Summary
Regulated cell adhesion and motility have important roles during growth, development and tissue homeostasis. Consequently, great efforts have been made to identify genes that control these processes. One candidate is Rap1, as it has been implicated in the regulation of adhesion and motility in cell culture. To further study the role of Rap1 during multicellular development, we generated a mutant in a potential Rap1 GTPase activating protein (RapGAPB) in Dictyostelium. rapGAPB– cells have increased levels of active Rap1 compared with wild-type cells, indicating that RapGAPB regulates Rap1 activity. Furthermore, rapGAPB– cells exhibit hallmark phenotypes of other known mutants with hyperactivated Rap1, including increased substrate adhesion and abnormal F-actin distribution. However, unlike these other mutants, rapGAPB– cells do not exhibit impaired motility or chemotaxis, indicating that RapGAPB might only regulate specific roles of Rap1. Importantly, we also found that RapGAPB regulates Rap1 activity during multicellular development and is required for normal morphogenesis. First, streams of aggregating rapGAPB– cells break up as a result of decreased cell-cell adhesion. Second, rapGAPB– cells exhibit cell-autonomous defects in prestalk cell patterning. Using cell-type-specific markers, we demonstrate that RapGAPB is required for the correct sorting behaviour of different cell types. Finally, we show that inactivation of RapGAPB affects prestalk and prespore cell adhesion. We therefore propose that a possible mechanism for RapGAPB-regulated cell sorting is through differential adhesion.
Keywords: Differential cell adhesion, Morphogenesis, Pattern formation, Rap1, RapGAP
Introduction
Cell interactions and changes in cellular behaviour, such as cell adhesion and motility, are required for growth, development and tissue homeostasis of multicellular organisms. The social amoeba Dictyostelium discoideum provides a model system to study the regulation of these processes. Dictyostelium normally live as single-celled amoeba feeding on bacteria in the soil. In response to starvation, the cells enter a developmental cycle, which leads to the aggregation of several thousand amoebae to make a multicellular mound. Aggregation is driven by chemotaxis towards waves of cAMP (Kessin, 2001). Within the mound, cells differentiate into either prestalk or prespore cells. Both cell types are initially found in a `salt and pepper' distribution, but then subsequently sort out into discreet tissues during a period of morphogenesis that results in the formation of a migratory slug (Maeda et al., 2003; Maruo et al., 2004; Thompson et al., 2004; Williams et al., 1989; Yamada et al., 2005). Finally, after a period of migration, further complex morphogenetic movements result in the formation of a fruiting body consisting of terminally differentiated stalk and spore cells, as well as ancillary supporting structures (Kessin, 2001).
A key question in this developing system is how distinct tissues along the anterior-posterior axis arise from initially scattered populations of cells. This process of cell sorting is thought to be driven by a combination of differential chemotaxis and cell adhesion. For example, separated prestalk and prespore cells exhibit different rates of chemotaxis towards cAMP (Early et al., 1995; Matsukuma and Durston, 1979) as well as differential adhesion following disaggregation (Lam et al., 1981). Despite this, a relatively small number of genes have been implicated in the regulation of cell sorting and tissue establishment. The identification of further genes will therefore be crucial for our understanding of the regulation of morphogenesis in Dictyostelium, as well as other organisms.
One candidate molecule for the regulation of cell type patterning is the Ras subfamily small GTPase Rap1, because it has been implicated in the regulation of cell adhesion and cell motility in mammalian cells (Bos, 2005; Fujita et al., 2006; Lorenowicz et al., 2006; McLeod et al., 2004; Shimonaka et al., 2003; Zhang et al., 2005) and vegetative Dictyostelium amoebae (Bosgraaf et al., 2005; Jeon et al., 2007b). Rap1 was first identified as a protein that could antagonise Ras signaling (Kitayama et al., 1989), but it is now widely recognised that its effects are mediated by signaling pathways that are distinct from those of the Ras pathway (Bos, 1998). Rap1 activity is dependent on whether it is in a GDP-bound (inactive) or GTP-bound (active) state. Rap1 activity can therefore be tightly controlled by factors that influence whether it is GTP-bound or GDP-bound. Guanine nucleotide exchange factors (GEFs) activate Rap1 activity by promoting exchange of GDP for GTP, whereas GTPase-activating proteins (GAPs) promote GTP hydrolysis.
The Dictyostelium genome encodes only one Rap gene, rapA (Robbins et al., 1990; Weeks et al., 2005), whereas there are a large number of predicted RapGAPs and several presumptive RapGEFs, thus raising the possibility that some of these might regulate specific aspects of Rap1 function. To date, one specific RapGEF (GbpD) and one specific RapGAP (RapGAP1) have been characterised (Bosgraaf et al., 2005; Jeon et al., 2007a; Kortholt et al., 2006). In addition, a Rap1 kinase effector (Phg2), that exhibits specific binding to GTP-bound Rap1, has been identified (Gebbie et al., 2004; Kortholt et al., 2006). Simple studies of Rap1 function in Dictyostelium have however proved difficult, because attempts to disrupt the rapA gene have so far failed and it is likely that rapA-null cells are non-viable. Despite this, it has been possible to study the role of Rap1 through the expression of dominant-negative (Rap1S17N) and constitutively active (Rap1G12V) forms of Rap1. In addition, cells in which GbpD or RapGAP1 has been overexpressed or disrupted, and cells in which phg2 has been disrupted, have proved extremely informative. From these studies, it has emerged that Rap1 is required for the regulation of diverse vegetative cell processes, including stress response and phagocytosis, and this might explain why the disruption of the Rap1 gene would be lethal (Kang et al., 2002; Rebstein et al., 1997; Seastone et al., 1999). Importantly, these and other studies (Jeon et al., 2007a; Jeon et al., 2007b; Kortholt et al., 2006) reveal that the regulation of Rap1 in Dictyostelium, as in mammalian cells, is required for cytoskeletal organisation and adhesion.
The role of Dictyostelium Rap1 is not confined to growing vegetative cells. Recent studies demonstrate that chemotaxis of starved cells towards cAMP involves Rap1. For example, Rap1 hyperactivation due to the expression of constitutively activated Rap1 (Rap1G12V) or the overexpression of GbpD in wild-type cells severely impairs the ability of cells to perform chemotaxis (Jeon et al., 2007a; Jeon et al., 2007b; Kortholt et al., 2006). cAMP stimulation also induces rapid and transient activation of Rap1 (Jeon et al., 2007b). Active Rap1 is localised to the leading edge of migrating cells where it is thought to promote the recruitment of Phg2, which in turn promotes moyosin II phosphorylation and disassembly at the leading edge (Jeon et al., 2007b). The role played by Rap1 during multicellular development is, however, poorly understood.
In this study, we have addressed this problem using a strain in which a gene encoding a potential RapGAP (rapGAPB) was disrupted (rapGAPB–). We show biochemically and phenotypically that RapGAPB is required to regulate the levels of Rap1. rapGAPB is expressed during multicellular development and is enriched in prestalk cells, thus suggesting a role in cell-type regulation. Consistent with this idea, we find that expression of rapGAPB and the regulation of Rap1 activity are required for normal morphogenesis and the correct patterning of specific subtypes of prestalk cells. To determine how Rap1 exerts its effects, we compared the adhesive properties of the different cell types in wild-type and mutant structures. Relative cell adhesion of both prestalk and prespore cells was affected. We therefore propose that Rap1-regulated differential cell adhesion is required for normal cell-type patterning in Dictyostelium.
Results
Identification of the gene encoding RapGAPB and generation of a rapGAPB– mutant
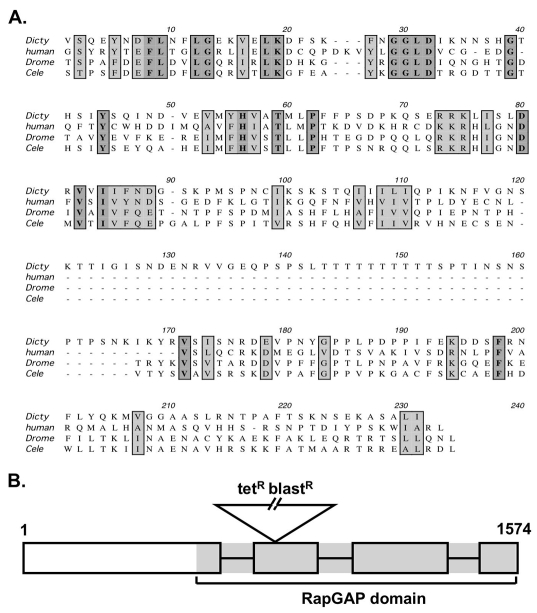
During the course of a genetic selection for patterning mutants (unpublished results), we isolated a mutant with a disruption within a gene predicted to encode a RapGAP (DDB0205291) (Fig. 1A). To ensure that the mutation affected protein function, we generated a new allele in which a 3 kb drug-resistance cassette was inserted within the RapGAP domain (Fig. 1B). This mutant very likely represents a null allele. First, no transcripts could be detected in the mutant by quantitative PCR with primers upstream of the insertion. Furthermore, we have generated a deletion allele in which the entire coding sequence was replaced by a blasticidin cassette and this resulted in an indistinguishable mutant phenotype (supplementary material Fig. S1).
Fig. 1.
Identification of a Dictyostelium RapGAP. (A) Alignment of the potential RapGAP domain of Dictyostelium RapGAPB (Dicty) with RapGAPs from human (human), Drosophila (Drome) and C. elegans (Cele). Identical residues are highlighted in dark grey and conserved residues in light grey. Dictyostelium RapGAPB shows approximately 20% conservation with the human RapGAP1 sequence. (B) Structure of the Dictyostelium rapGAPB gene to illustrate the position of the insertion cassette. Numbers indicate base pairs, grey area encodes the predicted rapGAP domain.
rapGAPB– cells show many phenotypic similarities to cells with hyperactivated Rap1, but chemotaxis is unaffected
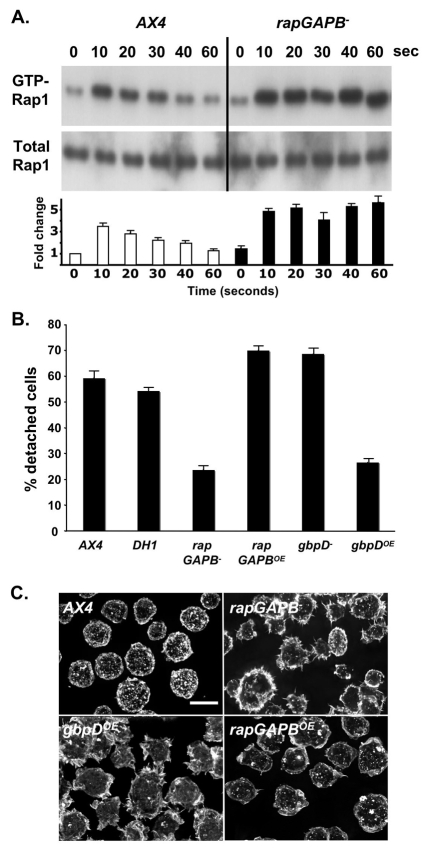
Since RapGAPB also exhibits sequence similarity with other GAPs, especially RanGAPs, it was important to first establish that it regulates Rap1 activity. Two clear predictions can be made for the effects of a RapGAP mutation. (1) It would be expected to exhibit elevated levels of Rap1. (2) It should show some phenotypic similarity to strains with artificially elevated Rap1 levels, such as the RapGEF overexpression strain gbpDOE. We therefore first measured the levels of activated Rap1 in response to cAMP stimulation. For this, we utilised an assay in which the Rap1-GTP binding domain of Byr2 can be used to pull down activated Rap1 from Dictyostelium cell extracts (Bolourani et al., 2008; Kang et al., 2002). Rap1 levels were compared in wild-type and rapGAPB– cells. Under these conditions, the levels of activated Rap1 normally rise dramatically within 10 seconds, before falling back to the unstimulated levels within 1 minute (Jeon et al., 2007b) (Fig. 2A). We found that although the basal levels of activated Rap1 were indistinguishable in unstimulated wild-type and rapGAPB– cells, clear differences could be seen in response to cAMP stimulation (Fig. 2A). First, the maximal level of Rap1 activation was greater in rapGAPB– cells than in wild-type cells. Second, the time course of deactivation was dramatically altered. Activated Rap1 levels barely decreased in rapGAPB– cells even after 1 minute (Fig. 2A). To ensure this was not due to a general defect in cAMP response kinetics, we measured the activation response of a related protein, RasG, under the same conditions. Activation and deactivation kinetics of RasG in response to cAMP were comparable in wild-type and mutant cells (data not shown). These findings are consistent with the idea that RapGAPB is specifically required to inactivate Rap1.
Fig. 2.
RapGAPB is required to regulate Rap1 activity. (A) Western blot showing levels of activated Rap1 (top panel) and total Rap1 (bottom panel). In wild-type cells, maximal increase in levels of active Rap1 was observed after 10 seconds of cAMP stimulation, followed by a steady decrease over 60 seconds of stimulation. In rapGAPB– cells, the maximal increase after 10 seconds of stimulation is slightly higher than that of wild-type cells. In addition, the level of active Rap1 does not decrease over 60 seconds, instead the maximal level is maintained. The level of total Rap1 is equal in all cell lysates. The graph shows fold change in amount of GTP-Rap1 relative to AX4 cells at 0 seconds. (B) Cell-substrate adhesion was measured by counting the number of vegetative cells that detached from a plate after shaking for 20 minutes. Approximately 60% of wild-type cells (AX4 and DH1) detached. Significantly fewer rapGAPB– and gbpDOE cells detached. By contrast, more rapGAPBOE and gbpD– cells detached. (C) Cells were stained with FITC conjugated phalloidin in order to observe F-actin distribution and viewed by deconvolution microscopy. rapGAPB– and gbpDOE cells exhibit a clearly different F-actin distribution and cell morphology compared with wild-type AX4 cells, whereas the rapGAPBOE cells appeared more like the wild type. Scale bar: 10 μm.
When Rap1 levels are altered in Dictyostelium, several hallmark phenotypes have been described (Jeon et al., 2007a; Jeon et al., 2007b; Kortholt et al., 2006). We therefore tested whether rapGAPB– cells also showed similar defects. First, we found that vegetative rapGAPB– cells attached more strongly to the substratum than wild-type control cells (Fig. 2B). The extent of the increase was similar to that seen with gbpDOE cells. This phenotype was efficiently rescued when a GFP-tagged version of RapGAPB (RapGAPBOE) was expressed in rapGAPB– mutant cells (Fig. 2B). In fact, these cells actually attached slightly less than wild-type cells (Fig. 2B), most likely because this line expresses ∼twofold higher levels of rapGAPB transcripts than the wild type (data not shown). We also tested whether rapGAPB– cells showed defects in morphology and actin distribution consistent with increased levels of Rap1 activation. When rapGAPB– or gbpDOE cells were stained with fluorescent phalloidin, both showed a characteristic spiky appearance with actin decorating the periphery of cells within filopodia, a noticeably different morphology to that of the wild type (Fig. 2C). Again, this phenotype was efficiently rescued by RapGAPBOE expression in rapGAPB– mutant cells (Fig. 2C). Taken together, these data strongly suggest that RapGAPB is required for the normal regulation of Rap1 activation.
Several recent studies provide evidence that the localised regulation of Rap1 activation is important for chemotaxis of Dictyostelium cells towards cAMP (Jeon et al., 2007a; Jeon et al., 2007b; Kortholt et al., 2006). Surprisingly, however, we could find no defect in cell movement in rapGAPB– cells. Mutant cells behaved like wild-type cells when both random movement and movement toward a chemoattractant were measured (Table 1).
Table 1.
Random movement and chemotaxis
|
Aggregation-competent cells
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
Vegetative cells Random movement
|
Random movement
|
Chemotaxis
|
||||||||
| Speed | Persistence | n | Speed | Persistence | n | Speed | Persistence | Directionality | n | |
| AX4 | 1.68±0.45 | –0.01±0.22 | 205 | 3.22±1.64 | 0.41±0.31 | 94 | 5.24±1.46 | 0.79±0.15 | 0.85±0.10 | 26 |
| RapGAPB– | 1.66±0.52 | 0.03±0.26 | 181 | 3.25±1.65 | 0.32±0.31 | 75 | 6.04±3.02 | 0.73±0.20 | 0.83±0.15 | 33 |
RapGAPB expression is required for developmentally regulated Rap1 activity and for normal morphogenesis
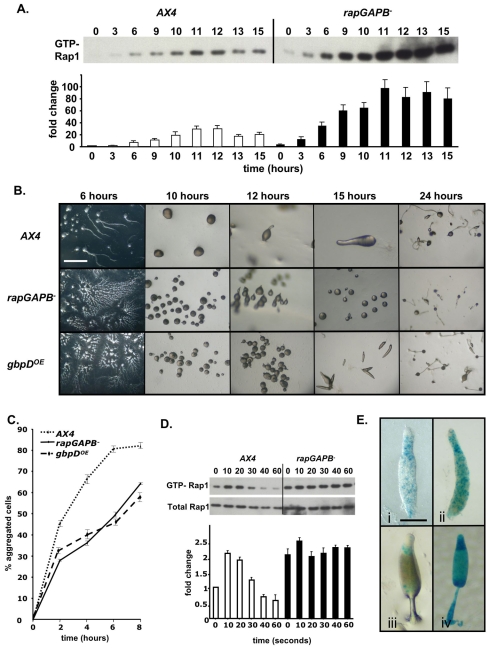
Although Rap1 levels have been shown to remain constant at all developmental stages, we tested whether levels of activated Rap1 exhibit developmental regulation. We found that in wild-type cells, levels of activated Rap1 are lowest in vegetative cells, but gradually increase during the first 12 hours of development. Levels are highest around the tipped mound stage then decrease slightly (Fig. 3A). Several dramatic differences were observed in the rapGAPB– mutant. First, levels of activated Rap1 were higher in vegetative cells and at all developmental stages. Second, levels did not decrease during the later stages of development, but instead remained at peak levels (Fig. 3A). These findings demonstrate that Rap1 activity is regulated during multicellular development, and that RapGAPB is required for its regulation.
Fig. 3.
RapGAPB expression is required for developmentally regulated Rap1 activity and for normal morphogenesis. (A) Developmental regulation of activated Rap1 levels. In wild-type cells, levels of GTP-Rap1 peak after 12 hours of development before decreasing. In rapGAPB– cells, GTP-Rap1 levels are higher at all time points and do not decrease during later developmental stages. The graph shows relative amounts of GTP-Rap1 compared with levels in AX4 cells at time 0. (B) Development of wild-type AX4, rapGAPB– and gbpDOE cells. Developing rapGAPB– and gbpDOE cells both formed streams that broke up, resulting in many small mounds. rapGAPB– and gbpDOE cells formed abnormal tip mound structures. The development of rapGAPB– cells temporarily halted at the mound stage before eventually forming small culminants. The small mounds formed by the gbpDOE cells made small slugs that culminated normally to form culminants. Scale bar: 2 mm. (C) Cell-cell adhesion of streaming cells was measured by disaggregating cells and counting the number of cells that reaggregated at different time points. rapGAPB– and gbpDOE cells are less adhesive because fewer cells reaggregated at all time points. (D) cAMP-induced activation of Rap1 in 12 hour developed cells. Western blot shows levels of activated Rap1 (top panel) and total Rap1 (bottom panel). In wild-type cells, levels of active Rap1 peaked after 10 seconds of cAMP stimulation, followed by a steady decrease. In rapGAPB– cells, the level in unstimulated cells was similar to the maximal level in wild-type cells at all time points. Graph shows relative levels of GTP-Rap1 compared with that in AX4 cells at 0 seconds. (E) To determine the expression pattern of rapGAPB, the rapGAPB promoter was placed upstream of lacZ and transformed into wild-type cells. When stained for a short time, expression is enriched in the collar region of slugs (i) and in the upper and lower cups of culminants (iii). After longer periods of staining, stained cells were predominantly found in the collar region, but also found scattered throughout the entire slug (ii) and in the apical tip, upper and lower cups and spores of culminants (iv). Scale bar: 1 mm.
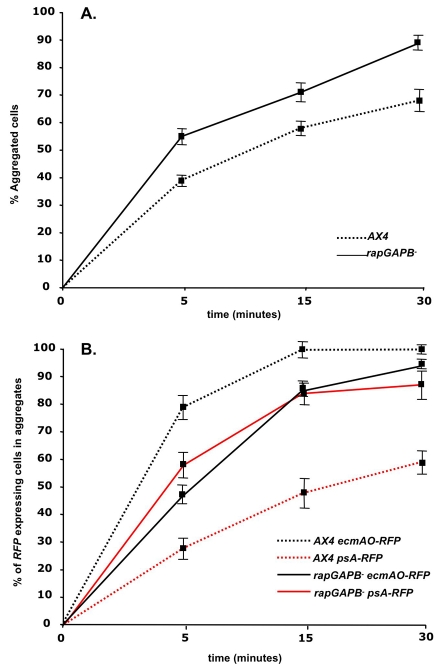
To determine whether regulation of Rap1 activity is required for normal multicellular development, we compared the development of wild-type and rapGAPB– cells. Mutant streams tended to break up, resulting in the formation of large numbers of small mounds. This defect was phenocopied when gbpDOE cells were allowed to develop (Fig. 3B). It has previously been shown theoretically and experimentally that the break up of streams can be triggered by increased cell movement and/or decreased cell adhesion (Gomer et al., 2002; Roisin-Bouffay et al., 2000). As we had been unable to detect any defect in cell motility in rapGAPB– mutant cells, we tested whether altered cell-cell adhesion might result in the break up of streams. Consistent with this idea, both rapGAPB– and gbpDOE cells exhibited much reduced cell-cell adhesion than wild-type cells in an assay in which streaming cells are dissociated and the extent of reaggregation measured (Fig. 3C).
Once aggregation has taken place, and resulted in the formation of small mounds, further development of rapGAPB– and gbpDOE mutant cells was also extremely aberrant (Fig. 3B). Most notably, both mutants formed large numbers of oddly shaped tipped mound structures. From this stage, rapGAPB– cells appeared to be unable to make migratory slugs, although small twisted fruiting bodies eventually formed. Unlike rapGAPB– cells, gbpDOE cells did eventually overcome the block at the mound stage and proceeded to the slug and fruiting body stages (Fig. 3B). This is probably because the actin15 promoter used to drive GbpD expression is expressed at lower levels as development proceeds (Knecht et al., 1986). Despite this, the similarities in phenotype between developing rapGAPB– and gbpDOE cells indicates that these defects are also probably due to misregulation of Rap1 activity.
As the major defect in morphogenesis of the rapGAPB– mutant occurs around the mound-slug transition, we tested whether activation of Rap1 in response to cAMP is also affected at this stage. In wild-type mounds, levels of activated Rap1 were found to peak within 10 seconds before decreasing rapidly. By contrast, cAMP stimulation had little affect on mound stage mutant cells. In fact, the basal level of activated Rap1 in rapGAPB– cells was already similar to the maximum level of stimulated wild-type cells, and did not decrease over the time course tested (Fig. 3D).
To better understand the role of RapGAPB in multicellular development, we also elucidated the rapGAPB expression pattern. Interestingly, rapGAPB mRNA is not uniformly expressed in all cell types. When a region 1 kb upstream of the rapGAPB gene was used to drive the expression of a lacZ reporter gene, β-galactosidase activity was most strongly expressed in the prestalk O (pstO) region of slugs after short periods of staining (Fig. 3E). After longer periods of staining, stained cells were also found scattered throughout the entire anterior and posterior regions of the slug, but still showed highest expression in the prestalk O region (Fig. 3E). Consistent with RapGAPB expression being the most highly expressed in prestalk cells, stained cells were found in the upper and lower cups of culminants after short periods of staining. However, after longer periods of staining, stained cells were also found in the apical tip, upper and lower cups of culminants, as well as the stalk and spores (Fig. 3E). Therefore, although rapGAPB is most strongly expressed in pstO cells, its expression is not exclusive to this cell type. This gene expression pattern presumably reflects that of the endogenous gene, because expression of rapGAPB driven by this promoter fragment completely rescues the developmental phenotype of the mutant (supplementary material Fig. S1).
rapGAPB– cells exhibit defects in prestalk cell patterning
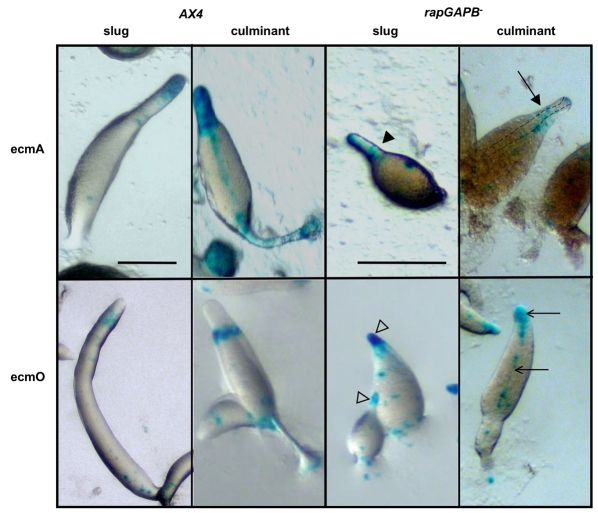
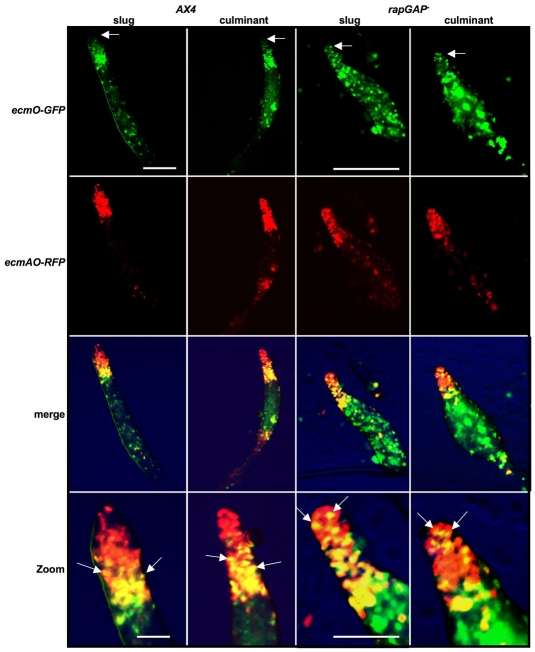
To elucidate whether the mutant phenotypes are due to defects in cell-type differentiation and/or pattern formation, rapGAPB– mutant cells were transformed with cell-type-specific reporter genes. ecmAO-lacZ (pstA and pstO) and ecmB-lacZ (pstAB and pstB) were used to label the various prestalk cell types, together with psA-lacZ as a general marker of prespore cell differentiation. ecmAO-lacZ, ecmB-lacZ and psA-lacZ expression was normal in rapGAPB– mutant cells (supplementary material Fig. S2). It was perhaps most surprising to find that ecmAO-lacZ expression was normal when one considers that rapGAPB is most strongly expressed in these cells. We did, however, find that this initial impression was somewhat misleading when gene expression was examined using more-specific cell-type markers. For this, we used constructs derived from different regions of the ecmAO promoter that drive expression specifically in pstA (ecmA-lacZ) and pstO (ecmO-lacZ) cell populations (Early et al., 1995; Early et al., 1993). When ecmA-lacZ and ecmO-lacZ expression was examined in developing wild-type and mutant structures, clear differences were seen. First, in mutant slugs and culminants, expression of ecmA-lacZ was not confined to the extreme tip but also appeared to occupy the collar and upper cup normally only occupied by pstO cells (Fig. 4). Second, ecmO-lacZ expression was strikingly aberrant as it was found in regions normally only occupied by pstA cells, namely the tip of slugs, as well as the apical tip and stalk of culminants (Fig. 4). These findings suggested that pstO and pstA cells are intermingled. To test this idea directly, we generated co-transformants in which RFP was driven by the ecmAO promoter and GFP by the ecmO promoter. In these strains, pstO cells appear yellow (due to expression of both RFP and GFP) and pstA cells appear red. In mutant slugs, pstO cells were not confined to the collar but also appeared to occupy the extreme tip normally only occupied by pstA cells (Fig. 5). Furthermore, pstO cells were found in the apical tip as well as the upper cup of mutant culminants. We therefore believe that in developing rapGAPB– mutant structures, the normally spatially distinct pstA and pstO cell populations are intermingled. The sum of this expression leads to an apparently normal pattern of ecmAO-lacZ expression.
Fig. 4.
PrestalkA and prestalkO markers are misexpressed in the rapGAPB– mutant. AX4 or rapGAPB– cells were transformed with cell-specific reporter genes. Expression was examined at both slug and early culminant stages. Clear differences were found in the expression of both ecmA-lacZ and ecmO-lacZ. In developing AX4 cells, ecmA-lacZ expression was found mainly in the front of the prestalk region at the slug stage and in the apical tip and stalk of culminants. By contrast, in developing rapGAPB– cells, ecmA-lacZ expression was found in the tip and collar (filled arrowhead) of the equivalent slug stage and mainly in the upper cups (filled arrow) of culminants, with less staining in the apical tip. Dotted line shows stalk tube. In developing AX4 cells, ecmO-lacZ expression was found mainly in the collar region at the slug stage and in the upper cup of culminants. By contrast, in developing rapGAPB– cells, ecmO-lacZ expression was strongly expressed in the tip (open arrowhead) of the equivalent slug stage and mainly in the apical tip (open arrow) and stalk (open arrow) of culminants. Scale bars: 1 mm.
Fig. 5.
Prestalk A and prestalk O cells are intermingled in the rapGAPB– mutant. AX4 and rapGAPB– cells expressing both ecmO-GFP and ecmAO-RFP were developed to the slug and early culminant stages. In developing AX4 cells, ecmO-GFP expression (green) was found mainly in the collar region of slugs and was mostly absent from the tip (arrowed). ecmAO-RFP expression (red) was found throughout the prestalk region of slugs. As a result, the merged image shows orange-yellow cells only in the collar (arrowed in the magnified image) and only red cells in tips of slugs. By contrast, in developing rapGAPB– cells, ecmO-GFP-expressing cells were found in both the collar and tip regions of the slug (arrowed) and ecmAO-RFP expression was found throughout the entire prestalk region. The resulting merged image shows orange-yellow cells in both the collar and tip of the slug (arrowed on the magnified image). A similar defect was observed at the early culminant stage. In AX4 culminants, ecmO-GFP expression was highest in the upper cup and largely absent from the apical tip (arrowed), whereas ecmAO-RFP expression was found in both the collar and apical tip. The merged image shows orange-yellow cells in only the upper cup (arrowed on the magnified image) and only red cells in the apical tip. However, in rapGAPB– culminants, ecmO-GFP and ecmAO-RFP expression were both found in the upper cup and apical tip (arrowed). The merged image shows orange-yellow cells in both the upper cup and apical tip (arrowed in the magnified image). Scale bars: 1 mm and 0.3 mm (zoom panels).
Importantly, our studies demonstrate that all cell types are present in developing rapGAPB– mutant cells, even though the pattern of expression of pstO and pstA markers is aberrant. One simple interpretation of these data is that RapGAPB is not required for cell-type determination, but instead for the control of the behaviour of the different cell types following differentiation, specifically the pstA and pstO cells. To further test this idea, we generated a chimera between labeled and unlabeled wild-type and rapGAPB– cells. In a control homotypic chimera, labeled cells were evenly distributed throughout slugs and culminants. However, when a minority of labeled rapGAPB– mutant cells were mixed with wild-type cells, labeled cells were seldom found in the anterior prestalk region of slugs or upper and lower cups of the culminant (Fig. 6A). By contrast, when mixed with a majority of unlabelled rapGAPB– mutant cells, wild-type cells were found predominantly within the anterior prestalk region of slugs and strongly labeled stalk and ancillary structures of culminants (Fig. 6A). Furthermore, when Rap1 levels were specifically increased in prestalk cells by placing GbpD or Rap1G12V under the control of a prestalk-specific promoter, these cells were also under-represented within anterior regions of the slug and in the prestalk-derived structures of the culminant (Fig. 6B).
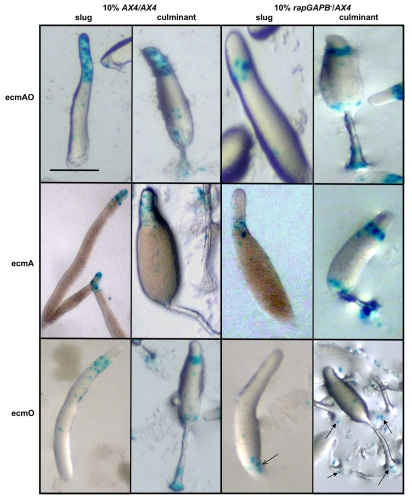
Fig. 6.
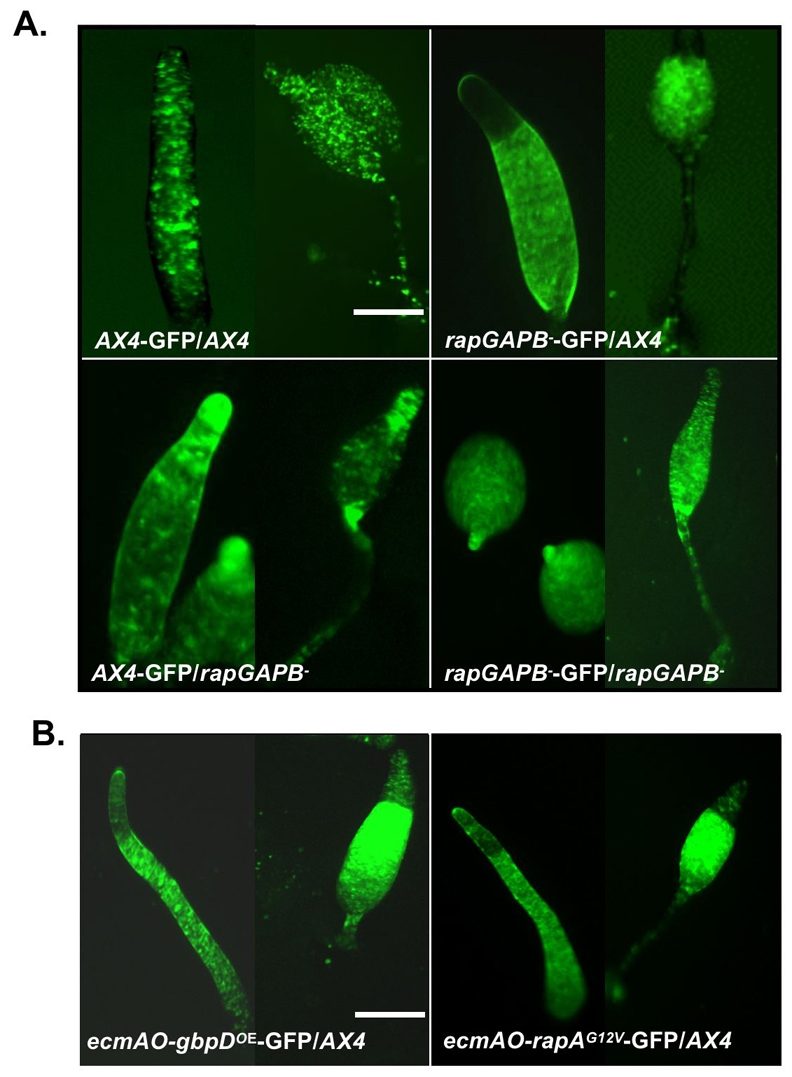
RapGAPB is required for prestalk cell patterning in chimeric development. (A) 10% GFP-labeled cells were mixed in chimera with 90% unlabeled cells and allowed to develop. In control homotypic mixes, GFP-labeled cells were evenly distributed throughout the slug and culminant. However, when GFP-labeled rapGAPB– cells were mixed with AX4 cells, the labeled cells were absent from the tip of slugs and from the upper and lower cups and stalk of culminants. The opposite pattern was observed when GFP labeled AX4 cells were mixed with rapGAPB– cells. (B) When GFP-labeled ecmAO-gbpDOE or ecmAO-rapAG12V cells were mixed with AX4 cells, the labeled cells were absent from the prestalk region of slugs and from the upper and lower cups and stalk of culminants. Scale bars: 1 mm.
Although Rap1 activation affects prestalk cell behaviour in chimeras, two questions arise. First, is this defect due to a failure to differentiate into prestalk cells in chimeras or due to misplacement of mutant cells? Second, does this reflect a general defect in prestalk cell differentiation or is it specific to certain prestalk subtypes? To address these questions, we examined the behaviour of wild-type and rapGAPB– mutant cells expressing prestalk and prespore markers in chimeras. Although no defect in psA or ecmB expression could be detected (supplementary material Fig. S3), chimeras with ecmAO-lacZ, ecmA-lacZ and ecmO-lacZ revealed clear defects. In control mixes, ecmAO-lacZ-labelled cells were found throughout the entire prestalk zone of slugs as well as the upper cup and apical tip of culminants (Fig. 7). By contrast, when ecmAO-lacZ transformed rapGAPB– cells were mixed with wild-type cells, labeled mutant cells were only present in the pstO collar of slugs and upper cup in culminants (Fig. 7). Despite an absence of mutant pstA cells from the tip of slugs and culminants in chimera, staining of ecmA-lacZ revealed that mutant pstA cells do differentiate but are misplaced to the region normally occupied by pstO cells (Fig. 7). Similarly, although mutant pstO cells differentiated in chimeras, their localisation was also aberrant. ecmO-lacZ-expressing mutant cells were largely found at the rear of chimeric slugs and scattered around the basal disc of culminants (Fig. 7). In fact, these cells appeared to be lost during the later stages of development. It is important to emphasise that stained cells are never found on the substratum in wild-type controls. Furthermore, there is not a general failure of rapGAPB– mutant cells to participate in chimera with wild-type cells. This defect is therefore highly specific to those cells differentiating as pstO cells. Taken together, these findings provide further strong support for the idea that that the regulation of Rap1 activity is crucial for the normal behaviour of pstA and pstO cells in pattern formation, but not pstA or pstO cell differentiation itself.
Fig. 7.
rapGAPB– mutant cells show defects in ecmAO-lacZ, ecmA-lacZ and ecmO-lacZ expression in chimeras. AX4 or rapGAPB– cells were transformed with cell-specific markers. 10% of cells expressing these markers were mixed in chimeras with 90% unlabeled cells and developed. Expression of the markers was observed at both the slug and early culminant stages of development. Clear defects were observed in the expression of ecmAO-lacZ, ecmA-lacZ and ecmO-lacZ in chimeras. When AX4 cells expressing ecmAO-lacZ were mixed with AX4 cells, ecmAO expression was found in the entire prestalk region of slugs and in the apical tip, upper cup and lower cup of culminants. However, when rapGAPB– cells expressing ecmAO-lacZ were mixed with AX4 cells, ecmAO expression was absent from the tip of slugs and only present in the collar region. In addition, in the culminant expression was absent from the apical tip. When AX4 cells expressing ecmA-lacZ were mixed with AX4 cells, ecmA expression was found in tip of slugs and in the apical tip and stalk of culminants. However, when rapGAPB– cells expressing ecmA-lacZ were mixed with AX4 cells, ecmA expression was absent from the tip of slugs and instead present in the collar region. In addition, expression was absent from the apical tip and found instead in the upper and lower cups of culminants. When AX4 cells expressing ecmO-lacZ were mixed with AX4 cells, ecmO expression was found in the collar region of slugs and in the upper cup and lower cups of culminants. However, when rapGAPB– cells expressing ecmO-lacZ were mixed with AX4 cells, ecmO expression was largely found at the rear of the slug and in the slime trail (open arrow). In addition, expressing cells were found mainly in the basal disk of the culminant and scattered on the agar (filled arrows). Scale bar: 1 mm.
rapGAPB– cells exhibit defects in cell-type-specific cell-cell adhesion
Differential cell adhesion has been proposed to have a role in tissue patterning in Dictyostelium. As rapGAPB– cells exhibit defects in cell-cell and substrate adhesion during aggregation and vegetative growth, we therefore tested whether defects in cell adhesion might also provide an explanation for the observed patterning defects. Cells were dissociated at the finger stage of development and the kinetics of reaggregation was used as a measure of cell-cell adhesion. Finger-stage rapGAPB– cells were found to be more adhesive than wild-type cells at the same stage of development (Fig. 8A). We next sought to determine whether the observed defect was due to a general increase in adhesion of all cell types. For this, wild-type and rapGAPB– mutant cells were transformed with ecmAO-RFP and psA-RFP reporter genes, which allowed the re-aggregation of prestalk and prespore cells to be followed. psA-RFP-expressing prespore cells were found to be more adhesive in the rapGAPB– mutant than the wild type. By contrast, ecmAO-RFP-expressing prestalk cells were less adhesive in the rapGAPB– mutant than in the wild type (Fig. 8B). Consequently, the relative adhesion of the two cell types is altered in the mutant. In fact, the adhesiveness of mutant prestalk and prespore cells was very similar, whereas wild-type prestalk cells were significantly more adhesive than prespore cells (Fig. 8B). These findings suggest that RapGAPB is required to regulate differential adhesion of both the prestalk and prespore populations. We propose that this defect might provide an explanation for the patterning defects observed in the developing rapGAPB– mutant.
Fig. 8.
rapGAPB– mutant cells show defects in prestalk and prespore cell-cell adhesion at the finger stage of development. Structures were disaggregated and the percentage of cells that aggregated after different times counted. (A) Cell-cell adhesion of wild-type (dotted line) and rapGAPB– cells (solid line). (B) Measurement of overall cell-cell adhesion. rapGAPB– cells are more adhesive than the wild type. Measurement of prespore and prestalk cell adhesion (psA-RFP or ecmAO-RFP expressing cells, respectively). rapGAPB– prespore cells are more adhesive than wild-type prespore cells and rapGAPB– prestalk cells are less adhesive than wild-type prestalk cells. As a consequence rapGAPB– prestalk and prespore cells show similar adhesion, whereas wild-type prestalk cells are more adhesive than wild-type prespore cells.
Discussion
We generated a mutant in a gene encoding a Dictyostelium RapGAP and used this mutant to further elucidate the role of Rap1. Measurements of the levels of activated Rap1 in the rapGAPB– mutant demonstrate that RapGAPB is required to switch Rap1 from its GTP-bound state to an inactive GDP-bound state. Our findings provide confirmation of some aspects of the role of Rap1 suggested by earlier overexpression studies. Importantly, they also provide novel insights into the role and complexity of regulation of Rap1 activity. Finally, we have been able to use this mutant to establish a role for Rap1 in the regulation of differential cell adhesion, cell type specific patterning and morphogenesis.
RapGAPB and chemotaxis
We found one important difference between the phenotype of rapGAPB– cells and other mutants with hyperactivated Rap1 levels. Although rapAG12V and gbpDOE cells exhibit severely reduced chemotaxis towards cAMP (Jeon et al., 2007b; Kortholt et al., 2006), chemotactic cell movement is unaffected in rapGAPB– cells. This is supported by the fact that gbpDOE cells are unable to aggregate on a bacterial lawn (Kortholt et al., 2006), whereas rapGAPB– cells readily form multicellular structures (supplementary material Fig. S1). One possible explanation is that another RapGAP also downregulates Rap1 activity and is required for chemotaxis. Indeed, downregulation of Rap1 by RapGAP1 results in slightly impaired chemotaxis, although the defect is not as severe as seen with cells expressing constitutively active Rap1 or overexpressing GbpD (Jeon et al., 2007a). Furthermore, the Dictyostelium genome encodes ten more putative RapGAPs. It will therefore be of great interest to study the phenotypes of mutants in these genes. It is also possible that any differences in chemotactic activity could be explained by the fact that the gbpDOE mutant was created in the DH1 parental strain, whereas the rapGAP1– and rapGAPB– mutants were created in AX3 and AX4 parental strains, respectively. Alternatively, the phenotypic difference could be due to differences in the levels of activated Rap1 between the strains. For example, in unstimulated aggregation-competent cells, although the basal levels of Rap1 were identical in rapGAPB– and wild-type cells, they are already at maximally stimulated levels in rapG12V cells (Jeon et al., 2007b). Consistent with this idea, rapGAP1– cells only show mild chemotaxis defects and exhibit wild-type levels of activated Rap1 in aggregation-competent cells (Jeon et al., 2007a). This difference in Rap1 activity between rapGAPB–, rapAG12V, gbpDOE and rapGAP1– cells could therefore underlie the difference in the ability of cells to chemotax towards cAMP. It is therefore possible that in aggregation-competent rapAG12V and gbpDOE cells, there is actually little if any response to cAMP when measured in terms of changes in levels of Rap1 activation. If this change were required for cells to efficiently sense and move towards a directional stimulus, then it could explain the inability of rapAG12V and gbpDOE cells to undergo chemotaxis, whereas rapGAPB– cells are unaffected. It is important to note, however, that the Rap1 activation response of rapGAPB– cells to cAMP is affected, despite the fact that we could not detect any phenotypic effect. It will be of great interest to determine whether rapGAPB– cells exhibit more subtle chemotactic defects under other conditions.
Rap1 regulation is required for normal pstO and pstA cell behaviour
Most studies have focused on the role of Rap1 signalling in vegetative cells and in the regulation of chemotaxis. However, it has previously been demonstrated that activated Rap1 can also affect development, albeit when expressed in a strain already compromised by expression of constitutively activated RasDG12T (Louis et al., 1997). Our study provides an important advance in understanding the role of Rap1 in cell-type differentiation and patterning. We have shown that Rap1 activity is developmentally regulated and that inactivation of RapGAPB leads to the misregulation of Rap1 during development. Consequently, the development of the rapGAPB– mutant is severely impaired. In the most severe case, rapGAPB– cells are barely able to complete development, and only after a prolonged arrest at the tip mound stage. Furthermore, we have shown that inactivation of RapGAPB, as well as overexpression of Rap1G12V or GbpDOE leads to common developmental defects, providing further evidence to support the idea that these defects are due to hyperactivation of Rap1.
As rapGAPB transcripts are enriched in prestalk cells, this suggested a requirement for Rap1 inactivation within prestalk cells. This idea is supported by the finding that both pstA and pstO cells are mislocalised in the rapGAPB– mutant when developed clonally. Furthermore, all strains tested with excess active Rap1 exhibited defects in prestalk cell sorting when mixed in chimeras with wild-type cells. Using cell-type-specific markers, we have shown that this is due to defects in both pstA and pstO cell behaviour. Most importantly, we found that the differentiation of pstA and pstO cells, as measured by expression of cell-type-specific markers, is not affected. Instead, the subsequent behaviour of these cells is perturbed. When rapGAPB– cells develop, pstA and pstO cells differentiate, even in chimeras with wild-type cells, but are unable to occupy their normal positions.
It has previously been proposed that cell motility and adhesion are required for the correct patterning of the different cell types during multicellular development in Dictyostelium (Chisholm and Firtel, 2004). We have been unable to detect any defect in chemotaxis of isolated rapGAPB– cells. However, it is impossible to rule out a role for RapGAPB in cell motility, because movement during multicellular development takes place in three dimensions and is therefore likely to represent a more testing environment in which cell motility and cell adhesion are tightly interlinked. Despite this, we believe that changes in cell adhesion may underlie the sorting defects observed, because rapGAPB– cells exhibit defects in cell-substrate adhesion and in the differential adhesive properties of prestalk and prespore cells. In addition, it has previously been shown that clonal inactivation of Rap1 during Drosophila wing disc development results in the dispersal of mutant cells into wild-type tissue (Knox and Brown, 2002). It is thought that this is due to the misplacement of adherens junctions within these cells. This idea is supported by the finding that altered levels of E-cadherin, a major adherens junction component, affects the sorting of dissociated embryonic zebrafish cells and mouse fibroblasts (Krieg et al., 2008; Steinberg and Takeichi, 1994). It does, however, seem that Rap1 probably plays a more widespread role in the regulation of cell adhesion (Bos, 2005; Kooistra et al., 2007). For example, Rap1 has also been shown to affect integrin-mediated adhesion of mammalian leukocytes (Reedquist et al., 2000).
To date, several different adhesion systems have been characterised through gene knockout and overexpression studies in Dictyostelium (Coates et al., 2002; Cornillon et al., 2006; Ginger et al., 1998; Harloff et al., 1989; Wang et al., 2000; Wong et al., 2002). Although these include cadherin and adherens junction systems, none of these appear to explain the defects we describe. For example, although adherens junctions have been shown to be required for normal culmination, they are not thought to be present at earlier stages (Grimson et al., 2000). Furthermore, gene knockouts of other adhesion systems, including the Dictyostelium cadherin homologue, cadA, show few similarities to the developmental defects resulting from Rap1 hyperactivation (Coates et al., 2002; Cornillon et al., 2006; Ginger et al., 1998; Harloff et al., 1989; Wang et al., 2000; Wong et al., 2002). Consequently, it will be of great interest to elucidate the molecular nature of the Rap1-regulated cell adhesion system in Dictyostelium because it might reveal novel modes of Rap1 regulation in other organisms. Furthermore, our studies show that the differentiation and behaviour of the prestalk cell population in Dictyostelium provides a model system to study the role of Rap1 in the developmental regulation of cell adhesion and pattern formation.
Materials and Methods
Strains, culture and maintenance
Dictyostelium strains were grown and maintained in association with Klebsiella aerogenes or in HL5 axenic medium (Sussman, 1987). The gbpD– cell line and gbpDOE plasmid were kindly provided by Peter Van Haastert. Transformants were selected in 10 μg/ml blasticidin or 20 μg/ml G418. For development, cells in exponential growth phase were harvested and washed before plating at a density of 6.4×106 cells/cm2 on KK2 (16.1 mM KH2PO4, 3.7 mM K2HPO4) plates in 1.5% purified agar.
Plasmid construction
For the disruption of the rapGAPB gene, a 3.5 kb genomic fragment from the rapGAPB locus was amplified from wild-type AX4 genomic DNA. A tetracycline-blasticidin (Tetr-Bsr) resistance cassette was inserted by in vitro transposition (Abe et al., 2003). Diagnostic PCR confirmed insertion of the cassette to be within the second exon, approximately 800 bp downstream from the start codon. For deletion of the rapGAPB gene, flanking genomic fragments were cloned into a floxed blasiticin cassette. Linearised constructs were transformed into AX4 cells by electroporation followed by blasticidin selection and confirmation of gene disruption or deletion by PCR. For overexpression construct generation, coding sequences were cloned as a translational fusion into pTX-GFP and transformed into AX4 cells. Clonal lines with equally strong GFP fluorescence were selected for further analysis. For cell-type-specific expression of constitutively active Rap1 and GbpD; rapAG12V was amplified from the Rap1-G12V plasmid, and a 4.4 kb gbpD genomic fragment were amplified by PCR from AX4 gDNA. Each fragment was cloned as a translational fusion into the ecmAO-GFP plasmid based upon pDdGFP. Cell-type-specific RFP and GFP constructs were generated by replacing lacZ with RFP or GFP.
Measurement of activated Rap1
The Ras-binding domain (RBD) of Byr2 was expressed in Escherichia coli as a GST fusion protein and purified as described previously (Kae et al., 2004). The purified GST-Byr2-RBD was used for the detection of activated Rap1. Cells were stimulated with 15 μM cAMP. Cells were lysed by mixing with an equal volume of 2× lysis buffer (20 mM sodium phosphate, pH 7.2, 2% Triton X-100, 20% Glycerol, 300 mM NaCl, 20 mM MgCl2, 2 mM EDTA, 2 mM Na3VO4, 10 mM NaF, containing two tablets of protease inhibitor (Roche Complete per 50 ml buffer). 400 μg protein was incubated with 100 μg GST-Byr2-RBD on glutathione-Sepharose beads (Amersham Biosciences) at 4°C for 1 hour. 50 μl of 1× SDS gel loading buffer was added to pelleted beads and the samples subjected to SDS-PAGE and western blot analysis with anti-Rap1 antibody.
F-actin staining
Cells in exponential growth phase were deposited onto glass coverslips in KK2. Cells were fixed in KK2 containing 0.1% Triton X-100 and 1% glutaraldehyde for 10 minutes. Cells were then stained with FITC-conjugated phalloidin for 30 minutes in the dark before mounting.
Adhesion assays
Cell substrate adhesion was measured using an adaptation of a published method (Fey et al., 2002). Briefly, 5×105 cells were grown on 3 cm tissue culture plates for 12 hours until confluent. The medium was replaced and plates were incubated for a further hour. Plates were then shaken at 120 r.p.m. for 20 minutes, after which the medium was removed and collected. The remaining adhering cells were detached from plates by repeated pipetting and collected. The number of cells in each of the two samples was determined in triplicate using a haemoctyometer. To quantify cell-cell adhesion, cells were developed on KK2 agar plates. Cells were harvested from plates and dissociated in a 21G needle in 1.0 ml KK2. 5×105 cells/ml were placed in a 1.5 ml tube and shaken at 120 r.p.m. for various times. The number of single cells in the sample was counted in triplicate using a haemocytometer. For quantification of prestalk and prespore cell-cell adhesion, cells expressing ecmAO-RFP or psA-RFP were used. The percentage of single RFP expressing cells was calculated out of the total number of RFP expressing cells in the sample.
Chemotaxis and random movement assay
To make cells aggregation competent, they were starved at 2×107 cell/ml. Cells were pulsed with cAMP for 5.5 hours. For analysis of chemotaxis, cells were challenged with a micropipette containing 1-2 μM cAMP. Track data were extracted using the `Manual Tracking' plug-in of ImageJ (Wayne Rasband, NIH) and motility parameters calculated. Speed was calculated from the positions of successive movements of the centroid of the cell, as was the persistence (the cosine of the angle of the path between centroids).
Supplementary Material
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/3/335/DC1
We thank Peter Van Haastert, Arjan Kortholt and the DictyBase Stock Centre for kindly providing strains and plasmids used in this study. We also thank Adam Hurlstone for his help with deconvolution microscopy. This work was supported by grants from the Medical Research Council, the Wellcome Trust and the Lister Institute of Preventive Medicine. Deposited in PMC for release after 6 months.
References
- Abe, T., Langenick, J. and Williams, J. G. (2003). Rapid generation of gene disruption constructs by in vitro transposition and identification of a Dictyostelium protein kinase that regulates its rate of growth and development. Nucleic Acids Res. 31, 107-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolourani, P., Spiegelman, G. B. and Weeks, G. (2008). Rap1 activation in response to cAMP occurs downstream of Ras activation during Dictyostelium aggregation. J. Biol. Chem. 283, 10232-10240. [DOI] [PubMed] [Google Scholar]
- Bos, J. L. (1998). All in the family? New insights and questions regarding interconnectivity of Ras, Rap1 and Ral. EMBO J. 17, 6776-6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos, J. L. (2005). Linking Rap to cell adhesion. Curr. Opin. Cell Biol. 17, 123-128. [DOI] [PubMed] [Google Scholar]
- Bosgraaf, L., Waijer, A., Engel, R., Visser, A. J. W. G., Wesseis, D., Soll, D. and van Haastert, P. J. M. (2005). RasGEF-containing proteins GbpC and GbpD have differential effects on cell polarity and chemotaxis in Dictyostelium. J. Cell Sci. 118, 1899-1910. [DOI] [PubMed] [Google Scholar]
- Chisholm, R. L. and Firtel, R. A. (2004). Insights into morphogenesis from a simple developmental system. Nat. Rev. Mol. Cell. Biol. 5, 531-541. [DOI] [PubMed] [Google Scholar]
- Coates, J. C., Grimson, M. J., Williams, R. S., Bergman, W., Blanton, R. L. and Harwood, A. J. (2002). Loss of the beta-catenin homologue aardvark causes ectopic stalk formation in Dictyostelium. Mech. Dev. 116, 117-127. [DOI] [PubMed] [Google Scholar]
- Cornillon, S., Gebbie, L., Benghezal, M., Nair, P., Keller, S., Wehrle-Haller, B., Charette, S. J., Bruckert, F., Letourneur, F. and Cosson, P. (2006). An adhesion molecule in free-living Dictyostelium amoebae with integrin beta features. EMBO Rep. 7, 617-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early, A. E., Gaskell, M. J., Traynor, D. and Williams, J. G. (1993). Two distinct populations of prestalk cells within the tip of the migratory Dictyostelium slug with differing fates at culmination. Development 118, 353-362. [DOI] [PubMed] [Google Scholar]
- Early, A., Abe, T. and Williams, J. (1995). Evidence for positional differentiation of prestalk cells and for a morphogenetic gradient in Dictyostelium. Cell 83, 91-99. [DOI] [PubMed] [Google Scholar]
- Fey, P., Stephens, S., Titus, M. A. and Chisholm, R. L. (2002). SadA, a novel adhesion receptor in Dictyostelium. J. Cell Biol. 159, 1109-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, Y., Hogan, C. and Braga, V. M. M. (2006). Regulation of cell-cell adhesion by Rap1. Methods Enzymol. 407, 359-372. [DOI] [PubMed] [Google Scholar]
- Gebbie, L., Benghezal, M., Cornillon, S., Froquet, R., Cherix, N., Malbouyres, M., Lefkir, Y., Grangeasse, C., Fache, S., Dalous, J. et al. (2004). Phg2, a kinase involved in adhesion and focal site modeling in Dictyostelium. Mol. Biol. Cell 15, 3915-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginger, R. S., Drury, L., Baader, C., Zhukovskaya, N. V. and Williams, J. G. (1998). A novel Dictyostelium cell surface protein important for both cell adhesion and cell sorting. Development 125, 3343-3352. [DOI] [PubMed] [Google Scholar]
- Gomer, R., Gao, T., Tang, Y. T., Knecht, D. and Titus, M. A. (2002). Cell motility mediates tissue size regulation in Dictyostelium. J. Muscle Res. Cell Motil. 23, 809-815. [DOI] [PubMed] [Google Scholar]
- Grimson, M. J., Coates, J. C., Reynolds, J. P., Shipman, M., Blanton, R. L. and Harwood, A. J. (2000). Adherens junctions and beta-catenin-mediated cell signalling in a non-metazoan organism. Nature 408, 727-731. [DOI] [PubMed] [Google Scholar]
- Harloff, C., Gerisch, G. and Noegel, A. A. (1989). Selective elimination of the contact site A protein of Dictyostelium discoideum by gene disruption. Genes Dev. 3, 2011-2019. [DOI] [PubMed] [Google Scholar]
- Jeon, T. J., Lee, D. J., Lee, S., Weeks, G. and Firtel, R. A. (2007a). Regulation of Rap1 activity by RapGAP1 controls cell adhesion at the front of chemotaxing cells. J. Cell Biol. 179, 833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon, T. J., Lee, D. J., Merlot, S., Weeks, G. and Firtel, R. A. (2007b). Rap1 controls cell adhesion and cell motility through the regulation of myosin II. J. Cell Biol. 176, 1021-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kae, H., Lim, C. J., Spiegelman, G. B. and Weeks, G. (2004). Chemoattractant-induced Ras activation during Dictyostelium aggregation. EMBO Rep. 5, 602-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, R. J., Kae, H., Ip, H., Spiegelman, G. B. and Weeks, G. (2002). Evidence for a role for the Dictyostelium Rap1 in cell viability and the response to osmotic stress. J. Cell Sci. 115, 3675-3682. [DOI] [PubMed] [Google Scholar]
- Kessin, R. H. (2001). Dictyostelium. Cambridge: Cambridge University Press.
- Kitayama, H., Sugimoto, Y., Matsuzaki, T., Ikawa, Y. and Noda, M. (1989). A Ras-related gene with transformation suppressor activity. Cell 56, 77-84. [DOI] [PubMed] [Google Scholar]
- Knecht, D. A., Cohen, S. M., Loomis, W. F. and Lodish, H. F. (1986). Developmental regulation of Dictyostelium discoideum actin gene fusions carried on low-copy and high-copy transformation vectors. Mol. Cell. Biol. 6, 3973-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox, A. L. and Brown, N. H. (2002). Rap1 GTPase regulation of adherens junction positioning and cell adhesion. Science 295, 1285-1288. [DOI] [PubMed] [Google Scholar]
- Kooistra, M. R., Dube, N. and Bos, J. L. (2007). Rap1: a key regulator in cell-cell junction formation. J. Cell Sci. 120, 17-22. [DOI] [PubMed] [Google Scholar]
- Kortholt, A., Rehmann, H., Kae, H., Bosgraaf, L., Keizer-Gunnink, I., Weeks, G., Wittinghofer, A. and Van Haastert, P. J. M. (2006). Characterization of the GbpD-activated Rap1 pathway regulating adhesion and cell polarity in Dictyostelium discoideum. J. Biol. Chem. 281, 23367-23376. [DOI] [PubMed] [Google Scholar]
- Krieg, M., Arboleda-Estudillo, Y., Puech, P. H., Kafer, J., Graner, F., Muller, D. J. and Heisenberg, C. P. (2008). Tensile forces govern germ-layer organization in zebrafish. Nat. Cell Biol. 10, 429-436. [DOI] [PubMed] [Google Scholar]
- Lam, T. Y., Pickering, G., Geltosky, J. and Siu, C. H. (1981). Differential cell cohesiveness expressed by prespore and prestalk cells of Dictyostelium discoideum. Differentiation 20, 22-28. [DOI] [PubMed] [Google Scholar]
- Lorenowicz, M. J., van Gils, J., de Boer, M., Hordijk, P. L. and Fernandez-Borja, M. (2006). Epac1-Rap1 signaling regulates monocyte adhesion and chemotaxis. J. Leukoc. Biol. 80, 1542-1552. [DOI] [PubMed] [Google Scholar]
- Louis, S. A., Weeks, G. and Spiegelman, G. B. (1997). Rap1 overexpression reveals that activated RasD induces separable defects during Dictyostelium development. Dev. Biol. 190, 273-283. [DOI] [PubMed] [Google Scholar]
- Maeda, M., Haruyo, S., Maruo, T., Ogihara, S., Iranfar, N., Fuller, D., Morio, T., Urushihara, H., Tanaka, T. and Loomis, W. F. (2003). Changing patterns of gene expression in prestalk cell subtypes of Dictyostelium recognised by in situ hybridisation with genes from microarray analyses. Eukaryotic Cell 2, 627-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruo, T., Sakamoto, H., Iranfar, N., Fuller, D., Morio, T., Urushihara, H., Tanaka, Y., Maeda, M. and Loomis, W. F. (2004). Control of cell type proportioning in Dictyostelium discoideum by differentiation-inducing factor as determined by in situ hybridization. Eukaryotic Cell 3, 124-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukuma, S. and Durston, A. J. (1979). Chemotactic cell sorting in Dictyostelium discoideum. J. Embryol. Exp. Morphol. 50, 243-251. [PubMed] [Google Scholar]
- McLeod, S. J., Shum, A. J., Lee, R. L., Takei, F. and Gold, M. R. (2004). The Rap GTPases regulate integrin-mediated adhesion, cell spreading, actin polymerization, and Pyk2 tyrosine phosphorylation in B lymphocytes. J. Biol. Chem. 279, 12009-12019. [DOI] [PubMed] [Google Scholar]
- Rebstein, P. J., Cardelli, J., Weeks, G. and Spiegelman, G. B. (1997). Mutational analysis of the role of Rap1 in regulating cytoskeletal function in Dictyostelium. Exp. Cell Res. 231, 276-283. [DOI] [PubMed] [Google Scholar]
- Reedquist, K. A., Ross, E., Koop, E. A., Wolthuis, R. M., Zwartkruis, F. J., van Kooyk, Y., Salmon, M., Buckley, C. D. and Bos, J. L. (2000). The small GTPase, Rap1, mediates CD31-induced integrin adhesion. J. Cell Biol. 148, 1151-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins, S. M., Suttorp, V. V., Weeks, G. and Spiegelman, G. B. (1990). A Ras-related gene from the lower eukaryote dictyostelium that is highly conserved relative to the human Rap genes. Nucleic Acids Res. 18, 5265-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roisin-Bouffay, C., Jang, W., Caprette, D. R. and Gomer, R. H. (2000). A precise group size in Dictyostelium is generated by a cell-counting factor modulating cell-cell adhesion. Mol. Cell 6, 953-959. [PubMed] [Google Scholar]
- Seastone, D. J., Zhang, L., Buczynski, G., Rebstein, P., Weeks, G., Spiegelman, G. and Cardelli, J. (1999). The small Mr Ras-like GTPase Rap1 and the phospholipase C pathway act to regulate phagocytosis in Dictyostelium discoideum. Mol. Biol. Cell 10, 393-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimonaka, M., Katagiri, K., Nakayama, T., Fujita, N., Tsuruo, T., Yoshie, O. and Kinashi, T. (2003). Rap1 translates chemokine signals to integrin activation, cell polarization, and motility across vascular endothelium under flow. J. Cell Biol. 161, 417-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg, M. S. and Takeichi, M. (1994). Experimental specification of cell sorting, tissue spreading, and specific spatial patterning by quantitative differences in cadherin expression. Proc. Natl. Acad. Sci. USA 91, 206-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman, M. (1987). Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol. 28, 9-29. [DOI] [PubMed] [Google Scholar]
- Thompson, C. R. L., Reichelt, S. and Kay, R. R. (2004). A demonstration of pattern formation without positional information in Dictyostelium. Dev. Growth Differ. 46, 363-369. [DOI] [PubMed] [Google Scholar]
- Wang, J., Hou, L., Awrey, D., Loomis, W. F., Firtel, R. A. and Siu, C. H. (2000). The membrane glycoprotein gp150 is encoded by the lagC gene and mediates cell-cell adhesion by heterophilic binding during Dictyostelium development. Dev. Biol. 227, 734-745. [DOI] [PubMed] [Google Scholar]
- Weeks, G., Gaudet, P. and Insall, R. (2005). The small GTPase superfamily. In Dictyostelium Genomics (ed. W. F. Loomis and A. Kuspa), pp. 173-210. Norfolk: Horizon Bioscience.
- Williams, J. G., Duffy, K. T., Lane, D. P., Mcrobbie, S. J., Harwood, A. J., Traynor, D., Kay, R. R. and Jermyn, K. A. (1989). Origins of the prestalk-prespore pattern in Dictyostelium development. Cell 59, 1157-1163. [DOI] [PubMed] [Google Scholar]
- Wong, E., Yang, C., Wang, J., Fuller, D., Loomis, W. F. and Siu, C. H. (2002). Disruption of the gene encoding the cell adhesion molecule DdCAD-1 leads to aberrant cell sorting and cell-type proportioning during Dictyostelium development. Development 129, 3839-3850. [DOI] [PubMed] [Google Scholar]
- Yamada, Y., Sakamoto, S., Ogihara, S. and Maeda, Y. (2005). Novel patterns of the gene expression regulation in the prestalk region along the antero-posterior axis during multicellular development of Dictyostelium. Gene Expr. Patterns 6, 63-68. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. C., Rehmann, H., Price, L. S., Riedl, J. and Bos, J. L. (2005). AF6 negatively regulates Rap1-induced cell adhesion. J. Biol. Chem. 280, 33200-33205. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.