Abstract
Pyrosequencing-based 16S rRNA gene surveys are increasingly utilized to study highly diverse bacterial communities, with special emphasis on utilizing the large number of sequences obtained (tens to hundreds of thousands) for species richness estimation. However, it is not yet clear how the number of operational taxonomic units (OTUs) and, hence, species richness estimates determined using shorter fragments at different taxonomic cutoffs correlates with the number of OTUs assigned using longer, nearly complete 16S rRNA gene fragments. We constructed a 16S rRNA clone library from an undisturbed tallgrass prairie soil (1,132 clones) and used it to compare species richness estimates obtained using eight pyrosequencing candidate fragments (99 to 361 bp in length) and the nearly full-length fragment. Fragments encompassing the V1 and V2 (V1+V2) region and the V6 region (generated using primer pairs 8F-338R and 967F-1046R) overestimated species richness; fragments encompassing the V3, V7, and V7+V8 hypervariable regions (generated using primer pairs 338F-530R, 1046F-1220R, and 1046F-1392R) underestimated species richness; and fragments encompassing the V4, V5+V6, and V6+V7 regions (generated using primer pairs 530F-805R, 805F-1046R, and 967F-1220R) provided estimates comparable to those obtained with the nearly full-length fragment. These patterns were observed regardless of the alignment method utilized or the parameter used to gauge comparative levels of species richness (number of OTUs observed, slope of scatter plots of pairwise distance values for short and nearly complete fragments, and nonparametric and parametric species richness estimates). Similar results were obtained when analyzing three other datasets derived from soil, adult Zebrafish gut, and basaltic formations in the East Pacific Rise. Regression analysis indicated that these observed discrepancies in species richness estimates within various regions could readily be explained by the proportions of hypervariable, variable, and conserved base pairs within an examined fragment.
Culture-independent 16S rRNA gene surveys are now routinely utilized to examine the microbial diversity in various environmental habitats. However, in surveys of highly diverse ecosystems, the size of clone libraries typically constructed (100 to 500 clones) allows for the identification only of members of the community that are present in high abundance (2, 13, 14, 17, 24, 51). In addition to the failure to detect the rare members of the ecosystem, these relatively small datasets provide inaccurate estimates when used for computing species richness within an ecosystem. Regardless of the approach utilized to estimate species richness, the estimates obtained are highly dependent on sample size, and smaller datasets typically result in the underestimation of species richness (14, 44, 47, 55).
The use of a pyrosequencing-based approach (40) in 16S gene-based diversity surveys promises to overcome both of the above-mentioned problems associated with inadequate sampling. The large number of 16S rRNA gene sequences produced (hundreds of thousands) allows access to rare members of the community (25; J. M. Tiedje, presented at the 108th General Meeting of the American Society for Microbiology, Boston, MA, 2008), as well as a relatively more accurate estimation of species richness. However, with the introduction of this new technology, it is necessary to correlate the results obtained from newer pyrosequencing-based surveys to the extensive collection of longer, capillary sequence-generated 16S rRNA gene sequences that has been deposited in public databases during the last 2 decades. Several recent studies have examined the utility of pyrosequencing fragments in providing an accurate survey of overall community structure (36) and investigated the ability of various fragments spanning the 16S rRNA gene to accurately predict the phylogenetic affiliation of pyrosequencing-generated fragments at various taxonomic cutoffs (35, 54). As such, these admirable efforts gave useful insights into the advantages and limitations of the pyrosequencing approach in 16S-based community surveys, pinpointed specific regions that provide better phylogenetic resolution than other pyrosequencing-generated regions, and provided a quantitative assessment of binning accuracy at various empirical cutoffs.
However, while issues regarding correlating phylogenies of shorter and longer fragments are actively being addressed, efforts to calibrate species richness data obtained from various pyrosequencing fragments at various taxonomic cutoffs to estimates obtained using longer 16S rRNA gene fragments are still lacking. It is unclear how pairwise distances and, hence, operational taxonomic unit (OTU) assignments and species richness estimates computed using various shorter fragments spanning various regions of the 16S rRNA gene will correlate to pairwise distances computed using the nearly complete 16S rRNA gene. Elucidating such differences between shorter and nearly complete fragments, as well as between shorter fragments representing different regions in the 16S rRNA gene, is absolutely necessary for accurate meta-analysis of species richness in previously published and future datasets constructed using various sequencing approaches.
Here, we constructed, sequenced, and analyzed a 16S rRNA library of 1,132 clones generated from an undisturbed tallgrass prairie soil in central Oklahoma and compared the numbers of OTUs and species richness values obtained using the full-length data sets (with and without the application of the Lane mask filter that excludes hypervariable regions from the phylogenetic analysis) (32) and fragments simulating pyrosequencing output generated by clipping where known conserved bacterial primers are encountered in the 16S rRNA gene. The lengths of the chosen simulated-pyrosequencing fragments represent amplicons that have been generated using the original GS20 pyrosequencing platform (≈100 bp) (25, 44, 48), similar to those currently being generated using the GS FLX pyrosequencing platform (≈250 bp) (1, 20, 35) or amplicons produced using the anticipated increase in the new GS XLR pyrosequencing platform (>250 bp). We show that the choice of the pyrosequenced fragment could indeed impact the number of OTUs calculated at different taxonomic cutoffs, with some fragments underestimating and others overestimating such parameters compared to the results with longer, nearly complete 16S rRNA gene fragments. We also show that even more marked differences could be encountered when comparing two pyrosequencing fragments within the same molecule. Further, we established a regression analysis that explains the nature of the observed discrepancies using the proportions of the hypervariable, variable, and conserved bases within fragments.
MATERIALS AND METHODS
Sampling and library construction.
A clone library, designated Soil-Okla-A, was constructed from undisturbed tallgrass prairie soil in central Oklahoma. The area is part of the Kessler Farm Field Laboratory research station in McClain County, OK (34° 58′ 31.74" N, 97° 31′ 18.05" W). Details of the soil structure and chemistry have previously been reported in a separate report that examined novelty and uniqueness patterns in Kessler Farm soil using a different 16S rRNA clone library (18). Samples were collected from the top 5 cm of soil in April 2005 and stored on dry ice while being transferred to the laboratory, where they were stored at −20°C upon arrival. DNA was extracted by using a FastDNA spin kit for soil (MP Biomedicals, Solon, OH), and the nearly complete 16S rRNA gene was amplified using primers 8F (5′ AGAGTTTGATCMTGGCTCAG 3′) and 1492R (5′ ACCTTGTTACGACTT 3′) in a 50-μl reaction mixture containing (final concentration) 2 μl of extracted DNA, 1× PCR buffer (Invitrogen), 2.5 mM MgSO4, 0.2 mM deoxynucleoside triphosphate mixture, 2.5 U platinum Taq DNA polymerase (Invitrogen), and 10 μM each of the forward and reverse primers. PCR amplification was carried out according to the following protocol: initial denaturation for 5 min at 95°C, followed by 20 cycles of denaturation at 95°C for 45 s, annealing at 52°C for 45 s, and elongation at 72°C for 1.5 min, with a final elongation step at 72°C for 15 min. The PCR products obtained were cloned into a TOPO-TA cloning vector according to the manufacturer's instructions (Invitrogen Corp., Carlsbad, CA). Sequencing of cloned inserts was conducted at the Advanced Center for Genome Technology (Norman, OK) as previously described (17). Chimeric sequences were identified by using Chimera check with the Bellerophon (version 3) function on the Greengenes web server (11). Twenty-nine sequences were identified as potential chimeras and removed from the data set.
Alignments and phylogenetic classification.
The nonchimeric sequences (1,132 clones) were aligned by utilizing a pairwise alignment approach using the ClustalX program (50). ClustalX performs a progressive pairwise alignment that is generally regarded as a more accurate approach than aligning sequences to a preexisting alignment database, e.g., Greengenes NAST aligner and the RDP database (8, 10). Nevertheless, pairwise alignment of the large number of fragments generated by pyrosequencing is computationally a daunting task. For example, for a 100,000-sequence data set, pairwise alignment will necessitate the generation of ∼1010 pairs of alignments. Pyrosequencing studies currently rely on aligning sequences against core sets of sequences in public databases. Therefore, in addition to ClustalX alignments, we generated a NAST alignment (10) in the Greengenes web server to compare the number of OTUs generated using a more accurate approach to the number generated by a more commonly used approach. The NAST aligned sequences were also used for determining the phylum-level affiliations of clones according to Hugenholtz taxonomy framework in the Greengenes classifier (11).
Clipping of shorter fragments to simulate pyrosequencing.
The aligned nearly full-length sequence file was imported to the Jalview program (7) to clip short sequences representing candidate regions for pyrosequencing using known conserved bacterial primers and with a suitable size for past, current, and near-future pyrosequencing platform capabilities (read length of 99 to 361 bp). These theoretical amplicons contain one or more of the variable regions V1 to V8 of the 16S rRNA molecule (see reference 41 for a canonical representation of the prokaryotic 16S rRNA gene molecule and the location of each of these variable regions within it). The forward and reverse primers used, length, and variable regions encompassed are shown in Table 1 for each of the eight short simulated fragments studied, as well as for the nearly full-length fragment.
TABLE 1.
Sequences of forward and reverse primers, variable sites encompassed, and base composition for the short, pyrosequencing-simulating regions studied and the nearly full-length fragment
| Bases | Forward primer | Reverse primer | Region | % of bases that are:
|
References | ||
|---|---|---|---|---|---|---|---|
| V | HV | C | |||||
| 27-355 | AGAGTTTGATCMTGGCTCAG | ACTCCTACGGGAGGCAGC | V1+V2 | 47 | 18 | 35 | 4, 38, 39 |
| 338-548 | GCTGCCTCCCGTAGGAGT | AATACGGAGGGTGCAAGCGT | V3 | 44 | 14 | 42 | 4, 38 |
| 530-826 | ACGCTTGCACCCTCCGTATT | GGATTAGATACCCTGGTAGTC | V4 | 57 | 5 | 38 | 4, 38 |
| 805-1065 | GACTACCAGGGTATCTAATCC | AGGTGCTGCATGGCTGT | V5+V6 | 49 | 10 | 41 | 21, 30, 38 |
| 967-1065 | CAACGCGAAGAACCTTACC | AGGTGCTGCATGGCTGT | V6 | 45 | 19 | 36 | 21, 30 |
| 967-1238 | CAACGCGAAGAACCTTACC | GTAGCRCGTGTGTMGCCC | V6+V7 | 44 | 9 | 47 | 21, 30, 31 |
| 1046-1238 | ACAGCCATGCAGCACCT | GTAGCRCGTGTGTMGCCC | V7 | 40 | 3 | 57 | 21, 30, 31 |
| 1046-1406 | ACAGCCATGCAGCACCT | GACGGGCGGTGWGTRCA | V7+V8 | 43 | 5 | 52 | 21, 30, 39 |
| 8-1492a | AGAGTTTGATCMTGGCTCAG | ACCTTGTTACGACTT | 51 | 10 | 39 | 34, 39, 53 | |
Nearly full-length fragment.
Distance matrix generation and OTU assignments.
Both the ClustalX and NAST-aligned files for the nearly full-length fragments, as well as the eight short fragments, were imported to Paup (Sinauer Associates, Inc., Sunderland, MA) to create full-distance matrices by using the “Create distance matrix” function within the program. The distance matrix was generated using the following parameters: Distance setting with F84 correction and a transition/transversion ratio of 2:1. Distance matrices generated using slightly different parameters (e.g., Distance setting with Jukes-Cantor correction, Kimura-2 correction, or Maximum likelihood correction) provided slightly different pairwise distance values and, hence, slightly different numbers of OTUs at different taxonomic cutoffs. However, the percent difference in OTU numbers obtained using each of the above-mentioned methods never exceeded 6.9% (range, 0.17% to 6.9%) and so the values presented satisfactorily represent pairwise distances of longer fragments.
The ratio of the number of OTUs obtained with any short region to the number of OTUs obtained with the nearly full-length region (referred to as the OTU ratio) was used as an indicator of whether a specific region overestimated or underestimated the number of OTUs compared to the results for the nearly full-length sequences. In addition, the distance matrix obtained was used to construct scatter plots between pairwise distance values of all possible pairs in the nearly full-length datasets (x axis) and the corresponding distances in each of the short regions (y axis), and the slopes of regression lines forced through the origin were calculated (see Fig. S1 in the supplemental material as an example of such plots). The values of the slopes for different regions indicate whether a specific region would over- or underestimate species richness compared to the results for the nearly full-length fragment. A slope value of around 1 suggests that the pyrosequencing fragment examined would give estimates of OTU numbers and, hence, species richness comparable to those obtained with the full-length fragment. A value greater than 1 suggests that a pyrosequencing fragment overestimates diversity, while a slope value of less than 1 suggests the opposite.
Species richness estimates using parametric and nonparametric approaches.
Since pyrosequencing-generated datasets are commonly used for species richness estimation (25, 44, 48), we sought to investigate whether the observed effect on the number of OTUs and the slopes of scatter plots of using the short pyrosequencing-simulating fragments would also apply to estimates of species richness. We estimated species richness using various approaches and compared the estimates obtained using each of the short fragments to those obtained using the nearly full-length fragment. We used DOTUR (46) to estimate species richness in all fragments examined using the nonparametric Chao and ACE estimators (6). We also used six different parametric distributions to fit the frequency data by the method of maximum likelihood, using a downloadable program (http://www.stat.cornell.edu/∼bunge/software.html) according to the criteria described previously (55). As previously observed (23, 29, 49, 55), the parametric model of mixture of two exponentials-mixed Poisson was the model that best fit the frequency data in all the datasets (the eight short pyrosequencing-simulating datasets, as well as the nearly full-length data set) and, hence, was used to estimate species richness. The choice of the most appropriate approach for species richness estimation is an issue hotly debated by microbial ecologists (29, 46), as well as macro ecologists (22, 42). Further, we understand that the sample size analyzed in this study is far from adequate to estimate the “true” species richness in soil. However, our goal is not to accurately determine the true species richness in the habitats examined but to provide a comparative tool for datasets that contain the same numbers of taxa. Under this scenario, we applied both parametric and nonparametric methods for estimating species richness as a comparative tool, an approach that is widely accepted in ecological studies (23, 26, 29, 49).
OTU assignments and species richness determinations for other environments.
We sought to determine whether the trends observed in the Soil-Okla-A clone library are unique to the Kessler Farm soil bacterial community, whether they represent a general trend characteristic of other soil ecosystems, or whether these trends will hold true in datasets derived from multiple habitats. To this end, using three previously reported datasets derived from another soil ecosystem (33), the digestive tract of conventionally raised adult zebrafish (43), and an endolithic and epilethic microbial community on basalt formations on the ocean floor (45), we determined the numbers of OTUs, slopes of scatter plots, and various species richness estimates using nearly full-length fragments and each of the eight candidate shorter pyrosequencing fragments as described above for the Soil-Okla-A clone library. All three datasets were generated using the same primer pair (8F and 1492R) that was also used to generate the Soil-Okla-A clone library, thus eliminating possible primer base bias or bias due to variations in the lengths of the amplicons produced.
Regression analysis.
As elaborated by Baker et al. (3) and Van de Peer et al. (52), the 16S rRNA molecule encompasses variable regions with bases that are, depending on the specific nucleotide substitution rate (vi) (3, 52), either totally conserved (vi = 0), conserved (vi = 10−0.925 to 10−0.425), variable (vi = 10−0.425 to 100.575), highly variable (vi ≥ 100.575), or >75% variable (present in Escherichia coli but absent in >75% of other bacteria). We hypothesized that the differences in the values of the slopes of pairwise distance scatter plots of different regions are due to differences in the frequency of base variability within each region. For each region, we determined the number of highly conserved and conserved bases (C), the number of variable bases (V), and the number of highly variable and >75%-variable bases (HV) as outlined by Baker et al. in reference 3. The percentages of C, V, and HV bases are shown in Table 1 for all the regions. To pinpoint the effect of base variability on the values of the slope, we calculated the Pearson correlation coefficient between the slope values and the numbers of bases (C, V, and HV), their ratios relative to the total (C/total, V/total, and HV/total), and all possible ratios (i.e., C/V, C/HV, V/C, V/HV, HV/C, and HV/V). We also calculated the Pearson correlation coefficient between the slope value and the length of the region. Since none of the above correlation coefficients exceeded 0.85, we sought to use multiple regression to correlate several variables to the values of the slope. Combinations of two or more of the above ratios were used in MS Excel using the function “Linest.” Multiple regression models were evaluated by comparing the values of slopes predicted by the model to the actual values. The model that best described the variations in the slope was the one that showed the highest Pearson correlation coefficient between actual and predicted slope values.
Nucleotide sequence accession numbers.
Sequences generated in this study were deposited in GenBank under accession numbers FJ478473 to FJ479604.
RESULTS
Composition of Soil-Okla-A clone library.
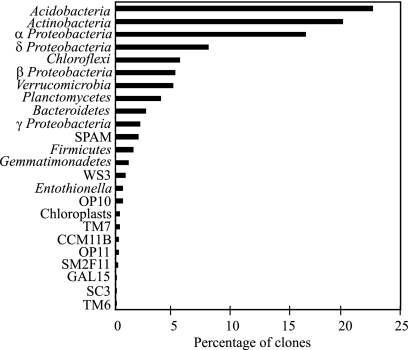
An examination of the 1,132 nearly full-length clones indicated that the Soil-Okla-A library displays a phylum-level community structure that is fairly typical of soils, with the nine major phyla often encountered in soil (Proteobacteria, Actinobacteria, Acidobacteria, Chloroflexi, Verrucomicrobia, Bacteroidetes, Planctomycetes, Gemmatimonadetes, and Firmicutes [28]) representing 95.2% of the clones in the library (Fig. 1). Compared to a previously published meta-analysis of multiple soil clone libraries (28), as well as a recently published quantitative PCR quantification of six different major bacterial phyla in 71 unique soils using group-specific quantitative PCR primers (19), the Soil-Okla-A clone library appears to be comparatively rich in Gammaproteobacteria, Alphaproteobacteria, and members of candidate division SPAM (Spring Alpine Meadow) and comparatively poor in members of the Firmicutes.
FIG. 1.
Phylum-level composition of the Soil-Okla-A clone library.
Comparison of numbers of OTUs obtained using nearly complete and shorter fragments.
Table 2 shows the number of OTUs obtained when the nearly full-length and the eight short pyrosequencing-simulating sequences were aligned using both pairwise alignment (using ClustalX) and multiple alignment (using the NAST alignment tool on the Greengenes web server). In general, the numbers of OTUs obtained following pairwise alignment were slightly higher than those obtained using NAST alignment (with the exception of the region encompassing V1 and V2 [the V1+V2 region]). However, the percent difference in the number of OTUs following either alignment approach was within 15% in most cases (Table 2). More importantly, the OTU ratios were comparable for long and short fragments following either method of alignment (see Table S1 in the supplemental material). Therefore, while pairwise alignment of sequences is known to give more accurate results than multiple alignment since each possible pair of sequences is aligned separately, the effect of the method of alignment on OTU assignment seems to be minimal. Since multiple alignments (e.g., NAST alignment) are becoming the method of choice with the increase in the number of sequences generated using pyrosequencing, we used the NAST-aligned file for further analysis.
TABLE 2.
Numbers of OTUs obtained using different methods of alignment at different taxonomic cutoffs with long and short sequences from the Soil-Okla-A clone librarya
| Regionb | Alignment method | No. of OTUs at indicated taxonomic cutoff (%)
|
Slopec | ||||
|---|---|---|---|---|---|---|---|
| 3 | 6 | 8 | 10 | 15 | |||
| V1+V2 | NAST | 652 | 499 | 407 | 345 | 233 | 1.2 |
| Clustal | 609 | 452 | 376 | 323 | 216 | ||
| V3 | NAST | 495 | 331 | 261 | 203 | 122 | 0.88 |
| Clustal | 499 | 359 | 259 | 223 | 132 | ||
| V4 | NAST | 619 | 414 | 327 | 267 | 158 | 0.97 |
| Clustal | 616 | 423 | 340 | 281 | 183 | ||
| V5+V6 | NAST | 570 | 393 | 328 | 267 | 166 | 1 |
| Clustal | 573 | 403 | 334 | 282 | 191 | ||
| V6 | NAST | 584 | 479 | 418 | 370 | 283 | 1.67 |
| Clustal | 545 | 500 | 438 | 371 | 231 | ||
| V6+V7 | NAST | 514 | 375 | 300 | 241 | 142 | 0.98 |
| Clustal | 575 | 391 | 312 | 278 | 172 | ||
| V7 | NAST | 340 | 194 | 149 | 112 | 61 | 0.6 |
| Clustal | 387 | 205 | 156 | 99 | 75 | ||
| V7+V8 | NAST | 412 | 241 | 187 | 143 | 73 | 0.65 |
| Clustal | 478 | 257 | 198 | 156 | 87 | ||
| NFL | NAST | 639 | 397 | 306 | 242 | 135 | NA |
| Clustal | 610 | 415 | 300 | 244 | 140 | NA | |
| LM | NAST | 424 | 263 | 195 | 157 | 84 | ND |
The results of the Clustal and NAST alignment methods at five different taxonomic cutoffs were compared using the nearly full-length sequences with and without the Lane mask filter and each of the eight studied short, pyrosequencing-simulating regions from the Soil-Okla-A clone library.
NFL, nearly full-length sequence (primer pair 8F-1492R); LM, nearly full-length region with the Lane mask applied.
Slopes of regression lines constructed between pairwise distances in the short, pyrosequencing-simulating regions versus the nearly full-length regions and forced through the origin. NA, not applicable; ND, not determined.
The OTU ratios, as well as the slopes of scatter plots between pairwise distances in each of the eight shorter regions and the nearly full-length fragment, were in general agreement in describing the relationship between long and short fragments. Following NAST alignment, the number of OTUs obtained using short simulated fragments ranged from 0.44 (V7-encompassing fragment at 15% cutoff) to 2.10 (V6-encompassing fragment at 15% cutoff) times the values obtained using the entire nearly complete longer 16S rRNA amplicon. This suggests that pyrosequencing could change our estimates of OTU numbers more than twofold in either direction. An even larger disparity could be detected when comparing the results for two pyrosequencing fragments with each other, a very plausible scenario with the anticipated future accumulation of pyrosequencing data. The numbers of OTUs obtained under this scenario differed as much as 4.64-fold (difference between the V6-encompassing fragment and the V7-encompassing fragment at the 15% taxonomic cutoff).
The comparison of the OTU estimates for each of the simulated fragments to those for the nearly full-length fragments at different taxonomic cutoffs, as well as the slopes of scatter plots (Table 2), indicates that, in general, fragments encompassing the V1+V2 regions and fragments encompassing the V6 region overestimated the number of OTUs at all taxonomic cutoffs (Table 2). On the other hand, fragments encompassing the V3, V7, and V7+V8 regions underestimated OTU numbers at all taxonomic cutoffs (Table 2). Fragments encompassing the V4, V5+V6, and V6+V7 (a composite of an overestimating fragment and an underestimating fragment) regions gave OTU numbers that were, in general, comparable to those obtained with the full sequence, as further evidenced by slope values of 0.97, 1, and 0.98, respectively, as well as by the fact that many of the OTU numbers at multiple cutoffs (12 out of 15) were within 15% of the OTU numbers obtained using the nearly full-length sequence (see Table S1 in the supplemental material).
It is important to note that these results were obtained by comparing the result for a specific pyrosequencing fragment to the result for the nearly complete 16S rRNA gene sequence with no filtration of hypervariable regions conducted prior to OTU assignments. Filtering hypervariable regions in nearly complete fragments prior to the generation of a distance matrix will significantly alter the pairwise distance values and, hence, the OTU assignments obtained. Examples of such filters include the Lane mask (32) and the Position variability by parsimony mask, available in the ARB software package (37). An examination of previously published 16S rRNA gene surveys showed that some studies did not apply filters for distance matrix generation or tree construction (15, 16), some applied filters only for the construction of phylogenetic trees but not for distance matrix generation (9, 12, 13, 17), and some applied filters for both distance matrix generation and, subsequently, tree construction (5, 18, 27), a process that is gaining popularity with the introduction of the “Create distance matrix” option in the Greengenes web server (11).
Therefore, in addition to comparison to the results for the unfiltered nearly complete 16S rRNA gene sequences described above, we determined the number of OTUs obtained with our nearly full-length sequence data set upon application of the Lane mask and compared it to estimates obtained with no mask, as well as estimates obtained with the eight short fragments. The numbers of OTUs obtained with the Lane mask were only 0.62 to 0.70 times the original number obtained with no Lane mask (Table 2). The estimates obtained using all short fragments were higher than those obtained with the Lane mask, by as much as 3.37-fold (compared to the V6 region at the 15% cutoff), except for fragments encompassing the V7 region, which gave estimates that were 0.71 to 0.80 times those of the nearly full-length fragment with the Lane mask, and the V7+V8 region, which gave estimates that were very similar (0.87- to 0.99-fold) to those of the nearly full-length fragment with the Lane mask (Table 2).
Comparing species richness estimates with short and long fragments at various taxonomic cutoffs in the Soil-Okla-A clone library.
Both parametric and nonparametric methods were used to estimate species richness for the nearly full-length data set, as well as for each of the eight short pyrosequencing-simulating datasets. As shown in Table 3, the results of all three species richness estimation methods were in general agreement with each other, as well as with the results obtained from OTU assignments, in describing the relationship between long and short fragments. Fragments encompassing the V1+V2 regions and fragments encompassing the V6 region overestimated species richness at all taxonomic cutoffs (Table 3); fragments encompassing the V3, V7, and V7+V8 regions underestimated diversities at all taxonomic cutoffs (Table 3); and fragments encompassing the V4, V5+V6, and V6+V7 regions gave numbers that were, in general, comparable to those obtained with the full sequence.
TABLE 3.
Species richness estimates obtained using long and short sequences from the Soil-Okla-A clone library at different taxonomic cutoffsa
| Regionb | Species richness estimation method | Species richness (mean ± SD) at indicated taxonomic cutoff (%)
|
||||
|---|---|---|---|---|---|---|
| 3 | 6 | 8 | 10 | 15 | ||
| V1+V2 | Chao | 1,874 ± 198 | 1,256 ± 151 | 802 ± 89 | 682 ± 86 | 397 ± 57 |
| ACE | 2,041 ± 209 | 1,169 ± 105 | 797 ± 68 | 649 ± 60 | 380 ± 38 | |
| Parametric | 4,589 ± 178 | 2,360 ± 94 | 1,616 ± 70 | 1,275 ± 59 | 737 ± 41 | |
| V3 | Chao | 1,050 ± 107 | 602 ± 72 | 418 ± 52 | 303 ± 41 | 145 ± 17 |
| ACE | 1,098 ± 94 | 562 ± 46 | 387 ± 31 | 286 ± 25 | 147 ± 13 | |
| Parametric | 2,273 ± 91 | 1,397 ± 68 | 686 ± 34 | 627 ± 37 | 190 ± 11 | |
| V4 | Chao | 1,704 ± 182 | 908 ± 110 | 611 ± 74 | 464 ± 60 | 211 ± 28 |
| ACE | 1,848 ± 171 | 913 ± 86 | 624 ± 60 | 455 ± 44 | 206 ± 19 | |
| Parametric | 3,428 ± 125 | 2,223 ± 100 | 1,034 ± 48 | 792 ± 40 | 305 ± 17 | |
| V5+V6 | Chao | 1,479 ± 161 | 784 ± 91 | 674 ± 95 | 546 ± 91 | 225 ± 30 |
| ACE | 1,576 ± 147 | 785 ± 69 | 594 ± 54 | 451 ± 44 | 219 ± 19 | |
| Parametric | 2,759 ± 125 | 1,539 ± 68 | 1,129 ± 53 | 904 ± 46 | 324 ± 17 | |
| V6 | Chao | 1,503 ± 158 | 1,031 ± 109 | 806 ± 85 | 736 ± 89 | 481 ± 60 |
| ACE | 1,728 ± 164 | 1,062 ± 94 | 827 ± 70 | 716 ± 64 | 454 ± 39 | |
| Parametric | 3,179 ± 121 | 2,302 ± 95 | 1,488 ± 62 | 1,407 ± 63 | 845 ± 41 | |
| V6+V7 | Chao | 1,246 ± 139 | 669 ± 69 | 494 ± 57 | 390 ± 52 | 187 ± 25 |
| ACE | 1,264 ± 115 | 704 ± 60 | 479 ± 39 | 370 ± 33 | 193 ± 20 | |
| Parametric | 2,688 ± 108 | 1,684 ± 79 | 1,023 ± 50 | 865 ± 48 | 292 ± 19 | |
| V7 | Chao | 660 ± 82 | 329 ± 56 | 209 ± 32 | 149 ± 25 | 76 ± 20 |
| ACE | 665 ± 63 | 298 ± 31 | 200 ± 20 | 148 ± 18 | 75 ± 12 | |
| Parametric | 1,209 ± 57 | 547 ± 32 | 343 ± 22 | 207 ± 13 | 91 ± 7 | |
| V7+V8 | Chao | 796 ± 85 | 373 ± 44 | 266 ± 33 | 206 ± 32 | 94 ± 22 |
| ACE | 807 ± 68 | 381 ± 36 | 276 ± 28 | 206 ± 25 | 91 ± 13 | |
| Parametric | 1,959 ± 87 | 706 ± 38 | 383 ± 20 | 288 ± 17 | 123 ± 10 | |
| NFL | Chao | 1,631 ± 161 | 805 ± 95 | 510 ± 57 | 429 ± 63 | 197 ± 34 |
| ACE | 1,833 ± 179 | 752 ± 64 | 517 ± 45 | 411 ± 42 | 192 ± 23 | |
| Parametric | 2,819 ± 98 | 1,790 ± 80 | 1,036 ± 51 | 912 ± 52 | 347 ± 24 | |
| LM | Chao | 868 ± 99 | 455 ± 60 | 310 ± 46 | 230 ± 35 | 125 ± 35 |
| ACE | 840 ± 70 | 448 ± 43 | 299 ± 31 | 229 ± 25 | 114 ± 18 | |
| Parametric | 2,183 ± 96 | 953 ± 52 | 463 ± 25 | 382 ± 24 | 187 ± 16 | |
Species richness estimates were obtained using the nearly full-length sequences with and without the Lane mask filter and each of the eight studied short, pyrosequencing-simulating regions from the Soil-Okla-A clone library at five different taxonomic cutoffs.
NFL, nearly full-length region (8F-1492R); LM, nearly full-length region with the Lane mask applied.
Estimates of species richness (average Chao, ACE, and parametric estimates) obtained using short simulated fragments ranged from 0.31 (V7-encompassing fragment at 10% cutoff) to 2.41 (V6-encompassing fragment at 15% cutoff) times the values obtained using the entire nearly complete longer 16S rRNA amplicon. Estimates obtained using two different pyrosequencing fragments differed by as much as 6.89-fold (between the V6-encompassing fragment and the V7-encompassing fragment at the 15% taxonomic cutoff). With very few exceptions, the standard deviations of the ratios Chaox,y/Chaoz,y, ACEx,y/ACEz,y, and Parametric species richness estimatex,y/Parametric species richness estimatez,y, where x,y denotes the short fragment x at cutoff y and z,y denotes the full-length fragment at cutoff y, were within 15% (see Table S2 in the supplemental material). Therefore, while the choice of the best species richness estimation method is critical when the “true” species richness is being sought, it seems to have minimal effect when the goal is to compare estimates. In addition, the results indicate that the use of the numbers of OTUs and slopes of scatter plots is sufficient and comparable to species richness estimates as a means to elucidate diversity discrepancies between long and short fragments.
Comparing OTU estimates and slopes of scatter plots of short and long fragments in libraries derived from other ecosystems.
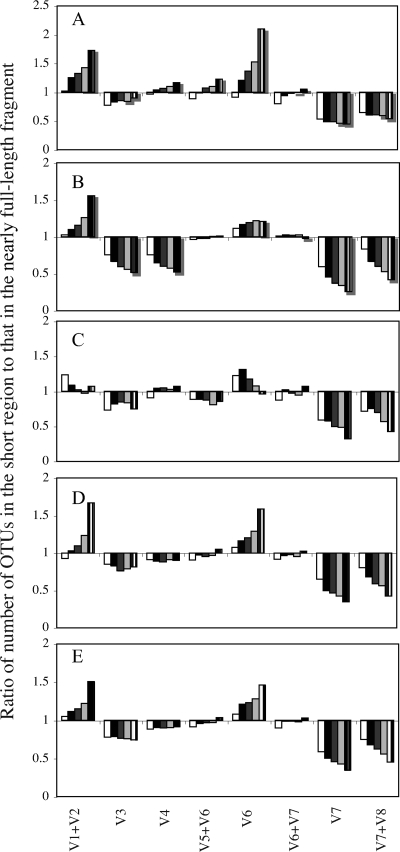
We sought to determine whether the trends observed in the Soil-Okla-A clone library are unique to the Kessler Farm soil bacterial community; whether they represent a general trend characteristic of various soil ecosystems; or whether they hold true in multiple datasets from global, diverse ecosystems. To this end, we repeated the calculations outlined above using three previously reported datasets from three distinct ecosystems with extremely different phylum-level community composition (see Table S4 in the supplemental material): (i) a soil with trembling aspen, paper birch, and sugar maple trees (1,152 clones) (33), (ii) digestive tract contents of conventionally raised adult zebrafish (612 clones) (library JFR0503 in reference 43), and (iii) endolithic and epilithic microbial communities inhabiting the ocean crust in the ocean floor in the East Pacific Rise (902 clones) (45). The trends obtained from OTU determinations and scatter plot slopes (Table 4) are strikingly similar to those observed with the Soil-Okla-A clone library (Table 2). The OTU ratios ranged between 0.26 (V7-encompassing fragment at the 15% cutoff) and 1.56 (V1+V2-encompassing fragment at the 15% cutoff) times the values obtained using the entire amplicon in aspen soil clone libraries, 0.32 to 1.31 times the values for the entire amplicon in zebrafish gut clone libraries, and 0.35 to 1.67 times the values for the entire amplicon in ocean floor communities, values that are comparable to those obtained with the Soil-Okla-A data set. The results for regions that overestimated diversity (V1+V2 and V6), underestimated diversity (V3, V7, and V7+V8), or provided comparable estimates (V4, V5+V6, and V6+V7) were the same in the four environments (except for the V4 region in the aspen soil and the V5+V6 region in the zebrafish gut, where they underestimated diversity) (Table 4, Fig. 2). Further, the slope values obtained from pairwise scatter plots were also strikingly similar in all four environments (Table 4). Finally, comparison of species richness estimate values for these three environments (see Table S3 in the supplemental material) mirrored the trends obtained using OTU numbers.
TABLE 4.
Numbers of OTUs and slopes of pairwise distance values obtained using short and long sequences at different taxonomic cutoffs for three different clone librariesa
| Environment | Regionb | No. of OTUs at indicated taxonomic cutoff (%)
|
Slopec | ||||
|---|---|---|---|---|---|---|---|
| 3 | 6 | 8 | 10 | 15 | |||
| Trembling aspen soil | V1+V2 | 702 | 616 | 568 | 527 | 436 | 1.23 |
| V3 | 516 | 374 | 292 | 235 | 144 | 0.87 | |
| V4 | 516 | 364 | 295 | 241 | 146 | 0.97 | |
| V5+V6 | 658 | 550 | 480 | 420 | 283 | 1.05 | |
| V6 | 761 | 656 | 585 | 511 | 339 | 1.8 | |
| V6+V7 | 690 | 575 | 501 | 429 | 274 | 1.07 | |
| V7 | 406 | 255 | 183 | 143 | 73 | 0.68 | |
| V7+V8 | 569 | 374 | 294 | 221 | 118 | 0.73 | |
| NFL | 682 | 561 | 490 | 418 | 280 | NA | |
| Zebrafish gut | V1+V2 | 78 | 49 | 41 | 36 | 30 | 1.27 |
| V3 | 46 | 37 | 34 | 31 | 21 | 0.72 | |
| V4 | 57 | 47 | 42 | 38 | 30 | 0.94 | |
| V5+V6 | 56 | 40 | 35 | 30 | 24 | 1.02 | |
| V6 | 77 | 59 | 47 | 40 | 27 | 1.35 | |
| V6+V7 | 55 | 46 | 39 | 35 | 30 | 0.96 | |
| V7 | 37 | 26 | 20 | 18 | 9 | 0.56 | |
| V7+V8 | 45 | 34 | 28 | 21 | 12 | 0.65 | |
| NFL | 63 | 45 | 40 | 37 | 28 | NA | |
| Basalt oceanic floor | V1+V2 | 450 | 389 | 355 | 326 | 259 | 1.24 |
| V3 | 414 | 313 | 247 | 208 | 126 | 0.86 | |
| V4 | 443 | 337 | 287 | 240 | 140 | 0.96 | |
| V5+V6 | 440 | 368 | 309 | 256 | 163 | 1.02 | |
| V6 | 521 | 440 | 390 | 341 | 246 | 1.74 | |
| V6+V7 | 445 | 365 | 318 | 251 | 159 | 0.94 | |
| V7 | 316 | 189 | 152 | 112 | 54 | 0.56 | |
| V7+V8 | 390 | 258 | 191 | 148 | 66 | 0.66 | |
| NFL | 486 | 378 | 325 | 264 | 155 | NA | |
Numbers of OTUs and slopes obtained using the nearly full-length sequences and each of the eight studied short, pyrosequencing-simulating regions at five different taxonomic cutoffs for three different clone libraries derived from soil, zebrafish gut, and ocean floor.
NFL, nearly full-length sequences (8F-1492R).
Slopes of regression lines constructed between pairwise distances in the short, pyrosequencing-simulating regions versus the nearly full-length regions and forced through the origin. NA, not applicable.
FIG. 2.
(A to D) Ratios of OTU numbers for the short pyrosequencing-simulating regions studied and OTU numbers for the nearly full-length fragment at the 3% (white), 6% (black), 8% (dark gray), 10% (light gray), and 15% (gray gridded) cutoffs in the Soil-Okla-A clone library (A), a clone library from trembling aspen soil (33) (B), a clone library from zebrafish gut (43) (C), and a composite clone library from the endolithic and epilithic communities inhabiting the ocean crust in the ocean floor in the East Pacific Ridge (45) (D). (E) Averages of the OTU ratios for the four clone libraries at the same taxonomic cutoffs.
Effect of taxonomic cutoff on bias associated with the use of shorter fragments in species richness determination.
We examined whether a specific short fragment would provide comparable performance (i.e., comparable levels of over- or underestimation of OTUs and species richness) at different putative phylogenetic cutoffs or whether such performance will be affected by the cutoff utilized. To this end, we calculated the OTU ratio for the four environments at each of the cutoffs of 3, 6, 8, 10, and 15% (Fig. 2). The results indicate that for specific fragments (V1+V2 and V6), these ratios change significantly, with a general trend of having higher ratios at higher taxonomic cutoffs. However, this is not the case for other fragments, where this ratio did not change significantly at all cutoffs utilized (V3 and V4).
Elucidation of factors behind discrepancies between pairwise distances for short and nearly full-length sequences.
While the results presented quantify the accuracy of specific regions for use in pyrosequencing studies at various taxonomic cutoffs, they do not explain the underlying reasons for the patterns observed. Since length has been shown to be an important factor in accurate determinations and utility, we sought to determine whether amplicon length affects the pattern of the species richness estimates (overestimation, underestimation, or comparable estimates). A weak correlation (r = 0.42) was observed between amplicon length and the slopes of pairwise distance graphs, taken as a measure of the species richness pattern (data not shown).
We hypothesized that since the 16S rRNA molecule has sites with various levels of evolutionary conservation (3), the proportion of these sites in a specific amplicon would impact the pairwise distance values obtained in the data set. A fragment with a higher proportion of variable regions than the full-length sequence would have higher pairwise distance values, and these differences will be reflected in a higher number of OTUs at any given taxonomic cutoff than will be found for other fragments with lower proportions of variable regions and vice versa.
To this end, we used the classification put forward by the reviews of Baker et al. (3) and Van de Peer et al. (52). In these reviews, all base pairs in the 16S rRNA gene of E. coli have been classified into highly conserved and conserved (C) base pairs, variable (V) base pairs, and highly variable and more than 75% variable (HV) base pairs. Using this classification, we determined the percentages of C, V, and HV base pairs in each of the pyrosequencing fragments and compared them to the percentages in the nearly full-length fragment. An initial inspection of these values among all different fragments under consideration showed a strong negative correlation (r = −0.81) between the slope values and the proportions of conserved bases (C/total). On the other hand, a strong positive correlation was observed between the slope values and the ratio HV/total (r = 0.83). The ratio V/total, on the other hand, correlated very weakly to the slope values (r = 0.17). The Pearson correlation coefficient between slope values and all possible base ratios ranged between −0.7 and −0.74 on the negative side and 0.64 to 0.83 on the positive side. Since none of the ratios or percentages had a correlation coefficient higher than 0.83, we sought to use multiple regression to better explain variability in slope values. We tested all possible combinations and chose the model that gave predicted slope values closest to the actual values (r = 0.93). The best model equation obtained was m (slope) = 30.5(C/total) + 11.5(HV/V) − 27.9(HV/total) − 8.5(C/V) + 5.25(HV/C) − 0.001(length) − 4.79.
DISCUSSION
This study provides an objective evaluation of the validity of species richness estimates produced using pyrosequencing-based approaches and their correlations to estimates produced using longer, nearly complete 16S rRNA gene fragments. With the utility and potential of pyrosequencing, it is anticipated that the number of pyrosequencing-based 16S rRNA environmental surveys will greatly increase within the next few years. Therefore, such an evaluation, together with the results of other studies that have investigated the utility of various fragments for phylogenetic binning (35, 36, 54), are extremely timely for comparison of the results obtained to those of Sanger sequencing-based 16S surveys, as well as for comparing pyrosequencing-produced datasets that utilized different regions within the 16S rRNA gene.
Four main conclusions arise from this study. (i) Various candidate pyrosequencing regions provide divergent estimates of OTUs and species richness that could be overestimates (e.g., the V1+V2-encompassing fragment, as well as the V6-encompassing fragment), underestimates (the V3-, V7-, and V7+V8-encompassing fragments), or comparable estimates (the V4-, V5+V6-, and V6+V7-encompassing fragments) relative to the results for nearly full-length fragments. (ii) The observed patterns of discrepancy observed for the Soil-Okla-A library hold true for the various environments tested. (iii) The level of bias in the estimates (i.e., the percentage of divergence from the value for the nearly full-length sequence) could differ within specific fragments (mainly the V1+V2- and the V6-encompassing regions) depending on the taxonomic cutoff used. (iv) The bias in species richness estimates could readily be explained and predicted by the proportion of hypervariable, variable, and conserved bases in the pyrosequencing fragment being utilized.
An examination of several 16S rRNA gene-based pyrosequencing surveys currently available indicates that several regions of the 16S molecule have already been utilized in such surveys. In their surveys of various marine ecosystems (meso- and bathypelagic layers of the North Atlantic Ocean and Pacific hydrothermal vents), Sogin et al. (48) and Huber et al. (25) utilized a 100-bp amplicon encompassing the V6 region. We show here that this region overestimates species richness compared to nearly full-length 16S rRNA gene fragments, especially at higher taxonomic cutoffs (Tables 3 and 4 and Fig. 2E). This level of overestimation would even be higher compared to the nearly full-length datasets that used the Lane mask prior to distance matrix calculation. Hamady et al. (20) used a fragment encompassing the V1+V2 region by using primer pair 8F-338R in their amplifications of the 16S rRNA gene from 286 different samples of human lung, river water, hypersaline microbial mats, air, and water from a hot spring. We show that, similar to the V6-encompassing fragment used in the previously mentioned marine studies, the use of the V1+V2-encompassing fragment would also result in the overestimation of species richness compared to nearly full-length 16S rRNA gene fragments, especially at higher taxonomic cutoffs (Tables 3 and 4 and Fig. 2E). On the other hand, the study by Andersson et al. used a V5+V6-encompassing fragment to study the bacterial diversity within various regions of the human gut using pyrosequencing (1). Such a fragment appears to provide estimates comparable to those produced using nearly full-length fragments for species richness estimation (Tables 3 and 4 and Fig. 2E).
As previously outlined, the choice of the most suitable regions within the 16S rRNA gene molecule should be tailored to provide not only accurate estimates of species richness but also accurate taxonomic assignments compared to the results for the nearly full-length 16S rRNA gene molecule. Using available collections of type strains and environmental survey 16S rRNA gene sequences, Wang et al. (54) determined the rate of accuracy of phylogenetic classification of 100-bp fragments along the entire length of the 16S rRNA gene molecule, moving 25 bp at a time. The results of this study suggested that regions around the V2 and V4 hypervariable region provide maximum accuracy for phylogenetic determination. In another study, using three large 16S rRNA gene datasets, Liu et al. clipped 100-, 250-, and 400-bp fragments upstream or downstream of conserved 16S rRNA bacterial primers and determined the recovery and coverage percentages obtained with these shorter fragments using five different methods for assigning taxonomy (35). The study recommends using fragments of 250 bp or more around the V1+V2, V3, and V4 regions. In comparison to these studies, our evaluation of species richness suggests that the V4, V5+V6, and V6+V7 regions provide estimates that are closest to the estimates obtained using longer fragments. Collectively, the V4-encompassing region (generated using primers 530F and 805R) appears to provide the best choice for both phylogenetic assignments and richness estimates.
The use of two regions in pyrosequencing surveys, one for phylogenetic identification and one for richness estimates for maximum phylogenetic and estimation accuracy, might be a viable but computationally and economically costly alternative. However, we believe that detecting discrepancies in species richness estimates should not automatically exclude a specific fragment from being considered for pyrosequencing, especially if such a fragment appears to provide accurate and reliable phylogenetic assignment. Therefore, quantifying the level of bias associated with each fragment, as well as understanding the factors contributing to the observed bias, is absolutely necessary for an objective evaluation of the species richness estimates obtained with such fragments. We show herein that the level of bias for a specific fragment could be readily predicted by a multiple regression model that incorporates the number of HV, V, and C regions within the fragment, as defined by the equation 30.5(C/total) + 11.5(HV/V) − 27.9(HV/total) − 8.5(C/V) + 5.25(HV/C) − 0.001(length) − 4.79. Such a predictor should prove useful when examining a new combination of primers targeting the domain Bacteria (5, 18, and 27) or using group-specific primers to target certain lineages.
The findings presented in this study are based on a data set of 1,132 sequences. As such, the size of the data set is much smaller than a typical pyrosequencing-generated data set. However, we believe that our experimental design is superior to the alternative of conducting a pyrosequencing run using the same KFS soil DNA and comparing the results to those obtained using Sanger sequencing for multiple reasons. First, for comparative purposes, you need two datasets (one long fragment and one short fragment). Therefore, the study must generate a similar number of Sanger-sequenced sequences for comparison to pyrosequencing-generated sequences, which is currently a daunting, nearly impossible task. Second, a real pyrosequencing run will allow us to compare only one shorter fragment to the nearly full-length fragment, while our current design allows us to compare all possible different candidate pyrosequencing fragments to the nearly full-length 16S rRNA molecule. Finally, conducting a pyrosequencing run could potentially introduce an unquantifiable level of primer bias, something the current experimental design avoids.
In addition, the results of regression analysis clearly suggest that the reasons behind such bias are intrinsic to the structure of the 16S rRNA molecule, i.e., the fact that some target regions for pyrosequencing have a higher or lower proportion of hypervariable and variable bases than the nearly full-length molecule. This fundamental property is present in each 16S rRNA gene molecule and, hence, is not likely to be dependent on the size of the data set. Therefore, we believe that the size of our data set will not affect the validity of the conclusions reached in this study.
Based on this study, we recommend the use of specific fragments (V4, V5+V6, and V6+V7) for pyrosequencing studies concerned with species richness determination in microbial communities. If other factors necessitate the use of other fragments (e.g., improving phylogenetic resolution or unsuitability of these regions for targeting a specific bacterial lineage), then the use of a regression study or the implementation of a pilot Sanger-sequencing prestudy is recommended for prior calibration of the results obtained.
Supplementary Material
Acknowledgments
This work has been supported by the National Science Foundation microbial observatories program (grant EF0801858) and financing from the Oklahoma State University start-up fund to M.S.E.
Footnotes
Published ahead of print on 26 June 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Andersson, A. F., M. Lindberg, H. Jakobsson, F. Backhed, P. Myren, and L. Engstrand. 2008. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3:e2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Axelrood, P. E., M. L. Chow, C. C. Radomski, J. M. McDermott, and J. Davies. 2002. Molecular characterization of bacterial diversity from British Columbia forest soils subjected to disturbance. Can. J. Microbiol. 48:655-674. [DOI] [PubMed] [Google Scholar]
- 3.Baker, G. C., J. J. Smith, and D. A. Cowan. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55:541-555. [DOI] [PubMed] [Google Scholar]
- 4.Boon, N., W. Windt, W. Verstraete, and E. M. Top. 2002. Evaluation of nested PCR-DGGE (denaturing gradient gel electrophoresis) with group-specific 16S rRNA primers for the analysis of bacterial communities from different wastewater treatment plants. FEMS Microbiol. Ecol. 39:101-112. [DOI] [PubMed] [Google Scholar]
- 5.Brodie, E. L., T. Z. DeSantis, D. C. Joyner, S. M. Baek, J. T. Larsen, G. L. Andersen, T. C. Hazen, P. M. Richardson, D. J. Herman, T. K. Tokunaga, J. M. Wan, and M. K. Firestone. 2006. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl. Environ. Microbiol. 72:6288-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 7.Clamp, M., J. Cuff, S. M. Searle, and G. J. Barton. 2004. The Jalview Java alignment editor. Bioinformatics 20:426-427. [DOI] [PubMed] [Google Scholar]
- 8.Cole, J. R., R. J. Chai, R. J. Farris, Q. Wang, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, A. M. Bandela, E. Cardenas, G. M. Garrity, and J. M. Tiedje. 2007. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35:D169-D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de la Torre, J. R., B. M. Goebel, E. I. Friedmann, and N. R. Pace. 2003. Microbial diversity of cryptoendolithic communities from the McMurdo dry valleys, Antarctica. Appl. Environ. Microbiol. 69:3858-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeSantis, T. Z., Jr., P. Hugenholtz, K. Keller, E. L. Brodie, N. Larsen, Y. M. Piceno, R. Phan, and G. L. Andersen. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:W394-W399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Anderson. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Souza, M. P., A. Amini, M. A. Dojka, I. J. Pickering, S. C. Dawson, N. R. Pace, and N. Terry. 2001. Identification and characterization of bacteria in a selenium-contaminated hypersaline evaporation pond. Appl. Environ. Microbiol. 67:3785-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunbar, J., S. M. Barns, L. O. Ticknor, and C. R. Kuske. 2002. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 68:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eder, W., L. L. Jahnke, M. Schmidt, and R. Huber. 2001. Microbial diversity of the brine-seawater interface of the Kebrit Deep, Red Sea, studied via 16S rRNA gene sequences and cultivation methods. Appl. Environ. Microbiol. 67:3077-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elshahed, M. S., F. Z. Najar, M. Aycock, C. Qu, B. A. Roe, and L. R. Krumholz. 2005. Metagenomic analysis of the microbial community at Zodletone Spring (Oklahoma): insights into the genome of a member of the novel candidate division OD1. Appl. Environ. Microbiol. 71:7598-7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elshahed, M. S., J. M. Senko, F. Z. Najar, S. M. Kenton, B. A. Roe, T. A. Dewers, J. R. Spear, and L. R. Krumholz. 2003. Bacterial diversity and sulfur cycling in a mesophilic sulfide-rich spring. Appl. Environ. Microbiol. 69:5609-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elshahed, M. S., N. H. Youssef, A. M. Spain, C. Sheik, F. Z. Najar, L. O. Sukharnikov, B. A. Roe, J. P. Davis, P. D. Schloss, and L. R. Krumholz. 2008. Novelty and uniqueness patterns of rare members of the soil biosphere. Appl. Environ. Microbiol. 74:5422-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fierer, N., M. A. Bradford, and R. B. Jackson. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354-1364. [DOI] [PubMed] [Google Scholar]
- 20.Hamady, M., J. J. Walker, J. K. Harris, N. J. Gold, and R. Knight. 2008. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods 5:235-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heuer, H., K. Hartung, G. Wieland, I. Kramer, and K. Smalla. 1999. Polynucleotide probes that target a hypervariable region of 16S rRNA genes to identify bacterial isolates corresponding to bands of community fingerprints. Appl. Environ. Microbiol. 65:1045-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill, T. C. J., K. A. Walsh, J. A. Harris, and B. F. Moffett. 2003. Using ecological diversity measure with bacterial communities. FEMS Microbiol. Ecol. 43:1-11. [DOI] [PubMed] [Google Scholar]
- 23.Hong, S.-H., J. Bunge, S.-O. Jeon, and S. S. Epstein. 2006. Predicting microbial species richness. Proc. Natl. Acad. Sci. USA 103:117-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huber, J. A., D. A. Butterfield, and J. A. Baross. 2003. Bacterial diversity in a subseafloor habitat following a deep-sea volcanic eruption. FEMS Microbiol. Ecol. 43:393-409. [DOI] [PubMed] [Google Scholar]
- 25.Huber, J. A., D. B. M. Welch, H. G. Morrison, S. M. Huse, P. R. Neal, D. A. Butterfield, and M. L. Sogin. 2007. Microbial population structures in the deep marine biosphere. Science 318:97-100. [DOI] [PubMed] [Google Scholar]
- 26.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isenbarger, T. A., M. Finney, C. Rios-Velazquez, J. Handelsman, and G. Ruvkun. 2008. Miniprimer PCR, a new lens for viewing the microbial world. Appl. Environ. Microbiol. 74:840-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janssen, P. H. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeon, S.-O., J. Bunge, T. Stoeck, K. J.-A. Barger, S.-H. Hong, and S. S. Epstein. 2006. Synthetic statistical approach reveals a high degree of richness of microbial eukaryotes in an anoxic water column. Appl. Environ. Microbiol. 72:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jonasson, J., M. Olofsson, and H.-J. Monstein. 2002. Classification, identification and subtyping of bacteria based on pyrosequencing and signature matching of 16S rDNA fragments. Acta Pathol. Microbiol. Immunol. Scand. 110:263-272. [DOI] [PubMed] [Google Scholar]
- 31.Kim, D., Y.-S. Kim, S.-K. Kim, S. W. Kim, G. J. Zylstra, Y. M. Kim, and E. Kim. 2002. Monocyclic aromatic hydrocarbon degradation by Rhodococcus sp. strain DK17. Appl. Environ. Microbiol. 68:3270-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-174. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 33.Lesaulnier, C., D. Papamichail, S. McCorkle, B. Olivier, S. Sliena, S. Taghavi, D. Zak, and D. van der Lelie. 2008. Elevated atmospheric CO2 affects soil microbial diversity associated with trembling aspen. Environ. Microbiol. 10:926-941. [DOI] [PubMed] [Google Scholar]
- 34.Lin, C., and D. A. Stahl. 1995. Taxon-specific probes for the cellulolytic genus Fibrobacter reveal abundant and novel equine-associated populations. Appl. Environ. Microbiol. 61:1348-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, Z., T. Z. DeSantis, G. L. Anderson, and R. Knight. 2008. Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res. 36:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, Z., C. Lozupone, M. Hamady, F. D. Bushman, and R. Knight. 2007. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 35:120-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Y. kumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Makemson, J. C., N. R. Fulayfil, W. Landry, L. M. Van Ert, C. F. Wimpee, E. A. Widder, and J. F. Case. 1997. Shewanella woodyi sp. nov., an exclusively respiratory luminous bacterium isolated from the Alboran Sea. Int. J. Syst. Bacteriol. 47:1034-1039. [DOI] [PubMed] [Google Scholar]
- 39.Marchesi, J. R., T. Sato, A. J. Weightman, T. A. Martin, J. C. Fry, S. J. Hiom, and W. G. Wade. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64:795-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Margulies, M., M. Egholm, W. E. Altman, S. Attiya, J. S. Bader, L. A. Bemben, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neefs, J.-M., Y. V. D. Peer, L. Hendriks, and R. D. Wachter. 1990. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 18:2237-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Hara, R. B. 2005. Species richness estimators: how many species can dance on the head of a pin? J. Animal Ecol. 74:375-386. [Google Scholar]
- 43.Rawls, J. F., M. Mahowald, R. E. Ley, and J. I. Gordon. 2006. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127:423-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roesch, L. F. W., R. R. Fulthorps, A. Riva, G. Casella, A. K. M. Hadwin, A. D. Kent, S. M. Daroub, F. A. O. Camargo, W. G. Farmerie, and E. W. Triplett. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1:283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santelli, C. M., B. N. Orcutt, E. Banning, W. Bach, C. L. Moyer, M. L. Sogin, H. Staudigel, and K. J. Edwards. 2008. Abundance and diversity of microbial life in ocean crust. Nature 453:653-656. [DOI] [PubMed] [Google Scholar]
- 46.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schloss, P. D., and J. Handelsman. 2006. Toward a census of bacteria in soil. PLoS Comput. Biol. 2:786-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sogin, M. L., H. G. Morrison, J. A. Huber, D. M. Welch, S. M. Huse, P. R. Neal, J. A. Arrieta, and G. H. Herndl. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. USA 103:12115-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoeck, T., J. Kasper, J. Bunge, C. Leslin, V. Ilyin, and S. Epstein. 2007. Protistan diversity in the arctic: a case of paleoclimate shaping modern biodiversity? PLoS One 8:e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX interface: flexible strategies for multiple sequence alignment aided by quality analysis tool. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Urbach, E., K. L. Vergin, L. Young, and A. Morse. 2001. Unusual bacterioplankton community structure in ultra-oligotrophic Crater lake. Limnol. Oceanogr. 46:557-572. [Google Scholar]
- 52.Van de Peer, Y., S. Chapelle, and R. De Wachter. 1996. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 24:3381-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vickerman, M. M., K. A. Brossard, D. B. Funk, A. M. Jesionowski, and S. R. Gill. 2007. Phylogenetic analysis of bacterial and archaeal species in symptomatic and asymptomatic endodontic infections. J. Med. Microbiol. 56:110-118. [DOI] [PubMed] [Google Scholar]
- 54.Wang, Q., G. M. Garrity, J. M. Tiedje, and J. R. Cole. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Youssef, N. H., and M. S. Elshahed. 2008. Species richness in soil bacterial communities: a proposed approach to overcome sample size bias. J. Microbiol. Methods 75:86-91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.