Abstract
OBJECTIVE
Germline mutations in three genes have been found in familial cases of cerebral cavernous malformations (CCM). We previously discovered somatic and germline truncating mutations in the KRIT1 gene supporting the “two-hit” mechanism of CCM lesion formation in a single lesion. The purpose of this study was to screen for somatic, nonheritable, mutations in three more lesions from different patients and identify the cell type(s) in which somatic mutations occur.
METHODS
Somatic mutations were sought in DNA from three surgically excised, fresh-frozen CCM lesions by cloning and screening PCR products generated from KRIT1 or PDCD10 coding regions. Laser capture microdissection (LCM) was used to isolated endothelial and nonendothelial cells in order to determine if somatic mutations were found in endothelial cells.
RESULTS
A CCM lesion harbored somatic and germline KRIT1 mutations on different chromosomes and are therefore biallelic. Both mutations are predicted to truncate the protein. The KRIT1 somatic mutations (novel c.1800delG mutation and previously identified 34 nucleotide deletion) in CCMs from two different patients were only found in the vascular endothelial cells lining caverns. No obvious somatic mutations were identified in the two other lesions; however, the results were inconclusive possibly due to the technical limitations or the fact that these specimens had a small proportion of vascular endothelial cells lining pristine caverns.
CONCLUSION
The “two-hit” mechanism occurs in vascular endothelial cells lining CCM caverns from two patients with somatic and Hispanic-American KRIT1 germline mutations. Methods for somatic mutation detection should focus on vascular endothelial cells lining pristine caverns.
Keywords: Genetics, Hemorrhagic, Mutation, Stroke, Vascular malformations
Introduction
CCM lesions have been found in 0.5% of the population(12). Autosomal dominant CCM (OMIM #116860) can be caused by germline, heritable mutations in KRIT1 (alias CCM1)(Laberge-le Couteulx, et al. 1999; Sahoo, et al. 1999), CCM2 (alias MGC4607)(6, 21) (21–23, 29) or PDCD10 (alias CCM3)(4, 23, 36) genes. The ancestral c.1363C>T KRIT1 mutation has been identified in most Hispanic-American patients(33). Findings of multiple lesions in familial CCM cases and single lesions in sporadic cases(12) are consistent with a “two-hit” mechanism(20) of CCM lesion formation and somatic mutation may be a common cause of CCM lesion genesis. In addition, radiotherapy has been implicated in the genesis of CCM, with multiple spinal cavernous malformations confined to the field of radiotherapy being observed in a patient, possibly resulting from induced somatic mutations(18). The “two-hit” hypothesis of CCM lesion formation has been tested by multiple groups(1, 13, 19, 26, 30–32) with conflicting results. Somatic mutations have been reported in six cases(1, 13, 19), including our previous study in which we identified a 34-base pair (bp) deletion (c.1465_1498delGAAATACTTGCTGAATTGACTAATCTGGATCCTC), that was found in 0–42% of cells, depending on the section of tissue examined from specimen 354. We hypothesized that the somatic mutation was restricted to a specific cell type such as the endothelial cells that have been shown to have defective cell-cell junctions(14, 17, 40). In this report, 3 additional CCM lesions were screened for somatic mutations. Patients 354, 356 and 410 had the germline Hispanic-American KRIT1 mutation (c1363C>T). Patient 332 had a PDCD10 germline mutation (c.608T>G). A novel somatic KRIT1 mutation (c.1300delG) in CCM specimen 410 was identified. Vascular endothelial cells lining pristine caverns harbored the novel and 34-bp deletion somatic mutations.
Materials and Methods
Tissue Samples/Germline mutations
Four patients consented to participation in this institutional-approved study. Diagnosis of CCM was based on characteristic gradient echo MRI findings, and pathological examination of the surgically removed lesion (Figure 1). Patient 354 and 356 each had four and five other affected family members, respectively. Patients 410 and 332 had no known family history, but each had a germline mutation in a CCM gene (Table 1). The germline mutation c.1363C>T KRIT1 denotes that at nucleotide 1363, numbered from the ATG start of translation, is a C that is changed to at T (coding region of Genbank reference sequence NM_194455).
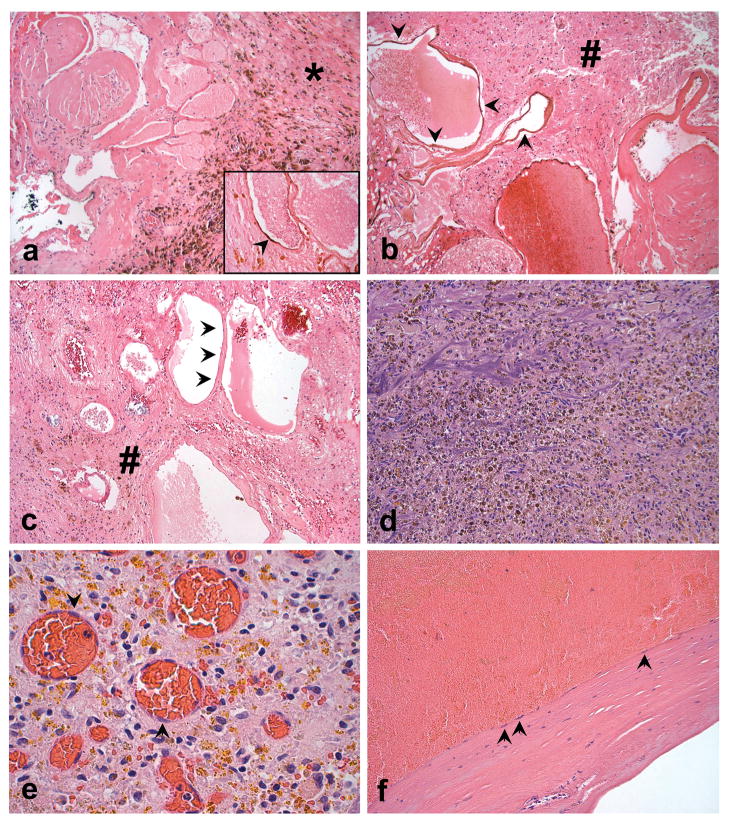
Figure 1. CCM Histopathology.
a. Low power photomicrograph of the CCM from sample 354, showing abundant hemosiderin pigment deposition in dense fibrotic tissue (*) adjacent to the caverns. (Hematoxylin and eosin (H&E), 100X). CD31 immunoreactivity definitively identifies the attenuated endothelial cells (arrowhead) typical of what was selected for LCM and genetic studies (inset, CD31 immunostaining with eosin counterstain, 600X). b. and c. Photomicrographs illustrating the numerous non-thrombosed caverns in CCMs from samples 410 and 356, respectively; note abundant adjacent brain tissue in these samples (#). The lining of the caverns (arrowheads) was typical of what was selected for LCM analysis (H&E, 200x). d. Specimen 332 showed a remotely ruptured vascular malformation, with only poorly preserved fragments of vessel wall and abundant hemosiderin pigment deposition (H&E, 200X). e. On higher power magnification, the resected 332 tissue contained the typical reparative response, with loose fibroblastic tissue, extensive golden-brown refractile hemosiderin pigment deposits, and delicate new capillary formation (H&E, 600X). The latter contained endothelial cells (arrowheads) that were selected for LCM and genetically analyzed, but these vessels were not part of the original CCM and are indicative of infiltrating new vessel. f. Also within the resection specimen was a thickened hyalinized vessel wall fragment that could be recognized to be part of either the CCM itself, or possibly a large draining altered vein; arrows indicate endothelial cells selected for LCM and genetic analyses (H&E, 200X).
Table I.
Clinical, Lesion and Genetic Summary
| KRIT1 Mutations | |||||||
|---|---|---|---|---|---|---|---|
| Patient | Age | Symptom | Location | Size (mm) | Total CCMs | Germline | Somatic |
| 354 | 54 | hemorrhage | Right posterior fossa | 38×32 | >12 | c.1363C>T | −34-bp |
| exon 14 | exon 15 | ||||||
| 410 | 16 | seizure | Left frontal lobe | 18×20 | 2 | c.1363C>T | c.1300delG |
| exon 14 | exon 14 | ||||||
| 356 | 15 | none | Right Parietal | 14×7 | 1 | c.1363C>T | Not conclusive |
| exon 14 | |||||||
| 332 | 3 | Hemorrhage | Right Cerebellar | 14×8 | 6 | c.608T>G | Not conclusive |
| PDCD10 | |||||||
| exon 10 | |||||||
Somatic Point Mutation Screening
DNA was extracted from blood and from three regions of the fresh-frozen CCM tissues as previously described(13). CCMs 410 and 356 had c.1363C>T KRIT1 germline mutations and were screened for somatic mutations in KRIT1. CCM 332 had a c.608T>G PDCD10 germline mutation and was screened for PDCD10 somatic mutations. Genomic DNA coding regions and consensus splice junctions were PCR-cloned (TOPO TA Cloning Kit, Invitrogen, Carlsbad, CA) and 46 clones for each exon were screened for mutations using denaturing high-pressure liquid chromatography (dHPLC) (9). Clones with aberrant patterns were sequenced (Figure 2A). Mutations that would obviously disrupt protein translation or were found in more than one clone were pursued with further verification analyses.
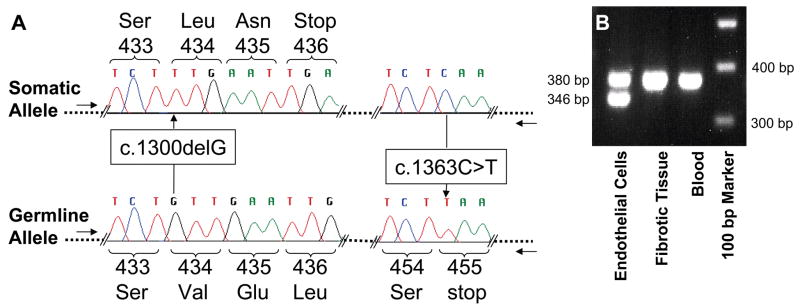
Figure 2. Genetic and LCM analyses.
A. Sequence analysis of KRIT1 exon 14 clones generated from DNA isolated from individual 410’s lesion using exon 14 genomic primers. The allele with the somatic G deletion located 1300 bp from the ATG start includes wt sequence (C) at base 1363 and is biallelic to the germline T mutation at base 1363. B. The 34-bp deletion was found only in the vascular endothelial cells (CD31+) LCM isolated from pristine caverns and not found in surrounding fibrotic tissue or from individual 354’s blood using exon 15 genomic primers (F- AAGCAGTTAACCACAAATTGG and R-GAAACTCAACAGATTTTGTGC).
Verification of Somatic Mutations
Verification of putative somatic mutations required analysis of PCR clones generated with a forward primer outside the original set, with an allele-specific reverse primer with wildtype (wt) sequence at the site of the germline mutation (CCM 410, GAAATATTGCTGAGTTTCTTG specific to 1363C wt allele; CCM 332, CACAGTTTTGAAGGTCTGAAGTATTT specific to the 608T wt allele). The somatic and germline CCM 410 mutations were both in exon 14, allowing genomic DNA analysis of alleles. For CCM 332, the putative somatic mutation was in exon 6 and the germline mutation was in exon 10; therefore, RNA was analyzed for verification.
Somatic Mutation Cell Type Mapping
LCM using a PixCell IIe microscope (Arcturus Engineering, Mountain View, CA) was employed to isolate non-glial dense fibrotic tissue (354), brain tissue (410) and vascular endothelial cells lining pristine caverns from both CCMs. Formalin-fixed paraffin-embedded (FFPE) CCM tissues were sliced (10 μM) and stained with eosin and CD31 (354) or eosin only (410). DNA was isolated using Pico Pure™ DNA Extraction Kit (Arcturus, Mountain View, CA).
For sample 354, 20–30 CD31+ endothelial cells lining roughly 10 caverns were collected in triplicate. Then, three densely fibrotic regions of 1 mm2 were collected in triplicate, which included some remote thrombus. The 354 endothelial cells and dense fibrotic non-glial tissue were PCR-amplified twice. The first PCR was with primers flanking the 34-bp deletion(13) (primary F-GGCTATAATGGAATTAAGGATTTCA and R-TTTCTGCCTCTAGTGCTTAC). The second PCR included 8 μl of the primary reaction and nested primers (secondary F-AAAGCACATGAAGTTGAAGG and R-TTCTACCAACCCACTCCC) in a 25 μl reaction volume. Products were analyzed on agarose gel (Figure 2B), bands were cut out of the gel, purified using QIAEXRII Gel Extraction Kit (Qiagen, Valencia, CA), and sequenced.
For sample 410, 20–30 endothelial cells lining roughly ten medium-sized caverns were collected in triplicate. Then, three regions of 1 mm2 surrounding brain were collected in triplicate. The 410 endothelial cells and brain were amplified three times with nested primers (primary, F-AAAGCACATGAAGTTGAAGG and R-TTCTACCAACCCACTCCC; secondary F-AAAGCACATGAAGTTGAAGGA and R-TTCTACCAACCCACTCCCA; tertiary F-AAAGCACATGAAGTTGAAGGA and R-GAAATATTGCTGAGTTTCTTG) and PCR-cloned separately using the 1363C allele-specific primer, and clones were sequenced. For sample 332, the caverns were relatively small in size and roughly only twenty endothelial cells lining twenty caverns were able to be collected in triplicate.
Results
Clinical Phenotypes (Table 1)
Patient 354 harbored greater than twelve CCM lesions. She had experienced at least three clinically significant hemorrhages over 1–2 years, resulting in disabling and progressive neurological deficits. MRI analysis was consistent with hemorrhage occurring in the CCM lesion that was removed and used in this genetic study.
Patient 410 had two CCM lesions. He had experienced seizures that were controlled with phenytoin. Troubling auras continued and at the time of surgery, it was unclear which of the two lesions was causing symptoms. Follow-up confirmed that his seizures resolved after removal of the left frontal lobe CCM that was used in this genetic study. Patient 356, a female, had a single asymptomatic lesion visible on MRI which was removed and used in this genetic study.
Patient 332 had six lesions. She had experienced multiple hemorrhages from different CCM lesions and had undergone three surgeries prior to the removal of the CCM used in this genetic study. The CCM used in this analysis was thought to have bled three months and one month prior to removal, based on symptoms and MRI changes.
Histopathology
Relative to the originally published specimen 354, specimens 410 and 356 contained fewer caverns with organizing thromboses, less fibrotic tissue and 40–60% surrounding normal brain tissue, based on selected tissue sections (Figure 1a, b, c). Specimen 332 included abundant hemosiderin with only fragments of remaining identified CCM vessel wall (Figure 1d) and extensive organization of the clot with granulation tissue-type response and likely new vessel formation (Figure 1e). Portions of a large caliber vessel wall, shown in Figure 1f, were also included in the specimen and were consistent with part of the ruptured CCM or the draining vein of the CCM. Vascular endothelial cells surrounding pristine caverns were not found in specimen 332 due to the extensive obliteration by previous hemorrhage.
KRIT1 Mutations
Specimens 354, 410 and 356 had germline c.1363C>T KRIT1 Hispanic-American mutations in blood and CCM DNA. In CCM 410 DNA, the germline mutation was biallelic to a novel somatic G deletion at nucleotide 1300 in exon 14 (Figure 2A). Clones in the primary screen were generated with intron primers flanking both somatic and germline mutations (intron primers flanking exon 14; F-AAAGCACATGAAGTTGAAGG and R-TTCTACCAACCCACTCCC). PCR-introduced errors were on average 5.6% for the entire 410 screen. Forty-six exon 14 clones were screened and the two clones with the G deletion were also wildtype, 1363C, showing the somatic mutation and germline mutations were biallelic. The c.1300delG deletion was not found in 94 subsequent clones with intron primers outside the original set (F-ATATCACCAACAGATTCTCAC and R-TTTCAGCAGTTTGAACACTAG), then verified in 5/176 allele-specific clones (F-AAAGCACATGAAGTTGAAGG and R-GAAATATTGCTGAGTTTCTTG) from frozen tissue, and found again in 96/192 of allele-specific clones from LCM endothelial cells, but not in 192 clones from LCM surrounding brain from FFPE tissue.
The somatic 34-bp deletion previously identified in exon 15 in CCM 354 was also localized to the DNA from vascular endothelial cells lining surrounding caverns but not in DNA from dense fibrotic regions surrounding the caverns, using allele-specific PCR analysis (Figure 2B). The two PCR bands were of variable intensity relative to each other in two independent experiments and roughly equal intensity when combined in the experiment pictured in Figure 2B, suggesting LCM and subsequent PCR amplifications were not quantitative.
In CCM 356, possible somatic mutations that would obviously disrupt the KRIT1 protein translation, were not found in 48 clones. Two possible somatic T deletion mutations were identified in intronic mononucleotide repeats. These mutations were at similar levels in both LCM endothelial cells and non-endothelial cell captured and therefore thought to be PCR-introduced errors. An 8% estimate of the PCR-cloning error rate was made by counting up the total number of clones with aberrant dHPLC patterns over the total number of clones with examined by dHPLC. Allele-specific RTPCR analysis was not employed with this sample and additional CCM genes were not screened due to insufficient lesion quantity.
PDCD10 Mutations
Sample 332 has a germline c.608T>G mutation that is predicted to truncate the PDCD10 protein by 10 amino acids. A potential somatic deletion of an A at position 205 from the ATG start in exon 6 was found in a mononucleotide repeat of 7 A’s. The frequency of PCR-generated deletions at a mononucleotide repeat was similar to the frequency of the possible somatic mutation. Aberrant clones were found at 4.8% in this screen. Therefore, it could not be determined conclusively if the A deletion was a somatic mutation in specimen 332 or only an artifact generated by the procedure.
Discussion
In this study, a novel somatic G deletion was identified that was biallelic to a germline mutation. While germline mutations are generally present in every cell type of the individual, a somatic mutation could be restricted to a cell type. Within the cell type(s) harboring the somatic mutation, each copy of the KRIT1 gene was predicted to result in a truncated KRIT1 protein from sample 410.
Methodological Interpretations
Several groups have failed to find somatic mutations possibly due to limitations in the methods employed(13, 19, 26, 31, 32). It is therefore, important to recognize the advantages and limitations of the methods used in this study. When RNA was isolated from three pieces of the surgically excised CCM (410), the somatic G deletion was initially found at very low proportions similar to what was expected for PCR-generated errors. However, this was the only mutation that was predicted to truncate the KRIT1 gene so it was sought in additional cell types. The frequency of the somatic mutation was significantly higher in vascular endothelial cells lining pristine caverns. Unlike sample 354, the somatic mutation in 410 DNA required cloning in order to be detected likely due to the fact that specimen 410 included a large amount of brain tissue surrounding the pristine caverns. Use of an allele-specific primer and especially LCM endothelial cells enhanced detection of the somatic G deletion to 50% of clones (96/192). The enrichment for 410 was not 100%. However, it should be stressed that the LCM PCR was not quantitative and equal intensities of both PCRed alleles were found in specimen 354 endothelial cells only after combining two independent experiments. Clones generated from endothelial cells without the G deletion may be due to mispriming of the allele-specific primer to the germline mutation allele, the capture of some nonendothelial cells, the capture of some endothelial cells lining normal vessel and/or caverns that are mosaic with both wildtype and mutant vascular endothelial cells in the caverns. The fact that some wildtype endothelial clones were found suggests that analyses for loss of heterozygosity in CCM endothelial cells may be complicated. Screening for mutations in the endothelial cells lining pristine caverns may enhance the ability to detect somatic mutations.
Analysis of CCM specimens 356 and 332 were inconclusive possibly due to methodological limitations as well as limitations in the specimens themselves which included low numbers (356) or no pristine caverns (332) in the slices examined. Candidate somatic mutations were found in mononucleotide repeats and may be due to PCR introduced errors(5). It is possible that PDCD10 c205delA somatic deletion in sample 332 could not be verified because the tissue that remained was without pristine caverns. Somatic mutations in specimens 356 and 332 could have been missed due to the low proportion of pristine caverns in the tissue, or because only putative somatic mutations that either obviously disrupted the coding sequence, or that were found in more than one clone, were followed. In addition, large-scale genomic deletions/duplication were not sought in this study and have been reported in KRIT1 (1/25 KRIT1 probands(22); 2/6 KRIT1 probands(10)) and in PDCD10 (1/8 PDCD10 probands (4)). The 3 CCM genes were not screened for somatic mutations due to insufficient sample and transheterozygosity may occur as has been shown for diseases like polycystic kidney disease that can be caused by a “two-hit” mechanism (38).
Finally, some types of CCM germline mutations have yet to be discovered. After screening the coding regions of the 3 CCM genes, mutations were found in 78–84% of CCM families and CCM cases with multiple lesions(7). Likewise, somatic mutations may not be limited to the coding region of the 3 CCM genes and mutations outside the coding region would have been missed in this study.
KRIT1
KRIT1 knockouts where both germline copies of the gene are disrupted in every cell of the organism are lethal at the embryonic stage in mice(39) and zebra fish(25) suggesting KRIT1 protein is essential to endothelial cell morphology and vessel morphogenesis in the heart (17). Our finding that some CCMs harbour somatic mutations is consistent with the hypothesis that “two-hits” in the same CCM gene are responsible for lesion genesis. Patients in this study inherited the first hit and occurrence of a second hit effectively rendered both copies of the KRIT1 gene unable to produce full-length protein in vascular endothelial cells within the CCM. Mapping of the somatic mutations to vascular endothelial cells correlates with the primary CCM pathology of endothelial cell proliferation and aberrancy in tight junctions(35, 40).
The CCM Pathway
The CCM proteins interact in the same pathway as scaffold or adaptor proteins that regulate trafficking and the cytoskeleton(16). The CCM2 protein physically interacts with both KRIT1 and CCM3 proteins(16, 37). KRIT1 protein binds to Rap1 and integrin cytoplasmic domain-associated protein, (ICAP-1α) suggesting KRIT1 is part of the integrin signaling pathway, cell adhesion, cell-cell junctions and regulation of proliferation (3, 14, 34, 41–43). The CCM2 protein has a phosphotyrosine-binding domain just as ICAP1α does, consistent with the assertion that it acts in the same pathway apparently binding the NPxY motif (41, 43) of KRIT1 protein. The phosphotyrosine binding (PTB) domain of the CCM3 protein (programmed cell death, PDCD10) binds KRIT1 protein through its NPxY motifs (16). CCM3 appears to be part of an apoptotic pathway (24) and the protein phosphatase 2A pathway that controls cell growth, proliferation and differentiation(15). Although multiple other proteins have been identified in the CCM pathway, the CCM genes have an essential non-redundant role in developing and maintaining blood vessels in the central nervous system.
Clinical Implications
The clinical course of CCM lesions is highly unpredictable. Within families and even in the same individual, CCM lesions have different propensities to bleed. This fact is consistent with an influence from genetic factors that are not inherited such as somatic mutations. In the “two-hit” mechanism CCM lesions are thought to be multiclonal and arise from independent events and are not due to metastasis. Therefore, individual lesions from the same patient would harbor unique somatic mutations similar to different somatic mutations contributing to the multiclonal nature of benign neurofibromatosis type 2(11) and polycystic kidney disease (PKD)(8). Intra-familial phenotypic variability has been primarily attributed to the “second-hit” in families with PKD(8) and more work needs to be done in order to determine if the somatic mutations contribute to the variable CCM phenotype.
Preventing the occurrence of somatic mutations is difficult and somatic mutations occur as part of normal development and aging(2, 27). Some of the suggested cancer prevention guidelines may be appropriate for individuals with multiple CCMs such as not smoking. Carcinogens from smoke that are absorbed into the blood would expose brain and spinal cord endothelial cells. Patients should limit their exposure to mutagens like radiation(18) that can cause somatic mutations in endothelial cells.
Many characteristics of CCMs make them potentially good candidates for gene therapy once gene therapy methods are proven successful (28). The “two-hit” mechanism is consistent with the hypothesis that introduction of the corresponding normal full-length gene might be sufficient to correct pathogenic endothelial cells. Gene therapy could be readily delivered to vascular endothelial cells through the blood stream. It would be important to determine the consequence of changing highly attenuated endothelial cells into cells with a more normal morphology and whether these changes could result in leakage or rupture. One primary limitation of gene therapy treatment of CCM would be the efficiency of gene transfer. Abnormal cells that were not successfully targeted would likely go on to regenerate the CCM.
Conclusions
Somatic mutations in KRIT1 were identified in vascular endothelial cells lining CCM caverns examined in two patients. The two somatic truncating mutations were biallelic, with germline Hispanic-American KRIT1, mutations suggesting the “two-hit” mechanism of disease may be responsible for lesion genesis in these two patients and that the primary defect lies in the vascular endothelial cells. Methods of somatic mutation detection should focus on vascular endothelial cells lining pristine caverns. Further investigations are warranted to determine how common the “two-hit” mechanism is found in CCM lesions.
Acknowledgments
Financial Disclosure: Financial support has not been received in conjunction with the generation of this submission.
Grant Information: This work was supported in part by Dr. Gault’s NINDS R21, 1 R21 NS053939-01A2
We thank the patients for participating in this study. We thank Mark P Burgoon, Kathryn Beauchamp, Carlyne D. Cool, Quoc-An Nguyen and Ruby Vianzon for technical support.
Footnotes
Disclosure of funding: Dr. Gault has received an NINDS R21, 1 R21 NS053939-01A2, starting July 1, 2008 for continuation of this work.
Author Justification Statement
Judith Gault, was responsible for writing the paper the experimental design and interpretation.
Issam Awad was responsible for facilitating patient enrollment, confirming diagnosis and writing the paper.
Peter Recksiek was responsible for enrolling patients, carrying out the experiments and technical interpretation of the results.
Robert Shenkar was responsible for manuscript preparation, patient enrollment and interpretation of results.
Robert Breeze was responsible for patient diagnosis, enrollment and clinical interpretation of the patient’s disease course.
Michael Handler was responsible for patient diagnosis, enrollment and clinical interpretation of the patient’s disease course.
B. K. Kleinschmidt-DeMasters was responsible for histopathological diagnosis and interpretation of the CCM lesions.
References
- 1.Akers AL, Johnson E, Steinberg GK, Zabramski JM, Marchuk DA. Biallelic somatic and germline mutations in cerebral cavernous malformations (CCMs): evidence for a two-hit mechanism of CCM pathogenesis. Hum Mol Genet. 2009;18:919–930. doi: 10.1093/hmg/ddn430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Araten DJ, Golde DW, Zhang RH, Thaler HT, Gargiulo L, Notaro R, Luzzatto L. A quantitative measurement of the human somatic mutation rate. Cancer Res. 2005;65:8111–8117. doi: 10.1158/0008-5472.CAN-04-1198. [DOI] [PubMed] [Google Scholar]
- 3.Beraud-Dufour S, Gautier R, Albiges-Rizo C, Chardin P, Faurobert E. Krit 1 interactions with microtubules and membranes are regulated by Rap1 and integrin cytoplasmic domain associated protein-1. Febs J. 2007;274:5518–5532. doi: 10.1111/j.1742-4658.2007.06068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergametti F, Denier C, Labauge P, Arnoult M, Boetto S, Clanet M, Coubes P, Echenne B, Ibrahim R, Irthum B, Jacquet G, Lonjon M, Moreau JJ, Neau JP, Parker F, Tremoulet M, Tournier-Lasserve E. Mutations within the Programmed Cell Death 10 Gene Cause Cerebral Cavernous Malformations. Am J Hum Genet. 2005;76:42–51. doi: 10.1086/426952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke LA, Rebelo CS, Goncalves J, Boavida MG, Jordan P. PCR amplification introduces errors into mononucleotide and dinucleotide repeat sequences. Mol Pathol. 2001;54:351–353. doi: 10.1136/mp.54.5.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denier C, Goutagny S, Labauge P, Krivosic V, Arnoult M, Cousin A, Benabid AL, Comoy J, Frerebeau P, Gilbert B, Houtteville JP, Jan M, Lapierre F, Loiseau H, Menei P, Mercier P, Moreau JJ, Nivelon-Chevallier A, Parker F, Redondo AM, Scarabin JM, Tremoulet M, Zerah M, Maciazek J, Tournier-Lasserve E. Mutations within the MGC4607 Gene Cause Cerebral Cavernous Malformations. Am J Hum Genet. 2004;74:326–337. doi: 10.1086/381718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denier C, Labauge P, Bergametti F, Marchelli F, Riant F, Arnoult M, Maciazek J, Vicaut E, Brunereau L, Tournier-Lasserve E. Genotype-phenotype correlations in cerebral cavernous malformations patients. Ann Neurol. 2006;60:550–556. doi: 10.1002/ana.20947. [DOI] [PubMed] [Google Scholar]
- 8.Devuyst O. [Variable progression of autosomal dominant polycystic kidney disease: genetic and molecular counterparts] Nephrol Ther. 2006;2 (Suppl 2):S104–108. [PubMed] [Google Scholar]
- 9.Donohoe E. Denaturing high-performance liquid chromatography using the WAVE DNA fragment analysis system. Methods Mol Med. 2005;108:173–187. doi: 10.1385/1-59259-850-1:173. [DOI] [PubMed] [Google Scholar]
- 10.Felbor U, Gaetzner S, Verlaan DJ, Vijzelaar R, Rouleau GA, Siegel AM. Large germline deletions and duplication in isolated cerebral cavernous malformation patients. Neurogenetics. 2007;8:149–153. doi: 10.1007/s10048-006-0076-7. [DOI] [PubMed] [Google Scholar]
- 11.Friedman JM, Fialkow PJ, Greene CL, Weinberg MN. Probable clonal origin of neurofibrosarcoma in a patient with hereditary neurofibromatosis. J Natl Cancer Inst. 1982;69:1289–1292. [PubMed] [Google Scholar]
- 12.Gault J, Sarin H, Awadallah NA, Shenkar R, Awad IA. Pathobiology of human cerebrovascular malformations: basic mechanisms and clinical relevance. Neurosurgery. 2004;55:1–16. discussion 16–17. [PubMed] [Google Scholar]
- 13.Gault J, Shenkar R, Recksiek P, Awad IA. Biallelic somatic and germ line CCM1 truncating mutations in a cerebral cavernous malformation lesion. Stroke. 2005;36:872–874. doi: 10.1161/01.STR.0000157586.20479.fd. [DOI] [PubMed] [Google Scholar]
- 14.Glading A, Han J, Stockton RA, Ginsberg MH. KRIT-1/CCM1 is a Rap1 effector that regulates endothelial cell cell junctions. J Cell Biol. 2007;179:247–254. doi: 10.1083/jcb.200705175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goudreault M, D’Ambrosio LM, Kean MJ, Mullin M, Larsen BG, Sanchez A, Chaudhry S, Chen GI, Sicheri F, Nesvizhskii AI, Aebersold R, Raught B, Gingras AC. A PP2A phosphatase high-density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol Cell Proteomics. 2008 doi: 10.1074/mcp.M800266-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hilder TL, Malone MH, Bencharit S, Colicelli J, Haystead TA, Johnson GL, Wu CC. Proteomic identification of the cerebral cavernous malformation signaling complex. J Proteome Res. 2007;6:4343–4355. doi: 10.1021/pr0704276. [DOI] [PubMed] [Google Scholar]
- 17.Hogan BM, Bussmann J, Wolburg H, Schulte-Merker S. ccm1 cell autonomously regulates endothelial cellular morphogenesis and vascular tubulogenesis in zebrafish. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn142. [DOI] [PubMed] [Google Scholar]
- 18.Jabbour P, Gault J, Murk SE, Awad IA. Multiple spinal cavernous malformations with atypical phenotype after prior irradiation: case report. Neurosurgery. 2004;55:1431. [PubMed] [Google Scholar]
- 19.Kehrer-Sawatzki H, Wilda M, Braun VM, Richter HP, Hameister H. Mutation and expression analysis of the KRIT1 gene associated with cerebral cavernous malformations (CCM1) Acta Neuropathol (Berl) 2002;104:231–240. doi: 10.1007/s00401-002-0552-6. [DOI] [PubMed] [Google Scholar]
- 20.Knudson AJ. Mutation and Cancer: Statistical Study of Retinoblastoma. Proc Natl Acad Sci U S A. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liquori CL, Berg MJ, Siegel AM, Huang E, Zawistowski JS, Stoffer T, Verlaan D, Balogun F, Hughes L, Leedom TP, Plummer NW, Cannella M, Maglione V, Squitieri F, Johnson EW, Rouleau GA, Ptacek L, Marchuk DA. Mutations in a gene encoding a novel protein containing a phosphotyrosine-binding domain cause type 2 cerebral cavernous malformations. Am J Hum Genet. 2003;73:1459–1464. doi: 10.1086/380314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liquori CL, Berg MJ, Squitieri F, Leedom TP, Ptacek L, Johnson EW, Marchuk DA. Deletions in CCM2 are a common cause of cerebral cavernous malformations. Am J Hum Genet. 2007;80:69–75. doi: 10.1086/510439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liquori CL, Berg MJ, Squitieri F, Ottenbacher M, Sorlie M, Leedom TP, Cannella M, Maglione V, Ptacek L, Johnson EW, Marchuk DA. Low frequency of PDCD10 mutations in a panel of CCM3 probands: potential for a fourth CCM locus. Hum Mutat. 2006;27:118. doi: 10.1002/humu.9389. [DOI] [PubMed] [Google Scholar]
- 24.Liu H, Wang Y, Zhang Y, Song Q, Di C, Chen G, Tang J, Ma D. TFAR19, a novel apoptosis-related gene cloned from human leukemia cell line TF-1, could enhance apoptosis of some tumor cells induced by growth factor withdrawal. Biochem Biophys Res Commun. 1999;254:203–210. doi: 10.1006/bbrc.1998.9893. [DOI] [PubMed] [Google Scholar]
- 25.Mably JD, Chuang LP, Serluca FC, Mohideen MA, Chen JN, Fishman MC. santa and valentine pattern concentric growth of cardiac myocardium in the zebrafish. Development. 2006;133:3139–3146. doi: 10.1242/dev.02469. [DOI] [PubMed] [Google Scholar]
- 26.Marini V, Ferrera L, Pigatto F, Origone P, Garre C, Dorcaratto A, Viale G, Alberti F, Mareni C. Search for loss of heterozygosity and mutation analysis of KRIT1 gene in CCM patients. Am J Med Genet A. 2004;130:98–101. doi: 10.1002/ajmg.a.30122. [DOI] [PubMed] [Google Scholar]
- 27.Marusyk A, DeGregori J. Declining cellular fitness with age promotes cancer initiation by selecting for adaptive oncogenic mutations. Biochim Biophys Acta. 2008;1785:1–11. doi: 10.1016/j.bbcan.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy SL, High KA. Gene therapy for haemophilia. Br J Haematol. 2008;140:479–487. doi: 10.1111/j.1365-2141.2007.06942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ortiz L, Costa AF, Bellido ML, Solano F, Garcia-Moreno JM, Gamero MA, Izquierdo G, Chadli A, Falcao F, Ferro J, Salas J, Alvarez-Cermeno JC, Montori M, Ramos-Arroyo MA, Palomino A, Pintado E, Lucas M. Study of cerebral cavernous malformation in Spain and Portugal: high prevalence of a 14 bp deletion in exon 5 of MGC4607 (CCM2 gene) J Neurol. 2007;254:322–326. doi: 10.1007/s00415-006-0359-9. [DOI] [PubMed] [Google Scholar]
- 30.Pagenstecher A, Stahl S, Sure U, Felbor U. A two-hit mechanism causes cerebral cavernous malformations: complete inactivation of CCM1, CCM2 or CCM3 in affected endothelial cells. Hum Mol Genet. 2009;18:911–918. doi: 10.1093/hmg/ddn420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plummer NW, Gallione CJ, Srinivasan S, Zawistowski JS, Louis DN, Marchuk DA. Loss of p53 sensitizes mice with a mutation in Ccm1 (KRIT1) to development of cerebral vascular malformations. Am J Pathol. 2004;165:1509–1518. doi: 10.1016/S0002-9440(10)63409-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reich P, Winkler J, Straube A, Steiger HJ, Peraud A. Molecular genetic investigations in the CCM1 gene in sporadic cerebral cavernomas. Neurology. 2003;60:1135–1138. doi: 10.1212/01.wnl.0000055470.62265.44. [DOI] [PubMed] [Google Scholar]
- 33.Sahoo T, Johnson EW, Thomas JW, Kuehl PM, Jones TL, Dokken CG, Touchman JW, Gallione CJ, Lee-Lin SQ, Kosofsky B, Kurth JH, Louis DN, Mettler G, Morrison L, Gil-Nagel A, Rich SS, Zabramski JM, Boguski MS, Green ED, Marchuk DA. Mutations in the gene encoding KRIT1, a Krev-1/rap1a binding protein, cause cerebral cavernous malformations (CCM1) Hum Mol Genet. 1999;8:2325–2333. doi: 10.1093/hmg/8.12.2325. [DOI] [PubMed] [Google Scholar]
- 34.Serebriiskii I, Estojak J, Sonoda G, Testa JR, Golemis EA. Association of Krev-1/rap1a with Krit1, a novel ankyrin repeat-containing protein encoded by a gene mapping to 7q21–22. Oncogene. 1997;15:1043–1049. doi: 10.1038/sj.onc.1201268. [DOI] [PubMed] [Google Scholar]
- 35.Tu J, Stoodley MA, Morgan MK, Storer KP. Ultrastructural characteristics of hemorrhagic, nonhemorrhagic, and recurrent cavernous malformations. J Neurosurg. 2005;103:903–909. doi: 10.3171/jns.2005.103.5.0903. [DOI] [PubMed] [Google Scholar]
- 36.Verlaan DJ, Roussel J, Laurent SB, Elger CE, Siegel AM, Rouleau GA. CCM3 mutations are uncommon in cerebral cavernous malformations. Neurology. 2005;65:1982–1983. doi: 10.1212/01.wnl.0000188903.75144.49. [DOI] [PubMed] [Google Scholar]
- 37.Voss K, Stahl S, Schleider E, Ullrich S, Nickel J, Mueller TD, Felbor U. CCM3 interacts with CCM2 indicating common pathogenesis for cerebral cavernous malformations. Neurogenetics. 2007 doi: 10.1007/s10048-007-0098-9. [DOI] [PubMed] [Google Scholar]
- 38.Watnick T, He N, Wang K, Liang Y, Parfrey P, Hefferton D, St George-Hyslop P, Germino G, Pei Y. Mutations of PKD1 in ADPKD2 cysts suggest a pathogenic effect of trans-heterozygous mutations. Nat Genet. 2000;25:143–144. doi: 10.1038/75981. [DOI] [PubMed] [Google Scholar]
- 39.Whitehead KJ, Plummer NW, Adams JA, Marchuk DA, Li DY. Ccm1 is required for arterial morphogenesis: implications for the etiology of human cavernous malformations. Development. 2004;131:1437–1448. doi: 10.1242/dev.01036. [DOI] [PubMed] [Google Scholar]
- 40.Wong JH, Awad IA, Kim JH. Ultrastructural pathological features of cerebrovascular malformations: a preliminary report. Neurosurgery. 2000;46:1454–1459. doi: 10.1097/00006123-200006000-00027. [DOI] [PubMed] [Google Scholar]
- 41.Zawistowski JS, Serebriiskii IG, Lee MF, Golemis EA, Marchuk DA. KRIT1 association with the integrin-binding protein ICAP-1: a new direction in the elucidation of cerebral cavernous malformations (CCM1) pathogenesis. Hum Mol Genet. 2002;11:389–396. doi: 10.1093/hmg/11.4.389. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J, Basu S, Rigamonti D, Dietz HC, Clatterbuck RE. krit1 modulates beta1-integrin-mediated endothelial cell proliferation. Neurosurgery. 2008;63:571–578. doi: 10.1227/01.NEU.0000325255.30268.B0. discussion 578. [DOI] [PubMed] [Google Scholar]
- 43.Zhang J, Clatterbuck RE, Rigamonti D, Chang DD, Dietz HC. Interaction between krit1 and icap1alpha infers perturbation of integrin beta1-mediated angiogenesis in the pathogenesis of cerebral cavernous malformation. Hum Mol Genet. 2001;10:2953–2960. doi: 10.1093/hmg/10.25.2953. [DOI] [PubMed] [Google Scholar]