Brucella abortus biovar 3 is an uncommon cause of human brucellosis in Spain, where Brucella melitensis has accounted for 97.5% of all cases. B. abortus biovar 3 was detected in specimens from four patients in Cáceres (west-central Spain) with a febrile syndrome associated with arthralgia and myalgia and from one patient in Lugo (northwestern Spain) with orchiepididymitis (March 2004 and April 2005). For tracing, these isolates were studied by biotyping, analysis of 16S rRNA genes, rpoB, gyrA, parC, and housekeeping genes, multilocus variable-number tandem-repeat analysis (MLVA-16), and analysis of hypervariable octameric oligonucleotide fingerprints (HOOF-prints).
Isolates were identified as B. abortus biovar 3 (3) by their CO2 requirements, H2S production, agglutination in anti-A-positive and anti-M-negative monospecific sera, and growth on thionin and basic fuchsin (20 μg/ml). They showed a single 16S rRNA gene and rpoB sequences (6, 9). The latter gene harbored substitutions (243-GAC, 268-ACT, and 340-GAA) with respect to the B. melitensis 16 M sequence under GenBank accession no. AE009516 (9). The replacement at codon 268 is thought to be a B. abortus marker because of its presence in reference biovars. The rpoB sequence showed a 100% match with the sequence of B. abortus biovar 7 strain 63/75 (GenBank accession no. DQ086138) (9). The authenticity of B. abortus biovar 7, however, has been questioned for many years (2, 5) because the reference strain was a mixture of B. abortus biovars 3 and 5. Further work showed the obtained rpoB gene differs at position 340 from the sequence of B. abortus biovar 7 reference strain Tulya (GenBank accession no. AY562180) (9). The Tulya strain and other African B. abortus biovar 3 isolates show distinct genetic patterns versus European isolates of the same biovar when analyzed with different genetic markers. The division of B. abortus biovar 3 into two groups has been proposed: 3a, containing the Tulya and African field isolates; and 3b, containing the European isolates (including those we analyzed) (10). Therefore, analyses of our five B. abortus biovar 3 isolates differed from those for the Tulya strain (9).
The gyrA and parC amplifications and sequencing were performed with the primers Bru_gyrA + 4651 (5′-TGCAGCGGTCTTATCTTGATT-3′), Bru_gyrA-5589 (5′-CAAACGAGGTCTGCAAAGG-3′), Bru_parC + 2040 (5′-CAAGCTGACCGAGCTTGAA-3′), and Bru_parC-2871 (5′-CACGAAGGCCGTCAGTATATC-3′) to provide polymorphisms related to the bacterial identification (7). All showed one gyrA (5285-T → C) mutation and one parC [2600-Ala(GCT) → Val(GTT)] mutation compared to B. melitensis 16 M. The gyrA change appears in B. abortus biovars and in Brucella suis, while the parC mutation is seen only in B. abortus biovars. The isolates showed the same sequence type, ST5 (gap-2 aroA-1 glk-1 dnaK-2 gyrB-1 trpE-4 cobQ-1 omp25-1 int-hyp-1) (11), identical to the corresponding sequence of the B. abortus biovar vaccine isolates S19 and RB51. The Spanish isolates differed at two (glk and trpE) and seven (gap, aroA, glk, dnaK, gyrB, trpE, and cobQ) of the nine housekeeping genes from those of the B. abortus biovar 3 5/93 strain and the Tulya strain, respectively (11).
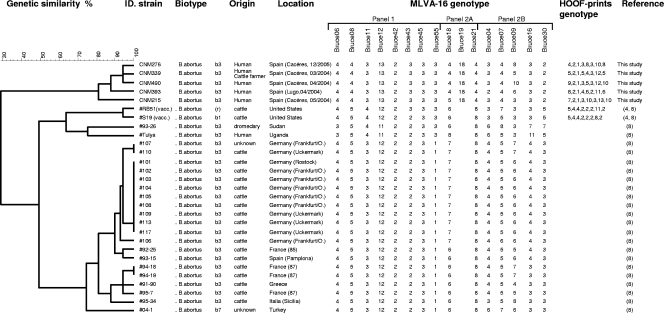
Five MLVA-16 (1) (genetic similarity range, 80% to 95%) and five HOOF-print (4) (35% to 65%) genotypes were obtained. In a comparison of those genotypes to other MLVA-15 genotypes (8), the Spanish cluster split from other European and African B. abortus biovar 3 isolates (similarity index, <40%), suggesting a different clonal lineage (Fig. 1).
FIG. 1.
Dendrogram of clustered MLVA-16 genotypes of the Spanish B. abortus biovar 3 human isolates (see the Brucella 2007 MLVA database [http://mlva.u-psud.fr]), the European B. abortus biovar 3 (3b) isolates, the African B. abortus biovar 3 (3a) Tulya strain, and the B. abortus biovar 1 RB51 and S19 vaccine isolates (8), generated using the categorical coefficient and UPGMA analysis (unweighted-pair group method using arithmetic averages). MLVA-16 information for Bruce09 was not available for these previously studied strains (8).
In summary, we described the first reported cases of B. abortus biovar 3 causing human brucellosis in Spain. MLVA and HOOF-print genotyping revealed a close genetic relationship between the detected emergent isolates; this is further supported by their identical 16S rRNA gene, rpoB, gyrA, parC, and housekeeping gene sequences.
Nucleotide sequence accession numbers.
GenBank has assigned the following accession numbers: for 16S rRNA, EF192470 to EF192474; for gyrA, EF420872; for parC, EF420873; and for rpoB, EU623594.
Acknowledgments
This work was supported by a grant from the Instituto de Salud Carlos III to A.N. and by project MPY 1116/07.
We thank S. Allix and M. Thiébaud (Afssa) for assistance in biotyping and Adrian Burton for checking the English version of the manuscript.
Footnotes
Published ahead of print on 24 June 2009.
REFERENCES
- 1.Al Dahouk, S., P. Le Flèche, K. Nöckler, I. Jacques, M. Grayon, H. C. Scholz, H. Tomaso, G. Vernaud, and H. Neubauer. 2007. Evaluation of Brucella MLVA typing for human brucellosis. J. Microbiol. Methods 69137-145. [DOI] [PubMed] [Google Scholar]
- 2.Allix, S., G. Le Carrou, M. Thiébaud, D. Albert, L. L. Perrett, C. E. Dawson, P. Groussaud, E. J. Stubberfield, M. Koylass, A. M. Whatmore, and B. Garin-Bastuji. Brucella abortus biovar 7: phenotypic and molecular evidence, p. 146. In Proc. Brucellosis 2008 Int. Res. Conf. (including 61st Brucellosis Res. Conf.), London, United Kingdom, 10 to 13 September 2008.
- 3.Alton, G. G., L. M. Jones, R. D. Angus, and J. M. Verger. 1988. Techniques for the brucellosis laboratory. Institut National de la Recherche Agronomique, Paris, France.
- 4.Bricker, B. J., D. R. Ewalt, and S. M. Halling. 2003. Brucella ‘HOOF-prints’: strain typing by multi-locus analysis of variable number tandem repeats (VNTRs). BMC Microbiol. 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gargani, G., and A. López-Merino. 2006. International Committee on Systematics of Prokaryotes—Subcommittee on the Taxonomy of Brucella--correspondence report (interim report) 1991-1993. Int. J. Syst. Evol. Microbiol. 561167-1168. [DOI] [PubMed] [Google Scholar]
- 6.Gee, J. E., B. K. De, P. N. Levett, A. M. Whitney, R. T Novak, and T. Popovic. 2004. Use of 16S rRNA gene sequencing for rapid confirmatory identification of Brucella isolates. J. Clin. Microbiol. 423649-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang, W. M. 1996. Bacterial genetic diversity based on type II DNA topoisomerase genes. Annu. Rev. Genet. 3079-107. [DOI] [PubMed] [Google Scholar]
- 8.Le Flèche, P., I. Jacques, M. Grayon, S. Al Dahouk, P. Bouchon, F. Denoeud, K. Nöckler, H. Neubauer, L. A. Guilloteau, and G. Vergnaud. 2006. Evaluation and selection of tandem repeat assay for a Brucella MLVA typing assay. BMC Microbiol. 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marianelli, C., F. Ciuchini, M. Tarantino, P. Pasquali, and R. Adone. 2006. Molecular characterization of the rpoB gene in Brucella species: new potential molecular markers for genotyping. Microb. Infect. 8860-865. [DOI] [PubMed] [Google Scholar]
- 10.Ocampo-Sosa, A. A., J. Aguëro-Balbín, and J. M. García-Lobo. 2005. Development of a new PCR assay to identify Brucella abortus biovars 5, 6 and 9 and the new subgroup 3b of biovar 3. Vet. Microbiol. 11041-51. [DOI] [PubMed] [Google Scholar]
- 11.Whatmore, A. M., L. L. Perret, and A. P. MacMillan. 2007. Characterisation of the genetic diversity of Brucella by multilocus sequencing. BMC Microbiol. 734. [DOI] [PMC free article] [PubMed] [Google Scholar]