Abstract
Protein Z-dependent protease inhibitor (ZPI) and antithrombin III (AT3) are members of the serpin superfamily of protease inhibitors that inhibit factor Xa (FXa) and other proteases in the coagulation pathway. While experimental structural information is available for the interaction of AT3 with FXa, at present there is no structural data regarding the interaction of ZPI with FXa, and the precise role of this interaction in the blood coagulation pathway is poorly understood. In an effort to gain a structural understanding of this system, we have built a solvent equilibrated three-dimensional structural model of the Michaelis complex of human ZPI/FXa using homology modeling, protein–protein docking and molecular dynamics simulation methods. Preliminary analysis of interactions at the complex interface from our simulations suggests that the interactions of the reactive center loop (RCL) and the exosite surface of ZPI with FXa are similar to those observed from X-ray crystal structure-based simulations of AT3/FXa. However, detailed comparison of our modeled structure of ZPI/FXa with that of AT3/FXa points to differences in interaction specificity at the reactive center and in the stability of the inhibitory complex, due to the presence of a tyrosine residue at the P1 position in ZPI, instead of the P1 arginine residue in AT3. The modeled structure also shows specific structural differences between AT3 and ZPI in the heparin-binding and flexible N-terminal tail regions. Our structural model of ZPI/FXa is also compatible with available experimental information regarding the importance for the inhibitory action of certain basic residues in FXa.
Keywords: Antithrombin III, Factor Xa, Homology modeling, Molecular dynamics simulation, Protein-protein docking, Protein Z-dependent protease inhibitor, Reactive center loop, Serpins
Introduction
Serpins (serine protease inhibitors) are involved in the regulation of several blood coagulation proteases and play a key role in the control of thrombosis and hemostasis [1, 2]. Protein Z-dependent protease inhibitor (ZPI) is a member of the serpin superfamily of protease inhibitors that inhibits factor Xa (FXa), factor IXa (FIXa) and factor XIa (FXIa) by different molecular mechanisms [3–7]. ZPI can produce rapid inhibition of FXa in the presence of the cofactor protein Z (PZ), negatively charged phospholipids and Ca2+ ions [3]. However, PZ is not absolutely required for inhibition, and ZPI by itself can inhibit FXa, although at a much slower rate (1,000-fold decrease in inhibition) [4]. ZPI therefore acts as an anticoagulant serpin. Low levels of either ZPI or PZ have been shown to be associated with ischemic stroke and central retinal vein or artery occlusion [8–10]. At the same time, ZPI is not the predominant physiological inhibitor of the coagulation pathway; the major function is thought to be dampening of the coagulation response prior to the formation of prothrombinase complex [5].
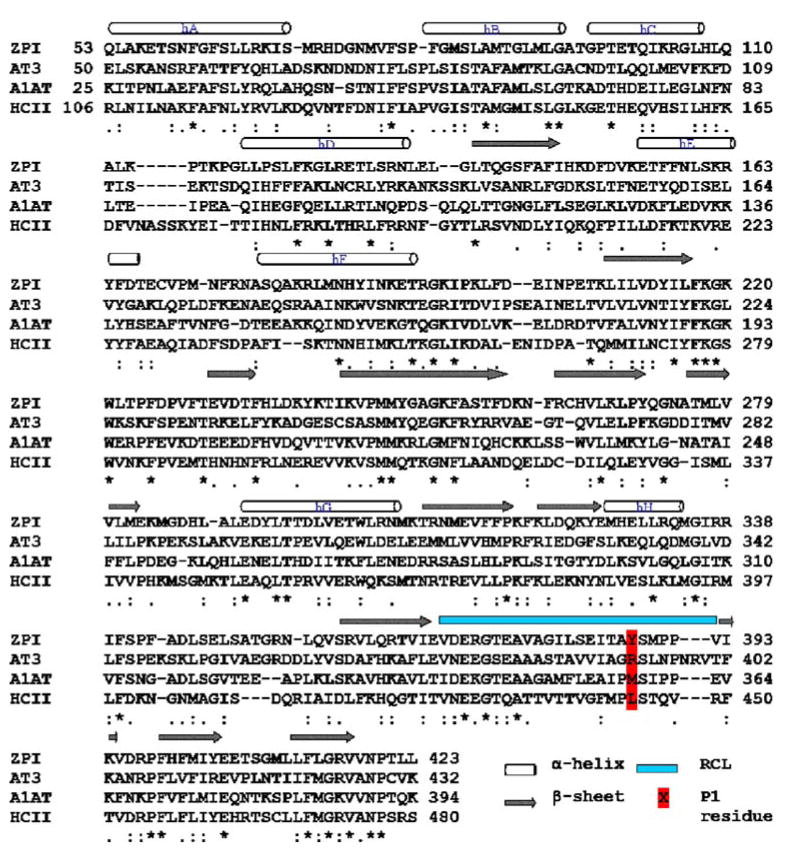
The protein structures of serpins are characterized by three β-sheets (A, B and C), 8–9 α-helices (hA–hI) and a reactive center loop (RCL) forming an extended, exposed conformation above the body of the serpin scaffold [2, 11]. From a structural point of view, ZPI can reasonably be considered to be similar to other serpins. Human ZPI is a single chain protein with 423 residues that have 26–31% sequence identity with the other serpins, including antithrombin III (AT3), α1-antitrypsin (A1AT) and heparin cofactor II (HCII) for which three-dimensional (3D) structures exist [3]. Sequence alignment of ZPI with other serpins (Fig. 1) predicts that the P1 residue at the reactive center of ZPI is a tyrosine. A study that constructed an altered form of ZPI with the P1 residue mutated to an alanine found that it lacked inhibitory activity against FXa [3].
Fig. 1.
Multiple sequence alignment of protein Z-dependent protease inhibitor (ZPI) with human antithrombin III (AT3), α1-antitrypsin (A1AT) and heparin cofactor II (HCII) using ClustalW. The N-terminal regions of the serpins share low homology and are not included in the figure. β-sheets (arrows) and α-helices (cylinders) are indicated above the alignment. Asterisks denote identity, colons strong similarity, and periods weak similarity. The reactive center loop (RCL) and the P1 residue at the reactive center are indicated
Despite the sequence similarity of ZPI with other serpins and its inhibitory activity against FXa, the structure of human ZPI is currently unresolved, with many unanswered questions about the role of ZPI in the coagulation pathway. What structural factors lead to the difference in inhibitory activity between ZPI and AT3? Is ZPI capable of binding to heparin like AT3 and HCII? How does PZ enhance the inhibitory activity of ZPI against FXa? We have recently proposed a solvent-equilibrated atomic structural model of PZ with bound Ca2+ ions [12]. In order to address the questions raised above, it would be desirable to build a realistic structural model of ZPI bound to FXa, to gain understanding of the atomic details of the interaction.
What is the nature of the PZ, ZPI and FXa interaction? We know that PZ and ZPI form a complex in plasma [13] with a Kd of ~ 7 nM [14], and we know the plasma concentration of PZ and ZPI to be 40 nM [15] and 53 nM [4], respectively. Indirect, but not structurally conclusive, evidence suggests a ternary complex on the membrane surface. If, however, we compute the plasma concentration of ZPI (53 nM) as molecules/Å3 (3.2×10−11 molecules/Å3) and compare this to the estimated surface concentration (0.5×10−7 molecules/Å3), we find that the concentration of ZPI at the surface is enhanced by a factor of ~1,600 over that in plasma. Here, we are estimating a PZ/ZPI complex to occupy 106 Å3 on the membrane surface, that approximately 10% of the surface is occupied by the complex (through the Gla domain of PZ), and that half of the PZ molecules at the surface will be bound to ZPI. These estimates are conservative so as not to bias the conclusion. Thus, following this reasoning, we are led to the simple result that a model of action of ZPI does not require a ternary complex of ZPI, PZ and FXa. Instead, a reasonable model is that PZ transports ZPI to the membrane surface, enhances ZPI concentration at the surface by more than 103-fold, where the ZPI can then bind membrane-bound FXa for its inhibitory action. Thus, we focus on a binary model of ZPI/FXa as the central inhibitory unit.
Is there good structural data on which to base a model of ZPI/FXa? Fortunately, a recent 3D X-ray crystal structure exists for a homologous system: the AT3 (S195A)/FXa/ pentasaccharide complex [16]. Although structural data for AT3 and HCII with thrombin (S195A) are also known, it is most reasonable to employ the AT3/FXa (S195A) as a primary modeling template due to thrombin versus FXa structural and sequence differences. The pentasaccharide moiety can be discarded since it is used as a heparin model in the X-ray work and the action of ZPI is known to not be enhanced by heparin [4]. Since we intend to employ a longtime molecular dynamics (MD) simulation on the initial modeled structure, significant conformational adjustments can occur to remove template bias and therefore accommodate the required structural changes in the ZPI model.
Using the sequence information for human ZPI, we have employed several computational modeling approaches to build a structural model of human ZPI in complex with FXa. Molecular dynamics simulations of both the X-ray crystal structure of the AT3/FXa complex [16] and our modeled structure of ZPI/FXa in explicitly solvated systems, followed by molecular mechanics Poisson-Boltzmann/surface area (MM-PBSA) calculations [17] were then used to estimate structural features potentially responsible for the differences in inhibitory activities between AT3 and ZPI. The derived structural model for ZPI/FXa provides new atomistic understanding of the functional role of ZPI.
Methods
Construction of ZPI/FXa model
Two different approaches were adopted to build a structural model of the ZPI/FXa complex. In the first approach, we built a model of the complex through homology modeling, starting from a multiple sequence alignment of ZPI with AT3, A1AT and HCII, and using the X-ray crystal structure of AT3/FXa (PDB code 2GD4) [16] as the single structural template. Multiple models of the complex were built using MODELLER 9v1 [18] and the best model was selected based on stereochemical and energetic evaluations. This model was then explicitly solvated and subjected to 25 ns MD simulation to obtain the final model of the complex.
In the second approach, we built a model of the ZPI/FXa complex using a protein–protein docking method, starting from a homology model of ZPI and an X-ray crystal structure of FXa. The rationale was that, although the flexible RCL region of ZPI probably interacts with the active site of FXa in a manner similar to AT3 and other serpins, the remainder of the interaction surface between ZPI and FXa could be different compared to AT3, owing to different surface complementarities and also to the slightly shorter length of the RCL, due to a three-amino-acid deletion in the C-terminal side of the reactive bond of ZPI (Fig. 1). In order to account for this possibility, protein–protein docking was used to explore ZPI binding to FXa.
Homology modeling
The amino acid sequence of human ZPI was retrieved from Swiss-Prot (UniProt entry Q9UK55) [19]. For the first approach, as mentioned above, a homology model of the ZPI/FXa complex was built using the X-ray crystal structure of AT3/FXa (PDB code 2GD4) as the single structural template. However for building a reliable sequence alignment, a multiple sequence alignment of ZPI with AT3, A1AT and HCII sequences was created with CLUSTALW [20], using the BLOSUM matrices for scoring the alignments, and this alignment (Fig. 1) was then used to build several structural models using MODELLER.
In order to build a structural model of ZPI alone for the protein–protein docking approach, a model of ZPI was built based on its homology to other serpins with known 3D structure. A multiple sequence alignment of ZPI with AT3, A1AT and HCII sequences was created with CLUSTALW (Fig. 1), and their corresponding X-ray structures (PDB entry 2ANT [21], 1QLP [22] and 1JMJ [23], respectively) were used as structural templates for the homology modeling. The main difference between the native and activated (bound) forms of serpins is in the conformation of the RCL region. The native form is usually in a compact conformation in which the N-terminal region of RCL is partially inserted into the top of β-sheet A, whereas the activated form is in an extended conformation in which the hinge region of RCL has been expelled from β-sheet A, with a release of constraints on the RCL [24]. Since our aim was to build a complex structure of activated ZPI with FXa, the RCL region of ZPI in this approach was modeled based on a proteinase-bound conformation. The structures of AT3 bound to thrombin (1TB6) [25] and FXa (2GD4) [16] in the presence of heparin were used to model the RCL region, while the remainder of the structure was modeled using all three serpins.
The obtained alignments from CLUSTALW were checked for insertions and deletions in the structurally conserved regions, especially the RCL region between the P4–P4′ residues and also fine-tuned manually. The best alignment (Fig. 1) was selected based on both the alignment score as well as the reciprocal positions of the conserved residues in the RCL region. The resulting alignments, for each case, were used as input to construct homology models of ZPI and ZPI/FXa using MODELLER. Three-dimensional models were built based on a distance restraint algorithm imposed from the multiple sequence alignment of the target sequence with the template structures, applying the CHARMM force field [26]. An optimization method involving conjugate gradients and MD with simulated annealing, available in MODELLER, was employed to minimize violations of spatial restraints. For the model building process, default parameters included in the “automodel” class were used. To guarantee sufficient conformational sampling of each active site residue, an ensemble of 30 models was built, from which the best final model was selected based on evaluation of stereochemical values from PROCHECK [27], the objective function from MODELLER and by visual inspection. The models were also assessed using the Verify3D algorithm [28, 29] to identify any region of improper folding.
Protein–protein docking
Since the serpin mechanism of protease inhibition involves the formation of an encounter complex (Michaelis complex) where the RCL region is recognized by the protease as a substrate [2], a truncated region of RCL, namely P3-P1′ residues (Thr385–Ser388), from the homology model was kept in place in the catalytic site of FXa. The remainder of the ZPI structure, except the RCL, was then docked to FXa using a modified version of the docking program FTDOCK [30]. The docking approach was based on surface complementarity with knowledge-based distance constraints applied to improve the conformational search efficiency. Explicit distance constraints were imposed in ZPI between the P3 residue (Thr385) and the C-terminal residue to sheet A (Val369) and between P1′ residue (Ser388) and Val392, based on the distances between their Cα atoms obtained from our ZPI model. One final model was picked from the docking solutions based on surface complementarity and residue pair potential scores. To pick the final model, the docking solutions were clustered based on the central position of the mobile molecule with respect to the static molecule, and the solution with the best score from the largest cluster was picked. The remainder of the RCL loop was then built using loop modeling techniques implemented in SYBYL 7.2 (Tripos) to obtain a model of the ZPI/FXa complex.
MD simulations and MM-PBSA calculations
A stepwise structure refinement for the two homology models (ZPI by itself, and ZPI in complex with FXa) was performed through a MD simulation, to obtain solvent-equilibrated models and to remove bad contacts. All MD simulations were performed using PMEMD9 in the AMBER9 [31] suite of molecular modeling programs. Force field parameters used were taken from the ff99SB forcefield included in the AMBER9 MD package. Four different systems were set up for final MD simulations: the two ZPI/FXa complex structures (one built from the homology approach and the other from the docking approach), the crystal structure of AT3/FXa complex (PDB code 2GD4)— modified to include missing N-terminal regions and to mutate the active site Ala195 of FXa back to serine—and additionally a modified version of the AT3/FXa complex structure, in which the P1 arginine in AT3 was mutated to an alanine. All structures were solvated in a 12.5 Å layer of TIP3P water molecules [32] and neutralized using Na+ or Cl− ions. Prior to structural equilibration, the systems were subjected to several stages of energy minimization and relaxation. In the first step, belly dynamics was performed on all the water molecules and counterions for 25 ps. This involved allowing motional freedom to the water molecules and the counterions to relax their positions, while the protein atoms were kept fixed. This was followed by an energy minimization of all the water molecules and counterions in 20,000 conjugate gradient steps to remove steric clashes, while the protein was held fixed. The whole system was then subjected to minimization in 1,000 steps of steepest descent, followed by 50,000 conjugate gradient steps. A stepwise, constant volume heating procedure was implemented over a 70 ps period to bring the system to 300 K, and then held at 300 K for another 100 ps, before starting constant pressure simulations for density equilibration. Equilibration times varied for different systems, and were followed by an unconstrained production run. Long-range electrostatic interactions were treated using the particle mesh Ewald (PME) method [33, 34], and a time step of 1.5 fs was used for all of the MD calculations. The ZPI/FXa complex built through the docking method was subjected to a self-guided MD simulation (SGMD) [35], to enhance conformational sampling efficiency. Since our aim was to examine whether the conformation of ZPI/FXa obtained through docking moved towards the conformation obtained through homology modeling or if it explored a different conformational path, we used SGMD to accelerate the systematic motions by applying a guiding force during the simulation. The motivation behind using accelerated conformational sampling was to obtain a qualitative idea, in a reasonable timescale, regarding the direction of movement of ZPI in the ZPI/FXa complex obtained through the docking method. A local sampling time of 2.0 ps and a guiding temperature of 1.0 K, which defines the strength of the guiding force in temperature units, were used for the SGMD method. The guiding force was applied to a part of the simulation system that included only the ZPI atoms. The stability of the system and the state of equilibration in each of the simulations were followed by monitoring the backbone root mean square deviation (RMSD) and the potential energy of the system.
The MM-PBSA method [17] was applied to estimate the free energies of binding from the molecular mechanical force field energy (EMM), and the solvation free energies (Gnonpolar + GPB) for the ZPI/FXa and AT3/FXa complex structures. This method is based on analysis of conformations obtained from MD simulations of the different systems and computing different energetic terms between the complex and both the serpin and the protease. Simulations of the serpin–protease complexes and the unbound proteins were used to extract the final 100 structural snapshots of the complexes and the unbound proteins after equilibration, which were then stripped of water molecules and counterions and the total free energy of the system is estimated as a sum of the following energy contributions,
where EMM is the forcefield energy computed from the snapshots in the MD simulation as the contribution from electrostatic (Eele) and van der Waals (EVDW) energetic interactions for the complex, Gnonpolar for the nonpolar contribution to the solvation free energy based on solvent accessible surface area (SASA) calculation, and GPB stands for the electrostatic part of the solvation free energy obtained by solving the Poisson-Boltzmann equation. Since an estimate of solute entropy is not included, the subsequent free energies are not absolute, but instead relative. Free energies of binding for the serpin–protease complex can be then estimated according to the following equation, where «X» corresponds to an average of a given value X over a sequence of snapshots taken from MD trajectories:
This procedure has been used successfully to estimate absolute [36, 37] and relative [38, 39] free energies of binding in protein–protein systems.
Results and discussion
Structural model of ZPI/FXa
The basic objective of our work was to build a 3D structural model of the ZPI/FXa complex, making use of homology information from previously resolved protein crystal structures. In one approach, the crystal structure of the AT3/FXa complex was used as the template, as ZPI shares significant homology with AT3 and also inhibits FXa through a similar serpin inhibitory mechanism [3]. The amino acid sequence alignment of human ZPI and AT3 showed 28% sequence identity (~50% similarity), when the non-conserved N-terminal tail region (residue 1–52) was excluded. All serpins, despite relatively low pairwise identity of primary structures (sometimes as low as 25%), share an extensive, common fold [2]. The sequence identities between AT3 and other blood coagulation serpins with known structure, namely HCII and A1AT are only 29% and 31%, respectively (excluding the N-terminal tail region), and yet they share significant structural similarity in their protein core. The protein core here is defined as the entire structure of the serpin excluding the peripheral elements of secondary structure, namely the N-terminal tail preceding the helix A and the approximately six residue long C-terminal tail region following sheet B. The RMSDs of the backbone atoms of AT3 with HCII and A1AT in their core regions are 1.14 Å and 1.41 Å, respectively. So, while it is generally held that greater than 30% identity between the target and template sequence is desirable for comparative modeling, in the case of serpins, a sequence identity of 28% between ZPI and AT3 offers a reasonable starting point for homology modeling.
The second model of the ZPI/FXa complex was built by first constructing a structural model of ZPI alone, using a multiple sequence alignment with AT3, HCII and A1AT (28, 29 and 32% sequence identity, respectively). The best solvent-equilibrated structural model of ZPI was then docked to FXa using protein–protein docking. Although this second model of ZPI presented a different interaction surface to FXa, and had a significantly different orientation of ZPI with respect to the first model, it was observed that, after a long self-guided MD simulation (26 ns), ZPI reoriented extensively with respect to FXa in this model and moved in the direction of the ZPI orientation in the first model, built through the homology approach from AT3/FXa. Therefore, the best model of the ZPI/FXa complex built through the first approach was used for structural refinement and analyses. It must be emphasized that there were no constraints imposed to effect this movement, and the SGMD movement in the second model could in principle have been in any direction. PROCHECK was used to calculate the ϕ–ψ angles for the Ramachandran plot, which shows that most of the ϕ–ψangles (83.0%) were located in the core region of the plot, and only 1.0% were in the disallowed region for the starting ZPI/FXa model (Fig. S1). For comparison, the same assessment was applied to the X-ray crystal structure of AT3/FXa (2GD4); it was found that 84.7% of ϕ–ψangles were located in the core regions and 0.3% were in the disallowed region. Thus, the fraction of residues in the modeled structure deviating from expected ϕ–ψRamachandran regions is comparable to that for the X-ray crystal structure of AT3/FXa. The improper geometries in the starting model were further refined using MD simulation (discussed below). The model was also assessed using the Verify3D algorithm (Fig. S2) that shows the 3D to one-dimensional (1D) scores of our model are similar to those obtained with the template structure of AT3. The values are mostly positive, except for the few residues around the P1 region that directly interact with FXa, which indicate that the structure has properly folded.
MD simulation of ZPI/FXa
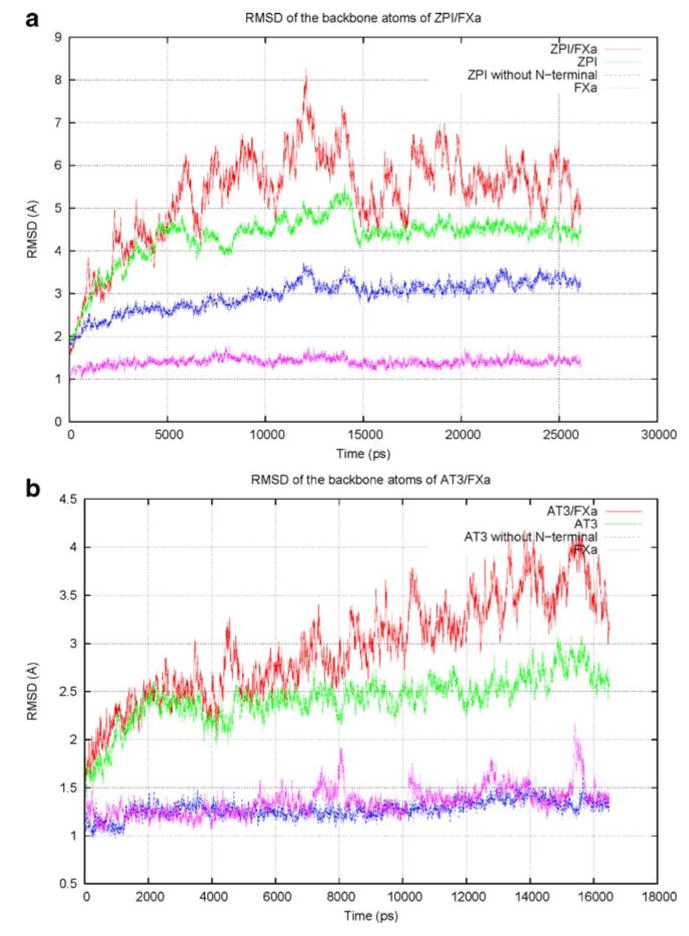
The quality of the initial model was improved by subjecting it to several stages of energy minimization and relaxation, followed by unconstrained MD production run in an aqueous environment until stable conformations with near constant RMSD values were reached. The steric clashes and improper geometries in the initial structure were removed after the MD simulation, to obtain a model with correct bond lengths and bond angles, and where individual atoms were clash-free. All of the residues in the disallowed regions of the ϕ–ψmap migrated into the allowed regions. The RMSD values comparing a series of structural snapshots of ZPI/FXa, taken at 3 ps intervals during the course of a 25 ns simulation, to the first ZPI/FXa snapshot are shown in Fig. 2a. The figure shows the backbone RMSD versus time for different structural regions of ZPI/FXa complex, calculated during the simulation. The plot shows that, after 15 ns of simulation time, the structures of ZPI and FXa have equilibrated and their backbone RMSDs were around 4.5 Å and 1.5 Å, respectively, compared to the starting structure. The high RMSD for ZPI is due to the flexible N-terminal tail region; when the N-terminal region was not included, the RMSD is approximately 3 Å from the starting structure. The N-terminal tail region is relatively unconstrained by packing, with few secondary structural features, leading to its increased flexibility. The RMSD for the overall serpin–protease complex, however, fluctuated between 4.5 and 7 Å due to the heightened mobility at the complex interface; a similar phenomenon is observed in a 17 ns control simulation of the solvated AT3/FXa X-ray crystal structure (Fig. 2b).
Fig. 2.
Plot of backbone root mean square deviation (RMSD) of a ZPI/factor Xa (FXa) over 25 ns molecular dynamics (MD) simulation using the best homology model as a reference, and b AT3/FXa over 17 ns simulation using the X-ray crystal structure (2GD4) as a reference. The crystal structure of AT3 was modified to include missing regions and to mutate the active site Ala195 of FXa back to serine
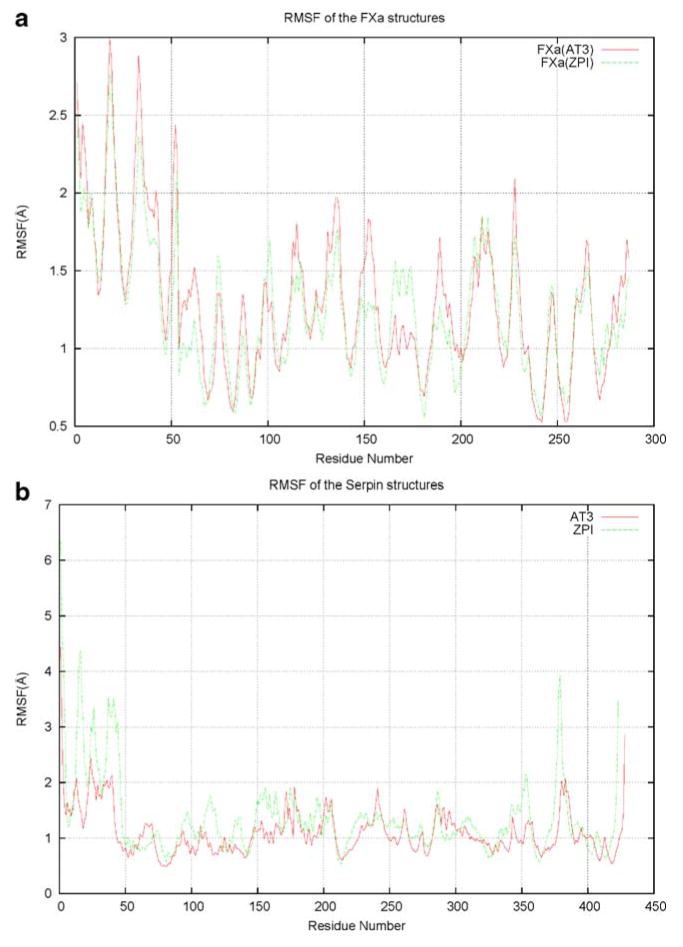
To identify the mobile structural elements, the atomic positional fluctuations for all the ZPI/FXa backbone atoms were monitored during the simulation time. A residue-based description of the local flexibility was obtained by calculating root mean square fluctuation (RMSF) values. The RMSF values describe the atomic positional fluctuations of the structure after fitting it to a reference frame, which in this case was the starting structure of the selected part of the trajectory. The difference between RMSD and RMSF values is that in the latter, the average is taken over time, giving a value for each residue (or atom) i, whereas with RMSD the average is taken over the residues, giving time-specific values. The backbone RMSF values calculated over the final 3 ns of the trajectories and averaged over each residue for the FXa (both AT3 and ZPI bound) and serpin (AT3 and ZPI) structures are shown in Fig. 3a, b.
Fig. 3.
Root mean square fluctuation (RMSF) of the backbone atoms averaged over each residue for a the FXa regions bound to ZPI and AT3, and b the serpins, ZPI and AT3
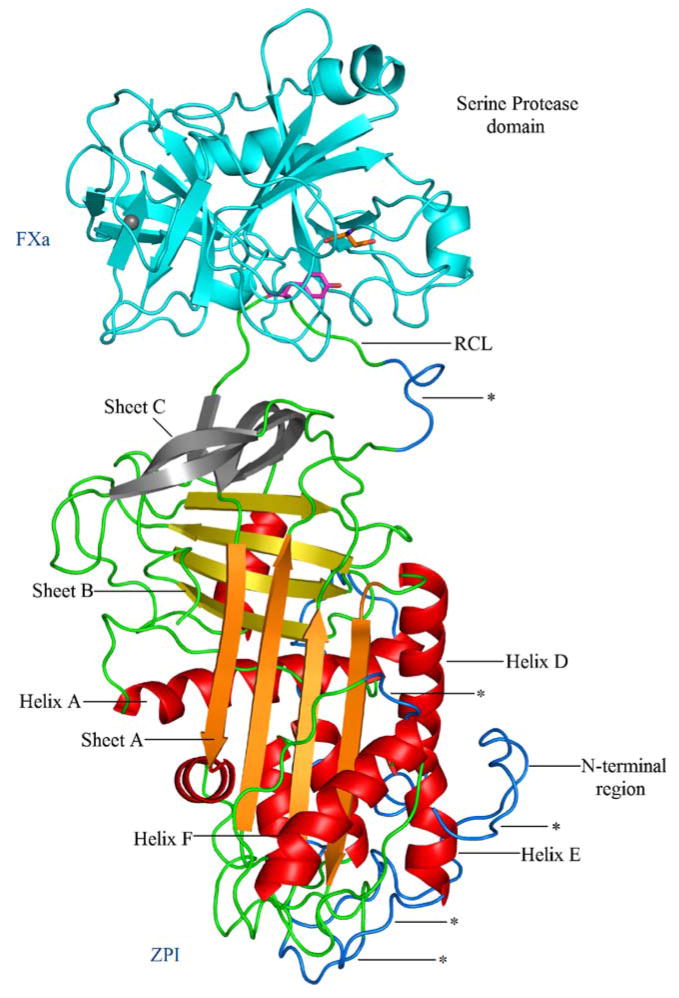
There is a significant level of similarity in the RMSF values for FXa, bound to both AT3 and ZPI (Fig. 3a). Plots of the RMSF data indicated that the largest atomic positional motions of ZPI/FXa and AT3/FXa complexes occurred in the approximately 50 amino acid residue long N-terminal tail region of the serpins (Fig. 3b). The fluctuation in ZPI for that region reaches a value of around 6.5 Å; in the same region of AT3, the fluctuation is smaller, the corresponding RMSF value being around 4.5 Å. Overall, the values of fluctuation in ZPI are slightly higher than for the AT3 structure, but the overall plot is generally similar between the two structures. The higher atomic fluctuation in ZPI is as expected, because of the absence of three disulfide bonds in ZPI compared to AT3 (at positions 8–128, 21–95 and 247–430). Other regions in ZPI that had higher RMSF values compared to their corresponding regions in AT3 include certain loop regions connecting the secondary structural elements (residues 110–118 preceding helix D, residues 149–154 located before helix E, residues 193–195 following helix F, residues 351–355 below sheet A) and residues 376–382 located N-terminal to the reactive site in the flexible RCL region. The regions in ZPI that are more flexible compared to AT3 are highlighted in Fig. 4.
Fig. 4.
Cartoon representation of the solvent-equilibrated structural model of ZPI/FXa complex. Structural elements colored in blue (also denoted by asterisks) highlight the regions of ZPI that are comparatively more flexible than in AT3, as indicated by RMSF values obtained from the simulation. The key secondary structural elements are labeled; P1 and the S1 residues are shown in stick representation
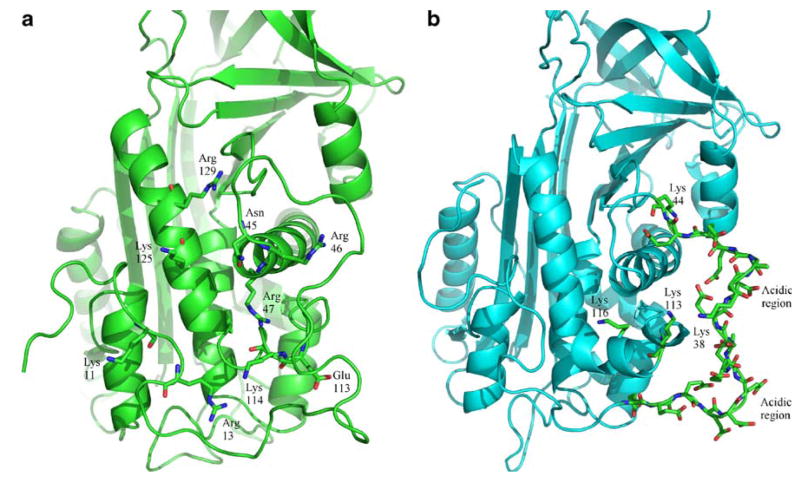
With the exception of protein C inhibitor, the heparin-binding serpins appear to use a structurally homologous region (helix D) as part of the positively charged heparin binding site [2, 40]. It was observed in a previous study that heparin did not significantly affect the ZPI inhibition of FXa in the presence of PZ, and increased the inhibitory activity to only a minor extent in the absence of PZ [4]. Comparing the heparin-binding region of AT3 to the corresponding region of ZPI shows that ZPI does not possess as many positively charged residues near helix D as in AT3 and the key residues involved in the interaction of heparin pentasaccharide with AT3 (PDB code 2GD4) are poorly conserved [41]. The surrounding region in ZPI, which contains residues 26–43, is in fact highly negatively charged and shows little homology to AT3 (Fig. 5). The acidic tail region shows some similarity to HCII, which also contains several acidic amino acid residues [22]. It is therefore likely that the weak effect of heparin on ZPI inhibition of FXa [4] is probably due to the inability of heparin to bind with significant affinity to this region.
Fig. 5.
a,b Heparin binding region. a Residues involved in binding to heparin in AT3. b Residues in the corresponding region of ZPI
The role of the flexible N-terminal tail region in ZPI is yet to be defined. The absence of two corresponding disulfide bonds in ZPI in this region compared to AT3 gives significant mobility and, unlike AT3, the movement of this flexible region is not coupled with helix D. The same two disulfide bonds are also absent in HCII, conferring significant mobility to the tail region in HCII compared to AT3. In the native structure of HCII, the N-terminal tail is sequestered through an interaction with the highly basic heparin binding region [42]. Binding of heparin releases the tail region, thus making it available for interaction with an exosite region in thrombin. In the crystal structure of the HCII-thrombin Michaelis complex, the acidic N-terminal tail forms ionic and hydrophobic contacts with exosite regions on thrombin through an allosteric mechanism [22]. In our 25 ns simulation of solvated ZPI/FXa, although there were significant atomic fluctuations and mobility in the N-terminal tail region, the tail did not move so as to interact directly with FXa. It is not clear whether this is due to inadequate sampling in our simulation or because the tail region is involved in extended intramolecular interactions with several positively charged residues in ZPI (Lys104, Arg105, Lys125, Arg133, and Arg163) that restricts its movement. Deletion of the N-terminal region in HCII abolished most of the glycosaminoglycans (GAG)-induced enhancement in inhibiting thrombin, indicating the importance of this region [43, 44]. Constructing and expressing a truncated form of ZPI without the N-terminal tail (tail deleted variant of ZPI) would similarly help to understand the function of this flexible region in inhibition of FXa and its potential role in binding to PZ.
MM-PBSA calculations
The ensembles of structural snapshots obtained from the MD simulations of ZPI/FXa and AT3/FXa were used to perform MM-PBSA calculations in order to estimate binding free energies of FXa to both ZPI and AT3. In addition, two separate MD simulations of AT3/FXa in which the P1 arginine residue in AT3 was mutated to alanine or tyrosine were performed and included in the calculations, to estimate the binding contribution from the P1 residue. These calculations resulted in a ΔGbinding of −276.66±11.30 kcal mol−1 for FXa binding to wild-type AT3, and a ΔGbinding of −262.48±11.59 kcal mol−1 for binding to wild-type ZPI. Since these calculations do not include solute entropic corrections, they provide only relative free energies. The MM-PBSA calculations in this study are used primarily to estimate a relative measure of the affinities of binding for the different complex structures, and are not intended as estimates of true binding energies. The calculations predict a stronger binding of AT3 to FXa than ZPI, with ΔGbinding of −14.18 kcal mol−1. Table 1 shows all the energy contributions for the different complexes along with the standard deviation values, as deduced from the MM-PBSA analysis. In order to further explore the difference in the relative binding affinity between AT3 and ZPI to FXa, similar calculations were performed on R393A and R393Y AT3/FXa complexes. These calculations resulted in a ΔGbinding of −243.46± 10.17 kcal mol−1 for FXa binding to R393A AT3, and a Δ3Gbinding of −264.60±10.22 kcal mol−1 for binding to R393Y AT3. The difference in the binding free energy between wild type AT3 and the P1 Ala substitution mutant (Δ3Gbinding of −33.2 kcal mol−1) is consistent with the critical role played by the Arg393 residue in the interaction with FXa. When the P1 residue in AT3 was mutated to a tyrosine (R393Y), the relative binding energy of the complex (3Gbinding of −264.60±10.22 kcal mol −1) is very close to the values observed in the ZPI/FXa complex (ΔGbinding of −262.48±11.59 kcal mol−1). These MM-PBSA results indicate that the presence of an arginine residue at the P1 position in AT3 creates a significant stabilization of the complex through an ionic interaction with the S1 residue (Asp189) of FXa, and produces a higher binding affinity than if alanine is present at the same position, or the P1 tyrosine residue present in ZPI.
Table 1.
Contribution of different energy terms and binding free energy (kcal mol−1) for wild type (wt) protein Z-dependent protease inhibitor/factor Xa (ZPI/FXa), wt human antithrombin III (AT3)/FXa, R393A AT3/FXa and R393Y AT3/FXa complexes along with their standard deviations (SD), obtained from molecular mechanics Poisson-Boltzmann/surface area (MM-PBSA) calculations
| Component | wt ZPI/FXa |
wt AT3/FXa |
R393A AT3/FXa |
R393Y AT3/FXa |
||||
|---|---|---|---|---|---|---|---|---|
| kcal/mol | SD | kcal/mol | SD | kcal/mol | SD | kcal/mol | SD | |
| ΔEele | −350.20 | ±13.63 | −378.07 | ±15.48 | −244.64 | ±13.14 | −356.32 | ±11.74 |
| ΔEVDW | −110.12 | ±5.33 | −137.40 | ±5.78 | −100.21 | ±5.49 | −114.69 | ±5.55 |
| ΔEMM = ΔEele + ΔEVDW | −460.32 | ±17.80 | −515.47 | ±20.31 | −344.85 | ±13.87 | −471.00 | ±13.55 |
| ΔGnonpolar | −206.58 | ±1.38 | −194.13 | ±1.18 | −195.72 | ±0.74 | −199.00 | ±1.00 |
| ΔGPB | 404.42 | ±11.50 | 432.94 | ±14.14 | 297.11 | ±11.48 | 405.40 | ±9.13 |
| ΔGbinding=ΔEMM + ΔGnonpolar + ΔGPB | −262.48 | ±11.59 | −276.66 | ±11.30 | −243.46 | ±10.17 | −264.60 | ±10.22 |
Specific side chain interactions
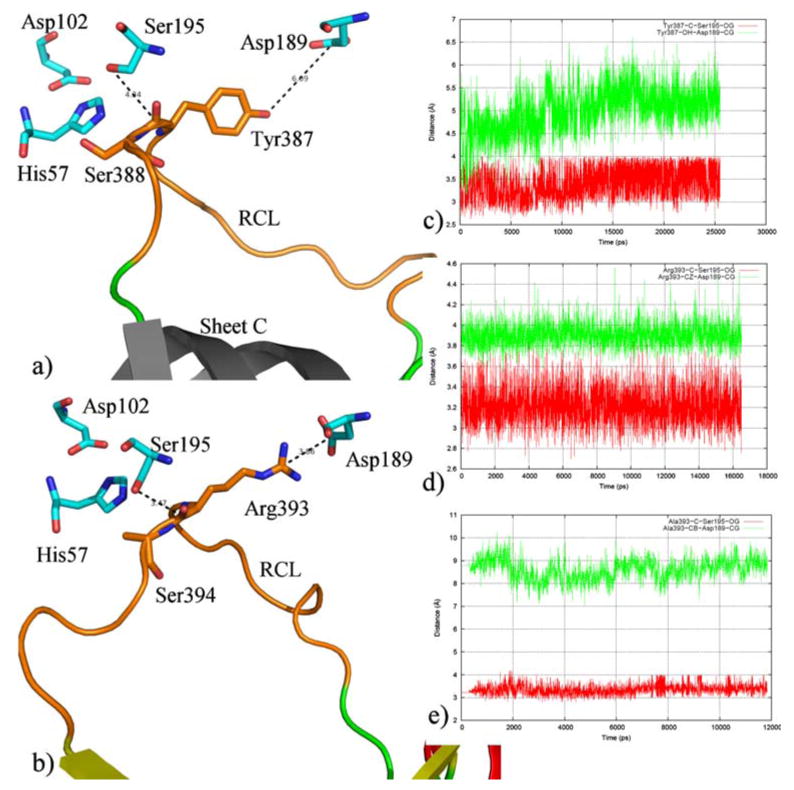
The ability to form a productive non-covalent Michaelis complex depends primarily on specific interactions between the RCL of the serpin and the active site region of the protease. But other exosite interactions with the body of the serpin have also been observed in many serpin–protease pairs. The extent of interactions between ZPI and FXa can be divided into two broad regions: the RCL region and the sheet C region of ZPI. The RCL region is responsible for the initial recognition of ZPI by FXa. In the non-covalent complex, our model has the P1 tyrosine (Tyr387) side chain positioned in the S1 specificity pocket of FXa, and the RCL is involved in extensive contacts with FXa. The contacts in the RCL region extend from residues P10 to P3′ (residues Ala378–Pro390); these form complementary interactions with subsites in and around the active site of FXa. In the case of AT3, a slightly larger set of residues (P9–P8′) in the RCL region are involved in interactions with FXa (based on solvent-equilibrated MD structure of the complex). Comparison of interaction between the critical P1 residue in ZPI and AT3 with residues in the active site of FXa, shows some differences. In ZPI, while the P1 Tyr387 is involved in several hydrophobic, main chain and side chain hydrogen-bonding interactions with FXa residues, it does not form an ionic interaction with the S1 residue (Asp189) as seen in AT3, where the P1 Arg393 forms a salt bridge with the S1 residue. In addition to this salt bridge interaction, Arg393 in AT3 is also involved in an aromatic interaction with Tyr228 in FXa; this interaction is absent in ZPI. As a consequence, the side chain–side chain distance between the P1 and S1 residues is larger in ZPI/FXa compared to AT3/FXa (Fig. 6). This is consistent with experimental results that show that ZPI does not form an SDS-stable complex with FXa, suggesting that, unlike the AT3/FXa complex, the ZPI/FXa complex has a larger dissociation constant, probably owing to weaker active site–RCL recognition interactions [3, 4]. The P1–S1 side chain distance, however, does not affect the distance between the active site Ser195 of FXa and the reactive bond in ZPI, i.e., ZPI is capable of forming the covalent acyl ester linkage that results in the cleavage of the peptide bond between Tyr387 and Ser388. The distance between Ser195 γO and the backbone carbon atom of Tyr387 of ZPI is around 3.5–4.0 Å, which is similar to the distance observed in AT3. The simulation of R393A AT3, for which the P1 arginine residue was mutated to an alanine, shows similar structural characteristics, wherein the Ala393 side-chain lacks the salt bridge interaction with the S1 Asp189 of FXa but still has the reactive bond of the serpin within 3.5 Å of the active site Ser195. This observation indicates that while the P1–S1 specificity interaction is important for the stability and the binding affinity of the complex as shown from the MM-PBSA calculations, the extended interactions between the RCL of the serpin and the active site of FXa contribute towards maintaining the non-covalent complex between the enzyme and the substrate (or inhibitor). This facilitates the subsequent formation of the covalent complex and the serpin cleavage. In other words, the effectiveness of the serpin inhibition, which depends on the formation of the covalent complex would be the same for ZPI and AT3, whereas the specificity of the interaction with FXa would be more favorable for AT3 due to the presence of an arginine at the P1 position compared to a tyrosine in ZPI. Table 2 shows a comparison of the specific FXa residues involved in interaction with the RCL region of ZPI and AT3, based on the solvent-equilibrated structural models obtained from our simulations.
Fig. 6.
Distance between P1 and S1 residues in a ZPI/FXa and b AT3/FXa. ZPI and AT3 are shown in cartoon representation with the portion of RCL region directly interacting with FXa colored orange. The P1 and P1′ residues are shown in stick representation. The catalytic triad residues and the S1 residue in FXa are also shown in stick representation (cyan) and colored by atom type. The right-hand panel shows a plot of the distances between P1 and S1 and P1 and Ser195 over the simulation time for c ZPI/FXa, d AT3/FXa, and e R393A AT3/FXa. The plots show that the P1–Ser195 distance is comparable between all three complexes, irrespective of their P1–S1 distance
Table 2.
Comparison of specific residues in FXa involved in interaction with the reactive center loop (RCL) region of ZPI and AT3. The interaction data is based on complete solvent-equilibrated structural model of the complexes
| Interaction between FXa and the RCL loop of the serpins | ||||
|---|---|---|---|---|
| Fxa | ZPI | RCL Position | AT3 | FXa |
| Arg222 | Ala378 | P10 | ||
| Arg222 | Gly379 | P9 | Ser385 | Arg222 |
| Arg222 | Ile380 | P8 | Thr386 | Gly218 |
| Arg222 | Leu381 | P7 | Ala387 | Glu217, Arg222 |
| Glu217 | Ser382 | P6 | Val388 | - |
| Phe174 | Glu383 | P5 | Val389 | Glu217 |
| Tyr99, Phe174, Trp215 | Ile384 | P4 | Ile390 | Tyr99, Phe174, Trp215 |
| Trp215, Gly216 | Thr385 | P3 | Ala391 | Trp215, Gly216 |
| His57, Ser214, Trp215 | Ala386 | P2 | Gly392 | Tyr99, Trp215 |
| His57, Ala190, Cys191, Gly193, Ser195, Val213, Ser214, Trp215, Gly216, Gly218 | Tyr387 | P1 | Arg393 | Asp189, Ala190, Gly193, Asp194, Ser195, Ser214, Gly218, Tyr228 |
| His57, Gly193, Ser195 | Ser388 | P″ | Ser394 | His57, Gly193, Ser195 |
| Phe41, Gly193 | Met389 | P″ | Leu395 | Phe41, Gly193 |
| Arg143 | Pro390 | P″ | Asn396 | Phe41 |
| P″ | Pro397 | Phe41, Ala61A | ||
| P″ | Asn398 | Asn35 | ||
| P″ | Arg399 | Asn35, Glu37 | ||
| P″ | Val400 | Glu37 | ||
| P″ | Thr401 | Glu37 | ||
In the canonical view (Fig. 4) of the non-covalent serpin complex, FXa is positioned atop the sheet C region of ZPI. So, in addition to the RCL interactions, residues in the sheet C region of ZPI, specifically residues Pro224, Phe229, Tyr249, Ala251, Gly252, Asn311, Glu313 and Phe315, are involved in interactions with FXa in this model. This is similar to interactions found in AT3, for which several residues in the sheet C region (Glu232, Asn233, Arg235, Glu237, Tyr253, Glu255 and His319) are involved in interactions with complementary residues in FXa. Similar exosite interactions are also observed in the HCII-thrombin complex, where residues in the sheet C region are involved in thrombin interactions.
Comparison with available experimental data, and location of known polymorphisms in the structural model
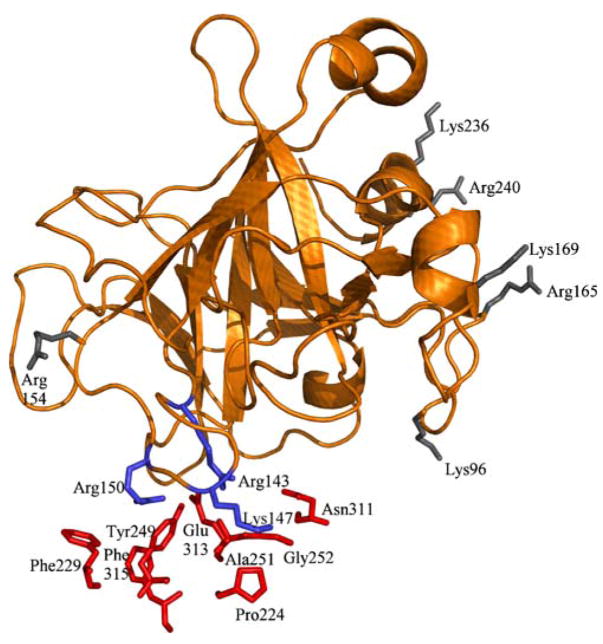
Comparison of our structural predictions with available experimental information regarding FXa residues predicted to be important for ZPI inhibitory activity [45] shows that, while our structural model agrees with some of the basic residues identified in FXa by the experiments as directly interacting with ZPI (Arg143, Lys147 and Arg150), most of the other basic residues identified in that study are present on a surface of FXa that is distant from the ZPI interaction surface in our model (Fig. 7). Therefore the significance of these other basic residues in binding to ZPI is unclear. One possibility is that, although these residues are not involved directly in interaction with the main body of the serpin, the flexible N-terminal tail region could still potentially interact with these basic residues. The N-terminal region in ZPI, which is composed of several acidic residues, is similar to the corresponding region in HCII and is unlike AT3, as it lacks the two disulfide bonds that are present in AT3 in this region. This confers additional mobility to the tail region in ZPI and could potentially enable its interactions with the basic FXa residues through an allosteric mechanism. These basic residues in FXa (Arg93, Lys96, Arg165, Lys169, Lys236 and Arg240) have also been implicated in heparin binding and are thought to be important for bridging FXa with AT3 [46]. If the N-terminal tail of ZPI is in fact involved in interactions with these basic residues of FXa, it could also explain why heparin has only minimal effect on ZPI inhibitory activity [4], since the tail region would overlap the heparin binding site on FXa. Another possibility is that, since the residues identified as important through the experimental study are based on the reaction rate constants for ZPI inhibition following their mutation, they do not correspond directly to the binding affinity of those mutants for ZPI. These basic residues in FXa could also possibly play a role in its interaction with the cofactor PZ. Table 3 lists the specific residues in ZPI and AT3 involved in interaction with the basic exosite residues of FXa, based on data obtained from the solvent-equilibrated structural model of the complexes. Experimental studies that explore the effect of mutations of these residues to their binding with PZ could shed light on the nature of binding interactions between FXa, ZPI and its non-proteinase ligand, PZ [47]. Our model thus identifies specific residues predicted to be directly involved in the recognition and binding for testing by mutagenesis experiments.
Fig. 7.
Mapping of FXa basic residues predicted to be important for ZPI inhibition [43]. FXa is shown in cartoon representation, with ZPI residues in stick representation and colored red. The residues in FXa (stick representation and colored blue) make direct contacts with ZPI residues (red), while the FXa residues colored gray are on a surface distant from ZPI
Table 3.
Residues in ZPI and AT3 involved in interaction with the basic exosite residues of FXa. The interaction data is based on complete solvent-equilibrated structural model of the complexes
| Interaction between serpins and the basic exosite residues of FXa | ||
|---|---|---|
| ZPI | FXa basic residues | AT3 |
| - | Lys96 | - |
| Glu313, Tyr249 | Arg143 | Glu255, Tyr253 |
| Pro224, Ala251, Glu375 | Lys147 | Glu232 |
| Phe229, Tyr249, Glu313, Phe315 | Arg150 | Glu237, Tyr253, His319, Asn233 |
| - | Arg154 | - |
| - | Arg165 | - |
| - | Lys169 | - |
| - | Lys236 | - |
| - | Arg240 | - |
Our structural model of ZPI/FXa also provides us the opportunity to compare with a list of mutations/polymorphisms found in a cohort of patients with venous thromboembolic diseases [48]. Of the mutations, two (R67X and W303X) are stop mutations and have been considered in subsequent studies [7, 49–51]. However, an analysis of the location of the remaining mutations shows that K25R and S40G are surface-exposed and located on the unstructured N-terminal tail, G75S is also surface-exposed and located in the loop region between helix A and helix B, S122T is surface-exposed and F124L is pointed towards the interior with both located in helix D, L137Q is surface-exposed on a loop between helix D and the first strand of sheet A, T140S is surface-exposed and is located on the first strand of sheet A, G250S is located in sheet C at the interface region between ZPI and FXa, and Q363R is located on the third strand of sheet A and is pointed towards the interior. Of these, G250S and Q363R appear to be located in structurally important regions in our model. A recent study has revealed a charge–change R67Q mutation (rather than a stop mutation) [52]; this is located surface-exposed on helix A in our structural model.
Summary
Comparative model building and solvent-equilibrated MD simulation study of the ZPI/FXa non-covalent complex points to the overall similarities and specific differences between AT3/FXa and ZPI/FXa Michaelis complexes. The atomistic structural model of ZPI/FXa gives a structural rationale as to how ZPI acts as a serpin and inhibits FXa by binding to its active site. Specific differences in the RCL and the sheet C region determine the relative specificity and stability of the ZPI/FXa non-covalent complex. The role of the flexible N-terminal region and the precise binding interaction of ZPI with PZ remain unclear. The binding interactions identified from our solvent-equilibrated structural model offer starting points for experimental verification of our model and for better understanding the interaction of ZPI with FXa.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Health (HL-06350), The intramural Program of NIEHS (Z01-ES043010-23), and National Science Foundation (FRG DMR 084549). We acknowledge the use of the computational resources provided by ITS at UNC-CH and the Biomedical Unit of the Pittsburgh Supercomputing Center. We thank our colleagues at UNC-CH for helpful conversations.
Footnotes
Conflict of interest statement The authors state that they have no conflict of interest.
Electronic supplementary material The online version of this article (doi:10.1007/s00894-008-0444-3) contains supplementary material, which is available to authorized users.
References
- 1.Rau JC, Beaulieu LM, Huntington JA, Church FC. Serpins in thrombosis, hemostasis and fibrinolysis. J Thromb Haemost. 2007;5:102–115. doi: 10.1111/j.1538-7836.2007.02516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gettins PG. Serpin structure, mechanism, and function. Chem Rev. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 3.Han X, Huang Z-F, Fiehler R, Broze GJ., Jr The protein Z-dependent protease inhibitor is a serpin. Biochemistry. 1999;38:11073–11078. doi: 10.1021/bi990641a. [DOI] [PubMed] [Google Scholar]
- 4.Han X, Fiehler R, Broze GJ., Jr Characterization of the protein Z-dependent protease inhibitor. Blood. 2000;96:3049–3055. [PubMed] [Google Scholar]
- 5.Heeb MJ, Cabral KM, Ruan L. Down-regulation of factor IXa in the factor Xase complex by protein Z-dependent protease inhibitor. J Biol Chem. 2005;280:33819–33825. doi: 10.1074/jbc.M506502200. [DOI] [PubMed] [Google Scholar]
- 6.Broze GJ., Jr Protein Z-dependent regulation of coagulation. Thromb Haemost. 2001;86:8–13. [PubMed] [Google Scholar]
- 7.Corral J, Gonzalez-Conejero R, Hernandez-Espinosa D, Vicente V. Protein Z/Z-dependent protease inhibitor (PZ/ZPI) anticoagulant system and thrombosis. Br J Haematol. 2007;137:99–108. doi: 10.1111/j.1365-2141.2007.06548.x. [DOI] [PubMed] [Google Scholar]
- 8.Vasse M, Guegan-Massardier E, Borg J-Y, Woimant F, Soria C. Frequency of protein Z deficiency in patients with ischaemic stroke. Lancet. 2001;357:933–934. doi: 10.1016/S0140-6736(00)04218-5. [DOI] [PubMed] [Google Scholar]
- 9.Heeb MJ, Paganini-Hill A, Griffin JH, Fisher M. Low protein Z levels and risk of ischemic stroke: differences by diabetic status and gender. Blood Cells Mol Dis. 2002;29:139–144. doi: 10.1006/bcmd.2002.0549. [DOI] [PubMed] [Google Scholar]
- 10.Koren-Michowitz M, Eting E, Rahimi-Levenne N, Gerach-Jehoshua O, Volcheck Y, Kornberg A. Protein Z levels and central retinal vein or artery occlusion. Eur J Haematol. 2002;75:401–405. doi: 10.1111/j.1600-0609.2005.00529.x. [DOI] [PubMed] [Google Scholar]
- 11.Law RH, Zhang Q, McGowan S, Buckle AM, Silverman GA, Wong W, Rosado CJ, Langendorf CG, Pike RN, Bird PI, Whisstock JC. An overview of the serpin superfamily. Genome Biol. 2006;7:216. doi: 10.1186/gb-2006-7-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee CJ, Chandrasekaran V, Duke RE, Perera L, Pedersen LG. A proposed structural model of human protein Z (PZ) J Thromb Haemost. 2007;5:1558–1561. doi: 10.1111/j.1538-7836.2007.02597.x. [DOI] [PubMed] [Google Scholar]
- 13.Tabatabai A, Fiehler R, Broze GJ., Jr Protein Z circulates in plasma in a complex with Protein Z-dependent protease inhibitor. Thromb Haemost. 2001;85:655–660. [PubMed] [Google Scholar]
- 14.Rezaie AR, Bae J-S, Manithody C, Qureshi SH, Yang L. Protein Z-dependent protease inhibitor binds to the C-terminal domain of protein. Z. J Biol Chem. 2008;283:19922–19926. doi: 10.1074/jbc.M802639200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miletich JP, Broze GJ., Jr Human plasma protein Z antigen: range in normal subjects and effect of warfarin therapy. Blood. 1987;69:1580–1586. [PubMed] [Google Scholar]
- 16.Johnson DJ, Li W, Adams TE, Huntington JA. Antithrombin-S195A factor Xa-heparin structure reveals the allosteric mechanism of Antithrombin activation. EMBO J. 2006;25:2029–2037. doi: 10.1038/sj.emboj.7601089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srinivasan J, Miller J, Kollman P, Case DA. Continuum solvent studies of stability of RNA hairpin loops and helices. J Biomol Struct Dyn. 1998;16:671–682. doi: 10.1080/07391102.1998.10508279. [DOI] [PubMed] [Google Scholar]
- 18.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 19.Bairoch A, Apweiler R. The SWISS-PROT protein sequence data bank and its supplement TrEMBL. Nucleic Acids Res. 1997;25:31–36. doi: 10.1093/nar/25.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignments through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skinner R, Abrahams JP, Whisstock JC, Lesk AM, Carrell RW, Wardell MR. The 2.6 Å structure of Antithrombin indicates a conformational change at the heparin binding site. J Mol Biol. 1997;266:601–609. doi: 10.1006/jmbi.1996.0798. [DOI] [PubMed] [Google Scholar]
- 22.Baglin TP, Carrell RW, Church FC, Esmon CT, Huntington JA. Crystal structures of native and thrombin-complexed heparin cofactor II reveal a multistep allosteric mechanism. Proc Natl Acad Sci USA. 2002;99:11079–11084. doi: 10.1073/pnas.162232399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elliott PR, Pei XY, Dafforn TR, Lomas DA. Topography of a 2.0 Å structure of α1-antitrypsin reveals targets for rational drug design to prevent conformational disease. Prot Sci. 2000;9:1274–1281. doi: 10.1110/ps.9.7.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cabrita LD, Bottomley SP. How do proteins avoid becoming too stable? Biophysical studies into metastable proteins. Eur Biophys J. 2004;33:83–88. doi: 10.1007/s00249-003-0356-1. [DOI] [PubMed] [Google Scholar]
- 25.Li W, Johnson DJ, Esmon CT, Huntington JA. Structure of the antithrombin-thrombin-heparin ternary complex reveals the antithrombotic mechanism of heparin. Nat Struct Mol Biol. 2004;11:857–862. doi: 10.1038/nsmb811. [DOI] [PubMed] [Google Scholar]
- 26.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMm: a program for macromolecular energy minimization and dynamics calculations. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 27.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 28.Bowie JU, Luethy R, Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253:164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- 29.Luethy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 30.Gabb HA, Jackson RM, Sternberg MJE. Modelling protein docking using shape complementarity, electrostatics and biochemical information. J Mol Biol. 1997;272:106–120. doi: 10.1006/jmbi.1997.1203. [DOI] [PubMed] [Google Scholar]
- 31.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA. AMBER. University of California; San Francisco: 2006. p. 9. [Google Scholar]
- 32.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 33.Darden T, York D, Pedersen L. Particle mesh Ewald: an N. log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 34.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 35.Wu X, Wang S. Self-guided molecular dynamics simulation for efficient conformational search. J Phys Chem B. 1998;102:7238–7250. [Google Scholar]
- 36.Gohlke H, Case DA. Converging free energy estimates: MM-PB(GB)SA studies on the protein-protein complex Ras-Raf. J Comput Chem. 2004;25:238–250. doi: 10.1002/jcc.10379. [DOI] [PubMed] [Google Scholar]
- 37.Almlöf M, Aqvist J, Smalas AO, Brandsdal BO. Probing the effect of point mutations at protein–protein interfaces with free energy calculations. Biophys J. 2006;90:433–442. doi: 10.1529/biophysj.105.073239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marinelli L, Cosconati S, Steinbrecher T, Limongelli V, Bertamino A, Novellino E, Case DA. Homology modeling of NR2B modulatory domain of NMDA receptor and analysis of Ifenprodil binding. Chem Med Chem. 2007;2:1498–1510. doi: 10.1002/cmdc.200700091. [DOI] [PubMed] [Google Scholar]
- 39.Brown SP, Muchmore SW. Rapid estimation of relative protein-ligand binding affinities using a high-throughput version of MM-PBSA. J Chem Inf Model. 2007;47:1493–1503. doi: 10.1021/ci700041j. [DOI] [PubMed] [Google Scholar]
- 40.Kuhn LA, Griffin JH, Fisher CL, Greengard JS, Bouma BN, Espana F, Tainer JA. Elucidating the structural chemistry of glycosaminoglycan recognition by protein C inhibitor. Proc Natl Acad Sci USA. 1990;87:8506–8510. doi: 10.1073/pnas.87.21.8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whisstock JC, Pike RN, Jin L, Skinner R, Pei XY, Carrell RW, Lesk AM. Conformational changes in serpins: II. The mechanism of activation of Antithrombin by heparin. J Mol Biol. 2000;301:1287–1305. doi: 10.1006/jmbi.2000.3982. [DOI] [PubMed] [Google Scholar]
- 42.Brinkmeyer S, Eckert R, Ragg H. Reformable intramolecular cross-linking of the N-terminal domain of heparin cofactor II: effects on enzyme inhibition. Eur J Biochem. 2004;271:4275–4283. doi: 10.1111/j.1432-1033.2004.04367.x. [DOI] [PubMed] [Google Scholar]
- 43.Ragg H, Ulshofer T, Gerewitz J. Glycosaminoglycan-mediated leuserpin-2/thrombin interaction. Structure-function relationships. J Biol Chem. 1990;265:22386–22391. [PubMed] [Google Scholar]
- 44.Van Deerlin VM, Tollefsen DM. The N-terminal acidic domain of heparin cofactor II mediates the inhibition of alpha-thrombin in the presence of glycosaminoglycans. J Biol Chem. 1991;266:20223–202231. [PubMed] [Google Scholar]
- 45.Rezaie AR, Manithody C, Yang L. Identification of Factor Xa Residues Critical for Interaction with Protein Z-dependent Protease Inhibitor. J Biol Chem. 2005;280:32722–32728. doi: 10.1074/jbc.M505517200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rezaie AR. Identification of basic residues in the heparin-binding exosite of factor Xa critical for heparin and factor Va binding. J Biol Chem. 2000;275:3320–3327. doi: 10.1074/jbc.275.5.3320. [DOI] [PubMed] [Google Scholar]
- 47.Patston PA, Church FC, Olson ST. Serpin-ligand interactions. Methods. 2004;32:93–109. doi: 10.1016/s1046-2023(03)00201-9. [DOI] [PubMed] [Google Scholar]
- 48.Van de Water N, Tan T, Ashton F, O’Grady A, Day T, Browett P, Ockelford P, Harper P. Mutations within the protein Z-dependent protease inhibitor gene are associated with venous thromboembolic disease: a new form of thrombophilia. Br J Haematol. 2004;127:190–194. doi: 10.1111/j.1365-2141.2004.05189.x. [DOI] [PubMed] [Google Scholar]
- 49.Razzari C, Martinelli I, Bucciarelli P, Viscardi Y, Biguzzi E. Polymorphisms of the protein Z-dependent protease inhibitor (ZPI) gene and the risk of venous thromboembolism. Thromb Haemost. 2006;95:909–910. [PubMed] [Google Scholar]
- 50.Fabbro D, Barillari G, Damante G. Mutations R67X and W303X of the protein Z-dependent protease inhibitor gene and venous thromboembolic disease: a case-control study in Italian subjects. J Thromb Thrombolysis. 2007;23:77–78. doi: 10.1007/s11239-006-9003-x. [DOI] [PubMed] [Google Scholar]
- 51.Folsom AR, Cushman M, Rasmussen-Torvik LJ, Heckbert SR, Tsai MY. Prospective study of polymorphisms of the protein Z-dependent protease inhibitor and risk of venous thromboembolism. Thromb Haemost. 2007;97:493–494. [PubMed] [Google Scholar]
- 52.Burggraf S, Dörhöfer B, Olgemöller B. Hybridization probe genotyping of the R67X nonsense polymorphism in the protein Z-dependent protease inhibitor reveals a new R67Q mutation. Clin Chem. 2007;53:1385–1387. doi: 10.1373/clinchem.2007.086421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.