Abstract
The high degree of specificity displayed by antibodies often results in varying potencies against antigen orthologs, which can affect the efficacy of these molecules in different animal models of disease. We have used a computational design strategy to improve the species cross-reactivity of an antibody-based inhibitor of the cancer-associated serine protease MT-SP1. In silico predictions were tested in vitro, and the most effective mutation, T98R, was shown to improve antibody affinity for the mouse ortholog of the enzyme 14-fold, resulting in an inhibitor with a KI of 340 pM. This improved affinity will be valuable in exploring the role of MT-SP1 in mouse models of cancer, and the strategy outlined here could be useful in fine-tuning antibody specificity.
The ability to engineer and biochemically manipulate antibodies is critical to their utility, and has helped usher in a new wave of biological therapeutics. While bacteriophage, ribosome, and yeast display techniques have revolutionized our ability to quickly raise antibodies to a specific target, these methods are all limited by their functional library size. Considerable effort has therefore gone into improving binding beyond the initial lead antibody hit from these libraries. Iterative rounds of combinatorial techniques such as CDR walking1, error-prone PCR2, or random or site directed mutagenesis3 have led to antibodies that regularly bind their antigens with KD’s in the picomolar range4. Structure-based and computational protein design techniques have also been used to streamline the antibody maturation process5–7.
Antibodies make outstanding therapeutic agents in part due to the high degree of specificity an antibody has for its antigen. Monoclonal antibodies often bind non-linear epitopes that depend on the precise three-dimensional arrangement of a constellation of amino acids. The specificity afforded by a monoclonal antibody has a downside, though, as it can hinder efforts to use a single antibody against species homologs. Antibodies raised against a specific human antigen often do not cross-react with nonhuman versions of that antigen, which can affect the choice and efficacy of pre-clinical animal models8, 9. The same combinatorial techniques used to improve antibody affinity have been used to modify the cross-reactivity of recombinant antibodies9–11; here we report that computational design can be used to improve the species cross-reactivity of an antibody.
We have previously described an antibody that inhibits the extracellular serine protease membrane-type serine protease 1 (MT-SP1/matriptase). The biology of MT-SP1 is complex, but it is posited to play a role in a number of biological processes, and dysregulation of the enzyme has been shown to play a role in tumor growth and metastasis (see reviews12, 13). The antibody, E2, was identified in a bacteriophage-displayed antibody library14, and is a potent inhibitor of MT-SP1, with a KI of 12 pM15. Though MT-SP1 and its mouse ortholog epithin are 87% identical, E2 is a 300-fold less potent inhibitor of the mouse version of the enzyme. The basis of this difference is not apparent from biochemical and structural analyses of the MT-SP1/E2 complex. There are only three residues on the protease surface that both make contacts with the antibody and are different between the human and mouse versions of the enzyme: Ile60 is a glutamine in the mouse ortholog, Arg60c a lysine, and Tyr146 is a glutamic acid, but these residues have been shown to not be critical for inhibition15. In order to better understand the role MT-SP1/epithin plays in tumor progression and metastasis, mouse models of cancer16 need to be utilized. In order to develop a version of E2 suitable for mouse experiments, computational design has been used to suggest mutations predicted to improve inhibition of the mouse version of the enzyme.
Designing a species cross-reactive antibody
In silico design strategies have been used to modify protein-protein specificity17, 18, and have successfully guided or aided in the understanding of the maturation of an antibody for an antigen6, 7, 19, 20. As is frequently the case in antibody-antigen interactions, hydrogen bonding and electrostatic interactions are important contributors to the binding affinity and specificity of the interaction between MT-SP1/epithin and the antibody considered here. The MT-SP1/epithin active site prefers positively charged substrates, and the heavy and light chain CDR3 loops of E2 are positively charged 14. To account for these electrostatic interactions, we used a molecular mechanics-based energy function, in conjunction with an implicit solvent model to treat the effects of water, to predict the effect on binding of mutations at the protease-antibody interface. This type of energy function has previously been shown to perform well in predicting mutations to increase binding affinity of antibodies6, 7, in studying specificity of enzymes for charged metabolites21, and in designing enzyme active-sites22, 23.
An estimate of the change in binding free energy upon mutation (ΔΔGmut) was calculated using the Protein Local Optimization Program (PLOP) by subtracting the calculated energies of unbound E2mutated and epithin species from the calculated energy of the E2mutated/epithin complex (Figure 1a). PLOP estimates free energy using the Optimized Potential for Liquid Simulations all atom (OPLS-AA) force field24–26, the Surface Generalized Born model27 of polar solvation, an estimator for the nonpolar component of the solvation free energy28, and a number of correction terms as detailed in Ghosh et al.27 and Jacobson et al.24 Computationally estimating changes in protein conformational entropy upon binding is extremely difficult, and we do not attempt to do so. For this reason, the calculated ΔΔGmut cannot be interpreted as a direct surrogate for the experimental change in binding affinity. The calculated values are much too large, because they do not include the entropy losses upon binding. Rather, as in previous work on specificity of proteins for small molecule substrates21 and inhibitors29, the calculated values are a qualitative measure, in this case used to identify mutations that may change binding affinity and specificity.
Figure 1.
(a) Predicted change in binding free energy for a point mutation is estimated in silico by the calculated change in free energy upon mutation of the E2/epithin complex minus the calculated change in free energy upon mutation of the unbound E2 antibody minus the calculated free energy of epithin. (b) Details of the methods used to calculate each free energy component shown in (a) (see text).
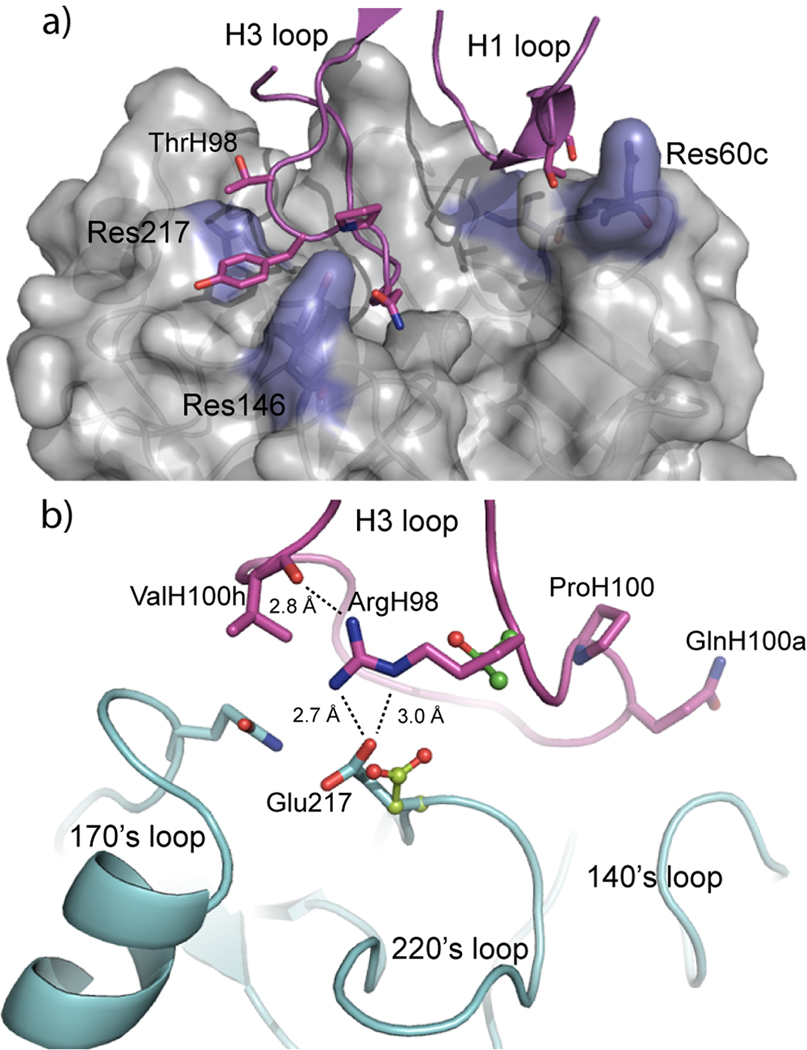
There is currently no available structure of the epithin/E2 complex, so a homology model of epithin was created with PLOP30 using the available MT-SP1 structure as a template. As part of the homology modeling process in PLOP, side chain rotamers are optimized24, 31 for all residues that differ between the two proteins. The epithin homology model was substituted for the protease in the MT-SP1/E2 crystal structure32, E2 was truncated to Fv length (ending at IleL106 and ValH111) and hydrogens were added and energy minimized33. A residue was selected for in silico mutation on the E2 heavy chain if at least one of its atoms was < 5 Å from any atom in the three residues that differ between MT-SP1 and epithin (Gln60, Lys60c, and Glu146, Figure 2a). The six chosen residues were ThrH28, SerH30, ThrH98, TyrH99, ProH100, and GlnH100a (Figure 2a). The computational workflow is outlined in Figure 1b and is described as follows: (1) each of the six residue side chains were mutated to the other 18 possible amino acid side chains (excluding cysteine); (2) the side chain rotamers of residues in the interface of the complex were optimized24, 31 and then energy minimized33; (3) the same interface residues were energy minimized33 in the unbound, mutated E2 and unbound epithin structures; and (4) ΔΔGmut was calculated as described above. Most of the changes were predicted to be neutral or worsen the antibody-antigen interaction, but 8 mutations were predicted to lower the free energy of the epithin-E2 complex by varying amounts and were tested experimentally.
Figure 2.
(a) The 6 residues (sticks) on the H1 and H3 CDR loops of E2 (magenta) that contact residues which are different between the human and mouse orthologs (blue) of the protease were chosen for in silico mutation. (b) Predicted effect of T98R mutation of the E2 H3 loop (magenta) on epithin (cyan) inhibition. The arginine side chain makes an intramolecular H-bond with the backbone carbonyl oxygen of ValH100h, and a hydrogen bond with the Glu217 of epithin. ThrH98 and Asp217 of MT-SP1 from the E2/MT-SP1 crystal structure are shown as ball-and-sticks. The model suggests that the T98R substitution does not affect MT-SP1 inhibition, because the Asp217 side chain cannot reach the guanidine group of ArgH98, which is locked in position by the intramolecular H-bond.
Testing the computational predictions
Fab constructs of the eight point mutants predicted to improve antibody binding were cloned via site-directed mutagenesis, expressed in E. coli, and purified as previously described15. The IC50’s of each point mutant were measured against epithin and MT-SP1, and relative KI’s were determined to normalize the IC50 with respect to the protease/substrate interaction15. The majority of the mutations had little effect on protease inhibition (Table 1). The heavy chain P100H mutant, predicted to improve binding significantly, was deleterious to protease inhibition. This is likely because eliminating proline significantly increases the flexibility of the backbone, an effect that is not captured in the modeling protocol. The T98R substitution, which was predicted to have the greatest effect on protease inhibition, improved the KI of E2 for epithin from 4.8 nM to 340 pM – a 14-fold improvement. The MT-SP1/E2 crystal structure32 provides a rationale for this improvement. The Thr98 side chain makes no contacts with the protease, and the Oγ2 atom is solvent exposed. The model of the epithin-E2 complex suggests the T98R mutation enables the arginine side chain to extend back towards the protease, make an intramolecular H-bond with the backbone carbonyl oxygen of ValH100h, and an intermolecular H-bond with the carboxylate group of Glu217 on the protease (Figure 2b). The 14-fold improvement in KI shown by this construct corresponds to a free energy gain of 1.6 kcal/mol, which is roughly the strength of one strong or two moderately strong H-bonds. This model also suggests why the T98R mutation has little effect on MT-SP1 inhibition. The human ortholog has an Asp residue at position 217, which, because its side chain is one atom shorter, cannot extend far enough to make this interaction. As a result, the threonine to arginine substitution allows a side chain that plays a minimal role in protease inhibition to be modified to broaden the species cross-reactivity of the E2 antibody.
Table 1.
Predicted and experimental antibody mutations
| Epithin Ki (nM) |
Fold Improvement2 |
ΔΔGexp1 (kcal/mol) |
ΔΔGpredicted (kcal/mol) |
MT-SP1 Ki (pM) |
Fold Improvement2 |
|
|---|---|---|---|---|---|---|
| E2 | 4.8 | 22.4 | ||||
| T28R | 3.2 | 1.5 | −0.24 | −13.2 | 39.3 | 0.6 |
| S30G | 14.0 | 0.3 | 0.63 | −17.6 | 165.9 | 0.1 |
| S30N | 14.7 | 0.3 | 0.66 | −11.1 | 155.1 | 0.1 |
| T98Q | 3.7 | 1.3 | −0.16 | −9.9 | 21.6 | 1.0 |
| T98R | 0.34 | 14.1 | −1.57 | −21.8 | 20.3 | 1.1 |
| Y99S | 11.5 | 0.4 | 0.52 | 0.12 | 48.6 | 0.5 |
| P100H | 78.5 | 0.1 | 1.66 | −16.3 | 1346.6 | 0.0 |
| Q100aV | 10.3 | 0.5 | 0.45 | −0.07 | 176.7 | 0.1 |
Relative KI’s were calculated from IC50 values according to: KI = IC50/(1 + [S] KM ) . Substrate KM’s were 21 µM for epithin and 41 µM for MT-SP1. See Ref. 15 for more details.
Fold improvement was determined by dividing the experimental (KI [E2] / KI [Mutant])
Our broadening of the species specificity of E2 highlights a number of reasons the antibody-antigen interaction is ideally suited for modification by computational design. When bound to proteins, antibodies often have more than 800 square angstroms of buried surface area, and are dominated by binding ‘hot-spots’34, which allows for the possibility of optimization at a number of discrete locations without destroying binding. Mutational studies on the MT-SP1/E2 complex suggested the antibody-antigen complex was dependent on interactions at ArgH100b and portions of the H2 and H3 loops that ‘grabbed’ the 90’s loop on the protease15, so we did not computationally screen these areas. The energetics of antibody-antigen complexes are dependent on side chain-side chain interactions more than main chain interactions35. Side-chain interactions can be easier to model, particularly when the protein backbone is relatively rigid. This is illustrated by our success modeling the effects of mutating side chain interactions at residue Thr98, and the difficulty experienced modeling Pro99, the only residue that was modified which significantly reduced inhibition. Also, the shape complementarity (as defined by the geometric match at a protein-protein interface) of antibody-antigen interactions tends to be worse than other well characterized protein-protein interactions36, and antibody CDR sequences are biased towards specific residues such as tyrosine, tryptophan, and aspartic acid37. This is likely a function of the inherent flexibility antibodies must have to bind to new and different antigens, and is clearly an efficient way to rapidly develop binders to many different targets, but likely leaves a portion of sequence and conformational space underexplored, and suggests there are a number of opportunities for computational modeling to predict mutations that could fine-tune the interaction.
In conclusion, computational design was used to predict a suite of mutations that could improve the species cross-specificity of an inhibitory antibody of the cancer associated serine protease MT-SP1. The substitution of an arginine for ThrH98 improved the affinity of the antibody for epithin by an order of magnitude, and had no effect on inhibition of the human ortholog. In conjunction with other similar published studies6, 7, the results presented here suggest that computational strategies can help guide the fine-tuning of antibody-antigen interactions, and streamline the process by which therapeutic antibodies are investigated in different animal models.
Acknowledgements
This work was supported by grants from the Sandler Program in the Basic Sciences (to MPJ), and NIH grants CA072006 and GM082250 (CSC and CJF). The work of BDS was supported in part by the Genentech Scholars Program. MPJ is a member of the Scientific Advisory Board of Schrodinger Inc. Molecular images were produced using PyMOL38.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barbas CF, 3rd, Hu D, Dunlop N, Sawyer L, Cababa D, Hendry RM, Nara PL, Burton DR. Proc Natl Acad Sci U S A. 1994;91(9):3809–3813. doi: 10.1073/pnas.91.9.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Razai A, Garcia-Rodriguez C, Lou J, Geren IN, Forsyth CM, Robles Y, Tsai R, Smith TJ, Smith LA, Siegel RW, Feldhaus M, Marks JD. J Mol Biol. 2005;351(1):158–169. doi: 10.1016/j.jmb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Ho M, Kreitman RJ, Onda M, Pastan I. J Biol Chem. 2005;280(1):607–617. doi: 10.1074/jbc.M409783200. [DOI] [PubMed] [Google Scholar]
- 4.Luginbuhl B, Kanyo Z, Jones RM, Fletterick RJ, Prusiner SB, Cohen FE, Williamson RA, Burton DR, Pluckthun A. J Mol Biol. 2006;363(1):75–97. doi: 10.1016/j.jmb.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 5.Barderas R, Desmet J, Timmerman P, Meloen R, Casal JI. Proc Natl Acad Sci U S A. 2008;105(26):9029–9034. doi: 10.1073/pnas.0801221105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark LA, Boriack-Sjodin PA, Eldredge J, Fitch C, Friedman B, Hanf KJ, Jarpe M, Liparoto SF, Li Y, Lugovskoy A, Miller S, Rushe M, Sherman W, Simon K, Van Vlijmen H. Protein Sci. 2006;15(5):949–960. doi: 10.1110/ps.052030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lippow SM, Wittrup KD, Tidor B. Nat Biotechnol. 2007;25(10):1171–1176. doi: 10.1038/nbt1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pegram M, Ngo D. Adv Drug Deliv Rev. 2006;58(5–6):723–734. doi: 10.1016/j.addr.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Werther WA, Gonzalez TN, O'Connor SJ, McCabe S, Chan B, Hotaling T, Champe M, Fox JA, Jardieu PM, Berman PW, Presta LG. J Immunol. 1996;157(11):4986–4995. [PubMed] [Google Scholar]
- 10.Garcia-Rodriguez C, Levy R, Arndt JW, Forsyth CM, Razai A, Lou J, Geren I, Stevens RC, Marks JD. Nat Biotechnol. 2007;25(1):107–116. doi: 10.1038/nbt1269. [DOI] [PubMed] [Google Scholar]
- 11.Liang WC, Wu X, Peale FV, Lee CV, Meng YG, Gutierrez J, Fu L, Malik AK, Gerber HP, Ferrara N, Fuh G. J Biol Chem. 2006;281(2):951–961. doi: 10.1074/jbc.M508199200. [DOI] [PubMed] [Google Scholar]
- 12.Bugge TH, List K, Szabo R. Front Biosci. 2007;12:5060–5070. doi: 10.2741/2448. [DOI] [PubMed] [Google Scholar]
- 13.Darragh MR, Bhatt AS, Craik CS. Front Biosci. 2008;13:528–539. doi: 10.2741/2698. [DOI] [PubMed] [Google Scholar]
- 14.Sun J, Pons J, Craik CS. Biochemistry. 2003;42(4):892–900. doi: 10.1021/bi026878f. [DOI] [PubMed] [Google Scholar]
- 15.Farady CJ, Sun J, Darragh MR, Miller SM, Craik CS. J Mol Biol. 2007;369(4):1041–1051. doi: 10.1016/j.jmb.2007.03.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welm AL, Sneddon JB, Taylor C, Nuyten DS, van de Vijver MJ, Hasegawa BH, Bishop JM. Proc Natl Acad Sci U S A. 2007;104(18):7570–7575. doi: 10.1073/pnas.0702095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joachimiak LA, Kortemme T, Stoddard BL, Baker D. J Mol Biol. 2006;361(1):195–208. doi: 10.1016/j.jmb.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 18.Shifman JM, Mayo SL. Proc Natl Acad Sci U S A. 2003;100(23):13274–13279. doi: 10.1073/pnas.2234277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chong LT, Duan Y, Wang L, Massova I, Kollman PA. Proc Natl Acad Sci U S A. 1999;96(25):14330–14335. doi: 10.1073/pnas.96.25.14330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sivasubramanian A, Maynard JA, Gray JJ. Proteins. 2008;70(1):218–230. doi: 10.1002/prot.21595. [DOI] [PubMed] [Google Scholar]
- 21.Kalyanaraman C, Bernacki K, Jacobson MP. Biochemistry. 2005;44(6):2059–2071. doi: 10.1021/bi0481186. [DOI] [PubMed] [Google Scholar]
- 22.Chakrabarti R, Klibanov AM, Friesner RA. Proc Natl Acad Sci U S A. 2005;102(34):12035–12040. doi: 10.1073/pnas.0505397102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakrabarti R, Klibanov AM, Friesner RA. Proc Natl Acad Sci U S A. 2005;102(29):10153–10158. doi: 10.1073/pnas.0504023102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobson MP, Kaminski GA, Friesner RA, Rapp CS. J Phys Chem B. 2002;106:11673–11680. [Google Scholar]
- 25.Jorgensen WL, Maxwell DS, Tirado-Rives J. Q ReV Biophys. 1993;26:49–61. [Google Scholar]
- 26.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. J Phys Chem B. 2001;105:6474–6487. [Google Scholar]
- 27.Ghosh A, Rapp CS, Friesner RA. J Phys Chem B. 1998;102:10983–10990. [Google Scholar]
- 28.Gallicchio E, Zhang LY, Levy RM. j Comp Chem. 2002;23(5):517–529. doi: 10.1002/jcc.10045. [DOI] [PubMed] [Google Scholar]
- 29.Huang N, Kalyanaraman C, Bernacki K, Jacobson MP. Phys Chem Chem Phys. 2006;8:5166–5177. doi: 10.1039/b608269f. [DOI] [PubMed] [Google Scholar]
- 30.Kenyon V, Chorny I, Carvajal WJ, Holman TR, Jacobson MP. J Med Chem. 2006;49(4):1356–1363. doi: 10.1021/jm050639j. [DOI] [PubMed] [Google Scholar]
- 31.Jacobson MP, Friesner RA, Xiang Z, Honig B. J Mol Biol. 2002;320(3):597–608. doi: 10.1016/s0022-2836(02)00470-9. [DOI] [PubMed] [Google Scholar]
- 32.Farady CJ, Egea PF, Schneider EL, Darragh MR, Craik CS. J Mol Biol. 2008;380(2):351–360. doi: 10.1016/j.jmb.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu K, Shirts MR, Friesner RA, Jacobson MP. J Chem Theory Comput. 2007;3:640–648. doi: 10.1021/ct600129f. [DOI] [PubMed] [Google Scholar]
- 34.Dall'Acqua W, Goldman ER, Eisenstein E, Mariuzza RA. Biochemistry. 1996;35(30):9667–9676. doi: 10.1021/bi960819i. [DOI] [PubMed] [Google Scholar]
- 35.Jackson RM. Protein Sci. 1999;8(3):603–613. doi: 10.1110/ps.8.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawrence MC, Colman PM. J Mol Biol. 1993;234(4):946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 37.Fellouse FA, Wiesmann C, Sidhu SS. Proc Natl Acad Sci U S A. 2004;101(34):12467–12472. doi: 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA. USA: DeLano Scientific; 2002. [Google Scholar]