Abstract
The African trypanosome, Trypanosoma brucei, has been shown to undergo genetic exchange in the laboratory, but controversy exists as to the role of genetic exchange in natural populations. Much of the analysis to date has been derived from isoenzyme or randomly amplified polymorphic DNA data with parasite material from a range of hosts and geographical locations. These markers fail to distinguish between the human infective (T. b. rhodesiense) and nonhuman infective (T. b. brucei) “subspecies” so that parasites derived from hosts other than humans potentially contain both subspecies. To overcome some of the inherent problems with the use of such markers and diverse populations, we have analyzed a well-defined population from a discrete geographical location (Busoga, Uganda) using three recently described minisatellite markers. The parasites were primarily isolated from humans and cattle with the latter isolates further characterized by their ability to resist lysis by human serum (equivalent to human infectivity). The minisatellite markers show high levels of polymorphism, and from the data obtained we conclude that T. b. rhodesiense is genetically isolated from T. b. brucei and can be unambiguously identified by its multilocus genotype. Analysis of the genotype frequencies in the separated T. b. brucei and T. b. rhodesiense populations shows the former has an epidemic population structure whereas the latter is clonal. This finding suggests that the strong linkage disequilibrium observed in previous analyses, where human and nonhuman infective trypanosomes were not distinguished, results from the treatment of two genetically isolated populations as a single population.
Human African sleeping sickness in East Africa is caused by the trypanosome Trypanosoma brucei rhodesiense, which occurs in a series of discrete historical foci of disease distributed throughout the range of the insect vector of the parasite, the tsetse fly. The disease is epidemic in nature with recurrent outbreaks in which large numbers of humans are infected interspersed with periods of relatively low incidence of the disease (1). The disease is a zoonosis with cattle and wild game harboring human infective trypanosomes (T. b. rhodesiense) as well as nonhuman infective T. b. brucei.
T. brucei stocks undergo sexual recombination in the laboratory (3–5), but there is considerable controversy as to the extent to which mating takes place in natural trypanosome populations (2, 6, 7, 8). There is an urgent need to resolve this controversy because of the resurgence of sleeping sickness, the frequent observation of drug resistance (9), and the finding that while all strains are infective for reservoir hosts of this zoonotic parasite, only certain strains can infect humans (1). A high level of sexual recombination would increase the diversity within a population (thus creating a wide range of genotypes) and so provide the means for the organism to adapt to changes in its environment. Significant levels of genetic exchange also would mean that traits such as human infectivity and drug resistance could be spread through a population of nonhuman infective trypanosomes.
Analysis of the population genetic structure of T. brucei, using a variety of different markers, has generated conflicting results and interpretations. Three different and opposing genetic structures have been proposed: Tait has proposed a panmictic population structure in which sexual recombination is frequent (6), Tibayrenc et al. (7) have proposed a clonal theory of population genetics where sexual recombination is not frequent enough to break the prevalent pattern of clonal population structure, whereas Maynard-Smith et al. (8) suggested an epidemic population structure where there is a background level of frequent sexual recombination with the occasional clonal expansion of a few particular genotypes.
An important complication of the analysis of isolates from tsetse and domestic animals in East Africa is that there are two subspecies of T. brucei (1), the human infective T. b. rhodesiense and the nonhuman infective T. b. brucei, and these hosts may well contain more than one subspecies. It is unknown whether these subspecies are genetically isolated but, if they are, this would potentially distort the conclusions that have been reached by treating both subspecies as a single population (7, 10). In the most detailed analysis to date, Hide et al. (2) analyzed trypanosomes isolated from cattle and humans whereby each isolate was analyzed for resistance to human serum, a method which distinguishes between human infective (T. b. rhodesiense) and nonhuman infective (T. b. brucei) trypanosomes. Analysis of isoenzyme variation revealed that human infective genotypes appeared to have an epidemic population structure whereas the nonhuman infective stocks appeared to be randomly mating. A cluster analysis, using restriction fragment length polymorphism patterns of repetitive sequences, showed that the human infective stocks formed a distinct homogeneous group separate from the T. b. brucei isolates. The markers used in this analysis suffered from two disadvantages. First, because of the repetitive nature of the DNA probes, the molecular fingerprint data cannot be interpreted genetically, and second, the parasites need to be amplified in mice and purified before marker analysis. To overcome these disadvantages, we have analyzed the population structure of T. brucei by using hypervariable minisatellite markers applied to a substantial set of isolates from a single geographical region, including those used by Hide et al. (2). The analysis thus provides a direct comparison between marker systems and independently tests the conclusions of Hide et al. about the population structure.
The data obtained have enabled us to resolve the key questions required to improve our understanding of the nature of sleeping sickness epidemics and the sources of human infection, first, the role of genetic exchange in determining population structure and second whether T. b. rhodesiense and T. b. brucei are genetically distinct. Two further critical questions were addressed, namely: how stable are the human infective genotypes in time and how widely they are distributed in different foci of human sleeping sickness? The results obtained validate the use of minisatellite markers for the analysis of T. brucei populations and provide insights into the population genetics of T. brucei.
Materials and Methods
Parasite Isolates.
Seventy-six Ugandan samples (isolated between 1988 and 1990) and tested for their ability to resist the lytic effect of human serum have been described in Hide et al. (2). Five Ugandan human samples isolated between 1960 and 1982 (10) and 11 samples isolated from the salivary glands of tsetse flies in 1969–1970 (11) also were examined for comparison but were not included in subsequent analysis. The above samples all were collected from the same region (Busoga) of Uganda. The 19 Zambian stocks, isolated between 1981 and 1983, have been described in Godfrey et al. (10). The 24 human isolates from Nyanza were collected by the East African Trypanosomiasis Research Organization in 1961, some of which have been described in Tait et al. (12). A full list of isolates used in this study is available as Table 3, which is published as supplemental data on the PNAS web site, www.pnas.org.
Single-Locus Minisatellite Analysis.
Primers and PCR conditions for the amplification of the minisatellite loci, cysteine-rich acidic integral membrane (CRAM), 292, and MS42 are as described (13). Allele band sizes for CRAM and 292 were determined by using Kodak 1D IMAGE ANALYSIS software, on the basis of mobilities relative to a reference standard lane (restriction fragments of λ HindIII and φx174 HaeIII). Repeated measurements of the size of the same allele were used to calculate the mean value and the standard deviation; two standard deviations corresponded to 2.3% of the estimated allele size. This measurement then was taken as the window size for every band measurement (±2.3% band size), and alleles were considered as identical if they were within this window. MS42 alleles were unequivocally identified by using minisatellite variant repeat mapping (A.M., unpublished work), based on the method described for human minisatellites (14). The genotypes for each minisatellite marker of each isolate are available in Table 3.
Analysis of Genetic Diversity.
Measures of genetic distance, agreement with Hardy-Weinberg equilibrium and linkage disequilibrium were calculated by using the genetic data analysis program (http://chee.unm.edu/gda/). The index of association (IA) was calculated as described by Maynard-Smith et al. (8). For this analysis 9 of 10 isolates that had not been tested for human serum resistance were included in the human serum-sensitive (HSS) group based on genotype analysis and the remaining isolate was included in the human serum-resistant (HSR) group as it contained genotype M5, which is solely associated with the HSR genotype in typed isolates.
Results
Allele and Genotype Analysis.
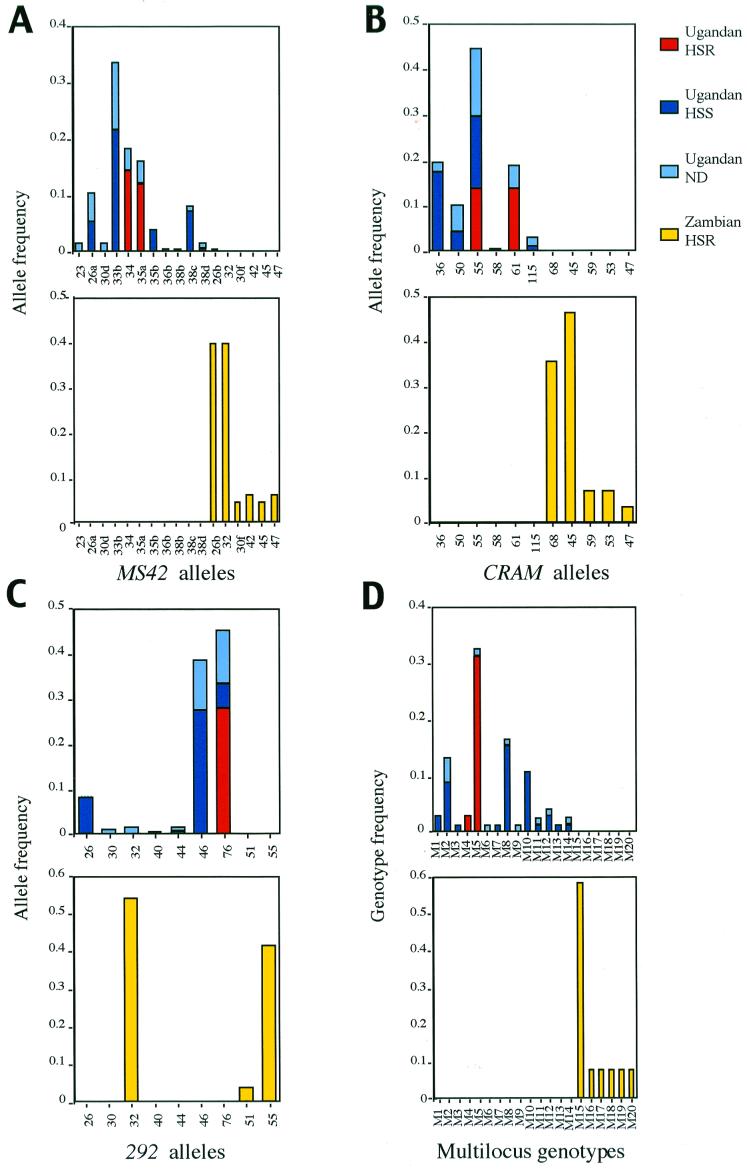
Each isolate was typed for the minisatellite markers, CRAM and 292, as described in MacLeod et al. (13), and allele size estimates revealed 16 different alleles for both minisatellites. Unequivocal allele identification for minisatellite MS42 was performed by using the minisatellite variant repeat mapping technique. In the isolates analyzed, 23 different MS42 alleles were identified by using minisatellite variant repeat-PCR compared with 17 alleles, which were identified by band size measurements alone. The number of alleles and the frequency of each was determined for the three collections of isolates (Zambian human; Ugandan human and HSR cattle isolates, T. b. rhodesiense; Ugandan HSS cattle isolates, T. b. brucei), and the results are presented in Fig. 1 A–C. Although the majority of the cattle isolates had been tested for human serum resistance, a minority had not. The Zambian population show a high frequency of two alleles at each locus and, in the case of CRAM and MS42, these alleles are exclusive to this population (Fig. 1 A and B). Comparison of the human and nonhuman infective isolates from Uganda (Busoga) show that the former only contain one or two alleles at each locus and, in the case of MS42, these alleles are exclusive and not shared with those of the nonhuman infective isolates (Fig. 1A). Combining the data for alleles at each of the three loci, for each isolate in turn, led to the definition of a series of multilocus genotypes and the results of this analysis are illustrated in Fig. 1D. Comparing the Zambian isolates with those from Uganda shows that the multilocus genotypes are exclusive to each geographical region, indicating that the two populations were totally distinct, and that the Zambian isolates primarily comprise a single multilocus genotype. Comparison of the human infective (T. b. rhodesiense) with the nonhuman infective isolates in a single geographical region (Busoga) shows that the two types can be distinguished with T. b. rhodesiense comprising of two discrete multilocus genotypes with one of these occurring at high frequency. In contrast, the nonhuman infective cattle isolates (T. b. brucei) are much more diverse, consisting of 12 different multilocus genotypes of varying frequency such that the two subspecies can be unambiguously differentiated. On this basis, the cattle isolates that have not been typed for human serum resistance can be assigned as either T. b. brucei or T. b. rhodesiense.
Figure 1.
The frequency of (A) MS42, (B) CRAM, and (C) 292 alleles in the Ugandan and Zambian populations. ND, not determined. (D) The frequency of multilocus genotypes for each population. Multilocus genotypes are given in Table 3.
Mixed T. brucei Infections.
All three minisatellite markers used in this study have heterozygosities of approximately 80%. Thus for the majority of stocks two alleles can be identified for each marker. In a significant proportion, nine of 50 (18%), of the 1988–1990 cattle isolates from Busoga three or more alleles were detected for at least one marker, indicating that at least two genetically distinct trypanosomes coexisted in the animal's bloodstream. A similar high level of mixed genotype infections has been previously detected by using these minisatellite markers in tsetse flies (13). Three mixed human isolates also have been detected; however, the possibility of laboratory contamination cannot be excluded for these isolates. All mixed samples were excluded from the population genetic analysis.
Population Structures.
Genotype frequency data were analyzed by standard techniques to determine deviation from Hardy–Weinberg equilibrium by using the genetic data analysis program (http://chee.unm.edu/gda/), and the results are presented in Table 1. It is clear that both the Zambian and Ugandan T. b. rhodesiense populations and the Ugandan T. b. brucei population show deviation from the genotype frequencies expected if each population was randomly mating. These data cannot be reconciled with the proposal of a panmictic population structure (6) but are unable to distinguish between clonal and epidemic structures.
Table 1.
Hardy–Weinberg and IA analysis for each population
| Population | n(ET) | Probability of
agreement with Hardy–Weinberg equilibrium
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
MS42
|
292
|
CRAM
|
IA
|
||||||
| All | ETs | All | ETs | All | ETs | All | ETs | ||
| Zambia, HSR | 19 (6) | 0.00 | 0.00 | 0.02 | 0.64 | 0.00 | 0.02 | 1.447 | −0.489 |
| Uganda, HSR | 23 (2) | 0.00 | NA | 1.00 | NA | 0.00 | NA | 1.539 | NA |
| Uganda, HSS | 41 (13) | 0.04 | 0.55 | 0.00 | 0.00 | 0.00 | 0.29 | 1.129 | −0.229 |
| Uganda, total | 64 (15) | 0.00 | 0.31 | 0.00 | 0.00 | 0.00 | 0.36 | 1.340 | −0.548 |
For this analysis nine of 10 isolates that had not been tested for human serum resistance were included in the HSS group based on genotype analysis. The remaining isolate was included in the HSR group as it contained the HSR genotype (M5). n, Number of isolates; NA, not applicable. Bold figures indicate agreement with Hardy–Weinberg equilibrium, i.e., probability of >0.05 or negative values of IA.
If deviation from Hardy–Weinberg equilibrium in the T. b. rhodesiense and T. b. brucei populations was caused by an epidemic population structure (2, 8), the removal of the most frequent genotypes from the analysis can be used to reveal the underlying population structure (8). To investigate this, the minisatellite data were reanalyzed by treating isolates with the same multilocus genotype as one individual or electrophoretic type (ET). For the Ugandan T. b. rhodesiense population only two highly related genotypes are present (see Table 2), unequivocally indicating that the population is clonal and so does not require any further analysis (Fig. 1D). Examination of the Ugandan T. b. brucei populations using ETs only, revealed, for two of three markers, that the population was in agreement with Hardy–Weinberg predictions, thus indicating that genetic exchange and recombination could be occurring in this population, but that the population structure was distorted by an epidemic spread of one or two genotypes (Table 1). By inspection of the data, in terms of the numbers of distinct alleles and their frequencies, this is clearly because of a predominance in the population of three common genotypes (M2, M8, and M10; Fig. 1D). Similarly, examination of ETs for the Zambian T. b. rhodesiense population indicated that one marker, 292, was found to be in agreement with Hardy–Weinberg expectations, although markers CRAM and MS42 remained in disagreement with the expected values, suggesting a clonal population structure. Indeed examination of the histogram of multilocus genotypes reveals that one major genotype dominates this population (genotype M15; Fig. 1D).
Table 2.
Genotypes of human infective Ugandan (Busoga) samples
| Year in which genotype was detected | Genotypes
|
|||
|---|---|---|---|---|
| MS42 | CRAM | 292 | Multilocus | |
| 1959, 1960, 1981, 1982 | 35a /34 | 61 /55 | 76 /44 | M21 |
| 1976 | 35d /34 | 61 /61 | 76 /44 | M22 |
| 1989 | 34 /34 | 61 /55 | 76 /76 | M4 |
| 1988, 1989, 1990 | 35a /34 | 61 /55 | 76 /76 | M5 |
The CRAM and 292 genotypes for human infective trypanosomes are given as estimates of the number of repeats in each allele. For minisatellite MS42 the genotypes are based on the number of repeats and the minisatellite variant repeat internal maps of each allele (A.M., unpublished work). The multilocus genotype, i.e. the combined results from the three minisatellites, are given. Each genotype was assigned an arbitrary number, prefixed M.
As an alternative approach to distinguish different population structures, the detection of linkage disequilibrium was calculated for each population by using the IA as described by Maynard-Smith et al. (8). The IA measures the association of alleles at different loci and has a predicted value of zero (or a negative value) for populations that are randomly mating, whereas if recombination is rare or absent the IA is large with a value significantly different from zero. The IA values for all populations are presented in Table 1, with the Zambian and Ugandan T. b. rhodesiense (HSR) populations and the Ugandan T. b. brucei population having large positive IA values, indicating that they are not randomly mating populations.
To investigate whether the large IA values for the Zambian T. b. rhodesiense population and the Ugandan T. b. brucei population were caused by an epidemic population structure, the minisatellite data were reanalyzed by using ETs only. The results revealed that the IA value for the Zambian T. b. rhodesiense population was greatly reduced, becoming negative, thus indicating that the population structure was distorted by an epidemic spread of one genotype (Table 1). Re-examination of the IA using ETs for the Ugandan T. b. brucei population also resulted in a negative value, indicating an epidemic population structure.
The use of ETs reduces the number of individuals in the analysis substantially, and coupled with the use of only three markers means that it is difficult to totally exclude the null hypothesis that these populations are unaffected by recombination. However, two additional points support our conclusion. First, it is difficult to explain the combinations of alleles on the null hypothesis (limited or no recombination), and second, similar conclusions were reached by Hide et al. (2) using independent loci.
T. b. rhodesiense Is Clonal.
It is clear from the genotype frequencies displayed in Fig. 1D that the Ugandan T. b. rhodesiense population consists of only two distinct genotypes (genotypes M4 and M5) with genotype M4 being detected twice whereas genotype M5 was detected 20 times. Five samples previously isolated from humans from the same area in Uganda between 1960 and 1982 were of highly related genotypes (genotypes M21 and M22), sharing the same basic complement of alleles. The different genotypes found in the Ugandan human infective stocks from different time points are presented in Table 2. The relationship between these genotypes suggests that they could have originated by self-fertilization; for example, genotype M21, which was first detected in this area in 1959, is heterozygous for alleles at MS42 and 292 could have self-fertilized to produce genotype M4, which is homozygous at both loci, and genotype M5, which is homozygous at minisatellite locus 292 (the major genotype detected between 1988 and 1990). Self-fertilization has been described in T. brucei (15, 16). It would appear that for the Busoga (Ugandan) focus at least, human infectivity is associated with one genotype and its predicted self-fertilization products, implying little or no recombination between the human infective and nonhuman infective stocks. The results presented here suggest that human infective stocks are clonal in origin, with stocks from just one lineage being able to infect humans, perhaps suggesting that human infectivity is an acquired attribute, which has not as yet, or cannot, spread through the rest of the T. brucei population by sexual recombination and that T. b. rhodesiense and T. b. brucei are completely distinct.
A T. b. rhodesiense Specific Marker for the Ugandan focus.
Results from this analysis indicate that the human infective samples from Uganda are very homogeneous, with 20 of 22 stocks isolated in 1988–1990 having the MS42 genotype 35a/34 (Table 2). The remaining two human infective stocks were homozygous for the 34 allele. One other stock isolated in 1976 had the genotype 35d/34, the 35d allele being only one repeat unit different from the 35a allele (A.M., unpublished work). It is clear from these data that there is an association between the presence of allele 34 in the MS42 genotype and human infectivity, because no nonhuman infective stocks had this allele (see Table 3). This suggests that the human infective Busoga samples can be defined by the presence of this allele i.e., this allele constitutes a Ugandan T. b. rhodesiense-specific marker.
Comparison of Ugandan and Zambian Samples.
A direct comparison of human isolates from Zambia with human isolates from Uganda does not identify any alleles in common (Fig. 1). These data suggest that the human isolates from the Zambian population are distinct from the human isolates from the Ugandan population. This indicates that T. b. rhodesiense samples from different areas may not be closely related, in contradiction to their classification in the same subspecies. Indeed, the T. b. rhodesiense Busoga samples appear to be more closely related to (although still distinct from) T. b. brucei Busoga samples than to the Zambian T. b. rhodesiense samples. This was confirmed by measurement of the genetic distance (17) between the populations (Ugandan T. b. rhodesiense/T. b. brucei 0.541; Ugandan T. b. rhodesiense/Zambian T. b. rhodesiense 0.787).
How Widespread and Persistent Is the T. b. rhodesiense Genotype?
The majority of samples were isolated from the Busoga region between 1988 and 1990, with the exception of five samples that were isolated from humans at earlier time points (1959, 1960, 1976, 1981, and 1982). Two multilocus genotypes were identified from the earlier samples (genotype M21 and M22, see Table 2), neither of which were detected in the large collection of samples from 1988–1990. The allelic relationship between the genotypes suggests that genotype M21 is the ancestral human infective genotype from which the other genotypes were derived, giving rise to genotypes M4 and M5 (probably by self-fertilization), which are the only human infective genotypes detected in the 1988–1990 Busoga samples.
Analysis of a collection of 24 samples, isolated from humans during an outbreak of sleeping sickness in 1961 in Nyanza in Kenya, approximately 100 km from Busoga in Uganda, revealed that T. b. rhodesiense genotype M21 was the main genotype prevalent in this focus with 20 of 24 human infective stocks being of this genotype (see Table 3). The data suggest that in the early 1960s genotype M21 predominated and was common to both foci. This genotype also was detected in a series of tsetse samples isolated from the Busoga region in 1969–1970 (see Table 3). Subsequently in the Busoga epidemic of the late 1980s, the predominant genotype was the highly related genotype M5, which only differs from genotype M21 by being homozygous for allele 76 at the 292 locus. This indicates that the common T. b. rhodesiense genotype (genotype M21 or the highly related genotype M4) have persisted in Southern Uganda and Kenya for at least 30 years, fulfilling one of the criteria for a clonal population (7).
Discussion
T. b. rhodesiense Is Distinct from T. b. brucei.
This study has revealed that, contrary to previous reports (18–20), Ugandan T. b. rhodesiense samples are distinct from T. b. brucei and can be defined by the use of three minisatellite markers in a simple genotype assay that allows the unambiguous distinction between T. b. rhodesiense and T. b. brucei. Although previous work by Hide et al. (2), using cluster analysis of restriction fragment length polymorphism patterns of repetitive sequences, has shown that human infective stocks form a distinct homogeneous group separate from the T. b. brucei isolates, the results described here contradict the findings of Hide et al. for two HSS isolates, Tira 68 and Tira 17, which were grouped with the HSR genotypes. The minisatellite data clearly identifies these stocks as not being of the HSR genotype. The highly discriminating minisatellite genotyping system, unlike the typing system based on repetitive probes, can be readily interpreted genetically and, as previous data using these minisatellites has shown that lysates of infected blood can be genotyped (13) a system for identifying Ugandan T. b. rhodesiense without the need for growth or purification of parasites is feasible. The lack of T. b. brucei samples from Zambia precludes the verification that the human infective genotypes identified here are specific for Zambian T. b. rhodesiense although they are completely distinct from the Ugandan T. b. rhodesiense. The large genetic distance between Zambian and Ugandan T. b. rhodesiense supports earlier findings that these populations are not closely related (20) and raises the question as to whether the ability to infect humans is a trait that has evolved on two separate occasions in different parts of Africa (A.M., unpublished work).
Mixed Infections.
In this study, 18% of isolates from cattle appeared to be of mixed genotype. The high prevalence of mixed T. brucei infections detected is in contrast to the isoenzyme data presented by Godfrey et al. (10), who examined a large number of trypanosome isolates from across Africa, but only detected mixed infections in approximately 2% in a range of different animals. This suggests that the prevalence of mixed genotype T. brucei infections is far higher than previously proposed. Indeed 12 mixed infected isolates were detected in the collection of T. brucei samples, which were not detected in the previous analysis by Hide et al. (2). The use of the repetitive probe restriction fragment length polymorphism patterns would misidentify such mixed isolates as containing novel genotypes. Clearly it is essential for population studies to identify any mixed isolates for subsequent analysis. The significant prevalence of mixed genotypes within cattle would suggest that tsetse could be readily infected with mixtures of different trypanosomes providing one of the prerequisites for genetic exchange. To formally demonstrate that mixed genotypes were present in these samples, trypanosome clones would need to be generated from the original stabilate of parasite material, in a similar fashion to the analysis of mixed tsetse infection described previously (13).
Population Structures.
The two different subspecies of T. brucei appear to have different population structures. T. b. rhodesiense is clonal, whereas the T. b. brucei population examined here has an epidemic population structure in which a background level of genetic recombination is obscured by the clonal expansion of a few genotypes. It would appear that host selection is an important determinant of the population structure of T. brucei, as particular genotypes of trypanosome are better adapted to survival within different mammalian hosts, and so the full level of T. brucei diversity is only apparent when tsetse flies are examined. These findings can explain the conflicting results of previous workers who examined populations comprising of both subspecies of T. brucei. For example, Tait examined stocks that were isolated from tsetse flies caught in the Lugala region of Busoga (6). A parsimonious interpretation of these data are that the predominant hosts were likely to have been wild game and cattle populations and thus the predominant subspecies sampled is likely to have been T. b. brucei. Hence the conclusion from this study was that this population was panmictic, albeit not statistically excluding lack of panmixia (21). In 1990, Tibayrenc examined samples isolated from humans (i.e., T. b. rhodesiense) and found evidence for a clonal population structure (7), an interpretation in agreement with the results presented here for T. b. rhodesiense. In 1993 Maynard-Smith examined samples from animals and humans (i.e., a mixture of both T. b. rhodesiense and T. b. brucei samples) from Lambwe Valley in Kenya and found evidence for an epidemic population structure (8). The high level of linkage disequilibrium identified in this study could have been compounded by treating the two genetically discrete populations of T. b. brucei and T. b. rhodesiense as a single population. Clearly, by using the markers described here this conclusion could be tested. In 1994 Hide et al. (2) examined a large proportion of the same stocks used in the current analysis using isoenzymes and concluded that T. b. rhodesiense and T. b. brucei did have different population structures, based on showing that the IA for T. b. brucei was negative whereas that for T. b. rhodesiense was positive but became negative when only ETs were considered, thus supporting an epidemic population structure for the latter. However, the consideration of only ETs in the T. b. rhodesiense population reduces the sample size drastically, making it difficult to determine whether the conclusion is statistically valid. In the current study, using the more informative minisatellite marker system, a similar result has been obtained. However, the ability to genetically interpret the minisatellite genotypes allowed the inspection of the genotypes concerned and revealed that they are highly related. Based on this analysis we have concluded that the Ugandan population of T. b. rhodesiense is clonal. However, it must be remembered that this may not necessarily be the case for other foci, but may be a unique feature of Ugandan trypanosomiasis. It is clear that the type of analysis described here could be extended to examine human infective and nonhuman infective isolates from other foci and thus begin to define the population structure of this parasite across its full geographical range.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Wellcome Trust to A. Tait and C.M.R.T.
Abbreviations
- CRAM
cysteine-rich acidic integral membrane
- IA
index of association
- HSR
human serum resistant
- HSS
human serum sensitive
- ET
electophoretic type
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.230434097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.230434097
References
- 1.Hoare C A. The Trypanosomes of Mammals. Oxford: Blackwell; 1972. [Google Scholar]
- 2.Hide G, Welburn S C, Tait A, Maudlin I. Parasitology. 1994;109:95–111. doi: 10.1017/s0031182000077805. [DOI] [PubMed] [Google Scholar]
- 3.Jenni L, Marti S, Schweizer J, Betschart B, Le Page R W F, Wells J M, Tait A, Paindavoine P, Pays E, Steinert A. Nature (London) 1986;322:173–175. doi: 10.1038/322173a0. [DOI] [PubMed] [Google Scholar]
- 4.Turner C M R, Sternberg J, Buchanan N, Smith E, Hide G, Tait A. Parasitology. 1990;101:377–386. doi: 10.1017/s0031182000060571. [DOI] [PubMed] [Google Scholar]
- 5.Gibson W C, Bailey M. Mol Biochem Parasitol. 1994;64:241–252. doi: 10.1016/0166-6851(94)00017-4. [DOI] [PubMed] [Google Scholar]
- 6.Tait A. Nature (London) 1980;287:536–538. doi: 10.1038/287536a0. [DOI] [PubMed] [Google Scholar]
- 7.Tibayrenc M, Kjellberg F, Ayala F J. Proc Natl Acad Sci USA. 1990;87:2414–2418. doi: 10.1073/pnas.87.7.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maynard-Smith J, Smith N H, O'Rourke M, Spratt B G. Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrett M P. Lancet. 1999;353:1113–1114. doi: 10.1016/S0140-6736(98)00416-4. [DOI] [PubMed] [Google Scholar]
- 10.Godfrey D G, Baker R D, Rickman L R, Mehlitz D. Adv Parasitol. 1990;29:1–74. doi: 10.1016/s0065-308x(08)60104-9. [DOI] [PubMed] [Google Scholar]
- 11.Goebloed E, Ligthart G S, Minter D M, Wilson A J, Dar F K, Paris J. Ann Trop Med Parasitol. 1973;67:31–43. [PubMed] [Google Scholar]
- 12.Tait A, Barry J D, Wink R, Sanderson A, Crowe J S. Parasitology. 1985;90:89–100. doi: 10.1017/s0031182000049040. [DOI] [PubMed] [Google Scholar]
- 13.MacLeod A, Turner C M R, Tait A. Mol Biochem Parasitol. 1999;102:237–248. doi: 10.1016/s0166-6851(99)00101-2. [DOI] [PubMed] [Google Scholar]
- 14.Jeffreys A J, MacLeod A, Tamaki K, Neil D L, Monckton D G. Nature (London) 1991;354:204–209. doi: 10.1038/354204a0. [DOI] [PubMed] [Google Scholar]
- 15.Tait A, Buchanan N, Hide G, Turner C M R. Mol Biochem Parasitol. 1996;76:31–42. doi: 10.1016/0166-6851(95)02528-6. [DOI] [PubMed] [Google Scholar]
- 16.Gibson W, Winters K, Mizen G, Kearns J, Bailey M. Microbiology. 1997;143:909–920. doi: 10.1099/00221287-143-3-909. [DOI] [PubMed] [Google Scholar]
- 17.Nei M. Genetics. 1978;23:341–369. doi: 10.1007/BF01908190. [DOI] [PubMed] [Google Scholar]
- 18.Borst P, Fase-Flowler F, Gibson W C. Mol Biochem Parasitol. 1981;3:117–131. doi: 10.1016/0166-6851(81)90011-6. [DOI] [PubMed] [Google Scholar]
- 19.Gibson W C, Marshall T F D C, Godfrey D G. Adv Parasitol. 1980;18:175–246. doi: 10.1016/s0065-308x(08)60400-5. [DOI] [PubMed] [Google Scholar]
- 20.Hide G, Buchanan N, Welburn S, Maudlin I, Barry J D, Tait A. Exp Parasitol. 1991;72:430–439. doi: 10.1016/0014-4894(91)90089-f. [DOI] [PubMed] [Google Scholar]
- 21.Cibulskis R E. Parasitology. 1988;96:303–322. doi: 10.1017/s0031182000058315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.