Abstract
A single-molecule detection setup based on total internal reflection fluorescence (TIRF) microscopy has been used to investigate association and dissociation kinetics of unlabeled 30mer DNA strands. Single-molecule sensitivity was accomplished by letting unlabeled DNA target strands mediate the binding of DNA-modified and fluorescently labeled liposomes to a DNA-modified surface. The liposomes, acting as signal enhancer elements, enabled the number of binding events as well as the residence time for high affinity binders (Kd < 1 nM, koff < 0.01 s−1) to be collected under equilibrium conditions at low pM concentrations. The mismatch discrimination obtained from the residence time data was shown to be concentration and temperature independent in intervals of 1–100 pM and 23–46°C, respectively. This suggests the method as a robust means for detection of point mutations at low target concentrations in, for example, single nucleotide polymorphism (SNP) analysis.
INTRODUCTION
Bulk-based methods such as differential-scanning and isothermal-mixing calorimetry (1,2) combined with theoretical representations have enabled predictions of the thermodynamic stability of arbitrary DNA duplexes of up to 40 bp (3). Similarly, kinetic studies of DNA-hybridization in bulk have provided information on the rate constants characterizing the hybridization and dissociation reactions (4). The introduction of DNA microarrays in the late 1980s, where spot-size miniaturization and advanced imaging techniques today provide multiplexed readout of 105–106 targets (5), has increased the throughput and applicability of DNA analysis enormously. However, with few exceptions, these assays rely on fluorescence read-out and end-point measurements, which generally exclude information on hybridization kinetics. In this context, surface-sensitive techniques that provide information on hybridization kinetics in the absence of fluorescent labels have emerged as attractive alternatives (6,7). Information on hybridization kinetics is also relevant for detection of point mutations (SNP analysis), since single mismatches have been shown to have clearly measurable influences on hybridization dynamics (8,9). However, most surface-sensitive techniques compatible with label-free read-out of hybridization dynamics, such as impedance spectroscopy (10), quartz crystal microbalance (QCM) (6) and surface plasmon resonance (SPR) (7,11), where in the latter case parallel analysis of up to 400 different probe sequences has been reported using imaging SPR (12), suffer from relatively low sensitivity. This, in turn, yields limits of detection (LOD) in the nM regime at best. Furthermore, the relatively low LOD of most label-free techniques implies that in order to achieve detectable signals, a high density of surface-immobilized DNA probes is required (typically >10 pmol/cm2). This, in turn, may influence the hybridization reaction negatively through steric hindrance or electrostatic repulsion (13), as evident from comparisons with related data recorded in bulk (14). In addition, high surface-probe densities increase the risk of mass-transport limitations (15,16), adding requirements on analysis using multi-component kinetic models causing additional uncertainties (16,17). Hence, sufficiently sensitive sensors concepts compatible with low probe densities (preferably <0.1 pmol/cm2) but still capable of providing information on binding/unbinding kinetics without having to directly label the analyzed DNA targets, are of great interest.
A number of concepts providing detection of unlabeled DNA (as well as protein) targets with impressive sensitivity have recently been introduced, demonstrating LODs reaching down to the fM concentration regime (18–22). However, most of these methods rely on some kind of signal enhancement through, for example, silver development (19) or enzymatic amplification (20). This, in turn, limits their applicability to end-point measurements. The semiconductor nanowire-based sensor concept introduced by Lieber et al., was proven capable of detecting DNA hybridization at fM concentrations without further signal enhancement (23). However, using this principle, target concentrations in the nM regime were needed to perform kinetic measurements under physiological conditions (24). A potentially promising label-free technique with single-molecule sensitivity was developed by Howorka and co-workers (25). With an oligonucleotide covalently attached inside a genetically engineered protein pore residing in a black lipid membrane, they successfully monitored hybridization and dissociation events of single 7 and 8 bases long DNA targets using single-channel current recording. However, this approach is based on sequential measurements of single binding events, which so far restricted its applicability to high target concentrations (µM) and low affinity binders (equal to or less than eight bases).
Imaging techniques provide a solution to the problem connected with sequential readout, but in order to reach single-molecule sensitivity, the target molecules must generally be fluorescently labeled. Due to rapid photobleaching, this restricts the detection time window to a few seconds (26,27) and can thus only be applied to low affinity binders (koff > 0.1 s−1). Using a sandwich format similar to that utilized by Cao et al. for detection of DNA and RNA targets (28), we recently demonstrated that TIRF microscopy can provide parallel detection of high affinity (koff < 0.01 s−1) single hybridization events with a LOD in the low fM regime. This was accomplished by letting unlabeled DNA targets mediate the binding of fluorescently labeled liposomes to a DNA-modified surface (29). In the present work, we extend the concept (schematically illustrated in Figure 1) by performing an analysis in equilibrium of single binding events. In this way, determination of the equilibrium dissociation constant, Kd, and a thermodynamic analysis of the dissociation rate constant, koff, are shown possible from a single injection of DNA targets at low pM concentrations. Operation under equilibrium binding conditions reduces problems related to mass-transport limitations, and the single-molecule sensitivity makes the concept compatible with low surface-probe densities (<0.1 pmol/cm2). Particular focus is put on the capacity of the assay to discriminate fully complementary from single mismatch sequences. The potential of single-mismatch discrimination was evaluated in a range of target concentrations (1–100 pM) and in a broad temperature interval (23–46°C). We also propose an alternative kinetic and thermodynamic analysis of this type of data, which relies on the ratio between the number of detaching liposomes and the integrated number of bound liposomes at equilibrium binding conditions.
Figure 1.
(A) Schematic illustration of the single-molecule sensing template. An unlabeled DNA target mediates the binding of rhodamine-labeled liposomes (Ø ∼100 nm) modified with, on average, one DNA duplex with a single stranded sticky-end (15 bases) to a TIR-illuminated surface modified with single stranded DNA (15 bases). The residence time of the liposomes on the surface is monitored for the kinetic analysis. The silicon dioxide surface is modified by self-assembly of biotinylated copolymer (PLL-g-PEG/PLL-g-PEGbiotin) followed by binding of Neutravidin conjugated to biotinylated single stranded DNA. (B) Time trace of binding and dissociation of a single liposome on the surface. The purple and yellow arrows mark the binding and dissociation event, respectively. (C) Microscopy snapshot of a subsection of the illuminated area during analysis. Green crosses indicate bound liposomes. Yellow rings show liposomes detaching from the surface in the subsequent frame. Purple crosses indicate new liposomes binding in the current frame. Field of view of the subsection is 40 × 40 µm.
MATERIALS AND METHODS
Preparation of DNA-modified liposomes
Liposomes were prepared by extrusion. 99 wt% egg-PC (l-α-phosphatidylcholine, Avanti Polar Lipids, USA) and 1 wt% rhodamine-DHPE (Lissamine™ rhodamine B 1,2-dihexadecanoyl-sn-glycero-3-phosphatidylethanolamine, Invitrogen, USA) with a total lipid mass of 2.5 mg were dissolved in 1 ml chloroform in a small round beaker and subsequently dried in a nitrogen environment for at least one hour. The lipids were rehydrated in 1 ml HEPES buffer (100 mM NaCl, 10 mM HEPES, pH 7.4) and were vortexed for about 3 min. The suspension was then extruded at least 21 times through a 100 nm polycarbonate membrane (Whatman, UK) and stored at 4°C. Prior to measurements, the liposomes were modified with DNA by incubating the liposome suspension with hybridized pairs of cholesterol-terminated DNA strands (3′-cholesterol CCC AGG CAG CAC GGA ATA AAG ACT ACA GGT-5′ and 5′-cholesterol-CCC TCC GTC GTG CCT-3′, MedProbe, Norway) for at least 1 h (1:1 molar ratio). This leads to irreversible self incorporation of the cholesterol-DNA construct (30). Prior to injection, the target sequence was added to the DNA-modified liposome solution (30 pM liposomes). The protocol used to modify liposomes with DNA was kindly provided by LayerLab AB, Gothenburg, Sweden.
Surface preparation
Prior to all surface modifications, the substrate (quartz cover glass) was cleaned using piranha solution (3:1 mixture of concentrated sulfuric acid and 30% hydrogen peroxide) (Caution: piranha solution reacts very violently and should be handled with great care) followed by extensive rinsing in Milli-Q™ water (Millipore, USA) and drying under nitrogen gas. The clean substrate was placed in a custom-made flow cell, equipped with a thermocouple and a temperature probe, after which the flow cell was filled with HEPES buffer. A 10 µg/ml solution of PLL-g-PEG/PLL-g-PEGbiotin {PLL(20)-g[3.5]-PEG(2), PLL(20)-g[3.5]-PEG(2)/PEG(3.4)-Biotin (50%), Surface Solutions, Switzerland} was injected (10:1 molar ratio of PLL-g-PEG/PLL-g-PEGbiotin) and adsorbed to the surface for >30 min. After subsequent rinsing, biotinylated DNA (5′-biotin-ACG TCA GTC TCA CCC-3′) conjugated to Neutravidin (150 nM, 1:1 molar ratio, Sigma-Aldrich, Germany) was incubated for >40 min. The modified surface was subsequently exposed to egg-PC liposomes without both fluorescent lipids and cholesterol-DNA for additional prevention of unspecific binding. Rhodamine-labeled, DNA-modified liposomes were finally added together with the target DNA strands (PM: 5′-TAT TTC TGA TGT CCA GGG TGA GAC TGA CGT-3′, MM: 5′-TAT TTC TAA TGT CCA GGG TGA GAC TGA CGT-3′, MedProbe, Norway). For the measurements of non-specific interactions, no target DNA was added. HEPES buffer (100 mM NaCl, 10 mM HEPES, pH 7.4) was used in all experiments.
TIRF setup and temperature control
TIRF excitation at 530 nm was obtained using a Kr-Ar mixed-gas ion laser, coupled to the substrate using a prism and refractive index-matched immersion oil. An inverted microscope (Nikon Eclipse TE2000-U, Nikon Corporation, Japan) with a 60× water immersion objective (NA = 1.00), TRITC filter set and shutter (MAC 5000, Ludl, USA), was coupled to a cooled CCD camera (iXon, Andor Technology, Northern Ireland) to collect the fluorescence micrographs. Time-laps images were acquired for 1 h at a rate of one image every 20 s. The temperature was controlled using a Peltier element (ELFA, Sweden) and the temperature was allowed to stabilize for at least 20 min before each measurement.
Image analysis
A liposome is counted as bound if its intensity exceeds the detection threshold (Figure 1B). If the intensity of a bound liposome suddenly drops below the dissociation threshold it is considered dissociated. Since all immobilized liposomes will be slightly affected by bleaching, the drop in intensity between two subsequent frames, must be larger than a specified value (typically half of the detection threshold) to be counted as a detached liposome. A small fraction (typically a few percent) of the total population of liposomes will reach intensities below the bleaching threshold due to bleaching, rather than release. Such liposomes are considered bleached and are not included in the analysis of the dissociation events, nor in the estimation of koff,eff (see below).
Due to the finite time-span of the measurement, liposomes that bind in the late part of a measurement will have less time to dissociate from the surface than liposomes that bind at earlier stages. This is accounted for by dividing the total measurement time into two parts of equal duration. Only liposomes that bind during the first half are studied. The number of these liposomes that subsequently lose contact with the surface at various times after the binding event, up to half the total measurement time, is determined. However, for the estimation of koff,eff (see below), binding during the entire measurement interval (1 h) is used in the analysis. Note, that the kinetic extraction is independent of at which point in time the actual measurement starts, since only new liposomes that bind during the measurement are included in the analysis.
RESULTS AND DISCUSSION
Two important parameters must be considered in order to successfully monitor residence times of single molecule binding events using fluorescence imaging techniques. First, the signal-to-noise ratio must be sufficiently high and consistent throughout the entire measurement to avoid false counts. TIRF-based illumination of bright surface-bound liposomes, each containing ∼1000 (∼1%) fluorescently labeled lipids, provides a convenient solution to this problem. The high amount of fluorophores per liposome in combination with time-laps imaging also reduces bleaching and enables monitoring of single molecule residence times for up to hours. This stands in contrast to most other single-molecule assays, which are usually limited to a few seconds, or, in the case of quantum dots, weaker signals and problems connected with emission blinking. Second, each interaction should correspond to a single-molecule binding event, which in the context of the assay used in this work means that a single molecular interaction should be responsible for each liposome binding event. This can be controlled using low densities of surface-immobilized DNA probes and/or by ensuring that each liposome carries no more than one DNA probe. However, in order to observe a sufficient number of binding events to produce reliable statistics we used in this work a 1:1 ratio of DNA and liposomes and a surface–probe density sufficiently high to ensure that the maximum liposome coverage, nmax, corresponds to the jamming limit (more than one surface immobilized DNA probe per projected liposome area). Although the 1:1 ratio implies that a significant fraction of the vesicles carry more than one cholesterol–DNA, single molecular interactions will still dominate at a sufficiently low target DNA concentration. With a Kd of a few nM for a typical 15-bp DNA helix used in this work (9), the coverage of DNA targets at a bulk concentration in the low pM range is less than 1% of the maximum coverage. Hence, the number of liposomes bound with more than one DNA tether to the substrate (less than <0.5% of the total number of bound liposomes) is expected to be negligible under the current experimental conditions (see further below).
Determination of the dissociation rate constants
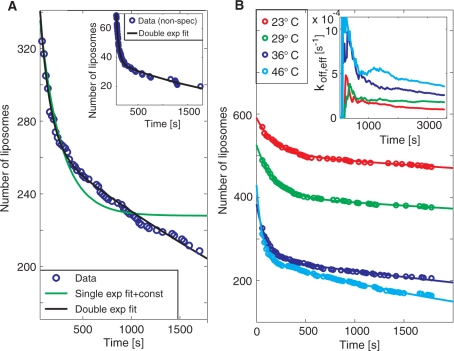
Kinetic information was extracted by following the residence time of individual DNA-target mediated liposome binding events. Figure 2A shows the number of DNA-modified liposomes that remain bound to the surface after a certain residence time in the presence of a 30–mer fully complementary target sequence at a concentration of 30 pM. Note that the difference in the number of liposomes between each time interval corresponds to the difference in magnitude of subsequent bars in a traditional residence-time histogram. Hence, although the analysis is made at equilibrium binding conditions, the data represent dissociation traces typically obtained by following the release upon rinsing in conventional affinity sensors. Note also that only liposomes that bind during the measurement time are accounted for in the analysis. In this way, liposomes that bound before the start of the measurement as well as low intensity liposomes that are below the threshold of detection (see the unmarked, low intensity, liposomes in the inset in Figure 1A) do not impair with the analysis. Figure 2B shows the same type of data generated upon subsequent heating from 23°C via 29°C and 36°C to 46°C at a single injection of DNA targets.
Figure 2.
(A) A typical dissociation trend for the fully complementary target at 36°C, showing the number of individual liposomes which still remain bound to the surface after a certain residence time (see also Figure 1B). To avoid influence from unspecific binding, liposomes that remained at the same position within the evanescent field for less than three frames (60 s) are not accounted for in the analysis. The dissociation rate was fitted with two (black line) and one, and an irreversible fraction, (green line) dissociation rate constants. From the double exponential fit ( f(t) = A1exp(−koff,1t) + A2exp(−koff,2t)) typical values for koff,1 and koff,2 were 0.007 s−1 and 1 × 10−4 s−1, respectively, at 36°C. Inset: Dissociation trend for non–specific interactions of liposomes at 23°C with koff,1 = 0.02 s−1 and koff,2 = 4 × 10−4 s−1. (B) The same type of data as in A for 23, 29, 36 and 46°C. Inset: Accumulative representation of the number of liposomes that has bound and subsequently lost contact with the surface divided with the total number of liposomes that has bound to the surface [Equation (1)], for the different temperatures.
From Figure 2A it is clear that the dissociation rate is better represented by two, rather than one dissociation rate constant, suggesting a deviation from the simple Langmuir binding model. To interpret the bi-exponential dissociation behavior, which is commonly observed in DNA kinetic studies using surface-based methods (7,9), several factors must be considered. Even though two independent hybridization events are required for target detection, release occurs as soon as one of the hybridized regions dissociates. This means that a single exponential dissociation trace is still expected, given that each of the two dissociation processes follows a single exponential trend and/or if non-specific liposome binding is sufficiently low. Non-specific binding was evaluated by adding liposomes in the absence of target DNA (inset in Figure 2A), demonstrating that the total number of binding events was less than 10% of the binding events for the complimentary target at 23°C (red curve in Figure 2B), and the dissociation rate was around 10 times faster (see figure legend). It is therefore reasonable to assume that the bi-exponential kinetics reflects either a distribution of several dissociation rates or some complex inherent process dynamics. To investigate the latter aspect, we performed kinetic Monte Carlo (MC) simulations using a coarse-grained lattice model representing the dissociation of two complementary DNA strands, when one of the strands is attached to the surface and the other strand is attached to a slowly diffusing bead (Zhdanov et al., submitted for publication). The conclusion from these simulations is that the distribution of the dissociation rates is indeed exponential. This, combined with the low density of surface-immobilized targets, suggests that the observed bi-exponential dissociation is likely due to heterogeneity in accessibility of the immobilized probe DNA molecules to bind the DNA targets. Indeed, from an evaluation of literature data, Svitel and co-workers report remarkably few surface-based systems that display binding kinetics that can be represented by an ideal binding process described by the Langmuir model (17).
Single mismatch discrimination
Even if the absolute dissociation rate constant is possibly influenced by the fact that the DNA probes are surface immobilized, the assay is still capable of discriminating a single mismatch (MM), from a perfect match (PM) (29). With the target strand being simultaneously bound to a surface-immobilized probe (with a melting point of ∼53°C) and a liposome-immobilized probe (with a melting point of ∼43°C), introduction of a single mismatch to the weaker of the two binding regions resulted in an increase of the fast and slow rate constants by a factor of 2 and 3, respectively, as summarized in Table 1.
Table 1.
Dissociation rate constants at ambient temperature (23°C) from curve fits of the residence times, koff,1 and koff,2, and the effective dissociation rate constant, koff,eff, including ± one standard deviation
| koff,1 (×10−3 s−1) | koff,2 (×10−5 s−1) | koff,eff (×10−4 s−1) | |
|---|---|---|---|
| MM | 6 ± 1 | 9 ± 0.9 | 3 ± 1 |
| PM | 3 ± 0.6 | 3 ± 0.8 | 0.9 ± 0.1 |
Although in good agreement with literature values on DNA kinetics on surfaces (6,9), the inherent complexity of this as well as most other surface-based assays call for simplified means to evaluate binding and dissociation kinetics. By analyzing single-molecule binding events in imaging mode, both the fast and slow dissociation rates can be converted into an efficient dissociation rate constant, koff,eff, by simply dividing the number of detaching liposomes from the start of the measurement at t = 0, n–(t), with an integral over the total observation time of the number of bound liposomes at the time t, n+(t), (see Supplementary Data for details):
| 1 |
Note that this expression describes an effective dissociation rate constant, koff,eff, that corresponds to (i) the sum of the individual dissociation rate constants along a chain of reversible bindings (note that this alternative way of determining an effective desorption rate constant, koff,eff is independent of the number of molecules in the chain and the expression will reduce to a normal one-step Langmuir model if only one reversible binding exist) as well as (ii) a bi-exponential process, in our case represented by koff,1 and koff,2. The temperature dependence of koff,eff is shown in the inset in Figure 2B, showing convergence towards a constant value as more statistics is collected. In agreement with both koff,1 and koff,2, koff,eff increases with increasing temperature. The absolute value, however, is somewhat larger than koff,2 but significantly lower than koff,1, and the introduction of a single mismatch leads to a reduction of koff,eff by a factor of three (Table 1). These observations suggest that koff,eff is primarily determined by the slower dissociation (koff,2). Most importantly however, this analysis shows that the single mismatch discrimination can be made from the image analysis directly, without time-consuming and sometimes uncertain fitting procedures.
Thermodynamic characterization of the dissociation reactions
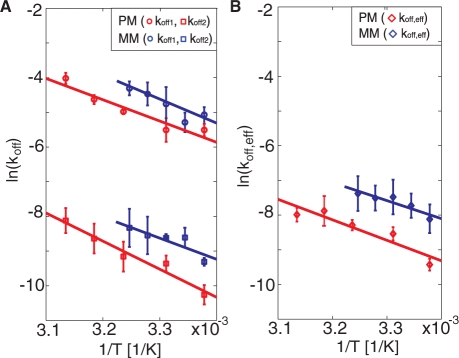
To evaluate the sensitivity of the mismatch discrimination to temperature variations, the thermodynamic parameters that govern the dissociation process was evaluated. Figure 3 shows the temperature dependence of the dissociation rate constants for the fully complementary and mismatch sequences. Since the analysis was made under equilibrium conditions, the temperature could be varied in a single experiment without addition of new DNA targets. In Figure 3, the temperature dependencies of the dissociation rate constants are displayed to facilitate an analysis using the Arrhenius expression
| 2 |
where A is the pre-exponential factor, R is the gas constant and Ea the activation energy.
Figure 3.
(A) The natural logarithm of koff,1 (circles) and koff,2 (squares) versus 1/T [Equation (2)] for the two target sequences: PM (red) and MM (blue). (B) The natural logarithm of koff,eff for the same target sequences as in A. No values are reported above 35°C for MM since at these temperatures the hybridization is too weak for sufficient statistics. Multiple temperature sweeps were performed for each DNA target. The error bars represent ± one standard deviation in dissociation rate constant from three independent measurements at each temperature.
In agreement with literature data (31), both koff,1 and koff,2 increase with increasing temperature. The temperature dependence of koff,2 of the fully complementary sequence displays a slightly larger slope than that of the single mismatch, while no significant difference in the temperature dependence was observed for koff,1. As summarized in Table 2, the corresponding activation energies vary between 52 and 81 kJ/mol. A similar trend was observed also for the effective dissociation constant, koff,eff, [Equation (1)]. Both the relative differences and the temperature dependence of koff,eff for the two target sequences are in agreement with those observed for koff,2.
Table 2.
The activation energy, Ea, determined from linear fits to the temperature dependence of the dissociation rate constants (koff,1 and koff,2) and from the effective rate constant (koff,eff), including ± one standard deviation (Figure 3)
| Ea for koff,1 (kJ/mol) | Ea for koff,2 (kJ/mol) | Ea for koff,eff (kJ/mol) | |
|---|---|---|---|
| MM | 69 ± 15 | 61 ± 17 | 52 ± 20 |
| PM | 61 ± 7 | 81 ± 10 | 59 ± 9 |
These results show that the single-mismatch and the fully complementary sequences reveal no significant differences in activation energies (Table 2), suggesting that the difference in the absolute dissociation rates has an entropic origin (31,32). Although in agreement with previous data using surface-based methods (7,31), the absolute activation energies are approximately a factor of eight lower than expected for a 15-bp duplex measured in bulk solution (33). Together with the observation of a bi-exponential dissociation behavior, this suggests that the underlying polymer support, and possibly also the liposomes, used as enhancer elements, do influence the kinetics of the dissociation reactions. However, these effects do not prohibit efficient single-mismatch discrimination in a wide temperature range.
Determination of equilibrium dissociation constants
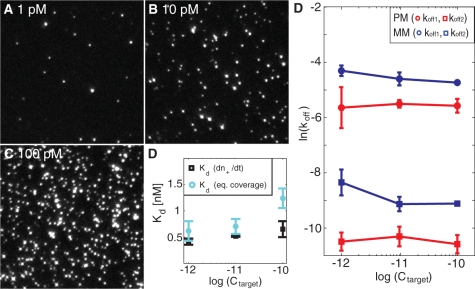
The single-particle imaging mode also provides a means to determine an effective equilibrium dissociation constant, Kd. This was accomplished by either quantifying the surface coverage at equilibrium conditions or by measuring the number of binding events per unit time, (dn+(t)/dt)eq, again from equilibrium binding conditions. Figure 4A–C show snapshots of the equilibrium coverage of liposomes at three different target concentrations (1, 10 and 100 pM), illustrating an essentially linear increase in the equilibrium coverage with a Kd of 1 ± 0.4 nM (Supplementary Data) for the fully complementary sequence (Figure 4D). Also (dn+(t)/dt)eq was observed to increase linearly with target concentration, as expected from its linear dependence on Ctarget (Supplementary Data):
| 3 |
where Cliposome is the concentration of liposome:cholesterol–DNA (30 pM), and Kd,A and Kd,B are the equilibrium dissociation constants for hybridization of the DNA target to the surface- and liposome-immobilized DNA probes, respectively. With nmax determined as the jamming limit of randomly adsorbed spheres (∼54% surface coverage) with a diameter of 100 nm, nmax becomes 69 μm−2. Under the assumption that Kd,A and Kd,B are approximately equal (Kd,A = Kd,B = Kd), the resulting equilibrium dissociation constant, Kd, becomes 0.6 ± 0.2 nM (Figure 4D) using koff,eff = 1 × 10−4 s−1 (Table 1). These values of Kd are in good agreement with literature data for a 15-mer DNA duplex (9). We stress, however, that since overlapping liposomes at high coverage lead to an underestimation of the counted number of liposomes – as illustrated by the overestimation of Kd at higher target concentrations (Figure 4D) − the uncertainty in the determination of Kd is larger than the uncertainty in the determination of koff − which is unaffected by the total fraction of monitored liposomes. This is particularly relevant with respect to mismatch discrimination, in which case the determination of Kd (and kon = koff/Kd) requires that the target concentration is known, while the determination of koff is independent of the target concentration. This is illustrated in Figure 4E, which demonstrates only a weak dependence on koff,1 and koff,2 on target concentration. If a significant fraction of the liposomes was mediated by more than one DNA target, a strong concentration dependence of the dissociation constant is expected (34). The weak concentration dependence of koff over two orders of magnitude thus strongly supports the assumption that each liposome-binding event is controlled by single DNA targets. Furthermore, in many practical situations aiming at, for example, single mismatch discrimination, such as in SNP diagnostics (35), the target concentrations are unknown. The proposed discrimination based on dissociation rate constants, shown to be independent of target concentration, may thus offer a reliable means for such analysis.
Figure 4.
(A–C) Microscopy snapshots at equilibrium coverage at three different target concentrations for the perfect match. The field of view is 50 × 50 μm yielding a coverage at equilibrium, neq, of 0.0088 μm−2, 0.057 μm−2 and 0.169 μm−2 at 1 pM, 10 pM and 100 pM, respectively. The intensity variation of the spots reflects the size distribution of the vesicles. (D) Equilibrium dissociation constant, Kd, determined from the number of bound liposomes per unit time, [dn+(t)/dt] (black) or the equilibrium coverage, neq (cyan), at different target concentrations. (E) The natural logarithm of koff,1 (circles) and koff,2 (squares) at the three different concentrations for the perfect match, PM (red), and the single mismatch, MM (blue).
CONCLUSION
We have utilized a single molecule assay to examine the kinetic and thermodynamic behavior of DNA binding and dissociation events. Kinetic information was extracted at low picomolar target concentrations, which are orders of magnitude lower than conventional surface-based instruments used for label-free kinetic measurements based on QCM or SPR. Although hundreds of individual binding events were used to produce sufficient statistics, LOD in the fM range can be reached without losing the possibility to analyze the kinetic and thermodynamic nature of the binding and release events. The assay is also compatible with array formats providing multiplexing capabilities through either bio-barcode identification or conventional spotting. The method may thus be an attractive alternative to commonly used assays for SNP analysis, such as microarrays (36) or fluorescent-bead assays (37). These methods are both sensitive to cross hybridization due to small differences in sequence for single SNPs. Hence, multiplex SNP analysis requires highly redundant probe sets for each SNP (38,39) and, in the case of fluorescent-bead assays, allele specific primer extension (ASPE) is generally used (40). SNP analysis based on difference in the rate of dissociation must not necessarily rely on enzymatic discrimination and since the target concentration must not be known a priori, there is in principle no need of more than one ‘spot’ per SNP.
We also emphasize the generic nature of the method. Liposomes can be readily modified with not only DNA, but also water-soluble proteins, and are ideally suited as hosts for proteins that naturally reside in the cell membrane of living cells. This, in combination with the capability of the method to permit measurements of dissociation kinetics at very low target concentrations for both high and low affinity interactions, points towards applicability of the method in studies of a multitude of biological processes. This holds especially true since the single molecule analysis in principle also permits investigations of the heterogeneity of individual binding sites. Although beyond the scope of the present work, which focuses on high affinity binders, this feature can be used to unravel whether deviations from a single exponential dissociation behavior, generally observed using surface-based formats, originates from surface heterogeneity or fluctuations at each specific binding site on the surface.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NanoBioMaps; INGVAR program to SSF. Funding for open access charge: Swedish Research Council for Engineering Sciences, contract numbers 2005-3140 and the INGVAR grant from the Strategic Research Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank LayerLab AB, Göteborg, Sweden, for the help with the design of the cholesterol-modified oligonucleotides.
REFERENCES
- 1.Rice SA, Doty P. The thermal denaturation of desoxyribose nucleic acid. J. Am. Chem. Soc. 1957;79:3937–3947. [Google Scholar]
- 2.Cheng YK, Pettitt BM. Stabilities of double-strand and triple-strand helical nucleic-acids. Prog. Biophys. Mol. Biol. 1992;58:225–257. doi: 10.1016/0079-6107(92)90007-s. [DOI] [PubMed] [Google Scholar]
- 3.Breslauer KJ, Frank R, Blocker H, Marky LA. Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. USA. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porschke D, Uhlenbec.Oc., Martin FH. Thermodynamics and kinetics of helix-coil transition of oligomers containing Gc base pairs. Biopolymers. 1973;12:1313–1335. [Google Scholar]
- 5.Matsuzaki H, Dong SL, Loi H, Di XJ, Liu GY, Hubbell E, Law J, Berntsen T, Chadha M, Hui H, et al. Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nat. Methods. 2004;1:109–111. doi: 10.1038/nmeth718. [DOI] [PubMed] [Google Scholar]
- 6.Okahata Y, Kawase M, Niikura K, Ohtake F, Furusawa H, Ebara Y. Kinetic measurements of DNA hybridisation an an oligonucleotide-immobilized 27-MHz quartz crystal microbalance. Anal. Chem. 1998;70:1288–1296. doi: 10.1021/ac970584w. [DOI] [PubMed] [Google Scholar]
- 7.Fiche JB, Buhot A, Calemczuk R, Livache T. Temperature effects on DNA chip experiments from surface plasmon resonance imaging: isotherms and melting curves. Biophys. J. 2007;92:935–946. doi: 10.1529/biophysj.106.097790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiche JB, Fuchs J, Buhot A, Calemczuk R, Livache T. Point mutation detection by surface plasmon resonance imaging coupled with a temperature scan method in a model system. Anal. Chem. 2008;80:1049–1057. doi: 10.1021/ac7019877. [DOI] [PubMed] [Google Scholar]
- 9.Tawa K, Knoll W. Mismatching base-pair dependence of the kinetics of DNA-DNA hybridization studied by surface plasmon fluorescence spectroscopy. Nucleic Acids Res. 2004;32:2372–2377. doi: 10.1093/nar/gkh572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao ZQ, Agarwal A, Trigg AD, Singh N, Fang C, Tung CH, Fan Y, Buddharaju KD, Kong JM. Silicon nanowire arrays for label-free detection of DNA. Anal. Chem. 2007;79:3291–3297. doi: 10.1021/ac061808q. [DOI] [PubMed] [Google Scholar]
- 11.Persson B, Stenhag K, Nilsson P, Larsson A, Uhlen M, Nygren PA. Analysis of oligonucleotide probe affinities using surface plasmon resonance: a means for mutational scanning. Anal. Biochem. 1997;246:34–44. doi: 10.1006/abio.1996.9988. [DOI] [PubMed] [Google Scholar]
- 12.Usui-Aoki K, Shimada K, Nagano M, Kawai M, Koga H. A novel approach to protein expression profiling using antibody microarrays combined with surface plasmon resonance technology. Proteomics. 2005;5:2396–2401. doi: 10.1002/pmic.200401171. [DOI] [PubMed] [Google Scholar]
- 13.Peterson AW, Heaton RJ, Georgiadis RM. The effect of surface probe density on DNA hybridization. Nucleic Acids Res. 2001;29:5163–5168. doi: 10.1093/nar/29.24.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levicky R, Horgan A. Physicochemical perspectives on DNA microarray and biosensor technologies. Trends Biotechnol. 2005;23:143–149. doi: 10.1016/j.tibtech.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Schuck P, Minton AP. Kinetic analysis of biosensor data: elementary tests for self consistency. Trends Biochem. Sci. 1996;21:458–460. doi: 10.1016/s0968-0004(96)20025-8. [DOI] [PubMed] [Google Scholar]
- 16.Schuck P, Minton AP. Analysis of mass transport-limited binding kinetics in evanescent wave biosensors. Anal. Biochem. 1996;240:262–272. doi: 10.1006/abio.1996.0356. [DOI] [PubMed] [Google Scholar]
- 17.Svitel J, Boukari H, Van Ryk D, Willson RC, Schuck P. Probing the functional heterogeneity of surface binding sites by analysis of experimental binding traces and the effect of mass transport limitation. Biophys. J. 2007;92:1742–1758. doi: 10.1529/biophysj.106.094615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao XJ, Tapec-Dytioco R, Tan WH. Ultrasensitive DNA detection using highly fluorescent bioconjugated nanoparticles. J. Am. Chem. Soc. 2003;125:11474–11475. doi: 10.1021/ja0358854. [DOI] [PubMed] [Google Scholar]
- 19.Nam JM, Thaxton CS, Mirkin CA. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 20.Goodrich TT, Lee HJ, Corn RM. Direct detection of genomic DNA by enzymatically amplified SPR imaging measurements of RNA microarrays. J. Am. Chem. Soc. 2004;126:4086–4087. doi: 10.1021/ja039823p. [DOI] [PubMed] [Google Scholar]
- 21.Zhang CY, Yeh HC, Kuroki MT, Wang TH. Single-quantum-dot-based DNA nanosensor. Nat. Mater. 2005;4:826–831. doi: 10.1038/nmat1508. [DOI] [PubMed] [Google Scholar]
- 22.Horejsh D, Martini F, Poccia F, Ippolito G, Di Caro A, Capobianchi MR. A molecular beacon, bead-based assay for the detection of nucleic acids by flow cytometry. Nucleic Acids Res. 2005;33:e13. doi: 10.1093/nar/gni015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hahm J, Lieber CM. Direct ultrasensitive electrical detection of DNA and DNA sequence variations using nanowire nanosensors. Nano Lett. 2004;4:51–54. [Google Scholar]
- 24.Bunimovich YL, Shin YS, Yeo WS, Amori M, Kwong G, Heath JR. Quantitative real-time measurements of DNA hybridization with alkylated nonoxidized silicon nanowires in electrolyte solution. J. Am. Chem. Soc. 2006;128:16323–16331. doi: 10.1021/ja065923u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howorka S, Movileanu L, Braha O, Bayley H. Kinetics of duplex formation for individual DNA strands within a single protein nanopore. Proc. Natl Acad. Sci. USA. 2001;98:12996–13001. doi: 10.1073/pnas.231434698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang SH, Shortreed MR, Yeung ES. Real-time dynamics of single-DNA molecules undergoing adsorption and desorption at liquid-solid interfaces. Anal. Chem. 2001;73:1091–1099. doi: 10.1021/ac0013599. [DOI] [PubMed] [Google Scholar]
- 27.Yao G, Fang XH, Yokota H, Yanagida T, Tan WH. Monitoring molecular beacon DNA probe hybridization at the single-molecule level. Chem. Eur. J. 2003;9:5686–5692. doi: 10.1002/chem.200304977. [DOI] [PubMed] [Google Scholar]
- 28.Cao YWC, Jin RC, Mirkin CA. Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science. 2002;297:1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 29.Gunnarsson A, Jonsson P, Marie R, Tegenfeldt JO, Hook F. Single-molecule detection and mismatch discrimination of unlabeled DNA targets. Nano Lett. 2008;8:183–188. doi: 10.1021/nl072401j. [DOI] [PubMed] [Google Scholar]
- 30.Pfeiffer I, Hook F. Bivalent cholesterol-based coupling of oligonucletides to lipid membrane assemblies. J. Am. Chem. Soc. 2004;126:10224–10225. doi: 10.1021/ja048514b. [DOI] [PubMed] [Google Scholar]
- 31.Ikuta S, Takagi K, Wallace RB, Itakura K. Dissociation kinetics of 19 base paired oligonucleotide-DNA duplexes containing different single mismatched base-pairs. Nucleic Acids Res. 1987;15:797–811. doi: 10.1093/nar/15.2.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pozhitkov AE, Stedtfeld RD, Hashsham SA, Noble PA. Revision of the nonequilibrium thermal dissociation and stringent washing approaches for identification of mixed nucleic acid targets by microarrays. Nucleic Acids Res. 2007;35:e70. doi: 10.1093/nar/gkm154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen CL, Wang WJ, Wang Z, Wei F, Zhao XS. Influence of secondary structure on kinetics and reaction mechanism of DNA hybridization. Nucleic Acids Res. 2007;35:2875–2884. doi: 10.1093/nar/gkm177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mammen M, Choi SK, Whitesides GM. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. 1998;37:2755–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 35.Schork NJ, Fallin D, Lanchbury JS. Single nucleotide polymorphisms and the future of genetic epidemiology. Clin. Genet. 2000;58:250–264. doi: 10.1034/j.1399-0004.2000.580402.x. [DOI] [PubMed] [Google Scholar]
- 36.Chee M, Yang R, Hubbell E, Berno A, Huang XC, Stern D, Winkler J, Lockhart DJ, Morris MS, Fodor SPA. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- 37.Lawrie GA, Robinson J, Corrie S, Ford K, Battersby BJ, Trau M. Multiplexed microsphere diagnostic tools in gene expression applications: factors and futures. Int. J. Nanomed. 2006;1:195–201. doi: 10.2147/nano.2006.1.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sapolsky RJ, Hsie L, Berno A, Ghandour G, Mittmann M, Fan JB. High-throughput polymorphism screening and genotyping with high-density oligonucleotide arrays. Genet. Anal. Biomol. Eng. 1999;14:187–192. doi: 10.1016/s1050-3862(98)00026-6. [DOI] [PubMed] [Google Scholar]
- 39.Hacia JG, Fan JB, Ryder O, Jin L, Edgemon K, Ghandour G, Mayer RA, Sun B, Hsie L, Robbins CM, et al. Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nat. Genet. 1999;22:164–167. doi: 10.1038/9674. [DOI] [PubMed] [Google Scholar]
- 40.Bortolin S, Black M, Modi H, Boszko I, Kobler D, Fieldhouse D, Lopes E, Lacroix JM, Grimwood R, Wells P, et al. Analytical validation of the tag-it high-throughput microsphere-based universal array genotyping platform: application to the multiplex detection of a panel of thrombophilia-associated single-nucleotide polymorphisms. Clin. Chem. 2004;50:2028–2036. doi: 10.1373/clinchem.2004.035071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.