Abstract
African green monkeys (genus Chlorocebus) can be infected with SIVagm, but do not develop AIDS. This natural host of SIV, like sooty mangabeys, maintains high levels of SIV replication but has evolved to avoid immunodeficiency. Elucidating the mechanisms that allow the natural hosts to co-exist with SIV without overt disease may provide crucial information to understand AIDS pathogenesis. Here we show: (1) many CD4+ T cells from African green monkeys down-regulate CD4 in vivo as they enter the memory pool, (2) down regulation of CD4 by memory T cells is independent of SIV infection, (3) the CD4− memory T cells maintain functions which are normally attributed to CD4 T cells including production of IL-2, production of IL-17, expression of FoxP3 and expression of CD40L (4) loss of CD4 expression protects these T cells from infection by SIVagm in vivo, and (5) these CD4− T cells can maintain MHC-II restriction. These data demonstrate that the absence of SIV-induced disease progression in natural hosts species may be partially explained by preservation of a subset of T cells that maintain CD4 T cell function while being resistant to SIV-infection in vivo.
INTRODUCTION
Simian immunodeficiency viruses (SIV) belong to the group of lentiviruses that infect non-human primates (NHP). The lentiviruses that cause immunodeficiencies in humans and Asian macaques originated from cross-species transmission of viruses that naturally infect nonhuman primates in Africa1. Like HIV-1 and HIV-2, all known SIV subtypes use CD4 as a receptor and either CCR5, CXCR4, or, as for SIVrcm, CCR22–4 as a co-receptor. Moreover, both SIV infection of Asian macaques and HIV-1 infection of humans result in chronic infection and the majority of infected individuals progress to AIDS.
In contrast, after SIV infection natural hosts generally do not progress to AIDS. Since natural hosts of SIV have co-evolved with the virus to avoid disease progression, dissecting the mechanisms underlying the nonprogressive nature of natural SIV infection will lead to a better understanding of the aspects of HIV infection responsible for the progressive nature of the disease in humans5–7. Natural hosts do not avoid disease progression by immunological control of the virus since SIV-infected natural hosts maintain high levels of viremia8–12. Moreover, experimental depletion of CD8+ T cells does not affect plasma viremia13 and natural hosts do not exhibit superior cellular control of viremia compared to HIV-infected humans or SIV-infected rhesus macaques(RM)14. The lentiviruses that infect natural hosts can be pathogenic. SIV infections of AGM and Sooty mangabeys (SM) have been correlated with short life spans of infected cells in vivo15–17. Moreover, SIVagm, which naturally infects African green monkeys (AGM), can be used to infect pigtail macaques who subsequently manifest simian AIDS18,19. Isolates of SIVsmm can also cause progressive infection in RM20–23. One fundamental difference between progressive SIV/HIV infection and nonprogressive SIV infection is the absence of immune activation, which is associated with disease progression in HIV-infected individuals24, during the chronic phase of infection in natural hosts9,11,25–28.
Previous studies performed using AGM have reported very low frequencies of CD4+ T cells28. AGM, however, remain disease free despite having low numbers of CD4+ T cells. In two reports, Murayama et al, described low frequencies of CD4+ T cells and high frequencies of CD8dim T-cells in healthy adult AGM29,30. They also found the CD8dim T cells could induce antibody production from B-cells in vitro and suggested that the CD8dim T-cells might supplement for the lack of CD4+ T-cells29. Here we have studied the frequencies, functionalities and in vivo infection frequencies of lymphocyte subsets from 36 AGM, 10 naturally infected, 8 experimentally infected, 11 uninfected adults and 7 uninfected juveniles (Table 1). Our data describe a mechanism by which AGM are able to survive SIVagm infection without succumbing to AIDS.
Table 1.
Clinical information of African green monkeys.
| Animal | Virus Strain | Infection Dose (TCID) | Plasma Viral Load (/mL) | CD4 T-cells (/μl blood) |
|---|---|---|---|---|
| AG3 | Naturally Infected | NA | 29600 | 341 |
| AG4 | Naturally Infected | NA | 19600 | 252 |
| AG5 | Naturally Infected | NA | 26000 | 63 |
| AG6 | Naturally Infected | NA | 1400 | 314 |
| AG10 | Naturally Infected | NA | 8000 | 210 |
| AG11 | Naturally Infected | NA | Undetected | 156 |
| AG12 | Naturally Infected | NA | 23600 | 60 |
| AG15 | Naturally Infected | NA | Undetected | 495 |
| AG16 | Naturally Infected | NA | 61100 | 452 |
| AG17 | Naturally Infected | NA | 148000 | 235 |
| AG7 | SIVagm90 | 1000 | 55800 | 449 |
| AG8 | SIVagmVer1 | 50 | 1660 | 180 |
| AG13 | SIVagm90 | 1000 | 31500 | 308 |
| AG19 | SIVagmVer1 | 50 | 44300 | 427 |
| AG22 | SIVagm90 | 1000 | 2066100 | 185 |
| AG24 | SIVagm90 | 1000 | 11500 | 301 |
| AG302 | SIVagm90 | 1000 | 19700 | 170 |
| AG346 | SIVagm90 | 1000 | 2000 | 17 |
| AG9 | Uninfected | NA | NA | 289 |
| AG23 | Uninfected | NA | NA | 821 |
| AG731 | Uninfected | NA | NA | 129 |
| AG5339 | Uninfected | NA | NA | 165 |
| AG5387 | Uninfected | NA | NA | 147 |
| AG5419A | Uninfected | NA | NA | 35 |
| AG5419B | Uninfected | NA | NA | 67 |
| AG5431 | Uninfected | NA | NA | 62 |
| AG5441 | Uninfected | NA | NA | 114 |
| AG5504 | Uninfected | NA | NA | 55 |
| AG5506 | Uninfected | NA | NA | 99 |
| AG25 | Uninfected juvenile | NA | NA | 1625 |
| AG26 | Uninfected juvenile | NA | NA | 2593 |
| AG28 | Uninfected juvenile | NA | NA | 2791 |
| AG30 | Uninfected juvenile | NA | NA | 1531 |
| AG31 | Uninfected juvenile | NA | NA | 3214 |
| AG32 | Uninfected juvenile | NA | NA | 2121 |
| AG33 | Uninfected juvenile | NA | NA | 2798 |
| HK14 | Uninfected sabeus | NA | NA | 378 |
RESULTS
Decreased frequencies of CD4+ T-cells correlate with increased frequencies of CD4−CD8αdim T-cells
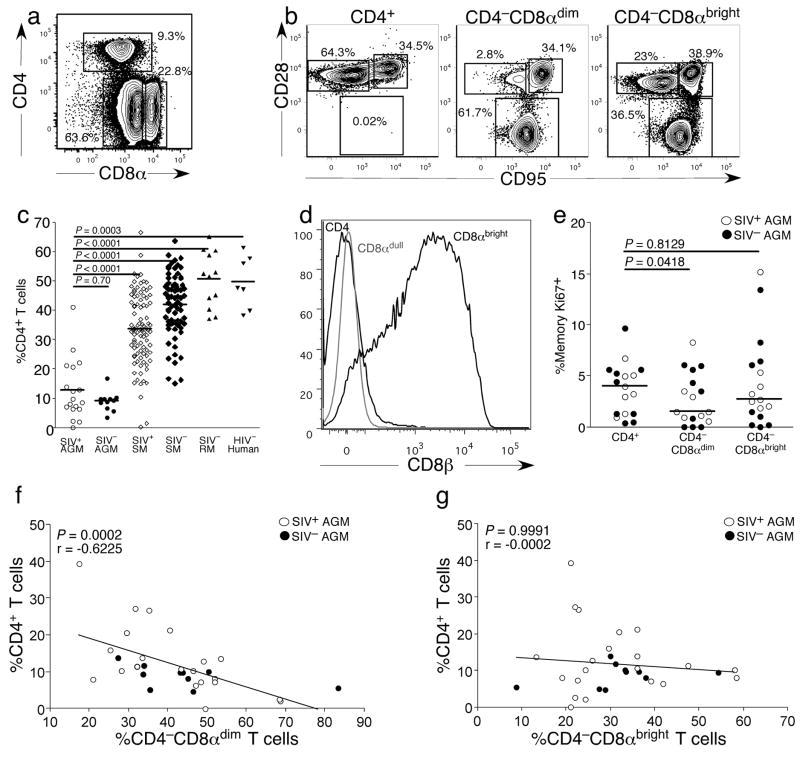
Loss of CD4+ T-cells is a hallmark of progression to AIDS in humans and Asian macaques. We therefore compared the frequency of CD4+ T-cells in SIVagm-infected and uninfected vervet AGM (one of four subspecies of AGM), HIV-uninfected humans, SIV-uninfected RM and SIVsmm-infected and uninfected SM. We observed three distinct T-cell populations in AGM based upon expression of CD4 and CD8α: CD4+ T-cells, CD4−CD8αdim T-cells and CD4−CD8αbright T-cells (Fig. 1a). We analyzed further the phenotypes of each subset based upon expression of CD28 and CD95 (Fig. 1b). The CD4+ and CD4−CD8αbright T-cells consisted of both memory and naive subsets (Fig. 1b). However, the CD4−CD8αdim T-cells consisted predominantly of memory T-cells (Fig. 1b). Moreover, adult AGM appeared to have surprisingly low frequencies of CD4+ T-cells and low CD4+ T-cell counts(Table 1). There were significantly lower frequencies of CD4+ T-cells in both SIVagm-infected and uninfected AGM compared to SIVsmm-infected or uninfected SM, SIV-uninfected RM or HIV-1-uninfected humans (Fig. 1c). In addition, the decrease in CD4+ T-cells was accompanied by an increase in the frequency of CD4−CD8αdim T-cells (Fig. 1a). Further phenotypic analysis showed that this CD4−CD8αdim T-cell population lacked expression of CD8β (Fig. 1d). In addition, the frequency of Ki67+ cells was significantly among CD4−CD8αdim T-cells compared to either the CD4+ or CD4−CD8αbright subsets, suggesting that the high frequency of CD4−CD8αdim T-cells was not due to preferential proliferation ofCD4−CD8αdim T-cells in vivo (Fig. 1e).
Figure 1. Phenotypic analysis of T-cell populations in vervet African green monkeys.
(a) Phenotype of T-cells in adult AGM. (b) Phenotype of individual subsets of T-cells in adult AGM. (c) Comparison of percent of CD3+ T-cells that express CD4 in peripheral blood in SIV+ and SIV− adult AGM, adult SIV+ and SIV− SM, adult SIV− RM, and HIV− adult humans. (d) Characterization of CD4−CD8αdim T-cells as CD8αβ− by analysis of CD8β expression. Cells were gated on live CD3+ small lymphocytes and analyzed for both CD4 and CD8β expression. (e) Ki67 expression by different subsets of memory T-cells. (f) Negative correlation between the frequencies of CD4+ T-cell and CD4−CD8αdim T-cells in adult AGM. (g) Correlation of the frequency of CD4+ T-cells and the frequency of CD4−CD8αbright T-cells in adult AGM. White circles represent SIV-infected AGM and black circles represent uninfected AGM. White diamonds represent SIV-infected SM and black diamonds prepresent uninfected SM. Black triangles represent SIV-uninfected RM and black squares represent HIV-uninfected humans. A Mann-Whitney U test was performed for c. A Spearman rank correlation was calculated for f and g.
The increased frequency of CD4−CD8αdim T-cells could be a reflection of a mathematical shift in percentages of T-cell populations due to the loss of CD4+ T-cells. However, we found that there was a significant negative correlation between the decrease in CD4+ T-cells and the increase in CD4−CD8αdim T-cells (Fig. 1f), but not for the CD4−CD8αbright T-cells (Fig. 1g). Therefore, the increase in the CD4−CD8αdim T-cell population was directly related to the decrease in CD4+ T-cells, suggesting that someCD4−CD8αdim T-cells might develop from CD4+ T-cells.
Some CD4−CD8αdim T-cells develop from CD4+ memory T-cells
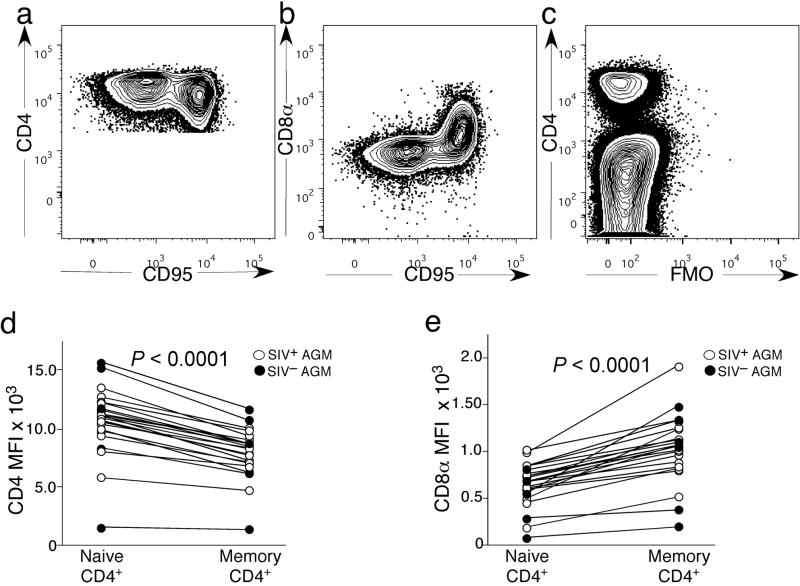
Since the frequencies of the CD4−CD8αdim T-cells negatively correlated with the frequencies of CD4+ T-cells (Fig. 1f), we hypothesized that some of the CD4−CD8αdim T-cells may have developed from CD4+ T-cells. Phenotypic analysis of the CD4+ T-cells from SIVagm-infected and uninfected AGM illustrated that the CD95+ memory CD4+ T-cells down-regulated CD4 (Fig. 2a) and up-regulated CD8α (Fig. 2b) compared to naive CD4+ T-cells. Fluorescence minus one analysis for CD8α was also performed to confirm that the CD4+ T-cells expressed CD8α (Fig 2c). The median fluorescent intensity (MFI) of CD4 was significantly lower in the memory subset of CD4+ T-cells compared to naive CD4+ T-cells (Fig. 2d) while the MFI of CD8α was significantly increased in the memory CD4+ T-cells over naive (Fig. 2e). Therefore, we hypothesized that upon stimulation CD4+ T-cells could transition to become CD4−CD8αdim T-cells.
Figure 2. CD8α and CD4 expression by naive and memory CD4+ T-cells.
(a) Representative staining of CD4+ memory and naive T-cells for CD4. (b) Representative staining of CD4+ memory and naive T-cells for CD8. (c) Fluorescence minus one control lacking antibodies against CD8α. (d) Median fluorescence intensity for CD4 expression in naive and memory CD4+ T-cells. (e) Median fluorescence intensity for CD8 expression in naive and memory CD4+ T-cells. A Wilcoxon matched pairs test was performed for 2d–e.
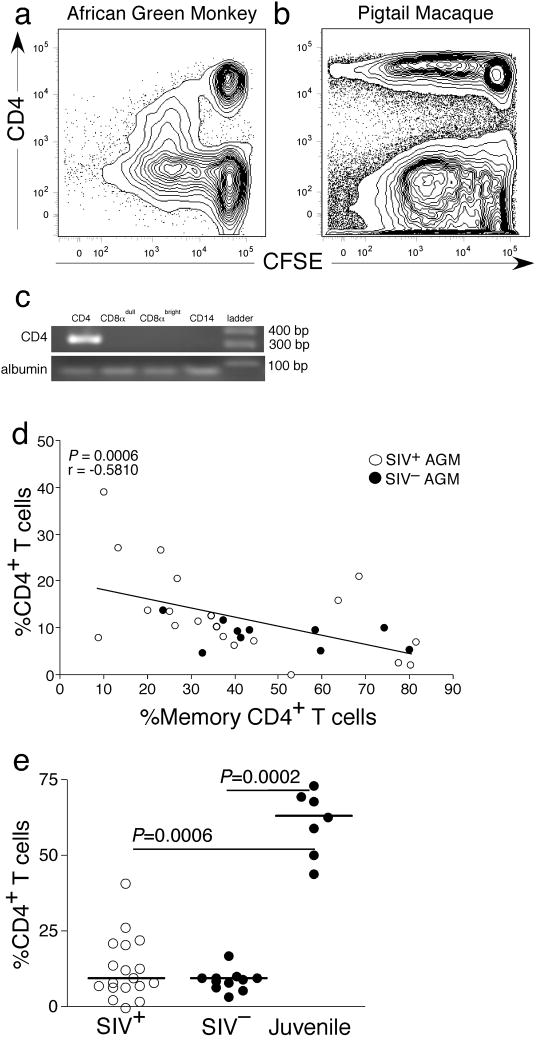
To study down-regulation of CD4 in vitro, we CFSE labeled and mitogenically stimulated peripheral blood mononucelear cells (PBMC) from several AGM and, for comparison, pigtail macaques. We then studied expression of CD4 and proliferation by flow cytometry after 5, 7, and 9 days in culture (Fig. 3a–b and Supplemental Fig. 1). We found that while AGM CD4+ T-cells lost CD4 expression, CD4+ T-cells from pigtail macaques maintained CD4 expression with cell division (Fig. 3a). It is a possibility that the CD4 down-regulation in AGM was transient, however, we did not see up-regulation of CD4 by day 9 (Supplemental Fig. 1). To confirm this finding, we also sorted naive CD4+ T-cells (>99% pure) prior to stimulation and found that after stimulation they became CD4−(Supplemental Fig. 1).
Figure 3. CD4−CD8αdim T-cells can develop from memory CD4+ T-cells.
(a) Down-regulation of CD4 by stimulated in vitro PBMC from AGM after 5 days of stimulation with SEB. (b) Maintenance of CD4 expression by stimulated PBMC from pigtail macaques in vitro after 5 days of stimulation with SEB. (c) CD4 mRNA expression in CD14+, CD4−CD8αdim, CD4−CD8αbright, or CD4+ lymphocyte subsets of an adult AGM. (d)Negative correlation between the frequency of memory CD4+ T-cells and the frequency of the total CD4+ T-cells in adult AGM. (e) Comparison of percent CD4+ T-cells in peripheral blood in SIV+ and SIV− adult and SIV− juvenile AGM. White circles represent SIV-infected AGM and black represent uninfected AGM. A Spearman rank correlation was calculated for 3c. A Mann Whitney U test was performed for 3e.
Analysis for CD4 mRNA within sorted CD4+, CD4−CD8αdim, CD4−CD8αbright T-cells and monocytes revealed that CD4 mRNA was only detected within CD4+ T-cells (Fig. 3c). Taken together, these data suggested that AGM memory CD4+ T-cells could become CD4−CD8αdim T-cells. If our hypothesis was correct, AGM would have concomitant decreases in the overall frequencies of total CD4+ T-cells with increases in memory CD4+ T-cells. Indeed, we found a significant negative correlation between the frequencies of memory CD4+ T-cells and the total frequency ofCD4+ T-cells (Fig. 3d). Hence as AGM accumulate memory CD4+ T-cells, the overall frequency of CD4+ T-cells decreases.
We reasoned that if some CD4+ T-cells were becoming CD4−CD8αdim T-cells upon stimulation into the memory pool, then animals with very little antigenic experience should have frequencies of CD4+ T-cells comparable to those of healthy RM and humans (Fig. 1c). Therefore, we measured the frequencies of T-cell subsets from PBMC obtained directly ex vivo from juvenile AGM (all less than 2 years old). In a total of seven juvenile AGM, the median frequency of CD4+ T-cells was 59% (39.6%–64.4%, Fig. 3e). These frequencies were comparable with those from non-immunocompromised RM and humans and were higher than the frequencies of CD4+ T-cells in adult AGM (P<0.0006). Furthermore, after we vaccinated the juvenile AGM with the standard influenza vaccine we observed an influenza-specific T-cell response from the CD4−CD8αdim T-cells to the MHC-II restricted antigen (Supplemental Fig. 2c). Taken together, these observations strongly suggest that many of the CD4−CD8αdim T-cells developed from memory CD4+ T-cells.
While we do not know the ages of many of the AGM, as they were imported from Tanzania, in the few animals from which we do have this information, there was no correlation between age and the frequency of CD4+ T-cells (data not shown). However, it is likely that age may be a factor in accumulation of CD4−CD8αdim T-cells as older animals are exposed to more antigens in vivo.
CD4−CD8αdim T-cells preserve CD4+ T-cell function while evading SIV infection
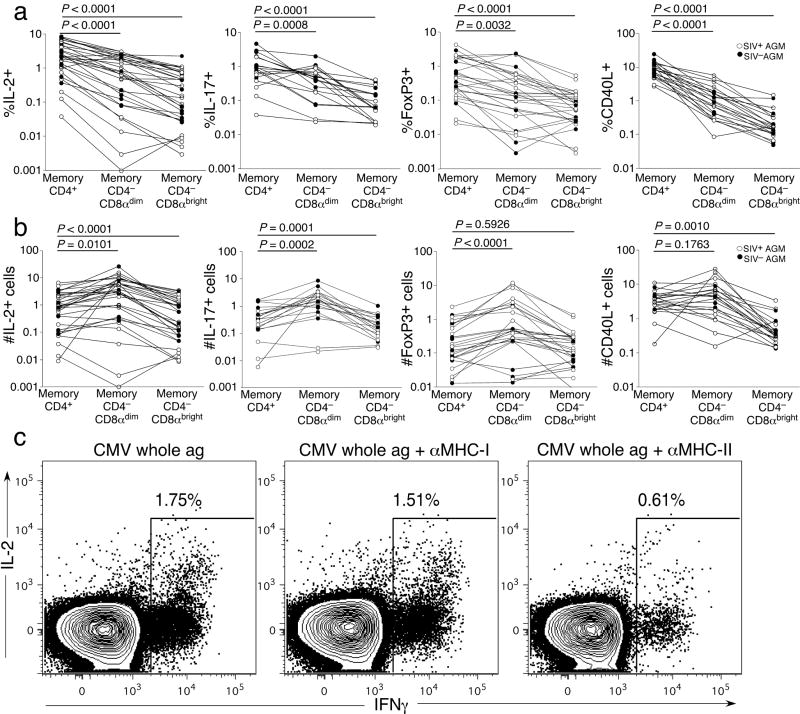
To elucidate the functions of the CD4−CD8αdim T-cells, we obtained PBMC from SIVagm-infected and uninfected adult AGM. Since it seemed apparent that many of the CD4−CD8αdim T-cells developed from CD4+ T-cells, we examined these T-cells for functions generally attributed to CD4+ T-cells. We found that upon stimulation with staphylococcal enterotoxin B (SEB), the CD4−CD8αdim T-cells could produceIL-2 and IL-17 (Supplemental Fig. 2 and Fig. 4a) and express CD40 ligand (CD40L) (Supplemental Fig. 2 and Fig. 4a). CD40L is typically expressed by activated CD4+ T-cells and results in enhancement of antigen presentation and induces B-cell class switching31. Additionally, a portion of the CD4−CD8αdim T-cells expressed the transcription factor FoxP3, thought to be predominantly expressed by regulatory CD4+ T-cells (Supplemental Fig. 2 and Fig. 4a)32. Hence the CD4−CD8αdim T-cell subset included T-cells that performed functions normally restricted to CD4+ T-cells. While the overall frequencies of memory CD4+ T-cells that could perform each function was significantly higher compared to the CD4−CD8αdim T-cells(Fig. 4a), the total frequency of CD4−CD8αdim T-cells was significantly greater than the overall frequency of CD4+ T-cells. We therefore calculated the relative number of cytokine+, CD40L+, and FoxP3+ T-cells for each T-cell subset (Fig. 4b). For IL-2, IL-17, and FoxP3 the relative number of CD4−CD8αdim T-cells was significantly greater than those of the CD4+ and CD4−CD8αbright T-cell subsets. The relative number of CD40L+ CD4−CD8αdim T-cells was equal to that of CD4+ T-cells. These observations suggest that there were actually greater numbers of T-cells performing CD4+ T-cell functions without actually expressing CD4.
Figure 4. CD4−CD8αdim T-cells can preserve CD4+ T-cell function.
(a) Comparison of the frequency of memory CD4+, CD4−CD8αdim, and CD4−CD8αbright T-cells performing various functions: IL-2 and IL-17 production, and FoxP3 and CD40L expression. (b) Comparison of the relative numbers of memory CD4+, CD4−CD8αdim, and CD4−CD8αbright T-cells from different memory T-cell subsets performing various functions: IL-2 and IL-17 production, and FoxP3 and CD40L expression (c) Responses of CD4−CD8αdim T-cells to CMV whole antigen in the presence and absence blocking antibodies to MHC-II or MHC-I. White circles represent SIV-infected AGM and black represent uninfected AGM. A Wilcoxon matched pairs test was performed for a–b.
CD4+ T-cells are restricted by MHC-II, while CD8+ T-cells are restricted to MHC-I. Therefore, to test further our hypothesis that the CD4−CD8αdim T-cells were acting as CD4+ T-cells, we screened PBMC from adult AGM for T-cell responses to a cytomegalovirus (CMV) whole antigen preparation. Presentation of this antigen requires processing through MHC-II33. We found one AGM that had T-cells responsive to CMV (AG731) and the entire CMV-specific T-cell response was restricted to CD4−CD8αdim T-cells(Fig. 4c and Supplemental Fig. 2b,c). To confirm that these CMV-specific CD4−CD8αdim T-cells were restricted by MHC-II we stimulated the T-cells in the presence of blocking antibodies specific to either MHC-II or MHC-I and measured production of cytokines. Blocking with anti-MHC class-II antibody decreased the frequency of responding CMV-specific CD4−CD8αdim T-cells by more than two-thirds, while blocking MHC-I had virtually no effect on the ability of CMV-specific CD4−CD8αdim T-cells to respond (Fig. 4c). Hence CD4−CD8αdim T-cells can be restricted by MHC-II in AGM.
Vervets are one of four subspecies of AGM along with sabeus, tantalus and grivets. To determine whether the phenomena which we observed were specific to vervet AGM, we examined T-cell populations in 6 adult sabeus AGM. Indeed, we found the presence of CD4−CD8αdim T-cells. Similar to the vervets, the CD4−CD8αdim T-cells from sabeus were able to perform CD4 functions (Supplementary Fig. 3). Confirmation of this phenomenon occurring in a second subspecies of AGM suggest that down-regulation of CD4 is characteristic of all AGM.
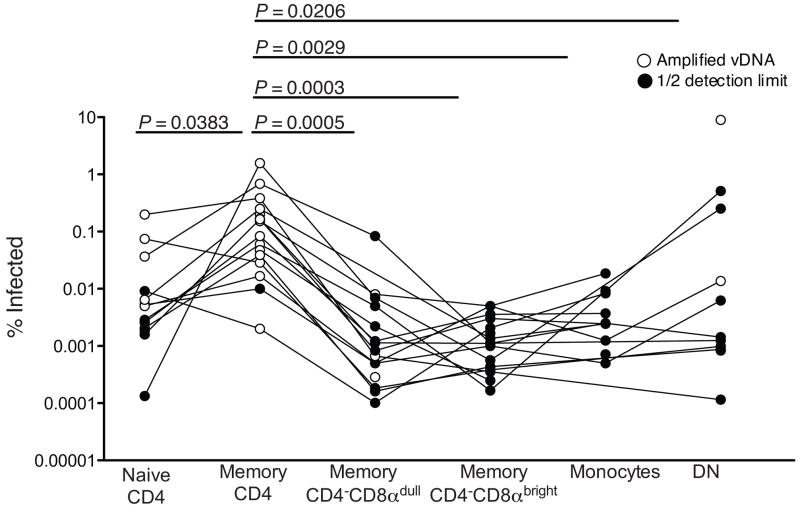
Since the CD4−CD8αdim T-cells have many functional characteristics of CD4+ T-cells, wenext determined which lymphocytes were infected by SIVagm in vivo. We flow cytometrically sorted individual subsets of lymphocytes from eighteen SIVagm-infected AGM and measured the in vivo infection frequency by quantitative real time PCR for SIVagm DNA. We sorted naive and memory CD4+ T-cells, memory CD4−CD8αdim T-cells, memory CD4−CD8αbright T-cells, memory CD4−CD8α− T-cells and monocytes. We found that memory CD4+ T-cells were the primary target for SIVagm in vivo (Fig. 5). Consistent with infection frequency patterns of HIV34, we also found that SIVagm could infect naive CD4+ T-cells, but memory CD4+ T-cells were preferentially infected. Importantly, CD4−CD8αdim T-cells, which we have shown can develop from CD4+ T-cells and maintain functions of CD4+ T-cells, were only very rarely, if ever, infected by SIVagm in vivo (Fig. 5). In 78% of the SIVagm-infected AGM we detected no viral DNA within the CD4−CD8αdim T-cell subset. Cell numbers were often limiting and it was conceivable that, in some cases, we did not detect any viral DNA due to a small number of sorted cells in each PCR. In samples in which no viral DNA was detected, we reported values calculated as 1/2 the lower limit of detection (closed circles), these values are based upon the number of sorted cells. In the few (22%) AGM where we did detect viral DNA within the CD4−CD8αdim T-cell subset the infection frequencies were very low (<0.01%). Taken together, these data suggest that CD4−CD8αdim T-cells preserve CD4+ T-cell function while evading SIV infection in vivo and in turn, these findings suggest a mechanism by which AGM are able remain disease free despite SIV infection.
Figure 5. Infection frequency of lymphocyte subsets.
Infection frequency of sorted lymphocyte subsets from SIVagm-infected adult AGM as determined by PCR. White circles represen T-cells with detectable viral DNA and black circles represent an undetectable infection frequency reported as one half the lower limit of detection based on the number of cells within each PCR reaction. A Wilcoxon matched pairs test was performed for this analysis.
DISCUSSION
We have shown that AGM, regardless of infection with SIVagm, have decreased numbers of total CD4+ T-cells that correlates with an increased population of CD4−CD8αdim T-cells. We demonstrated that some of these CD4−CD8αdim T-cells developed from memory CD4+ T-cells. This conclusion is strengthened by the observation that juvenile AGM, who have very low frequencies of memory T-cells in vivo, have high frequencies of CD4+ T-cells and low frequencies of CD4−CD8αdim T-cells and that in vitro stimulation of naive CD4+ T-cells results in down-regulation of CD4 and up-regulation of CD8α. In addition, we have demonstrated that in adult AGM some of these CD4−CD8αdim T-cells, upon stimulation, exhibit functions generally attributed to CD4+ T-cells. The CD4−CD8αdim T-cells can be restricted by MHC-II. Despite the fact many of the T-cells in this population likely developed from CD4+ T-cells and maintain the ability to perform functions attributed to CD4+ T-cells, they are able to evade infection by SIVagm. These data provide a mechanism by which AGM are able to survive chronic SIVagm infection without progression to simian AIDS.
Our proposed mechanism underlying the nonpathogenic nature of SIVagm infection could contribute to the lack of immune activation seen in the natural hosts of SIV. Indeed, preservation of CD4+ T-cell function may well contribute to the lack of immune activation in AGM and SM28,35,36. Additionally, there is a significantly lower frequency of CD4+CCR5+ T-cells in natural hosts of SIV when compared to non-natural hosts16,37, suggesting that the lack of CCR5 expression results in decreased homing to sites of inflammation, thereby preventing activation and inflammation. This decrease in activated T-cells could reduce the number targets for SIV. Taken together, these data may suggest that in natural hosts, the virus may be preferentially targeting macrophages. However, we have found no SIVagm DNA within highly purified monocytes, consistent with previous reports that SIVagm preferentially replicates in lymphocytes during chronic infection16,28. Also, previous comparative studies between pathogenic HIV infection of humans or SIV infection of Asian macaques and SIV infection of SM or AGM have shown that while both SM and AGM lose CD4+ T-cells from the gastrointestinal tract during the acute phase of infection, these animals do not manifest immune activation11,28,36. Microbial translocation, which causes immune activation in the chronic phase of HIV and SIV infection of Asian macaques38–45, does not occur in the chronic phase of SIV infection of either AGM or sooty mangabeys28,42. The functionality of mucosal CD4−CD8αdim T-cells, which is present at high frequencies in AGM28, should be assessed for a role in preventing microbial translocation and immune activation in SIVagm-infected AGM.
Many other cellular populations, such as NK cells, γδ T-cells and NKT-cells, express the CD8αα homodimer. Indeed, some of the CD4−CD8αdim T-cells from AGM express the γδ TCR (data not shown). Moreover, many of the CD28− memory T-cells within the CD4−CD8αdim T-cell subset express granzyme B (data not shown), not typically expressed by CD4+ T-cells. Therefore, it is clear that not all of the CD4−CD8αdim T-cells were originally CD4+ T-cells. While AGM have significantly higher frequencies of CD4−CD8αdim T-cells compared to humans, Bolassel et al, recently showed that slow progressing HIV-1-infected individuals have a significantly higher frequency of CD4−CD8αdim T-cells compared to chronically HIV-infected individuals46. This finding suggests a similar phenomenon to what we describe here could slow disease progression in HIV-infected humans.
While CD4−CD8αdim T-cells have previously been studied, it has not been demonstrated that memory CD4+ T-cells down-regulate CD4 and up-regulate CD8α. How these T-cells are able to perform functions without the CD4 molecule is unclear. Importantly, Rahemtulla et al, demonstrated normal development and function of lymphoid organs, B cells and CD8+ T-cells in CD4 knockout mice. However, antibody production, IL-2 secretion, and MHC-II restricted responses were significantly abrogated47. The authors subsequently showed that CD4−CD8− T-cells in CD4 knockout mice could partially compensate for the lack of CD4 T-cells48, similar to what we observe in AGM. CD4 has been shown to be important for initiating the downstream kinase signaling that results in T-cell activation49. It is possible that in AGM CD4−CD8αdim T-cells sufficient lck phosphorylation occurs, or alternative signals exist, but the actual mechanism(s) underlying the switch from CD4+ to CD4−CD8αdim remains unclear. Indeed, previous studies have reported genetic differences between certain regulatory elements from AGM compared to other primates50. One possibility is that a change may occur in the methylation states of the enhancer and silencer regions in the genome51. Also, differential expression of the MAZR protein, a protein suppressor of the CD8α enhancer region52, may also exist.
It is not unprecedented that changes in the frequencies of T-cells within individual subsets occur in lentiviral infections. For example, while SM generally maintain healthy CD4+ T-cell counts during chronic SIVsmm infection, infection with a dual tropic (CXCR4/CCR5) strain of SIVsmm resulted in the loss of CD4+ T-cells, but preservation CD4−CD8− T-cells53. Moreover, one report identified several naturally SIV-infected SM with very low frequencies of CD4+ T-cells26. The functions and ontogeny of T-cells in these SIVsmm-infected SM infected are currently under investigation but could represent a similar phenomenon.
Natural hosts have co-evolved with SIV to avoid disease progression, though the mechanisms by which this occurs may diverge since most SM maintain healthy frequencies of CD4+ T-cells. In AGM, we show that this co-evolution with SIVagm has occurred, in part, by the development of CD4−CD8αdim T-cells from memory CD4+ T-cells. Additionally, African lions, who remain disease free after infection with feline immunodeficiency virus, maintain a high frequency of CD8βdull T-cells regardless CD4+ T-cell loss54. Hence, down-regulation of CD4 may be associated with lack of disease progression in multiple immunodeficiency lentiviral infections.
We have identified and characterized a mechanism by which AGM are able to survive chronic SIVagm infection. In non-natural hosts of immunodeficiency lentiviruses depletion of CD4+ T-cells leads to AIDS. However, SIVagm-infected AGM maintain immune responses, remain healthy, and live normal life spans. We provide evidence for the conversion of CD4+ T-cells to CD4−CD8αdim T-cells, which likely plays a key role in the lack of clinical signs of AIDS in AGM. Once the mechanism by which the CD4+ T-cells are able to convert to CD4−CD8αdim T-cells has been understood, interventions aimed at mimicking this phenomenon could be developed for preventative and therapeutic trials.
METHODS
Animals
Eighteen SIVagm-infected vervet AGM (Chlorocebus pygerythrus), 11SIVagm-uninfected vervet AGM, 7SIVagm-uninfected juveline (less than 2 years old) vervet AGM, and 12 SIV-negative rhesus macaques (Macaca mulatta) were housed at Bioqual (Rockville, MD) and 6SIV-infected sabaeus AGM (C. sabeus) were housed at the Tulane National Primate Center (Convington, LA). SIVsmm-infected and uninfected SM were housed at the Yerkes National Primate Center (Atlanta, GA). All animals were housed in accordance with the National Research Council Guide for the Care and use of Laboratory Animals, and all protocols were approved by the relevant institutional animal care and use committees. Ten of the SIV+ AGM vervets, were infected in the wild and eight were experimentally infected with 50 or 1000 TCID50 of SIVagm90 intravenously (Table 1). Virus was isolated as previously described10. AG11 and AG15 are seropositive for SIVagm.
Human Subjects
Five HIV-1 uninfected subjects were recruited at the National Institutes of Health. The subjects all gave informed consent prior to entry into this study and all studies were approved by the institution’s IRB.
Flow cytometry
For intracellular cytokine staining (ICS), PBMC were incubated overnight at 37°C with media alone, 1μg of Staphylococcal enterotoxin B (SEB) (Sigma, St. Louis, MO), or 1μg of cytomegalovirus (CMV) whole antigen (Microbix Biosystems, Toronto, Ontario, Canada) in the presence of 0.5μg each of monoclonal antibodies to CD28 (CD28.2, Beckman Coulter, Fullerton, CA) and CD49d (9F10, BD Bioscience, San Jose, CA) and 1 μg/ml Brefeldin A (Sigma, St. Louis, MO). For some experiments PBMC were pretreated for 1 hour at 37°C with antibodies against MHC-I (G46-2.6, BD Bioscience) or MHC-II (TU39, BD Bioscience). After stimulation, cells were washed twice and incubated with Live/Dead fixable aqua dead cell stain (Invitrogen, Carlsbad, CA). Cells were then stained for the surface markers using monoclonal antibodies to CD3 (SP34-2, BD Bioscience), CD4 (L200, BD Bioscience), CD8 (RPA-T8, BD Bioscience), and CD95 (DX2, BD bioscience). Cells were washed and permeabilized with Cytofix/Cytoperm buffer (BD Bioscience). Cells were then intracellularly stained with fluorescent-conjugated monoclonal antibodies to IFNγ (4S.B3, BD Bioscience), IL-17 (eBio64DEC17, eBioscience, San Diego, CA), IL-2 (MQ1-17H12, BD Bioscience), CD40L (TRAP1, BD Bioscience), or Ki67 (B56, BD Bioscience). Cells were incubated at 4°C for 20 minutes. Cells were washed and then fixed with a 1% paraformaldehyde solution (Electron Microscopy Sciences, Hatfield, PA).
For proliferation, PBMC or flow cytometrically sorted naive CD4 T-cells were stained with 0.125 μM carboxyfluorescein succinimidyl ester (CFSE, Invitrogen) and then stimulated with 1μg/mL concanavalin A (Sigma), 1μg/mL staphylococcal enterotoxin B (Sigma), or anti-CD3/anti-CD28 microbeads at a 1:4 cell to bead ratio (generous gift from Joern E. Schmitz and Roland Zahn) for 5, 7, and 9 days. Cells were then labeled with fluorescent antibodies directed towards CD3, CD4, and CD8 (BD Bioscience).
For analysis of FoxP3 expression fresh PBMCs were surface stained and then permeabilized using FoxP3 permeabilization solution (eBioscience). Cells were intracellularly stained for FoxP3 (PCH101, eBioscience). Cells were washed and then fixed using a 1% PFA solution. All flow cytometry samples were run on a FACSAria (BD Bioscience) using FACSDiva software (BDBioscience) and data were analyzed using FlowJo (Tree Star, Ashland, OR).
Polymerase chain reaction
Quantitative real time PCR
Cell populations were sorted flow cytometrically and were lysed using 25 μL of a 1:100 dilution of proteinase K (Roche, Indianapolis, IN) in 10mM Tris buffer. Quantitative PCR was performed using 5 μL of cell lysates per reaction. Reaction conditions were as follows: 95°C holding stage for 5 minutes, and 40 cycles of 95°C for 15 seconds followed by 60°C for 1 minute. The Taq DNA polymerase kit (Invitrogen) was used. The sequence of the forward primer for SIVagm is GTCCAGTCTCAGCATTTACTTG. The reverse primer sequence is CGGGCATTGAGGTTTTTCAC. The probe sequence is CAGATGTTGAAGCTGACCATTTGGG. For cell number quantitation monkey albumin was measured as previously described55. The PCR machine used was the StepOne Plus (Applied Biosystems, Foster City, CA) and the analysis was performed using StepOne software (Applied Biosystems).
Reverse transcription PCR
Viable cell populations were sorted flow cytometrically and mRNA was isolated using Oligotex Direct mRNA Mini Kit (Qiagen, Valencia, CA) by following the manufacturer’s protocol. Following cDNA synthesis with random hexamers and Superscript II RNase H- reverse transcriptase transcripts of CD4 were amplified by PCR using the forward primer TCGGATTGACTGCCAACTCTG and reverse primer AAGGCGAGCGGGAAGGAGAA. Reaction conditions were as follows: 95°C holding stage for 5 minutes, and 40 cycles of 95°C for 15 seconds followed by 60°C for 1 minute. To control for the amount of mRNA, a second PCR was performed using primers for albumin (as above).
Statistics
All statistical analyses were performed using Prism software (GraphPad, La Jolla, CA). Statistically significance was based upon P values less than 0.05.
Supplementary Material
Acknowledgments
These studies were supported by the intramural NIAID, NIH program and by R01 AI064066 (IP), R01 AI065325 (CA) and RR-00168 (TNPRC). The authors would like to thank the Bad Boys of Cleveland for helpful discussions. We are grateful to Drs. Joern Schmitz and Roland Zahn for the kind donation of anti-CD3/CD28 microbeads for stimulation of T-cells from nonhuman primates. We are also appreciative to Dr. Bernard Lafont and Gary Mettler for their technical advice.
Bibliography
- 1.Hahn BH, Shaw GM, De Cock KM, Sharp PM. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 2.Ling B, Veazey RS, Marx PA. Nonpathogenic CCR2-tropic SIVrcm after serial passage and its effect on SIVmac infection of Indian rhesus macaques. Virology. 2008;379:38–44. doi: 10.1016/j.virol.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y, et al. Use of inhibitors to evaluate coreceptor usage by simian and simian/human immunodeficiency viruses and human immunodeficiency virus type 2 in primary cells. J Virol. 2000;74:6893–6910. doi: 10.1128/jvi.74.15.6893-6910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beer BE, et al. Characterization of novel simian immunodeficiency viruses from red-capped mangabeys from Nigeria (SIVrcmNG409 and -NG411) J Virol. 2001;75:12014–12027. doi: 10.1128/JVI.75.24.12014-12027.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pandrea I, Sodora DL, Silvestri G, Apetrei C. Into the wild: simian immunodeficiency virus (SIV) infection in natural hosts. Trends Immunol. 2008;29:419–428. doi: 10.1016/j.it.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silvestri G, Paiardini M, Pandrea I, Lederman MM, Sodora DL. Understanding the benign nature of SIV infection in natural hosts. J Clin Invest. 2007;117:3148–3154. doi: 10.1172/JCI33034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirsch VM. What can natural infection of African monkeys with simian immunodeficiency virus tell us about the pathogenesis of AIDS? AIDS Rev. 2004;6:40–53. [PubMed] [Google Scholar]
- 8.Goldstein S, et al. Comparison of simian immunodeficiency virus SIVagmVer replication and CD4+ T-cell dynamics in vervet and sabaeus African green monkeys. J Virol. 2006;80:4868–4877. doi: 10.1128/JVI.80.10.4868-4877.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandrea I, et al. Simian immunodeficiency virus SIVagm. sab infection of Caribbean African green monkeys: a new model for the study of SIV pathogenesis in natural hosts. J Virol. 2006;80:4858–4867. doi: 10.1128/JVI.80.10.4858-4867.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstein S, et al. Wide range of viral load in healthy african green monkeys naturally infected with simian immunodeficiency virus. J Virol. 2000;74:11744–11753. doi: 10.1128/jvi.74.24.11744-11753.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silvestri G, et al. Nonpathogenic SIV infection of sooty mangabeys is characterized by limited bystander immunopathology despite chronic high-level viremia. Immunity. 2003;18:441–452. doi: 10.1016/s1074-7613(03)00060-8. [DOI] [PubMed] [Google Scholar]
- 12.Pandrea I, et al. Simian immunodeficiency viruses replication dynamics in African non-human primate hosts: common patterns and species-specific differences. J Med Primatol. 2006;35:194–201. doi: 10.1111/j.1600-0684.2006.00168.x. [DOI] [PubMed] [Google Scholar]
- 13.Barry AP, et al. Depletion of CD8+ cells in sooty mangabey monkeys naturally infected with simian immunodeficiency virus reveals limited role for immune control of virus replication in a natural host species. J Immunol. 2007;178:8002–8012. doi: 10.4049/jimmunol.178.12.8002. [DOI] [PubMed] [Google Scholar]
- 14.Dunham R, et al. The AIDS resistance of naturally SIV-infected sooty mangabeys is independent of cellular immunity to the virus. Blood. 2006;108:209–217. doi: 10.1182/blood-2005-12-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klatt NR, et al. Availability of activated CD4+ T-cells dictates the level of viremia in naturally SIV-infected sooty mangabeys. J Clin Invest. 2008;118:2039–2049. doi: 10.1172/JCI33814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pandrea I, et al. Paucity of CD4+ CCR5+ T-cells may prevent transmission of simian immunodeficiency virus in natural nonhuman primate hosts by breast-feeding. J Virol. 2008;82:5501–5509. doi: 10.1128/JVI.02555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon SN, et al. Short-lived infected cells support virus replication in sooty mangabeys naturally infected with simian immunodeficiency virus: implications for AIDS pathogenesis. J Virol. 2008;82:3725–3735. doi: 10.1128/JVI.02408-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirsch VM, et al. Induction of AIDS by simian immunodeficiency virus from an African green monkey: species-specific variation in pathogenicity correlates with the extent of in vivo replication. J Virol. 1995;69:955–967. doi: 10.1128/jvi.69.2.955-967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstein S, et al. Plateau levels of viremia correlate with the degree of CD4+-T-cell loss in simian immunodeficiency virus SIVagm-infected pigtailed macaques: variable pathogenicity of natural SIVagm isolates. J Virol. 2005;79:5153–5162. doi: 10.1128/JVI.79.8.5153-5162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watson A, et al. Plasma viremia in macaques infected with simian immunodeficiency virus: plasma viral load early in infection predicts survival. J Virol. 1997;71:284–290. doi: 10.1128/jvi.71.1.284-290.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirsch V, et al. A molecularly cloned, pathogenic, neutralization-resistant simian immunodeficiency virus, SIVsmE543-3. J Virol. 1997;71:1608–1620. doi: 10.1128/jvi.71.2.1608-1620.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, et al. Complete nucleotide sequence, genome organization, and biological properties of human immunodeficiency virus type 1 in vivo: evidence for limited defectiveness and complementation. J Virol. 1992;66:6587–6600. doi: 10.1128/jvi.66.11.6587-6600.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fultz PN, McClure HM, Anderson DC, Switzer WM. Identification and biologic characterization of an acutely lethal variant of simian immunodeficiency virus from sooty mangabeys (SIV/SMM) AIDS Res Hum Retroviruses. 1989;5:397–409. doi: 10.1089/aid.1989.5.397. [DOI] [PubMed] [Google Scholar]
- 24.Giorgi JV, et al. Shorter survival in advanced human immunodeficiency virus type 1 infection is more closely associated with T lymphocyte activation than with plasma virus burden or virus chemokine coreceptor usage. J Infect Dis. 1999;179:859–870. doi: 10.1086/314660. [DOI] [PubMed] [Google Scholar]
- 25.Chakrabarti LA, et al. Normal T-cell turnover in sooty mangabeys harboring active simian immunodeficiency virus infection. J Virol. 2000;74:1209–1223. doi: 10.1128/jvi.74.3.1209-1223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sumpter B, et al. Correlates of preserved CD4(+) T-cell homeostasis during natural, nonpathogenic simian immunodeficiency virus infection of sooty mangabeys: implications for AIDS pathogenesis. J Immunol. 2007;178:1680–1691. doi: 10.4049/jimmunol.178.3.1680. [DOI] [PubMed] [Google Scholar]
- 27.Pandrea I, et al. High levels of SIVmnd-1 replication in chronically infected Mandrillus sphinx. Virology. 2003;317:119–127. doi: 10.1016/j.virol.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 28.Pandrea IV, et al. Acute Loss of Intestinal CD4+ T-cells Is Not Predictive of Simian Immunodeficiency Virus Virulence. J Immunol. 2007;179:3035–3046. doi: 10.4049/jimmunol.179.5.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murayama Y, Mukai R, Inoue-Murayama M, Yoshikawa Y. An African green monkey lacking peripheral CD4 lymphocytes that retains helper T-cell activity and coexists with SIVagm. Clin Exp Immunol. 1999;117:504–512. doi: 10.1046/j.1365-2249.1999.00999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murayama Y, et al. CD4 and CD8 expressions in African green monkey helper T lymphocytes: implication for resistance to SIV infection. Int Immunol. 1997;9:843–851. doi: 10.1093/intimm/9.6.843. [DOI] [PubMed] [Google Scholar]
- 31.Banchereau J, et al. The CD40 antigen and its ligand. Annu Rev Immunol. 1994;12:881–922. doi: 10.1146/annurev.iy.12.040194.004313. [DOI] [PubMed] [Google Scholar]
- 32.Karube K, et al. Expression of FoxP3, a key molecule in CD4CD25 regulatory T-cells, in adult T-cell leukaemia/lymphoma cells. Br J Haematol. 2004;126:81–84. doi: 10.1111/j.1365-2141.2004.04999.x. [DOI] [PubMed] [Google Scholar]
- 33.Pitcher CJ, et al. HIV-1-specific CD4+ T-cells are detectable in most individuals with active HIV-1 infection, but decline with prolonged viral suppression. Nat Med. 1999;5:518–525. doi: 10.1038/8400. [DOI] [PubMed] [Google Scholar]
- 34.Brenchley JM, et al. T-Cell Subsets That Harbor Human Immunodeficiency Virus (HIV) In Vivo: Implications for HIV Pathogenesis. J Virol. 2004;78:1160–1168. doi: 10.1128/JVI.78.3.1160-1168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenchley JM, et al. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood. 2008;112:2826–2835. doi: 10.1182/blood-2008-05-159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordon SN, et al. Severe Depletion of Mucosal CD4+ T-cells in AIDS-Free Simian Immunodeficiency Virus-Infected Sooty Mangabeys. J Immunol. 2007;179:3026–3034. doi: 10.4049/jimmunol.179.5.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pandrea I, et al. Paucity of CD4+CCR5+ T-cells is a typical feature of natural SIV hosts. Blood. 2007;109:1069–1076. doi: 10.1182/blood-2006-05-024364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hunt PW, et al. Relationship between T-cell Activation and CD4(+) T-cell Count in HIV-Seropositive Individuals with Undetectable Plasma HIV RNA Levels in the Absence of Therapy. J Infect Dis. 2008;197:126–133. doi: 10.1086/524143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang W, et al. Plasma Levels of Bacterial DNA Correlate with Immune Activation and the Magnitude of Immune Restoration in Persons with Antiretroviral-Treated HIV Infection. J Infect Dis. 2009;199:1177–1185. doi: 10.1086/597476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papasavvas E, et al. Delayed loss of control of plasma lipopolysaccharide levels after therapy interruption in chronically HIV-1-infected patients. AIDS. 2009;23:369–375. doi: 10.1097/QAD.0b013e32831e9c76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gregson JN, et al. Elevated plasma lipopolysaccharide is not sufficient to drive natural killer cell activation in HIV-1-infected individuals. AIDS. 2009;23:29–34. doi: 10.1097/QAD.0b013e3283199780. [DOI] [PubMed] [Google Scholar]
- 42.Brenchley JM, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 43.Ancuta P, et al. Microbial translocation is associated with increased monocyte activation and dementia in AIDS patients. PLoS ONE. 2008;3:e2516. doi: 10.1371/journal.pone.0002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balagopal A, et al. Human Immunodeficiency Virus-Related Microbial Translocation and Progression of Hepatitis C. Gastroenterology. 2008 doi: 10.1053/j.gastro.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marchetti G, et al. Microbial translocation is associated with sustained failure in CD4+ T-cell reconstitution in HIV-infected patients on long-term highly active antiretroviral therapy. AIDS. 2008;22:2035–2038. doi: 10.1097/QAD.0b013e3283112d29. [DOI] [PubMed] [Google Scholar]
- 46.Boulassel MR, Mercier F, Gilmore N, Routy JP. Immunophenotypic patterns of CD8+ T-cell subsets expressing CD8alphaalpha and IL-7Ralpha in viremic, aviremic and slow progressor HIV-1-infected subjects. Clin Immunol. 2007;124:149–157. doi: 10.1016/j.clim.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 47.Rahemtulla A, et al. Normal development and function of CD8+ cells but markedly decreased helper cell activity in mice lacking CD4. Nature. 1991;353:180–184. doi: 10.1038/353180a0. [DOI] [PubMed] [Google Scholar]
- 48.Rahemtulla A, et al. Class II major histocompatibility complex-restricted T-cell function in CD4-deficient mice. Eur J Immunol. 1994;24:2213–2218. doi: 10.1002/eji.1830240942. [DOI] [PubMed] [Google Scholar]
- 49.Barber EK, Dasgupta JD, Schlossman SF, Trevillyan JM, Rudd CE. The CD4 and CD8 antigens are coupled to a protein-tyrosine kinase (p56lck) that phosphorylates the CD3 complex. Proc Natl Acad Sci U S A. 1989;86:3277–3281. doi: 10.1073/pnas.86.9.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsunaga S, Mukai R, Inoue-Murayama M, Yoshikawa Y, Murayama Y. Sequence and functional properties of African green monkey CD4 silencer. Immunol Lett. 2000;75:47–53. doi: 10.1016/s0165-2478(00)00273-x. [DOI] [PubMed] [Google Scholar]
- 51.Kioussis D, Ellmeier W. Chromatin and CD4, CD8A and CD8B gene expression during thymic differentiation. Nat Rev Immunol. 2002;2:909–919. doi: 10.1038/nri952. [DOI] [PubMed] [Google Scholar]
- 52.Bilic I, et al. Negative regulation of CD8 expression via Cd8 enhancer-mediated recruitment of the zinc finger protein MAZR. Nat Immunol. 2006;7:392–400. doi: 10.1038/ni1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Milush JM, et al. Virally induced CD4+ T-cell depletion is not sufficient to induce AIDS in a natural host. J Immunol. 2007;179:3047–3056. doi: 10.4049/jimmunol.179.5.3047. [DOI] [PubMed] [Google Scholar]
- 54.Roelke ME, et al. T-lymphocyte profiles in FIV-infected wild lions and pumas reveal CD4 depletion. J Wildl Dis. 2006;42:234–248. doi: 10.7589/0090-3558-42.2.234. [DOI] [PubMed] [Google Scholar]
- 55.Mattapallil JJ, et al. Massive infection and loss of memory CD4+ T-cells in multiple tissues during acute SIV infection. Nature. 2005;434:1093–1097. doi: 10.1038/nature03501. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.