SUMMARY
Deregulated Myc triggers a variety of intrinsic tumor suppressor programs that serve to restrain Myc’s oncogenic potential. Since Myc activity is also required for normal cell proliferation, activation of intrinsic tumor suppression must be triggered only when Myc signaling is oncogenic. However, how cells discriminate between normal and oncogenic Myc is unknown. Here we show that distinct threshold levels of Myc govern its output in vivo: low levels of deregulated Myc are competent to drive ectopic proliferation of somatic cells and oncogenesis but activation of the apoptotic and ARF/p53 intrinsic tumor surveillance pathways requires Myc over-expression. The requirement to keep activated oncogenes at low level to avoid engaging tumor suppression is likely an important selective pressure governing the early stages of tumor microevolution.
SIGNIFICANCE
Cancers are prevented by the activation of intrinsic tumor suppression programs that either fix the damage in cells or ensure that the damaged cells cannot propagate. Cancers can only arise once these tumor suppressor pathways are abrogated. Importantly, activation of such tumor suppressor pathways must be restricted only by oncogenic, not normal, growth signals. Using a novel in vivo model of Myc-induced tumorigenesis in which Myc function is deregulated without concomitant over-expression, we show that tumor surveillance programs are triggered specifically by Myc over-expression, not deregulation. Nonetheless, low-level deregulated Myc remains potently oncogenic. These observations identify a novel mechanism by which the tumor suppressor defense mechanisms can be circumvented, with implications for our understanding of early stage neoplasia.
INTRODUCTION
The Myc oncoprotein is a pleiotropic transcription factor of the bHLH-LZip family with the essential role of engaging and coordinating expression of the diverse genes necessary for efficient and orderly proliferation of somatic cells. In normal cells, both c-Myc mRNA and protein expression are low and continuously dependent on mitogen signaling (Liu and Levens, 2006; Rabbitts et al., 1985; Ramsay et al., 1984). In most human cancers, by contrast, Myc expression is deregulated and/or elevated. Sometimes this is due to alterations in the myc gene itself that either disrupt its normal regulation (e.g. chromosomal translocation, retrovirus integration, gene amplification), increase Myc mRNA or protein stability, or abrogate Myc auto-repression (reviewed by (Nesbit et al., 1999; Popescu and Zimonjic, 2002; Spencer and Groudine, 1991). In most tumors, however, the c-myc gene appears normal and its elevated and persistent activity appears to be due to its relentless induction by upstream oncoproteins, such as oncogenic kinases, Ras or the Wnt/β-Catenin pathway.
The extent to which Myc deregulation versus Myc over-expression contributes to Myc oncogenic activity is unclear. Increasing Myc levels often correlate with the more advanced and aggressive variants of tumors, suggesting that over-expression plays some part in Myc-driven oncogenesis. Moreover, since over-expression drives novel interactions between Myc and low affinity promoter elements, some have suggested that Myc’s oncogenic actions arise from precocious recruitment of novel genes (Fernandez et al., 2003). On the other hand, deregulation of Myc without over-expression is sufficient to obviate the dependency of normal cell proliferation on mitogens and to block the response of cycling cells to anti-proliferative cues and is most likely a consequence of aberrantly sustained modulation of normal Myc target genes.
It is now generally accepted that spontaneous tumorigenesis is largely suppressed by the obligate coupling of proliferation to anti-oncogenic programs such as senescence and apoptosis (Evan and Littlewood, 1998; Lowe et al., 2004). Myc is a prototypical example of this phenomenon. Oncogenically activated Myc is a potent inducer of cell proliferation but also engages the ARF/p53 tumor suppressor pathway (Eischen et al., 1999; Kamijo et al., 1998; Schmitt et al., 1999; Zindy et al., 1998) and apoptosis (Askew et al., 1991; Evan et al., 1992), both tumor suppressor programs that antagonize cell expansion and restrict Myc’s oncogenic potential. However, since Myc mediates the proliferation of normal cells, it is clear that activation of ARF/p53 and apoptosis must be restricted to situations where Myc is oncogenic. How cells distinguish between Myc that is activated by mitogenic signals and Myc that is oncogenically activated is unknown, although candidates include the abnormal persistence of oncogenic Myc, its activation outside the normal context of other mitogenic signals, and its over-expression.
To address these issues, we have developed a novel Myc transgenic mouse in which latent expression of the reversibly switchable variant of Myc, MycERT2 is driven by the constitutive and ubiquitously active Rosa26 promoter. Overt MycERT2 expression is then triggered in any target tissue by the hit-and-run action of Cre recombinase. Due to the relative weakness of the Rosa26 promoter, the level of MycER expressed in tissues of such animals is very low and close to the physiological level of Myc following normal mitogen stimulation. We have used this model to define the oncogenic properties of Myc in vivo when deregulated but not significantly over-expressed. Our studies indicate that the level at which Myc is expressed plays an unforeseen and critical role in determining its oncogenic potential.
RESULTS
Generation of R26-lsl-MERT2 and R26-MERT2 mice
The switchable, 4-hydroxytamoxifen (4-OHT)-dependent variant of Myc was generated by fusing Myc to the modified hormone-binding domain of the modified estrogen receptor ERT2 (Indra et al., 1999). To direct expression conditionally of MycERT2 to target tissues, the cDNA was inserted downstream of the ubiquitously active Rosa26 locus, preceded by a strong translational termination sequence flanked by loxP recombination sites. Murine embryonic stem (ES) cell clones transfected with the resulting targeting vector were screened by Southern blotting to identify single insertions into the Rosa26 locus (Supplemental Fig. 1). Germ line transmission of the R26-lsl-MERT2 allele by chimeric mice was verified by PCR amplification across the Myc-ERT2 sequence junction. We refer to the resulting line of mice as R26-lsl-MERT2.
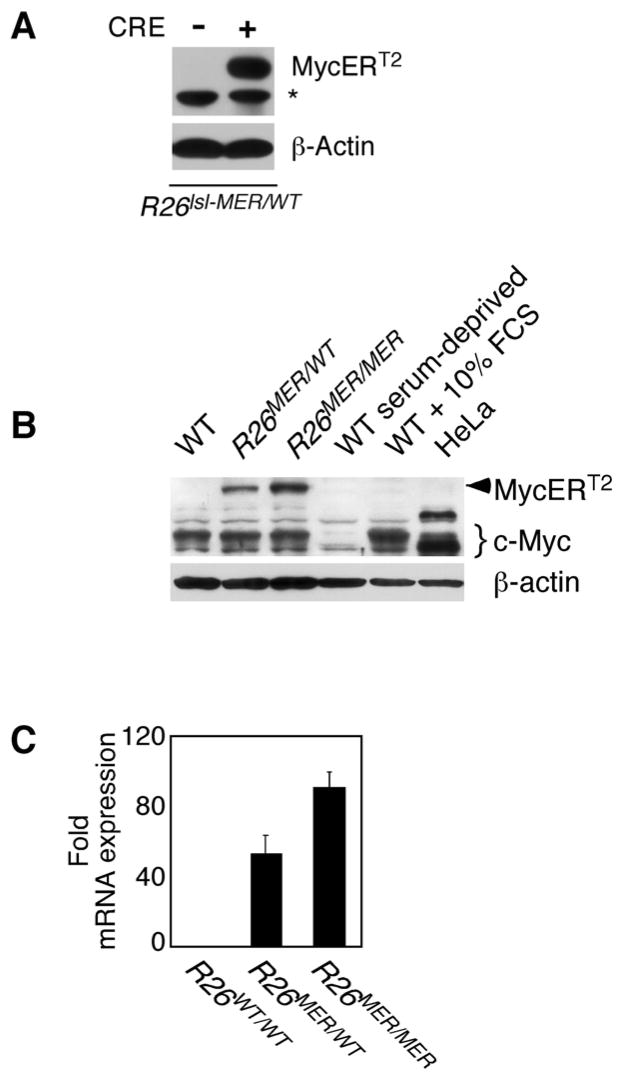
To verify that MycERT2 expression is dependent upon excision of the translational termination element, we expressed Cre recombinase in R26-lsl-MycERT2-derived embryonic fibroblasts (MEFs). Immunoblotting with anti-human ER antibody demonstrated expression of the predicted 92 kDa MycERT2 fusion protein only in MEFs expressing Cre (Fig. 1A). To compare directly in somatic cells and tissues the levels of MycERT2 expression driven from one or two copies of the Rosa26 promoter with those of endogenous Myc, R26-lsl-MycERT2 mice were crossed with the germ line deletor strain, ZP3-Cre (Lewandoski et al., 1997). Rosa26-driven MycERT2 is expressed in all somatic cells of such crosses, which we henceforth refer to as R26MER mice. Relative levels of endogenous Myc and ectopic MycERT2 proteins in log phase, serum-deprived and serum-stimulated WT MEFs, and in heterozygous R26MER/WT and homozygous R26MER/MER MEFs were then compared by immunoblotting cell extracts with SC42 anti-Myc antibody, which recognizes an epitope common to Myc and MycERT2. MycERT2, absent from WT MEFs, was expressed in homozygous R26MER/MER cells at approximately twice the level of MycERT2 present in heterozygous R26MER/WT cells (Figure 1B). This was confirmed at the mRNA level by real–time quantitative PCR (Figure 1C). The steady state level of MycERT2 in R26MER/WT MEFs was a little below, and the MycERT2 levels in R26MER/MER MEFs a little above, that of endogenous Myc in logarithmically growing WT MEFs, and far below the ~30,000 Myc molecules per cell present in HeLa cells (Moore et al., 1987). MycERT2 protein half-life was short (15–20′) (Suppl. Fig. 2A), essentially identical to that of endogenous Myc (Flinn et al., 1998; Ramsay et al., 1984). Unlike endogenous Myc, however, R26-driven MycERT2 expression was unaffected by serum status (Suppl. Fig. 2B). Above ~10,000 total Myc molecules per cell, Myc autoregulates, suppressing expression from its endogenous promoter (Penn et al., 1990). To ascertain whether MycERT2, when activated, is expressed at a level sufficient to suppress endogenous Myc, we repeated the analysis in Figure 1B using MEFs treated continuously with 4-OHT to activate MycERT2. We observed negligible down-regulation of endogenous Myc upon activation of MycERT2 in either R26MER/wt or R26MER/MER MEFs (Suppl. Fig. 2C), consistent with the very low levels of MycERT2 present in R26MER MEFs.
Figure 1. Characteristics of MycERT2 expression in R26lsl-MER MEFs.
A) Rosa26-driven MycERT2 expression in R26lsl-MER MEFs is dependent upon Cre recombinase-dependent and is transgene dose-dependent. R26lsl-MER/WT MEFs were infected with retrovirus driving Cre recombinase (+) or control vector (−) and selected with puromycin for 3 days. Western blotting of whole cell lysates with anti-human ERαantibody reveals expression of the predicted 92Kda MycERT2 fusion protein. The asterisk denotes a non-specific band.
B) Simultaneous comparison of levels of endogenous Myc and MycERT2 in wt, R26MER/wt and R26MER/MER MEFs. MEFs were isolated from embryos of each genotype and cultured in complete growth medium/10% FBS. Equal numbers of cell equivalents from each lysate were western blotted with SC42 anti-Myc antibody, detecting an epitope common to endogenous Myc and MycERT2. Extracts from serum-deprived wt MEFs and from wt MEFs 2 hrs after addition of fresh medium were probed alongside, together with an equivalent number of HeLa cells, which over express c-Myc (~30,000 molecules per cell) for comparison (Moore et al., 1987).
C) Dose dependent expression of MycERT2 mRNA driven from the Rosa26 locus. Q-PCR quantitation (mean + SEM) of MycERT2 mRNA isolated from in wt, R26MER/WT and R26MER/MER MEFs, all normalized to GUS (n=3).
We used Q-PCR with selective primers both to quantitate the relative levels of endogenous myc versus MycERT2 mRNA in tissues of tamoxifen-treated and untreated R26MER/MER mice and to assess whether endogenous Myc expression is suppressed by R26-driven MycERT2 in vivo. As expected, levels of endogenous Myc were extremely low in non-proliferating tissues. In spleen, where cell proliferation is substantial, MycERT2 mRNA levels were comparable with those of endogenous myc. We observed no significant suppression of endogenous Myc upon activation of MycERT2 in any tissue aside from colonic epithelium (Suppl. Fig. 3).
Deregulated expression of low-level Myc induces ectopic proliferation in multiple adult tissues
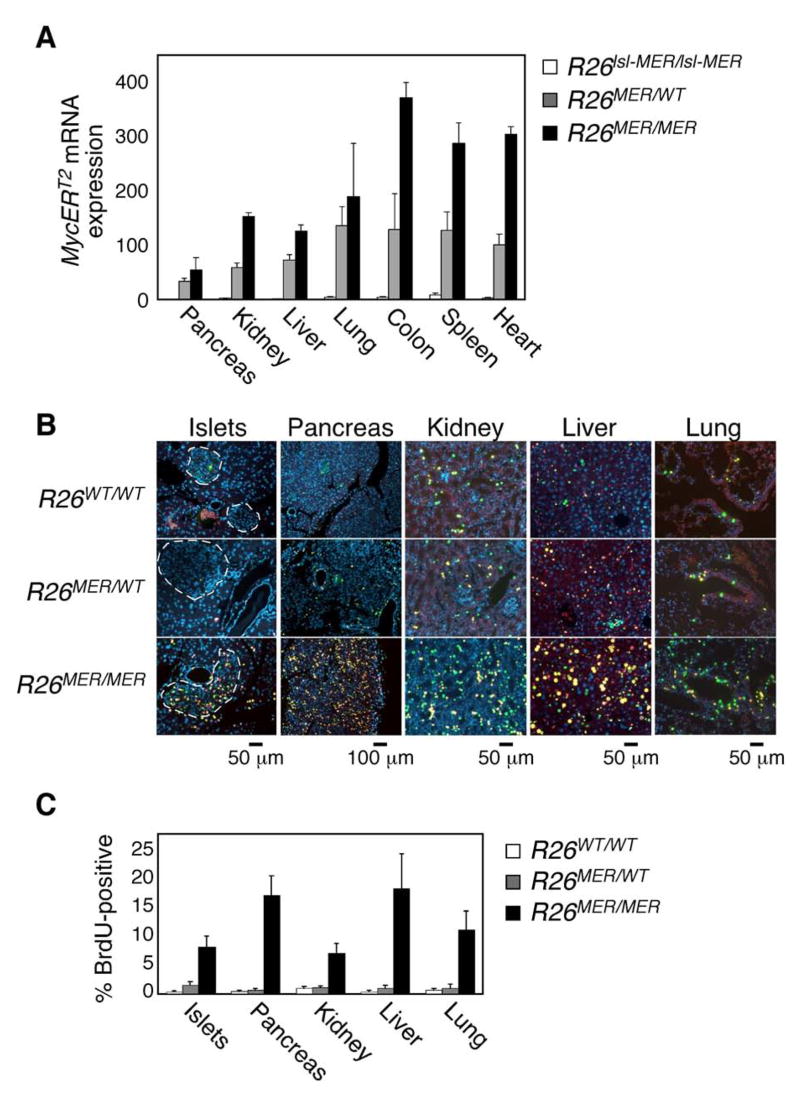
The relative contributions to oncogenesis made by Myc deregulation versus Myc over-expression remain unclear. The low level of deregulated MycERT2 expression in R26MER tissues therefore provided a unique opportunity to ascertain the impact of Myc deregulation, without overt over-expression, in adult mouse tissues. To confirm MycERT2 expression in tissues, RNA was extracted from multiple organs from adult (2-month old) homozygous R26-lsl-MERT2 and hetero R26MER/wt and R26MER/MER homozygous mice and MycERT2 transcript levels quantified by Q-PCR. MycERT2 mRNA was essentially undetectable in any organs of R26-lsl-MycERT2 mice but exhibited gene dose-dependent expression in all tested organs of R26MER animals (Fig. 2A).
Figure 2. Dose-dependent induction of proliferation by Rosa26-driven MycERT2 in Vivo.
A) Q-PCR analysis of MycERT2 mRNA levels (mean + SEM) in selected organs from untreated adult R26lsl-MER/lsl-MER (white bars), R26MER/WT (grey bars) and R26MER/MER (black bars) mice, normalized to GUS (n=3).
B) Representative images of 2-colour immunofluorescent detection of BrdU/IdU incorporation, indicating instances of S-phase progression in mice treated daily with tamoxifen for 6 days. Yellow-Orange: BrdU incorporation (S phase on day 3); Green: IdU incorporation (S phase on day 6).
C) Quantification (mean + SEM) of BrdU incorporation in tissues of control, R26MER/wt and R26MER/MER mice at day 6.
To activate MycERT2 acutely in all tissues, tamoxifen was daily administered systemically for 6 days to adult R26WT/WT, R26MER/WT and R26MER/MER mice. Proliferation in tissues was assessed by systemic administration of BrdU at day 3 followed by iododeoxyuridine (IdU) at day 6 to capture initial and delayed S phases (Burns and Kuan, 2005). BrdU and IdU incorporation in most tissues of tamoxifen-treated R26MER/WT (colonic epithelium was the exception – see below) was indistinguishable from that in R26WT/WT; by contrast, we saw a marked increase in incorporation of both BrdU and IdU in multiple tissues of homozygous R26MER/MER animals (Fig. 2B) - including exocrine pancreas, endocrine pancreas, kidney, liver and lung. Of note, no proliferation was observed in skeletal or cardiac muscle, or in brain aside from the epithelial choroid plexus (Suppl. Table 1). These data demonstrate that deregulation of low-level Myc is alone sufficient to drive sustained proliferation in a significant subset of tissues. However, MycERT2-induced proliferation was only evident in homozygous R26MER/MER tissues (Fig. 2C), indicating that a critical minimum threshold level of Myc is needed to drive proliferation in “permissive” tissues and this threshold is straddled by the levels of MycERT2 expression in hetero- and homozygous Rosa26-MycERT2 mice. Ipso facto, we conclude that the ectopic Myc activity in tissues of homozygous R26MER/MER mice is close to the minimum required to drive ectopic proliferation.
Different threshold levels of Myc trigger Myc-induced proliferation versus induction of apoptosis and ARF
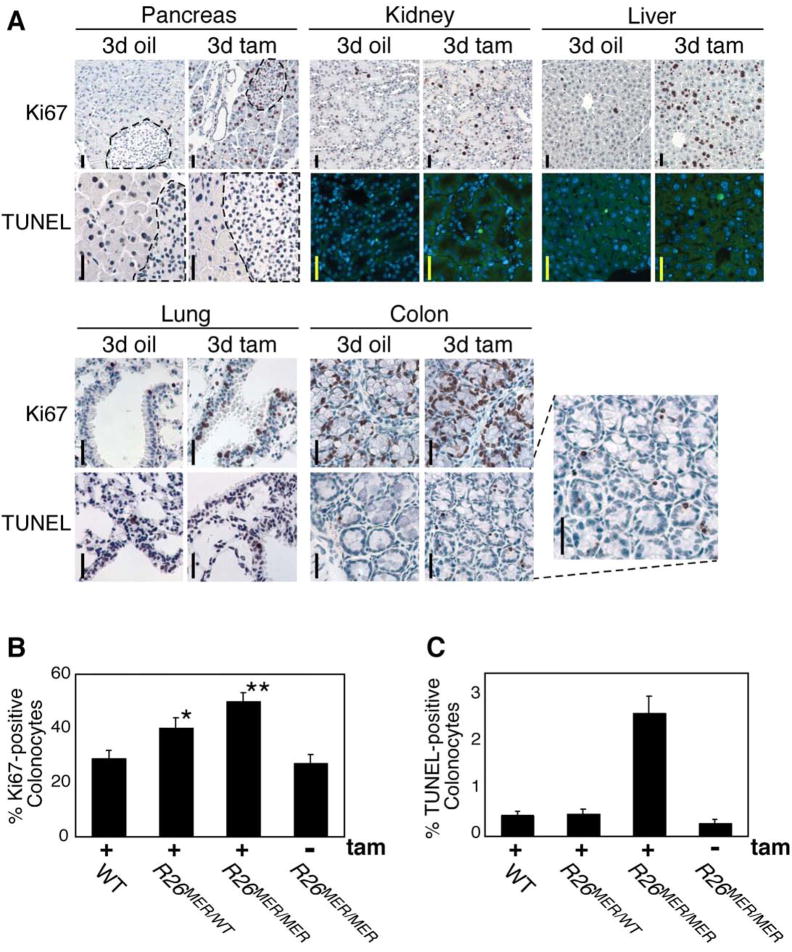
Myc-induced apoptosis is a key intrinsic tumor suppressor mechanism that limits Myc oncogenic potential. However, which factors determine whether proliferation or cell death is the predominant outcome of Myc activation remain unclear. To ascertain whether activation of low-level MycERT2 induces apoptosis in tissues in vivo, Myc was activated systemically in adult R26MER/MER mice for 0, 3 or 6 days and tissue sections from multiple organs then probed with Ki67-specific antibody to identify cells in cycle, and by TUNEL to identify apoptotic cells. As with our BrdU/IdU incorporation data, proliferation was robustly induced by 3 days of MycERT2 activation in the same proliferation permissive organs as above, remaining elevated at 6 days in pancreatic islets, kidney and lung (Fig. 3A and Suppl. Fig. 4; summarized in Suppl. Table 1). However, we observed no Myc-induced apoptosis in any tissues (with the exception of colonic epithelium - see below). We conclude that quasi-physiological levels of deregulated Myc are sufficient to drive proliferation in most permissive tissues without engaging concomitant apoptosis.
Figure 3. Widespread Myc-induced ectopic proliferation occurs without apoptosis in R26MER/MER mice.
A) Organs from adult R26MER/MER mice treated for 3 days with tamoxifen (tam) (n=7) or oil carrier (n=6), assessed for proliferation (Ki67 staining), and apoptosis (TUNEL assay). Activation of MycERT2 drives abundant ectopic cell cycle entry in multiple tissues (exocrine and endocrine pancreas, kidney, liver, lung and colon are presented). However, with the exception of the colon (inset), no tissue exhibited concomitant Myc-induced apoptosis. Scale bars = 25 μm.
B) Quantification (mean + SEM) of Ki67 staining in colon sections from tamoxifen-treated R26WT/WT (n=4), R26MER/WT (n=6), R26MER/MER mice (n=3), and from R26MER/MER mice treated with carrier (n=3). T Test analysis indicates that the increase in proliferation from R26WT/WT to R26MER/WT is significant (* P = 0.001) as is the increase from R26MER/WT to R26MER/MER (** P = 0.004).
C) Quantification (mean + SEM) of TUNEL positive cells in sections of colonic epithelium from same mice as B).
As mentioned, colonic epithelium of R26MER/MER mice was a notable exception to the above: Myc activation induced abundant apoptosis. Provocatively, out of all R26MER tissues tested, colonic epithelium exhibited the highest level of MycERT2 expression (Fig. 2A). Indeed, even the single allele of MycERT2 in heterozygous R26MER/WT mice drove sufficient Myc expression to trigger proliferation in colonic epithelium upon activation (P value = 0.001; Fig. 3B). In colonic epithelium of homozygous R26MER/MER animals (Fig. 3B) Myc-induced proliferation was even more marked: although accompanied by apoptosis (Fig. 3C). Together, these observations intimated that induction of apoptosis in vivo might require a higher level of Myc than proliferation.
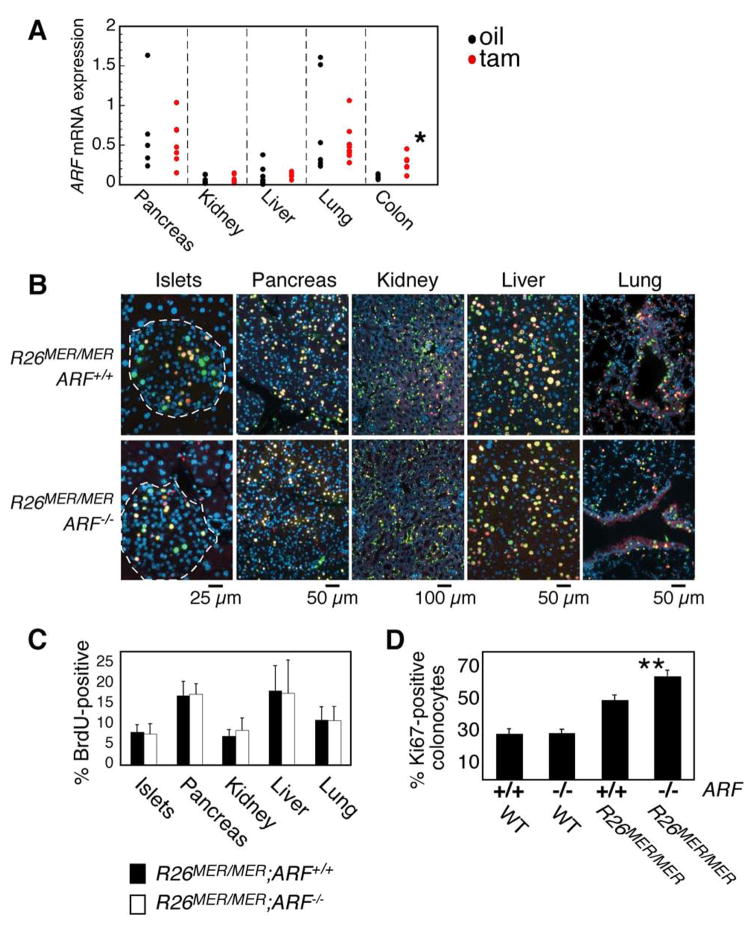
Like apoptosis, induction of p19ARF by oncogenic Myc is a critical restraint to Myc oncogenesis that, by activating p53 through inhibition of Mdm-2 (Kamijo et al., 1998; Weber et al., 1999; Zindy et al., 1998), can both drive apoptosis (Hermeking and Eick, 1994; Wagner et al., 1994; Zindy et al., 1998) and suppress proliferation (Finch et al., 2006). p19ARF may also exert p53-independent tumor suppressive functions (Lowe and Sherr, 2003), in part mediated by direct binding and functional inactivation of Myc (Datta et al., 2004; Qi et al., 2004). To ascertain whether the low, yet mitogenic, level of deregulated Myc in tissues of R26MER/MER mice is sufficient to induce ARF expression, we quantified p19ARF mRNA and protein expression following MycERT2 activation (Fig. 4A). Notwithstanding some variability in measured basal levels of ARF mRNA between tissues and individual isolates, we saw no measurable induction by Myc of ARF mRNA or protein in any tissue except, once again, colonic epithelium. Because it has recently been suggested that even extremely low levels of p19ARF may exert tumor suppressive activity (Bertwistle and Sherr, 2007), we also confirmed genetically that the Myc phenotypes we observe in vivo are p19ARF-independent by crossing the R26MER/MER mice into the ARF−/− background. ARF status had no impact on the extent, kinetics or duration of Myc-induced proliferation in most R26MER/MER mouse tissues (Fig. 4B, C). Together, these data show that the low level of deregulated Myc expressed in most proliferation-permissive tissues of R26MER/MER mice, while adequate to induce widespread proliferation, is insufficient to engage the p19ARF/p53 tumor suppressor pathway.
Figure 4. Activation of MycERT2 in most tissues of R26MER/MER does not induce the p19ARF tumor suppressor.
A) Q-PCR analysis for expression of ARF mRNA in organs from adult R26MER/MER mice treated for 3 days with tamoxifen (n=7) or oil carrier (n=7), normalized to GUS. No ARF is induced by MycERT2 activation in any tested tissue except for colonic epithelium, where it is statistically significant (* P value = 0.004).
B) Representative 2-colour immunoflourescence images of tissues from R26MER/MER;ARF+/+ (n=7) and R26MER/MER;ARF−/− (n=4) mice, injected daily with tamoxifen for 6 days. Mice were systemically pulsed with BrdU on day 2 and IdU on day 6, as per Figure 2, to identify cells in S phase at day 2 (Yellow-Orange) and day 6 (green).
C) Quantification (mean + SEM) of BrdU incorporation on day 6. Note that the data for ARF+/+ are the same as those presented in Figures 2C.
D) Quantification (mean + SEM) of Ki67 staining of colon sections from the indicated mice treated for 3 days with tamoxifen. Data for ARF+/+ samples are the same as those presented in Figure 3B. R26WT/WT;ARF−/− (n=3); R26MER/MER;ARF−/− (n=2).
R26MER/MER colonic epithelium was, again, an exception to the above: Myc activation induced a >2 fold induction of ARF mRNA (Fig. 4A; P value = 0.004) in colon and induction of p19ARF was confirmed by immunoblotting of colonocyte lysates prepared from fresh tissue (not shown). Furthermore, Myc-induced colonocyte proliferation was significantly enhanced in an ARF−/− background (Fig. 4D). As with apoptosis, this is consistent with the notion that induction of p19ARF requires higher levels of Myc than proliferation.
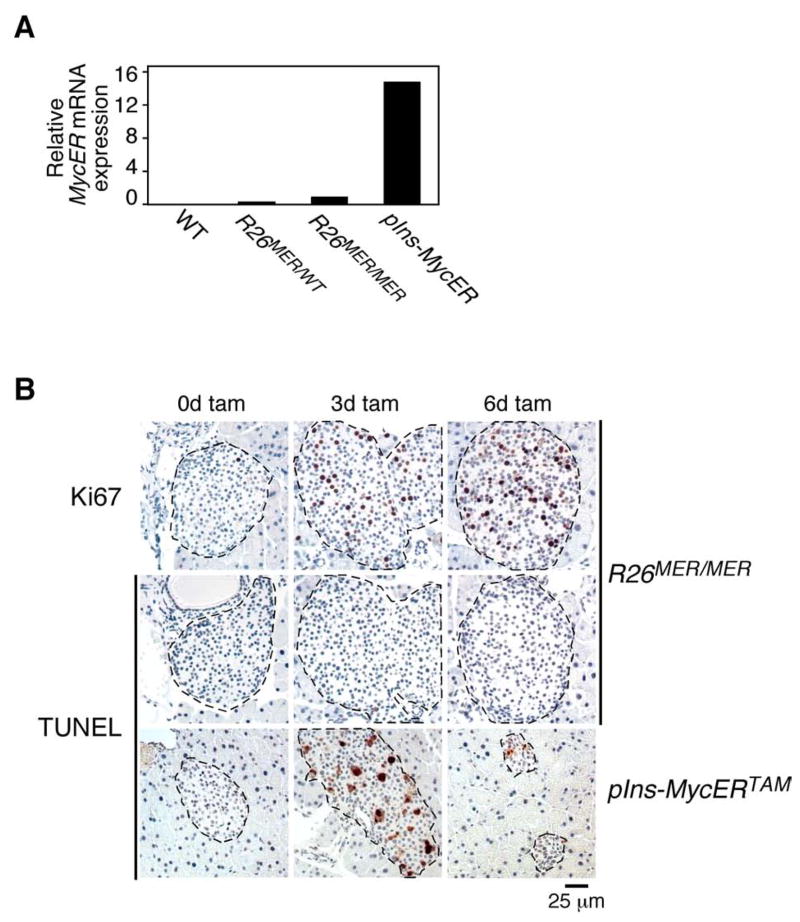
To ascertain directly whether the thresholds for Myc-induced proliferation versus tumor suppression (induction of apoptosis and ARF) are set at different levels of Myc expression in vivo, we used the pIns-MycERTAM β-cell mouse model in which the powerful rat insulin promoter drives high level expression of 4-OHT-dependent MycER in pancreatic β cells (Suppl. Fig. 5A). We previously showed that activation of Myc in this model efficiently triggers β cell proliferation but that this is rapidly curtailed by concomitant activation of both apoptosis and induction of nucleolar p19ARF (Suppl. Figs. 5A, 5B & 6) (Finch et al., 2006; Pelengaris et al., 2002). Q-PCR and immunoblot analyses of pancreatic islets isolated from pIns-MycERTAM, R26MER/WT and R26MER/MER mice indicated that MycERTAM mRNA and protein expression are ~15 fold higher in pIns-MycERTAM islets than those of MycERT2 in islets from homozygous R26MER/MER animals (Fig. 5A and Suppl. Fig. 5C). To compare directly the net outcome of acute Myc activation in pIns-MycERTAM versus Rosa26MER/MER islets we activated MycER in vivo for 3 and 6 days and then stained pancreatic tissue sections for markers of proliferation (Ki67) and apoptosis (TUNEL). As expected (Finch et al., 2006; Pelengaris et al., 2002), high levels of activated Myc in the pIns-MycERTAM islets induced proliferation together with both ARF induction and overwhelming β-cell apoptosis, resulting in rapid islet involution. By contrast, activation of low level Myc in islets of R26MER/MER mice drove proliferation without any attendant apoptosis or islet involution, resulting in progressive islet hyperplasia (Fig. 5B and Supp. Fig. 7). Likewise, activation of low-level MycERT2 in lung epithelium of R26MER/MER mice failed to elicit ARF expression whereas the high levels of MycERT2 delivered into lung epithelium by adenovirus vector potently induced ARF upon tamoxifen activation (Supp. Fig. 5D). Hence, in both β cells and lung in vivo, deregulated Myc is competent to activate intrinsic tumor suppression pathways so long as it is expressed at elevated levels.
Figure 5. Induction of apoptosis requires higher levels of deregulated Myc than induction of ectopic proliferation.
A) Q-PCR analysis of MycER mRNA expression in islets isolated from disaggregated pancreata of untreated designated mice, pooled according to genotype (n>2 per cohort). Expression was normalized to GUS and results are expressed relative to MycERT2 mRNA in R26MER/MER islets.
B) TUNEL staining of pancreatic islets from R26MER/MER mice, treated with tamoxifen for 3 (n=7) or 6 (n=6) days (center panels), with matched samples from similarly treated pIns-MycERTAM mice (lower panels). Ki67 staining of adjacent sections from R26MER/MER pancreata (upper panels) demonstrates tamoxifen-induced functionality of Rosa26-driven MycERT2 protein.
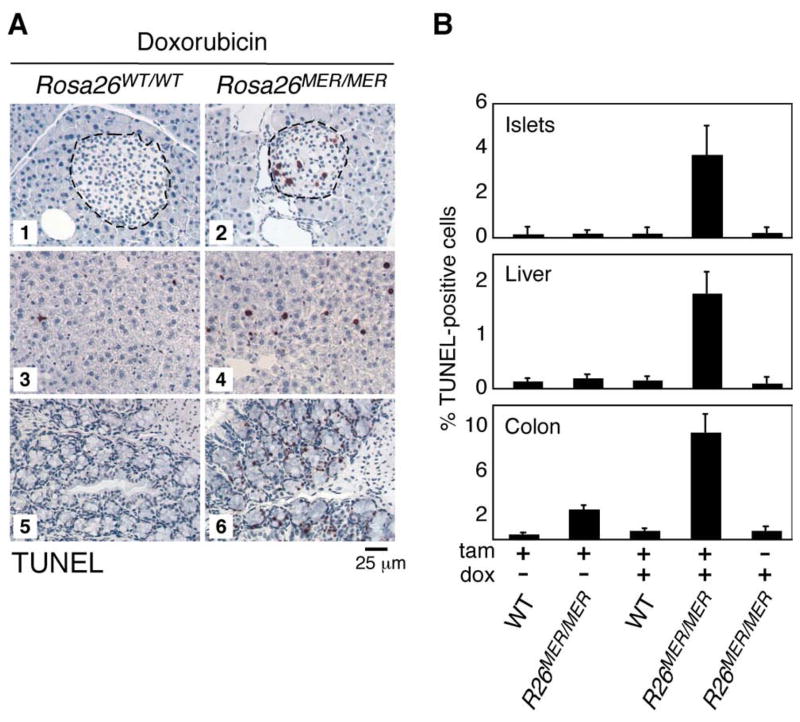
There are two plausible explanations for how different threshold levels of Myc might preferentially trigger proliferation versus apoptosis. It could be that low and high levels of Myc engage different sets of target gene sets, perhaps due to differing affinity Myc-binding promoter/enhancer elements (i.e. Myc discriminates) (Suppl. Fig. 8A). In this model, low-level Myc simply fails to activate the apoptotic transcriptional program. Alternatively, Myc might regulate the same genes in each case but to a greater extent when expressed at a higher level, and the differential Myc outputs arise from different thresholds at which the proliferative and apoptotic programs fire (i.e. downstream factors decide) (Suppl. Fig. 8B). In this case, low level Myc still activates the apoptotic program, but to a level insufficient to trigger it. We reasoned that if the second model holds, sub-apoptotic low-level Myc should, by priming the apoptotic machinery, sensitize cells to induction of apoptosis by other triggers. We therefore co-exposed R26MER/MER mice to both tamoxifen, to activate MycERT2 in tissues, and a sub-apoptotic dose of the cytotoxic agent doxorubicin. The combination of both sub-apoptotic stimuli triggered significant apoptosis in several tissues, including pancreatic islets and liver, and significantly exacerbated the extent of Myc-induced apoptosis in colonic epithelium (Fig. 6A & B). Thus, low level deregulated Myc, although insufficient to trigger apoptosis itself, nonetheless engages the Myc apoptotic program.
Figure 6. Sub-apoptotic Myc synergises with sub-apoptotic doxorubicin to induce apoptosis in multiple tissues.
A) TUNEL staining of pancreatic islets (panels 1, 2), liver (panels 3, 4), and colon (panels 5, 6) from R26WT/WT (n=2) and R26MER/MER (n=4) mice treated for 3 days with tamoxifen and overnight with 10 mg/kg doxorubicin.
B) Quantification (mean + SEM) of TUNEL staining in sections from pancreas (islets), liver and colon from the above mice treated with tamoxifen and/or doxorubicin. (n=2–4 mice per cohort).
Deregulation of low-level Myc is tumorigenic
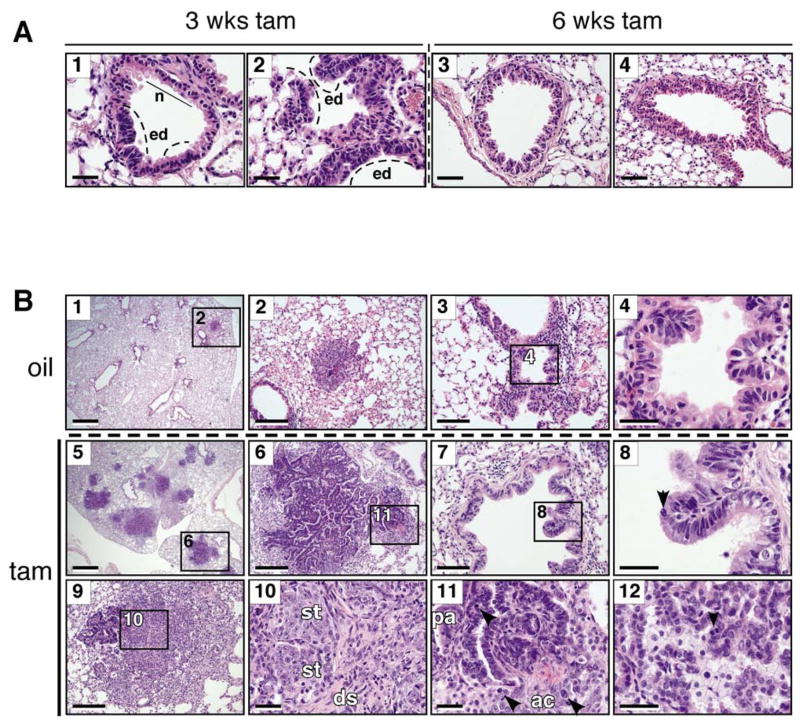
Although low-level deregulated Myc is sufficient to drive ectopic proliferation in tissues, it remained possible that actual tumorigenicity, like apoptosis and ARF induction, required higher-level Myc expression. To investigate this, we used intranasal delivery of recombinant Adenovirus-expressing Cre (Ad-Cre) to trigger sporadic expression of MycERT2 in lung epithelia of homozygous R26lsl-MER/lsl-MER mice. MycERT2 was then activated by daily systemic administration of tamoxifen for 3 (n=4) or 6 (n=5) weeks. After 3 weeks of sustained Myc activity, bronchiolar (Fig. 7A, panel 1) and bronchioalevolar junction regions (panel 2) of R26lsl-MER/lsl-MER lungs exhibited multiple hyperplastic and dysplastic epithelial foci with pseudostratification and occasional micropapillary tuft formation. After 6 weeks, more extensive micropapillary and papillary hyperplasia was clearly evident (panel 3), with several bronchioles exhibiting Clara cell hyperplasia and sloughing into the airway lumen (panel 4). These Myc-induced lesions closely resemble the early bronchiolaveolar lesions that develop 6–8 weeks after sporadic activation of oncogenic K-RasG12D driven from the endogenous KRas2 promoter (Jackson et al., 2005; Jackson et al., 2001; Johnson et al., 2001).
Figure 7. Low level deregulated Myc is tumorigenic in vivo.
A) Representative images of H & E-stained lungs from tamoxifen treated, Ad-Cre infected R26lsl-MER/lsl-MER mice showing multiple pre-neoplastic pulmonary lesions. Panels 1, 2: Localized epithelial dysplasia (ed) evident in airway epithelium (panel 1) and bronchioalveolar junctions (panel 2) after 3 weeks of sustained MycERT2 activation. Compare with normal bronchiolar epithelium (n). Scale bars=20 μm. Panels 3, 4: Micro-papillary invaginations (panel 3) and atypical Clara cell proliferation characterized by vertical stacking of bronchiolar epithelial cells with apical nuclei (panel 4) present after 6 weeks of sustained MycERT2 activation. Scale bars=40 μm.
B) H & E stained lungs from Ad-Cre infected LSL-K-RasG12D;R26lsl-MER/lsl-MER mice treated with oil carrier (n=3, panels 1–4) or tamoxifen (n=6, panels 5–12) for 6 weeks. Panels 1, 2, 5 & 6: Size comparison between tumors driven by K-RasG12D alone (1, 2) and K-RasG12D/MycERT2 combined (5, 6). Scale bars=0.4 □m (1, 5) and 100 μm (2, 6). The numbered rectangular regions within panels 1, 3, 5, 6, 7 and 9 are shown enlarged in panels 2, 4, 6, 11, 8, and 10 respectively. Panels 3, 4, 7 & 8: Micropapillary formation along the bronchioles driven by K-RasG12D alone (3, 4) and K-RasG12D/MycERT2 combined (7, 8). The arrowhead points to a mitotic figure in panel 8. Scale bars=50μm (3, 7) and 20 μm (4, 8). Panels 9–11: Progression to carcinoma observed after 6 weeks of combined KrasG12D/MycERT2 driven oncogenesis. Scale bars=100 μm (9) and 20 μm (10, 11, 12). Panel 10: Nests of solid tumor (st) and isolated tumor cells surrounded by desmoplastic stroma (ds). Panels 11, 12: Mitotic figures (arrowheads) present in combined K-RasG12D/MycERT2-driven papillary adenoma (pa), adenocarcinoma (ac; 11) and acinar adenocarcinoma.
Individually, Myc or Ras are usually insufficient to drive full tumor progression but together they exhibit potent oncogenic cooperation (Land et al., 1983). To ascertain whether low-level deregulated Myc cooperate with Ras oncogenesis in lung epithelium in vivo, we crossed homozygous R26lsl-MER/lsl-MER and heterozygous R26lsl-MER/wt mice into the conditional LSL-K-RasG12D model (Jackson et al., 2005; Jackson et al., 2001; Johnson et al., 2001). Sporadic co-expression of both oncogenes in lung epithelium was triggered by Ad-Cre, and then MycERT2 activated or not with tamoxifen. Induction of K-RasG12D alone elicited atypical adenomatous hyperplasias within 2 weeks, small adenomas by 6 weeks post infection and overt adenocarcinomas only after 16–26 weeks (Jackson et al., 2005; Jackson et al., 2001). Identical hyperplastic and dysplastic bronchiolar lesions arose with identical kinetics and efficiency in LSL-K-RasG12D;R26lsl-MER/lsl-MER mice never given tamoxifen (i.e. where only Ras is activated) (Fig. 7B, panels 1–4) and in control LSL-K-RasG12D-only mice treated with either oil (n=2) or tamoxifen (n=8; data not shown). By contrast, activation of MycERT2 together with K-RasG12D profoundly accelerated tumor progression over K-RasG12D alone in homozygous, but not heterozygous, Rosa26-lsl-MycERT2 animals. By 6 weeks post Ad-Cre infection, papillary adenomas were both more numerous and significantly larger (panels 5 and 6) in co-activated mice, some already having progressed to invasive adenocarcinoma (panels 9–11). Such adenocarcinomas were poorly differentiated, growing as small tumor clusters with individual tumor cells embedded within desmoplastic stroma (panel 10). Whereas few mitotic figures were evident in the lesions induced by K-RasG12D alone, they were readily detectable in the bronchiolar lesions (compare panels 4 and 8), adenomas (panel 11) and adenocarcinomas (panels 11 and 12) induced by K-RasG12D and MycERT2 together, indicating that low level expression of deregulated confers a significantly higher level of tumor cell proliferation and is possessed of potent tumorigenic activity.
DISCUSSION
Uncontrolled proliferation of somatic cells is an ever-present risk for large, long-lived organisms like vertebrates whose tissues harbor cells that proliferate throughout life. Hence, multiple surveillance mechanisms have evolved to suppress the emergence and propagation of tumor cells. One of these involves the tight coupling of tumor suppression to the programs that drive cell proliferation, a phenomenon dubbed “intrinsic tumor suppression” (Lowe et al., 2004). Such obligate coupling serves to annul the immediate growth advantage afforded by oncogenic activation of the cell’s proliferative machinery. The Myc transcription factor is a prototypical example of this. In normal cells, Myc coordinates and drives the diverse intra- and extra-cellular programs required for orderly expansion of normal somatic cells. However, oncogenic Myc triggers the ARF/p53 tumor suppressor pathway and apoptosis, two potent tumor suppressor programs that efficiently restrain Myc’s oncogenic potential (Evan and Littlewood, 1998; Lowe et al., 2004). However, for “intrinsic tumor suppression” to be compatible with the orderly proliferation in normal tissues, cells must accurately and reliably discriminate between normal mitogenic and oncogenic signaling. How they do this, however, is unclear. One potential distinctive hallmark of oncogenic signals is their aberrant context: in normal cells Myc is never activated alone but always in the context of collateral signaling pathways. Another possibility is the unusual persistence of oncogenic Myc - endogenous Myc activity is tightly mitogen-dependent and episodic. Finally, as a consequence of its mutation or its relentless induction by other oncogenes, Myc is usually present at significantly higher levels in tumors than in normal cells.
The undeniable importance of intrinsic tumor suppression is demonstrated by the profound acceleration of oncogenesis in Myc transgenic mouse models afforded by genetic lesions that block apoptosis or circumvent the ARF/p53 pathway. Unfortunately, classical transgenic Myc models employ highly active tissue-specific promoter/enhancer elements to drive transgenic Myc expression in the requisite target tissue. Hence, Myc is both deregulated and elevated, making it impossible to ascertain which specific aberrant feature of oncogenic Myc engages tumor suppression. To circumvent this shortcoming, we made use of the relatively weak, but ubiquitous, Rosa26 promoter to drive low-level deregulated expression of the switchable form of Myc, MycERT2, in target tissues. A MycERT2 open reading frame was inserted downstream of the endogenous Rosa26 promoter but its expression blocked by an intervening transcriptional STOP element flanked by loxP recombination sites. Hit-and-run excision of the STOP element by Cre recombinase triggers targeted expression of MycERT2; however, MycERT2 activity is dependent upon continuous provision of 4-OHT ligand.
To ascertain the level at which the Rosa26 promoter drives MycERT2 expression in adult tissues, we crossed mice harboring one or two copies of the Rosa26-LSL-MycERT2 into the ZP3-Cre background, which excises the floxed STOP element in the egg. The resulting R26MER animals express MycERT2 from the Rosa26 promoter in all tissues. In all tested R26MER tissues MycERT2 expression levels were broadly comparable, although somewhat higher in colonic epithelium. In every tissue examined, homozygous R26MER/MER mice expressed approximately twice as much MycERT2 mRNA and protein as their heterozygous R26MER/WT littermates.
Acute activation of MycERT2 in tissues of homozygous R26MER/MER mice triggered widespread ectopic proliferation in many tissues including endocrine and exocrine pancreas, liver, kidney epithelium, lung, skin and lymphoid organs. In most instances, proliferation was sustained at least out to 6 days of continuous Myc activation: analysis beyond this time was curtailed by precipitous onset of anemia and dehydration that coincided with intestinal hemorrhage and necessitated euthanasia. In contrast to homozygous R26MER/MER mice, activation of MycERT2 in heterozygous R26MER/wt animals elicited no increased proliferation in any tissues except colon and, weakly, spleen red pulp. Given there is only a two-fold difference in MycERT2 levels in heterozygous versus homozygous R26MER mice, this defines a sharp minimum threshold level of intracellular Myc required to elicit ectopic proliferation in vivo. It also indicates that the level of ectopic activated MycERT2 in homozygous R26MER/MER tissues is close to the minimum necessary to drive ectopic proliferation. It is interesting that the level of ectopic MycERT2 in heterozygous R26MER/WT tissues, while comparable to that of endogenous Myc in MEFs proliferating in response to serum, is insufficient to drive those tissues into cycle. We guess this reflects the fact that serum mitogens co-engage both Myc and multiple, additional synergistic signaling pathways. By contrast, Myc is the sole engine of proliferation in R26MER/WTcells. It is also possible that activated MycERT2 has reduced specific activity compared to wt Myc protein, leading to an overestimation of the level of functional Myc in R26MER tissues. Of note, while MycERT2 was expressed in all R26MER/MER tissues tested, some failed to proliferate when MycERT2 was activated. Why this should be is unclear but it is likely that some cell types are constitutionally unable to proliferate due to structural constraints or lack of critical components of the cell cycle machinery. Yet others may be competent to proliferate only in response to higher levels of Myc than are expressed in R26MER/MER tissues. An obvious example of this would be those many differentiated cell types that express appreciable levels of Mad proteins (Chin et al., 1995; Queva et al., 1998; Vastrik et al., 1995), which mitigate Myc action by competing with Myc for Max.
Not only is low-level constitutive expression of MycERT2 sufficient to induce and maintain aberrant proliferation in many normal R26MER/MER tissues in vivo but it is also oncogenic, efficiently driving early-stage tumorigenesis in lung epithelium and, in cooperation with activated Ras, full progression to invasive adenocarcinoma (Fig. 7). Nonetheless, even though Rosa26-driven Myc is oncogenic and both deregulated and active out of normal mitogenic context, it fails to induce either apoptosis or measurable ARF expression in most tissue types. This indicates a fundamental difference in how Myc engages its oncogenic and tumor suppressive outputs. The one notable exception to this is colonic epithelium and, since this is the R26MER tissue with the highest level of MycERT2 expression, we guessed that over-expression of Myc is the likely determining trigger of intrinsic tumor suppression. We confirmed this directly in pancreatic β cells and lung epithelium: in both tissues further elevation of Myc levels triggered both apoptosis and ARF. Therefore, we conclude that it is the aberrant intensity of oncogenic Myc, and not its abnormal persistence or context, that triggers intrinsic tumor suppression. Recently it was shown that induction of both ARF and replicative senescence by activated Ras in vivo also requires over-expression of the activated oncoprotein (Sarkisian et al., 2007). Hence, preternaturally high intensity appears to be the general metric by which cells discriminate between normal and oncogenic signals. However, we do not discount the possibility that even low levels of deregulated Myc may be sufficient, in some cell lineages, to engage apoptosis and/or ARF expression at some level that suppresses tumor initiation. Indeed, data from studies on transgenic Myc in the well-characterized Eμ-myc lymphoma model suggest that even very low Myc levels can, at some point in tumor evolution, contribute to the selective pressure to inactivate ARF or p53 (Bertwistle and Sherr, 2007).
Paradoxically, our data suggest that low-level deregulated Myc may be a more efficient initiator of oncogenesis than Myc that is over-expressed, since the latter can be tolerated only by cells that have already lost their tumor suppressor pathways. Indeed, high initial levels of Myc may even impede onset of tumorigenesis, a conclusion consistent with recent surprising findings that co-expression of Myc delays the mean latency of tumor onset in a transgenic RasG12D lung tumor model (Tran et al., 2008). This elegant study used the CC10 promoter to drive Doxycyclin-dependent Myc expression in Clara cells and alveolar type II pneumocytes at very high levels, at least 10 times the quasi-physiological level of MycERT2 in lung epithelium of R26MER/MER mice (Fig. 2A). As our data show, such high levels of Myc breach the ARF/apoptotic triggering threshold and, by engaging intrinsic tumor suppression, this has the immediate effect of staunching tumorigenesis. By contrast, oncogenic cooperation in the LSL-K-RasG12D;R26lsl-MER/lsl-MER mouse is efficient because it involves levels of oncogenic Myc (and presumably Ras) that are too low to trigger significant intrinsic tumor suppression. We surmise that low-level oncogene expression may be general characteristic of early stage spontaneous tumors and that most tumors evolve via a two stage selective process. Early on, when tumor suppressor pathways remain intact, selection strongly favors low-level oncogene activity and the relatively indolent clonal expansion it confers. Elevated oncogene activity, together with the increased aggressiveness it confers on tumors, becomes subject to positive selection only once the appropriate intrinsic tumor suppression pathways have been eroded by sporadic mutation.
How might different levels of Myc trigger such distinct biological outputs? One possibility is that Myc-induced ARF/p53 and apoptosis are mediated by a distinct set of target genes, perhaps those with lower affinity promoter/enhancer elements that respond only to elevated Myc levels (Supplemental Fig. 5A). Alternatively, Myc might regulate identical sets of gene targets in both cases, although to lesser or greater overall extents when Myc is low or high respectively, with net outcome determined by the different execution thresholds of each output program (Supplemental Fig. 5B). To distinguish between the two possibilities, we asked whether expression of Myc at levels that trigger no overt apoptosis nonetheless reduces the threshold for induction of apoptosis by other insults. MycERT2 was globally activated in R26MER/MER mice, which were then systemically exposed to the cytotoxic drug Doxorubicin at a dose that was itself sub-apoptotic. The combination of sub-lethal Myc and Doxorubicin induced apoptosis in several tissues. Myc-induced apoptosis is mediated through the intrinsic mitochondrial pathway (Juin et al., 1999) that is triggered when the anti-apoptotic buffering of the Bcl-2/Bcl-xL proteins is neutralized by the sum of activated BH3 proteins. Our data imply that low-level Myc still activates its requisite BH3 apoptotic effectors but to a level insufficient to neutralize Bcl-2/Bcl-xL without the cooperation of collateral apoptotic signals. Of note, we saw no correlation between Myc-induced proliferation and Myc-induced sensitivity to Doxyrubicin is different tissues, indicating that Myc-dependent Doxyrubicin sensitivity is not merely a consequence of that increased tissue’s proliferative status but reflects an inherent variability in the innate sensitivity to Myc-induced apoptosis among different cell types in vivo. Indeed, even in any one cell type the threshold for triggering Myc-induced apoptosis is highly dependent upon changes in relative levels of the Bcl-2 family proteins (Bissonnette et al., 1992; Fanidi et al., 1992; Juin et al., 2002; Wagner et al., 1993) and availability of survival signals (Harrington et al., 1994a; Harrington et al., 1994b). Hence, the level of Myc required to trigger apoptosis is not an innate, fixed property of a cell but exquisitely dependent upon the cell’s internal state and microenvironment. Importantly, such tissue-specific variations in the threshold set points for Myc oncogenic and apoptotic output mean that the innate susceptibilities of different tissues to Myc-driven oncogenesis will depend greatly on the level at which oncogenic Myc is expressed. Whether the operational threshold at which Myc triggers ARF is similarly subject to such protean, cell type-specific influences is unknown. One last implication of the different firing thresholds at which Myc’s distinct outputs (proliferation and tumor suppression) are set is that relatively subtle changes in overall Myc activity may suffice to shift above or below each specific threshold, thereby triggering dramatically different outcomes. Likewise, it is possible that mutations that merely augment or degrade overall Myc activity, by shifting the effective level of Myc above or below different output thresholds, could appear selective for specific Myc functions.
MATERIALS AND METHODS
Generation of Rosa26-MER targeting vector and mice
c-myc cDNA lacking a stop codon was excised from pBabe-MycERTAM (Littlewood et al., 1995) and annealed to T2 point-mutated ligand-binding-domain of human Estrogen Receptor from pCreERT2 (Indra et al., 1999), followed by the IRES-EGFP fragment (generously provided by Drs David Dankort and Martin McMahon, UCSF) generating c-MycERT2-IRES-EGFP (MIE). This MIE fragment was then incorporated into the previously described Rosa26 vector pBigT (Srinivas et al., 2001) then excised incorporating both the floxed STOP/neomycinR cassette and MIE. The final vector, pRosa26fsMIE, was linearized with SwaI, transfected into murine ES cells and Neomycin-resistant clones identified by Southern blotting. An Internal probe was generated by random priming from a gel-purified 1.6 Kb NcoI/HindIII fragment of pR26-PAS, further digested with AscI to separate it from co-migrating DNA. External probe was generated by random priming from a 330 bp NotI fragment of the Rosa26 promoter, generously provided by Dr Philippe Soriano (FHCRC). Chimeric mice were initially bred with C57/Bl6, until germ line transmission of the allele was observed. The consequent R26-lsl-MycERT2 mouse line was bred to C57/Bl6, then alternately to FVBN, and maintained on a mixed FVBN/C57/Bl6 background.
Other mouse strains, procedures and genotyping
All procedures involving mice were performed in accordance with protocol number AN 076148 (UCSF IACUC). Zp3-Cre (Lewandoski et al., 1997), LSL-K-RasG12D (Jackson et al., 2001), p19ARF−/− (Kamijo et al., 1999) and pIns-MycERTAM (Pelengaris et al., 2002) mice are described previously. Tamoxifen (Sigma), dissolved in peanut oil, was administered daily by intraperitoneal (IP) injection for a maximum of 6 weeks at a dose of 1 mg/20 g body mass per day. For BrdU/IdU double labeling, bromodeoxyuridine (Sigma) and Iododeoxyuridine (Sigma), dissolved in Tris buffered saline, were injected IP three times at 6 hr intervals on day 3, while IdU was injected once 5–6 hrs prior to harvest one day after the last tam injection (day 6). To deliver adenovirus-Cre recombinase (Ad-Cre), mice were anesthetized with 2.5% Avertin (250 μl/20 g body mass) and 5×107 pfu Ad-Cre was administered as previously described (Fasbender et al., 1998). Details of primer/probe sets used for genotyping and expression analysis can be found in the Supplementary Information.
Cell culture and immunoblotting
Embryonic fibroblasts were isolated from E13.5 R26-lsl-MycERT2 and R26-MycERT2 mice. Whole cell lysates were prepared by dissolving cells in Tween lysis buffer [150 mM NaCl; 50 mM HEPES, pH 7.5; 1 mM EDTA; 2.5 mM EGTA; 0.1% Tween 20 + CompleteTM protease inhibitor cocktail (PIs; Roche)] followed by sonication. Nuclear extracts were made in low salt buffer [20 mM KCl; 10 mM HEPES, pH 7.5; 1mM MgCl2; 1 mM CaCl2; 0.1% Triton x-100 + PIs]. For liver extracts, freshly isolated livers were homogenized in 10 vol/wt T-PerTM (Pierce) + PIs, fractionated by centrifugation at 1,000 x G and the pellets re-suspended in 1 volume T-Per + 0.2% SDS, followed by sonication. Islets were prepared from disaggregated pancreata as previously described (Lawlor et al., 2006) and lysed in Tween lysis buffer, sonicated and cleared by centrifugation at 12,000 x G prior to loading. For p19ARF analysis, cell lysates were loaded on glycerol/acrylamide gels and fractionated by discontinuous electrophoresis and electroblotted onto PVDF membranes. Anti-ERα (Santa Cruz, SC-543); anti-c-Myc (Santa Cruz, SC42 and 9E10); X-Myc1 (G.E.); anti-p19ARF 5-C3-1 (Bertwistle et al., 2004), generously provided by Dr Martine Roussel, St Jude Hospital); anti-Lamin A/C (Santa Cruz, SC7293); anti-α-Actin (Sigma) were used as primary antibodies. Secondary horseradish peroxidase-conjugated antibodies (Amersham) were detected by chemiluminescence.
Immunohistochemistry and immunoflourescence
5 μm paraffin embedded sections were probed with Ki67 antibody (SP6, Neomarkers: Fremont, CA) was used at 1:200 in 3% BSA overnight (o/n) at 4°C and detected with biotinylated goat anti-rabbit (Vector Labs) followed by Vectastain ABCTM detection (Vector Labs) using stable diamobenzidine (DAB) solution (Invitrogen). For BrdU/IdU double staining, mouse α-BrdU (1:20, Roche), which recognizes both BrdU and IdU, was used in conjunction with rat α-BrdU (1:200, Serotec) that recognizes BrdU only. Bound primary antibodies were detected using Alexa-488 conjugated α-mouse IgG and Alexa-568 conjugated α-rat IgG (Molecular Probes). For p19ARF immunohistochemistry, blocking was performed in 10% normal goat serum (NGS) with 0.1% Triton x-100. Primary antibody (5-C3-1) was diluted 1:10 in 3% NGS + 0.1% Triton x-100 and incubated overnight at 4°C. TUNEL staining was performed using the ApoptagTM peroxidase labeled kit (Chemicon) or ApoptagTM flourescein labeled kit (Chemicon) according to the manufacturers directions. Otherwise, tissue sections were blocked overnight in 3% BSA prior to addition of peroxidase-conjugated anti-digoxigenin.
Supplementary Material
Acknowledgments
We thank all members of the Evan laboratory for sage help in preparing this manuscript, most especially Laura Soucek. Special thanks to Fanya Rostker for animal husbandry. We are grateful to Drs Michael Fried, Martin McMahon, Courtney Broaddus, Doug Hanahan, Kevin Shannon, Doug Green, Francesca Mariani, David Dankort, Suzanne Schubbert, Scott Oakes, Jay Debnath, Scott Kogan and Bill Weiss for insightful comments. We thank all members of the Cancer Center core facilities (Dr. Nigel Kileen, Bill Hyun, Kirsten Copren, Langdon Smythe, Nataliya Korets and Eva Soliven). DJM was supported in part by a Ruth L. Kirschstein NRSA Fellowship (CA099363). MRJ is Enrique Cepero fellow of the Damon Runyon Cancer Research Foundation. Funded by the NIH NCI (R01-CA106526 to GIE).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES CITED
- Askew DS, Ashmun RA, Simmons BC, Cleveland JL. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene. 1991;6:1915–1922. [PubMed] [Google Scholar]
- Bertwistle D, Sherr CJ. Regulation of the ARF tumor suppressor in Eμ-Myc transgenic mice: longitudinal study of Myc-induced lymphomagenesis. Blood. 2007;109:792–794. doi: 10.1182/blood-2006-07-033985. [DOI] [PubMed] [Google Scholar]
- Bertwistle D, Zindy F, Sherr CJ, Roussel MF. Monoclonal antibodies to the mouse p19(Arf) tumor suppressor protein. Hybridomics. 2004;23:293–300. doi: 10.1089/hyb.2004.23.293. [DOI] [PubMed] [Google Scholar]
- Bissonnette R, Echeverri F, Mahboubi A, Green D. Apoptotic cell death induced by c-myc is inhibited by bcl-2. Nature. 1992;359:552–554. doi: 10.1038/359552a0. [DOI] [PubMed] [Google Scholar]
- Burns KA, Kuan CY. Low doses of bromo- and iododeoxyuridine produce near-saturation labeling of adult proliferative populations in the dentate gyrus. Eur J Neurosci. 2005;21:803–807. doi: 10.1111/j.1460-9568.2005.03907.x. [DOI] [PubMed] [Google Scholar]
- Chin L, Schreiber-Agus N, Pellicer I, Chen K, Lee HW, Dudast M, Cordon-Cardo C, DePinho RA. Contrasting roles for Myc and Mad proteins in cellular growth and differentiation. Proc Natl Acad Sci U S A. 1995;92:8488–8492. doi: 10.1073/pnas.92.18.8488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Nag A, Pan W, Hay N, Gartel AL, Colamonici O, Mori Y, Raychaudhuri P. Myc-ARF (alternate reading frame) interaction inhibits the functions of Myc. J Biol Chem. 2004;279:36698–36707. doi: 10.1074/jbc.M312305200. [DOI] [PubMed] [Google Scholar]
- Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan G, Littlewood T. A matter of life and cell death. Science. 1998;281:1317–1322. doi: 10.1126/science.281.5381.1317. [DOI] [PubMed] [Google Scholar]
- Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, Waters CM, Penn LZ, Hancock DC. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Fanidi A, Harrington EA, Evan GI. Cooperative interaction between c-myc and bcl-2 proto-oncogenes. Nature. 1992;359:554–556. doi: 10.1038/359554a0. [DOI] [PubMed] [Google Scholar]
- Fasbender A, Lee JH, Walters RW, Moninger TO, Zabner J, Welsh MJ. Incorporation of adenovirus in calcium phosphate precipitates enhances gene transfer to airway epithelia in vitro and in vivo. J Clin Invest. 1998;102:184–193. doi: 10.1172/JCI2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch A, Prescott J, Shchors K, Hunt A, Soucek L, Dansen TB, Swigart LB, Evan GI. Bcl-XL gain of function and p19ARF loss of function cooperate oncogenically with Myc in vivo by distinct mechanisms. Cancer Cell. 2006;10:113–120. doi: 10.1016/j.ccr.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Flinn EM, Busch CMC, Wright APH. Myc Boxes, which Are conserved in Myc family proteins, are signals for protein degradation via the Proteasome. Mol Cell Biol. 1998;18:5961–5969. doi: 10.1128/mcb.18.10.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington EA, Bennett MR, Fanidi A, Evan GI. c-Myc-induced apoptosis in fibroblasts is inhibited by specific cytokines. EMBO J. 1994a;13:3286–3295. doi: 10.1002/j.1460-2075.1994.tb06630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington EA, Fanidi A, Evan GI. Oncogenes and cell death. Curr Opin Genet Dev. 1994b;4:120–129. doi: 10.1016/0959-437x(94)90100-7. [DOI] [PubMed] [Google Scholar]
- Hermeking H, Eick D. Mediation of c-Myc-induced apoptosis by p53. Science. 1994;265:2091–2093. doi: 10.1126/science.8091232. [DOI] [PubMed] [Google Scholar]
- Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, Jacks T. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- Juin P, Hueber AO, Littlewood T, Evan G. c-Myc-induced sensitization to apoptosis is mediated through cytochrome c release. Genes Dev. 1999;13:1367–1381. doi: 10.1101/gad.13.11.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juin P, Hunt A, Littlewood T, Griffiths B, Swigart LB, Korsmeyer S, Evan G. c-Myc functionally cooperates with Bax to induce apoptosis. Mol Cell Biol. 2002;22:6158–6169. doi: 10.1128/MCB.22.17.6158-6169.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamijo T, van de Kamp E, Chong MJ, Zindy F, Diehl JA, Sherr CJ, McKinnon PJ. Loss of the ARF tumor suppressor reverses premature replicative arrest but not radiation hypersensitivity arising from disabled ATM function. Cancer Res. 1999;59:2464–2469. [PubMed] [Google Scholar]
- Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci U S A. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Lawlor ER, Soucek L, Brown-Swigart L, Shchors K, Bialucha CU, Evan GI. Reversible kinetic analysis of Myc targets in vivo provides novel insights into Myc-mediated tumorigenesis. Cancer Res. 2006;66:4591–4601. doi: 10.1158/0008-5472.CAN-05-3826. [DOI] [PubMed] [Google Scholar]
- Lewandoski M, Wassarman KM, Martin GR. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr Biol. 1997;7:148–151. doi: 10.1016/s0960-9822(06)00059-5. [DOI] [PubMed] [Google Scholar]
- Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Levens D. Making myc. Curr Top Microbiol Immunol. 2006;302:1–32. doi: 10.1007/3-540-32952-8_1. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr Opin Genet Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- Moore JP, Hancock DC, Littlewood TD, Evan GI. A sensitive and quantitative enzyme-linked immunosorbence assay for the c-myc and N-myc oncoproteins. Oncogene Res. 1987;2:65–80. [PubMed] [Google Scholar]
- Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002;109:321–334. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- Penn LJ, Brooks MW, Laufer EM, Land H. Negative autoregulation of c-myc transcription. EMBO Journal. 1990;9:1113–1121. doi: 10.1002/j.1460-2075.1990.tb08217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu NC, Zimonjic DB. Chromosome-mediated alterations of the MYC gene in human cancer. J Cell Mol Med. 2002;6:151–159. doi: 10.1111/j.1582-4934.2002.tb00183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Gregory MA, Li Z, Brousal JP, West K, Hann SR. p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature. 2004;431:712–717. doi: 10.1038/nature02958. [DOI] [PubMed] [Google Scholar]
- Queva C, Hurlin PJ, Foley KP, Eisenman RN. Sequential expression of the MAD family of transcriptional repressors during differentiation and development. Oncogene. 1998;16:967–977. doi: 10.1038/sj.onc.1201611. [DOI] [PubMed] [Google Scholar]
- Rabbitts PH, Watson JV, Lamond A, Forster A, Stinson MA, Evan G, Fischer W, Atherton E, Sheppard R, Rabbitts TH. Metabolism of c-myc gene products: c-myc mRNA and protein expression in the cell cycle. Embo J. 1985;4:2009–2015. doi: 10.1002/j.1460-2075.1985.tb03885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay G, Evan GI, Bishop JM. The protein encoded by the human proto-oncogene c-myc. Proc Natl Acad Sci USA. 1984;81:7742–7746. doi: 10.1073/pnas.81.24.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkisian CJ, Keister BA, Stairs DB, Boxer RB, Moody SE, Chodosh LA. Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis. Nat Cell Biol. 2007;9:493–505. doi: 10.1038/ncb1567. [DOI] [PubMed] [Google Scholar]
- Schmitt CA, McCurrach ME, de Stanchina E, Wallace-Brodeur RR, Lowe SW. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 1999;13:2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer CA, Groudine M. Control of c-myc regulation in normal and neoplastic cells. Adv Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastrik I, Kaipainen A, Penttila TL, Lymboussakis A, Alitalo R, Parvinen M, Alitalo K. Expression of the mad gene during cell differentiation in vivo and its inhibition of cell growth in vitro. J Cell Biol. 1995;128:1197–1208. doi: 10.1083/jcb.128.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AJ, Kokontis JM, Hay N. Myc-mediated apoptosis requires wild-type p53 in a manner independent of cell cycle arrest and the ability of p53 to induce p21waf1/cip1. Genes Dev. 1994;8:2817–2830. doi: 10.1101/gad.8.23.2817. [DOI] [PubMed] [Google Scholar]
- Wagner AJ, Small MB, Hay N. Myc-mediated apoptosis is blocked by ectopic expression of bcl-2. Mol Cell Biol. 1993;13:2432–2440. doi: 10.1128/mcb.13.4.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar ARF sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.