Abstract
Cell-penetrating peptides (CPPs) are small cationic peptides that cross the cell membrane while carrying macromolecular cargoes. We use solid-state NMR to investigate the structure and lipid interaction of two cationic residues, Arg10 and Lys13, in the CPP penetratin. 13C chemical shifts indicate that Arg10 adopts a rigid β-strand conformation in the liquid-crystalline state of anionic lipid membranes. This behavior contrasts with all other residues observed so far in this peptide, which adopt a dynamic β-turn conformation with coil-like chemical shifts at physiological temperature. Low-temperature 13C-31P distances between the peptide and the lipid phosphates indicate that both the Arg10 guanidinium Cζ and the Lys13 Cε lie in close proximity to the lipid 31P (4.0 - 4.2 Å), proving the existence of charge-charge interaction for both Arg10 and Lys13 in the gel-phase membrane. However, since lysine substitution in CPPs are known to reduce their translocation ability, we propose that low temperature stabilizes both lysine and arginine interactions with the phosphates, whereas at high temperature the lysine-phosphate interaction is much weaker than the arginine-phosphate interaction. This is supported by the unusually high rigidity of the Arg10 sidechain and its β-strand conformation at high temperature. The latter is proposed to be important for ion pair formation by allowing close approach of the lipid headgroups to guanidinium sidechains. 19F and 13C spin diffusion experiments indicate that penetratin is oligomerized into β-sheets in gel-phase membranes. These solid-state NMR data indicate that guanidinium-phosphate interactions exist in penetratin, and guanidinium groups play a stronger structural role than ammonium groups in the lipid-assisted translocation of CPPs across liquid-crystalline cell membranes.
Keywords: cell-penetrating peptide, lipid bilayer, guanidinium-phosphate interaction, 13C-31P REDOR, arginine, lysine
Cell-penetrating peptides (CPP) are arginine- and lysine-rich cationic peptides that can readily enter cells not only by themselves but also carrying other macromolecular cargos (1-3). Thus they are promising drug-delivery molecules. Many studies have established that the intracellular entry of CPPs is related to their strong affinity to lipid bilayers (4). The lipid membrane can be the plasma membrane of the cell or the endosomal membrane from which CPPs must escape after endocytosis (5). The fundamental biophysical question of interest is how these highly cationic peptides cross the hydrophobic part of the lipid bilayer against the free energy barrier, and doing so without causing permanent damage to the membrane, in contrast to another family of cationic membrane peptides, antimicrobial peptides (AMPs).
Several models have been proposed to explain the membrane translocation of CPPs. The inverse micelle model proposes that transient inverse micelles form in the membrane to trap the peptides from the outer leaflet and subsequently release them to the inner leaflet (6, 7). However, this model is inconsistent with the lipid 31P spectra (8), and the large rearrangement of lipids is difficult to achieve energetically. The electroporation model (9) posits that at low concentrations CPPs bind only to the outer leaflet of the bilayer, thus causing a transmembrane electric field. Above a threshold peptide concentration, the membrane is permeabilized in a electroporationlike manner, which creates transient defects that enable the peptides to distribute to both leaflets, thus relieving the membrane curvature stress (9-11). The third model posits that the guanidinium ions in these arginine-rich peptides associate with the lipid phosphate groups to neutralize the arginine residues and thus allow the peptides to cross the membrane without a high energy penalty. This model is supported by phase transfer experiments of oligoarginnines (12) and by molecular dynamics simulation of the HIV-1 Tat peptide, which showed transient association of arginine residues with the phosphate groups on both sides of the bilayer (13).
We recently investigated the depth of insertion and conformation of a CPP, penetratin, using solid-state 13C and 31P NMR. Penetratin is the first discovered CPP and is derived from the third helix (residues 43-58) of the Drosophilia Antennapedia homeodomain (14). Using Mn2+ paramagnetic relaxation enhancement (PRE) experiments, we showed that penetratin is bound to both leaflets of the lipid bilayer at both low and high concentrations (peptide-lipid molar ratios of 1:40 and 1:15) (15, 16). This data indicates that the electroporation model is unlikely for penetratin. No 31P peaks at the isotropic frequency or the hexagonal-phase frequency were observed, thus ruling out the inverse micelle model. In addition, we found that penetratin undergoes an interesting conformational change, as manifested by 13C chemical shifts, from a β-sheet structure in the gel-phase membrane to a coil-like conformation in the liquid-crystalline membrane (17). The coil-like conformation at high temperature has non-negligible residual order parameters of 0.23 - 0.52 (17), indicating that the peptide remains structured. We hypothesized that the high-temperature conformation is a β-turn that undergoes uniaxial rotation around the bilayer normal.
Given the above experimental evidence against the electroporation model and the inverse micelle model, we now test the validity of the guanidinium-phosphate complexation model for the membrane translocation of penetratin. For this purpose we measured 13C-31P distances between several peptide sidechains and lipid 31P atoms. As shown before for an arginine-rich antimicrobial peptide, strong associations with the lipid phosphates manifest as short 13C-31P distances (18, 19). We show here that the cationic Arg10 in penetratin indeed exhibits shorter distances to phosphate groups than hydrophobic residues. However, another cationic residue, Lys13, also exhibits short 13C-31P distances, despite the fact that Lys mutants of CPPs have much weaker translocation activities. We show that the answer to this puzzle lies not in the low-temperature structure and distances of the two residues, but in their high-temperature dynamic structures, which differ significantly. And it is the structure in the liquid-crystalline membrane that accounts for the distinct roles of Arg and Lys in CPP entry into the cell. Finally, we investigate the oligomeric structure of penetratin in gel-phase membranes using 19F and 13C spin diffusion NMR.
Materials and Methods
All lipids were purchased from Avanti Polar Lipids (Alabaster, AL) and used without further purification. Penetratin (RQIKI WFQNR RMKW KK), which contains three arginines and four lysines, was synthesized using standard Fmoc solid-phase peptide synthesis methods (20). Uniformly 13C, 15N-labeled arginine was purchased from Cambridge Isotope Laboratory and incorporated into Arg10 in the peptide. Ile3 and Lys13 were labeled in two other peptide samples as described before (17). 4-19F-Phe7 labeled penetratin was used for 19F experiments to determine the oligomeric structure. All peptide samples were purified by HPLC to > 95% purity.
Hydrated membrane samples were prepared using an aqueous-phase mixing method. Lipids were first codissolved in chloroform at the desired molar ratios and dried under a stream of N2 gas. After lyophilization in cyclohexane overnight, the dry lipid powder was suspended in water and freeze-thawed several times before the peptide solution was added. The solution was incubated overnight to facilitate binding, then centrifuged at 55,000 rpm for 3 hours to obtain a hydrated membrane pellet. For the Arg10 experiments, a hydrated DMPC/DMPG (8:7) membrane with a peptide : lipid molar ratio of 1:15 was used in most 2D correlation and distance measurements. The molar ratio was chosen to balance the positive charges of the peptide (+7) by the negatively charged PG lipids (-1). This DMPC/DMPG sample is supplemented by a hydrated POPC/POPG (8:7) sample for the conformation study, and by a trehalose-protected dry POPE/POPG (8:7) sample for the 13C-31P REDOR experiment. For distance measurements of other residues, several trehalose-protected dry DMPC/DMPG membranes were used to ensure that both the peptide and the lipid headgroups are completely immobilized at low temperature (21). Trehalose is known to protect the lamellar structure of the lipid bilayer in the absence of water (22). Below we refer to the non-trehalose containing membrane samples as hydrated samples to distinguish from the dry trehalose-containing samples.
Solid-state NMR experiments
All experiments were carried out on a Bruker DSX-400 (9.4 Tesla) spectrometer (Karlsruhe, Germany) at a resonance frequency of 100.7 MHz for 13C, 376.8 MHz for 19F and 162.1 MHz for 31P. Magic-angle-spinning (MAS) probes tuned to 1H/13C/31P and 1H/19F/X and equipped with 4 mm spinning modules were used for all experiments. Low temperature was achieved using a Kinetics Thermal System XR air-jet sample cooler (Stone Ridge, NY). The temperature of the sample was read from a thermocouple placed near the rotor and was not further calibrated. Typical 90° pulse lengths are 3.5-5.0 μs. 1H decoupling fields of 50-80 kHz were used. 13C, 31P and 19F chemical shifts were referenced externally to the α-Gly 13C' resonance at 176.49 ppm on the TMS scale, the hydroxyapatite 31P signal at 2.73 ppm and the Teflon 19F signal at -122 ppm, respectively.
13C cross polarization (CP) MAS experiments were conducted with a contact time of 0.5-1.0 ms at a typical Hartman-Hahn field strength of 50 kHz. For variable-temperature experiments, samples were stabilized for at least 20 min at each temperature before data acquisition. 2D 13C-13C dipolar assisted rotational resonance (DARR) experiments (23) were conducted under 5 kHz MAS and with a mixing time of 20 ms and 30 ms. A 1H-driven spin diffusion experiment with a longer mixing time of 50 ms was used to detect inter-residue cross peaks of penetratin in DMPC/DMPG membranes.
13C-1H and 15N-1H dipolar couplings were measured using DIPSHIFT (24, 25) experiments at 303 K under 3.401 and 3.000 kHz MAS, respectively. The MREV-8 sequence was used for 1H homonuclear decoupling (26), with an 105° 1H pulse length of 4.0 μs. In the C-H DIPSHIFT experiment, the 13C-13C dipolar coupling is removed by MAS while the 13C-13C scalar coupling has no effect on the t1-dependent intensity modulation due to the constant time nature of the evolution period. The normalized t1 intensities were fitted using a home-written Fortran program. The best-fit values were divided by the theoretical scaling factor of 0.47 for the MREV-8 sequence. For the doubled N-H DIPSHIFT experiment, the fit value was further divided by 2 to obtain the true couplings. The ratio between the true coupling and the rigid limit value gives the order parameter SXH. The rigid-limit coupling used was 22.7 kHz for C-H and 10.6 kHz for N-H dipolar couplings. Simulations took into account the difference between the XH and XH2 spin systems.
Frequency-selective rotational-echo double-resonance (REDOR) experiments were used to measure distances between peptide 13C and lipid 31P (27, 28). The experiments were conducted at ~230 K under 4 kHz MAS. A rotor-synchronized soft 13C Gaussian 180° pulse of 1000 μs was applied in the middle of the REDOR period to suppress the 13C-13C J-coupling between the on-resonance 13C and its directly bonded 13C spins. 31P 180° pulses of 9 μs were applied every half rotor period. The DMPC/DMPG sample were used to measure the Arg10 sidechain and CO distances to 31P, whereas the POPE/POPG sample was used to measure the Cα -31P distance, as the DMPC Cγ peak (54 ppm) overlaps with the Arg10 Cα signal.
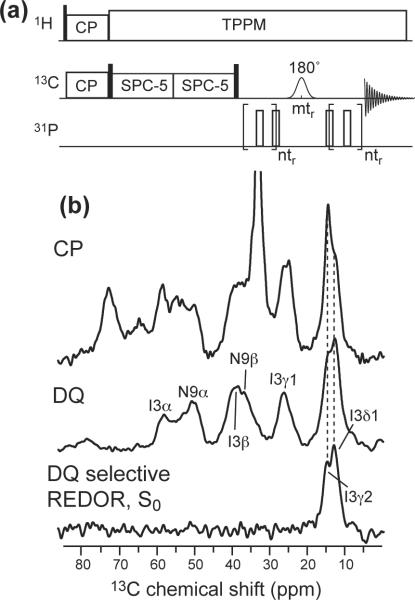
A double-quantum (DQ) selective REDOR experiment (Figure 5a) was designed to measure the 13C-31P distance of Ile3 sidechains, whose signals overlap extensively with the lipid 13C peaks. An SPC-5 pulse train (29) was used to create DQ coherence of the labeled 13C sites and suppress the natural abundance lipid 13C signals. The efficiency of the DQ-REDOR experiment is about 20% of the single-quantum selective REDOR experiment.
Figure 5.
(a) Pulse sequence for the DQ selective REDOR experiment. (b) 13C MAS spectra of Ile3, Asn9-labeled penetratin in trehalose-protected DMPC/DMPG bilayers. Top: CP spectrum, showing both the lipid and peptide signals. Middle: DQ-filtered spectrum, showing only peptide signals. Bottom: DQ selective REDOR S0 spectrum, showing only the Ile3 sidechain Cγ2 and Cδ1 signals.
For the 13CO-31P REDOR experiment, the DQ-REDOR experiment was not used due to the low sensitivity of the CO signal. Thus, the lipid natural-abundance contribution to the 13CO signal was corrected using the equation (S/S0)observed = 0.79(S/S0)peptide + 0.21(S/S0)lipid, where the weight fractions were obtained from the peptide-lipid molar ratio. At the low temperature used for the REDOR experiments, the lipid and peptide CO groups have very similar CP efficiencies, thus the natural abundance correction is relatively accurate.
All 13C-31P REDOR data were fit by two-spin simulations. As we showed before, for distances shorter than 5 Å, two-spin simulations are sufficient. For distances larger than 7 Å, the two-spin simulation only slightly overestimates the distances compared to the vertical distance from 13C to the 31P plane obtained from a multi-spin simulation (18).
The oligomeric structure of penetratin was determined using the 19F CODEX experiment (30, 31). The experiments were conducted at 233 K on a trehalose-protected DMPC/DMPG sample to freeze potential motion of the peptide. Two experiments were conducted for each mixing time: an exchange experiment (S) with the desired mixing time (τm) and a short z-filter (τz), and a reference experiment (S0) with interchanged τm and τz. The normalized intensity, S/S0, was measured as a function of the mixing time until it reached a plateau. The inverse of the equilibrium S/S0 value gives the minimum oligomeric number. Error bars were propagated from the signal-to-noise ratios of the isotropic peak and its sidebands. The τm-dependent CODEX curve was simulated as described before (32) to extract intermolecular distances.
Results
Arg10 conformation and dynamics in penetratin
In the present study, we focus on Arg10, one of the three arginine residues in penetratin, to understand whether cationic residues in general and arginine residues in particular play a special role in the membrane translocation of the peptide. We first investigate the conformation of Arg10. We recently reported the reversible conformational change of many penetratin residues in the lipid bilayer between a β-turn state at high temperature and a β-strand state at low temperature. This conformational change was manifested as chemical shift changes and was observed at Ile3, Ile5, Gln8, Asn9 and Lys13. The chemical shift change is independent of the membrane composition (POPC/POPG and DMPC/DMPG), anionic lipid fraction (PC/PG = 8:7 and 3:1), and peptide concentration (P/L = 1:15 and 1:30) (17).
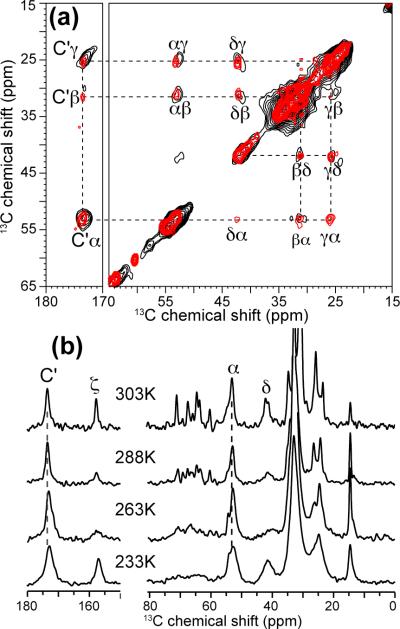
Figure 1a shows the 2D 13C-13C correlation spectra of Arg10-labeled penetratin in DMPC/DMPG bilayers at 303 K and 234 K. Most intra-residue cross peaks are seen, and show no frequency differences between high and low temperatures. Thus, in contrast to all other residues examined, Arg10 does not have temperature-induced conformational change. The difference of the experimental isotropic chemical shifts from the random coil values reflects the secondary structure of the protein (33). Based on the 13CO, 13Cα and 13Cβ 13C isotropic shifts (Supporting Information Table S1), we find Arg10 adopts β-strand conformation at both high and low temperatures.
Figure 1.
(a) 2D 13C-13C correlation spectra of Arg10 labeled penetratin in DMPC/DMPG (8:7) membranes at 303 K (red) and 234 K (black), with DARR mixing times of 30 ms and 20 ms, respectively. (b) 1D 13C CP-MAS spectra of Arg10-penetratin in the DMPC/DMPG membrane as a function of temperature. Lines guide the eye for Arg10 backbone signals and the lack of temperature-induced chemical shift changes.
One-dimensional 13C CP-MAS spectra scanned between 303 K and 233 K (Figure 1b) confirm the lack of chemical shift changes in DMPC/DMPG bilayers. Moreover, the 1D spectra show that the penetratin 13C lines are broader at low temperature. This phenomenon is common to many membrane peptides (34, 35), and can be attributed to conformational distribution of the peptide in the lipid membrane, which is averaged at high temperature but frozen in at low temperature. For the sidechain Cδ signal, the largest line broadening is observed between 288 K and 263 K, below which the lines sharpen again. This is a definitive signature of intermediatetimescale motion, which means that at 303 K the Arg10 sidechain undergoes fast torsional motion. The lack of exchange broadening for the Cα and CO signals indicate that the Arg10 backbone is already in the slow motional limit at 303 K.
We also measured the Arg10 chemical shifts in POPC/POPG (8:7) membranes (Supporting Information Figure S1) and similarly found only β-strand chemical shifts in a wide temperature range. Figure 2 plots the 13C secondary chemical shifts of Arg10 at two temperatures in two different lipid membranes. The temperature-independent β-strand conformation of Arg10 differs from all other residues examined so far in penetratin (17).
Figure 2.
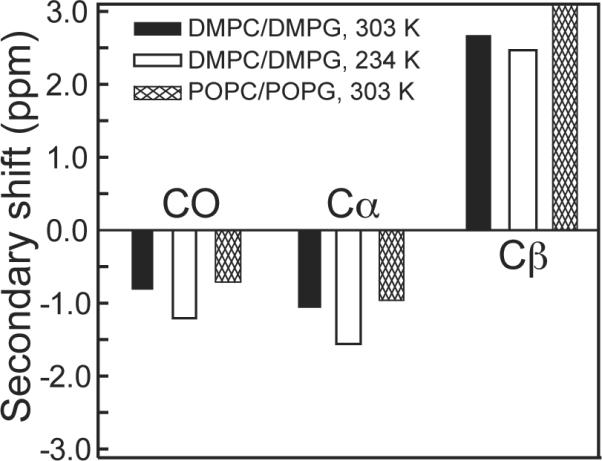
13C secondary chemical shifts of CO, Cα and Cβ of Arg10 of penetratin in two lipid membranes and at two temperatures.
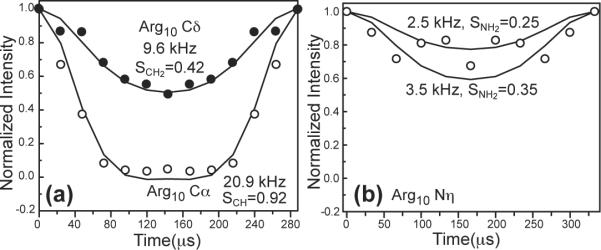
The β-strand conformation is usually more rigid than a coil or turn conformation due to hydrogen bond constraints, and thus should have order parameters close to 1 (17, 36). To verify this, we measured the C-H dipolar couplings of various Arg10 segments in DMPC/DMPG bilayers. Figure 3a shows the 13C-1H DIPSHIFT curves of Cα and Cδ at 303 K. The backbone Cα-Hα dipolar coupling is 20.9 kHz, corresponding to an SCH of 0.92, which translates to a small motional amplitude of 13° (37). This order parameter fits into the SCH range of 0.89-0.94 measured for the other five residues when in the β-strand conformation. In comparison, the sidechain Cδ has a SCH of 0.42. While this value is much lower than the backbone due to the many torsional motions of the sidechain, it is actually larger than all other measured sidechain order parameters, which range from 0.23 to 0.37 in the β-sheet conformation (17). For comparison, the Lys13 sidechain Cε was previously found to have an SCH order parameter of 0.33 (17). We also measured the N-H dipolar coupling of the guanidinium Nη group, and found an N-H dipolar coupling of 3.2 ± 0.5 kHz (Figure 3b). This translates to an SNH of 0.30±0.05, which is significant considering this segment is six bonds away from the backbone Cα.
Figure 3.
X-H DIPSHIFT time evolution of Arg10 sites. (a) C-H dipolar couplings of α (open circles) and Cδ (filled circles) at 303 K, measured under 3.401 kHz MAS. The best-fit true couplings are given along with the corresponding order parameters. (b) N-H dipolar coupling of Nη, measured under 3.000 kHz MAS. The scatter in the data is bracketed by two simulated curves, giving an SNH of 0.30 ± 0.05.
13C-31P distances between penetratin and lipid headgroups
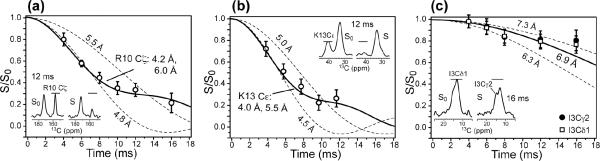
The main goal of the present study is to determine if the cationic residues in penetratin interact strongly with the negatively charged lipid phosphates. 13C-31P distance measurements can provide this information site-specifically. Figure 4 shows the 13C-31P REDOR data of Arg10, Lys13, and Ile3 sidechains in DMPC/DMPG membranes. The experiments were carried out at 233 K where the 31P chemical shift span is 195 - 198 ppm, corresponding to fully immobilized headgroups. The arginine Cζ and lysine Cεgroups directly neighbor the cationic amines and have well resolved chemical shifts of 157.0 ppm and 40.2 ppm, respectively, thus they are ideal reporters of the interaction of these sidechain ends with the lipid headgroups. Arg10 Cζ exhibits significant 13C-31P REDOR dephasing of S/S0 = 0.35 by 10 ms (Figure 4a), indicating relatively short distances to 31P. The time dependence of the REDOR intensities cannot be fit to a single distance due to the presence of a kink around 12 ms. Instead, a combination of a long distance of 6.0 Å and a short distance of 4.2 Å at a 1:1 ratio is found by a least-squares analysis to fit the data best (RMSD = 0.029). The short distance of 4.2 Å can only be satisfied if the guanidinium N-H groups are within hydrogen bonding distance with the O-P groups (18). Similarly, Lys13 Cε exhibits significant dephasing and the REDOR intensities are best fit by two distances of 4.0 Å and 5.5 Å (1:1) (RMSD = 0.036). Again, the short distance supports hydrogen bonding with the lipid phosphate groups. Details of the two-distance best fit for Arg10 and Lys13 are shown in Supporting Information Figure S2. Single-distance fitting of the Arg10 REDOR data indicate that the longer distance must be greater than 5.5 Å while the shorter distance must be smaller than 4.8 Å (Figure 4a). Further, the 13C-31P distances cannot be shorter than 3.6 Å due to steric constraints. Thus, two-distance REDOR curves were calculated using short distances of 3.6 - 4.8 Å with an increment of 0.2 Å and long distances of 5.4 - 7.2 Å with an increment of 0.3 Å, and the two contributions were averaged at a 1:1 ratio. The Lys13 data was analyzed similarly. The simulations indicate that the experimental uncertainties for these 13C-31P REDOR data are about ±0.2 Å for distances shorter than 5.5 Å and ±0.4 Å for distances longer than 5.5 Å.
Figure 4.
13C-31P REDOR data of penetratin sidechains in lipid membranes at 233 K. (a) Arg10 Cξ in hydrated DMPC/DMPG (8:7) bilayers. (b) Lys13 Cε in dry trehalose-protected DMPC/DMPG (8:7) bilayers. (c) Ile3 methyl groups in dry trehalose-protected DMPC/DMPG (8:7) bilayers. Representative REDOR S0 and S spectra are shown in the inset.
Are the short 13C-31P distances of Arg10 and Lys13 specific to the cationic sidechains or are they also true for hydrophobic residues in penetratin? To answer this question, we measured the 13Cγ2-31P and 13Cδ±-31P distances of the neutral hydrophobic residue Ile3. Its Cδ is three bonds away from Cα, which is similarly separated from the backbone as lysine Cε. To remove the lipid natural abundance 13C signals that overlap with the Ile Cγ2 and Cδ1 peaks between 8.9 and 19.0 ppm, we designed a DQ selective REDOR experiment, whose pulse sequence is shown in Figure 5a. The DQ-selected spectra of Ile3 Cγ2 and Cδ1 signals are shown in Figure 5b (middle and bottom spectra). The REDOR dephasing of Ile3 is shown in Figure 4c. Much less REDOR decay is observed, with S/S0 values of ~0.80 at 16 ms. The data is best fit to a distance of 6.9 Å for both Cγ2 and Cδ1, which is 2.7-2.9 Å longer than the Arg10 Cζ and Lys13 Cε. Thus, the short 13C-31P distances are specific to arginine and lysine sidechains instead of being true for all sidechains.
We also measured the 13C-31P distances of Arg10 backbone Cα and CO, which are 6.8 Å and 7.8 Å, respectively (Figure 6). These values fall into the range of 6.9-8.2 Å previously measured for other residues (15). The 13CO data was corrected for the lipid natural abundance signals, whose systematic uncertainty is much smaller than the random noise of the data. All 13C-31P distances are summarized in Table 1.
Figure 6.
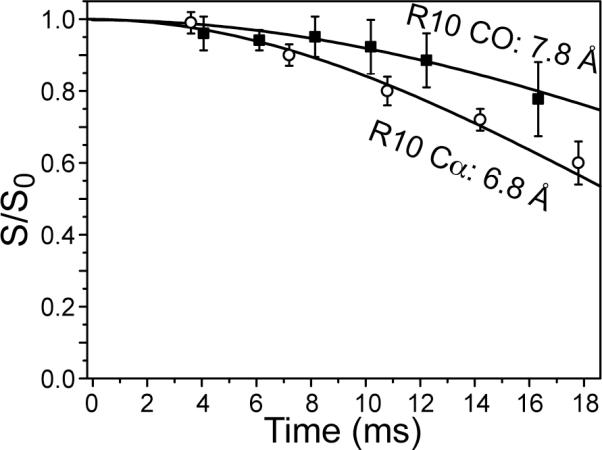
13C-31P REDOR of Arg10 α (open circles) and CO (filled squares) at 233 K. The α data were measured in dry trehalose-protected POPE/POPG (8:7) bilayers. The CO data were measured in frozen hydrated DMPC/DMPG (8:7) membranes.
Table 1.
13C-31 P distances of penetratin residues in lipid membranes at P/L =1:15 and 233 K. Cα and Lys13 Cα distances are obtained from ref (15).
| Residue | 13C-31P distances (Å) | ||
|---|---|---|---|
| Ile3 | Cα: 8.2, | Cγ2: 6.9, | Cδ1: 6.9 |
| Arg10 | CO: 7.8, | Cα: 6.8, | Cζ: 4.2, 6.0 |
| Lys13 | Cα: 6.9, | Cε: 4.0,5.5 | |
Oligomeric structure of penetratin in the lipid membrane
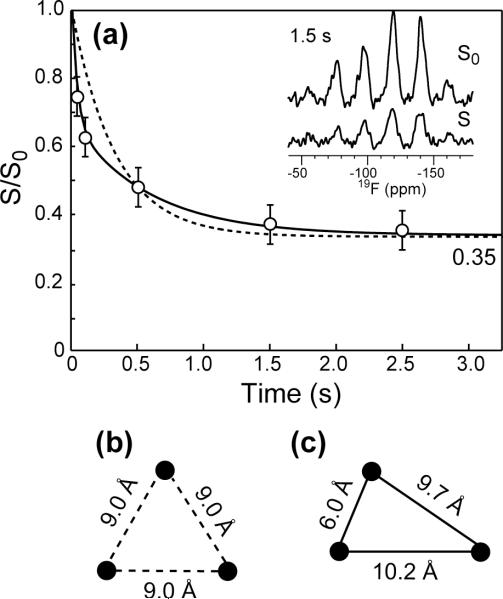
The 19F CODEX experiment was used to determine the oligomeric number and intermolecular distances of penetratin in gel-phase membranes. Figure 7a shows the normalized exchange intensities of 4-19F-Phe7 penetratin in trehalose-protected DMPC/DMPG bilayers. The CODEX intensities decay to an equilibrium value of 0.35 by 2.5 s, indicating three-spin clusters detectable by the 19F distance ruler. To fit the decay trajectory quantitatively, we first assumed an equilateral triangle geometry for the three 19F spins (Figure 7b). The best-fit possible under this assumption gives an internuclear distance of 9.0 Å for each side of the triangle; however, the fit curve (dashed line) does not capture the fast initial decay of the experimental data. To better fit the bi-exponential nature of the data, we then used a triangular geometry with one short distance of much less than 9 Å and two distances longer than or comparable to 9 Å. Modeling of penetratin as a trimer of antiparallel β-strands (see below) yielded one distance of 6.0 Å and two distances of about 10 Å (Figure 7c), which were found to give excellent fit to the experimental data.
Figure 7.
(a) Normalized CODEX intensities as a function of mixing time for 4-19F-Phe7 penetratin in trehalose-protected DMPC/DMPG (8:7) membrane. The data were collected under 8 kHz MAS and 233 K. Representative S0 and S spectra are shown. (b) The equilateral triangle geometry used to generate the dashed-line fit curve in (a). (c) The three-spin geometry with unequal distances used to obtain the solid-line best-fit curve in (a).
To further constrain the intermolecular packing of penetratin in the lipid membrane, we measured a 2D 1H-driven 13C spin diffusion spectrum with a mixing time of 50 ms. Figure 8 shows the 2D spectrum of Ile5, Gln8, Lys13-labeled penetratin in DMPC/DMPG bilayers at 249 K. Two inter-residue cross peaks were observed: I5α-K13α and Q8δ-I5α. Since the peptide adopts a β-strand conformation at this temperature, the intramolecular distances are ~27 Å and 11 Å for I5α-K13α and Q8δ-I5α, respectively, which are too long to be observed by 13C spin diffusion NMR. Thus, these cross peaks must result from intermolecular contacts, which are most likely less than 6 Å for the 50 ms mixing time used.
Figure 8.
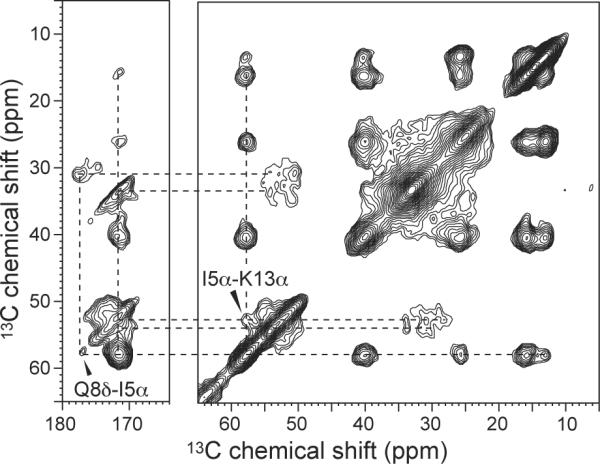
2D 13C-13C correlation spectrum of Ile5, Gln8, and Lys13-labeled penetratin in DMPC/DMPG (8:7) bilayers at 249 K. The spin diffusion mixing time was 50 ms. Two inter-residue cross peaks are detected and assigned.
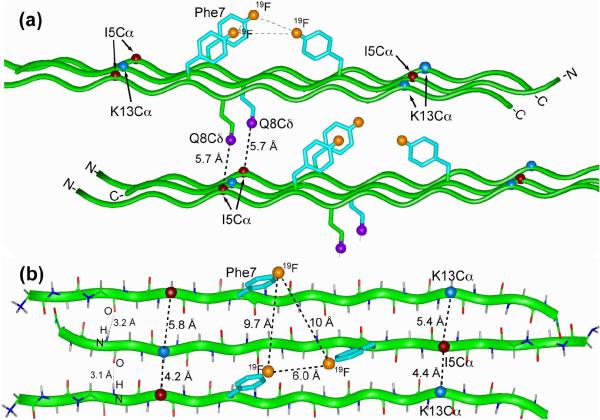
Using standard geometries for β-sheets, where inter-strand hydrogen bonds have RN-O distances of 2.8 - 3.4 Å and backbone torsion angles are ϕ = -139°, ψ = 135°, we built a β-sheet model for penetratin at low temperature that is consistent with the 19F and 13C spin diffusion data (38, 39). Three penetratin β-strands are arranged as a trimer, with the middle strand antiparallel to the two outer strands and shifted by one residue (Figure 9a). This arrangement gives inter-strand I5α-K13α distances of 4.4 - 5.8 Å, consistent with the 2D 13C spectrum. The three Phe7 rings point to the same side of the β-sheet, giving 19F-19F distances of 6.0 Å, 9.7 Å and 10 Å (Figure 9b). Short Q8δ-I5α distances cannot be satisfied within the same β-sheet, but requires two β-sheets stacked in parallel, with an inter-sheet distance of ~10 Å. This gives a Q8δ-I5α distance of 5.7 Å (Figure 9a), where the Q8 χ1 angle is -177°, which is the dominant rotamer of Gln in the β-sheet conformation (40).
Figure 9.
Oligomeric structure of penetratin in the gel-phase membrane. (a) Side view. (b) Top view. Constraints used to build the model include Phe7 19F-19F distances of 6.0, 9.7 and 10 Å, I5α-Q8δ and I5α-K13α distances of less than 6.0 Å, RN-O hydrogen-bond distances of 2.8 - 3.4 Å, and inter-sheet distances of ~10 Å. The β-strand backbone has uniform (ϕ, ψ) angles of (-139°, 135°). The χ1 angle is -177° for both Phe7 and Gln8.
Discussion
Interaction of charged sidechains in penetratin with lipid phosphates
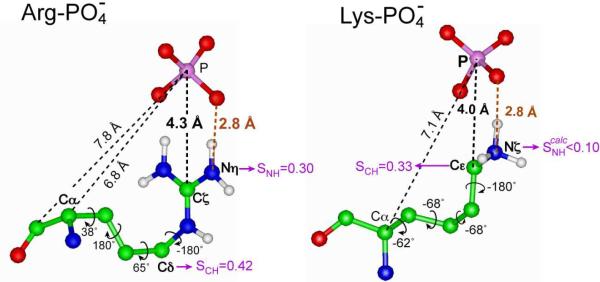
The main finding of the current study is that an arginine and a lysine sidechain in penetratin both form close contacts with the lipid phosphates at low temperature. The Arg10 Cζ-P distance of 4.2 Å and Lys13 Cε-P distance of 4.0 Å both indicate the formation of N-H…O-P hydrogen bonds. Figure 10 shows the sidechain conformations of Arg10 and Lys13 and the spatial arrangements with a phosphate group that satisfy the distances measured here. The N-O distances in both cases must be less than 3.0 Å to satisfy the experimental Cζ and Cε distances to 31P.
Figure 10.
Low-temperature sidechain conformation and phosphate-interaction of Arg10 and Lys in penetratin. The measured 13C-31P distances are indicated in black; the implied N-H…O hydrogen bond distances are indicated in brown. Sidechain torsion angles that satisfy both backbone and sidechain 13C-31P distances are indicated. At physiological temperature, sidechain order parameters of various segments indicate that the lysine-phosphate complex is significantly weakened whereas the arginine-phosphate complex remains.
The short distances of the Arg10 sidechain to the lipid 31P indicates that guanidinium-phosphate complexation occurs not only in antimicrobial peptides but also in cell-penetrating peptides, even though they differ in whether they cause permanent membrane damage. The similarity of lipid-peptide charge-charge attraction and hydrogen-bond formation suggests that penetratin, like some AMPs, also uses this interaction as the main mechanism for its function, which is crossing the lipid membrane. The fundamental driving force for the complex formation is the reduction in the free energy when a neutral species crosses the bilayer. The complexation entails that the peptide drags some lipid headgroups into the hydrophobic region of the membrane, thus causing membrane disorder. However, since no isotropic signal was observed in the 31P spectra of penetratin-containing POPC/POPG (8:7) membranes (15), the disorder is probably transient and not observable on the NMR timescale or under NMR experimental conditions. The 13C-31P distances must be measured at low temperature in the gel-phase membrane in order to freeze molecular motions that would average the dipolar couplings. At physiological temperature where motion is abundant and the penetratin structure is neither a canonical α-helix nor a β-strand (17), whether the 13C-31P distances remain short is not possible to determine, but can be inferred from the sidechain dynamics of the residues (see below).
More interestingly, we found that Arg10 and Lys13 sidechains both establish short distances to 31P at low temperature. This is at first puzzling, since it is well documented that CPP analogs where arginine residues were replaced by lysine have much weaker translocation abilities (41, 42). For penetratin, which contains three arginines and four lysines, cellular uptake efficiencies have been compared among the wild-type peptide, the all-arginine analog, and the all-lysine analog. The efficiency was found to be the highest for the all-arginine analog and the lowest for the all-lysine analog (43). Since both arginine and lysine bear a positive charge at neutral pH, the higher activity of arginine-rich peptides has been suggested to be due to more diffuse charge distribution of the guanidinium group, or the ability of guanidinium ions to form multiple hydrogen bonds with oxyanions in a spatially directed manner (44).
We propose that the similar 13C-31P distances of Arg10 and Lys13 is only true at low temperature at which the REDOR experiments are carried out. Low temperature stabilizes charge-charge interactions, and masks different lipid interactions between Arg and Lys at physiological temperature. Indeed, there is good evidence for a much weaker interaction of the lysine sidechain with phosphates at ambient temperature. First, whereas the Arg10 Nη-Hη order parameters could be measured, attempts to determine the Lys13 Nζ order parameter failed due to unstable 1H-15N cross polarization, which in itself indicates extensive dynamics of the amino group. Second, Arg10 Cδ and Nη have C-H and N-H order parameters of 0.42 and 0.30±0.05 at 303 K, which are relatively large considering that they are three and six bonds away from the backbone Cα. Further, the similar order parameters indicate that the guanidinium moiety as a whole is relatively rigid at physiological temperature, which should facilitate its complexation with the phosphate groups. In comparison, the Lys13 Nζ- Hζ order parameter, although not directly measurable, can be estimated as the product of the Cε SCH of 0.33 with an additional scaling factor of 0.33 due to the three-site jumps of the amino group. Thus, the maximum Lys13 Nζ-Hζ order parameter should be only 0.10, which is much smaller than the Arg10 Nη order parameter of 0.30. With this small order parameter, the Lys13 amino group is unlikely to form any long-lasting hydrogen bonds with lipid phosphates.
The fact that Arg10 adopts a β-sheet backbone conformation that is independent of the temperature or membrane composition, in contrast to all other residues seen so far in penetratin, further supports the unique interaction of arginine with lipid phosphates. Residues Ile3, Ile5, Gln8, Asn9, and Lys13 all exhibit coil-like chemical shifts at high temperature, which were assigned to a β-turn conformation (17). These data, together with the Arg10 chemical shifts measured here, suggest that the β-turn connect a short stretch of β-strand encompassing Arg10. β-turn residues separated by short β-strands are present in various naturally occurring proteins. For example, elastins have recurring (VPGVV)n sequences where the central PG residues adopt a β-turn conformation whereas the flanking Val residues have the β-strand conformation (45).
For penetratin, between the β-turn Asn9 and Lys13, there are two arginine residues and one Met (RRM). It is very likely that the conformational propensity of Arg10 is not unique to this residue but is also true for Arg11, because the peptide backbone may be forced into an extended structure in order to allow the lipid headgroups to approach the charged guanidinium moieties to form the guanidinium-phosphate complex. In other words, guanidinium-phosphate interactions may be the cause of the persistent β-sheet conformation of Arg10 at high and low temperatures. As a corollary, the fact that Lys13 adopts the β-turn instead of β-strand conformation at high temperature is yet another piece of evidence that the ammonium group has much weaker interactions with the phosphates in the liquid-crystalline membrane.
The β-sheet oligomeric structure of penetratin in the gel-phase membrane is energetically favorable. The establishment of intermolecular C=O … H-N hydrogen bonds reduces the free-energy cost of inserting the peptide into the membrane (46). As discussed above, the extended conformation (Figure 9) may facilitate the close approach of lipid phosphate groups with the cationic sidechains, thus allowing both arginine and lysine to interact with the phosphates and establish short 13C-31P distances (Figure 10).
In conclusion, we have shown that strong guanidinium-phosphate interactions exist in the cell-penetrating peptide penetratin, similar to antimicrobial peptides. Moreover, by considering not only low-temperature 13C-31P distances of Arg10 and Lys13, but also high-temperature order parameters of the two sidechains and the unique high-temperature β-strand conformation of Arg10, we deduce that the arginine sidechain interacts more strongly with lipid phosphates than the lysine sidechain at physiological temperature. Therefore, charge- and hydrogen-bond-stabilized guanidinium-phosphate interaction is not only responsible for membrane translocation of this cationic peptide, but also influences the conformation of the peptide.
Supplementary Material
Acknowledgment
We thank Professor Klaus Schmidt-Rohr for discussions of the 19F CODEX results.
Funding Information: This work is supported by the National Institutes of Health grants GM-066976 to M. H.
Abbreviations
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphatidylcholine
- DMPG
1,2-dimyristoyl-sn-glycero-3-phosphatidylglycerol
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine
- POPE
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- POPG
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylglycerol
Footnotes
Supporting information available 13C chemical shift assignments of Arg10-labeled penetratin in POPC/POPG membranes and RMSD analyses of REDOR data are provided. This supplemental material is free of charge online at http://pubs.acs.org.
References
- (1).Eguchi A, Akuta T, Okuyama H, Senda T, Yokoi H, Inokuchi H, Fujita S, et al. Protein transduction domain of HIV-1 Tat protein promotes efficient delivery of DNA into mammalian cells. J. Biol. Chem. 2001;276:26204–26210. doi: 10.1074/jbc.M010625200. [DOI] [PubMed] [Google Scholar]
- (2).Gratton JP, Yu J, Griffith JW, Babbitt RW, Scotland RS, Hickey R, Giordano FJ, et al. Cell-permeable peptides improve cellular uptake and therapeutic gene delivery of replication-deficient viruses in cells and in vivo. Nat. Med. 2003;9:357–362. doi: 10.1038/nm835. [DOI] [PubMed] [Google Scholar]
- (3).Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- (4).Fischer R, Fotin-Mleczek M, Hufnagel H, Brock R. Break on through to the other side-biophysics and cell biology shed light on cell-penetrating peptides. ChemBioChem. 2005;6:2126–2142. doi: 10.1002/cbic.200500044. [DOI] [PubMed] [Google Scholar]
- (5).Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ, Chernomordik LV, et al. Cell-penetrating peptides. A reevaluation of the mechanism of cellular uptake. J. Biol. Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- (6).Derossi D, Calvet S, Trembleau A, Brunissen A, Chassaing G, Prochiantz A. Cell internalization of the third helix of the Antennapedia homeodomain is receptor-independent. J. Biol. Chem. 1996;271:18188–18193. doi: 10.1074/jbc.271.30.18188. [DOI] [PubMed] [Google Scholar]
- (7).Berlose JP, Convert O, Derossi D, Brunissen A, Chassaing G. Conformational and associative behaviours of the third helix of antennapedia homeodomain in membrane-mimetic environments. Eur. J. Biochem. 1996;242:372–386. doi: 10.1111/j.1432-1033.1996.0372r.x. [DOI] [PubMed] [Google Scholar]
- (8).Prochiantz A. Getting hydrophilic compounds into cells: lessons from homeopeptides. Curr. Opin. Neurobiol. 1996;6:629–634. doi: 10.1016/s0959-4388(96)80095-x. [DOI] [PubMed] [Google Scholar]
- (9).Binder H, Lindblom G. Charge-dependent translocation of the Trojan peptide penetratin across lipid membranes. Biophys. J. 2003;85:982–995. doi: 10.1016/S0006-3495(03)74537-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Drin G, Cottin S, Blanc E, Rees AR, Temsamani J. Studies on the internalization mechanism of cationic cell-penetrating peptides. J. Biol. Chem. 2003;278:31192–31201. doi: 10.1074/jbc.M303938200. [DOI] [PubMed] [Google Scholar]
- (11).Zhang W, Smith SO. Mechanism of penetration of Antp(43-58) into membrane bilayers. Biochemistry. 2005;44:10110–10118. doi: 10.1021/bi050341v. [DOI] [PubMed] [Google Scholar]
- (12).Rothbard JB, Jessop TC, Lewis RS, Murray BA, Wender PA. Role of membrane potential and hydrogen bonding in the mechanism of translocation of guanidinium-rich peptides into cells. J. Am. Chem. Soc. 2004;126:9506–9507. doi: 10.1021/ja0482536. [DOI] [PubMed] [Google Scholar]
- (13).Herce HD, Garcia AE. Molecular dynamics simulations suggest a mechanism for translocation of the HIV-1 TAT peptide across lipid membranes. Proc. Natl. Acad. Sci. U. S. A. 2007;104:20805–20810. doi: 10.1073/pnas.0706574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Derossi D, Joliot AH, Chassaing G, Prochiantz A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J. Biol. Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- (15).Su Y, Mani R, Hong M. Asymmetric insertion of membrane proteins in lipid bilayers by solid-state NMR paramagnetic relaxation enhancement: a cell-penetrating Peptide example. J. Am. Chem. Soc. 2008;130:8856–8864. doi: 10.1021/ja802383t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Buffy JJ, Hong T, Yamaguchi S, Waring A, Lehrer RI, Hong M. Solid-State NMR Investigation of the Depth of Insertion of Protegin-1 in Lipid Bilayers Using Paramagnetic Mn2+ Biophys. J. 2003;85:2363–2373. doi: 10.1016/s0006-3495(03)74660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Su Y, Mani R, Doherty T, Waring AJ, Hong M. Reversible sheet-turn conformational change of a cell-penetrating peptide in lipid bilayers studied by solid-state NMR. J. Mol. Biol. 2008;381:1133–1144. doi: 10.1016/j.jmb.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Tang M, Waring AJ, Hong M. Phosphate-mediated arginine insertion into lipid membranes and pore formation by a cationic membrane peptide from solid-state NMR. J. Am. Chem. Soc. 2007;129:11438–11446. doi: 10.1021/ja072511s. [DOI] [PubMed] [Google Scholar]
- (19).Tang M, Waring AJ, Lehrer RI, Hong M. Effects of guanidinium-phosphate hydrogen bonding on the membrane-bound structure and activity of an arginine-rich membrane peptide from solid-state NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2008;47:3202–3205. doi: 10.1002/anie.200705993. [DOI] [PubMed] [Google Scholar]
- (20).Yamaguchi S, Hong T, Waring A, Lehrer RI, Hong M. Solid-state NMR investigations of peptide-lipid interaction and orientation of a beta-sheet antimicrobial peptide, protegrin. Biochemistry. 2002;41:9852–9862. doi: 10.1021/bi0257991. [DOI] [PubMed] [Google Scholar]
- (21).Tang M, Waring AJ, Hong M. Trehalose-protected lipid membranes for determining membrane protein structure and insertion. J. Magn. Reson. 2007;184:222–227. doi: 10.1016/j.jmr.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Crowe JH, Carpenter JF, Crowe LM. The role of vitrification in anhydrobiosis. Annu. Rev. Physiol. 1998;60:73–103. doi: 10.1146/annurev.physiol.60.1.73. [DOI] [PubMed] [Google Scholar]
- (23).Takegoshi K, Nakamura S, Terao T. C-13-H-1 dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 2001;344:631–637. [Google Scholar]
- (24).Munowitz MG, Griffin RG, Bodenhausen G, Huang TH. Two-dimensional rotational spin-echo nuclear magnetic resonance in solids: correlation of chemical shift and dipolar interactions. J. Am. Chem. Soc. 1981;103:2529–2533. [Google Scholar]
- (25).Hong M, Gross JD, Rienstra CM, Griffin RG, Kumashiro KK, Schmidt-Rohr K. Coupling amplification in 2D MAS NMR and its application to torsion angle determination in peptides. J. Magn. Reson. 1997;129:85–92. doi: 10.1006/jmre.1997.1242. [DOI] [PubMed] [Google Scholar]
- (26).Rhim WK, Elleman DD, Vaughan RW. Analysis of multiple-pulse NMR in solids. J. Chem. Phys. 1973;59:3740–3749. [Google Scholar]
- (27).Jaroniec CP, Tounge BA, Rienstra CM, Herzfeld J, Griffin RG. Measurement of 13C-15N distances in uniformly 13C labeled biomolecules: J-decoupled REDOR. J. Am. Chem. Soc. 1999;121:10237–10238. [Google Scholar]
- (28).Jaroniec CP, Tounge BA, Herzfeld J, Griffin RG. Frequency selective heteronuclear dipolar recoupling in rotating solids: accurate (13)C-(15)N distance measurements in uniformly (13)C,(15)N-labeled peptides. J. Am. Chem. Soc. 2001;123:3507–3519. doi: 10.1021/ja003266e. [DOI] [PubMed] [Google Scholar]
- (29).Hohwy M, Rienstra CM, Jaroniec CP, Griffin RG. Fivefold symmetric homonuclear dipolar recoupling in rotating solids: application to double-quantum spectroscopy. J. Chem. Phys. 1999;110:7983–7992. [Google Scholar]
- (30).deAzevedo ER, Bonagamba TJ, Hu W, Schmidt-Rohr K. Centerband-only detection of exchange: efficient analysis of dynamics in solids by NMR. J. Am. Chem. Soc. 1999;121:8411–8412. [Google Scholar]
- (31).Buffy JJ, Waring AJ, Hong M. Determination of Peptide Oligomerization in Lipid Membranes with Magic-Angle Spinning Spin Diffusion NMR. J. Am. Chem. Soc. 2005;127:4477–4483. doi: 10.1021/ja043621r. [DOI] [PubMed] [Google Scholar]
- (32).Luo W, Hong M. Determination of the oligomeric number and intermolecular distances of membrane protein assemblies by anisotropic 1H-driven spin diffusion NMR spectroscopy. J. Am. Chem. Soc. 2006;128:7242–7251. doi: 10.1021/ja0603406. [DOI] [PubMed] [Google Scholar]
- (33).Zhang H, Neal S, Wishart DS. RefDB: A database of uniformly referenced protein chemical shifts. J. Biomol. NMR. 2003;25:173–195. doi: 10.1023/a:1022836027055. [DOI] [PubMed] [Google Scholar]
- (34).Cady SD, Hong M. Amantadine-Induced Conformational and Dynamical Changes of the Influenza M2 Transmembrane Proton Channel. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1483–1488. doi: 10.1073/pnas.0711500105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Tang M, Waring AJ, Hong M. Arginine Dynamics in a Membrane-Bound Cationic Beta-Hairpin Peptide from Solid-State NMR. ChemBioChem. 2008;9:1487–1492. doi: 10.1002/cbic.200800005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Buffy JJ, Waring AJ, Lehrer RI, Hong M. Immobilization and aggregation of the antimicrobial peptide protegrin-1 in lipid bilayers investigated by solid-state NMR. Biochemistry. 2003;42:13725–13734. doi: 10.1021/bi035187w. [DOI] [PubMed] [Google Scholar]
- (37).Huster D, Xiao LS, Hong M. Solid-State NMR Investigation of the dynamics of colicin Ia channel-forming domain. Biochemistry. 2001;40:7662–7674. doi: 10.1021/bi0027231. [DOI] [PubMed] [Google Scholar]
- (38).Mani R, Cady SD, Tang M, Waring AJ, Lehrer RI, Hong M. Membrane-dependent oligomeric structure and pore formation of a b-hairpin antimicrobial peptide in lipid bilayers from solid-state NMR. Proc. Natl. Acad. Sci. USA. 2006;103:16242–16247. doi: 10.1073/pnas.0605079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Mani R, Tang M, Wu X, Buffy JJ, Waring AJ, Sherman MA, Hong M. Membrane-bound dimer structure of a b-hairpin antimicrobial peptide from rotationalecho double-resonance solid-state NMR. Biochemistry. 2006;45:8341–8349. doi: 10.1021/bi060305b. [DOI] [PubMed] [Google Scholar]
- (40).Lovell SC, Word JM, Richardson JS, Richardson DC. The penultimate rotamer library. Proteins: Struct., Funct., Genet. 2000;40:389–408. [PubMed] [Google Scholar]
- (41).Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L, Rothbard JB. The design, synthesis, and evaluation of molecules that enable or enhance cellular uptake: peptoid molecular transporters. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13003–13008. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Mitchell DJ, Kim DT, Steinman L, Fathman CG, Rothbard JB. Polyarginine enters cells more efficiently than other polycationic homopolymers. J. Pept. Res. 2000;56:318–325. doi: 10.1034/j.1399-3011.2000.00723.x. [DOI] [PubMed] [Google Scholar]
- (43).Amand HL, Fant K, Nordén B, Esbjörner EK. Stimulated endocytosis in penetratin uptake: effect of arginine and lysine. Biochem. Biophys. Res. Commun. 2008;371:621–625. doi: 10.1016/j.bbrc.2008.04.039. [DOI] [PubMed] [Google Scholar]
- (44).Jiang Y, Ruta V, Chen J, Lee A, MacKinnon R. The principle of gating charge movement in a voltage-dependent K+ channel. Nature. 2003;423:42–48. doi: 10.1038/nature01581. [DOI] [PubMed] [Google Scholar]
- (45).Yao XL, Hong M. Structural Distribution in an Elastin-Mimetic Peptide (VPGVG)3 Investigated by Solid-State NMR. J. Am. Chem. Soc. 2004;126:4199–4210. doi: 10.1021/ja036686n. [DOI] [PubMed] [Google Scholar]
- (46).White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.