Abstract
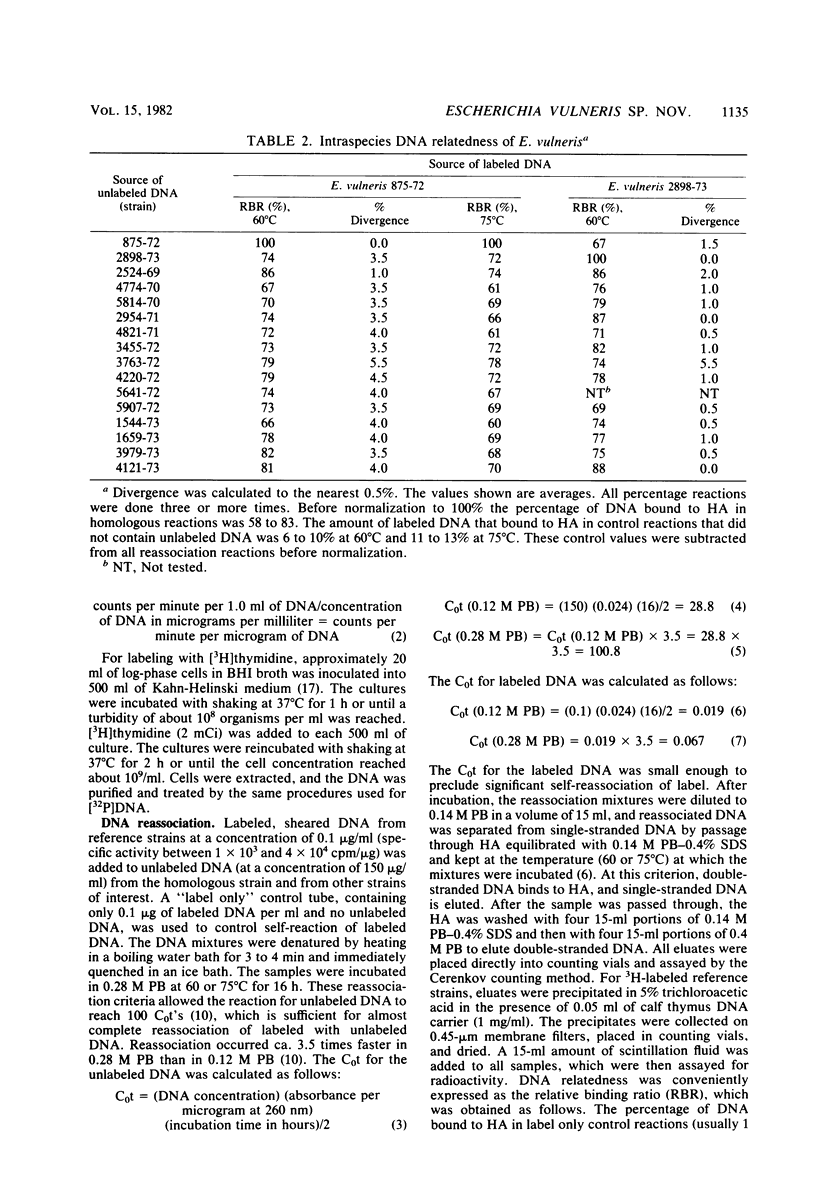
The name Escherichia vulneris sp. nov. (formerly called Alma group 1 and Enteric group 1 by the Centers for Disease Control and API group 2 by Analytab Products, Inc.) is proposed for a group of isolates from the United States and Canada, 74% of which were from human wounds. E. vulneris is a gram-negative, oxidase-negative, fermentative, motile rod with the characteristics of the family Enterobacteriaceae. Biochemical reactions characteristic of 61 E. vulneris strains were positive tests for methyl red, malonate, and lysine decarboxylase; a delayed positive test for arginine dihydrolase; acid production from d-mannitol, l-arabinose, raffinose, l-rhamnose, d-xylose, trehalose, cellobiose, and melibiose; negative tests for Voges-Proskauer, indole, urea, H2S, citrate, ornithine decarboxylase, phenylalanine deaminase, and DNase; and no acid from dulcitol, adonitol, myo-inositol, and d-sorbitol. Two-thirds of the strains produced yellow pigment. Most strains gave negative or delayed positive reactions in tests for lactose, sucrose, and KCN. The E. vulneris strains tested were resistant to penicillin and clindamycin, were resistant or showed intermediate zones of inhibition to carbenicillin and erythromycin, and were susceptible to 14 other antibiotics. DNA relatedness of 15 E. vulneris strains to the type strain averaged 75% in reactions at 60°C and 69% in reactions at 75°C, indicating that they comprise a separate species. DNA relatedness to other species in the family Enterobacteriaceae was 6 to 39%, an indication that this new species belongs in the family. E. vulneris showed the highest relatedness to species of Escherichia (25 to 39%) and Enterobacter (24 to 35%). On the basis of biochemical similarity, the new species was placed in the genus Escherichia. The type strain of E. vulneris is ATCC 33821 (CDC 875-72).
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BERNS K. I., THOMAS C. A., Jr ISOLATION OF HIGH MOLECULAR WEIGHT DNA FROM HEMOPHILUS INFLUENZAE. J Mol Biol. 1965 Mar;11:476–490. doi: 10.1016/s0022-2836(65)80004-3. [DOI] [PubMed] [Google Scholar]
- Bauer A. W., Kirby W. M., Sherris J. C., Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966 Apr;45(4):493–496. [PubMed] [Google Scholar]
- Bonner T. I., Brenner D. J., Neufeld B. R., Britten R. J. Reduction in the rate of DNA reassociation by sequence divergence. J Mol Biol. 1973 Dec 5;81(2):123–135. doi: 10.1016/0022-2836(73)90184-8. [DOI] [PubMed] [Google Scholar]
- Brenner D. J., Davis B. R., Steigerwalt A. G., Riddle C. F., McWhorter A. C., Allen S. D., Farmer J. J., 3rd, Saitoh Y., Fanning G. R. Atypical biogroups of Escherichia coli found in clinical specimens and description of Escherichia hermannii sp. nov. J Clin Microbiol. 1982 Apr;15(4):703–713. doi: 10.1128/jcm.15.4.703-713.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner D. J., Fanning G. R., Rake A. V., Johnson K. E. Batch procedure for thermal elution of DNA from hydroxyapatite. Anal Biochem. 1969 Apr 4;28(1):447–459. doi: 10.1016/0003-2697(69)90199-7. [DOI] [PubMed] [Google Scholar]
- Brenner D. J., Fanning G. R., Skerman F. J., Falkow S. Polynucleotide sequence divergence among strains of Escherichia coli and closely related organisms. J Bacteriol. 1972 Mar;109(3):953–965. doi: 10.1128/jb.109.3.953-965.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T. Measurement of 32P activity in a liquid scintillation counter without the use of scintillator. Anal Biochem. 1968 Jan;22(1):70–73. doi: 10.1016/0003-2697(68)90260-1. [DOI] [PubMed] [Google Scholar]
- FALKOW S., CITARELLA R. V. MOLECULAR HOMOLOGY OF F-MEROGENOTE DNA. J Mol Biol. 1965 May;12:138–151. doi: 10.1016/s0022-2836(65)80288-1. [DOI] [PubMed] [Google Scholar]
- Farmer J. J., 3rd, Fanning G. R., Huntley-Carter G. P., Holmes B., Hickman F. W., Richard C., Brenner D. J. Kluyvera, a new (redefined) genus in the family Enterobacteriaceae: identification of Kluyvera ascorbata sp. nov. and Kluyvera cryocrescens sp. nov. in clinical specimens. J Clin Microbiol. 1981 May;13(5):919–933. doi: 10.1128/jcm.13.5.919-933.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman F. W., Farmer J. J., 3rd Salmonella typhi: identification, antibiograms, serology, and bacteriophage typing. Am J Med Technol. 1978 Dec;44(12):1149–1159. [PubMed] [Google Scholar]
- KAHN P., HELINSKI D. R. RELATIONSHIP BETWEEN COLICINOGENIC FACTORS E1 AND V AND AN F FACTOR IN ESCHERICHIA COLI. J Bacteriol. 1964 Dec;88:1573–1579. doi: 10.1128/jb.88.6.1573-1579.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]