Abstract
In this work, we develop a fully automated method for the quality assessment prediction of protein structural models generated by structure prediction approaches such as fold recognition servers, or ab initio methods. The approach is based on fragment comparisons and a consensus Cα contact potential derived from the set of models to be assessed and was tested on CASP7 server models. The average Pearson linear correlation coefficient between predicted quality and model GDT-score per target is 0.83 for the 98 targets which is better than those of other quality assessment methods that participated in CASP7. Our method also outperforms the other methods by about 3% as assessed by the total GDT-score of the selected top models.
Keywords: model quality assessment prediction, TASSER, SP3
INTRODUCTION
Selecting the best quality models from a set of predicted structures is an essential part of protein structure prediction1,2. In the latest Community Wide Experiment on the Critical Assessment of Techniques for Protein Structure Prediction (CASP7)3(http://predictioncenter.org/casp7/), a new prediction category that judges the quality of models and the reliability of predicting certain residues in the structure was implemented. There are a number of methods that address this issue that can be conceptually divided into the following three categories: (1) statistical methods, (2) machine learning methods, and (3) energy function based methods. Examples of statistical methods include those based on clustering in ab initio structure prediction4,5 and the 3D-jury6,7 approach used for meta-servers. Their outcome depends on the population statistics of the models. Statistical methods do not depend on the details of the models whereas energy function based methods require a detailed molecular description. Such energy function based methods include physics-based and knowledge-based energies for discriminating the native structure from decoys, near native structure selection and for the assessment of protein models8–21. Some meta-servers use machine learning approaches to select models from individual servers22–24. Eramian et al. studied 24 assessment scores in the literature and used Support Vector Machine (SVM) regression to combine some of these machine learning approaches and energy function based approaches for predicting errors in protein structure models25. In additional to methods that predict the overall quality of protein structure models, there are also alternative methods that assign a local quality score to each residue. This could be useful for constructing hybrid protein structure models7,26,27.
In this work, we have developed a knowledge based energy function method which employs a score function based on fragment comparisons in combination with a statistical potential to predict the quality of protein models. The approach only uses the Cα coordinates of the models. We tested our method in CASP7 (submitted prediction under name TASSER-QA), where it was shown to have the best average Pearson linear correlation coefficient and was the top ranked among the participating methods in its ability to select the best structures from the CASP7 server models. The method was also used by the TASSER group for selecting models for submission28.
METHOD
Fragment library generation and fragment comparison
The SP3 threading method29,30 was used to generate the fragment library for fragment comparison. The details of SP3 were published elsewhere29. Here we re-optimized the parameters with a full grid search on the five dimensional parameter space. The new optimal solution (w0, w1, w2ndary, wstruc, sshift) is (3.5, 0.1, −1.50, 0.5, 0.7). This resulted in the one-to-one match alignment accuracy of 66.1% against the ProSup structure alignment benchmark31 compared to the original accuracy of 65.3%. Another change made to SP3 that increases its sensitivity is the inclusion of profiles generated by PSIBLAST32 with a looser e-value cutoff of 1.0. To the target sequence, the sequence profile is replaced by the average of two profiles with e-value cutoffs 0.001 and 1.0, and to the templates, the structurally derived profile is replaced by the average of original and the PSIBLAST profile with an e-value cutoff of 1.0.
We extend the SP3 threading method29 to compute local sequence similarity between query and template sequences by computing and recording the alignment score at each query sequence position aligned to each template during threading. The position dependent score is then smoothed by averaging over a 9 residue wide sliding window. For each position, 9 residue long fragments of the top 25 scoring templates are selected to form the fragment library used for subsequent fragment comparison. Fragment comparison is done in the following way: For each residue position in the model for the query sequence, a nine residue fragment with the given residue in the middle (less in the N or C-terminus, for example: the fragment for the first residue will be residues 1–5) is compared with the 25 corresponding fragments in the fragment library according to their pairwise root-mean-square-deviation (RMSD). The fragment comparison score Efrg is the average RMSD over the 25 fragments and over all model residue positions.
Consensus Cα contact potential
The consensus Cα contact potential is constructed from the set of models to be assessed using a similar procedure as was applied to TASSER33,34. For the set of models to be assessed (in practice the top scoring models of the CASP7 servers), a protein specific consensus Cα contact potential between Cαs is calculated as:
| (1) |
where Θ5(x) and Θ6(x) are step functions defined as
| (2) |
pij is the fraction of models that the ith residue Cα is in contact with the jth residue Cα in the models and rij is the Cα distance between residues i and j. p0 defines the minimal fraction threshold that two residues are in contact and 6 Å is the distance cutoff that defines whether a given pair of residues are in contact. In this work, pij is predicted from all the models to be assessed. For example, if the ith residue Cα is in contact with the jth residue Cα (distance < 6 Å) in n1 models of total n assessed models, then pij = n1/n. When pij > p0 =0.3, we consider residues i and j to be involved in a real contact in the native structure and the Θ5 terms are effective in Eq. 1. The first term in Eq. 1 favors pairs predicted as being in contact that are within 6 Å, whereas the secondary term penalizes predicted contact pairs that are farther apart than 6 Å when the total violation exceeds a threshold value of Ncp. The weights wr3, wr4 and Ncp are taken from TASSER33,34.
The score used for predicting model quality is a simple combination of Efrg and Econtact:
| (3) |
where wc is a relative weight of the two terms that will be determined from optimization on a training set (see below). Nr is the number of residues in the model. Because the value of Eq is not between 0 and 1, we transform it by the following logistic function so that the ranking score is in the range 0–1.
| (4) |
Training and testing datasets
The only free parameter in the current approach is wc. We optimized it on all the server models for the 40 easy targets (classified as those having a SP3 Z-score ≥ 5.6) in CASP630. The object function is the total TM-score35 of the selected top models with respect to their native structures. To mimic the real prediction situation, the template library used for generating the fragment library in the optimization was built from structures released before May 28, 2004 which was before the CASP6 prediction season. The optimized result for wc is 0.2.
We tested and compared our method with other methods on the server models of the 98 targets in CASP73. The fragment libraries for all the targets were generated during the CASP blind test. We only evaluated the first models from each server and only those models with full length structures (no missing residues). Predictions by other quality assessment methods were downloaded from the CASP7 website. We only examined predictions for tertiary structures (not for alignment models) so that we could also calculate the model GDT-score36 and TM-score35.
RESULTS
We examined the performance of our approach (we participated in CASP7 as the TASSER-QA, group ID 125) as well as several other methods that participated in CASP7 by using as structural similarity measure both the GDT-score36 and TM-score35 on the 98 targets. Because we submitted predictions in CASP7 with only ranking and no predicted quality scores, the official assessors excluded our method as well as several other methods from their assessment which mostly focused on correlation analysis rather than the quality of the selected top models. Here, we are able to use the predicted quality scores given by Eq. 4 for correlation analysis and compare TASSER-QA with other methods.
In Table 1, we compile the results of the first prediction (named with “Target ID” QA “group ID”_1) using the GDT-score36 as the structural similarity measure. The methods compared in Table 1 (also the following Tables) include only those that have predicted quality scores and have average correlation coefficients of both Pearson and Spearman rank greater than 0.5 in our analysis. Table 2 shows similar results when the TM-score35 is used as the structural similarity measure. In Table 1, TASSER-QA has the highest average (per target) Pearson linear correlation coefficients of 0.834; nevertheless, it is insignificantly different from Pcon’s results. TASSER-QA also has the highest total GDT-score 58.06 of the first ranked models which are about 3% better than the next best method ABIpro-h which has total GDT-score of 56.49. The Pcons and LEE methods have the best average Spearman rank correlation coefficients37 among all methods, with TASSER-QA ranking third. However, their differences are not statistically significant. TASSER-QA is only marginally (with an insignificant p-value of 0.07) better in total GDT-score than the best Zhang-Server whose models are used by TASSER-QA and other methods, and it is about 6% less than the best possible models as ranked by GDT-score (see Table 1).
Table 1.
Average correlation coefficients and total GDT-scores of the top quality assessment predictors in CASP7. The structural similarity of model to native is measured by GDT-score36.
| Method | # of targets | Pearson | Spearman | GDT-score | |||
|---|---|---|---|---|---|---|---|
| Avea | p-valueb | Avec | p-value | Total(ave.)d | p-value | ||
| 125(TASSER-QA)e | 98 | 0.834 | 0.734 | 58.06(0.592) | |||
| TASSER-QA-allf | 98 | 0.828 | 0.51 | 0.791 | 8.6×10−8 | 57.22(0.584) | 0.08 |
| 634(Pcons) | 98 | 0.811 | 0.16 | 0.757 | 0.18 | 55.05(0.562) | 3.7×10−7 |
| 556(LEE) | 96 | 0.797 | 0.049 | 0.747 | 0.67 | 53.86(0.561) | 2.4×10−6 |
| 713(Circle-QA) | 98 | 0.730 | 2.7×10−14 | 0.662 | 1.7×10−5 | 56.06(0.572) | 1.9×10−4 |
| 633(ProQ) | 98 | 0.719 | 1.1×10−12 | 0.597 | 6.4×10−12 | 54.15(0.553) | 4.5×10−6 |
| 038(GeneSilico) | 88 | 0.712 | 1.4×10−11 | 0.621 | 1.4×10−8 | 49.09(0.558) | 2.1×10−5 |
| 692(ProQlocal) | 98 | 0.711 | 2.5×10−11 | 0.591 | 8.1×10−12 | 54.03(0.551) | 3.1×10−6 |
| 178(Bilab) | 98 | 0.699 | 1.7×10−12 | 0.585 | 2.9×10−10 | 54.53(0.556) | 1.2×10−4 |
| 704(QA-ModFOLD) | 98 | 0.675 | 5.9×10−15 | 0.600 | 3.9×10−10 | 53.92(0.550) | 2.0×10−6 |
| 699(ABIpro-h) | 98 | 0.674 | 2.5×10−12 | 0.629 | 6.7×10−7 | 56.49(0.576) | 7.8×10−3 |
| 091(Ma-OPUS) | 97 | 0.662 | 1.6×10−17 | 0.608 | 5.5×10−9 | 53.25(0.549) | 5.1×10−5 |
| 013(Jones-UCL) | 81 | 0.647 | 2.3×10−16 | 0.544 | 7.7×10−16 | 45.28(0.559) | 3.5×10−7 |
| 703(QA-ModCHECK) | 71 | 0.624 | 1.9×10−13 | 0.575 | 2.7×10−8 | 30.61(0.431) | 1.4×10−8 |
| 717(CaspIta-FRST) | 90 | 0.586 | 1.4×10−25 | 0.518 | 3.9×10−21 | 48.49(0.539) | 6.6×10−10 |
| 025(Zhang-Server)g | 98 | - | - | 57.35(0.585) | 0.07 | ||
| Besth | 98 | - | - | 62.00(0.633) | |||
Pearson linear correlation coefficient. The results are the average per target. They may be slightly different from those in CASP7 website we did not include models where the alignments alone are given. We also calculate the GDT-scores based on target chains rather than domains.
P-values are given for the differences of results between TASSER-QA and other methods. Difference with p-value of < 0.05 is considered significant at 95% confidence level.
Spearman rank correlation coefficient. The results are the average per target.
Total GDT-score of the server models that are ranked the first by quality assessment prediction methods. Numbers in parenthesis are averages per target.
This work using only first models from servers. Descriptions of all other methods can be found at the CASP73 website http://predictioncenter.org/casp7/.
This work using all models from servers.
Zhang-Server is not a quality-assessment (QA) prediction method. It is the best server whose models were used by QA methods and used here as a baseline for comparison.
Ranked by GDT-score as another baseline.
Table 2.
Average correlation coefficients and total GDT-scores of the top quality assessment predictors in CASP7 for the 67 easy targets.
| Method | # of targets | Pearson | Spearman | GDT-score | |||
|---|---|---|---|---|---|---|---|
| Ave. | p-value | Ave. | p-value | Total(ave.) | p-value | ||
| 125(TASSER-QA) | 67 | 0.926 | 0.772 | 47.32(0.706) | |||
| 634(Pcons) | 67 | 0.895 | 0.04 | 0.796 | 0.18 | 45.12(0.673) | 9.5×10−6 |
| 556(LEE) | 65 | 0.954 | 0.01 | 0.842 | 2.9×10−4 | 44.50(0.685) | 4.0×10−4 |
| 713(Circle-QA) | 67 | 0.817 | 9.0×10−24 | 0.701 | 5.6×10−4 | 46.17(0.689) | 1.2×10−3 |
| 633(ProQ) | 67 | 0.802 | 1.2×10−13 | 0.618 | 2.2×10−9 | 44.33(0.662) | 1.7×10−5 |
| 038(GeneSilico) | 60 | 0.799 | 3.4×10−13 | 0.651 | 8.2×10−7 | 39.94(0.666) | 1.4×10−4 |
| 692(ProQlocal) | 67 | 0.801 | 9.0×10−14 | 0.618 | 2.1×10−9 | 44.35(0.662) | 2.0×10−5 |
| 178(Bilab) | 67 | 0.784 | 8.5×10−11 | 0.614 | 1.4×10−7 | 44.43(0.663) | 3.3×10−4 |
| 704(QA-ModFOLD) | 67 | 0.773 | 5.4×10−14 | 0.659 | 6.3×10−7 | 44.20(0.660) | 3.3×10−5 |
| 699(ABIpro-h) | 67 | 0.771 | 1.4×10−9 | 0.685 | 4.3×10−4 | 46.02(0.687) | 2.0×10−4 |
| 091(Ma-OPUS) | 66 | 0.747 | 3.2×10−17 | 0.654 | 4.7×10−6 | 44.13(0.669) | 1.7×10−3 |
| 013(Jones-UCL) | 56 | 0.734 | 1.9×10−16 | 0.586 | 7.3×10−12 | 37.72(0.674) | 1.8×10−6 |
| 703(QA-ModCHECK) | 47 | 0.673 | 3.8×10−15 | 0.587 | 5.4×10−7 | 24.09(0.512) | 3.1×10−7 |
| 717(CaspIta-FRST) | 61 | 0.662 | 1.8×10−22 | 0.550 | 2.2×10−15 | 39.55(0.648) | 2.7×10−8 |
| 025(Zhang-Server) | 67 | - | - | 47.14(0.704) | 0.34 | ||
We further examine the performance of the compared methods on easy, medium and hard targets as classified by our in-house 3D-jury approach28 (see http://cssb.biology.gatech.edu/skolnick/files/tasser-qa/ for classification list). Table 2 shows the results for the 67 easy targets, and Table 3 shows the results for the 31 medium/hard targets. In both Tables 2 and 3, the GDT-score is used as the structural similarity measure. For the easy targets, the LEE method has the best Pearson correlation coefficient and the best Spearman correlation coefficient, whereas TASSER-QA has the second and third best results, respectively, on the basis of the Pearson and Spearman correlation coefficients. TASSER-QA has a significantly higher total GDT-score than those of all other quality assessment methods. For the medium/hard targets, TASSER-QA surpasses the LEE method and has the highest Pearson correlation coefficient and the second highest Spearman correlation coefficient next only to that of Pcons (see table 3). The total GDT-score (10.74) of TASSER-QA is highest, but is almost the same as that of method ABIpro-h (10.47). The difference in total GDT-score between TASSER-QA and many other methods are statistically indistinguishable probably due to small data size of 31 targets.
Table 3.
Average correlation coefficients and total GDT-scores of the top quality assessment predictors in CASP7 for the 31 medium/hard targets.
| Method | # of targets | Pearson | Spearman | GDT-score | |||
|---|---|---|---|---|---|---|---|
| Ave. | p-value | Ave. | p-value | Total(ave.) | p-value | ||
| 125(TASSER-QA) | 31 | 0.636 | 0.651 | 10.74(0.346) | |||
| 634(Pcons) | 31 | 0.628 | 0.86 | 0.674 | 0.58 | 9.93(0.320) | 0.01 |
| 556(LEE) | 31 | 0.470 | 1.2×10−3 | 0.546 | 5.6×10−3 | 9.36(0.302) | 1.7×10−3 |
| 713(Circle-QA) | 31 | 0.543 | 9.8×10−3 | 0.579 | 0.012 | 9.89(0.319) | 0.04 |
| 633(ProQ) | 31 | 0.541 | 8.8×10−3 | 0.551 | 7.5×10−4 | 9.82(0.317) | 0.07 |
| 038(GeneSilico) | 28 | 0.527 | 0.01 | 0.558 | 6.1×10−3 | 9.15(0.327) | 0.06 |
| 692(ProQlocal) | 31 | 0.514 | 9.1×10−3 | 0.533 | 1.1×10−3 | 9.68(0.312) | 0.04 |
| 178(Bilab) | 31 | 0.515 | 1.7×10−3 | 0.522 | 6.6×10−4 | 10.09(0.326) | 0.15 |
| 704(QA-ModFOLD) | 31 | 0.464 | 3.2×10−4 | 0.473 | 1.4×10−4 | 9.72(0.314) | 0.02 |
| 699(ABIpro-h) | 31 | 0.463 | 3.0×10−4 | 0.508 | 3.7×10−4 | 10.47(0.338) | 0.58 |
| 091(Ma-OPUS) | 31 | 0.482 | 4.5×10−4 | 0.508 | 3.9×10−4 | 9.11(0.294) | 0.01 |
| 013(Jones-UCL) | 25 | 0.453 | 1.1×10−3 | 0.451 | 3.3×10−5 | 7.55(0.302) | 0.02 |
| 703(QA-ModCHECK) | 24 | 0.527 | 0.03 | 0.552 | 0.01 | 6.52(0.272) | 3.8×10−3 |
| 717(CaspIta-FRST) | 29 | 0.426 | 4.3×10−6 | 0.451 | 5.7×10−7 | 8.94(0.308) | 5.5×10−3 |
| 025(Zhang-Server) | 31 | - | - | 10.21(0.329) | 0.13 | ||
In Table 4, we list the numbers of targets for which server models are ranked the first by TASSER-QA and by actual quality GDT-score. The top three servers by TASSER-QA are the same as those by GDT-score. However, Zhang-Server and MetaTasser are over-selected by TASSER-QA: 33 by TASSER-QA vs. 24 by GDT-score for the Zhang-Server and 17 by TASSER-QA vs. 12 by the GDT-score for MetaTasser. Although Zhang-Server contributes one third of the top ranked models by TASSER-QA and it is significantly better than other servers in CASP7, the total GDT-score (57.64) by TASSER-QA changes only very little when models from Zhang-Server are eliminated from the selection process.
Table 4.
Numbers of targets for which the server models are ranked the first by TASSER-QA and by GDT-score.
| Servera | TASSER-QA | GDT-score |
|---|---|---|
| Zhang-Server | 33 | 24 |
| MetaTasser | 17 | 12 |
| Pmodeller6 | 6 | 6 |
| ROBETTA | 5 | 3 |
| HHpred3 | 5 | 5 |
| Bilab-ENABLE | 4 | 1 |
| SAM_T06_server | 3 | 3 |
| RAPTOR | 3 | 2 |
| BayesHH | 3 | 4 |
| SPARKS2 | 2 | 0 |
We refer to the CASP7 website http://predictioncenter.org/casp7/ for abstracts of individual server methods. Only top ten servers by TASSER-QA are presented.
Another analysis we carried out is the effects of the two terms in Eq. 3. When the contact potential term Econtact is set to zero, we get the result (Pearson, Spearman, total GDT-score) = (0.763, 0.648, 56.80). They are all significantly worse than those given by the full terms in Eq. 3 that are (0.834, 0.734, 58.06). If the fragment comparison term Efrg is set to zero, we get (0.831, 0.698, 56.60). The correlation coefficients change slightly, but the total GDT-score changes significantly. These results show that both Efrg and Econtact are important to the better performance of TASSER-QA method.
In CASP7, TASSER-QA only considered the first model given by each individual server (i.e. model names ending with “_TS1”). This is similar to the best-model-mode of the 3D-jury method6. It is of interest to know how much worse TASSER-QA will be if this strategy is not implemented or cannot be used because of the absence of ranking information from the individual servers. By including all models (model names ending with “_TS1” to “_TS5”) in the assessment prediction, TASSER-QA will have (Pearson, Spearman, total GDT-score) = (0.828, 0.791, 57.22) compared to the original (0.834, 0.734, 58.06) (see also Table 1). The Pearson correlation coefficient and total GDT-score are still the best of the compared methods. The Spearman rank correlation coefficient of TASSER-QA moves from the third to the top position. However, the strategy of pre-filtering possibly worse models other than the first models (ending with “_TS1”) by individual servers helps in the selection of good models by TASSER-QA as assessed by the total GDT-score. A similar result was also observed in the 3D-jury method6. That is because both TASSER-QA and 3D-jury use the similarity information between models which depends on the ensemble of models. This may also be true for other methods that depend only on the properties of individual models, e.g., Pcons will have (Pearson, Spearman, total GDT-score) = (0.820, 0.696, 55.62) if only first models by individual servers are used compared to (0.810, 0.757, 55.05) when all top five server models are assessed. The fact that TASSER-QA using all models is indistinguishable from using first models in Pearson’s correlation coefficient and total GDT-score (see Table 1) indicates that it can be reliably applied to models with no ranking information like those from CASP set-up.
From these results, a general trend for the correlation coefficients is also observed: the more models that are assessed, the better is the Spearman rank correlation coefficient and the worse is the Pearson correlation coefficient. This may be attributable to the intrinsic mathematical properties of the two kinds of coefficients and may have nothing to do with the properties of assessment prediction scores.
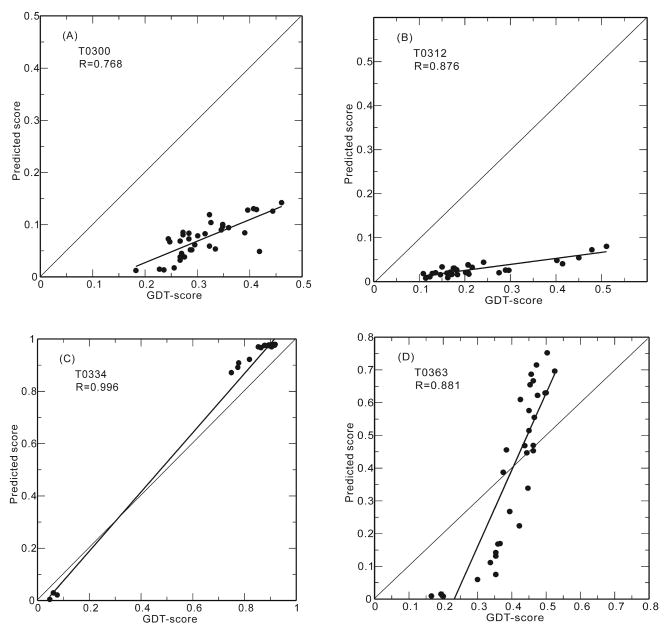
In Figure 1, we show some examples of the linear correlation between the TASSER-QA predicted scores and the actual GDT-scores of the models. The correlation coefficient can be as high as 0.996 (0.961 after excluding the three apparently outliers of very low GDT-scores) for target T0334. As shown in Table 2, the average linear correlation coefficient for easy targets is 0.926. It can also be very poor for some hard targets. For example, the correlation coefficient for target T0356 is −0.164 and for target T0361, it is 0.083 (not shown in Figure 1), even though TASSER-QA has the best average linear correlation coefficient of 0.636 for the 31 medium/hard targets.
Figure 1.
We next consider the application of TASSER-QA to situations not like a CASP set-up in which there are many servers and almost all of them provide internally ranked models for the same protein target. The first such application is one when there are only a few servers, with no internal ranking information. In Table 5, we show the results for models from only two moderately performing servers ROBETTA and SP3 in CASP7 and we assume no rank information among the 10 (5 from each server) models for each target. We have also assumed that the predictions of all other compared methods do not depend on the ensemble of models so that we can directly use their CASP7 predictions on the subset of models without having to re-do the predictions (although TASSER-QA’s dependence on the ensemble has been taken into account through re-calculating the consensus potential). TASSER-QA is applied using all 10 models from the two servers. We see in Table 5 that TASSER-QA is still the best for both correlation methods and total GDT-score. The selected top model quality by many methods is better than that of ROBETTA or SP3 but is more than 5% worse than that of the best models.
Table 5.
Average correlation coefficients and total GDT-scores of the top quality assessment predictors in CASP7 for models from ROBETTA and SP3 servers only.
| Method | # of targets | Pearson | Spearman | GDT-score | |||
|---|---|---|---|---|---|---|---|
| Ave. | p-value | Ave. | p-value | Total(ave.) | p-value | ||
| 125(TASSER-QA-all) | 98 | 0.677 | 0.609 | 55.11(0.562) | |||
| 634(Pcons) | 98 | 0.577 | 0.03 | 0.577 | 0.40 | 54.04(0.551) | 0.13 |
| 556(LEE) | 96 | 0.526 | 1.3×10−3 | 0.476 | 4.2×10−3 | 52.71(0.549) | 0.09 |
| 713(Circle-QA) | 98 | 0.569 | 4.4×10−3 | 0.527 | 0.02 | 54.42(0.555) | 0.22 |
| 633(ProQ) | 98 | 0.430 | 5.4×10−7 | 0.391 | 5.4×10−6 | 53.98(0.551) | 0.10 |
| 038(GeneSilico) | 80 | 0.491 | 1.4×10−3 | 0.454 | 3.3×10−3 | 44.94(0.562) | 0.16 |
| 692(ProQlocal) | 97 | 0.418 | 3.6×10−7 | 0.383 | 5.1×10−6 | 53.88(0.555) | 0.15 |
| 178(Bilab) | 98 | 0.537 | 5.7×10−7 | 0.489 | 9.2×10−4 | 54.38(0.555) | 0.15 |
| 704(QA-ModFOLD) | 98 | 0.555 | 1.4×10−3 | 0.513 | 5.4×10−3 | 54.20(0.553) | 0.19 |
| 699(ABIpro-h) | 98 | 0.568 | 1.9×10−3 | 0.526 | 6.8×10−3 | 54.14(0.552) | 0.07 |
| 091(Ma-OPUS) | 97 | 0.549 | 7.2×10−4 | 0.505 | 5.7×10−3 | 52.18(0.538) | 6.8×10−3 |
| 013(Jones-UCL) | 81 | 0.439 | 9.6×10−7 | 0.408 | 3.5×10−5 | 44.76(0.553) | 0.06 |
| 703(QA-ModCHECK) | 71 | 0.546 | 0.11 | 0.502 | 0.16 | 38.37(0.540) | 0.48 |
| 717(CaspIta-FRST) | 90 | 0.473 | 4.2×10−5 | 0.485 | 4.9×10−3 | 49.20(0.547) | 0.02 |
| ROBETTA | 98 | - | - | 52.87(0.539) | |||
| SP3 | 98 | - | - | 51.58(0.526) | |||
| Best | 98 | - | - | 58.01(0.592) | |||
The second application is on models generated by ab initio or refinement modeling methods such as ROSETTA and TASSER. In this situation, there are a large number models with no a priori ranking information and the usual way of selecting models is to use clustering methods such as SPICKER4. To show TASSER-QA also works well in this situation, we apply TASSER-QA on TASSER generated models for CASP7 targets with 16,000 models for each target and compare it with SPICKER on the same set of models. The results are compiled in Table 6. The p-values that characterize the statistical significances of the difference between the total GDT-scores of TASSER-QA and SPICKER selected models are all > 0.05 except for one case when the fragment comparison term is set to zero and best of top five selected models are used for GDT-score computation. That means TASSER-QA has comparable performance with SPICKER on TASSER generated models. Table 6 also shows that the fragment comparison term is slightly more transferable to this kind of models and the combination of fragment comparison and consensus potential does not give better results.
Table 6.
Comparison of TASSER-QA with SPICKER on TASSER-generated models for the 98 CASP7 targets.
| GDT-score of First modele | GDT-score of Best of top five | |
|---|---|---|
| 98 targets | ||
| SPICKERa | 53.93 | 55.31 |
| TASSER-QA-allb | 54.29 (0.17) | 55.23 (0.75) |
| TASSER-QA-all-frgc | 54.35 (0.18) | 55.74 (0.16) |
| TASSER-QA-all-cad | 53.81 (0.64) | 54.79 (0.04) |
| 67 easy targets | ||
| SPICKER | 45.31 | 45.96 |
| TASSER-QA-all | 45.31 (1.0) | 45.90 (0.73) |
| TASSER-QA-all-frg | 45.57 (0.21) | 46.31 (0.09) |
| TASSER-QA-all-ca | 45.19 (0.42) | 45.66 (0.08) |
| 31 medium/hard targets | ||
| SPICKER | 8.62 | 9.35 |
| TASSER-QA-all | 8.98 (0.09) | 9.32 (0.45) |
| TASSER-QA-all-frg | 8.78 (0.49) | 9.43 (0.74) |
| TASSER-QA-all-ca | 8.63 (0.97) | 9.13 (0.26) |
Clustering method in Ref 4.
This work using all models.
This work using all models with the consensus term in Eq. (3) set to zero.
This work using all models with the fragment comparison term in Eq. (3) set to zero.
Numbers in parenthesis are p-values for the differences between TASSER-QA and SPICKER.
CONCLUSIONS
In this work, we presented a simple but accurate model quality assessment prediction method that is comparable to or even better than the state-of-the-art methods available to date. The method combines fragment comparisons and a consensus Cα contact potential. The fragment library is obtained by an extension of the SP3 threading method. The consensus Cα contact potential is derived from the models to be assessed using the same approach as in TASSER33,34. This consensus Cα contact potential takes into account the effect of similarities among the models which behaves in some sense like 3D-jury6. Both terms in Eq. 3 are important for the current method to be successful. This approach is fully automated and is useful for selecting the best possible models from a set of structures provided by other methods. The resulting selected models can also be used as a starting point for further refinement. The current methodology was used by TASSER group in CASP7 28 in selecting the final models for submission. In practice, it can also be used by TASSER34 or other refinement methods in selecting initial models from other servers (e.g. all server models in CASP7) for refinement. The fact that this method has a very good Pearson correlation coefficient for easy targets makes it a suitable approach for near native structure selection. For medium/hard targets, although the Pearson correlation coefficient is worse, it is still better than other existing approaches.
We note that for most of the easy targets, the correlations between predicted quality score and actual GDT-score are very good, but the slopes of the linear correlations are not close to one. That means the prediction is good for the relative quality of the models within a given target, but not good enough for absolute quality of the models that can be compared between different targets. This is true for TASSER-QA as well as for other top performing methods.
The current method can also be extended to assign a local quality measure of individual residues by simply considering the contribution of individual residues to Eq. 4. How well the extended prediction works on identifying high quality regions of the predicted structure needs further investigation.
Acknowledgments
This research was supported in part by grant Nos. GM-37408 and GM-48835 of the Division of General Medical Sciences of the National Institutes of Health.
References
- 1.Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 2.Skolnick J, Fetrow J, Kolinski A. Structural genomics and its importance for gene function analysis. Nat Biotechnol. 2000;18:283–287. doi: 10.1038/73723. [DOI] [PubMed] [Google Scholar]
- 3.Moult J, Fidelis K, Hubbard T, Kryshtafovych A, Rost B, Tramontano A. 7th Critical Assessment of Techniques for Protein Structure Prediction [Google Scholar]
- 4.Zhang Y, Skolnick J. SPICKER: a clustering approach to identify near-native protein fold. J Comput Chem. 2004;25:865–871. doi: 10.1002/jcc.20011. [DOI] [PubMed] [Google Scholar]
- 5.Simons KT, Strauss C, Baker D. Prospects for ab initio protein structural genomics. J Mol Biol. 2001;306:1191–1199. doi: 10.1006/jmbi.2000.4459. [DOI] [PubMed] [Google Scholar]
- 6.Ginalski K, Elofsson A, Fischer D, Rychlewski L. 3D-jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- 7.Sasson I, Fischer D. Modeling three-dimensional protein structures for CASP5 using 3D-SHOTGUN meta-predictors. Proteins: Structure, function and Genetics. 2003;53:389–394. doi: 10.1002/prot.10544. [DOI] [PubMed] [Google Scholar]
- 8.Melo F, Sanchez R, Sali A. statistical potentials for fold assessment. Protein Sci. 2002;11:430–448. doi: 10.1002/pro.110430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou H, Zhou Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Science. 2002;11:2714–2726. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lazaridis T, Karplus M. Discrimination of the native from misfolded protein models with an energy function including implicit solvation. J Mol Biol. 1998;288:477–487. doi: 10.1006/jmbi.1999.2685. [DOI] [PubMed] [Google Scholar]
- 11.Melo F, Feytmans E. Assessing protein structures with a non-local atomic interaction energy. J Mol Biol. 1998;277:1141–1152. doi: 10.1006/jmbi.1998.1665. [DOI] [PubMed] [Google Scholar]
- 12.Lu H, Skolnick J. A distance-dependent atomic knowledge-based potential for improved protein structure selection. Proteins. 2001;44:223–232. doi: 10.1002/prot.1087. [DOI] [PubMed] [Google Scholar]
- 13.Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 14.Petrey D, Honig B. Free energy determinants of tertiary structure and the evaluation of protein models. Protein Sci. 2000;9:2181–2191. doi: 10.1110/ps.9.11.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia Y, Huang ES, Levitt M, Samudrala R. Ab initio construction of protein tertiary structures using a hierarchical approach. J Mol Biol. 2000;300:171–185. doi: 10.1006/jmbi.2000.3835. [DOI] [PubMed] [Google Scholar]
- 16.Dominy BN, Books CL. Identifying native-like protein structures using physics-based potentials. J Comput Chem. 2002;23:147–160. doi: 10.1002/jcc.10018. [DOI] [PubMed] [Google Scholar]
- 17.Tobi D, Elber R. Distance-dependent, pair potential for protein folding: results from linear optimization. Proteins. 2000;41:40–46. [PubMed] [Google Scholar]
- 18.Luthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 19.Eisenberg D, Luthy R, Bowie JU. VERIFY3D: Assessment of protein models with three-dimensional profiles. Methods in Enzym. 1997;277:396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- 20.Zhang C, Liu S, Zhou H, Zhou Y. An accurate residue-level pair potential of mean force for folding and binding based on the distance-scaled ideal-gas reference state. Protein Sci. 2004;13(400–411) doi: 10.1110/ps.03348304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang C, Liu S, Zhou Y. Accurate and efficient loop selections using DFIRE-based all-atom statistical potential. Protein Sci. 2004;13:391–399. doi: 10.1110/ps.03411904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lundsrom J, Rychlewski L, Bunnicki J, Elofsson A. Pcons: a neural-network-based consensus predictor that improves fold recognition. Protein Science. 2001;10:2354–2362. doi: 10.1110/ps.08501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wallner B, Fang H, Elosson A. Automatic consensus-based fold recognition using Pcons, ProQ, and Pmodeller. Proteins: Structure, Function and Genetics Suppl. 2003;6:534–541. doi: 10.1002/prot.10536. [DOI] [PubMed] [Google Scholar]
- 24.Wallner B, Elofsson A. Can correct protein models be identified? Protein Science. 2003;12:1073–1086. doi: 10.1110/ps.0236803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eramian D, Shen MY, Devos D, Melo F, Sali A, Marti-Renom MA. A composite score for predicting errors in protein structure models. Protein Science. 2006;15:1653–1666. doi: 10.1110/ps.062095806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wallner B, Elofsson A. Identification of correct regions in protein models using structural, alignment, and consensus information. Protein Sci. 2005;15:900–913. doi: 10.1110/ps.051799606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischer D. 3D-SHOTGUN: a novel, cooperative, fold-recognition meta-predictor. Proteins. 2003;51:434–441. doi: 10.1002/prot.10357. [DOI] [PubMed] [Google Scholar]
- 28.Zhou H, Pandit SB, Lee S, Borreguerro J, Chen H, Wroblewska L, Skolnick J. Analysis of TASSER based CASP7 protein structure prediction results. Proteins. 2007 doi: 10.1002/prot.21649. (submitted) [DOI] [PubMed] [Google Scholar]
- 29.Zhou H, Zhou Y. Fold recognition by combining sequence profiles derived from evolution and from depth-dependent structural alignment of fragments. Proteins. 2005;58:321–328. doi: 10.1002/prot.20308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou H, Zhou Y. SPARKS 2 and SP3 servers in CASP 6. Proteins(Supplement CASP issue) 2005;(suppl 7):152–156. doi: 10.1002/prot.20732. [DOI] [PubMed] [Google Scholar]
- 31.Domingues FS, Lackner P, Andreeva A, Sippl MJ. Structure-based evaluation of sequence comparison and fold recognition alignment accuracy. J Mol Biol. 2000;297:1003–1013. doi: 10.1006/jmbi.2000.3615. [DOI] [PubMed] [Google Scholar]
- 32.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman D-J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acid Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Kolinski A, Skolnick J. TOUCHSTONE II: A new approach to ab initio protein structure prediction. Biophysical Journal. 2003;85:1145–1164. doi: 10.1016/S0006-3495(03)74551-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Skolnick J. Automated structure prediction of weakly homologous proteins on genomic scale. Proc Natl Acad Sci (USA) 2004;101:7594–7599. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Skolnick J. A scoring function for the automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 36.Zemla A, Venclovas C, Moult J, Fidelis K. Processing and analysis of CASP3 protein structure predictions. Proteins. 1999;3:22–29. doi: 10.1002/(sici)1097-0134(1999)37:3+<22::aid-prot5>3.3.co;2-n. [DOI] [PubMed] [Google Scholar]
- 37.Press WH, Flannery BP, Teukolsky SA, Vetterling WT. Numerical Recipes: The Art of Scientific Computing. Cambridge University Press; 1989. [Google Scholar]