Abstract
This past January, in Cuernavaca Mexico, a conglomerate of scientists met to discuss the contemporary view of Bacterial Locomotion and Signal Transduction (BLAST). The BLAST meetings represent a field that has its roots in chemotaxis and the flagellum-based motility but now encompass all types of cellular movement and signalling. The topics varied from the interactions between molecules to the interactions between species. We heard about 3D reconstructions of transmembrane chemoreceptors within cells, new biophysical methods for understanding cellular engines, intricate phosphorelays, elaborate gene networks, new messenger molecules, and emerging behaviours within complex populations of cells. At BLAST X we gained an appreciation for the lifestyle choices bacteria make, how they get to where they are going, and the molecular mechanisms that underlie their decisions. Herein we review the highlights of the meeting.
Where should I go? What should I do? These are questions not only asked by the wayward college student, but also by the simple bacterial cell; and while it may a year abroad and a therapist to answer them in the former case, for the latter, the precise tools of molecular science can be brought to bear on the problem. Ultimately, the behaviour of an organism should be understandable in terms of its macromolecules, and their perpetual dance of changing partners in response to a changing world. The underlying switches, their logic, mechanisms, and consequences inspired the research reported at the 10th biennial meeting on Bacterial Locomotion and Signal Transduction (BLAST), chaired by David Zusman (UC Berkeley) and organized by John Sandy Parkinson (University of Utah), Joe Falke (University of Colorado), Philip Matsumura (University of Illinois) and Michael Manson (Texas A&M University).
Over the past 20 years, BLAST has served as a central congress for the field of bacterial signalling and motility. This is an area of study that began in the 1970s with the discovery of bacterial chemoreceptors and characterization of the flagellar motor. In recent years, our understanding of chemotaxis has undergone a renaissance of sorts, with the realization that chemosensory and motility machinery form complex assemblies that integrate with membranes, cell walls and are utilized for much more than just moving towards or away from a given chemoattractant. An onslaught of new genetic and structural data, as well as the application of in vivo quantification methods, have raised bacterial chemotaxis to arguably the best-understood signalling system in biology. Moreover, many other so-called “two-component systems” have been discovered that regulate a multitude of behaviours including stress responses, quorum sensing, sporulation, cell-growth and virulence. We are just beginning to comprehend how these systems couple with membranes and the cytoskeleton to coordinate different types of motility and drive adaptation to new environments. New investigations of biofilms bring with them interdisciplinary work aimed at understanding the signalling networks that underlie cell growth, differentiation, adhesion and cellular communities. Every two years, the BLAST meeting consolidates the efforts in the above areas of a very collegial group of microbiologists, bacterial physiologists, biochemists and biophysicists. This past January 18–29th, 2009, Blast X was held in Cuernavava Mexico. Here we summarize for those in attendance, and those not, what went on.
Chemoreceptor Structure and Function
The field of two-component signalling traces its origins to Julius Adler’s realization that the migration of bacteria up (or down) chemical gradients is a receptor-mediated processes (Adler, 1975). Over the past several decades the molecular players and their mechanisms, primarily in E. coli, have been worked out with increasing detail (Figure 1). At the same time, quantitative in vivo studies have revealed the remarkable properties of the sensory system in terms of its sensitivity, gain and dynamic range. Major breakthroughs in this area continue to be reported at BLAST. At the heart of the sensory apparatus is a high degree of molecular cooperativity, and this year, we viewed the structural outlines of its manifestation. Much previous work has suggested that chemoreceptors, the CheA kinase and coupling protein CheW cluster at the poles of cells within receptor-kinase arrays. CryoEM tomography from the labs of Sriram Subramaniam (National Institutes of Health) and Grant Jensen (California Institute of Technology) brought this home in striking fashion with high resolution images of the hexagonal arrays the chemoreceptors form in whole cells (Briegel et al., 2008, Khursigara et al., 2008a). In a technical tour-de-force, Ariane Briegel from the Jensen lab showed that these hexagonal lattices are remarkably conserved across a wide range of bacteria, from enteric proteobacteria to Thermotaga (Figure 2). All of the visualized arrays appear to be based on a trimeric assembly of chemoreceptors that had been anticipated by previous studies (Hazelbauer et al., 2008). Cezar Khursigara from the Subramaniam lab added that the receptor clusters in Caulobacter crescentus do not appear to be completely structured within the hexagonal framework, and this could be key for function (Khursigara et al., 2008a).
Figure 1.
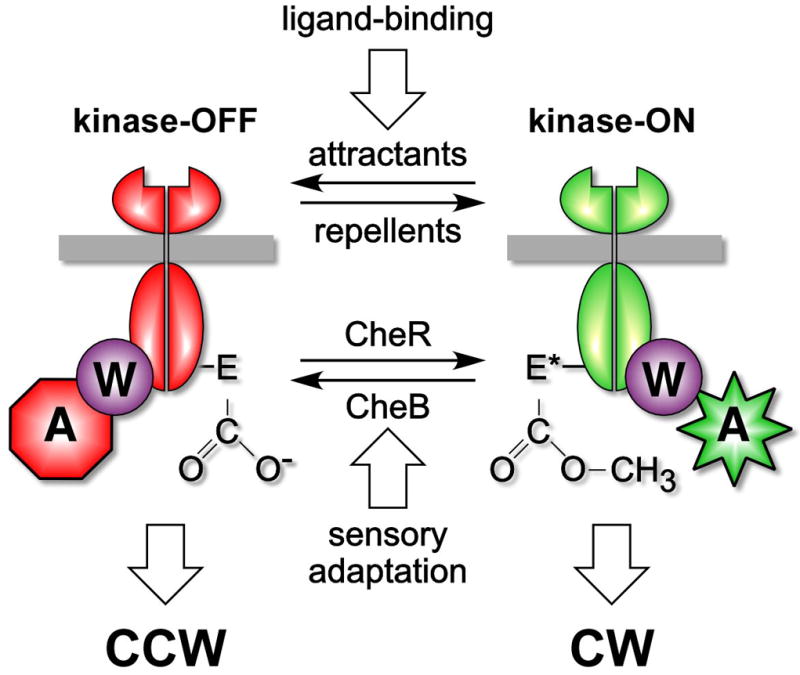
The two-state model of chemoreceptor signaling. Methyl-accepting chemotaxis proteins (MCPs) or chemoreceptors regulate the activity of the signal-transducing histidine kinase CheA, which is coupled to MCPs through CheW. The MCP receptors can be viewed as having two activity states, one that is inhibitory with respect to the kinase and one that is activating. Attractant binding promotes the inhibitory state (and ultimately counter clockwise (CCW) rotation of the flagellar motor), whereas cytoplasmic methylation favors the activating state (and ultimately clockwise (CW) flagellar rotation). Action by the CheR methyltransferase, is countered by the CheB methylesterase, which itself is regulated by CheA through phosphorylation. In this way, kinase activity is used to reset the receptor state equilibrium as the bacteria swim up a gradient of ligand. Courtesy of Sandy Parkinson.
Figure 2.
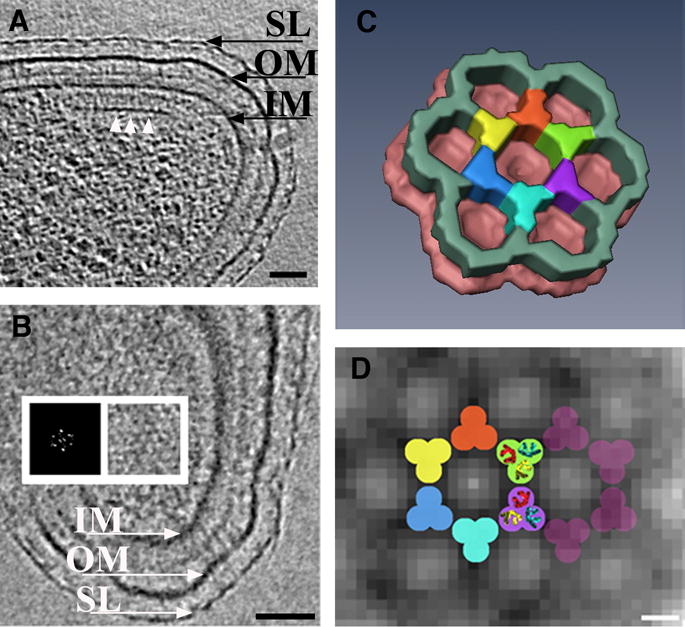
Wild-type chemoreceptor array architecture in Caulobacter crescentus (A) tomographic slice perpendicular to the chemoreceptor array. The base plate composed of CheA-CheW (white arrows) is visible running parallel to the inner membrane, at a distance of 31nm. Thin pillar-like densities connecting the base plate with the inner membrane are evident. (B) Slice through the tomogram parallel to the membranes directly above the base plate. The hexagonal lattice of the chemoreceptor array is visible. A power spectrum confirms this and demonstrates a center-to-center spacing of 12nm (inset). (C) Manually segmented three-dimensional surface representation of a unit cell of the region where the base plate and the chemoreceptor tips connect. Averaging and six-fold symmetry were applied. (D) Model for the arrangement of the receptors superimposed upon tomographic data. The dimensions and symmetry of the bottom half of the trimers-of-dimers from the crystal structure (PDB ID 1QU7) fit well in each intersection of the observed hexagonal lattice. SL: S-Layer, OM: outer membrane, IM: inner membrane. Scale bars: A and B: 50 nm, D: 5nm. Courtesy of Ariane Briegel and Grant Jensen.
Given these new data, the tantalizing question now is how the receptors function within these arrays. Fortunately, dissecting receptor mechanisms is old hat to the BLAST community. A new conceptual dimension, though, is that not all chemoreceptors appear to transmit signals in the same way. How far does what we know about the canonical Escherichia coli receptors Tar and Tsr carry to other systems? George Glekas working with George Ordal (University of Illinois) demonstrated that chemoreceptors from the Gram positive Bacillus subtilis (MCPB) have very different extracellular domains to those of the E. coli MCP proteins Tar and Tsr. Glekas went on to show through homology modelling, mutant studies and behavioural assays, that MCPB resembles the quorum-sensing receptor LuxQ (Neiditch et al., 2006) in that the structure comprises two PAS domains aligned along a central helix. The chemoattractant binds in the “upper” PAS domain and likely sends a signal via transmembrane 1 (TM1), instead of TM2, as in Tar and Tsr (Falke & Hazelbauer, 2001). Jim Remington working with Karen Guillemin (both at the University of Oregon) and Karen Ottemann (UC Santa Cruz) presented a crystal structure from a similar sensory receptor found in Helicobacter pylori. The H. pylori receptor also contains PAS domains in helical association, albeit one instead of two per subunit. The canonical view of signalling by Tar and Tsr, is that ligand binding causes displacement of TM2 in a “piston” like motion that affects the cytoplasmic domains. Support for a non-piston motion in other receptors was provided by work of Roger Draheim and Gunnar von Heijne (Stockholm University), using a combination of biochemical studies, some of which Draheim pioneered while working with Mike Manson (Texas A&M) (Draheim et al., 2006). Careful positioning and monitoring of TM2 in the sensor kinase EnvZ relative to the membrane indicates that a helix rotation rather than a displacement might be the key conformational switch.
An important revelation at the previous BLAST IX was the structure of the chemoreceptor HAMP domain (Hulko et al., 2006), which lies just inside the cytoplasmic membrane and transduces signals between the transmembrane helices and the adaptation and signalling domains. The structure suggested a number of mechanisms that are now being probed, including a rotational “gear-box” model (Hulko et al., 2006), and a membrane association model (Manson, 2008). Work from Gus Wright and Rachel Crowder in Michael Manson’s lab (Texas A&M) probed the effects of substitutions and length on the linker that connects transmembrane helix TM2 to HAMP. Although some substitutions and extensions are tolerated, changes that alter helicity are not, suggesting that HAMP connects to TM2 through a helix, whose length and flexibility can vary.
Although most chemoreceptors use HAMP domains, not all have extracellular ligand binding domains. For example the E. coli aerotaxis receptor (Aer) senses redox via an intracellular flavin-binding PAS domain (Taylor, 2007). Asharie Campbell working with Kylie Watts, Mark Johnson and Barry Taylor (Loma Linda University) and a co-winner of the Kadner Award for best student poster presentation, conducted a genetic screen to find residues in the Aer PAS domain critical for FAD binding and signal transduction. These studies revealed that conformational signals likely propagate from the FAD-binding pocket out to the Ncap (an N-terminal, variable region of PAS domains), where a structural state is unmasked to turn the receptor “on”. Through a set of cross-linking and accessibility studies, Kylie Watts went on to show that changes in the PAS Ncap motion could affect HAMP because the two domains directly interact in Aer. Furthermore, the data point to key changes at the base of the HAMP domain when the receptor is activated. Clearly, there is more than one way for signals to go into HAMP; it remains to be established whether there is only one way out.
Further on down the receptor, we find the most conserved regions of the molecule, those that undergo reversible methylation in the adaptation response, and the highly conserved tip that binds CheA and CheW. Kalin Swain working with Joe Falke (University of Colorado) and also a Kadner awardee, showed that differential stability of the adaptation region vs. the signalling tip is a key factor in kinase activation. Residue substitutions that destabilize packing in the tip activate CheA. On the other hand, destabilizing the adaptation region has the opposite effect. This is interesting because, as Rick Dahlquist (UC Santa Barbara) remarked, there are no known mutations in CheA that activate to the extent that receptors do. One could then reason that the receptors enforce a state of CheA that the molecule cannot otherwise achieve. How this is achieved by destabilizing the interaction domain of the receptor is puzzling. The potential importance of dynamics in receptor function was also apparent from a new crystal structure of a soluble signalling domain of chemoreceptor from T. maritima (Pollard et al., 2009) presented by Abiola Pollard (from the lab of Brian Crane, Cornell University). The structure revealed a new conformational state for the receptor in which a helical bulge in the adaptation regions propagates to the displacement of the receptor tip relative to the helical stalks. However, the most dramatic new insights into receptor conformational change were provided by 3D reconstructions of overproduced Tsr chemoreceptors by cryoEM tomography. Cezar Khursigara (from the Subramaniam lab, NIH) showed reconstructions of trimeric receptor dimers produced at abnormally high levels (Khursigara et al., 2008b). The objects in the images fall into two classes whose dimensions depend on the presence of attractant and receptor modification state (Figure 4). The expanded structures correlate with the inhibitory state (CheA off) and the compact structures correlate with an activated state (CheA on). In both cases, the conserved tips stay associated, but might move up or down as the more proximal membrane regions swell or contract. One caveat of the images is that, in the overproduced state, the receptor tips are locked in a “zipper” conformational that has been shown to correlate with inactivity (Khursigara et al., 2008b). Nonetheless, aspects of the observed conformational changes are likely relevant, and their significance can now be tested.
Figure 4.
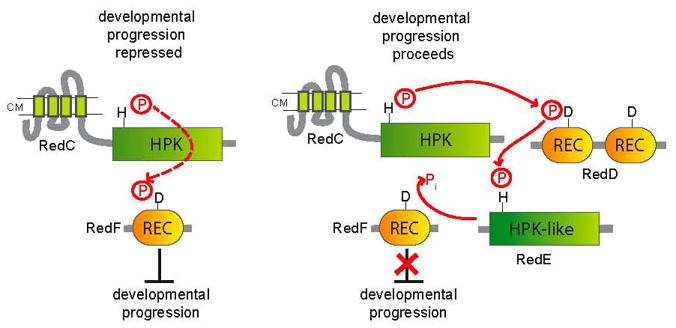
Model for the Red four component TCS mechanism controlling progression through the M. xanthus developmental program. Left panel: Developmental progression is repressed when RedC histidine protein kinase (HPK) phosphorylates the receiver (REC) domain of RedF, a stand-alone response regulator. Right panel: Development proceeds when RedC instead phosphorylates the first REC domain of RedD, a dual receiver response regulator. The phosphoryl group is then transferred to the HPK-like protein, RedE, and RedE acts as a phosphatase on RedF. Courtesy of Penelope Higgs.
Along these lines, Diego Massazza and Claudia Studdert (Universidad Nacional de Mar del Plata), together with Sandy Parkinson (University of Utah) are further extending the original cross-linking studies of Studdert and Parkinson (Studdert & Parkinson, 2004) that provided in vivo evidence for receptor trimers with the goal of finding conformationally specific reporter sites. Consistent with the tomograms, this work also indicates that the receptor timers expand when attractant is bound but that the dimers do not undergo vertical displacements relative to each other. If the right set of reporter sites can be found, the approach has great promise for revealing the details of how the receptors activate.
At the end of the day, it is CheA activity that really matters to the cell, and to understand in depth the chemotaxis system, we must grapple with how receptors regulate the kinase, ultimately in the context of the hexagonal arrays. Annette Erbse (from the Falke lab, University of Colorado) presented an in-depth biochemical study on reconstituted and ex-vivo isolate receptor-kinase complexes integrated in membranes and showed that they remain highly stable and active under many perturbing conditions. This suggests that once assembled, the arrays do not readily exchange components or recruit new factors, but rather function as one huge allosteric enzyme. The extreme stability of the reconstituted arrays conflicts somewhat with in vivo FRAP experiments from Victor Sourjik’s lab that demonstrate exchange of array components on a much faster times scale (Schulmeister et al., 2008). In cells, Erbse and Falke propose that a machinery for disassembling the ultrastable complexes should exist, to limit the size of the complexes and prevent them from filling the cell membrane. The in vivo and ex-vivo results would be reconciled if the disassembly machinery is lost when complexes are isolated from cells. Finally, some direct data on CheA, CheW and receptor interactions was provided by site-specific spin-labelling studies performed by Jaya Bhatnagar (from the groups of Jack Freed and Brian Crane, Cornell University). This method involves systematically modifying the components of a soluble complex of T. maritima chemoreceptor signalling domains, CheA and CheW with nitroxide spin labels and measuring distances of separation with pulsed ESR spectroscopy. Distance restraints among the components map out relative domain orientations and contacts within the complexes that are largely consistent with interfaces identified by other biochemical and genetic methods. Despite these advances, we still don’t have a good model for how CheA and CheW interact with the receptors in the hexagonal lattice, nor for how CheA activity is regulated by receptors. This brings back the important questions of how stable the arrays are, how different the structural states need to be to elicit activity changes, and what dynamic processes allow for interconversion. Discussions lead by Roger Alexander, who with Thierry Emonet (Yale University) is modelling array responses under a number of conditions, left us scratching our heads.
While the molecular mechanisms of chemotaxis were of great interest at BLAST X, so too where their consequences. Elegant FRET readout experiments pioneered by Victor Sourjik (University of Heidelberg) and Howard Berg (Harvard University) have tracked in vivo kinase activity with great accuracy and specificity(Sourjik & Berg, 2004), and allowed quantitative models of signal propagation and adaptation to be developed and validated. Tom Shimizu, who is now working with Berg, carried these methods forward to define the kinetics of receptor adaptation using time-varying attractant stimuli. Unlike findings from early work from Block and Berg, which followed flagellar output (Block et al., 1983), direct monitoring of CheA activity shows the adaptation feedback to be weak, consistent with slow recovery from small perturbations and a noisy steady state. No response thresholds were observed either to positive or negative ramps, so in contrast to the famous line by Berg that “E. coli is an optimist” – not bothering to try new directions when the going is good, Shimizu remarked that “the receptor:kinase complex is a realist” – because it reports on all that is there. Using similar methods, Silke Neumann from the Sourjik lab probed the dose response profile of the less abundant E. coli receptors Tap, Trg, and found that they do not sense with the same dynamic range and sensitivity as the more abundant receptors Tar and Tsr. Notably, these receptors bind their ligands through periplasmic binding proteins, and this primary binding event could dampen the receptor response. Nonetheless, the less abundant receptors can be described well by modification of the allosteric model of coupled receptor clusters that was developed by Ned Wingreen (Princeton University) for the major receptors (Skoge et al., 2006).
BLAST X showed that we have come a long way in understanding chemotaxis, but can we apply what we know to design something new? This challenge is being met in the labs of Bill Degrado and Mark Goulian (University of Pennsylvania). Postdoc Shalom Goldberg presented data from a creative study in which he engineered E. coli to move towards non-standard ligands by introducing an enzyme into the periplasm that converts an inert substrate that the bacteria does not sense into one that it does. This strategy can be extended to generate symbiotic behaviour between cell types, in which one strain provides the attractant to which the other can respond. Like married postdocs searching for academic positions, the two strains size up the world for each other and decide to move off together. Jerry Hazelbauer (University of Missouri) remarked that Julius Adler himself would have been proud of this concept.
Sensor Kinases and Phospho Relays
Although CheA and CheY might represent the archetypal two-component bacterial sensing system, they are really only the tip of the iceberg. Two-component signalling systems, composed of a sensor histidine kinase (HK) that phosphorylates a response regulator (RR) are widely used by microorganisms to sense and adapt to their environment in the broadest sense. In most cases, the RR regulates gene expression via its coupled DNA binding domain. At this meeting, researchers focused on some interesting departures from ‘classical’ two-component systems, as well as on new approaches for better understanding the molecular mechanisms underlying signal stability and protein-protein interactions. Our one-dimensional view of signal transduction has changed. A multi-dimensional network of interactions between diverse signalling chains has emerged in the past years. While several signals are often integrated into one regulatory pathway, it is not uncommon for one signal to regulate several individual pathways.
Sean Crosson (University of Chicago) reported on the LovK/LovR two-component system from Caulobacter crescentus as an example for how the integration of two environmental signals, visible light and oxidative/osmotic stress, coordinate to control cell envelope physiology. The photosensory kinase LovK, possesses a flavin-bound LOV domain. When C. crescentus is exposed to blue light, cell-cell adhesion is accentuated via LovK/LovR (Purcell et al., 2007). A second two-component system in C. crescentus, PhyK/PhyR, mediates the response to oxidative/osmotic stress by positively regulating the expression of the σT regulon. How are both systems interlinked? Phosphorylated LovR inhibits the kinase PhyK, which results in a down regulation of the σT regulon. This integration is a fine example of a feedback-regulated signalling network – an emerging theme in two-component signalling.
Another talk by Alla Kaserer (from the West laboratory, University of Oklahoma) also dealt with complex inputs, in this case, adaptation to osmotic, oxidative and other environmental stresses. Such environmental adaptation in the eukaryote Saccharomyces cerevisiae is mediated through a branched, multi-step phosphorelay system. This system, which is involved in stress response, consists of SLN1, a hybrid kinase, YPD1, a histidine-containing phosphotransfer protein, and two response regulators, SSK1 and SKN7. Kaserer investigated the effect of osmolytes on regulation of the central RR, SSK1. SLN1 is active during non-osmotic stress and shuttles phosphoryl groups to SSK1 via YPD1. Hyperosmotic shock produces a rapid efflux of water and changes in intracellular ion/solute concentration, which are compensated by increasing cellular glycerol levels. Kinetic data suggest that a modest increase in osmolyte concentration reduces the phosphorylated life-time of SSK1, thus facilitating activation of the downstream MAP kinase cascade and glycerol production. In contrast, the combinatory effect of high levels of NaCl and glycerol on rates of phosphotransfer favors phosphorylation of SSK1 and signal attenuation. These changes are an example of how the environmental milieu can directly influence rate constants of the phosphotransfer reactions underlying signaling.
An example of a multifunctional two-component regulatory mechanism is the essential WalK/WalR system that regulates cell wall homeostasis in Staphylococcus aureus, which was presented by Sarah Dubrac (from the Msadek laboratory, Institut Pasteur). WalK/WalR depletion arrests cell wall biosynthesis and turnover, leading to the formation of thicker cell walls with rougher outer surfaces and an abnormal distribution of division septa (Dubrac et al., 2008). The WalK/WalR regulon consists of more than 30 genes, including 10 genes involved in cell wall degradation. In addition to its role in cell wall homeostasis, WalK/WalR also positively regulates biofilm formation and suppresses cell-to-cell aggregation. Dubrac postulated that the multifunctionality of WalK/WalR and its global activation of cell wall degradation contribute to the ability of the bacteria to synchronize essential aspects of growth to the environment.
Amber Bible (from the Alexandre laboratory, University of Tennessee) discussed the multiple two-component pathways that mediate changes in motility and cellular morphology in Azospirillum brasilense. A. brasilense flocculates under high oxygen and limiting nitrogen concentrations. It possesses four chemotaxis operons, of which one, che1, regulates motility, cell-cell-aggregation and exopolysaccharide production associated with flocculation and cell length (Bible et al., 2008). Another chemotaxis operon, che4, was also found to regulate chemotaxis activity and cell length, but not flocculation. Thus, cross regulation between parallel chemotaxis pathways permits sets of cellular functions to be integrated into a complex network.
One of the most valuable organisms for studying prokaryotic multicellularity is Myxococcus xanthus. The progression through the developmental program of fruiting body formation in M. xanthus is regulated by an atypical two-component signal transduction system consisting of four components (RedC-F). RedC is a typical membrane-bound histidine kinase, RedD consists of two receiver domains, RedE is a soluble histidine-kinase-like protein, and RedF is a single receiver domain response regulator (Higgs et al., 2005). Penelope Higgs (Max-Planck-Institute for Terrestrial Microbiology) presented her current model of the temporal regulation of fruiting body formation by the Red-system. Cell aggregation is repressed when RedC phosphorylates RedF, and it is relieved when RedC phosphorylates RedD. However, there is complex crosstalk between RedD and RedF. Another player, RedE can accept phosphate from RedD and then act as a phosphatase for RedF (Figure 3). How this complex network of phosphoryl group transfer ensures the proper progression of development is unclear. This system offers some tantalizing questions such as the nature of the external signals regulating substrate specificity of RedC how the interaction between RedE and RedF is regulated.
Figure 3.
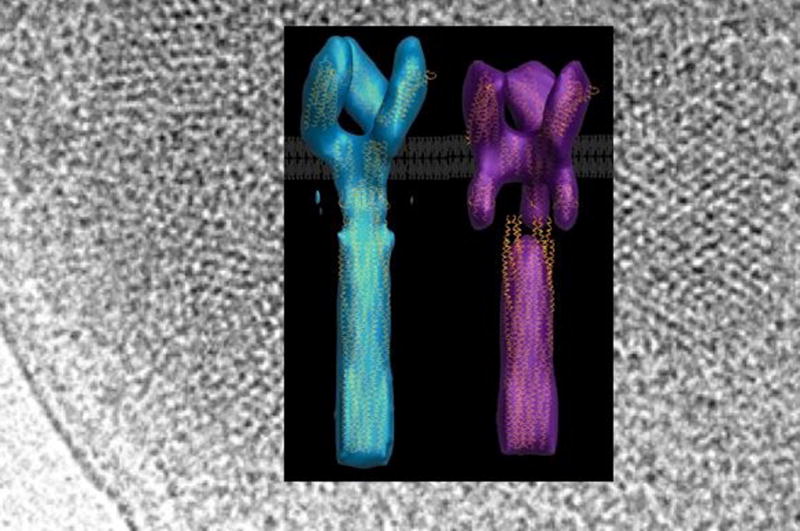
Electron tomography reconstructions of Tsr chemoreceptors overexpressed in whole cells. The chemoreceptor structures fall into two classes: one that is contracted in regions beneath the cytoplasmic membrane that map to the location of the HAMP domain (left) and another that is expanded (right). The proportion of molecules in the expanded state increases when attractant is added, whereas the proportion of molecules in the contracted state increases when modifications are made to the receptor to mimic methylation. The receptors are shown on a background of the hexagonal lattices they form at the poles of cells. Courtesy of Cezar Khursigara and Sriram Subramaniam.
M. xanthus exhibits positive gliding taxis towards phosphatidylethanolamine (PE). Both, the Dif and the Frz pathway are required for excitation and adaptation, respectively (Bonner et al., 2005). Zhaomin Yang (Virginia Tech) analyzed the crosstalk between these pathways. The MCP-homolog DifA senses PE but does not undergo methylation. Instead, signals sensed through DifA modulate methylation of another MCP homolog FrzCD. In addition, and independently of DifA, M. xanthus senses PE through FrzCD. The Dif-independent, Frz-dependent PE-sensing increases FrzCD methylation and subsequent adaptation, while Dif-dependent signalling suppresses FrzCD methylation in order to delay adaptation (Xu et al., 2008). Therefore, methylation of FrzCD is governed by opposing forces from Dif-dependent and Dif-independent sensing mechanisms. Why does excitation through Dif inhibit rather than promote adaptation? Yang reconciled the necessity for such a regulation with the low gliding speed of M. xanthus. In order to allow an appropriate ratio of net movement and adaptation, the latter mechanism is delayed by the interdependence of the Dif and Frz pathways.
Edith Diaz-Mireles (from the Bolam laboratory Newcastle University) received the Robert Macnab Award for the best poster presented by a postdoctoral scientist for her poster “Carbohydrate sensing by a human gut symbiont”. The work, which was carried out by Hongjun Zheng, identified a hybrid two-component protein that binds fructose and controls fructan utilization of Bacteroides thetaiotaomicron, a dominant member of the distal intestinal microbiota. This exciting study promises great steps towards understanding of the perception of complex polysaccharides by bacteria.
During the course of this meeting, it became apparent that we must revise the “one protein-one function” idea. Steven Porter (from the Armitage lab, University of Oxford) illustrated this point with studies of a bifunctional kinase-phosphatase from Rhodobacter sphaeroides (Porter et al., 2008a). R. sphaeroides has a complex chemosensory pathway with multiple histidine kinases and response regulators, but it lacks phosphatases such as CheZ, CheC, FliY or CheX. It has two chemosensory clusters, each harbouring a distinct set of chemotaxis proteins (Porter et al., 2008b). The four CheA histidine kinases of R. sphaeroides exhibit different domain organization. While CheA1 and CheA2 resemble E. coli CheA (domains P1-P5) and form homodimers, CheA3 and CheA4 together form a functional CheA. CheA4 consists of only the P3, P4 and P5 domains and phosphorylates CheA3, which contains a novel 794-residue domain between its P1 and P5 domains. Porter showed that CheA3 is both a principal phosphodonor of CheY1 and CheY6, and also a specific phosphatase for CheY6. Dephosphorylation of CheY6 is accelerated by a factor of 3 due to CheA3 activity. Therefore, the dephosphorylation half-life of CheY6 is 1.4 s, which correlates well with the 1 sec stimulus response time of R. sphaeroides. Porter then localized the phosphatase activity to a 200-aa segment in the novel phosphatase domain of CheA3. It will be interesting to learn more about the underlying molecular mechanism for response regulator dephosphorylation in this unique bifunctional kinase/phosphatase.
Autophosphorylation in homodimeric histidine kinases is usually envisaged as an intermolecular reaction (i.e. one subunit phosphorylates the other) (Swanson et al., 1993). Gabriela Peña-Sandoval from the Georgellis laboratory (Universidad Nacional Autonoma de Mexico) proved that this paradigm does not apply to the ArcB system. She applied complementation analysis and in vitro phosphorylation assays to show that the phosphorylation site and the kinase activity of ArcB must be present in the same subunit. The E. coli ArcB sensor kinase is a tripartite membrane-spanning sensor kinase that is activated during anoxic growth. ArcB/ArcA/RssB constitute a branched “three-component system”, with ArcA-P acting as an activator of gene expression (Malpica et al., 2006). ArcB is silenced under aerobic conditions, when quinines oxidize Cys residues in the cytoplasmic PAS domain to intermolecular disulfide bridges.
The necessity for a synchronized time scale for adaptive responses became apparent in the studies presented by Yang on the phosphatidylethanolamine taxis of M. xanthus, as well as in Porter’s report on the bifunctional kinase-phosphatase in R. sphaeroides. What molecular mechanisms allow the kinetics of signal transduction to match the timescale of life? Robert Bourret (University of North Carolina) tackled this particular question with a structural approach to response regulator autodephosphorylation. Mutations changing positions 14, 59 and 89 in E. coli CheY changed the dephosphorylation rate by a factor of 100 (Thomas et al., 2008). Bourret presented high-resolution X-ray crystal structures for five variant CheY proteins in complex with the phosphoryl mimic BeF3−. He then compared these structures with the corresponding wild-type CheY structure as well as with the structures of four different response regulators (ArcA, DctD, PhoB, and Spo0F) that exhibit much slower autodephosphorylation rates. The structural analysis showed that specific side chains occlude a nucleophilic water molecule from the phosphoryl group, thereby reducing the rate of autodephosphorylation. In addition, interactions between residues may function to delay conformational change from the active to the inactive conformation, which would reduce the rate of the autodephosphorylation reaction if the two events were coupled.
It is intriguing that phosphorelays generate a high degree of specificity and variation from the same building blocks: the HisKA and RR domains. How is such specificity achieved? The current availability of large sequence databases from bacterial genomes led Hendrik Szurmant with colleagues Jim Hoch (The Scripps Research Institute), M. Weight (Institute for Scientific Interchange Foundation, Italy) Ed. White and T. Hwa (UC San Diego) to mine specific protein interaction surfaces, merely from sequence data. Previous methods were successful in identifying residue positions on two interacting proteins that co-varied across a sequence family, but it had been difficult to differentiate indirect effects from those resulting from direct contacts (Szurmant et al., 2008). Szurmant and colleagues have now combined a co-variance based approach with global interference analysis to build up networks of interacting sites and thereby distinguish indirect interactions from direct ones. The new method was successful in application to a set of over 2500 representatives of bacterial two component systems and allowed identification of hetero-interactions between sensor kinases and response regulators as well as homo-interactions between response regulators (Weigt et al., 2009). Szurmant indicated that this new approach could allow the prediction of interaction surfaces important for the assembly of multi-protein complexes, even when only one copy of the partner is present per genome.
Overall, it became strikingly apparent during this meeting that our understanding of two-component signalling systems is being dramatically impacted by the realization of how bacteria adapt to different environments through the evolution of new sensory kinases and phosphorelay systems. With this in mind, a huge gain of knowledge can be expected from the study of two-component systems in pathogenic and symbiotic bacteria.
The Physical Control of Movement
A central player of the BLAST meeting is the bacterial flagellum. This fascinating rotary nanomachine has been the focus of our attention for over 30 years and still is able to deliver new insights into its function, assembly and how the cell regulates its uses. As well as dissecting just how the chemotaxis system processes environmental signals that dictate the rotary state of the flagellum, the most recent BLAST focused on two important aspects of flagellar biology – regulation and further insights into how the structure is rotated.
Flagellar-mediated motility is the primary mode of directed movement in the liquid environment of planktonic cells. This is in contrast to surface associated movement that utilises either flagella or type IV pili, as exemplified in M. xanthus. Recent advances in microscopy techniques are beginning to aid our research into how flagella promote surface movement (Sowa & Berry, 2008b), several posters focused on describing new insights into communal movement of E. coli using such techniques. A key leap forward in our appreciation of the flagellar motor has been achieved through recent studies into the temporal dynamics of specific structural components and the underlying gene regulation that coordinates flagellar assembly and gene expression.
Flagellar assembly is a temporal process that occurs from the base up. The structure is anchored into the bacterial membrane by the basal body, which is held in place by two outer rings that associate with the peptidoglycan and LPS of Gram-negative bacteria. The basal body is assembled from over 20 structural proteins (Berg, 2003). Three of these proteins form a cytoplasmic ring known as the C-ring that assembles at the cytoplasmic interface of the inner membrane anchor FliF. It is the C-ring that interacts with the motor force generators, otherwise known as the stators, and the chemotaxis machinery. While the interaction of the chemotaxis protein CheY dictates how the flagellum is rotated, the stators are responsible for the physical rotation of the flagellum. For many flagellated bacterial species, these stators comprise two proteins, MotA and MotB. MotAB functions as a H+ pump that generates the necessary proton motive force to promote the interactions between MotAB and the C-ring and rotate the flagellum (Kojima & Blair, 2004). However, we know of an ever-growing number of flagellar systems that use a Na+ pump in the place of MotAB. Na+-dependent stators are often found in bacteria that utilise two flagella systems, such as Vibrio alginolyticus where each system employs a different stator (McCarter, 2001). Therefore, it is not that surprising, but still intriguing, that there are systems that might use either a H+ or Na+ dependent stator to rotate the same flagellum. Kai Thormann (Max Planck Institute for Terrestrial Microbiology) presented the elegant story of how one such two stator/one flagellum system utilises both pumps (Paulick et al., 2009). He described the way Shewanella oneidensis utilises a H+ dependent stator at low NaCl concentrations and switches to the Na+ dependent stator at high NaCl concentration. Thus, S. oneidensis maintains the functionality of its flagellum by sensing the ionic concentration of the environment and adjusting the use of two stators accordingly. The next intriguing question to explore is how the sensing mechanism is coupled to stator utilisation.
Continuing on the stator theme, Seiji Kojima (Nagoya University) presented data on the structure of MotB from Salmonella enterica serovar Typhimurium.. The study of the E. coli and S. Typhimurium flagellar systems over the last three decades has resulted in the development of an intricate atomic-level picture of the flagellum structure. The stator proteins MotA and MotB interact with the flagellum in an A4B2 configuration. MotA is an integral inner membrane protein that acts as the proton pump and MotB is a partially periplasmic protein proposed regulate the activity of MotA. MotB possesses a characteristic C-terminal peptidoglycan-binding motif. A recently-proposed model based on the crystal structure of the H. pylori MotB homologue explains how this motif could interact with peptidoglycan (Roujeinikova, 2008). Kojima presented a larger structure of the S. Typhimurium MotB C-terminal domain than used by Roujeinikova. Assuming that the distance between the peptidoglycan layer and inner membrane is 100 Å, the new structural data suggests that a large conformational change is needed for MotB to activate the MotAB pump.
The stators of the flagellum interact with the C-ring, which is comprised of many copies of three proteins: FliG, FliM and FliN. MotA interacts with FliG, while FliM is the docking site for CheY-P, although all three components are required for torque generation (Sowa & Berry, 2008a). Other than the discussion on stator structure and function of the flagellum, BLAST X paid special attention to the development of techniques and models to investigate the switching mechanism of directional rotation directed by CheY-P:FliM interactions. Richard Branch (from the Berry group, Oxford University) described a mathematical model that quantifies the conformational spread mechanism proposed by Duke (Bray & Duke, 2004) using new high-resolution switching data. This approach validated further the use of conformational spread to model how CheY-P interacts within a given FliG:M:N of the ring to instigate a chain reaction through neighbouring complexes. The model and data presented by Branch suggests that incomplete switches will occur depending on the level of CheY-P interaction. Following from Branch’s talk, Peter Reuven from the Eisenbach group (Weizmann Institute, Israel) described an unstable CheY-YFP protein chimera that they have created to investigate whether the CheY-P switch demonstrates hysteresis. The unstable fusion will allow precise control over intracellular CheY-P concentration. In part, both studies have been motivated from the inability of the previous data of Cluzel to fit perfectly on a Hill curve of traditional cooperativity (Cluzel et al., 2000). The imperfect fits suggest that the interaction and disassociation of CheY-P within the Cring might follow more than one rate. The comparison of the CheY:C-ring interaction through fluorescence-calibrated correlation spectroscopy when CheY-YFP is either increasing or decreasing should help explain some of the previous observations regarding this key interaction.
Still on the subject of flagellar rotation, Mathieu Gauthier (from the lab of Simon Rainville, Laval University) discussed the use of lasers to punch holes in bacterial membranes and gain control of flagellar rotation. This project was inspired by earlier work in which Fung and Berg (Fung & Berg, 1995) used chemical damage to obtain access to the cytoplasm of immobilized E. coli cells. Following this strategy, Gauthier and colleagues immobilized a motile cell within in a microcapillary such that a free flagellum hung out from the tip of the tube. By ablating the cell membrane on the inner side with a finely tuned laser pulse, they could manipulate and monitor conditions on both sides of the membrane. For example, the ability to precisely control the proton motive force allows for very accurate correlations between driving force and flagellar motion. This method promises new insight into the detailed mechanism of torque generation.
Many of the studies mentioned so far focus on a single flagellum in a particular region of the cell surface. However, in reality, bacteria such as E. coli and S. typhimurium utilize multiple flagella orientated in a peritrichous manner over the cell surface (Macnab, 2003). There are between 4 and 10 flagella in these two bacteria (Jarrell & McBride, 2008, Macnab, 2003). As well as being the focus of attention for the biophysicists, the E. coli and S. typhimurium systems have for long entertained molecular geneticists interested in how the assembly process is regulated. We heard two talks on aspects of this regulatory network. Katsumi Imada (Osaka University) presented the structure of FliT and discussed its implications for the function of this protein and its relationship with other Type III secretion system chaperone structures. FliT is the chaperone for the filament cap protein FliD. However, it also plays a regulatory role during flagellar assembly, as it can negatively regulate the activity of FlhD4C2 through a direct protein-protein interaction (Yamamoto & Kutsukake, 2006).
The regulation of FlhD4C2 by FliT is only one regulatory circuit that modulates flagellar gene expression in response to flagellar assembly. In the enteric bacteria, an integrated regulatory network coordinates flagellar gene expression and assembly by modulating the activities of either FlhD4C2 or the flagellar specific sigma factors σ28 (See (Apel & Surette, 2008, McCarter, 2006) and (Aldridge & Hughes, 2002)for recent reviews). The main regulatory signals contributed through the FlgM, FliT and FliZ proteins. FlgM is an anti-σ28 factor that is secreted once the Hook Basal Body (HBB) is complete. This concept of utilising secretion to sense assembly is seen throughout all the regulatory circuits involved in regulating flagellar gene expression. It plays a critical role in activating σ28 dependent transcription during a Fla− to Fla+ transition (Chevance & Hughes, 2008). In contrast, FliT and FliZ have opposite effects upon FlhD4C2 activity, FliZ being a positive regulator of FlhD4C2 activity, in contrast to FliT. Chris Rao (University of Illinois) presented some recent work done in collaboration with the Aldridge lab on the fine-tuning of the rate of flagellar gene expression in response to changes in the secretion rate of growing flagella (Brown et al., 2008). This work asks how the system responds to the assembly of multiple flagella, rather than the first one to be built. Rao presented a mathematical model that focused on the response of the FlgM: σ28 regulatory circuit to changes in secretion rates. This model builds on dynamic gene expression data from studies in which the rate of FlgM secretion was reduced by using a FlgM chimera that is four times the size of FlgM itself (Brown et al., 2008).
In summary, BLAST X focused on the functional rotation of a flagellum and touched upon the regulation of the assembly process. Key to these talks were the use of new technologies to obtain data sets at previously unachievable resolution and with much more control over the systems being studied. It was clear that while the underlying principles of the function and assembly of a flagellum are well documented, there are still some fascinating mechanisms that remain unexplored. Like phosphorelays, one area that is gaining momentum is the study of central concepts involving a much wider range of bacterial species than has hitherto been possible, largely because of the ongoing accumulation of genomic data. These alternative systems will surely offer new and interesting opportunities for the next generations to build upon the truisms established by the pioneers the field.
Consequences of decision making – gene regulation
The goal of sensory systems is to perceive a given signal and utilise this information to dictate a response. The two outputs of sensory systems primarily discussed at BLAST X were the physical control of cellular processes by chemotaxis-like systems and the control of gene expression. The critical question for all signalling systems is what to do with the information once it has been passed through the sensory cascade to the output domains of the regulatory components. As discussed above, the classic output of two-component systems is activation of a RR-transcription factor. The net result will be either up-regulation or down-regulation of target gene expression often via changes in transcription. W heard about several new findings with respect to cross-talk between multiple regulatory pathways. Some examples include systems that coordinate subsets of genes within a regulatory network, the gene expression patterns of two alternative life styles, or the expression of different virulence factors at different stages of an infection.
In this latter respect, BLAST X discussed the use of regulatory systems during inter species and host-bacterial interactions. Two examples of such regulation were touched upon by talks from the Spiro group (University of Texas at Dallas) and the Bustamante group from Cuernavaca (Universidad Nacional Autonoma de Mexico). Jonathan Partridge from the Spiro group described some new data obtained from their recent ChIP-Chip experiments designed to identify genes regulated by the nitric oxide (NO) sensor NsrR (Rankin et al., 2008). In this talk data were presented that indicates NsrR regulates motility in response to NO challenge. Specifically, NsrR was suggested to be a negative regulator of flagellar gene expression based on its interaction with a number of FlhD4C2 dependent promoters. By this mechanism NO has the potential to stimulate motility; clearly further investigation will explain why such a response is employed. Interestingly, the response of E. coli observed by Partridge and Spiro is somewhat similar to what has been observed in P. aeruginosa, where NO exposure causes biofilm dispersal (Barraud et al., 2006).
Staying on the pathogenicity aspect Luary Martinez from Victor Bustamante’s group described their recent work on the control of Salmonella virulence gene expression by the BarA/SirA system. There has been a growing interest in how Salmonella controls the expression of the virulent type 3 secretions systems (SPI-1 and SPI-2) during pathogenesis. The Bustamante group have recently shown that the major transcriptional regulator HilD controls expression from both SPI-1 and SPI-2 (Bustamante et al., 2008). Martinez discussed how the two-component system BarA/SirA can influence HilD regulation of SPI-1 and SPI-2. She described data that confirmed previous findings suggesting that the regulation of hilD expression by SirA is not direct but through the csrABC system (Chevance & Hughes, 2008) (Fortune et al., 2006). However, their data, in addition to shedding more light on the intricacies of this regulatory network, has also posed more questions on how hilD expression responds to environmental signals.
Behavioural Responses: Individuality and Community Living
One of the major lifestyle decisions that two-component systems influence is whether to coordinate behaviour with other cells. For cells to interact with each other they must be physically close; not surprisingly, changes in motility are tightly coupled to such choices. At BLAST X, we heard about novel behaviours of cell populations and the regulatory switches that promote neighbourliness.
In addition to being a paradigm for multicellular development, Myxococcus xanthus is a long standing model system for studying coordinated motility. As part of their lifecycle, M. xanthus cells move together and coalesce into fruiting bodies of differentiated cells (Zusman et al., 2007). The associated motion requires so-called “gliding motility”, which is very different from flagellar motility but is controlled by sensory systems not unlike those found in E. coli chemotaxis. The M. xanthus Frz chemosensory system holds much interest as the central regulatory system for the cellular reversals that characterize gliding (Zusman et al., 2007). Emilia Mauriello (from the Zusman lab, UC Berkeley) is exploring intracellular localization of the MCP homolog FrzCD, which regulates the CheA homolog FrzE (Mauriello, 2009). FrzCD appears to decorate a helical filament with the same pitch as MreB, the actin homolog. FrzCD function and its capacity for adaptation (via methylation) is related to this clustering. Since Myxococci usually move in groups, an interesting question is what happens to FrzCD clusters when cells are together? Amazingly, FrzCD clusters align in neighbouring cells. Side-to-side contacts between cells correlate with FrzCD localization. Furthermore, it appears that the intercellular associations induce reversals. Thus, in order to coordinate their movement, M. xanthus regulates the positioning of intracellular structures controlling motility. These results suggest that FrzCD detects and responds to signals from a cell contact sensitive signaling system (Mauriello, 2009).
The lab of John Kirby (University of Iowa) has discovered an entirely new cooperative behaviour of M. xanthus – predataxis (Berleman et al., 2008). Jeb Berleman from the Kirby lab reported how M. xanthus colonies show a rippling, multicellular structure that only occurs when eating prey, such as E. coli (Figure 5). Myxococcus populations have a much different form when exploring new territory. On investigation of the involvement of the Frz system in this behaviour, Berleman found that mutations affecting both FrzF and FrzG, the methyltransferase and methylesterase that respectively modify FrzCD, are defective in predation. However, the frzG mutant is hyper-reversing while frzF does not reverse (note that these are opposite to the paradigm established for the analogous E. coli adaptation enzymes). The WT shows a prey cell concentration dependent rippling response such that wavelength decreases (due to increased reversal frequency of individual cells) when prey concentration increases. Importantly, the components that induce rippling are non-diffusible macromolecules (eg, peptidoglycan, DNA, cell debris) that must come in direct contact with predator cells.
Figure 5.
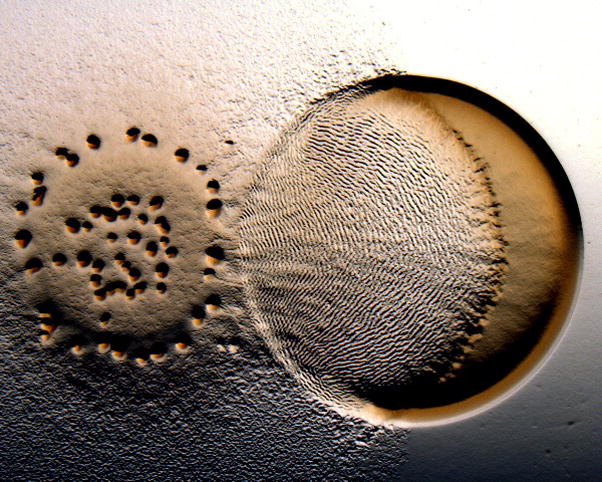
The Predataxis behavior of M. xanthus. When eating E. coli, M. xanthus forms an unusual rippling structure that results from the coordinated reversals of predatory cells. Courtesy of John Kirby.
M. xanthus is certainly not the only bacterial species that forms complex multicellular communities of differentiated cells. Bacillus subtilis can also differentiate into multiple cell types, including motile, matrix producing, and sporulating cells. B. subtilis biofilms produce aerial spore-forming structures that are similar to the fruiting bodies of M. xanthus with different cell types occupying different locations within the structures (Lopez et al., 2009). Spo0A, when phosphorylated, is a positive regulator of sporulation, whereas extracellular matrix production negatively regulates motility. Hera Vlamakis (from the Kolter Lab at Harvard Medical School) asked when and where B. subtilis express flagella, matrix, and sporulation specific genes within a biofilm (Vlamakis et al., 2008). Fluorescent reporters were used as markers of different cell fates (SspB – sporulation reporter; flagellin – motility reporter, and YqxM – matrix reporter) were examined in thin sections of the mature colonies (Figure 6). Flagellin expression peaked at the base of the biofilm and at the outer edge of the colony, sporulation gene expression was evident at the top of the aerial structures and at the centre of the colony, and the matrix reporter gene was expressed throughout the biofilm. Overall, the studies show that on solid surfaces matrix is produced, which in turn induces sporulation, matrix mutants do not sporulate in these conditions. Spo0A~P is a critical factor: at low concentrations, matrix production is induced and at high concentrations, sporulation is induced. Of the kinases known to regulate Spo0A, KinA and KinB stimulate high Spo0A~P levels, while KinC and KinD generate lower levels. Furthermore, a matrix/kinD double mutant restores sporulation on solid surfaces. Therefore, it appears that KinD acts like a phosphatase that is inhibited by matrix material (or a signal therein) that otherwise sets the level of Spo0A~P needed for sporulation.
Figure 6.
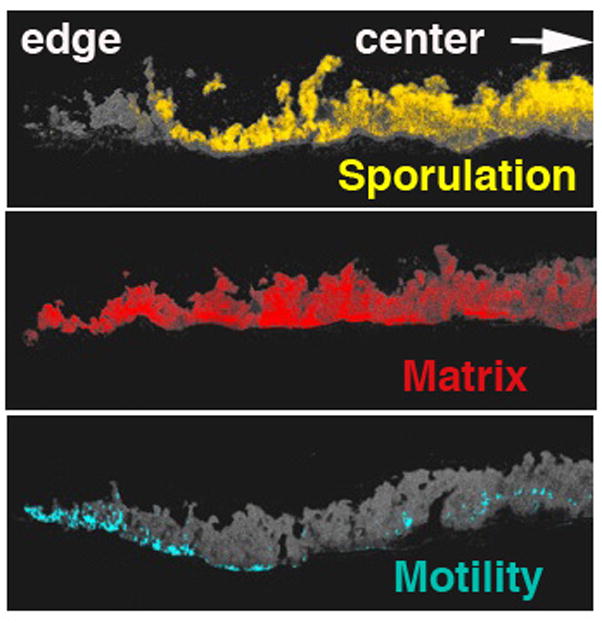
Differentiation within a B. subtilis biofilm. Fluorescent markers for cells expressing genes associated with flagella (blue), the extracellular matrix (red) and sporulation factors (yellow) localize to different positions of the colony, shown here in thin crosssection, with aerial structures above and solid substrate below. Courtesy of Hera Vlamakis and Roberto Kolter.
How exactly the sporulation switch in B. subtilis is thrown is not a simple question to answer. Spo0A is the master regulator for entry into sporulation. It is activated by sensor kinases (i.e. KinA) through the phosphorelay in which KinA phosphorylates Spo0F, which transfers phosphoryl groups to Spo0B and finally to Spo0A. Spo0A has early targets, which respond to low concentrations, and late targets, which respond to high concentrations, the last ones being critical for the entry into the sporulation process. Arnaud Chastanet (from the Losick lab, Harvard University) investigated why a sub-population does not enter the sporulation pathway and whether Spo0A can generate a bistable switch that could account for it. A bistable switch could have been operative because Spo0A upregulates itself and also increases Spo0F levels in a positive feedback loop. In wild-type populations a fraction of the cells (about 30%) do not generate asymmetric septa required for sporulation. However, overproduction of neither Spo0A nor Spo0F changed the sporulation level, while overproduction of KinA results in nearly 100% sporulation. Thus, the flux of phosphoryl groups through the system, not the total Spo0A concentration, may be the rate limiting step for throwing what is an unstable, rather than a bistable sporulation switch.
Robert Belas (University of Maryland) described a switch between planktonic and biofilm lifestyles in Rhodobacter-like bacteria. Roseobacter and Silicibacter are marine alpha proteobacteria that constitute nearly 20% of all prokaryotes in oceans. Silicibacter forms biofilms in symbiosis with host phytoplankton, and can live as a sessile cell (which forms multicellular rosettes) or as a flagellated swimmer (single cell) that chemotaxes toward phyto compounds. Genes regulating flagella biosynthesis and function reside in a 35 kb cluster similar in organization to the fla2 cluster in Rhodobacter sphaeroides. A transposon mutagenesis screen showed that FlaBCD are required for swimming. FlaC is a DNA binding domain protein and FlaD is a helix-turn-helix protein of the MarR transcription regulator family. Genetics and behavioural assays indicate that FlaC regulates the switch between motile and sessile forms of the cell. Lastly, FlaB might be a sensor kinase responsible for phosphorylation or phosphotransfer to FlaC, although what FlaB senses is currently unknown (Belas, 2009).
What to do with your flagella seems to be an important consideration in deciding to give up the “jet-set” planktonic life and settle down for the long haul in a biofilm. In E. coli, for example, the RcsABCD, functions to inhibit the master regulator of flagellar biosynthesis FlhDC, and regulate biofilm formation. In contrast to previous assumptions, data from Ricardo Oropeza (Universidad Nacional Autonoma de Mexico) indicates that RcsC (a hybrid sensor kinase) is required for biofilm formation in E. coli but that other genes in the operon encoding RcsB (a response regulator) and RcsD (which contains a phosphotransfer domain) are not. Previous models have proposed that RcsF transduces information through RcsCD to RcsBA and then to FlhDC. RcsBA then positively regulates genes involved in biofilm production: cps (colanic acid) and tubulin homologue gene ftsZ. However, RcsC still induced biofilms in an rcsBD background, which indicates that RcsB and RcsD are not required for biofilm formation.
In some cases, having flagella can be detrimental to colonizing certain environments. This conclusion was reached by Juan Gonzalez (University of Texas) in his work on the relationship between Sinorhizobium meliloti and alfalfa sprouts (Hoang et al., 2008, Gurich, 2009), in which bacteria enter root-derived nodules and differentiate into a new N2-fixing form. Bacterial population density increases around the root prior to invasion and gene regulation by the ExpR/Sin quorum sensing of S. meliloti takes place. Recent work showed that a mutant unable to make the quorum-sensing signal molecules (sinI) does not invade plants as efficiently as the wild-type strain, while a signal receptor mutant (expR) displays no invasion defects. Further analysis determined that at high cell density quorum sensing shuts down flagellar synthesis in the wild-type strain. On the other hand, motility and chemotaxis genes are highly expressed in the sinI mutant but repressed in the expR mutant during all phases of growth. These data indicated that the failure to inactivate flagellar synthesis at high cell population density might be detrimental to the successful invasion of the plant. Interestingly, a sinI mutant that is also defective in flagellin production can invade normally, which suggests that flagella themselves inhibit colonization and when quorum is reached. Flagella production is down-regulated to provide optimal conditions for the bacterial-host interactions.
Biofilms are more than just cells - the secreted macromolecules created by their denizens hold the communities together. Matt Chapman (University of Michigan) discovered that bacteria assemble amyloid fibres (curli fibres) (Barnhart & Chapman, 2006), not unlike those found in diseases involving protein misfolding, such as Alzheimer disease. The curli fibres bind exopolysaccharides (EPS) in E. coli and are required to make biofilms. These fibres are made from proteins rich in β-sheets (CsgA) and, as it turns out, cheap to make energetically due to their amino acid content (high Gly and Ser) (Wang & Chapman, 2008). Genetic screens for mutants affected in curli fibre production identified some interesting targets, an Na/H antiporter, and a number of transcriptional regulators. As Chapman remarked, the amyloid structures that cause so much trouble in neurodegenerative diseases might simply be a rendition of a very general type of fibrous protein structure found in many contexts throughout biology.
Some bacteria rely on rather unusual solid substrates for growth and their relationship to these materials depends on two-component signalling. Hoa Tran working with Robert Weis and Derek Lovely (University of Massachusetts) is exploring the importance of chemotaxis operons to the growth and metabolism of Geobacter sulfurreducens. Geobacter are anaerobic δ-proteobacteria that generate energy by reducing extracellular iron. However, G. sulfurreducens does not have flagella; in liquid medium, cells form conductive biofilms that can adhere directly to graphite electrodes. However, genome sequencing of G. sulfurreducens revealed a total of 70 che genes in 6 clusters (Tran et al., 2008). Knock-outs of some genes within the che5 cluster (cheR, cheA, cheB and cheW) led to altered production of extracellular compounds, including two key c-type cytochromes in the outer membrane (OmcS and OmcZ) involved in iron reduction. Thus, in Geobacter, the chemotaxis sensor system appears to have been co-opted to regulate important electron-transfer proteins involved in biofilm growth on oxidation substrates.
There is no doubt that studies of bacterial mobility and signal transduction have advanced our knowledge in many areas, but what your program officer wants to know is if bacterial motility matters for disease. Md Motaleb, in collaboration with Nyles Charon (University of West Virginia) and Patricia Rosa (RML, NIH) reported their efforts to determine the importance of motility to the pathogenesis of Borrelia burgdorferi, the causal agent of Lyme disease. B. burgdorferi must live both in tick and in mammalian hosts. Microarray data indicate that motility genes are upregulated during infection. Motaleb et al knocked out the gene for the major flagellin protein, flaB, and tested the mutants for virulence. However, the confounding problem here is that B. burgdorferi has 21 linear and circular plasmids, many of the plasmids are necessary for virulence and are very difficult to maintain under laboratory conditions. After heroic efforts, a flaB− strain was isolated that retained all of the plasmids necessary for virulence (20). Unfortunately, the complemented flaB mutant consistently lost a plasmid known to be necessary for virulence. Nevertheless, when a tick was inoculated with the WT or the flaB− mutant and then used to infect a mouse, only the WT was able to generate an infection. The result provides the strong evidence that motility is required for infection by B. burgdorferi.
Birgit Prüβ (North Dakota State University) tested the importance of motility in the pathogenicity of another bacterium, Yersinia enterocolitica - a facultative anaerobe that is transmitted orally and causes fever and diarrhoea in humans. Prüβ used a chicken embryo lethality assay to test whether the master regulator FlhDC, which induces flagellar gene expression, also affects virulence gene expression. Indeed, mutations in flhD and flhB reduced lethality. It was suggested that the effect of FlhDC may actually be mediated through type III secretion systems that are also under FlhDC control and secrete virulence factors. In another important human pathogen, H. pylori, which is responsible for ulcer disease, Karen Ottemann (UC Santa Cruz) is determining how many chemoreceptors the bacterium has, and what they do. Luckily, the number is a small one, at four, which may readily allow delineation of their role in infection.
Finally, as a slight departure from the traditional BLAST themes, Zemer Gitai (Princeton University) described his labs efforts to identify key factors in chromosome segregation with the possibility that new signalling transduction systems might well turn up. Chromosome segregation is one of the essential processes of cell division and, hence, lies at the crux of developmental decisions. Zemer’s lab carried out a high throughput fluorescence-based screen in C. crescentus to examine localization of ~3000 proteins. Many proteins exhibited filamentous localization patterns, including known filamentous proteins, such as ParA. ParA is an ATPase that participates in F episome segregation by interacting with the parS DNA sequence. Zemer’s follow-up studies suggested that C. crescentus chromosome segregation proceeds through two distinct phases and that the MreB actin homolog controls the slow early phase while ParA controls the fast late phase. By analogy to the mitotic spindle of eukaryotic cells, ParA is predicted to display dynamic instability, similar to that observed with tubulin.
Conclusion
From helix rotations, through gene networks, to multi-cellular behaviours, we come to understand our microbial partners like never before. Certainly, we look on their decisions with more than detached interest. At BLAST, the study of pathogenesis has not yet held central stage, but does have a foothold in our discussions. This is probably because the focus of BLAST for many years has been the basic understanding of the how bacteria sense their environment and the best experimental organisms are not necessarily the most infectious. However, we predict that this will change as increasing knowledge opens up new avenues for coercing our bacterial friends into decisions and lifestyles perhaps more beneficial for us then for them. We can’t wait to be surprised, amused and enlightened at BLAST XI.
References
- Adler J. Chemotaxis In Bacteria. Annu Rev Biochem. 1975;44:341–356. doi: 10.1146/annurev.bi.44.070175.002013. [DOI] [PubMed] [Google Scholar]
- Aldridge P, Hughes KT. Regulation of flagellar assembly. Curr Opin Microbiol. 2002;5:160–165. doi: 10.1016/s1369-5274(02)00302-8. [DOI] [PubMed] [Google Scholar]
- Apel D, Surette MG. Bringing order to a complex molecular machine: The assembly of the bacterial flagella. Biochim Biophys Acta-Biomembr. 2008;1778:1851–1858. doi: 10.1016/j.bbamem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud N, Hassett DJ, Hwang SH, Rice SA, Kjelleberg S, Webb JS. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol. 2006;188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belas RE, Horikawa SI, Aizawa Suvanasuthi R. Genetic determinants of Silicibacter sp. TM1040 motility. J Bacteriol. 2009 doi: 10.1128/JB.00429-09. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg HC. The Rotary Motor of Bacterial Flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- Berleman JE, Scott J, Chumley T, Kirby JR. Predataxis behavior in Myxococcus xanthus. Proc Natl Acad Sci U S A. 2008;105:17127–17132. doi: 10.1073/pnas.0804387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bible AN, Stephens BB, Ortega DR, Xie ZH, Alexandre G. Function of a chemotaxis-like signal transduction pathway in modulating motility, cell clumping, and cell length in the alphaproteobacterium Azospirillum brasilense. J Bacteriol. 2008;190:6365–6375. doi: 10.1128/JB.00734-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block SM, Segall JE, Berg HC. Adaptation Kinetics in Bacterial Chemotaxis. J Bacteriol. 1983;154:312–323. doi: 10.1128/jb.154.1.312-323.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner PJ, Xu Q, Black WP, Li Z, Yang ZM, Shimkets LJ. The Dif chemosensory pathway is directly involved in phosphatidylethanolamine sensory transduction in Myxococcus xanthus. Mol Microbiol. 2005;57:1499–1508. doi: 10.1111/j.1365-2958.2005.04785.x. [DOI] [PubMed] [Google Scholar]
- Bray D, Duke T. Conformational spread: the propagation of allosteric states in large multiprotein complexes. Annu Rev Biophys Biomol Struct. 2004;33:53–73. doi: 10.1146/annurev.biophys.33.110502.132703. [DOI] [PubMed] [Google Scholar]
- Briegel A, Ding HJ, Li Z, Werner J, Gitai Z, Dias DP, Jensen RB, Jensen GJ. Location and architecture of the Caulobacter crescentus chemoreceptor array. Mol Microbiol. 2008;69:30–41. doi: 10.1111/j.1365-2958.2008.06219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JD, Saini S, Aldridge C, Herbert J, Rao CV, Aldridge PD. The rate of protein secretion dictates the temporal dynamics of flagellar gene expression. Mol Microbiol. 2008;70:924–937. doi: 10.1111/j.1365-2958.2008.06455.x. [DOI] [PubMed] [Google Scholar]
- Bustamante VH, Martinez LC, Santana FJ, Knodler LA, Steele-Mortimer O, Puente JL. HilD-mediated transcriptional cross-talk between SPI-1 and SPI-2. Proc Natl Acad Sci U S A. 2008;105:14591–14596. doi: 10.1073/pnas.0801205105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluzel P, Surette M, Leibler S. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science. 2000;287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- Draheim RR, Bormans AF, Lai RZ, Manson MD. Tuning a bacterial chemoreceptor with protein-membrane interactions. Biochemistry. 2006;45:14655–14664. doi: 10.1021/bi061259i. [DOI] [PubMed] [Google Scholar]
- Dubrac S, Bisicchia P, Devine KM, Msadek T. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol Microbiol. 2008;70:1307–1322. doi: 10.1111/j.1365-2958.2008.06483.x. [DOI] [PubMed] [Google Scholar]
- Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune DR, Suyemoto M, Altier C. Identification of CsrC and characterization of its role in epithelial cell invasion in Salmonella enterica serovar Typhimurium. Infect Immun. 2006;74:331–339. doi: 10.1128/IAI.74.1.331-339.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung DC, Berg HC. POWERING THE FLAGELLAR MOTOR OF ESCHERICHIA-COLI WITH AN EXTERNAL VOLTAGE-SOURCE. Nature. 1995;375:809–812. doi: 10.1038/375809a0. [DOI] [PubMed] [Google Scholar]
- Gurich N, Gonzalez JE. The Role of Quorum Sensing in the Sinorhizobium meliloti-Alfalfa Symbiosis. J Bacteriol. 2009 doi: 10.1128/JB.00376-09. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends In Biochemical Sciences. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs PI, Cho KY, Whitworth DE, Evans LS, Zusman DR. Four unusual two-component signal transduction homologs, RedC to RedF, are necessary for timely development in Myxococcus xanthus. J Bacteriol. 2005;187:8191–8195. doi: 10.1128/JB.187.23.8191-8195.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang HH, Gurich N, Gonzalez JE. Regulation of motility by the ExpR/Sin quorum-sensing system in Sinorhizobium meliloti. J Bacteriol. 2008;190:861–871. doi: 10.1128/JB.01310-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, Coles M. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126:929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- Jarrell KF, McBride MJ. The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol. 2008;6:466–476. doi: 10.1038/nrmicro1900. [DOI] [PubMed] [Google Scholar]
- Khursigara CM, Wu XW, Subramaniam S. Chemoreceptors in Caulobacter crescentus: Trimers of receptor dimers in a partially ordered hexagonally packed array. J Bacteriol. 2008a;190:6805–6810. doi: 10.1128/JB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khursigara CM, Wu XW, Zhang PJ, Lefman J, Subramaniam S. Role of HAMP domains in chemotaxis signaling by bacterial chemoreceptors. Proc Natl Acad Sci U S A. 2008b;105:16555–16560. doi: 10.1073/pnas.0806401105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Blair DF. The bacterial flagellar motor: structure and function of a complex molecular machine. Int Rev Cytol. 2004;233:93–134. doi: 10.1016/S0074-7696(04)33003-2. [DOI] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Kolter R. Generation of multiple cell types in Bacillus subtilis. Fems Microbiol Rev. 2009;33:152–163. doi: 10.1111/j.1574-6976.2008.00148.x. [DOI] [PubMed] [Google Scholar]
- Macnab RM. How bacteria assemble flagella. Annu Rev Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- Malpica R, Sandoval GRP, Rodriguez C, Franco B, Georgellis D. Signaling by the arc two-component system provides a link between the redox state of the quinone pool and gene expression. Antioxid Redox Signal. 2006;8:781–795. doi: 10.1089/ars.2006.8.781. [DOI] [PubMed] [Google Scholar]
- Manson MD. The tie that binds the dynamic duo: the connector between AS1 and AS2 in the HAMP domain of the Escherichia coli Tsr chemoreceptor. J Bacteriol. 2008;190:6544–6547. doi: 10.1128/JB.00943-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauriello EMF, Astling DP, Sliuarsenko O, Zusman DR. Localization of a bacterial cytoplasmic receptor is dynamic and changes with cell-cell contacts. Proc Natl Acad Sci U S A. 2009;106:4852–4857. doi: 10.1073/pnas.0810583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter LL. Polar flagellar motility of the Vibrionaceae. Microbiol Mol Biol Rev. 2001;65:445–462. doi: 10.1128/MMBR.65.3.445-462.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter LL. Regulation of flagella. Curr Opin Microbiol. 2006;9:180–186. doi: 10.1016/j.mib.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Neiditch MB, Federle MJ, Pompeani AJ, Kelly RC, Swem DL, Jeffrey PD, Bassler BL, Hughson FM. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–1108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulick A, Koerdt A, Lassak J, Huntley S, Wilms I, Narberhaus F, Thormann KM. Two different stator systems drive a single polar flagellum in Shewanella oneidensis MR-1. Mol Microbiol. 2009;71:836–850. doi: 10.1111/j.1365-2958.2008.06570.x. [DOI] [PubMed] [Google Scholar]
- Pollard AM, Bilwes AM, Crane BR. The Structure of a Soluble Chemoreceptor Suggests a Mechanism for Propagating Conformational Signals. Biochemistry. 2009;48:1936–1944. doi: 10.1021/bi801727m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter SL, Roberts MAJ, Manning CS, Armitage JP. A bifunctional kinase-phosphatase in bacterial chemotaxis. Proc Natl Acad Sci U S A. 2008a;105:18531–18536. doi: 10.1073/pnas.0808010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter SL, Wadhams GH, Armitage JP. Rhodobacter sphaeroides: complexity in chemotactic signalling. Trends Microbiol. 2008b;16:251–260. doi: 10.1016/j.tim.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Purcell EB, Siegal-Gaskins D, Rawling DC, Fiebig A, Crosson S. A photosensory two-component system regulates bacterial cell attachment. Proc Natl Acad Sci U S A. 2007;104:18241–18246. doi: 10.1073/pnas.0705887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin LD, Bodenmiller DM, Partridge JD, Nishino SF, Spain JC, Spiro S. Escherichia coli NsrR regulates a pathway for the oxidation of 3-nitrotyramine to 4-hydroxy-3-nitrophenylacetate. J Bacteriol. 2008;190:6170–6177. doi: 10.1128/JB.00508-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roujeinikova A. Crystal structure of the cell wall anchor domain of MotB,, a stator component of the bacterial flagellar motor: Implications for peptidoglycan recognition. Proc Natl Acad Sci U S A. 2008;105:10348–10353. doi: 10.1073/pnas.0803039105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulmeister S, Ruttorf M, Thiem S, Kentner D, Lebiedz D, Sourjik V. Protein exchange dynamics at chemoreceptor clusters in Escherichia coli. Proc Natl Acad Sci U S A. 2008;105:6403–6408. doi: 10.1073/pnas.0710611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoge ML, Endres RG, Wingreen NS. Receptor-receptor coupling in bacterial chemotaxis: Evidence for strongly coupled clusters. Biophys J. 2006;90:4317–4326. doi: 10.1529/biophysj.105.079905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Berg HC. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- Sowa Y, Berry RM. Bacterial flagellar motor. Q Rev Biophys. 2008a;41:103–132. doi: 10.1017/S0033583508004691. [DOI] [PubMed] [Google Scholar]
- Sowa Y, Berry RM. Bacterial flagellar motor. Q Rev Biophys. 2008b;41:103–132. doi: 10.1017/S0033583508004691. [DOI] [PubMed] [Google Scholar]
- Studdert CA, Parkinson JS. Crosslinking snapshots of bacterial chemoreceptor squads. Proc Natl Acad Sci U S A. 2004;101:2117–2122. doi: 10.1073/pnas.0308622100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson RV, Bourret RB, Simon MI. Intermolecular complementation of the kinae activity of CheA. Mol Microbiol. 1993;8:435–441. doi: 10.1111/j.1365-2958.1993.tb01588.x. [DOI] [PubMed] [Google Scholar]
- Szurmant H, Bobay BG, White RA, Sullivan DM, Thompson RJ, Hwa T, Hoch JA, Cavanagh J. Co-evolving motions at protein-protein interfaces of two-component signaling systems identified by covariance analysis. Biochemistry. 2008;47:7782–7784. doi: 10.1021/bi8009604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BL. Aer on the inside looking out: paradigm for a PAS-HAMP role in sensing oxygen, redox and energy. Mol Microbiol. 2007;65:1415–1424. doi: 10.1111/j.1365-2958.2007.05889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SA, Brewster JA, Bourret RB. Two variable active site residues modulate response regulator phosphoryl group stability. Mol Microbiol. 2008;69:453–465. doi: 10.1111/j.1365-2958.2008.06296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HT, Krushkal J, Antommattei FM, Lovley DR, Weis RM. Comparative genomics of Geobacter chemotaxis genes reveals diverse signaling function. BMC Genomics. 2008:9. doi: 10.1186/1471-2164-9-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis H, Aguilar C, Losick R, Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008;22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chapman MR. Sequence determinants of bacterial amyloid formation. J Mol Biol. 2008;380:570–580. doi: 10.1016/j.jmb.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigt M, White RA, Szurmant H, Hoch JA, Hwa T. Identification of direct residue contacts in protein-protein interaction by message passing. Proc Natl Acad Sci U S A. 2009;106:67–72. doi: 10.1073/pnas.0805923106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Black WP, Cadieux CL, Yang ZM. Independence and interdependence of Dif and Frz chemosensory pathways in Myxococcus xanthus chemotaxis. Mol Microbiol. 2008;69:714–723. doi: 10.1111/j.1365-2958.2008.06322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Kutsukake K. FliT acts as an anti-FlhD2C2 factor in the transcriptional control of the flagellar regulon in Salmonella enterica serovar typhimurium. J Bacteriol. 2006;188:6703–6708. doi: 10.1128/JB.00799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusman DR, Scott AE, Yang Z, Kirby JR. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat Rev Microbiol. 2007;5:862–872. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]


