Abstract
Copper appears to be influencing the distribution and abundance of phytoplankton in marine environments, and cyanobacteria are thought to be the most sensitive of the phytoplankton groups to copper toxicity. By using growth assays of phylogenetically divergent clades, we found that coastal strains of marine Synechococcus species were more tolerant to copper shock than open-ocean strains. The global transcriptional response to two levels of copper shock were determined for both a coastal strain and an open-ocean strain of marine Synechococcus species using whole-genome expression microarrays. Both strains showed an osmoregulatory-like response, perhaps as a result of increasing membrane permeability. This could have implications for marine carbon cycling if copper shock leads to dissolved organic carbon leakage in Synechococcus species. The two strains additionally showed a common reduction in levels of photosynthesis-related gene transcripts. Contrastingly, the open-ocean strain showed a general stress response, whereas the coastal strain exhibited a more specifically oxidative or heavy-metal acclimation response that may be conferring tolerance. In addition, the coastal strain activated more regulatory elements and transporters, many of which are not conserved in other marine Synechococcus strains and may have been acquired by horizontal gene transfer. Thus, tolerance to copper shock in some marine Synechococcus strains may in part be a result of a generally increased ability to sense and respond in a more stress-specific manner.
Marine Synechococcus and Prochlorococcus species are together thought to be responsible for at least 20% of global carbon fixation (25). They play such a significant role in the carbon cycle that it is essential to understand to which environmental stresses they are susceptible and how they respond. Cyanobacteria have been shown to be particularly sensitive to copper, for example, compared to other marine phytoplanktons (9). As a group, cyanobacteria have various levels of copper tolerance, with Prochlorococcus species thought to be more sensitive to copper than Synechococcus species (27). Within Synechococcus species, strain tolerance has been shown to vary (9), but those experiments with equilibrated copper levels were done prior to a better understanding of Synechococcus phylogenetic diversity.
Applying a copper “shock” seems just as environmentally relevant as using equilibrated copper levels, since there are several mechanisms by which we could see sharp, rapid spikes of Cu2+ concentrations in the surface waters where marine Synechococcus strains are found. Although the Cu2+ concentration is generally low (10−14 M) in the surface waters of the open ocean, it can increase by 3 or 4 orders of magnitude below the euphotic zone (12), and a mixing event could thus cause a sharp increase in Cu2+ concentrations. Furthermore, in coastal environments, a sharp spike in free-copper levels can occur after a heavy rain due mostly to anthropogenic inputs (6). Finally, there is evidence that dust deposition can introduce copper into surface waters (35). Research suggests that cyanobacterial distribution is affected by free-copper levels both coastally and in the open ocean (27, 30), and culture studies have shown that in response to copper stress, marine Synechococcus species can produce strong extracellular L1-type ligands that bind Cu2+, perhaps as a defense mechanism (31). Although the biosynthetic pathways of these ligands are unknown, Cu-L1 masses have been determined (47). Considering the dynamic nature of marine free-copper levels and the known sensitivity of marine Synechococcus species to copper, it seems likely that Synechococcus species are experiencing copper shock and have adaptive responses to this shock. The transcriptional response to a quick increase of Cu2+ in already growing cultures has not been systematically tested in marine Synechococcus strains and seems a logical next step.
Since copper is not only a toxicant but a micronutrient as well, a study of responses to various levels of copper must take into account an organism's need to carefully balance the uptake and efflux of copper in order to maintain homeostasis between deprivation on one side and toxic effects on the other. In the freshwater cyanobacterium Synechocystis species, copper uptake and transport have been well defined and involve two copper-transporting P1-type ATPases and a copper metallochaperone, Atx1. Copper is imported via one ATPase at the plasma membrane, where is it then transferred to Atx1, which shuttles it to the thylakoid membrane and transfers it to the other ATPase (8). However, no homolog for Atx1 has been found in marine Synechococcus species genomes, although there is a putative homolog for CtaA, the plasma membrane copper-importing ATPase, that has not been functionally validated (33).
The toxicity of excess copper is thought to arise through several different mechanisms, although there is still debate as to which is dominant. First, cycling between Cu2+ and Cu+ triggers the production of hydroxyl radicals, which can cause damage to enzymes and membranes (37). Additionally, an excess of copper may cause it to competitively bind to other high-affinity metal transporters and inhibit or interfere with the uptake of other essential metals (42). Finally, excess copper can interfere with photosynthesis by inhibiting electron transport, especially in photosystem II (PSII) (3). The response to these toxic effects can be twofold, including both a primary copper efflux response as well as secondary response to combat the effects of toxicity. Global transcriptional responses to various copper levels have been studied in detail for other microorganisms such as diatoms, Escherichia coli, and Pseudomonas aeruginosa, to name a few (14, 24, 43). Bacterial copper efflux and its regulation are probably best described for E. coli and include a P-type ATPase, CopA; a multicopper oxidase, CueO; and a proton-driven copper efflux pump, CusCFBA, as well as a two-component signal transduction system and a MerR-like transcriptional activator that is induced by copper (38). There are additionally some previously identified copper tolerance proteins, CutABCDE, whose functions are less well understood (38). The transcriptional response to excess copper in E. coli results in the upregulation of some of these genes involved in copper export, such as CopA, as well as secondary responses that include the envelope and superoxide stress responses and iron homeostasis (24). However, although there is a putative homolog for CutA in marine Synechococcus species, none of the copper sensor proteins used by E. coli and other bacteria have been found in marine Synechococcus species genomes, and the five histidine kinases in Synechococcus sp. strain WH8102 (hereafter strain WH8102) seem to be involved mainly in energy, phosphate, or osmoregulatory sensing (34).
The recent sequencing of clade I coastal marine Synechococcus sp. strain CC9311 (hereafter strain CC9311) revealed a metal-intensive ecological strategy, as indicated by the many more genes involved in metal homeostasis as well as metal enzymes and sensor kinases that were not present in oligotrophic strains such as WH8102 (clade III) (34). Note that these two strains are quite different overall: of the 2,951 total genes in CC9311, 1,730 are shared with WH8102, which has 2,589 genes in total. There are several genes of particular relevance to the potential copper tolerance present in CC9311 and not in WH8102, including a cation-dependent facilitator (CDF) efflux transporter and five bacterial ferritin genes. The purpose of this study is to first clearly determine differences in tolerance to copper shock between several coastal and open-ocean strains of marine Synechococcus species, as indicated by growth responses, and then compare and contrast global transcriptional responses to copper shock between a sensitive strain and a more tolerant strain.
MATERIALS AND METHODS
Cell culture and growth assays.
Batch cultures of Synechococcus sp. strains WH8102, CC9311, CC9605, and CC9902, representing phylogenetic clades III, I, II, and IV, respectively (1), were grown in chelexed SOW medium, as previously described by Dupont et al. (16), with the exceptions that copper was not added to the medium in the trace-metal mix and that nickel was added at a final concentration of 50 nM. All cultures were grown axenically except CC9902. When in the early exponential phase, as measured by fluorescence-based growth curves and as validated later with flow cytometry, 20 ml was aliquoted into glass culture tubes. Cultures were then given approximately 24 h to adjust to the transfer, and Cu-EDTA was then added in triplicate at 1 μM, 100 nM, 10 nM, and 1 nM, as were controls with sterile milliQ water added. Using a MINEQL calculation (46), this corresponds approximately to equilibrium free-copper levels equivalent to pCu 9.1, pCu 10.1, pCu 11.1, and pCu 12.1 (pCu = −log10[Cu2+]). At fixed intervals for the following 48 h, fluorescence was measured with a Turner Designs 10-AU apparatus, and 1-ml samples were fixed with 1% glutaraldehyde and frozen at −80°C for later analysis using a FACsort flow cytometer.
Flow cytometry.
Samples were run as described previously by Collier and Palenik (13), with the exceptions that these samples were 1,000-fold diluted and run for 2 min and that a compensation matrix was not applied for the analysis. Briefly, samples were run on a Becton-Dickson FACsort flow cytometer, and all samples from a given growth assay were run on the same day. Each sample was run with a known concentration of 0.97-μm standard beads, and several bead-only controls were also run. The statistical significance of the difference between treatments in cell number was determined using a one-way analysis of variance and Dunnett's posttests for the final time points.
Microarray RNA extraction.
Duplicate 3-liter or 4-liter batch cultures of WH8102 and CC9311 were grown up to the early exponential phase, and half the culture was spun down at room temperature at 10,400 × g, resuspended in Trizol, and frozen at −80°C for eventual RNA extraction, taking care that the harvesting time stayed under 30 min. Cu-EDTA was added to the other half for Cu2+ levels of pCu 10.1 (high) and pCu 11.1 (moderate) for WH8102 and pCu 9.1 (high) and pCu 10.1 (moderate) for CC9311. After a 2-h incubation, these cells were harvested as outlined above. Total RNA was extracted using a Trizol-based method, followed by Qiagen RNeasy kit purification, both according to the manufacturer's specifications except that an extra 65°C incubation for 60 min was added at the onset to lyse cells.
DNA microarray design and construction.
A full-genome microarray for WH8102 consisting of a mixed population of PCR amplicons (2,142 genes) and 70-mer oligonucleotides (389 genes) was constructed. Unique PCR amplicons representing each gene were approximately 800 bp in size or smaller if the gene size was smaller. Unique 70-mer oligonucleotides were utilized for genes under 200 bp in size and for the two genes that we were not able to successfully PCR amplify. PCR products and oligonucleotides were suspended in 50% dimethyl sulfoxide (DMSO) and deposited onto aminosilane-coated UltraGAPS glass slides (Corning) using an Intelligent Automation Systems array spotter. Each slide included six replicates of each PCR amplicon or oligonucleotide, negative controls such as 50% DMSO-50% deionized water, and positive controls including a total mix of WH8102 PCR amplicons, Arabidopsis PCR amplicons, and 70-mer oligonucleotides as described previously by Hegde et al. (19). After printing, slides were air dried for 30 min, and DNA was irreversibly cross-linked to the surface by using a Stratalinker apparatus (Stratagene) to deliver 250 mJ/cm2 of short-wavelength UV energy and stored in desiccators at room temperature. CC9311 arrays were constructed from unique 70-mer oligonucleotides for each of 2,892 genes in a similar manner. Each gene was represented six times on an array. Negative controls were 50% DMSO-50% deionized water, and positive controls included Arabidopsis PCR amplicons and 70-mer oligonucleotides.
DNA microarray transcriptional profiling.
An indirect labeling method was used to label cDNA as previously described (36), where cDNA is synthesized in the presence of a nucleoside triphosphate analog containing a reactive aminoallyl group to which a fluorescent dye molecule, either cyanine 3 (Cy3) or cyanine 5 (Cy5), is coupled. Approximately 4 μg of total RNA was used for indirect labeling, leading to the production of approximately 4 μg of cDNA with approximately 200 pmol of dye molecules incorporated per microgram of cDNA synthesized. Prior to hybridization, labeled cDNA was scanned spectrophotometrically to ensure optimal dye incorporation per sample for adequate signal intensity. Each experiment typically included a minimum of two biological replicates, a minimum of three technical replicates for each biological sample, and at least one flip-dye experiment per biological replicate.
All hybridizations were performed as previously described (36), and slides were promptly scanned at a 10-μm resolution using an Axon 4000B scanner with GenePix 4.0 software. Processing of the tagged image file format images from hybridized arrays was performed using TIGR-Spotfinder (www.tigr.org/software), and the data sets were normalized by applying the LOWESS algorithm, using block mode and a smooth parameter of 0.33, which is available in the TIGR-MIDAS package (www.tigr.org/software). Statistical analysis was performed on the mean of log2-transformed signal ratios of replicate spots using the Statistical Analysis of Microarrays algorithms (45) with a false discovery rate of less than 1% and a cutoff of a 0.4-log2-fold/−0.4-log2-fold change. Gene function was predicted using KEGG (23) and in-house predictions. Comparisons between Synechococcus genomes were performed using analyses described previously by Dufresne et al. (15) to obtain orthologs from all 11 marine Synechococcus species genomes.
ArrayExpress.
All microarray data presented here are in accordance with the Microarray Gene Expression Data Society's recommendations regarding minimum information about a microarray experiment (10).
RT-PCR and semiquantitative real-time PCR validation.
Several micrograms of RNA from each microarray experiment was saved and used for quantitative PCR (QPCR) validation for three of the four microarray sets. For the fourth microarray set, WH8102 under conditions of high copper, the experiment was repeated in triplicate, and new RNA was extracted. For all sets, cDNA was synthesized using 750 ng of RNA per sample and an Invitrogen Superscript III cDNA kit according to kit instructions. Reverse transcriptase (RT) PCR negative controls (no-RT) were prepared by preparing duplicates of each sample but omitting the addition of Superscript III to one of the samples, which were then used to verify that there was no genomic DNA contaminating the samples. Two to three technical replicates of each reaction were done concurrently, and RT-PCR products were combined upon completion.
Semiquantitative PCR was performed using a Stratagene Mx3000P apparatus with Stratagene SYBR green master mix and primers designed using Primer3 (40). Reactions were set up according to Stratagene master mix specifications. Samples were run in triplicate, and a no-RT control was run for each sample to exclude the possibility of DNA contamination. A five-point standard curve was generated using WH8102 or CC9311 genomic DNA of a known concentration (measured using a NanoDrop spectrophotometer) in a serial 10-fold dilution. This curve was then used to convert the cycle number of the QPCR output to nanograms of cDNA. Several housekeeping genes were also run for each sample, and standard curves were generated for these genes as well. Similar conversions to nanograms of cDNA were performed for the housekeeping genes, and these values were then divided into nanograms of cDNA values for the genes of interest for each sample, thus generating values for each sample relative to a housekeeping gene. The change between values for control and copper-shocked conditions was then calculated by using a log2 transformation.
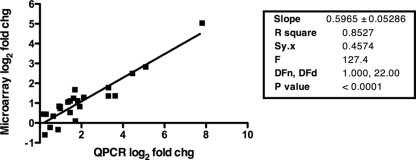
Quantitative RT-PCR results were found to significantly correlate with microarray results for the genes assayed (Fig. 1). For the majority of the genes, quantitative RT-PCR yielded higher computed changes than the microarray, excepting a couple of genes with very low, yet statistically significant, microarray changes, which is typical (44). Hence, the 0.4- and −0.4-log2-fold-change cutoff applied to the microarray data was considered to be appropriate.
FIG. 1.
QPCR correlation with microarray data. Shown are data for comparisons of log2 changes (chg) in expression obtained from the microarray results and QPCR validation. The line indicates regression analysis, and the box shows accompanying statistics, where Sy.x is the standard deviation of the residuals and DFn and DFd are the degrees of freedom accompanying the F test.
Microarray data accession number.
Descriptions of the microarray experiments, quantitation data, and array design have been deposited into the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) and have been assigned accession number GSE13910.
RESULTS
Copper shock growth assays.
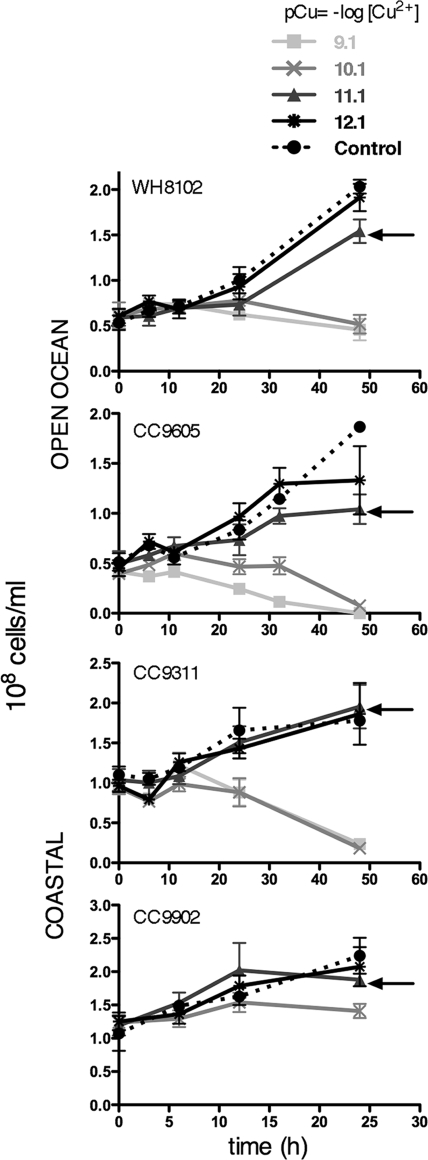
The flow cytometric cell counts revealed statistically significant decreases in the numbers of Synechococcus cells per ml relative to the growing control culture with the addition of the two largest amounts of copper (pCu 9.1 and pCu 10.1) in all four strains after 48 h, except CC9902, which had significant decreases in numbers of cells in response to pCu 10.1 after 24 h but was not tested at 48 h and was not tested at the pCu 9.1 level (Fig. 2 and Table 1). For the next-highest addition (pCu 11.1), only the two open-ocean strains, WH8102 and CC9605, showed a statistically significant decrease in cell numbers compared to the control. Finally, for the lowest addition (pCu 12.1), only CC9605 showed a statistically significant decrease in cell numbers compared to the control. Using flow cytometry, measurable decreases in fluorescence per cell were detectable as early as 12 h in both CC9311 and WH8102 at the highest copper levels, pCu 9.1 and pCu 10.1 (data not shown). This demonstrates that the physiological effects were occurring within this time frame.
FIG. 2.
Numbers of Synechococcus species cells/ml calculated from flow cytometric data over 24 or 48 h after treatment for all four strains. Error bars represent ±1 standard deviation. Arrows indicate the pCu 11 final time point, with pCu 11 being the level at which open-ocean strain cell numbers decreased significantly relative to the control and coastal strain cell numbers, which do not. For statistical results of these endpoint comparisons between treatments, see Table 1.
TABLE 1.
Synechococcus species copper shock growth assays and comparison of final time points for all copper treatments versus control
| Synechococcus sp. strain | Welch one-way ANOVA resultd | Avg no. of cells/ml (108)a
|
||||
|---|---|---|---|---|---|---|
| pCu 9 | pCu 10 | pCu 11 | pCu 12 | Control | ||
| CC9605 | F(4,b 3.20c) = 1198.5; P < 0.0001 | 0.0005* | 0.076* | 1.042* | 1.331* | 1.865 |
| WH8102 | F(4, 4.30) = 97.53; P = 0.0002 | 0.457* | 0.519* | 1.542* | 1.913 | 2.033 |
| CC9902 (24 h) | F(3, 4.01) = 13.5; P = 0.0145 | NA | 1.410* | 1.879 | 2.074 | 2.239 |
| CC9311 | F(4, 4.67) = 916.5; P < 0.0001 | 0.233* | 0.183* | 1.955 | 1.864 | 1.780 |
Asterisks indicate a statistically significant difference from the control, based on a Dunnett's post hoc test, with a P value of <0.05. NA, not applicable.
Degrees of freedom between groups.
Degrees of freedom within groups.
ANOVA, analysis of variance.
Microarray.
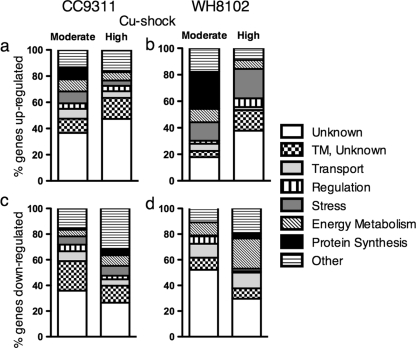
A total of four sets of global expression microarray experiments were conducted, two for coastal strain CC9311 and two for open-ocean strain WH8102. For each strain, both moderate- and high-copper shocks were applied, and sets of significantly regulated genes were determined. For CC9311, moderate shock yielded 120 upregulated and 78 downregulated genes out of a possible 2,893 total protein-coding genes, meaning that 6.8% of the protein-coding genes were differentially regulated. The high-copper shock yielded 317 upregulated and 528 downregulated genes (29.2% differentially regulated). The overlap of significant genes between these two treatments was fairly high, with 122 genes being differentially regulated under both shock conditions out of 198 possible genes (this being the total number of significant genes for the moderate-copper shock), which indicates a physiologically consistent response between the two treatments. For WH8102, the moderate-copper shock yielded 129 upregulated and 138 downregulated genes of a possible 2,528 total protein-coding genes, with 10.3% of all protein-coding genes being differentially regulated. The high-copper shock yielded 45 upregulated and 98 downregulated genes (5.5% differentially regulated). Again, the overlap of significant genes between the two treatments was high, with 54 genes of 143 possible genes being regulated with both treatments. The significant genes were grouped into general categories and subcategories based on predicted function using KEGG and in-house predictions (Fig. 3; see Tables S1 to S4 in the supplemental material).
FIG. 3.
All significantly upregulated and downregulated genes for Synechococcus sp. strains WH8102 (b and d) and CC9311 (a and c) in all four microarray experiments, grouped by functional categories assigned based on KEGG and in-house predictions. “Moderate” indicates conditions of moderate-copper shock, “High” indicates high-copper shock, and “TM, Unknown” represents genes of unknown function with transmembrane domains. Energy metabolism includes oxidative phosphorylation, photosynthesis, carbon fixation, and nitrogen and sulfur metabolism.
The overlap of orthologous genes between the two strains was modest, with 24 orthologous genes being significantly regulated under conditions of high-copper shock in both strains and 16 orthologous genes overlapping under conditions of moderate-copper shock (Table 2). However, there were also common types of responses not involving directly orthologous genes. For example, along with the putative osmoregulatory two-component response regulator that was upregulated in both strains (Table 2), there were also accompanying osmoregulatory responses in each strain. The open-ocean strain upregulated an osmolyte transporter, glycine betaine (SYNW0229), and the coastal strain upregulated a gene involved in osmolyte synthesis (glucosylglycerol-phosphate phosphatase [sync_1171]). Furthermore, both strains highly downregulated several porins under both shock conditions, although only one, sync_1542 (SYNW2224) (Table 2), was orthologous.
TABLE 2.
Between-strain overlap of orthologous genes that were significantly regulated under either high- and moderate-copper shock conditionsa
| Shock condition and CC9311 locus | Log2-fold change for CC9311 | WH8102
|
Description | |
|---|---|---|---|---|
| Locus | Log2 fold change | |||
| High copper | ||||
| Upregulated | ||||
| sync_1233 | 1.37 | SYNW0807 | 1.68 | Two-component sensor histidine kinase (osmoregulatory) |
| sync_1232 | 2.51 | SYNW0808 | 0.99 | Two-component response regulator (osmoregulatory) |
| sync_1231 | 2.41 | SYNW0809 | 0.82 | Conserved hypothetical protein (putative osmoregulatory) |
| sync_1903 | 1.20 | SYNW1510 | 0.78 | Conserved hypothetical protein |
| sync_1905 | 0.77 | SYNW1512 | 0.84 | Conserved hypothetical protein |
| sync_2523 | 1.36 | SYNW2176 | 1.78 | Possible serine protease |
| sync_1050 | 1.05 | SYNW0921 | 0.57 | Conserved hypothetical protein, metal binding domain? |
| sync_2710 | 3.11 | SYNW2330 | 0.80 | Conserved hypothetical protein |
| Downregulated | ||||
| sync_0562 | −0.54 | SYNW1960 | −0.46 | PSI subunit IV (PsaE) |
| sync_0133 | −0.48 | SYNW0144 | −0.59 | PSI iron-sulfur center subunit VII (PsaC) |
| sync_2496 | −1.16 | SYNW0334 | −0.62 | PSII PsbX protein |
| sync_2586 | −0.52 | SYNW2232 | −0.72 | PSII D2 protein |
| sync_1422 | −0.41 | SYNW1303 | −0.62 | Cytochrome b6/f complex subunit VIII |
| sync_1542 | −1.18 | SYNW2224 | −0.97 | Possible porin |
| sync_0059 | −0.84 | SYNW0058 | −0.74 | 50S ribosomal protein L35 |
| sync_2224 | −1.12 | SYNW0546 | −0.44 | 50S ribosomal protein L27 |
| sync_2274 | −0.44 | SYNW0521 | −0.42 | Putative ferredoxin-thioredoxin reductase variable chain |
| sync_2872 | −0.59 | SYNW2442 | −0.77 | Putative urea ABC transporter urea binding protein |
| sync_2789 | −0.43 | SYNW2375 | −0.52 | Putative glycine cleavage H protein |
| sync_0271 | −0.74 | SYNW0233 | −0.41 | Conserved hypothetical protein |
| sync_0950 | −0.47 | SYNW2103 | −0.86 | Conserved hypothetical protein |
| sync_1807 | −0.53 | SYNW1730 | −0.41 | Conserved hypothetical protein |
| sync_2027 | −0.44 | SYNW1778 | −0.41 | Conserved hypothetical protein |
| sync_2900 | −1.11 | SYNW2489 | −0.49 | Conserved hypothetical protein |
| Moderate copper | ||||
| Upregulated | ||||
| sync_2313 | 0.53 | SYNW0494 | 1.30 | ATP synthase subunit alpha |
| sync_2314 | 0.50 | SYNW0493 | 0.89 | ATP synthase delta chain |
| sync_1966 | 0.62 | SYNW1717 | 0.96 | Ribulose bisphosphate carboxylase small chain |
| sync_2725 | 0.68 | SYNW2341 | 1.06 | 50S ribosomal protein L10 |
| sync_2223 | 0.56 | SYNW0547 | 0.65 | 50S ribosomal protein L21 |
| sync_0433 | 0.54 | SYNW2071 | 1.08 | 30S ribosomal protein S19 |
| sync_2523 | 0.82 | SYNW2176 | 0.84 | Possible serine protease |
| sync_2862 | 0.80 | SYNW0194 | 0.64 | Similar to leukotoxin secretion protein |
| sync_1398 | 0.78 | SYNW1278 | 1.23 | Heat shock protein HtpG (stress) |
| sync_1569 | 0.66 | SYNW1073 | 0.53 | Glutamine synthetase glutamate-ammonia ligase |
| sync_1903 | 1.05 | SYNW1510 | 0.88 | Conserved hypothetical protein |
| sync_2099 | 1.67 | SYNW1886 | 0.57 | Conserved hypothetical protein |
| sync_2204 | 0.90 | SYNW0567 | 0.64 | Conserved hypothetical protein |
| sync_2881 | 0.65 | SYNW2456 | 0.42 | Conserved hypothetical protein |
| Downregulated | ||||
| sync_0497 | −0.44 | SYNW2011 | −0.84 | Bilin biosynthesis protein MpeU |
| sync_0014 | −0.95 | SYNW0014 | −0.42 | RNA binding region RNP-1 (RNA recognition motif) |
Putative operons are composed of sync_1233, sync_1232, and sync_1231; sync_1903 and sync_1905; and sync_2313 and sync_2314.
Aside from the shared osmoregulatory response, there was also a common photosynthetic response. Based on KEGG classifications, photosynthesis-related genes are grouped into several categories including PSI, PSII, the cytochrome b6f complex, photosynthetic electron transport, F-type ATPase, and antenna proteins. There are 87 of these genes in WH8102 and 81 in CC9311, and excluding the antenna proteins, which vary between the two strains, the other categories contain orthologous genes between strains except for one extra gene in the strain WH8102 in the electron transport category. Of the 37 total photosystem genes (PSI and PSII), 14 are downregulated in the coastal strain and 10 are downregulated in the open-ocean strain under conditions of high-copper shock. Neither strain upregulated any of the PSI, PSII, photosynthetic electron transport, or F-type ATPases under high-copper shock conditions. However, under conditions of moderate-copper shock, both strains upregulated F-type ATPase genes as well as genes involved in carbon fixation. The coastal strain upregulated photosynthetic antenna proteins (including phycoerythrin, phycocyanin, and allophycocyanin) and downregulated cytochrome b6/f complex genes, whereas WH8102 downregulated photosynthetic antenna proteins (for specific genes, see Tables S1 to S4 in the supplemental material).
Finally, there were also several conserved hypothetical proteins that were upregulated in both strains (Table 2), including one, SYNW0921 (sync_1050), with a signaling peptide and four conserved cysteines as some of the only conserved residues (Fig. 4), creating a possible metal binding domain. Other conserved hypothetical proteins, including SYNW1510 (sync_1903), appear to be stress related and are found under other shock conditions (data not shown).
FIG. 4.
Alignment of conserved hypothetical protein SYNW0921 in marine Synechococcus species. The signaling peptide portion of the protein is not included.
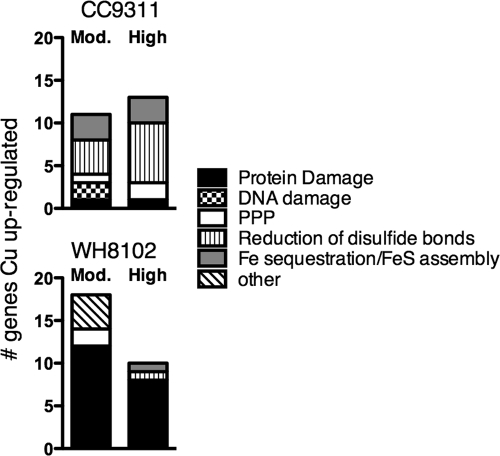
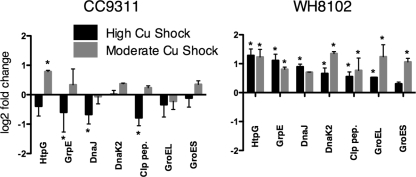
There were several clear differences in the responses between the two strains, the first being the type of stress response that was activated. Figure 5 compares the different types of stress genes activated in each strain based on KEGG classifications and in-house predictions. The activated genes of the coastal strain that have been linked to the oxidative stress response in Escherichia coli and Staphylococcus aureus (7, 21) include those coding for the upregulation of ferritin to sequester iron and keep it from redox cycling; iron-sulfur (FeS) assembly proteins to repair damaged FeS clusters, which are especially sensitive to oxidative stress; methionine sulfoxide reductase and ferredoxin thioredoxin reductase to reduce unwanted disulfide bonds created by oxidative stress; and components of the pentose phosphate pathway to generate reducing power (Fig. 5). Conversely, the open-ocean strain upregulated mostly general stress response genes, that is, protein-folding genes, chaperones, and proteases such as heat shock proteins, GroEL/GroES, Clp proteases, and DnaJ/DnaK (Fig. 6). Excepting one, HtpG, these genes were not upregulated at all in CC9311, and in fact, under conditions of high-copper shock, many of these genes were significantly downregulated (Fig. 6). However, CC9311 is known to upregulate these genes under other shock conditions such as mitomycin C shock (data not shown).
FIG. 5.
Breakdown of the general category of “stress” from Fig. 2 into subcategories based on predicted roles. Shown is a comparison of significantly upregulated genes in this general category for each of the four microarray experiments, where “Mod.” is moderate-copper shock, “High” is high-copper shock, and “PPP” stands for the pentose phosphate pathway.
FIG. 6.
Comparison of “stress” gene expressions between two strains, CC9311 (left) and WH8102 (right). Bars indicate log2 average changes in expression determined from microarray expression data, error bars represent 1 standard deviation, and asterisks indicate significant regulation with a 1% false discovery rate cutoff. Clp pep., endopeptidase Clp, ATP binding chain C.
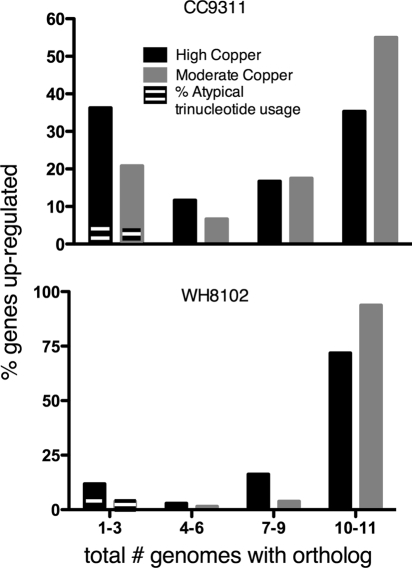
Another intriguing difference was the coastal strain's activation of many genes that are not conserved in the other 11 sequenced marine Synechococcus strains, compared to the open-ocean strain, which activated mostly conserved genes (Fig. 7). Of particular interest among the “unique” CC9311 genes were two operons activated under both shock conditions, which are listed in Table 3 along with other select “unique” CC9311 genes (for a full list, see Table S5 in the supplemental material). The first, consisting of sync_1213 to sync_1217, has an ATP-binding cassette (ABC) transporter, an efflux mediator, and a large protein, likely effluxed, with a divalent metal cation binding domain and a protein binding domain (Table 3). The entire operon is predicted to be horizontally acquired based on atypical trinucleotide usage, and the closest gene ortholog appears in another marine cyanobacterium, Cyanobium sp. strain PCC7001. The second operon, consisting of sync_1491 to sync_1494, includes a sigma-70 factor, a thioredoxin-like protein, a biogenesis protein, and a possible anti-sigma factor. The genes in this operon had the greatest fold-change values in the whole data set under conditions of both the high-copper and moderate-copper shock and have orthologs in only one other Synechococcus species strain, WH5701, but they do not have atypical trinucleotide usage (Table 3). This operon is also of interest, as it represents a difference in the regulatory responses between the two strains. CC9311 activated several sigma factors, whereas WH8102 did not activate any. Of the other “unique” CC9311 genes activated, there were several others with an atypical trinucleotide content, some of which have orthologs in marine bacteria (Table 3).
FIG. 7.
Comparison of conservations of upregulated genes in each strain. Eleven total marine Synechococcus species genomes (including the two strains from the microarray experiments) were probed for orthologs of genes in each microarray experiment. Synechococcus species genomes included were those of strains BL107, CC9902, CC9605, WH7803, WH7805, RS9916, RS9917, WH5701, RCC307, WH8102, and CC9311. Total numbers of upregulated genes for each experiment were as follows: 317 and 120 for CC9311 under conditions of high- and moderate-copper shock, respectively, and 45 and 129 for WH8102 under conditions of high- and moderate-copper shock, respectively. Percent atypical nucleotide usage was calculated based on the number of atypical genes as a percentage of the total number of upregulated genes for each treatment.
TABLE 3.
CC9311 copper-induced genes with low-level conservation in Synechococcus speciesa
| Locus | Gene | Log2-fold changeb | Closest BLASTp homolog (E value)c |
|---|---|---|---|
| sync_1217 | Structural toxin protein RtxA* | 0.93 | Cyanobium sp. strain PCC7001 (∼1e−120) |
| sync_1216 | LssD protein, putative* | 1.82 | Cyanobium sp. strain PCC7001 (∼1e−120) |
| sync_1215 | ABC transporter ATP binding/permease fusion* | 2.49 | Cyanobium sp. strain PCC7001 (∼1e−120) |
| sync_1214 | ABC transporter RzcB, putative* | 2.20 | Cyanobium sp. strain PCC7001 (∼1e−120) |
| sync_1213 | Hypothetical protein* | 1.28 | Synechococcus sp. strain 7805 (9e−21) |
| sync_1496 | Thioredoxin-like protein | 6.31 | Synechococcus sp. strain WH5701 (∼1e−60) |
| sync_1495 | Biogenesis protein | 6.07 | Synechococcus sp. strain WH5701 (∼1e−60) |
| sync_1494 | RNA polymerase sigma-70 factor family | 5.03 | Synechococcus sp. strain WH5701 (∼1e−60) |
| sync_1493 | Conserved hypothetical protein | 4.40 | Synechococcus sp. strain WH5701 (∼1e−60) |
| sync_0183 | Undecaprenyl-phosphate glucosephosphotransferase* | 1.05 | Staphylococcus aureus (4e−37) |
| sync_0182 | Posttranslational flagellin modification protein B* | 0.90 | Campylobacter coli (3e−38) |
| sync_0172 | Imidazole glycerol phosphate synthase glutamine amidotransferase subunit* | 0.84 | Hoeflea phototrophica (5e−50) |
| sync_1515 | Long-chain fatty acid coenzyme A ligase* | 0.82 | Synechococcus sp. strain WH5701 (3e−60) |
| sync_1447 | Hypothetical protein* | 0.88 | Leuconostoc mesenteroides (0.24) |
| sync_2420 | Hypothetical protein* | 0.90 | Uncultured marine microorganism (3e−11) |
| sync_1504 | Hypothetical protein* | 0.87 | Anabaena variabilis (1e−52) |
Partial list. Asterisks indicate atypical trinucleotide usage.
From high-copper-shock microarrays.
BLASTp search against the nonredundant database done in March 2009.
Other differences included the expression of transport and membrane proteins. First, there is a contrasting use of multidrug and protein efflux transporters between the two strains. We predicted the use of CDF transporters and a P-type ATPase that is homologous to a copper importer in Synechocystis species, CtaA, but none of these were upregulated in either strain, contrary to our predictions. Instead, the open-ocean strain upregulated one major facilitator superfamily multidrug efflux ABC transporter (SYNW0800) under conditions of high-copper shock and another (SYNW0193) under conditions of moderate-copper shock. The open-ocean strain also upregulated Sec system genes involved in protein export and an ortholog of CutA, an E. coli periplasmic copper tolerance protein, which were not upregulated in the coastal strain. The coastal strain upregulated several nonorthologous multidrug efflux transporters including a resistance-nodulation-cell division (RND) family transporter (sync_2767) and several major facilitator superfamily transporters (sync_0936 and sync_1126).
There were also differences in the two strains in genes involved in ion transport and membrane permeability. As mentioned above, both strains downregulated porins, but unique to the coastal strain was the upregulation of two small mechanosensitive ion channels (sync_2818 and sync_2824), neither of which has an ortholog in the genome of strain WH8102, and the downregulation of large conductance-mechanosensitive ion channels (sync_0202 and sync_2281). Also unique to the coastal strain was the upregulation of a fatty acid desaturase (sync_2793) and the downregulation of both potassium and chloride ion channels (sync_2515, sync_0823, and sync_1314).
There was also generally more downregulation of genes with transmembrane domains in CC9311. Under conditions of moderate-copper shock in CC9311, 35% of all the significantly downregulated genes had transmembrane domains, a majority of which have no known function. Conversely, only 18% of the downregulated genes under conditions of moderate-copper shock in WH8102 had transmembrane domains (Fig. 3). Even considering total percentages of genes with transmembrane domains in the genome of each strain (22.58% in CC9311 and 19.03% in WH8102), this still appears to be a disproportionate downregulation of proteins with membrane domains for the coastal strain.
Lastly, there were 12 genes with modified expression levels in the coastal strain that had orthologs only in the genomes of Synechococcus species strains with known copper resistance, as shown here, that is, CC9311 and CC9902, and that were not present in the genomes of Synechococcus sp. strain CC9605 or WH8102 (see asterisks in Tables S3 and S4 in the supplemental material). These genes included a modified small mechanosensitive ion channel and several genes of unknown function.
DISCUSSION
The two coastal strains of marine Synechococcus species from two divergent clades (clades I and IV) are able to cope with free-copper levels in the range of pCu 11 better than the two open-oceans strains (clades II and III). This range is thought to be an ecologically relevant one (12). The higher tolerance of CC9311 also confirms the predictions from the sequenced genome that this strain would be more metal tolerant (34). Coastal strains may in general be better adapted to a quick-changing environment, including stresses other than copper. Of course, a monospecific laboratory culture of Synechococcus species may respond very differently to copper shock than when it is found with the consortia of microbes present in the marine environment. In the future, it will be important to test whether the gene expression changes that we found can be replicated with environmental samples.
The transcriptional response to copper stress in Synechococcus appears to be distinct from those of other bacteria such as E. coli and P. aeruginosa, for the most part. Based on the transcriptional responses of E. coli and P. aeruginosa to copper shock, we expected to see the upregulation of copper efflux-related genes such as CDF transporters, P-type ATPases, and RND transporters as well as an oxidative stress response including an upregulation of superoxide dismutase, an envelope stress response including the downregulation of porins (in P. aeruginosa), and an iron homeostasis response (24, 43). While there are not very many clear orthologs for the copper-induced genes identified in these studies, we can look at response types for similarities. Contrary to our predictions, genes related to active copper efflux, including CDF transporters and P-type ATPases, did not appear to be upregulated, although one RND transporter in the coastal strain was induced. In the coastal strain, there was evidence that both an oxidative stress response and an iron homeostasis response were induced, with some common genes like ferritin being induced; however, superoxide dismutase genes were not upregulated. One similarity to data from previous work was the downregulation of porins and an outer membrane permeability response in P. aeruginosa, which was also seen for both strains of Synechococcus.
In fact, taken as a whole, the common transcriptional response between the strains suggests that membrane damage and changes in membrane permeability may be some of the primary toxic effects of copper that both coastal and open-ocean Synechococcus strains combat, perhaps with an osmoregulatory-like response. The first indication of this, as mentioned above, is the downregulation of porins, which is not surprising considering that an E. coli mutant lacking porins was found to be more copper resistant, and thus, copper is thought to diffuse through porins into the periplasm (26). The osmoregulatory response can be seen in the activation of a two-component response regulator that is likely involved in the osmoregulatory response considering both its sequence similarity to another known and well-studied osmosensor pair, OmpR and EnvZ in Escherichia coli (22, 18), and its upregulation under conditions of osmotic shock in WH8102 (our unpublished data). The osmoregulatory-like response can also be seen in the upregulation of osmolyte transport (glycine betaine) and synthesis as well as the regulation of many membrane-associated proteins and, in the coastal strain, the regulation of mechanosensitive ion channels, which are activated by increased membrane tension and help prevent cell lysis (20). Finally, a previously reported study of copper toxicity in Saccharomyces cerevisiae found that tolerance is dependent on the fatty acid composition of the plasma membrane (2), and a fatty acid desaturase was upregulated in the coastal strain under conditions of moderate-copper shock.
In the marine environment, a loss of membrane integrity due to copper shock in Synechococcus species could have significant implications for ecosystem function and carbon flux through the system. Phytoplankton excretion of dissolved organic carbon has been studied for many years (17) and has even been measured in strains of Prochlorococcus species, the sister group of Synechococcus species (4). The percentage of photosynthate that is excreted is highly variable, perhaps as a result of unclear environmental factors, potentially including copper toxicity. These “leaky” phytoplankton cells are thought to contribute significantly to the cycling of carbon by way of the microbial loop. These results suggest that copper could be one of the factors contributing to dissolved organic carbon excretion, and further investigation into this would be valuable.
Another common response between the two strains is that of the unknown common copper-responsive genes, of which there were several. We can only speculate as to their function, and more work regarding their copper specificity needs to be done, but perhaps most interesting was the copper-responsive gene family with four conserved cysteines, two of which are only one amino acid apart and the other two of which were surrounding the pair. Cysteine residues are often involved in binding metals, copper in particular, in plastocyanin, for example. Additionally, a recurring CxC motif is a defining feature of metallothioneins (39). This family also has a predicted signaling peptide, and we hypothesize that this protein is a secreted copper binding protein.
The final common response between the two strains, the downregulation of PSI and PSII genes under conditions of high-copper shock, reveals that copper in marine Synechococcus species may directly affect PSII or also inhibit electron transport, as it was previously shown to do in other systems (5, 32). Conversely, this response could be explained by an oxidative stress mechanism. The possible inhibition of photosynthesis and, thus, growth in response to copper shock, while not surprising, does serve to underscore the clear toxicity that these cultures are experiencing in response to levels of copper that can be found in marine systems. Under conditions of moderate-copper shock, there is a slightly different response in that both strains upregulate carbon fixation and F-type ATPase genes, possibly attempting to combat the inhibition of these processes instead of slowing growth. Perhaps more intriguing were the differences in responses regarding the iron-containing cytochrome b6/f complex genes and photosynthetic antenna proteins. The tolerant strain decreases transcription of cytochrome b6/f genes, and it is known that some cyanobacteria can switch to copper-containing plastocyanin under conditions of iron limitation (48). Although the microarray results do not show an increase in plastocyanin transcript levels, it is possible that the coastal strain is switching to the copper-containing equivalent protein. The tolerant strain also increases levels of expression of the antenna proteins, indicating that it is still trying to put resources into photosynthetic activity, while the sensitive strain is ramping down all growth-related activity and putting all its resources into repair and recovery.
From the response of the tolerant strain, it seems likely that the higher tolerance comes from an increased ability to sense and respond with more specialized regulatory pathways and transporters to excess copper or perhaps, more generally, oxidative stress. The open-ocean strain's transcriptional response, especially under conditions of high-copper shock, can best be described as a general stress response. Although there is surely a copper-specific component, most of the upregulated genes would be activated under any number of shock conditions. On the other hand, the expression patterns of the coastal strain can be best defined by its unique, and perhaps copper-specific, response. The genes with the highest changes in levels of expression are transporters, uncharacterized membrane proteins, and sigma factors, many of which are not conserved in marine Synechococcus species and could be copper specific. Even the genes that could be characterized as stress response genes, such as the iron sequestration and disulfide bond reduction genes, constitute a more specialized response to oxidative or heavy-metal stress than the protein chaperones and proteases that the open-ocean strain upregulates. Copper shock appears to trigger additional transcriptional regulators, perhaps using as-yet-undefined copper sensor proteins. The copper specificity of these genes, especially the ambiguous transporters such as the operon composed of sync_1213 to sync_1217, which could be effluxing a copper binding protein, needs to be tested further in the future.
It is especially intriguing that many of the specialized genes that the coastal strain is upregulating are not generally conserved in marine Synechococcus species, and some have atypical trinucleotide usage, suggesting that they may have been horizontally transferred. Several of these genes also have putative homologs in marine bacteria, and one operon in particular, sync_1214 to sync_1217, has a high level of homology with an operon present in another cyanobacterium, Cyanobium sp. strain PCC7001, which was isolated from a coastal marine location. Interestingly, in Cyanobium species, the genes in the operon all have a typical trinucleotide content excepting the gene at the top of the operon, the ABC transporter RzcB. If the operon was horizontally acquired, this furthermore implies that these recent gene acquisitions have already been integrated into the transcriptional regulatory network of the Synechococcus strain. Horizontal gene transfer has been shown to be important for heavy-metal tolerance in the Bacteria, especially in sediment systems, and copper resistance plasmids, in Ralstonia metallidurans, for example, were described previously (28, 29). Apart from plasmid-borne resistance, there is an example of a set of efflux genes found in an environmental Xanthomonas strain that confers copper resistance and is predicted to be horizontally transferred (41). However, in oceans, the acquisition of heavy-metal tolerance through horizontal gene transfer has not been shown. If these recently acquired genes do contribute to the copper tolerance of this strain of Synechococcus species, it would be yet another indication that Synechococcus species may be able to adapt to a microenvironment by acquiring and using novel genes when necessary.
The copper concentration in the marine environment is increasingly understood to affect phytoplankton distributions because of both its toxicity and its necessity to life. This study is a step toward a better understanding of how different phylogenetic and ecological groups of Synechococcus species respond to copper toxicity, which will eventually help us to better predict and explain distribution patterns of Synechococcus species as well as to better understand the adaptive mechanisms that Synechococcus species can employ in the wake of environmental disturbances.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation (grants NSF:MCB08-17775 and NSF:RR166-585/3505738).
Footnotes
Published ahead of print on 5 June 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ahlgren, N. A., and G. Rocap. 2006. Culture isolation and culture-independent clone libraries reveal new marine Synechococcus ecotypes with distinctive light and N physiologies. Appl. Environ. Microbiol. 72:7193-7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avery, S. V., N. G. Howlett, and S. Radice. 1996. Copper toxicity towards Saccharomyces cerevisiae: dependence on plasma membrane fatty acid composition. Appl. Environ. Microbiol. 62:3960-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron, M., J. B. Arellano, and J. L. Gorge. 1995. Copper and photosystem-II—a controversial relationship. Physiol. Plant. 94:174-180. [Google Scholar]
- 4.Bertilsson, S., O. Berglund, M. J. Pullin, and S. W. Chisholm. 2005. Release of dissolved organic matter by Prochlorococcus. Vie Milieu Life Environ. 55:225-231. [Google Scholar]
- 5.Bhargava, P., Y. Mishra, A. K. Srivastava, O. P. Narayan, and L. C. Rai. 2008. Excess copper induces anoxygenic photosynthesis in Anabaena doliolum: a homology based proteomic assessment of its survival strategy. Photosynth. Res. 96:61-74. [DOI] [PubMed] [Google Scholar]
- 6.Blake, A. C., D. B. Chadwick, A. Zirino, and I. Rivera-Duarte. 2004. Spatial and temporal variations in copper speciation in San Diego Bay. Estuaries 27:437-447. [Google Scholar]
- 7.Bore, E., S. Langsrud, O. Langsrud, T. M. Rode, and A. Holck. 2007. Acid-shock responses in Staphylococcus aureus investigated by global gene expression analysis. Microbiology 153:2289-2303. [DOI] [PubMed] [Google Scholar]
- 8.Borrelly, G. P. M., C. A. Blindauer, R. Schmid, C. S. Butler, C. E. Cooper, I. Harvey, P. J. Sadler, and N. J. Robinson. 2004. A novel copper site in a cyanobacterial metallochaperone. Biochem. J. 378:293-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brand, L. E., W. G. Sunda, and R. R. L. Guillard. 1986. Reduction of marine-phytoplankton reproduction rates by copper and cadmium. J. Exp. Mar. Biol. Ecol. 96:225-250. [Google Scholar]
- 10.Brazma, A., P. Hingamp, J. Quackenbush, G. Sherlock, P. Spellman, C. Stoeckert, J. Aach, W. Ansorge, C. A. Ball, H. C. Causton, T. Gaasterland, P. Glenisson, F. C. P. Holstege, I. F. Kim, V. Markowitz, J. C. Matese, H. Parkinson, A. Robinson, U. Sarkans, S. Schulze-Kremer, J. Stewart, R. Taylor, J. Vilo, and M. Vingron. 2001. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat. Genet. 29:365-371. [DOI] [PubMed] [Google Scholar]
- 11.Reference deleted.
- 12.Coale, K. H., and K. W. Bruland. 1990. Spatial and temporal variability in copper complexation in the North Pacific. Deep Sea Res. A 37:317-336. [Google Scholar]
- 13.Collier, J. L., and B. Palenik. 2003. Phycoerythrin-containing picoplankton in the Southern California Bight. Deep Sea Res. II 50:2405-2422. [Google Scholar]
- 14.Davis, A. K., M. Hildebrand, and B. Palenik. 2006. Gene expression induced by copper stress in the diatom Thalassiosira pseudonana. Eukaryot. Cell 5:1157-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dufresne, A., M. Ostrowski, D. J. Scanlan, L. Garczarek, S. Mazard, B. P. Palenik, I. T. Paulsen, N. T. de Marsac, P. Wincker, C. Dossat, S. Ferriera, J. Johnson, A. F. Post, W. R. Hess, and F. Partensky. 2008. Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 9:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupont, C. L., K. Barbeau, and B. Palenik. 2008. Ni uptake and limitation in marine Synechococcus strains. Appl. Environ. Microbiol. 74:23-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fogg, G. E. 1983. The ecological significance of extracellular products of phytoplankton photosynthesis. Bot. Mar. 26:3-14. [Google Scholar]
- 18.Forst, S., D. Comeau, S. Norioka, and M. Inouye. 1987. Localization and membrane topology of EnvZ, a protein involved in osmoregulation of OmpF and PmpC in Escherichia coli. J. Biol. Chem. 262:16433-16438. [PubMed] [Google Scholar]
- 19.Hegde, P., R. Qi, K. Abernathy, C. Gay, S. Dharap, R. Gaspard, J. E. Hughes, E. Snesrud, N. Lee, and J. Quackenbush. 2000. A concise guide to cDNA microarray analysis. BioTechniques 29:548-550, 552-554, 556. [DOI] [PubMed] [Google Scholar]
- 20.Hurst, A. C., E. Petrov, A. Kloda, T. Nguyen, L. Hool, and B. Martinac. 2008. MscS, the bacterial mechanosensitive channel of small conductance. Int. J. Biochem. Cell Biol. 40:581-585. [DOI] [PubMed] [Google Scholar]
- 21.Imlay, J. A. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77:755-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jo, Y. L., F. Nara, S. Ichihara, T. Mizuno, and S. Mizushima. 1986. Purification and characterization of the OmpR protein, a positive regulator involved in osmoregulatory expression of the ompF and ompC genes in Escherichia coli. J. Biol. Chem. 261:5252-5256. [PubMed] [Google Scholar]
- 23.Kanehisa, M., and S. Goto. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28:27-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kershaw, C. J., N. L. Brown, C. Constantinidou, M. D. Patel, and J. L. Hobman. 2005. The expression profile of Escherichia coli K-12 in response to minimal, optimal and excess copper concentrations. Microbiology 151:1187-1198. [DOI] [PubMed] [Google Scholar]
- 25.Li, W. K. W. 1994. Primary production of prochlorophytes, cyanobacteria, and eukaryotic ultraphytoplankton—measurements from flow cytometric sorting. Limnol. Oceanogr. 39:169-175. [Google Scholar]
- 26.Lutkenhaus, J. F. 1977. Role of a major outer membrane protein in Escherichia coli. J. Bacteriol. 131:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mann, E. L., N. Ahlgren, J. W. Moffett, and S. W. Chisholm. 2002. Copper toxicity and cyanobacteria ecology in the Sargasso Sea. Limnol. Oceanogr. 47:976-988. [Google Scholar]
- 28.Martinez, R. J., Y. L. Wang, M. A. Raimondo, J. M. Coombs, T. Barkay, and P. A. Sobecky. 2006. Horizontal gene transfer of P-IB-type ATPases among bacteria isolated from radionuclide- and metal-contaminated subsurface soils. Appl. Environ. Microbiol. 72:3111-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mergeay, M., S. Monchy, T. Vallaeys, V. Auquier, A. Benotmane, P. Bertin, S. Taghavi, J. Dunn, D. van der Lelie, and R. Wattiez. 2003. Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: towards a catalogue of metal-responsive genes. FEMS Microbiol. Rev. 27:385-410. [DOI] [PubMed] [Google Scholar]
- 30.Moffett, J. W., L. E. Brand, P. L. Croot, and K. A. Barbeau. 1997. Cu speciation and cyanobacterial distribution in harbors subject to anthropogenic Cu inputs. Limnol. Oceanogr. 42:789-799. [Google Scholar]
- 31.Moffett, J. W., and L. E. Brand. 1996. Production of strong, extracellular Cu chelators by marine cyanobacteria in response to Cu stress. Limnol. Oceanogr. 41:388-395. [Google Scholar]
- 32.Mohanty, N., I. Vass, and S. Demeter. 1989. Copper toxicity affects photosystem-II electron-transport at the secondary quinone acceptor, QB. Plant Physiol. 90:175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palenik, B., B. Brahamsha, F. W. Larimer, M. Land, L. Hauser, P. Chain, J. Lamerdin, W. Regala, E. E. Allen, J. McCarren, I. Paulsen, A. Dufresne, F. Partensky, E. A. Webb, and J. Waterbury. 2003. The genome of a motile marine Synechococcus. Nature 424:1037-1042. [DOI] [PubMed] [Google Scholar]
- 34.Palenik, B., Q. H. Ren, C. L. Dupont, G. S. Myers, J. F. Heidelberg, J. H. Badger, R. Madupu, W. C. Nelson, L. M. Brinkac, R. J. Dodson, A. S. Durkin, S. C. Daugherty, S. A. Sullivan, H. Khouri, Y. Mohamoud, R. Halpin, and I. T. Paulsen. 2006. Genome sequence of Synechococcus CC9311: insights into adaptation to a coastal environment. Proc. Natl. Acad. Sci. USA 103:13555-13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paytan, A., K. R. M. Mackey, Y. Chen, I. D. Lima, S. C. Doney, N. Mahowald, R. Labiosa, and A. F. Post. 2009. Toxicity of atmospheric aerosols on marine phytoplankton. Proc. Natl. Acad. Sci. USA 106:4601-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peterson, S. N., C. K. Sung, R. Cline, B. V. Desai, E. C. Snesrud, P. Luo, J. Walling, H. Y. Li, M. Mintz, G. Tsegaye, P. C. Burr, Y. Do, S. Ahn, J. Gilbert, R. D. Fleischmann, and D. A. Morrison. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51:1051-1070. [DOI] [PubMed] [Google Scholar]
- 37.Pinto, E., T. C. S. Sigaud-Kutner, M. A. S. Leitao, O. K. Okamoto, D. Morse, and P. Colepicolo. 2003. Heavy metal-induced oxidative stress in algae. J. Phycol. 39:1008-1018. [Google Scholar]
- 38.Rensing, C., and G. Grass. 2003. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 27:197-213. [DOI] [PubMed] [Google Scholar]
- 39.Romero-Isart, N., and M. Vasak. 2002. Advances in the structure and chemistry of metallothioneins. J. Inorg. Biochem. 88:388-396. [DOI] [PubMed] [Google Scholar]
- 40.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 41.Ryan, R. P., D. J. Ryan, Y. C. Sun, F. M. Li, Y. P. Wang, and D. N. Dowling. 2007. An acquired efflux system is responsible for copper resistance in Xanthomonas strain IG-8 isolated from China. FEMS Microbiol. Lett. 268:40-46. [DOI] [PubMed] [Google Scholar]
- 42.Sunda, W. G., and S. A. Huntsman. 1998. Interactions among Cu2+, Zn2+, and Mn2+ in controlling cellular Mn, Zn, and growth rate in the coastal alga Chlamydomonas. Limnol. Oceanogr. 43:1055-1064. [Google Scholar]
- 43.Teitzel, G. M., A. Geddie, S. K. De Long, M. J. Kirisits, M. Whiteley, and M. R. Parsek. 2006. Survival and growth in the presence of elevated copper: transcriptional profiling of copper-stressed Pseudomonas aeruginosa. J. Bacteriol. 188:7242-7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tetu, S. G., B. Brahamsha, D. A. Johnson, V. Tai, K. Phillippy, B. Palenik, and I. T. Paulsen. 2 April 2009. Microarray analysis of phosphate regulation in the marine cyanobacterium Synechococcus sp. WH8102. ISME J. [Epub ahead of print.] doi: 10.1038/ismej.2009.31. [DOI] [PubMed]
- 45.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Westall, J. C., J. L. Zachary, and F. M. M. Morel. 1976. MINEQL—general algorithm for computation of chemical-equilibrium in aqueous systems. Abstr. Pap. Am. Chem. Soc. 172:8. [Google Scholar]
- 47.Wiramanaden, C. I. E., J. T. Cullen, A. R. S. Ross, and K. J. Orians. 2008. Cyanobacterial copper-binding ligands isolated from artificial seawater cultures. Mar. Chem. 110:28-41. [Google Scholar]
- 48.Zhang, L., B. McSpadden, H. B. Pakrasi, and J. Whitmarsh. 1992. Copper-mediated regulation of cytochrome c553 and plastocyanin in the cyanobacterium Synechocystis 6803. J. Biol. Chem. 267:19054-19059. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.