Abstract
Drosophila melanogaster offers many unique advantages for deciphering the complexities of glycan biosynthesis and function. The completion of the Drosophila genome sequencing project as well as the comprehensive catalogue of existing mutations and phenotypes have lead to a prolific database where many of the genes involved in glycan synthesis, assembly, modification, and recognition have been identified and characterized. Recent biochemical and molecular studies have elucidated the structure of the glycans present in Drosophila. Powerful genetic approaches have uncovered a number of critical biological roles for glycans during development that impact on our understanding of their function during mammalian development. Here, we summarize key recent findings and provide evidence for the usefulness of this model organism in unraveling the complexities of glycobiology across many species.
Keywords: Development, Drosophila, glycosylation
Introduction
The complexities of glycan biosynthesis and structure necessitate the use of a system that affords the advantages of sophisticated genetics as well as reduced genome redundancy. The Drosophila melanogaster genome is inherently less redundant (∼14,000 genes in Drosophila versus ∼25,000 in humans) (Rubin et al. 2000; Stein 2004; Hahn et al. 2007) and can therefore circumvent the functional redundancy present in many glycosyltransferase families. Additionally, the elegant genetic strategies employed in Drosophila have been used historically to uncover and elucidate many complex biological events. Random mutagenesis coupled with sophisticated mapping techniques has yielded the identification of many novel genes responsible for a plethora of evolutionarily conserved regulatory events. Comprehensive databases cataloguing alleles, phenotypes, expression patterns, and interacting partners of the many genes previously characterized further provide an invaluable resource for rapidly deciphering the function of additional newly discovered genes. Continuing efforts to mutagenize every gene in the genome through transposon targeting techniques have provided a wealth of reagents to interrogate the function of a large number of previously uncharacterized genes. Whole genome RNA interference (RNAi) in insect cell culture provides a system for rapidly cataloguing the function of genes in any cellular process for which a screen has been developed (e.g., viability, growth, morphological changes, cell signaling, cell division, cell adhesion) (Kiger et al. 2003; Boutros et al. 2004). Additionally, techniques for tissue- and stage-specific knockdown of gene expression via RNAi in the fly can address the role of specific genes in specific developmental processes. The recent construction of a genome-wide transgenic RNAi library in the fly will enable researchers to rapidly interrogate the developmental consequences of almost any gene of interest (Dietzl et al. 2007).
In recent years, much progress has been made using Drosophila melanogaster to study many diverse aspects of glycobiology. In this review, we will summarize recent work elucidating glycan function using the fly as a model system.
Glycan function in Drosophila
The role of proteoglycans during Drosophila development
The importance of glycans during Drosophila development was first discovered while dissecting the developmental impact of the components of proteoglycans, which consist of a glycosaminoglycan (GAG) chain attached to serine residues of core proteins. While a detailed account of the extensive advances in this field is beyond the scope of this review, we will highlight key points elucidated in the fly. Both secreted and cell-surface proteoglycans exist in the fly. Cell surface proteoglycans consist of two classes: glypicans, which are membrane-bound via a GPI anchor and are modified with heparan sulfate (HS); and syndecans, which are transmembrane proteins modified with heparan sulfate or chondroitin sulfate (CS). Studies in the fly and fly cell culture systems have led to a number of different models describing the role of proteoglycans in hedgehog (Hh), wingless (Wg), fibroblast growth factor (FGF), and decapentaplegic/TGF-β (Dpp) signaling pathways during development (reviewed in Nybakken and Perrimon 2002 and Hacker et al. 2005). These models include: (1) proteoglycans acting as coreceptors to enhance the receptor–ligand interaction; (2) proteoglycans aiding in requisite ligand dimerization for efficient receptor binding; and (3) proteoglycans influencing ligand/morphogen gradient formation by regulating ligand diffusion, transport, stability, secretion, or endocytosis. While initial information defining the role of proteoglycans came from analysis of mutations in genes encoding the core proteins to which the GAG chains are attached, genetic screens have revealed (and continue to reveal) the importance of genes responsible for GAG biosynthesis and modification (Figure 1).
Fig. 1.
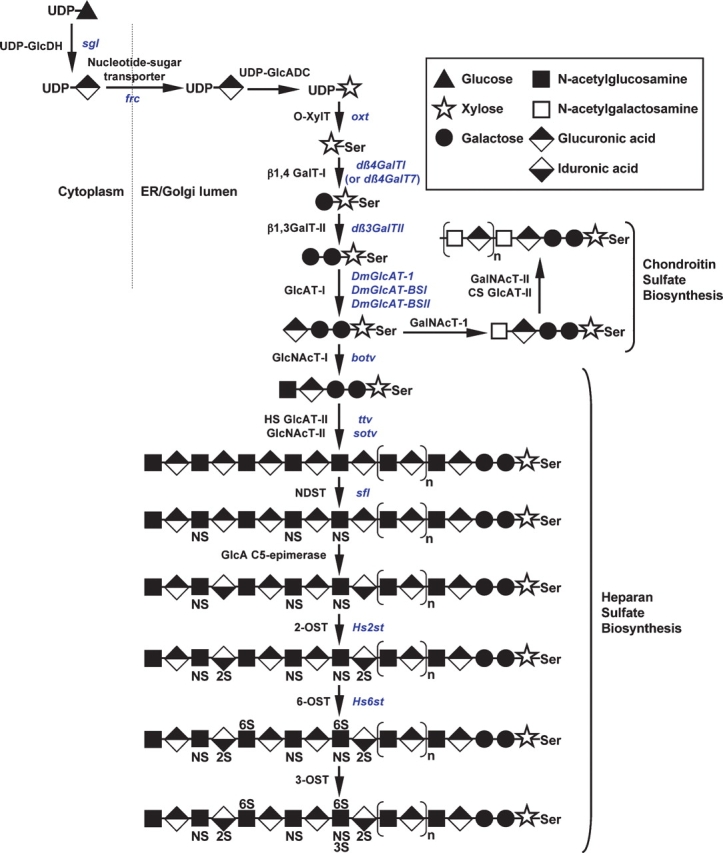
Biosynthesis of glycosaminoglycans. The initiation of chondroitin sulfate (CS) and the complete synthesis of heparan sulfate (HS) are shown. Enzymes responsible for catalyzing each step are shown in black and the corresponding Drosophila genes are shown in blue. Enzyme abbreviations are as follows: UDP-GlcDH, UDP-glucose dehydrogenase; UDP-GlcADC, UDP-glucuronic acid decarboxylase; O-XylT, polypeptide O-xylosyltransferase; β1,4GalT-I, xylose-β1,4-galactosyltransferase; β1,3GalT-II, galactose-β1,3-galactosyltransferase; GlcAT-I, galactose-β1,3-glucuronyltransferase; GalNAcT-I, glucuronic acid-β1,4-N-acetylgalactosaminyltransferase; CS GlcAT-II, chondroitin sulfate GalNAc-β1,3-glucuronyltransferase; GalNAcT-II, glucuronic acid-β1, 4-N-acetylgalactosaminyltransferase; GlcNAcT-I, glucuronic acid-α1,4-N-acetylglucosaminyltransferase; HS GlcAT-II, heparan sulfate GlcNAc-β1, 4-glucuronyltransferase; GlcNAcT-II, glucuronic acid α1,4-N-acetylglucosaminyltransferase; NDST, N-deacetylase/N-sulfotransferase; 2-OST, 2-O-sulfotransferase; 6-OST, 6-O-sulfotransferase; 3-OST, 3-O-sulfotransferase.
GAG synthesis is initiated by the addition of a xylose to the peptide backbone of proteins destined to become proteoglycans (Figure 1). Drosophila has one xylosyltransferase (encoded by the oxt gene) responsible for catalyzing this addition (Wilson 2002; Brunner et al. 2006). Defects in this gene have not yet been characterized. The core xylose is then further extended by the addition of galactose through the action of a single β1,4-galactosyltransferase (β1,4 GalT-I) encoded by the gene dβ4GalT7 (also known as dβ4GalTI) (Nakamura et al. 2002; Takemae et al. 2003). RNAi to dβ4GalT7 in Drosophila impaired HS and CS biosynthesis and resulted in abnormal wing and leg morphology, phenocopying defects in Hh and Dpp signaling (Nakamura et al. 2002; Takemae et al. 2003). The next step in the pathway is catalyzed by a proteoglycan β1,3-galactosyltransferase (encoded by the dβ3GalTII gene; Figure 1), which transfers galactose to the Galβ1-4Xyl disaccharide core. RNAi to dβ3GalTII resulted in decreased levels of heparan sulfate proteoglycans (HSPGs) and decreased levels of extracellular Wg (Ueyama et al. 2008). The fly also expresses three glucuronyltransferases, encoded by the genes DmGlcAT-I, DmGlcAT-BSI, and DmGlcAT-BSII (BS stands for “broad specificity”). DmGlcAT-I demonstrated specificity for transferring GlcA to the linkage region trisaccharide (Galß1-3Galß1-4Xyl) of proteoglycans (Kim et al. 2003). In contrast, DmGlcAT-BSI and -BSII transferred GlcA to a wide range of substrates, including proteoglycans, glycolipids, and glycoproteins, suggesting their potential involvement in the synthesis and extension of a variety of glycans in the fly (Kim et al. 2003). Interestingly, DmGlcAT-BSI and DmGlcAT-BSII are widely expressed during development, suggesting that GlcA may serve as a major negatively charged sugar in the fly, as sialic acid addition appears to be much more restricted (see section on N-Linked glycosylation).
The next steps of GAG biosynthesis are catalyzed by members of the EXT (hereditary multiple exostosin) gene family. Brother of tout velu (botv) encodes an N-acetylglucosoaminyltransferase that adds GlcNAc to the tetrasaccharide core of HSPGs and may also contribute to chain elongation (Han et al. 2004; Izumikawa et al. 2006). Tout velu (ttv) and sister of tout velu (sotv) encode enzymes which form a complex responsible for the sequential, repeating addition of GlcA and GlcNAc to elongate the HS chains (Han et al. 2004; Izumikawa et al. 2006). Mutations in ttv, sotv, or botv result in impared Wg, Hh and Dpp signaling as well as reduced or abrogated HS synthesis, indicating the crucial role of GAGs in morphogen gradient formation (Toyoda et al. 2000; Takei et al. 2003; Han et al. 2004; Dasgupta et al. 2007). Additionally, mutations in the sugarless (sgl) or sulfateless (sfl) genes (encoding the enzymes UDP-glucose dehydrogenase and N-deacetylase/N-sulfotransferase, respectively) (Figure 1), displayed aberrant tracheal morphogenesis, similar to defects in FGFR signaling (Lin et al. 1999; Selleck 2000; Toyoda et al. 2000;). Recent studies have further demonstrated strict temporal control of GAG synthesis during Drosophila embryonic development. The mechanism, which involves developmentally regulated translational control of ttv and sgl, is thought to be an evolutionarily conserved means of modulating growth factor activity and morphogen gradient formation at specific times during development by regulating GAG biosynthesis (Bornemann et al. 2008).
Additional genes involved in GAG biosynthesis and modification include two encoding sulfotransferases (Hs2st and Hs6st). While mutation of either gene alone did not result in significant defects due to compensatory sulfation, mutation of both genes resulted in severe defects in FGF signaling, indicating the importance of overall GAG sulfation levels as opposed to specific sulfation patterns for signaling in certain developmental contexts (Kamimura et al. 2006; Xu et al. 2007). Also, the nucleotide sugar transporter, Fringe connection (encoded by the frc gene) transports UDP-GlcA (and other nucleotide sugars) into the ER/Golgi, further influencing proteoglycan biosynthesis, as well as the synthesis of other glycans (Goto et al. 2001; Selva et al. 2001). From these studies, it has become widely appreciated that the GAG component of proteoglycans and the enzymes regulating their biosynthesis function in many diverse aspects of development.
N-Linked glycosylation in Drosophila
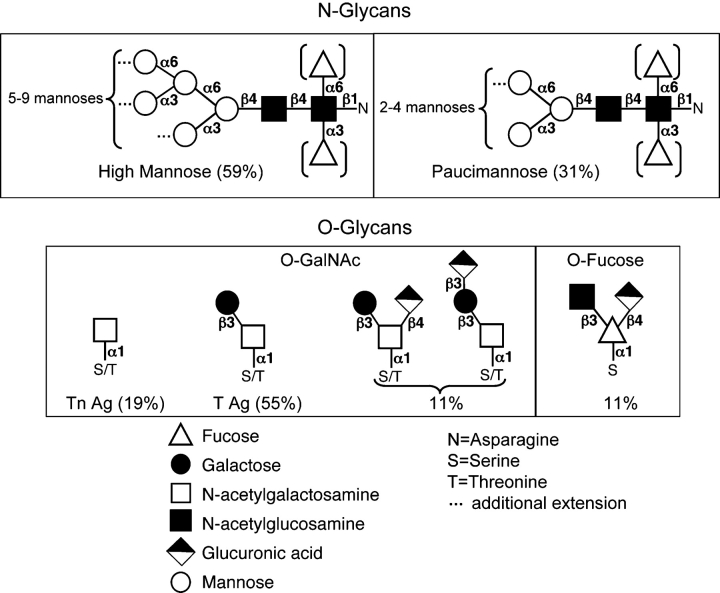
Recent mass spectrometry studies by North et al. (2006) and Aoki et al. (2007) have defined the structure of N-linked glycans present on protein substrates during Drosophila embryonic development (Figure 2). The predominant N-glycan structures found consist of high mannose and paucimannose and may be fucosylated at the chitobiose core. Only minor amounts of hybrid, bi- and tri-antennary complex glycans were observed, with some containing α2,6-linked sialic acid (North et al. 2006; Aoki et al. 2007). This composition differs considerably from that seen in mammals, where N-glycans are predominantly hybrid and complex, with abundant sialylation (Gagneux and Varki 1999). However, Drosophila N-glycans appear to be more similar to mammalian N-glycans than those from another model organism, Caenorhabditis elegans, which contain unique high fucose, phosphorylcholine, and methylated structures (Cipollo et al. 2005; Hanneman et al. 2006; Paschinger et al. 2008). Interestingly, the N-glycan profile of the fly changes as development proceeds, suggesting specific regulation of the glycosylation machinery and roles for certain glycan structures during different stages of development (Aoki et al. 2007).
Fig. 2.
N- and O-linked glycan structures present in Drosophila melanogaster. Shown are the major types of N-linked and O-linked glycans found in Drosophila and their relative abundance. N-Glycans consist primarily of high mannose (59%) and paucimannose (31%); hybrid (7%) and complex (3%) structures are much less abundant. Brackets indicate that an α1,3 fucose or α1,6 fucose may also be present in these structures. Mucin-type O-linked glycans are predominantly comprised of the Tn antigen (Tn Ag) and T antigen (T Ag or Core 1) structures. The only detectable protein O-fucose glycan in Drosophila is a glucuronyl trisaccharide (Aoki et al. 2008).
A number of genes involved in N-glycan biosynthesis in Drosophila have recently been characterized. Wollknauel (wol) encodes a UDP-glucose:dolichyl-phosphate glucosyltransferase responsible for dolichol-sugar synthesis. Mutations in this gene result in an unfolded protein response and disruption of embryonic patterning (Haecker et al. 2008). Mutations in other genes involved in later stages of N-glycan synthesis appear to primarily affect nervous system development and function. The abundant paucimannose N-linked glycans in the fly are attributed to the gene fused lobes (fdl), which encodes an N-acetylglucosaminidase that is responsible for cleaving the N-acetylglucosamine (GlcNAc) of hybrid N-glycans to form paucimannose N-glycans (Leonard et al. 2006). Mutations in this gene cause altered central nervous system (CNS) development resulting in fusion of the mushroom body β lobes of the brain (Boquet et al. 2000). Paradoxically, mutations in the Drosophila Mgat1 orthologue, which transfers GlcNAc to the paucimannose N-glycans to produce hybrid N-glycans, result in a similar “fused lobes” phenotype (Sarkar et al. 2006), suggesting key roles for both hybrid and paucimannose N-glycans in CNS development. In addition to the fused lobe phenotype, Drosophila Mgat1 mutants display reduced locomotory activity, reduced lifespan, and sterility in males, indicating roles for β1,2-N-acetylglucosaminyltransferase I-dependent N-glycans in reproduction and homeostasis in the adult fly (Sarkar et al. 2006).
In contrast to mammals, which have dozens of genes encoding sialyltransferases, the Drosophila genome has only one sialyltansferase gene (SiaT) (Koles et al. 2004). The Drosophila SiaT encodes an α2,6 sialyltransferase that is evolutionarily related to the vertebrate ST6 sialyltransferase (Koles et al. 2004). This enzyme acts on oligosaccharides and glycoproteins but not on glycolipids in vitro. SiaT is expressed in a very tissue- and stage-specific fashion, being found only in a subset of cells in the developing CNS (Koles et al. 2004). Indeed, mutations in SiaT result in adults with reduced lifespan, reduced tolerance to heat, and reduced locomotory activity, supporting a role for sialic acid modification of glycans in nervous system function (Koles et al. 2004). To date, additional genes involved in N-glycan biosynthesis have not yet been characterized in the fly but ongoing work by many groups centers around investigating the role of these glycans in protein structure, function, processing, and stability influencing fly development and homeostasis.
O-Linked glycosylation in Drosophila
O-GlcNAc
O-Linked GlcNAc was the first O-glycan to be detected in Drosophila. Early studies using lectin staining as well as radiolabeling, identified O-GlcNAc in distinct banding patterns along polytene chromosomes (Kelly and Hart 1989), providing evidence for the presence of this glycan on nuclear and chromatin-associated proteins in Drosophila. However, most of our current understanding of the O-GlcNAc modification has come from studies in mammalian systems, although a number of groups are now analyzing O-GlcNAc function in the fly.
O-Mannose
The role of O-linked mannose on protein substrates is of great interest as mutations in the glycosyltransferases responsible for this modification in humans result in muscular dystrophies (Muntoni et al. 2004). Drosophila has two genes, rotated abdomen (rt) and twisted (tw), encoding protein O-mannosyltransferases that are orthologues of the vertebrate POMT1 and POMT2 O-mannosyltransferases (Martin-Blanco and Garcia-Bellido 1996; Willer et al. 2003; Ichimiya et al. 2004; Lyalin et al. 2006). Similar to vertebrates, RT and TW appear to be required simultaneously, perhaps acting as a heterocomplex responsible for the transfer of mannose to protein substrates. Mutations in either gene result in defects in muscle development leading to a rotated abdomen phenotype in adults (Martin-Blanco and Garcia-Bellido 1996; Lyalin et al. 2006). In mammals, the major substrate of the O-mannosyltransferases is α-dystroglycan. Drosophila also has a dystroglycan (Dg) that, when mutated, results in muscle phenotypes similar, but not identical to those seen in tw and rt mutants (Haines et al. 2007). These enzymatic and phenotypic similarities between the fly and mammals suggest that Drosophila will be a valuable model system for deciphering the mechanistic role of O-linked mannose in muscle development and function. Additionally, the orthologue of the mammalian POMGnT-I (which extends the core mannose through the addition of GlcNAc) has not been found in Drosophila, suggesting that O-mannose glycans in the fly may be simpler in structure and thus more amenable to experimental analysis.
O-Linked GalNAc (Mucin-type O-glycosylation)
Mucin-type O-linked glycosylation and the family of UDP-N-acetyl- galactosamine:polypeptide N-acetylgalactosaminyltransferases (PGANTs) that initiates it are conserved in Drosophila. However, mucin-type O-glycans in flies are less extended relative to their mammalian counterparts, consisting primarily of the core 1 structure (T Ag; Galβ1-3GalNAcα1-O-S/T) (North et al. 2006), the Tn antigen (Tn Ag; GalNAcα1-O-S/T), and the core 1 structure modified with GlcA attached to either the Gal or GalNAc (Aoki et al. 2008; Breloy et al. 2008; M. Tiemeyer, personal communication) (Figure 2). No evidence for sialylated O-glycans has been found.
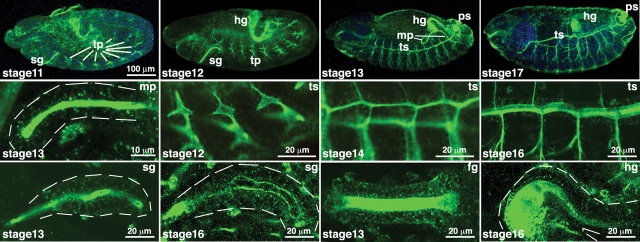
The fly has 12 putative genes encoding PGANTs, 9 of which have demonstrated biochemical activity in vitro (Schwientek et al. 2002; Ten Hagen and Tran 2002; Ten Hagen et al. 2003) (Table I). These genes display dynamic spatial and temporal regulation, suggesting that their coordinated expression determines what proteins (and what regions within those proteins) acquire O-glycans at various stages during development (Table I) (Tian and Ten Hagen 2006). Indeed, tissue staining using lectins and antibodies has illustrated the diversity of cells and organs expressing O-glycans at specific stages of development (Fredieu and Mahowald 1994; D’Amico and Jacobs 1995; Tian and Ten Hagen 2007a; Figure 3). Of note, is the unique spatial expression of mucin-type O-glycans along the presumptive apical and luminal regions of developing tubular tissues (Figure 3), which may play key roles in proper tube formation (see below).
Table I.
Summary of Drosophila pgants
| Name | CG no. | Activity | Expressed ina |
|---|---|---|---|
| pgant1 | CG8182 | Peptide/glycopeptide transferase | Embryonic gut, salivary glands, antennomaxillary complex, and posterior spiracles; third instar larval wing, eye-antennal, leg and haltere imaginal disks; adult male and female |
| pgant2 | CG3254 | Peptide/glycopeptide transferase | Embryonic brain and trachea; third instar larval wing and eye-antennal imaginal disks; adult male and female heads |
| pgant3 | CG4445 | Peptide/glycopeptide transferase | Embryonic gut, posterior spiracles, pharynx, esophagus, and epidermis; third instar larval wing, eye-antennal, leg and haltere imaginal disks; adult male and female |
| pgant4 | CG31956 | Glycopeptide transferase | Embryonic gut and proventriculus; third instar larval wing, eye-antennal, leg and haltere imaginal disks; adult male and female bodies |
| pgant5 | CG31651 | Peptide/glycopeptide transferase | Embryonic gut, salivary glands, antennomaxillary complex, posterior spiracles, and epidermis; third instar larval wing, eye-antennal, leg and haltere imaginal disks; adult male and female |
| pgant6 | CG2103 | Glycopeptide transferase | Embryonic gut, salivary glands, antennomaxillary complex, and epidermis; third instar larval wing, eye-antennal, leg and haltere imaginal disks; adult male and female |
| pgant7 | CG6394 | Glycopeptide transferase | Embryonic gut, salivary glands, antennomaxillary complex, and epidermis; third instar larval wing, eye-antennal, leg and haltere imaginal disks; adult male and female |
| pgant8 | CG7297 | Peptide/glycopeptide transferase | Embryonic gut; adult male and female |
| pgant35A | CG7480 | Peptide/glycopeptide transferase | Embryonic gut, salivary glands, trachea, and posterior spiracles; third instar larval wing, eye-antennal, leg and haltere imaginal disks; adult male and female |
| NA | CG30463b | ND | Embryonic amnioserosa, gut, and salivary glands; third instar larval wing, eye-antennal, leg and haltere imaginal disks |
| NA | CG10000b | ND | Embryonic gut |
| NA | CG31776b | ND | Embryonic gut and antennomaxillary complex; third instar larval wing, eye-antennal, leg and haltere imaginal disks |
Fig. 3.
Mucin-type O-linked glycan expression is found throughout Drosophila embryogenesis. Tn Ag (GalNAcα1-S/T) was detected by immunofluorescence and confocal imaging using antibodies directed against this glycan (described in Tian and Ten Hagen 2007a). Embryos at various stages of development (shown in the bottom-left corner of each image) are shown across the top row. The bottom and middle rows show enlarged images of developing tubular structures (denoted in the top-right corner of each image). O-Glycans are abundant along the apical and luminal regions of the developing organs shown. Dashed white lines are included to illustrate the outer boundaries of certain organs. fg, foregut; hg, hindgut; mp, malpighian tubules; ps, posterior spiracles; sg, salivary gland; tp, tracheal placodes; ts, tracheal system. Adapted from Glycobiology, 17, 820-827 (2007) by copyright permission of Oxford University Press.
Biochemical studies have revealed a hierarchy of action within the PGANT family, with certain members acting on previously unmodified peptides as initiating transferases (peptide transferases) and others acting to further modify previously glycosylated substrates (glycopeptide transferases), further supporting the coordinated action of PGANTs in dictating O-glycosyaltion patterns. Biochemical analyses also demonstrated that fly and mammalian orthologues have similar substrate preferences and preferred sites of GalNAc addition within those substrates (Ten Hagen et al. 2003; Gerken et al. 2008). This functional conservation may indicate conserved biological roles for members of this glycosyltransferase family that have been maintained over the course of evolution.
While other extending transferases have not been characterized in the fly, genes encoding core 1 β3-galactosyltransferases (core 1 β3-GalTs) have been identified (Muller et al. 2005). In contrast to mammals, which have one core 1 β3-GalT whose activity requires a chaperone (Ju et al. 2002; Ju and Cummings 2002), in vitro studies in the fly suggest that there may be as many as four functional core 1 β3-GalTs that do not appear to require a chaperone for activity (Muller et al. 2005). It is of note that the majority of mucin-type O-linked glycans in the fly consist of the unmodified core 1 structure (North et al. 2006; Aoki et al. 2008), suggesting that evolutionary pressures may have favored expansion of core 1 β3-GalTs in the fly. Regulation of core 1 β3-galactosyltransferase activity appears to be under different genetic controls in flies versus mammals; the multiple fly core 1 β3-GalT genes exhibit unique spatial expression patterns (Muller et al. 2005) whereas the single mammalian core 1 β3-GalT gene is ubiquitously expressed (Ju et al. 2002; Ju and Cummings 2002). Additionally, the activity of the mammalian transferase is influenced by expression of its chaperone, and possibly other competing transferases.
Crucial roles for mucin-type O-glycosylation were first demonstrated in Drosophila, where one member of the family (pgant35A) was found to be recessive lethal (Schwientek et al. 2002; Ten Hagen and Tran 2002). This was the first demonstration that mucin-type O-glycosylation was required for viability in any organism. Additional work on this enzyme has recently revealed a role during tracheal tube formation (Tian and Ten Hagen 2007b), consistent with the abundant presence of O-glycans in this organ (Figure 3). Mucin-type O-glycans normally present along the apical and luminal surfaces of the developing respiratory system in the fly were absent in pgant35A maternal/zygotic mutants. Proteins normally seen along the apical and luminal regions of the tracheal system were found in cytoplasmic vesicles, and septate junction (tight junction) proteins normally found along the lateral regions of cells comprising the tracheal tubes were mislocalized to more apical positions, indicating defects in apicobasal polarity. The resultant tracheal tubes were irregular in shape and diameter and lacked an intact diffusion barrier. These results suggest a role for mucin-type O-glycans in proper formation of the apical and luminal surfaces of the tracheal system, possibly by influencing trafficking/maintenance of proteins destined for those surfaces. Roles for mucin-type O-glycans in mammalian organ formation and tubulogenesis were also seen in mice deficient for the core 1 β3-GalT, where mice displayed defective vasculature formation and died embryonically from fatal brain hemorrhages (Xia et al. 2004). Additionally, hypomorphic mutations in core1 β3-GalT resulted in defective glomeruli and proximal tubules in the kidney, adding additional support for the role of mucin-type O-glycans in tubulogenesis across diverse species (Alexander et al. 2006).
O-Linked Fucose and Glucose in Conserved Signaling Pathways
The identification of the role of glycans in the evolutionarily conserved Notch signaling pathway has generated tremendous interest (reviewed in Stanley 2007). Genetic screens in Drosophila initially identified the gene fringe (fng) as a key regulator of Notch signaling during development (Cohen et al. 1997; Fleming et al. 1997; Panin et al. 1997; Klein and Arias 1998). fng expression was shown to enhance the activation of Notch signaling by the ligand Delta while inhibiting activation by the ligand, Serrate (Fleming et al. 1997; Panin et al. 1997). Biochemical and genetic studies revealed that fng encodes a glycosyltransferase responsible for the addition of GlcNAc in a β1,3-linkage to O-linked fucose present on protein substrates, such as the epidermal growth factor (EGF) repeats of Notch and its ligands, Delta and Serrate (Bruckner et al. 2000; Moloney et al. 2000; Panin et al. 2002). These studies provided the first evidence for a regulatory role of this type of glycan in a highly conserved signaling pathway. Cell culture studies demonstrated that the presence of this glycan affects receptor/ligand binding and may also affect subsequent signaling events (Lei et al. 2003; Okajima et al. 2003). Subsequent studies in mammalian systems have verified the role of these glycans in Notch signaling (reviewed in Stanley 2007), illustrating the utility of Drosophila to decipher glycan function across species.
The identification of the GlcNAc-Fuc disaccharide on Notch and its receptors lead to a search for the glycosyltransferase that is responsible for the addition of the core fucose. Two protein O-fucosyltransferases (OFUT1 and OFUT2) exist in the fly; OFUT1 is primarily responsible for adding fucose to the EGF repeats of Notch, Delta, and Serrate (Okajima and Irvine 2002; Panin et al. 2002; Okajima et al. 2003) while OFUT2 adds fucose to thrombospondin type I repeats (TSRs), but not EGF repeats (Luo, Koles, et al. 2006; Luo, Nita-Lazar, et al. 2006;). Genetic studies in the fly indicate that the role of the OFUT1 fucosyltransferase in Notch signaling is quite complex and not solely a function of the addition of O-linked fucose. In support of this, it has been shown that certain Notch signaling defects in ofut1 mutants can be rescued by a catalytically inactive form of OFUT1 (Okajima et al. 2005). This is the case during Drosophila embryonic neurogenesis, where catalytically inactive OFUT1 can restore Notch signaling during nervous system development (Okajima et al. 2008). This fucosyltransferase-independent function of OFUT1 is further supported by the observation that mutants in the GDP-fucose transporter (Gmd) (which fail to transport GDP-fucose into the ER/Golgi and are defective in fucosylation) do not display Notch-related nervous system defects, indicating that fucose addition to Notch per se is not required for nervous system development even though the OFUT1 protein is required (Okajima et al. 2008). Based on Notch localization studies, the authors propose that the OFUT1 protein has a chaperone activity responsible for proper folding, secretion, and/or cell-surface expression of Notch that is independent of its enzymatic activity (Okajima et al. 2005; reviewed in Stanley 2007).
A number of Notch signaling defects do, however, appear to depend on the fucosyltransferase activity of OFUT1 and are mimicked in Gmd mutants (Ishikawa et al. 2005; Okajima et al. 2005). These defects phenocopy fng defects, suggesting that the addition of O-fucose by OFUT1 in this instance is necessary to form the substrate for the Fng glycosyltransferase. Additional work also indicates that the removal of an O-fucose site from Notch affects ligand binding in the absence of Fng, suggesting that the O-fucose may function in receptor–ligand interactions independent of serving as a substrate for GlcNAc addition by Fng (Lei et al. 2003). Thus, it appears that OFUT1 performs different functions in different developmental contexts, some of which are dependent upon fucosyltransferase activity and others that involve a chaperone function independent of enzymatic activity. While the mammalian orthologue of OFUT1 (Pofut1) does not appear to have a similar chaperone activity, it is clear that genetic studies in the fly have led to significant insights into the roles of O-fucose glycans in a major, conserved signaling pathway in higher eukaryotes.
Most recently, genetic studies in the fly identified yet another glycan involved in Notch signaling (Acar et al. 2008). The protein O-glucosyltransferase, Rumi, is responsible for the addition of O-linked glucose to serine residues in certain EGF repeats of Notch, serving to regulate Notch folding and/or trafficking. Unlike OFUT1, the influence of Rumi on Notch signaling is dependent on its glycosyltransferase activity. Continued work in the fly will no doubt shed more light on the mechanistic role of these glycans and others in the regulation of Notch signaling during eukaryotic development.
Glycosphingolipid (GSL) Function in Drosophila
Glycosphingolipids (GSLs) in Drosophila consist primarily of the Manβ1-4Glcβ1-ceramide core (arthro-series), which can be elongated by additional sugars (such as GalNAc, GlcNAc, and Gal) or phosphoethanolamine (reviewed in Seppo and Tiemeyer 2000). The GSL core structure (Glc-ceramide) is synthesized by glucosyl ceramide synthase (DGlcT-1) in flies, which transfers glucose from UDP-glucose to ceramide (Cer) (Kohyama-Koganeya et al. 2004). RNAi to DGlcT-1 resulted in increased apoptosis, possibly due to increased ceramide levels, which are known to be pro-apoptotic (Kohyama-Koganeya et al. 2004). The next step in GSLs synthesis is controlled by the egghead (egh) gene, which encodes a GDP-mannose:βGlc β1,4-mannosyltransferase responsible for forming Manβ1-4Glcβ1-Cer (Wandall et al. 2003, 2005). The brainiac (brn) gene, encoding a UDP-GlcNAc:βMan β1,3-GlcNAc transferase, then adds GlcNAc to form GlcNAcβ1-3Manβ1-4Glcβ1-Cer (Muller et al. 2002; Wandall et al. 2005). Mutations in egh or brn cause loss of apicobasal polarity in the follicular epithelium, indicating a role for these genes in epithelial maintenance and cell adhesion (Goode, Melnick, et al. 1996; Goode, Morgan, et al. 1996). Additionally, these mutants displayed certain neurogenic phenotypes, such as loss of ventral and cephalic epidermal cells and hypertrophy in certain regions of the nervous system. These phenotypes mimicked those seen for certain other signaling molecules (Notch and EGF), suggesting that GSLs synthesized by egh and brn may be involved in regulating receptor–ligand interactions by altering occupancy in lipid rafts.
GSL chains are further modified by the addition of neutral sugars. To that end, two members of the β1,4-N-acetylgalact- osyltransferase enzyme family (β1,4 GalNAcTs) have been identified in Drosophila (β4GalNAcTA and β4GalNAcTB) (Haines and Irvine 2005). Recent work indicates that β4GalNAcTA and β4GalNAcTB modify GSLs by the addition of GalNAc to the GlcNAcβ1-3Manβ1-4Glcβ1-Cer structure (Chen et al. 2007; Stolz et al. 2008). Analysis of GSL structures in β4GalNAcTA and β4GalNAcTB single mutants indicates that β4GalNAcTB is the major enzyme responsible for GSL modification (Stolz et al. 2008). While mutations in β4GalNAcTB produced epithelial defects in ovarian follicle cells in a small proportion of animals (Chen et al. 2007), β4GalNAcTA mutants displayed altered behavioral (Haines and Irvine 2005) nerve and muscle phenotypes (Haines and Stewart 2007), suggesting that these enzymes are not functionally redundant, but rather have unique roles in vivo. Collectively, these studies demonstrate that two genes with presumably the same enzymatic activity and expression patterns have unique roles in GSL biosynthesis. This raises the possibility that one or both may also be involved in modifying other glycans (Sasaki et al. 2007) or that one may require a cofactor, adaptor, or chaperone that modulates its activity (Chen et al. 2007). However, no effects on viability or fertility were noted, even in animals doubly mutant for both genes, indicating that extension of the GSLs by these enzymes is not required for survival.
Conclusions
The studies summarized herein highlight the recent progress that has been made in defining the genes responsible for glycan biosynthesis in Drosophila as well as their unique biological functions through the use of powerful genetic and molecular techniques unique to this organism. Taking advantage of genome-wide RNAi screens both in cell culture as well as in the fly itself will further aid our fundamental understanding of the biological role of glycans and the enzymes that form them. Future studies defining the repertoire of proteins that are glycosylated as well as the glycan binding molecules with which they interact will shed light on the mechanistic role of glycans in many conserved aspects of biology. Recently 205 glycoproteins carrying N-linked glycans were identified in the Drosophila brain (Koles et al. 2007). The repertoire of proteins carrying this modification is very diverse, including extracellular matrix proteins, cell adhesion proteins, transporters, cell surface receptors, proteases, ion channel components, and enzymes involved in a wide variety of metabolism and cellular functions. Other recent studies have begun to define proteins modified by mucin-type O-linked glycosylation in Drosophila cells, including those comprising the extracellular matrix, as well as pathogen recognition proteins, stress response proteins, secreted proteases, and protease inhibitors (Schwientek et al. 2007). While a great deal has been discovered in the fly, it only serves to highlight that we are still firmly on the tip of the iceberg in terms of understanding the complex roles of glycans during all stages of eukaryotic development. Ongoing efforts by many talented groups will help to unravel the complexities of glycobiology in many diverse developmental systems and organisms.
Acknowledgments
We would like to thank the many members of the community who have contributed to the work mentioned herein. We would like to thank Drs. Lawrence A. Tabak, Jaya Raman, Yu Guan, and Hazuki Miwa for carefully reading this manuscript. This research was supported by the Intramural Research Program of the National Institute of Dental and Craniofacial Research, National Institutes of Health.
Glossary
Abbreviations
- Cer
ceramide
- CNS
central nervous system
- EGF
epidermal growth factor
- GAG
glycosaminoglycan
- Gal
galactose
- GalNAc
N-acetylgalactosamine
- GlcA
glucuronic acid
- GlcNAc
N-acetylglucosamine
- GSL
glycosphingolipid
- Fuc
fucose
- ppGaNTase or ppGalNAcT or pgant or PGANT
UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase
- Man
mannose
References
- Acar M, Jafar-Nejad H, Takeuchi H, Rajan A, Ibrani D, Rana NA, Pan H, Haltiwanger RS, Bellen HJ. Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell. 2008;132:247–258. doi: 10.1016/j.cell.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander W, Viney EM, Zhang J, Metcalf D, Kauppi M, Hyland CD, Carpinelli MR, Stevenson W, Croker BA, Hilton AA, et al. Thrombocytopenia and kidney disease in mice with a mutation in the C1galt1 gene. Proc Natl Acad Sci USA. 2006;103:16442–16447. doi: 10.1073/pnas.0607872103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Perlman M, Lim J, Cantu R, Wells L, Tiemeyer M. Dynamic developmental elaboration of N-linked glycan complexity in the Drosophila melanogaster embryo. J Biol Chem. 2007;282:9127–9142. doi: 10.1074/jbc.M606711200. [DOI] [PubMed] [Google Scholar]
- Aoki K, Porterfield M, Lee SS, Dong B, Nguyen K, McGlamry KH, Tiemeyer M. The diversity of O-linked glycans expressed during Drosophila melanogaster development reflects stage- and tissue-specific requirements for cell signaling. J Biol Chem. 2008 doi: 10.1074/jbc.M804925200. In press. PMID: 18725413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boquet I, Hitier R, Dumas M, Chaminade M, Préat T. Central brain postembryonic development in Drosophila: Implication of genes expressed at the interhemispheric junction. J Neurobiol. 2000;42:33–48. [PubMed] [Google Scholar]
- Bornemann DJ, Park S, Phin S, Warrior R. A translational block to HSPG synthesis permits BMP signaling in the early Drosophila embryo. Development. 2008;135:1039–1047. doi: 10.1242/dev.017061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Paro R, Perrimon N. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- Breloy I, Schwientek T, Lehr S, Hanisch FG. Glucuronic acid can extend O-linked core 1 glycans, but it contributes only weakly to the negative surface charge of Drosophila melanogaster Schneider-2 cells. FEBS Lett. 2008;582:1593–1598. doi: 10.1016/j.febslet.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Bruckner K, Perez L, Clausen H, Cohen S. Glycosyltransferase activity of Fringe modulates Notch–Delta interactions. Nature. 2000;406:411–415. doi: 10.1038/35019075. [DOI] [PubMed] [Google Scholar]
- Brunner A, Kolarich D, Voglmeir J, Paschinger K, Wilson IB. Comparative characterisation of recombinant invertebrate and vertebrate peptide O-xylosyltransferases. Glycoconj J. 2006;23:543–554. doi: 10.1007/s10719-006-7633-z. [DOI] [PubMed] [Google Scholar]
- Chen YW, Pedersen JW, Wandall HH, Levery SB, Pizette S, Clausen H, Cohen SM. Glycosphingolipids with extended sugar chain have specialized functions in development and behavior of Drosophila. Dev Biol. 2007;306:736–749. doi: 10.1016/j.ydbio.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Cipollo JF, Awad AM, Costello CE, Hirschberg CB. N-Glycans of Caenorhabditis elegans are specific to developmental stages. J Biol Chem. 2005;280:26063–26072. doi: 10.1074/jbc.M503828200. [DOI] [PubMed] [Google Scholar]
- Cohen B, Bashirullah A, Dagnino L, Campbell C, Fisher WW, Leow CC, Whiting E, Ryan D, Zinyk D, Boulianne G, Hui CC, et al. Fringe boundaries coincide with Notch-dependent patterning centres in mammals and alter Notch-dependent development in Drosophila. Nat Genet. 1997;16:283–288. doi: 10.1038/ng0797-283. [DOI] [PubMed] [Google Scholar]
- D’Amico P, Jacobs JR. Lectin histochemistry of the Drosophila embryo. Tissue Cell. 1995;27:23–30. doi: 10.1016/s0040-8166(95)80005-0. [DOI] [PubMed] [Google Scholar]
- Dasgupta U, Dixit BL, Rusch M, Selleck S, The I. Functional conservation of the human EXT1 tumor suppressor gene and its Drosophila homolog tout velu. Dev Genes Evol. 2007;217:555–561. doi: 10.1007/s00427-007-0163-2. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Fleming RJ, Gu Y, Hukriede NA. Serrate-mediated activation of Notch is specifically blocked by the product of the gene fringe in the dorsal compartment of the Drosophila wing imaginal disc. Development. 1997;124:2973–2981. doi: 10.1242/dev.124.15.2973. [DOI] [PubMed] [Google Scholar]
- Fredieu JR, Mahowald AP. Glycoconjugate expression during Drosophila embryogenesis. Acta Anat. 1994;149:89–99. doi: 10.1159/000147562. [DOI] [PubMed] [Google Scholar]
- Gagneux P, Varki A. Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology. 1999;9:747–755. doi: 10.1093/glycob/9.8.747. [DOI] [PubMed] [Google Scholar]
- Gerken TA, Ten Hagen KG, Jamison O. Conservation of peptide acceptor preferences between Drosophila and mammalian polypeptide-GalNAc transferase orthologue pairs. Glycobiology. 2008;18:861–870. doi: 10.1093/glycob/cwn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode S, Melnick M, Chou TB, Perrimon N. The neurogenic genes egghead and brainiac define a novel signaling pathway essential for epithelial morphogenesis during Drosophila oogenesis. Development. 1996;122:3863–3879. doi: 10.1242/dev.122.12.3863. [DOI] [PubMed] [Google Scholar]
- Goode S, Morgan M, Liang YP, Mahowald AP. Brainiac encodes a novel, putative secreted protein that cooperates with Grk TGF alpha in the genesis of the follicular epithelium. Dev Biol. 1996;178:35–50. doi: 10.1006/dbio.1996.0196. [DOI] [PubMed] [Google Scholar]
- Goto S, Taniguchi M, Muraoka M, Toyoda H, Sado Y, Kawakita M, Hayashi S. UDP-sugar transporter implicated in glycosylation and processing of Notch. Nat Cell Biol. 2001;3:816–822. doi: 10.1038/ncb0901-816. [DOI] [PubMed] [Google Scholar]
- Hacker U, Nybakken K, Perrimon N. Heparan sulphate proteoglycans: The sweet side of development. Nat Rev Mol Cell Biol. 2005;6:530–541. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- Haecker A, Bergman M, Neupert C, Moussian B, Luschnig S, Aebi M, Mannervik M. Wollknauel is required for embryo patterning and encodes the Drosophila ALG5 UDP-glucose:dolichyl-phosphate glucosyltransferase. Development. 2008;135:1745–1749. doi: 10.1242/dev.020891. [DOI] [PubMed] [Google Scholar]
- Hahn M, Han MV, Han S-G. Gene family evolution across 12 Drosophila genomes. PLoS Genetics. 2007;3(11):2135–2146. doi: 10.1371/journal.pgen.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines N, Irvine KD. Functional analysis of Drosophila beta1,4-N-acetlygalactosaminyltransferases. Glycobiology. 2005;15:335–346. doi: 10.1093/glycob/cwi017. [DOI] [PubMed] [Google Scholar]
- Haines N, Seabrooke S, Stewart BA. Dystroglycan and protein O-mannosyltransferases 1 and 2 are required to maintain integrity of Drosophila larval muscles. Mol Biol Cell. 2007;18:4721–4730. doi: 10.1091/mbc.E07-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines N, Stewart BA. Functional roles for beta1,4-N-acetlygalactosaminyltransferase-A in Drosophila larval neurons and muscles. Genetics. 2007;175:671–979. doi: 10.1534/genetics.106.065565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Belenkaya TY, Khodoun M, Tauchi M, Lin X, Lin X. Distinct and collaborative roles of Drosophila EXT family proteins in morphogen signalling and gradient formation. Development. 2004;131:1563–1575. doi: 10.1242/dev.01051. [DOI] [PubMed] [Google Scholar]
- Hanneman AJ, Rosa JC, Ashline D, Reinhold VN. Isomer and glycomer complexities of core GlcNAcs in Caenorhabditis elegans. Glycobiology. 2006;16:874–890. doi: 10.1093/glycob/cwl011. [DOI] [PubMed] [Google Scholar]
- Ichimiya T, Manya H, Ohmae Y, Yoshida H, Takahashi K, Ueda R, Endo T, Nishihara S. The twisted abdomen phenotype of Drosophila POMT1 and POMT2 mutants coincides with their heterophilic protein O-mannosyltransferase activity. J Biol Chem. 2004;279:42638–42647. doi: 10.1074/jbc.M404900200. [DOI] [PubMed] [Google Scholar]
- Ishikawa HO, Higashi S, Ayukawa T, Sasamura T, Kitagawa M, Harigaya K, Aoki K, Ishida N, Sanai Y, Matsuno K. Notch deficiency implicated in the pathogenesis of congenital disorder of glycosylation IIc. Proc Natl Acad Sci USA. 2005;102:18532–18537. doi: 10.1073/pnas.0504115102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumikawa T, Egusa N, Taniguchi F, Sugahara K, Kitagawa H. Heparan sulfate polymerization in Drosophila. J Biol Chem. 2006;281:1929–1934. doi: 10.1074/jbc.M509138200. [DOI] [PubMed] [Google Scholar]
- Ju T, Brewer K, D'Souza A, Cummings RD, Canfield WM. Cloning and expression of human core 1 β1,3-galactosyltransferase. J Biol Chem. 2002;277:178–186. doi: 10.1074/jbc.M109060200. [DOI] [PubMed] [Google Scholar]
- Ju T, Cummings RD. A unique molecular chaperone Cosmc required for activity of the mammalian core 1 beta 3-galactosyltransferase. Proc Natl Acad Sci USA. 2002;99:16613–16618. doi: 10.1073/pnas.262438199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura K, Koyama T, Habuchi H, Ueda R, Masu M, Kimata K, Nakato H. Specific and flexible roles of heparan sulfate modifications in Drosophila FGF signaling. J Cell Biol. 2006;174:773–778. doi: 10.1083/jcb.200603129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Hart GW. Glycosylation of chromosomal proteins: Localization of O-linked N-acetylglucosamine in Drosophila chromatin. Cell. 1989;57:243–251. doi: 10.1016/0092-8674(89)90962-8. [DOI] [PubMed] [Google Scholar]
- Kiger AA, Baum B, Jones S, Jones MR, Coulson A, Echeverri C, Perrimon N. A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2003;2:27. doi: 10.1186/1475-4924-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BT, Tsuchida K, Lincecum J, Kitagawa H, Bernfield M, Sugahara K. Identification and characterization of three Drosophila melanogaster glucuronyltransferases responsible for the synthesis of the conserved glycosaminoglycan-protein linkage region of proteoglycans. Two novel homologs exhibit broad specificity toward oligosaccharides from proteoglycans, glycoproteins, and glycosphingolipids. J Biol Chem. 2003;278:9116–9124. doi: 10.1074/jbc.M209344200. [DOI] [PubMed] [Google Scholar]
- Klein T, Arias AM. Interactions among Delta, Serrate and Fringe modulate Notch activity during Drosophila wing development. Development. 1998;125:2951–2962. doi: 10.1242/dev.125.15.2951. [DOI] [PubMed] [Google Scholar]
- Kohyama-Koganeya A, Sasamura T, Oshima E, Suzuki E, Nishihara S, Ueda R, Hirabayashi Y. Drosophila glucosylceramide synthase: A negative regulator of cell death mediated by proapoptotic factors. J Biol Chem. 2004;279:35995–36002. doi: 10.1074/jbc.M400444200. [DOI] [PubMed] [Google Scholar]
- Koles K, Irvine KD, Panin VM. Functional characterization of Drosophila sialyltransferase. J Biol Chem. 2004;279:4346–4357. doi: 10.1074/jbc.M309912200. [DOI] [PubMed] [Google Scholar]
- Koles K, Lim JM, Aoki K, Porterfield M, Tiemeyer M, Wells L, Panin V. Identification of N-glycosylated proteins from the central nervous system of Drosophila melanogaster. Glycobiology. 2007;17:1388–1403. doi: 10.1093/glycob/cwm097. [DOI] [PubMed] [Google Scholar]
- Lei L, Xu A, Panin VM, Irvine KD. An O-fucose site in the ligand binding domain inhibits Notch activation. Development. 2003;130:6411–6421. doi: 10.1242/dev.00883. [DOI] [PubMed] [Google Scholar]
- Léonard R, Rendic D, Rabouille C, Wilson IB, Préat T, Altmann F. The Drosophila fused lobes gene encodes an N-acetylglucosaminidase involved in N-glycan processing. J Biol Chem. 2006;281:4867–4875. doi: 10.1074/jbc.M511023200. [DOI] [PubMed] [Google Scholar]
- Lin X, Buff EM, Perrimon N, Michelson AM. Heparan sulfate proteoglycans are essential for FGF receptor signaling during Drosophila embryonic development. Development. 1999;126:3715–3723. doi: 10.1242/dev.126.17.3715. [DOI] [PubMed] [Google Scholar]
- Luo Y, Koles K, Vorndam W, Haltiwanger RS, Panin VM. Protein O-fucosyltransferase 2 adds O-fucose to thrombospondin type 1 repeats. J Biol Chem. 2006;281:9393–9399. doi: 10.1074/jbc.M511975200. [DOI] [PubMed] [Google Scholar]
- Luo Y, Nita-Lazar A, Haltiwanger RS. Two distinct pathways for O-fucosylation of epidermal growth factor-like or thrombospondin type 1 repeats. J Biol Chem. 2006;281:9385–9392. doi: 10.1074/jbc.M511974200. [DOI] [PubMed] [Google Scholar]
- Lyalin D, Koles K, Roosendaal SD, Repnikova E, Van Wechel L, Panin VM. The twisted gene encodes Drosophila protein O-mannosyltransferase 2 and genetically interacts with the rotated abdomen gene encoding Drosophila protein O-mannosyltransferase 1. Genetics. 2006;172:343–353. doi: 10.1534/genetics.105.049650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Blanco E, García-Bellido A. Mutations in the rotated abdomen locus affect muscle development and reveal an intrinsic asymmetry in Drosophila. Proc Natl Acad Sci USA. 1996;93:6048–6052. doi: 10.1073/pnas.93.12.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine KD, Haltiwanger RS, et al. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- Muller R, Altmann F, Zhou D, Hennet T. The Drosophila melanogaster brainiac protein is a glycolipid-specific beta 1,3-N-acetylglucosaminyltransferase. J Biol Chem. 2002;277:32417–32420. doi: 10.1074/jbc.C200381200. [DOI] [PubMed] [Google Scholar]
- Muller R, Hulsmeier AJ, Altmann F, Ten Hagen KG, Tiemeyer M, Hennet T. Characterization of mucin-type core-1 β1-3-galactosyltransferase homologous enzymes in Drosophila melanogaster. FEBS J. 2005;272:4295–4305. doi: 10.1111/j.1742-4658.2005.04838.x. [DOI] [PubMed] [Google Scholar]
- Muntoni F, Brockington M, Torelli S, Brown SC. Defective glycosylation in congenital muscular dystrophies. Curr Opin Neurol. 2004;17:205–209. doi: 10.1097/00019052-200404000-00020. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Haines N, Chen J, Okajima T, Furukawa K, Urano T, Stanley P, Irvine KD, Furukawa K. Identification of a Drosophila gene encoding xylosylprotein beta4-galactosyltransferase that is essential for the synthesis of glycosaminoglycans and for morphogenesis. J Biol Chem. 2002;277:46280–46288. doi: 10.1074/jbc.M203873200. [DOI] [PubMed] [Google Scholar]
- North SJ, Koles K, Hembd C, Morris HR, Dell A, Panin VM, Haslam SM. Glycomics studies of Drosophila melanogaster embryos. Glycoconj J. 2006;23:345–354. doi: 10.1007/s10719-006-6693-4. [DOI] [PubMed] [Google Scholar]
- Nybakken K, Perrimon N. Heparan sulfate proteoglycan modulation of developmental signaling in Drosophila. Biochim Biophys Acta. 2002;1573:280–291. doi: 10.1016/s0304-4165(02)00395-1. [DOI] [PubMed] [Google Scholar]
- Okajima T, Irvine KD. Regulation of notch signaling by O-linked fucose. Cell. 2002;111:893–904. doi: 10.1016/s0092-8674(02)01114-5. [DOI] [PubMed] [Google Scholar]
- Okajima T, Reddy B, Matsuda T, Irvine KD. Contributions of chaperone and glycosyltransferase activities of O-fucosyltransferase 1 to Notch signaling. BMC Biol. 2008;6:1–10. doi: 10.1186/1741-7007-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okajima T, Xu A, Irvine KD. Modulation of notch-ligand binding by protein O-fucosyltransferase 1 and fringe. J Biol Chem. 2003;278:42340–42345. doi: 10.1074/jbc.M308687200. [DOI] [PubMed] [Google Scholar]
- Okajima T, Xu A, Lei L, Irvine KD. Chaperone activity of protein O-fucosyltransferase 1 promotes notch receptor folding. Science. 2005;307:1599–1603. doi: 10.1126/science.1108995. [DOI] [PubMed] [Google Scholar]
- Panin VM, Papayannopoulos V, Wilson R, Irvine KD. Fringe modulates Notch–ligand interactions. Nature. 1997;387:908–912. doi: 10.1038/43191. [DOI] [PubMed] [Google Scholar]
- Panin VM, Shao L, Lei L, Moloney DJ, Irvine KD, Haltiwanger RS. Notch ligands are substrates for protein O-fucosyltransferase-1 and Fringe. J Biol Chem. 2002;277:29945–29952. doi: 10.1074/jbc.M204445200. [DOI] [PubMed] [Google Scholar]
- Paschinger K, Gutternigg M, Rendić D, Wilson IB. The N-glycosylation pattern of Caenorhabditis elegans. Carb Res. 2008;343:2041–2049. doi: 10.1016/j.carres.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar M, Leventis PA, Silvescu CI, Reinhold VN, Schachter H, Boulianne GL. Null mutations in Drosophila N-acetylglucosaminyltransferase I produce defects in locomotion and a reduced life span. J Biol Chem. 2006;281:12776–12785. doi: 10.1074/jbc.M512769200. [DOI] [PubMed] [Google Scholar]
- Sasaki N, Yoshida H, Fuwa TJ, Kinoshita-Toyoda A, Toyoda H, Hirabayashi Y, Ishida H, Ueda R, Nishihara S. Drosophila beta 1,4-N-acetylgalactosaminyltransferase-A synthesizes the LacdiNAc structures on several glycoproteins and glycosphingolipids. Biochem Biophys Res Commun. 2007;354:522–527. doi: 10.1016/j.bbrc.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Schwientek T, Bennett EP, Flores C, Thacker J, Hollmann M, Reis CA, Behrens J, Mandel U, Keck B, Schafer MA, et al. Functional conservation of subfamilies of putative UDP-N-acetylgalactosaminyltransferases in Drosophila, Caenorhabditis elegans, and mammals. J Biol Chem. 2002;277:22623–22638. doi: 10.1074/jbc.M202684200. [DOI] [PubMed] [Google Scholar]
- Schwientek T, Mandel U, Roth U, Muller S, Hanisch FG. A serial lectin approach to the mucin-type O-glycoproteome of Drosophila melanogaster S2 cells. Proteomics. 2007;7:3264–3277. doi: 10.1002/pmic.200600793. [DOI] [PubMed] [Google Scholar]
- Selleck SB. Proteoglycans and pattern formation: Sugar biochemistry meets developmental genetics. Trends Genet. 2000;16:206–212. doi: 10.1016/s0168-9525(00)01997-1. [DOI] [PubMed] [Google Scholar]
- Selva EM, Hong K, Baeg GH, Beverley SM, Turco SJ, Perrimon N, Häcker U. Dual role of the fringe connection gene in both heparan sulphate and fringe-dependent signalling events. Nat Cell Biol. 2001;3:809–815. doi: 10.1038/ncb0901-809. [DOI] [PubMed] [Google Scholar]
- Seppo A, Tiemeyer M. Function and structure of Drosophila glycans. Glycobiology. 2000;10:751–760. doi: 10.1093/glycob/10.8.751. [DOI] [PubMed] [Google Scholar]
- Stanley P. Regulation of Notch signaling by glycosylation. Curr Opin Struct Biol. 2007;17:530–535. doi: 10.1016/j.sbi.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD. Human genome: End of the beginning. Nature. 2004;431:915–916. doi: 10.1038/431915a. [DOI] [PubMed] [Google Scholar]
- Stolz A, Haines N, Pich A, Irvine KD, Hokke CH, Deelder AM, Gerardy-Schahn R, Wuhrer M, Bakker H. Distinct contributions of beta 4GalNAcTA and beta 4GalNAcTB to Drosophila glycosphingolipid biosynthesis. Glycoconj J. 2008;25:167–175. doi: 10.1007/s10719-007-9069-5. [DOI] [PubMed] [Google Scholar]
- Takei Y, Ozawa Y, Sato M, Watanabe A, Tabata T. Three Drosophila EXT genes shape morphogen gradients through synthesis of heparan sulfate proteoglycans. Development. 2003;131:73–82. doi: 10.1242/dev.00913. [DOI] [PubMed] [Google Scholar]
- Takemae H, Ueda R, Okubo R, Nakato H, Izumi S, Saigo K, Nishihara S. Proteoglycan UDP-galactose:beta-xylose beta 1,4-galactosyltransferase I is essential for viability in Drosophila melanogaster. J Biol Chem. 2003;278:15571–15578. doi: 10.1074/jbc.M301123200. [DOI] [PubMed] [Google Scholar]
- Ten Hagen KG, Tran DT. A UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase is essential for viability in Drosophila melanogaster. J Biol Chem. 2002;277:22616–22622. doi: 10.1074/jbc.M201807200. [DOI] [PubMed] [Google Scholar]
- Ten Hagen KG, Tran DT, Gerken TA, Stein DS, Zhang Z. Functional characterization and expression analysis of members of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family from Drosophila melanogaster. J Biol Chem. 2003;278:35039–35048. doi: 10.1074/jbc.M303836200. [DOI] [PubMed] [Google Scholar]
- Tian E, Ten Hagen KG. Expression of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family is spatially and temporally regulated during Drosophila development. Glycobiology. 2006;16:83–95. doi: 10.1093/glycob/cwj051. [DOI] [PubMed] [Google Scholar]
- Tian E, Ten Hagen KG. O-linked glycan expression during Drosophila development. Glycobiology. 2007;17:820–827. doi: 10.1093/glycob/cwm056. [DOI] [PubMed] [Google Scholar]
- Tian E, Ten Hagen KG. A UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase is required for epithelial tube formation. J Biol Chem. 2007;282:606–614. doi: 10.1074/jbc.M606268200. [DOI] [PubMed] [Google Scholar]
- Toyoda H, Kinoshita-Toyoda A, Fox B, Selleck SB. Structural analysis of glycosaminoglycans in animals bearing mutations in sugarless, sulfateless, and tout-velu. Drosophila homologues of vertebrate genes encoding glycosaminoglycan biosynthetic enzymes. J Biol Chem. 2000;275:21856–21861. doi: 10.1074/jbc.M003540200. [DOI] [PubMed] [Google Scholar]
- Ueyama M, Takemae H, Ohmae Y, Yoshida H, Toyoda H, Ueda R, Nishihara S. Functional analysis of proteoglycan galactosyltransferase II RNA interference mutant flies. J Biol Chem. 2008;283:6076–6084. doi: 10.1074/jbc.M709189200. [DOI] [PubMed] [Google Scholar]
- Wandall HH, Pedersen JW, Park C, Levery SB, Pizette S, Cohen SM, Schwientek T, Clausen H. Drosophila egghead encodes a beta 1,4-mannosyltransferase predicted to form the immediate precursor glycosphingolipid substrate for brainiac. J Biol Chem. 2003;278:1411–1414. doi: 10.1074/jbc.C200619200. [DOI] [PubMed] [Google Scholar]
- Wandall HH, Pizette S, Pedersen JW, Eichert H, Levery SB, Mandel U, Cohen SM, Clausen H. Egghead and brainiac are essential for glycosphingolipid biosynthesis in vivo. J Biol Chem. 2005;280:4858–4863. doi: 10.1074/jbc.C400571200. [DOI] [PubMed] [Google Scholar]
- Willer T, Valero MC, Tanner W, Cruces J, Strahl S. O-Mannosyl glycans: From yeast to novel associations with human disease. Curr Opin Struct Biol. 2003;13:621–630. doi: 10.1016/j.sbi.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Wilson IB. Functional characterization of Drosophila melanogaster peptide O-xylosyltransferase, the key enzyme for proteoglycan chain initiation and member of the core 2/I N-acetylglucosaminyltransferase family. J Biol Chem. 2002;277:21207–21212. doi: 10.1074/jbc.M201634200. [DOI] [PubMed] [Google Scholar]
- Xia L, Ju T, Westmuckett A, An G, Ivanciu L, McDaniel JM, Lupu F, Cummings RD, McEver RP. Defective angiogenesis and fatal embryonic hemorrhage in mice lacking core 1-derived O-glycans. J Cell Biol. 2004;164:451–459. doi: 10.1083/jcb.200311112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Song D, Pedersen LC, Liu J. Mutational study of heparan sulfate 2-O-sulfotransferase and chondroitin sulfate 2-O-sulfotransferase. J Biol Chem. 2007;282:8356–8367. doi: 10.1074/jbc.M608062200. [DOI] [PubMed] [Google Scholar]