Abstract
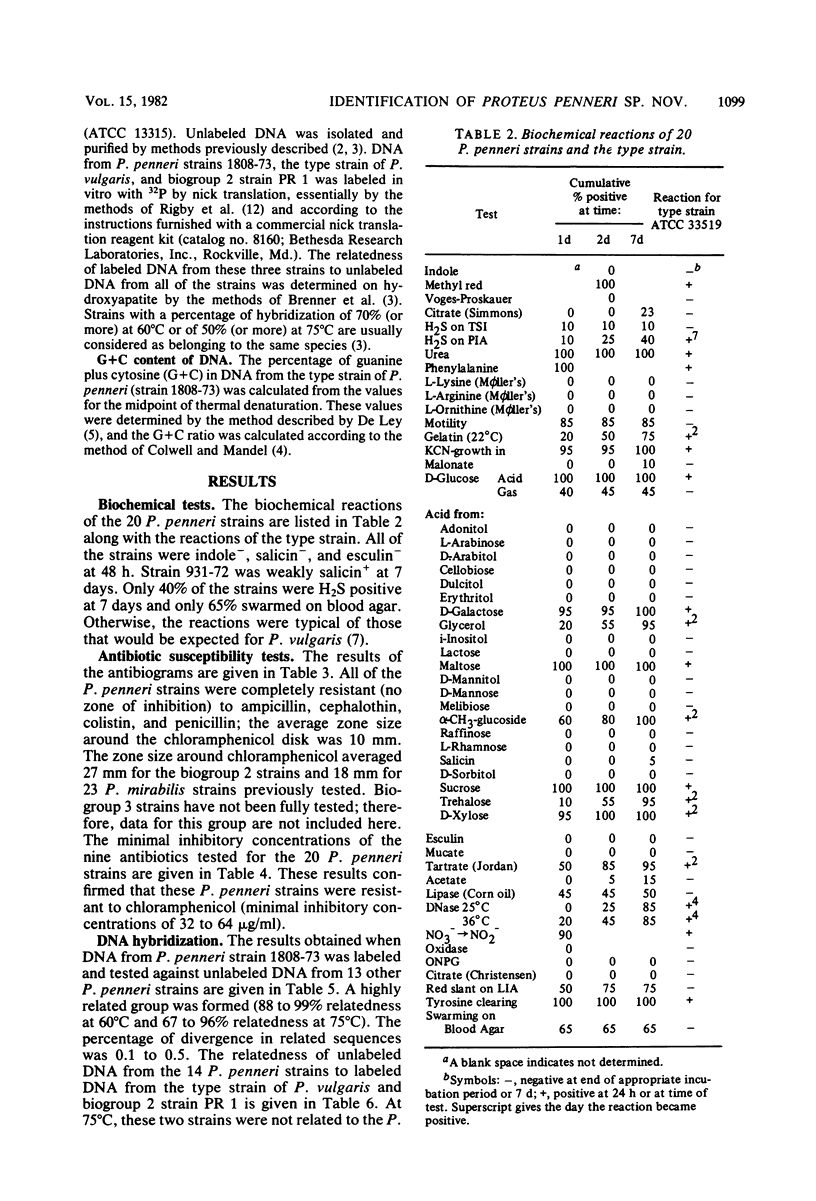
The name Proteus penneri sp. nov. is proposed for a group of organisms previously called Proteus vulgaris indole negative or P. vulgaris biogroup 1. All of these strains were salicin negative, esculin negative, and chloramphenicol resistant (zone size, less than 14 mm). DNA relatedness studies indicated that when DNA from P. penneri strain 1808-73 was labeled and tested against unlabeled DNA from 13 other P penneri strains, a highly related group was formed (88 to 99% relatedness at 60 degrees C and 67 to 99% relatedness at 75 degrees C). Strain 1808-73 (ATCC 33519) is proposed as the type strain of P. penneri. In this study, two distinct groups of indole-positive P. vulgaris strains were also apparent. The first group (defined as P. vulgaris biogroup 2) was indole positive, salicin positive, and esculin positive, and the second group (defined as P. vulgaris biogroup 3) was indole positive, salicin negative, and esculin negative. The current type strain of P. vulgaris (ATCC 13315) belongs to biogroup 3. The DNA from P. penneri strains was not highly related to labeled DNA from the type strain of P. vulgaris (14 to 30% relatedness at 75 degrees C) or from P. vulgaris strain PR 1 (ATCC 29905), which belongs to biogroup 2 (27 to 33% relatedness at 75 degrees C). Strains of biogroup 2 were sensitive to chloramphenicol (zone size, greater than 19mm), and 10 of these strains formed a highly related group by DNA hybridization when DNA from PR 1 was labeled (64 to 100% relatedness at 60 degrees C and 70 to 100% relatedness at 75 degrees C), but they were not highly relatedness to the type strain of P. vulgaris (51 to 68% relatedness at 60 degrees C and 14 to 44% relatedness at 75 degrees C). Further DNA relatedness studies are needed on strains of biogroup 3 before a definitive taxonomic proposal can be made for these two indole-positive biogroups.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bauer A. W., Kirby W. M., Sherris J. C., Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966 Apr;45(4):493–496. [PubMed] [Google Scholar]
- Brenner D. J., Fanning G. R., Johnson K. E., Citarella R. V., Falkow S. Polynucleotide sequence relationships among members of Enterobacteriaceae. J Bacteriol. 1969 May;98(2):637–650. doi: 10.1128/jb.98.2.637-650.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COLWELL R. R., MANDEL M. ADANSONIAN ANALYSIS AND DEOXYRIBONUCLEIC ACID BASE COMPOSITION OF SOME GRAM-NEGATIVE BACTERIA. J Bacteriol. 1964 Jun;87:1412–1422. doi: 10.1128/jb.87.6.1412-1422.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ley J. Reexamination of the association between melting point, buoyant density, and chemical base composition of deoxyribonucleic acid. J Bacteriol. 1970 Mar;101(3):738–754. doi: 10.1128/jb.101.3.738-754.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman F. W., Farmer J. J., 3rd Salmonella typhi: identification, antibiograms, serology, and bacteriophage typing. Am J Med Technol. 1978 Dec;44(12):1149–1159. [PubMed] [Google Scholar]
- Hickman F. W., Framer J. J., 3rd, Steigerwalt A. G., Brenner D. J. Unusual groups of Morganella ("Proteus") morganii isolated from clinical specimens: lysine-positive and ornithine-negative biogroups. J Clin Microbiol. 1980 Jul;12(1):88–94. doi: 10.1128/jcm.12.1.88-94.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis D. G., Hickman F. W., Fanning G. R., Farmer J. J., 3rd, Weaver R. E., Brenner D. J. Tatumella ptyseos gen. nov., sp. nov., a member of the family Enterobacteriaceae found in clinical specimens. J Clin Microbiol. 1981 Jul;14(1):79–88. doi: 10.1128/jcm.14.1.79-88.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKell J., Jones D. A numerical taxonomic study of Proteus-Providence bacteria. J Appl Bacteriol. 1976 Aug;41(1):143–161. doi: 10.1111/j.1365-2672.1976.tb00614.x. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- von Graevenitz A., Nourbakhsh M. Antimicrobial resistance of the genera Proteus, Providencia and Serratia with special reference to multiple resistance patterns. Med Microbiol Immunol. 1972;157(2):142–148. doi: 10.1007/BF02124474. [DOI] [PubMed] [Google Scholar]


