing the avrBs2 promoter and ORF was cloned into the BamHI site in pDD62. To construct pDD62(avrBs2-HA), PCR was used to amplify the DNA region 3′ of the avrBs2 ClaI site in p81533 to introduce a HA epitope, an in-frame stop codon, and a BamHI site followed by a SalI site. This ClaI-SalI fragment replaced the ClaI-SalI fragment in p81533 (3) creating p81533(avrBs2-HA). The HindIII-SalI fragment of p81533(avrBs2-HA) was then used to replace the HindIII-XhoI fragment in pDD62(avrBs2) creating pDD62(avrBs2-HA). The HindIII-BamHI fragment of p81533(avrBs2-HA) was cloned into pVSP61 creating pVSP61(promoterless avrBs297–714-HA). 3′ avrBs2 deletions were made by digesting p81533b with ScaI, PvuII, NcoI, or StuI, adding a BamHI linker to the cleaved DNA, and cloning the subsequent BamHI fragment into pDD62 to create pDD62(avrBs21–417), pDD62(avrBs21–497), pDD62(avrBs21–519), and pDD62(avrBs21–574), respectively. To engineer 5′ avrBs2 deletions, the promoter and coding region of interest (codon 1–97 at the HindIII site) were amplified separately and used as DNA template for a subsequent overlap PCR reaction. Once cloned into pCR4Blunt-TOPO, HindIII fragments were cloned into the HindIII site of pVSP61(promoterless avrBs297–714-HA), creating pVSP61(avrBs219–714-HA), pVSP61(avrBs251–714-HA), pVSP61(avrBs262–714-HA), and pVSP61(avrBs297–714-HA), all possessing the avrBs2 promoter. For avrBs2-avrRpt2 fusions, the 3′ end of avrRpt280–255 was amplified by using pRSR0 (28) as DNA template and cloned into pDD62(avrBs2) as a HindIII-XhoI fragment replacing the 3′ avrBs2 sequence creating pDD62(avrBs21–97+avrRpt280–255). Smaller regions of avrBs2 were amplified and cloned into the BamHI-HindIII site of pDD62(avrBs21–97+ avrRpt280–255), creating pDD62(avrBs21–58+ avrRpt280–255), pDD62(avrBs21–41+ avrRpt280–255), pDD62(avrBs21–28+ avrRpt280–255), and pDD62(avrBs21+avrRpt280-255), all possessing the avrBs2 promoter. By using overlap PCR, frameshifts (fs) were constructed by inserting a G nucleotide(s) (+1 and +2) after the AUG start codon of avrBs2. Reciprocal changes after avrBs2 codon 18 were made creating pVSP61(avrBs2+1fs-HA) and pVSP61(avrBs2+2fs-HA).
For transient expression by using A. tumefaciens, constructs were made in pMD1 (4) or a derivative pMDD1, where the XbaI-SacI fragment was replaced with a XbaI-SacI linker containing a BamHI site and a XhoI site followed by stop codons in all three reading frames. BamHI fragments in pDD62(avrBs2) and pDD62(avrBs2-HA) were cloned into the BamHI site of pMD1 creating pMD1(avrBs2) and pMD1(avrBs2-HA), respectively. XbaI-BamHI fragments in pVSP61(avrBs219–714-HA), pVSP61(avrBs251–714-HA), pVSP61(avrBs262–714-HA), pVSP61(avrBs297–714-HA), pVSP61(avrBs2+1fsHA), and pVSP61(avrBs2+2 fsHA) were cloned into the corresponding sites in pMD1 creating pMD1(avrBs219–714-HA), pMD1(avrBs251–714-HA), pMD1(avrBs262–714-HA), pMD1(avrBs297–714-HA), pMD1(avrBs2+1fs-HA), and pMD1(avrBs2+2fs-HA), respectively. BamHI fragments in pDD62(avrBs21–417), pDD62(avrBs21–497), pDD62(avrBs21–519), and pDD62(avrBs21–574) were cloned into the BamHI site of pMDD1, creating pMDD1(avrBs21–417), pMDD1(avrBs21–497), pMDD1(avrBs21–519), and pMDD1(avrBs21–574), respectively.
Secretion Assay.
Xcv 85* and 85* ΔhrcV strains grown overnight at 28°C on NYGA were suspended in secretion media, pH 7 (19), containing rif 5 μg/ml and kan 12 μg/ml. Ten-milliliter cultures (1 × 108 cells ml−1) were shaken for 12 h at 28°C and collected at 2,500 × g for 5 min at room temperature. Cells were washed in 1 mM MgCl2, repelleted, and resuspended in MgCl2. Bacteria were diluted to 4 × 108 cells ml−1 in 4 ml of secretion media, pH 5.4, containing rif 5 μg/ml, and 50 μg/ml BSA. Cultures were shaken for 4.5 h at 28°C. Cellular lysate fractions were obtained by precipitating 125 μl of each culture with 10% trichloroacetic acid on ice for 30 min. Protein was collected by centrifugation at 14,000 × g for 30 min, washed with 100% ethanol, and resuspended with 40 μl of sample buffer (20). Culture fluid fractions were obtained by removing cells from the remaining culture by centrifugation and then directly filtering supernatants through a 0.45-μm filter (HT Tuffryn, Gelman). Filtrates were precipitated and resuspended in 60 μl of sample buffer. Twenty microliters of the cellular lysate fraction and 30 μl of the culture fluid fraction were analyzed.
Protein Gels and Immunoblot Analysis.
Protein from Xc strains and A. tumefaciens-infected leaves was extracted from frozen samples by homogenization by using a Kontes pestle (Fisher) in a microfuge tube. Samples were suspended with buffer (8 M urea/0.1 M NaPO4/0.01 M Tris⋅HCl, pH 8.0), rocked for 30 min at room temperature, and then centrifuged at 14,000 × g for 15 min to obtain the supernatant identified as the total cellular protein. Protein fractions were mixed with sample buffer, boiled for 5 min, and then analyzed in a 10% gel by SDS/PAGE. For immunoblot analysis, proteins were transferred from gels to nitropure nitrocellulose (Osmonics) by electroblotting at 0.3 amps for 1 h in transfer buffer containing 10 mM 3-[cyclohexylamino]-1-propane sulfonic acid, pH 11.0, 10% (vol/vol) methanol. AvrBs2, AvrRpt2, and NPT II were detected by using rabbit polyclonal antisera at 1:2,000 (2), 1:2,000 (29), and 1:1,000 (5 Prime→3 Prime), respectively, in 20 mM Tris⋅HCl, pH 7.5/0.5 M NaCl/0.05% Tween 20 buffer containing 5% nonfat milk, followed by horseradish peroxidase-conjugated secondary antibodies and chemiluminescence.
Plant Growth and Bacterial Inoculation.
Pepper cultivars (cv.) Early Calwonder (ECW; bs2,bs2) and the near-isogenic cv. ECW-20R (Bs2,Bs2) were used for Xcv and A. tumefaciens inoculations. Arabidopsis thaliana ecotype Col-0 wild-type (RPS2,RPS2) and rps2–201 mutant (rps2, rps2) plants (30) were used for Xcc inoculations. A. thaliana plants were grown in chambers at 22°C under 8 h photoperiod. Pepper plants were grown under greenhouse conditions. Bacteria were hand infiltrated into plant leaves through a small wound by using a 1-cc syringe.
Agrobacterium-Mediated Transient Expression Assay.
A. tumefaciens was grown overnight at 28°C on Luria agar medium containing rif 100 μg/ml, tet 5 μg/ml, and kan 35 μg/ml. Bacteria were collected and incubated in inducing media (10 mM Mes, pH 5.6/10 mM MgCl2/and 150 μM acetosyringone) for 2 h before inoculation.
Results
Type III-Dependent Secretion of AvrBs2 Protein from Xcv.
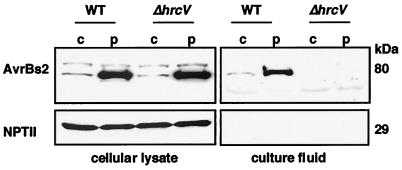
To dissect the targeting of AvrBs2 protein to host cells, we first determined whether AvrBs2 is secreted by the type III pathway. We used the Xcv strain 85* that contains a chromosomal copy of avrBs2 and constitutively expresses hrp genes (locus encoding the type III apparatus) because of a mutation in the regulatory gene hrpG (25). We also used Xcv strain 85* ΔhrcV, a secretion defective strain, to test hrp-dependent secretion of AvrBs2. Conditions previously established to assay for Xcv secretion (19) were used to detect AvrBs2 secretion. Immunoblot analysis shows that AvrBs2 protein is present in cellular lysate from Xcv 85* and Xcv 85* ΔhrcV strains when avrBs2 is expressed chromosomally or by pDD62 (Fig. 1). Importantly, the secretion competent Xcv strains released AvrBs2 into the culture fluid (Fig. 1). Robust AvrBs2 expression and secretion was observed only when avrBs2 was expressed from pDD62. To confirm that AvrBs2 protein in the Xcv 85* culture fluids was not because of cell lysis, immunoblot analysis was repeated by using antisera for NPT II, a cytoplasmic protein encoded by the plasmid pDD62. NPTII was detected only in the cellular lysate and not in the culture fluid (Fig. 1.). These data demonstrate that Xcv secretes AvrBs2 by the type III pathway and that secretion is hrp dependent. Considering that secretion of AvrBs2 from Xcv strain 85* pDD62(avrBs2) was more robust, we subsequently expressed all avrBs2 constructs from pDD62.
Figure 1.
Type III-dependent secretion of AvrBs2 from Xcv. Immunoblot analysis of AvrBs2 and NPT II protein in cellular lysate and culture fluid isolated from Xcv strain 85* pDD62(avrBs2) expressing AvrBs2 (80 kDa) and NPT II (29 kDa). wt; wild-type secretion strain. ΔhrcV; secretion mutant. c; chromosomal avrBs2. p; plasmid avrBs2.
Identification of AvrBs2's in Planta Effector Domain by Deletion Analysis.
We next performed mutagenesis of avrBs2 to determine the effector domain of AvrBs2 required for avrBs2-dependent localized cell death in Bs2 pepper plants. AvrBs2 N-terminal deletions were constructed introducing an ATG before AvrBs2 codon 19, 51, 62, and 97 (3). Xcv GM98–38 strains carrying pDD62(avrBs2-HA), pVSP61(avrBs219–714-HA), pVSP61(avrBs251–714-HA), pVSP61(avrBs262–714-HA), or pVSP61(avrBs297–714-HA) all expressed the expected AvrBs2-specific polypeptides (Table 1). However, when hand inoculated into Bs2 pepper leaves, only Xcv expressing AvrBs2-HA was able to induce avrBs2-dependent cell death (Table 1). To determine whether the N-terminal deletions were affecting the delivery of AvrBs2 or Bs2-dependent recognition (25) directly, all of the N-terminal AvrBs2 deletions were cloned into pMD1 and then expressed in planta by using the Agrobacterium-mediated transient expression assay. AvrBs2-specific cell death was induced by all of the N-terminal deletion constructs, except AvrBs297–714-HA (Table 1). These data clearly show that the N terminus of AvrBs2 is not required to trigger a Bs2-dependent response in planta, suggesting that these truncated proteins are instead defective in type III targeting. Moreover, in planta Bs2-dependent recognition requires the N-terminal AvrBs2 domain between codon 62 and 97.
Table 1.
Phenotypes of AvrBs2 deletion polypeptides expressed by Xcv and by Agrobacterium-mediated transformation in pepper Bs2 leaves
| AvrBs2 protein* |
Xcv
expression
|
A. tumefaciens expression in
planta
|
||
|---|---|---|---|---|
| Phenotype† | Protein | Phenotype† | Protein | |
| Mature protein | ||||
| 1-714 | HR | Yes | HR | Yes |
| 1-714-HA‡ | HR | Yes | HR | Yes |
| N-terminal deletions | ||||
| 19-714-HA | NS | Yes | HR | Yes |
| 51-714-HA | NS | Yes | HR | Yes |
| 62-714-HA | NS | Yes | HR | Yes |
| 97-714-HA | NS | Yes | NS | Yes |
| C-terminal deletions | ||||
| 1-417 | NS | No | NS | No |
| 1-497 | w-HR | Yes | HR | Yes |
| 1-519 | HR | Yes | HR | Yes |
Codon numbering is based on the predicted protein for ORF1 (3).
HR, hypersensitive cell death response; NS, no symptoms, w-HR, weak HR.
HA, hemagglutinin epitope.
We also deleted the C-terminal region of AvrBs2 to define the effector domain required for avrBs2-dependent localized cell death in Bs2 pepper plants. Xcv GM98–38 carrying pDD62(avrBs2), pDD62(avrBs21–417), pDD62(avrBs21–497), pDD62(avrBs21–519), or pDD62(avrBs21–574) were inoculated into Bs2 pepper leaves. All of the C-terminal deleted AvrBs2 polypeptides induced cell death except for AvrBs21–417 (Table 1). However, AvrBs21–417 protein could not be detected in Xcv. Failure to detect this polypeptide may reflect antibody specificity and/or protein stability. These results demonstrate that the C-terminal 217 codons of AvrBs2 are dispensable for in planta Bs2-dependent recognition of AvrBs2.
Identification of AvrBs2 Type III Secretion Signals By Using a AvrRpt2 Reporter.
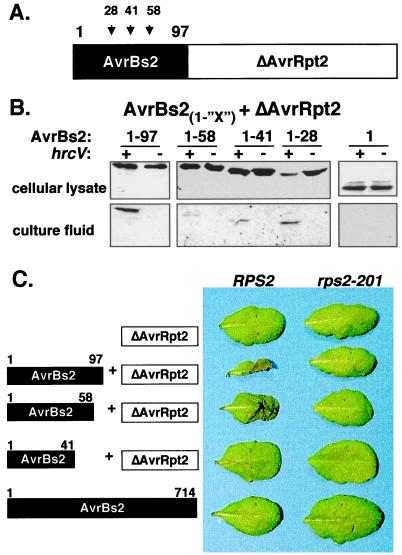
To identify the type III signals in AvrBs2 required for secretion and translocation to host plants, we fused the N-terminal coding region of AvrBs2 (codons 1–97) to the C-terminal coding region of AvrRpt2 (codons 80–255), a P. syringae type III effector (20). We predicted that the N-terminal 97 codons of AvrBs2 fused to AvrRpt280–255 (devoid of type signals, Fig. 2A) would be sufficient to secrete the fusion protein through the Xcv type III apparatus. Immunoblot analysis shows that Xcv strain 85* carrying pDD62(avrBs21–97+avrRpt80–255) expressed the predicted 30-kDa fusion protein in cell lysate and then secreted it into the culture fluid (Fig. 2B). No AvrBs21–97+AvrRpt280–255 protein was detected in the culture fluid of Xcv strain 85* ΔhrcV carrying pDD62(avrBs21–97+avrRpt80–255) demonstrating that secretion was hrp dependent. The intracellular NPT II marker was not detected in the culture fluid of any strain presented in Fig. 2B (data not shown), confirming that the presence of all detected fusion proteins in the culture fluid was not a result of Xcv lysis. These data show that the first 97 codons of AvrBs2 contain the AvrBs2 type III secretion signal, which is sufficient to target the P. syringae truncated AvrRpt280–255 protein through the Xcv type III apparatus.
Figure 2.
(A) Schematic of chimeric AvrBs2-AvrRpt2 fusion proteins. AvrBs2 codons 1–28, 1–41, 1–58, and 1–97 were independently fused to AvrRpt2 codons 80–255 (ΔAvrRpt2). (B) AvrBs2 N-terminal codons secrete the AvrRpt280–255 effector via the Xcv type III pathway. Immunoblot analysis of cellular lysate and culture fluid isolated from Xcv strain 85* expressing AvrBs21-“X”+AvrRpt280–255 in pDD62 by using AvrRpt2 antisera. Numbers refer to the respective codon positions of AvrBs2 fused to the reporter AvrRpt280–255 protein. +, wild-type secretion strain. −, ΔhrcV secretion mutant. (C) AvrRpt2-dependent cell death in resistant A. thaliana RPS2 leaves inoculated with Xcc strains expressing the AvrBs2-AvrRpt2 fusion proteins. Resistant RPS2 and susceptible rps2–201 A. thaliana leaves were inoculated with a 2.5 × 108 cells ml−1 suspension of bacteria and then photographed 24 h later. Schematic of protein expressed in Xcc shown on left. Symptoms of Xcc in leaves shown on right.
Next, we deleted codons within the AvrBs2 domain of the AvrBs21–97+AvrRpt280–255 fusion protein to identify the minimal region of AvrBs2 required for in vitro type III secretion in Xcv. Xcv 85* and 85* ΔhrcV strains expressing AvrBs2 codons 1–58, 1–41, and 1–28 fused to AvrRpt280–255 (Fig. 2A) were tested for secretion into culture fluid. Immunoblot analysis shows that all of the fusion proteins were expressed in Xcv cellular lysate and secreted into the culture fluid in a hrp-dependent manner (Fig. 2B.). A fusion protein containing the first 16 codons of AvrBs2 was poorly expressed in Xcv; therefore, its secretion could not be confirmed (data not shown). Thus, we have defined the first 28 codons of AvrBs2 to be the minimal signal sufficient for detectable in vitro secretion of AvrRpt280–255 into Xcv culture fluid.
Identification of AvrBs2 Type III Signals Required for Translocation to a Host By Using a AvrRpt2 Reporter.
To determine whether codons 1–97 of AvrBs2 are sufficient for directing the secretion and translocation of the AvrRpt2 reporter to host cells, we inoculated Xc pv. campestris (Xcc) strains expressing the AvrBs21–97+AvrRpt280–255 fusion protein into A. thaliana leaves. We chose Xcc to express the fusion protein because Xcc does not inherently induce localized cell death in A. thaliana (data not shown). Furthermore, resistant RPS2 A. thaliana plants specifically recognize AvrRpt280–255 when it is expressed in planta (20). Thus, if codons 1–97 of AvrBs2 are sufficient for type III translocation of the AvrRpt2 reporter, then Xcc expressing the AvrBs21–97+AvrRpt280–255 fusion protein should induce AvrRpt2-dependent localized cell death on resistant RPS2 A. thaliana plants. Resistant RPS2 A. thaliana leaves infected with Xcc expressing AvrBs21–97+AvrRpt280–255 induced AvrRpt2-dependent cell death (Fig. 2C). No symptoms were observed on rps2–201 leaves, an A. thaliana mutant lacking a functional RPS2 allele. Conversely, avrRpt2-dependent cell death was not induced in RPS2 A. thaliana leaves infected with Xcc expressing AvrRpt280–255 or mature AvrBs2 (Fig. 2C). This data demonstrates that the first 97 codons of AvrBs2 are required for Xanthomonas to translocate AvrRpt280–255 to RPS2 A. thaliana plants.
To identify the minimal region of AvrBs2 required for type III translocation, the AvrBs2-AvrRpt2 fusion proteins used to dissect AvrBs2-specific type III secretion signals (Fig. 2B) were expressed in Xcc. The resulting Xcc strains were used to inoculate A. thaliana plants. Only resistant RPS2 A. thaliana plants infected with Xcc expressing AvrBs21–97+AvrRpt280–255 and AvrBs21–58+AvrRpt280–255 induced avrRpt2-dependent cell death in RPS2 A. thaliana plants (Fig. 2C). No symptoms were observed in rps2–201 A. thaliana plants. Conversely, Xcc expressing AvrBs21–41+AvrRpt280–255 was unable to translocate AvrRpt280–255 to RPS2 plants (Fig. 2C), even though it is targeted for secretion (Fig. 2B). All fusion proteins were equally expressed in Xcc (data not shown); therefore, type III translocation deficiency was not because of protein instability. We have thus found that AvrBs2 codons 1–58 are required for Xanthomonas to translocate the AvrRpt2 reporter protein to A. thaliana.
We next explored the possibility that the type III apparatus in P. syringae might recognize the AvrBs2 type III signals used in Xanthomonas. P. syringae pv. tomato DC3000 strains expressing the AvrBs21–97+AvrRpt280–255 fusion protein were inoculated onto A. thaliana plants. No symptoms were observed in these plants (data not shown). This indicates that the type III apparatus in P. syringae pv. tomato cannot recognize AvrBs2-specific type III signal sequences for the delivery of the Pseudomonas-specific AvrRpt2 reporter.
Identification of an mRNA Secretion Signal in AvrBs2.
Some bacterial effectors possess mRNA secretion signals (16, 22), suggesting that type III apparatuses are capable of recognizing signals that couple mRNA translation to polypeptide secretion. To determine whether AvrBs2 possesses an mRNA secretion signal, we constructed frameshift mutations in avrBs2 by inserting nucleotides immediately after the AUG start codon. The correct reading frame was restored after codon 18 by reciprocal nucleotide changes. We chose to introduce the frameshifts within the first 28 codons of AvrBs2 because this is the region required for the secretion of AvrRpt280–255 in Xcv (Fig. 2B). Because changes made within the first 28 codons of AvrBs2 affected protein stability in Xcv (data not shown), the AvrBs2 frameshift proteins, AvrBs2+1fs-HA and AvrBs2+2fs-HA, were not assayed for in vitro type III secretion. Instead, we analyzed the frameshift proteins for biological activity in sensitive Xcv infection assays.
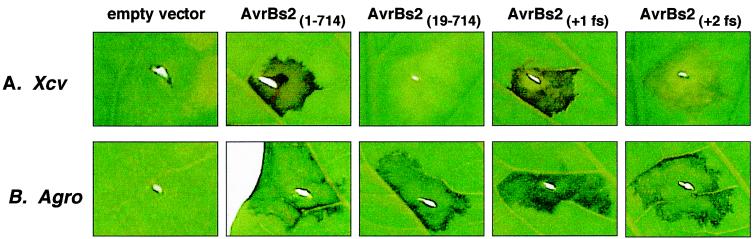
Resistant Bs2 pepper plants infected with Xcv GM98–38 expressing wild-type AvrBs2 induced Bs2-dependent localized cell death (Fig. 3A), whereas Xcv GM98–38 alone displayed no symptoms (Fig. 3A). Leaves infected with Xcv GM98–38 expressing AvrBs219–714 also displayed no symptoms (Fig. 3A), indicating that the first 18 codons of AvrBs2 are required for the effective delivery of the AvrBs2 signal to Bs2 pepper plants. However, if AvrBs219–714 is expressed transiently in planta, it is recognized by Bs2 plants and cell death is induced (Fig. 3B). These data support our type III secretion and translocation assays demonstrating that the region within the first 28 codons of AvrBs2 is essential for type III delivery to the host (Fig. 2 B and C). Interestingly, resistant Bs2 pepper plants infected with Xcv GM98–38 strains expressing either AvrBs2+1fs-HA or AvrBs2+2fs-HA induce Bs2-dependent cell death (Fig. 3A). Although the AvrBs2+1fs-HA effector protein was more potent in inducing Bs2-dependent cell death in Xcv infections, both frameshift proteins were equally effective in inducing cell death when transiently expressed in planta compared with wild type AvrBs2 (Fig. 3B). The reading frames (+1 and +2) resulting from mutagenesis did not encode peptides with physical properties distinctly different from that encoded by the correct AvrBs2 reading frame. Considering that the frameshift proteins retain AvrBs2-specific biological activity when expressed by Xcv, we have identified a possible mRNA signal within the first 18 codons of AvrBs2.
Figure 3.
avrBs2-dependent localized cell death in resistant Bs2 pepper is mediated via an mRNA signal. (A) Xcv symptoms on resistant Bs2 pepper leaves. Plants were inoculated with a 2 × 108 cells ml−1 suspension of Xcv strain GM98–38 carrying pDD62 (empty vector), pDD62(avrBs21–714), pVSP61(avrBs219–714-HA), pVSP61(avrBs2+1fs-HA), or pVSP61(avrBs2+2fs-HA). Symptoms were photographed 48 h after inoculation. (B) Agrobacterium-mediated transient expression of avrBs2 genes in resistant Bs2 pepper leaves. Plants were inoculated with a 6 × 108 cells ml-1 suspension of A. tumefaciens (Agro) strain C58C1 (pCH32) carrying pMD1 (empty vector), pMD1(avrBs21–714), pMD1(avrBs219–714-HA), pMD1(avrBs2+1fs-HA), or pMD1(avrBs2+2fs-HA). Symptoms were photographed 72 h after inoculation.
Discussion
In this study, we provide evidence supporting type III-dependent secretion of AvrBs2. We show that the phytopathogenic bacterium Xcv secretes mature AvrBs2 protein into culture fluid, and that secretion requires a functional type III secretion system. Mutation of hrcV, a predicted inner membrane structural component of the type III apparatus in Xcv (31, 32), prevented the secretion of AvrBs2 from Xcv and the induction of localized cell death in resistant Bs2 pepper plants (data not shown). These results show that secretion of AvrBs2 through the type III apparatus is one event essential for the molecular recognition of this effector in planta.
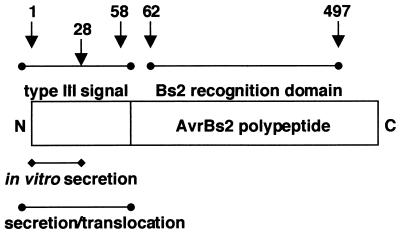
In addition, we have characterized the molecular signals required for the type III secretion and translocation of AvrBs2 from Xcv to pepper plants (summarized in Fig. 4). The use of AvrRpt2, a P. syringae type III effector protein, as a reporter in Xcv enabled us to localize key regions in AvrBs2 required for type III-dependent secretion and translocation. We show that the truncated AvrRpt280–255 protein, a region devoid of type III signals, was an ideal reporter for the mapping of AvrBs2 type III signals in Xcv. Consistent with our hypothesis, the N-terminal 97 codons of AvrBs2 fused to AvrRpt280–255 was sufficient to target AvrRpt280–255 through the Xcv type III pathway. AvrBs21–97+AvrRpt280–255 was secreted into Xcv culture fluid in a type III-dependent manner. Moreover, the same fusion protein was able to induce avrRpt2-dependent cell death in Xcc infected RPS2 A. thaliana plants, indirectly demonstrating translocation of the AvrRpt280–255 protein. By using the AvrRpt280–255 reporter, the minimal region of AvrBs2 required for efficient type III secretion into Xcv culture fluids was mapped to the first 28 codons of AvrBs2. Yet the first 58 codons of AvrBs2 were required for phenotypic expression of the AvrRpt280–255 reporter in planta. We have thus identified two modular domains required for type III delivery, one for in vitro secretion and another for secretion and translocation. The spatial distribution of AvrBs2's secretion and translocation domains resembles those identified for YopE and YopH (15, 17, 33). Curiously, the translocation domain of YopE has been recently shown to be involved in the inhibition of YopE type III targeting in the absence of its SycE chaperone (34). We are further investigating the functional role of the secretion and translocation domains encompassing AvrBs2's type III signal, as well as the possibility of a chaperone.
Figure 4.
Schematic of AvrBs2 type III signals and Bs2 in planta recognition domain. Numbers refer to the codons in AvrBs2.
Type III effectors from Yersinia and P. syringae have been recently shown to possess mRNA targeting signals, which are recognized by the type III secretion apparatus in Yersinia enterocolitica (16, 22). The existence of mRNA signals in type III effectors suggests a chaperone-independent mechanism for protein targeting via this pathway. We explored the possibility that an mRNA targeting signal may be present within the defined type III secretion domain (codons 1–28) identified for AvrBs2. Sensitive Xcv infection assays in Bs2 pepper plants revealed that frameshift mutations within the first 18 codons of AvrBs2 are tolerated and still enable phenotypic expression of avrBs2 in the host. This suggests that an mRNA targeting domain resides within first 18 codons of this effector, and that the type III secretion pathway in Xcv is also capable of recognizing an mRNA signal sequence. Our work suggests that type III secretion in Xcv, as in Y. enterocolitica (16), is governed by signals that couple mRNA translation to polypeptide targeting.
We have also identified the effector domain of AvrBs2 that is sufficient to initiate Bs2-specific disease resistance-mediated cell death in planta. Our localized cell death assays have mapped the recognition domain of AvrBs2 to codons 62 and 497 of the mature polypeptide. Although the biological function of AvrBs2 remains unclear, it shares homology to agrocinopine synthase (ACS) of A. tumefaciens and the glycerophosphoryl diester phosphodiesterase UgpQ of E. coli (3). The effector domain of AvrBs2 mapped herein resides within the region of highest homology to ACS and UgpQ (2, 3). Interestingly, all natural field strains of Xcv overcoming Bs2 plant resistance possess molecular lesions within codons 62 and 497 of the avrBs2 gene (2).
In conclusion, this work has identified the structural domains in AvrBs2 required for Xcv type III delivery to Bs2 pepper plants. We are using this information to design experiments to formally demonstrate the direct translocation of AvrBs2 into plant cells. Phytopathogenic bacteria residing in the apoplastic fluid of plant mesophyll cells have to traverse the cell wall to deliver effector proteins. Thus, although plant and animal pathogens may share similar type III secretion signals, we expect plant pathogens to use unique strategies to invade the cell wall barrier. Progress in understanding how Xcv presents this key virulence determinant to host cells will provide new insight in to strategies used by the pathogen to avoid host recognition and to cause disease.
Acknowledgments
We thank Peter Repetti and Mike Axtell for constructive comments regarding this manuscript. This work was supported by Novartis Agricultural Discovery Institute (San Diego, CA).
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.230450797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.230450797
References
- 1.Kearney B, Staskawicz B J. Nature (London) 1990;346:385–386. doi: 10.1038/346385a0. [DOI] [PubMed] [Google Scholar]
- 2.Gassmann, W., Dahlbeck, D., Chesnokova, O., Minsavage, G. V., Jones, J. B. & Staskawicz, B. J. (2000) J. Bacteriol.182, in press. [DOI] [PMC free article] [PubMed]
- 3.Swords K M M, Dahlbeck D, Kearney B, Roy M, Staskawicz B J. J Bacteriol. 1996;178:4661–4669. doi: 10.1128/jb.178.15.4661-4669.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tai T H, Dahlbeck D, Clark E T, Gajiwala P, Pasion R, Whalen M C, Stall R E, Staskawicz B J. Proc Natl Acad Sci USA. 1999;96:14153–14158. doi: 10.1073/pnas.96.24.14153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minsavage G V, Dahlbeck D, Whalen M C, Kearney B, Bonas U, Staskawicz B J, Stall R E. Mol Plant–Microbe Inter. 1990;3:41–47. [Google Scholar]
- 6.Meyers B C, Dickerman A W, Michelmore R W, Sivaramakrishnan S, Sobral B W, Young N D. Plant J. 1999;20:317–332. doi: 10.1046/j.1365-313x.1999.t01-1-00606.x. [DOI] [PubMed] [Google Scholar]
- 7.Hammond-Kosack K E, Jones J D G. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:575–607. doi: 10.1146/annurev.arplant.48.1.575. [DOI] [PubMed] [Google Scholar]
- 8.Kousik C S, Ritchie D F. Plant Dis. 1998;82:181–186. doi: 10.1094/PDIS.1998.82.2.181. [DOI] [PubMed] [Google Scholar]
- 9.Kousik C S, Ritchie D F. Phytopathology. 1996;86:1336–1343. [Google Scholar]
- 10.Bonas U. Mol Plant–Microbe Inter. 1991;4:81–88. doi: 10.1094/mpmi-4-628. [DOI] [PubMed] [Google Scholar]
- 11.Kjemtrup S, Nimchuk Z, Dangl J L. Curr Opin Microbiol. 2000;3:73–78. doi: 10.1016/s1369-5274(99)00054-5. [DOI] [PubMed] [Google Scholar]
- 12.Galan J E, Collmer A. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 13.Hueck C J. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornelis G R. Proc Natl Acad Sci USA. 2000;97:8778–8783. doi: 10.1073/pnas.97.16.8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sory M P, Boland A, Lambermont I, Cornelis G R. Proc Natl Acad Sci USA. 1995;92:11998–2002. doi: 10.1073/pnas.92.26.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson D, Schneewind O. Science. 1997;278:1140–1143. doi: 10.1126/science.278.5340.1140. [DOI] [PubMed] [Google Scholar]
- 17.Schesser K, Frithz-Lindsten E, Wolf-Watz H. J Bacteriol. 1996;178:7227–7233. doi: 10.1128/jb.178.24.7227-7233.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wattiau P, Woestyn S, Cornelis G R. Mol Microbiol. 1996;20:255–262. doi: 10.1111/j.1365-2958.1996.tb02614.x. [DOI] [PubMed] [Google Scholar]
- 19.Rossier O, Wengelnik K, Hahn K, Bonas U. Proc Natl Acad Sci USA. 1999;96:9368–9373. doi: 10.1073/pnas.96.16.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mudgett M B, Staskawicz B J. Mol Microbiol. 1999;32:927–941. doi: 10.1046/j.1365-2958.1999.01403.x. [DOI] [PubMed] [Google Scholar]
- 21.van Dijk K, Fouts D E, Rehm A H, Hill A R, Collmer A, Alfano J R. J Bacteriol. 1999;181:4790–4797. doi: 10.1128/jb.181.16.4790-4797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson D M, Fouts D E, Collmer A, Schneewind O. Proc Natl Acad Sci USA. 1999;96:12839–12843. doi: 10.1073/pnas.96.22.12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alfano J R, Kim H S, Delaney T P, Collmer A. Mol Plant–Microbe Inter. 1997;10:580–588. doi: 10.1094/MPMI.1997.10.5.580. [DOI] [PubMed] [Google Scholar]
- 24.Ham J H, Bauer D W, Fouts D E, Collmer A. Proc Natl Acad Sci USA. 1998;95:10206–10211. doi: 10.1073/pnas.95.17.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wengelnik K, Rossier O, Bonas U. J Bacteriol. 1999;181:6828–6831. doi: 10.1128/jb.181.21.6828-6831.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 27.Turner P, Barber C, Daniels M. Mol Gen Genet. 1984;195:101–107. [Google Scholar]
- 28.Innes R W, Bent A F, Kunkel B N, Bisgrove S R, Staskawicz B J. J Bacteriol. 1993;175:4859–4869. doi: 10.1128/jb.175.15.4859-4869.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McNellis T W, Mudgett M B, Li K, Aoyama T, Horvath D, Chua N-H, Staskawicz B J. Plant J. 1998;14:247–257. doi: 10.1046/j.1365-313x.1998.00106.x. [DOI] [PubMed] [Google Scholar]
- 30.Kunkel B N, Bent A F, Dahlbeck D, Innes R W, Staskawicz B J. Plant Cell. 1993;5:865–875. doi: 10.1105/tpc.5.8.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bogdanove A J, Beer S V, Bonas U, Boucher C A, Collmer A, Coplin D L, Cornelis G R, Huang H-C, Hutchineson S W, Panopoulos N J, et al. Mol Microbiol. 1996;20:681–683. doi: 10.1046/j.1365-2958.1996.5731077.x. [DOI] [PubMed] [Google Scholar]
- 32.Fenselau S, Balbo I, Bonas U. Mol Plant–Microbe Inter. 1992;5:390–396. doi: 10.1094/mpmi-5-390. [DOI] [PubMed] [Google Scholar]
- 33.Boland A, Sory M-P, Iriarte M, Kerbourch C, Wattiau P, Cornelis G R. EMBO J. 1996;15:5191–5201. [PMC free article] [PubMed] [Google Scholar]
- 34.Boyd A P, Lambermont I, Cornelis G R. J Bacteriol. 2000;182:4811–4821. doi: 10.1128/jb.182.17.4811-4821.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]