Abstract
During estrogen-induced proliferation, c-Myc and cyclin D1 initiate independent pathways that activate cyclin E1-Cdk2 by sequestration and/or downregulation of the CDK inhibitor p21Waf1/Cip1, without significant increases in cyclin E1 protein levels. In contrast, cyclin E2 undergoes a marked increase in expression, which occurs within 9 to 12 h of estrogen treatment of antiestrogen-pretreated MCF-7 breast cancer cells. Both E cyclins are important to estrogen action, as small interfering RNA (siRNA)-mediated knockdown of either cyclin E1 or cyclin E2 attenuated estrogen-mediated proliferation. Inducible expression of cyclin D1 upregulated cyclin E2, while siRNA-mediated knockdown of cyclin D1 attenuated estrogen effects on cyclin E2. However, manipulation of c-Myc levels did not profoundly affect cyclin E2. Cyclin E2 induction by estrogen was accompanied by recruitment of E2F1 to the cyclin E1 and E2 promoters, and cyclin D1 induction was sufficient for E2F1 recruitment. siRNA-mediated knockdown of the chromatin remodelling factor CHD8 prevented cyclin E2 upregulation. Together, these data indicate that cyclin E2-Cdk2 activation by estrogen occurs via E2F- and CHD8-mediated transcription of cyclin E2 downstream of cyclin D1. This contrasts with the predominant regulation of cyclin E1-Cdk2 activity via CDK inhibitor association downstream of both c-Myc and cyclin D1 and indicates that cyclins E1 and E2 are not always coordinately regulated.
In mammalian cells, there are two E-type cyclins, cyclins E1 and E2 (collectively referred to as cyclin E), that activate Cdk2 in late G1 phase. The E-type cyclins are encoded by separate genes, located at chromosomes 19q12 (CCNE1) and 8q22.1 (CCNE2) in humans, but share substantial sequence identity and functional redundancy. The predominant function of cyclin E is believed to be the activation of Cdk2 and consequent effects on cell cycle progression and DNA replication, since both E-type cyclins accelerate the G1-to-S-phase transition and most cyclin E-Cdk2 substrates identified to date have roles in cell cycle progression and DNA replication (34). Cyclin E also has crucial, and possibly CDK-independent, functions in the initiation of DNA replication and centrosome duplication (25, 26, 42), and its deregulation may promote oncogenesis via genomic instability (63) arising through centrosome amplification (35, 49, 63) or alterations in replication complex assembly (19, 45).
Overexpression of cyclin E1 in the mouse mammary gland leads to tumor formation at a low frequency (∼12%) and long latency (8 to 13 months) (4), although tumorigenesis may be augmented by cooperation with other oncogenic events, particularly p53 inactivation (61). High expression of cyclin E1 protein in breast cancers is strongly correlated with proliferative markers such as elevated Ki67 levels and an elevated mitotic index (6), suggesting that cyclin E1 promotes tumor cell proliferation. This provides good evidence for an association between cyclin E1 and breast oncogenesis, but cyclin E2 has not been independently examined in models of tumorigenesis. In contrast with the apparent redundancy between the two E-type cyclins in many experimental models, clinical data suggest that cyclins E1 and E2 may have distinct roles in breast cancer, with cyclin E2 being particularly important in estrogen receptor (ER)-positive cancers. Two quantitative reverse transcription-PCR studies have identified cyclin E1 and cyclin E2 mRNA levels as significant, independent prognostic indicators in multivariate models (15, 60), and in one study they remained significant predictors of metastasis-free and overall survival when they were included in the same multivariate model, indicating that they make distinct contributions to patient outcome (60). In both studies, the prognostic value of cyclin E2 was particularly affected by either ER status or endocrine therapy: Sieuwerts et al. (60) found that cyclin E2 mRNA was associated with poor outcome only in ER-positive cancers but that cyclin E1 mRNA was prognostic in both ER-positive and ER-negative cancers, while Desmedt et al. (15) showed that cyclin E2 mRNA was significantly associated with outcome in systemically untreated patients but not in tamoxifen-treated patients. Thus, there is strong evidence that a high level of cyclin E2 is a predictor of poor outcome in breast cancer, especially in the context of estrogen action. Moreover, cyclin E2 is the only common gene in three major gene expression signatures that predict reduced survival in breast cancer (62, 69, 73), further emphasizing the need to investigate cyclin E2 independently of cyclin E1.
The proliferative effects of estrogen are thought to be responsible for its role as a causative agent in breast cancer (31), and the ability of estrogen to promote S-phase entry depends on Cdk2 activation (55, 70). In order to define the targets of estrogen-induced mitogenesis, we have used an in vitro model in which MCF-7 breast cancer cells are pretreated with the pure antiestrogen ICI 182780 and hence arrested in a quiescent state (12), from which they can be induced to reenter the cell cycle by treatment with estrogen (56). c-Myc and cyclin D1 are ER target genes that are important mediators of estrogen stimulation of cell cycle progression (7, 21). Estrogen-responsive regulatory sequences have been identified in the proximal promoters of both these genes (13, 17), and more recently, ER binding to enhancer sequences in the region of the MYC and CCND1 (cyclin D1) genes has been identified (11, 18). Once transcriptionally activated by estrogen, c-Myc and cyclin D1 initiate independent pathways that converge at or before cyclin E1-Cdk2 activation (55). Following estrogen treatment of antiestrogen-pretreated cells, cyclin E1-Cdk2 complexes are activated without significant increases in cyclin E1 protein abundance. Instead, cyclin E1-Cdk2 activation occurs as a consequence of (i) sequestration of p21Waf1/Cip1 into newly formed cyclin D1-Cdk4 complexes at the expense of p21Waf1/Cip1's association with cyclin E1-Cdk2 (52, 54-56); (ii) c-Myc-mediated inhibition of p21Waf1/Cip1 transcription, which allows cyclin E1 to form active complexes with Cdk2 (43, 52, 54, 56); and (iii) Cdc25-mediated activation of Cdk2 (22).
Microarray analysis by laboratories, including our own, has identified cyclin E2 as a strongly estrogen-responsive gene (14, 48). Since cyclin E2 has not been characterized in any detail in the context of estrogen regulation and the clinical studies summarized above indicate a potential role for cyclin E2 in hormone-responsive breast cancer that is distinct from that of cyclin E1, we have examined cyclin E2 regulation following estrogen treatment of breast cancer cells, in comparison with cyclin E1 regulation. In this model, cyclin E1-Cdk2 is activated by both c-Myc and cyclin D1. However, the upregulation of cyclin E2 was induced primarily through cyclin D1, required the chromatin remodelling factor chromodomain helicase DNA binding protein 8 (CHD8), and was associated with changes in the association of E2F transcription factors with the cyclin E2 gene promoter. Thus, cyclin E2-Cdk2 is under distinct regulation by estrogen compared to cyclin E1-Cdk2. These data may shed light on why cyclins E1 and E2 have discrete relationships with ER positivity and endocrine therapy in breast cancer.
MATERIALS AND METHODS
Cell lines and culture.
MCF-7 and T-47D cells were cultured in RPMI 1640 medium supplemented with 5% fetal calf serum and insulin (10 μg/ml). Cyclin D1 clone 13 and c-Myc zinc-inducible cell lines are described in references 55 and 48. Further cyclin D1-inducible cell lines were derived as follows. pΔMT (46) was engineered to be compatible with Gateway technology (pΔMT-GW; Invitrogen, Mount Waverley, VIC, Australia) and to contain a selectable marker (neomycin). Cyclin D1 was amplified by PCR from MCF-7 cDNA and inserted into pDONR221 (Invitrogen) by using BP Gateway recombination (Invitrogen). The cyclin D1 “kinase-dead” mutant (cyclin D1 K112E) (24) was generated by site-directed mutagenesis of pDONR221-wild-type cyclin D1 (cyclin D1 WT) using the Phusion site-directed mutagenesis kit (Finnzymes, Espoo, Finland) according to the manufacturer's instructions. cDNAs were then inserted into pΔMT-GW by LR Gateway recombination (Invitrogen).
Cyclin D1 WT and cyclin D1 K112E cell lines were generated by transfecting MCF-7 cells with the pΔMT-GW empty vector, pΔMT-GW cyclin D1 WT, or pΔMT-GW cyclin D1 K112E using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Forty-eight hours after transfection, cells were selected with Geneticin (G418, 400 μg/ml; Gibco, Invitrogen) for 15 days, and the resulting G418-resistant colonies were expanded and tested for expression of cyclin D1 WT and cyclin D1 K112E.
Steroid hormone treatments.
Exponentially proliferating cells were treated for 48 h with 1 × 10−8 M ICI 182780 (a kind gift of Alan Wakeling, Astra-Zeneca Pharmaceuticals, Alderly Park, Cheshire, United Kingdom) to induce quiescence. Cells were subsequently stimulated with 1 × 10−7 M 17β-estradiol [3,17 β-dihydroxy-1,3,5(10)-estratriene; Sigma, Castle Hill, NSW, Australia] and collected as indicated below. Steroid hormones and antagonists were each dissolved in ethanol (EtOH) at a 1,000-fold final concentration, and control cultures received EtOH vehicle to the same final concentration.
siRNA transfection.
Gene-specific small interfering RNAs (siRNAs) (cyclin D1 smart pool [catalog no. L-003210-00], cyclin D1-15 [J-003210-15], cyclin D1-18 [J-003210-18], cyclin E1 [L-003213-00], cyclin E2 [L-003214-00], c-Myc-17 [D-003282-17]) and the controls On-Target Plus siCONTROL pool (D-001810-10), siGENOME nontargeting siRNA 2 (D-001210-02), and On-Target Plus siCONTROL individual siRNAs (D-001810-1-4)) were purchased from Dharmacon (Lafayette, CO). Silencer Select individual siRNAs to CHD8 (catalog no. s33581 and s33582) were purchased from Ambion (Applied Biosystems, Scoresby, Victoria, Australia) and pooled. Transfections were performed at 5 to 100 nM with Lipofectamine 2000 (Invitrogen). Experiments were performed with either siGenome On-Target, On-Target-Plus control siRNAs (Dharmacon), or Silencer Select negative-control 1 and 2 (Ambion), which gave rise to essentially the same results.
Western blot analysis.
Protein lysates were harvested as described in reference 55, and 10 to 30 μg of lysate was separated using NuPage polyacrylamide gels (Invitrogen) prior to transfer of the lysate to polyvinyl difluoride membranes. The membranes were incubated with the following primary antibodies: Cdc6 (180.2), Cdk2 (M2), cyclin E1 (HE12), E2F1 (KH95), and c-Myc (9E10) from Santa Cruz Biotechnology (Santa Cruz, CA); cyclin D1 (DCS6; Novocastra, Newcastle-upon-Tyne, United Kingdom); cyclin E2 (Cell Signaling, Danvers, MA); p21Waf1/Cip1 and p27Kip1 (610233 and 610241, respectively; BD Transduction Laboratories, Lexington, KY); β-actin (AC-15, Sigma); and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 4300; Ambion, Austin, TX). The secondary antibodies were horseradish peroxidase-conjugated sheep anti-mouse or donkey anti-rabbit antibodies (Amersham, Rydelmere, NSW, Australia), and specific proteins were visualized by chemiluminescence (Perkin-Elmer, Rowville, VIC, Australia). Densitometry was performed using the software ImageJ (57).
We performed siRNA and immunoprecipitation experiments to confirm antibody specificity to cyclins E1 and E2. Two cyclin E2-specific antibodies were used for immunoprecipitation: H-140 (Santa Cruz) and I1775 (Epitomics). These antibodies immunoprecipitated a protein that was detectable only by a cyclin E2 antibody in Western blotting and not by a cyclin E1 antibody (see Fig. 2C). Conversely, immunoprecipitation with a second cyclin E1 antibody (C-19; Santa Cruz) gave rise to a 52-kDa band that was detectable only by cyclin E1 Western blotting (see Fig. 2C). Only the protein detected by the cyclin E1 antibody (HE12; Santa Cruz) was reduced by cyclin E1 siRNA treatment, and similarly, cyclin E2 siRNA reduced only cyclin E2 protein (Cell Signaling) (see Fig. 3A).
FIG. 2.
Estrogen stimulation of MCF-7 cells leads to a redistribution of cyclin/Cdk/CDK inhibitor complexes and an increase in cyclin E1-Cdk2 and cyclin E2-Cdk2 kinase activity. (A) The kinase activities of cyclin E1 and E2 immunoprecipitates toward the histone H1 substrate were determined across the time course of estrogen stimulation. (B) Relative kinase activities were quantitated by densitometry, and even loadings of samples were confirmed by Coomassie blue staining of histone proteins (not shown). (C) Arrested cells were treated with estrogen or vehicle (EtOH) for 16 h, and lysates were immunoprecipitated with antibodies to p21Waf1/Cip1, p27Kip1, and cyclins E1 and E2. Immunoprecipitates were subjected to Western blotting for p21Waf1/Cip1, p27Kip1, cyclin D1, cyclin E1, cyclin E2, and Cdk2. Ab, antibody; α, anti; IgG, immunoglobulin G.
FIG. 3.
siRNA-mediated knockdown of cyclins E1 and E2 reduces estrogen-induced proliferation following estrogen rescue. (A) Cell lysates were collected from proliferating MCF-7 cells either treated with 20 nM pooled cyclin E1 siRNA (CycE1), pooled cyclin E2 siRNA (CycE2), or nontargeting pooled siRNA (NTPool) or mock transfected (mock) and immunoprobed for cyclin E1, cyclin E2, and β-actin. (B) Growth-arrested MCF-7 cells were transfected with 20 nM pooled cyclin E1 siRNA, pooled cyclin E2 siRNA, or nontargeting pooled siRNA or mock transfected (mock). Cells were then stimulated with estrogen for 0, 3, and 5 days or a vehicle control. Cell lysates were subjected to Western blotting for cyclin E1, cyclin E2, and β-actin. (C) Cell number was determined by hemacytometer cell counting. Data presented are the means from duplicate experiments, where duplicate counts were performed on triplicate treatments. Error bars represent SEM. Statistical analyses were performed using t tests (day 3, for cyclin E1 siRNA versus controls, P < 0.03; for cyclin E2 siRNA versus controls, P < 0.02; day 5, for cyclin E1 siRNA versus controls, P < 0.009; for cyclin E2 siRNA versus controls, P < 0.006).
Immunoprecipitation and kinase assays.
Immunoprecipitation was performed against 100 to 500 μg of cell lysate using the Trueblot system (eBioscience, San Diego, CA). The following antibodies were used: p21Waf1/Cip1 (M-19), p27Kip1 (C-19), cyclin E1 (C-19), cyclin E2 (H-140), cyclin A (C-19), and Cdk2 (C-19) from Santa Cruz Biotechnology or cyclin E2 from Epitomics (Burlingame, CA). To determine kinase activity, immunoprecipitates of cyclin E1, cyclin E2, cyclin A, and Cdk2 were collected with protein A-Sepharose beads (Zymed, Invitrogen), and the activity of each toward histone H1 substrates was determined as previously described (12).
Proliferation assays.
Proliferation was assessed by hemacytometer cell counting or by flow cytometry using either propidium iodide or bromodeoxyuridine-propidium iodide staining, as previously described (46, 67). Nocodazole [methyl-[5-(2-thienyl-carbonyl)-1H-benzimidazol-2-yl] carbamate] (Sigma) was prepared in dimethyl sulfoxide and used at a final concentration of 50 ng/ml.
RNA extraction and quantitation of mRNA expression.
Total RNA was extracted using the RNeasy minikit (Qiagen, Doncaster, VIC, Australia) as described in the manufacturer's instructions. Reverse transcription was performed as instructed by the manufacturer on 0.5 to 1 μg of RNA using a Promega (Annandale, NSW, Australia) reverse transcription system, with the following amendments. Each reaction mixture was incubated for 60 min at 42 to 55°C, before inactivation at 95°C for 5 min. cDNA was diluted to 200 to 500 μl with H2O and stored at −20°C.
Quantitative reverse transcription-PCR was performed on an Applied Biosystems ABI Prism 7900HT system. All reagents and specific gene expression assays were purchased from Applied Biosystems. Gene expression assays were performed with CHD8 (catalog no. Hs00394229_m1), cyclin D1 (Hs00277039_m1), cyclin E1 (Hs00180319_m1), cyclin E2 (Hs00180319_m1), β-2-microglobulin (Hs99999907_m1), c-Myc (HS00153408_m1), and human RPLPO (large ribosomal protein; 4326314E). For each reaction, 4.5 μl of diluted cDNA was mixed with 5 μl TaqMan universal PCR master mix and 0.5 μl of the gene expression assay mixture. Each reaction mixture was pipetted in triplicate using the EpMotion 5070 automated pipetting system (Eppendorf, North Ryde, NSW, Australia), before amplification using standard gene expression assay conditions. Standard curves of each mRNA were created using serial dilutions of MCF-7 cDNA, and relative expression was determined using the ΔΔCT method, where CT is the threshold cycle, with the RPLPO or β-2-microglobulin levels as internal controls.
ChIP.
Cell lysates were fixed with formalin, and chromatin immunoprecipitation (ChIP) was performed as described previously (10), using antibodies to E2F1 (sc-193; Santa Cruz) and E2F4 (sc-1082; Santa Cruz). Immunoprecipitated DNA was amplified by quantitative PCR using the Applied Biosystems SYBR green master mix. The following primers were used to amplify regions from the cyclin E1 and cyclin E2 gene promoters: for cyclin E1, TCTTCAGAGAGCCAGGAAGG (forward) and TTGGGTCGTTCATTCATTCA (reverse), and for cyclin E2 AAATCCAGGAGTTGCAGTGG (forward) and ACTCTCAGGGGCTCCTTCTC (reverse). Each ChIP experiment was performed at least in duplicate, and each PCR mixture was amplified in triplicate.
RESULTS
Estrogen regulation of cyclin E1 and cyclin E2 expression and activity.
Both cyclins E1 and E2 have been identified in microarray studies as upregulated by estrogen at single time points (14, 48). We therefore examined the dynamics of cyclin E1 and E2 expression and activity following estrogen stimulation in initial experiments. After induction of quiescence with 10 nM ICI 182780 treatment for 48 h, MCF-7 cells were treated with 100 nM 17β-estradiol and cell lysates collected at intervals up to 24 h after estrogen rescue. In this model, essentially the entire cell population reinitiates cell cycle progression following estrogen treatment, reaching S phase between 12 and 16 h and G2 phase at ∼24 h and dividing after ∼30 h (Fig. 1A) (48, 55, 56). Analyses of time courses of estrogen treatment identified a small increase in cyclin E1 mRNA (2.4-fold) that peaked 12 h after estrogen treatment but little change in cyclin E1 protein level (Fig. 1B and E), consistent with previous data (56). Unlike cyclin E1, cyclin E2 was strongly upregulated, with both mRNA and protein reaching ≥10-fold-higher levels 12 to 20 h after estrogen treatment (Fig. 1B and E). Estrogen-mediated upregulation of cyclin E2 was apparent over a wide range of estrogen concentrations (Fig. 1C). Thus, cyclin E2 is induced to much higher levels than cyclin E1 during estrogen rescue, and the induction of cyclin E2 appears to be slightly delayed compared with cyclin E1 induction in this model system since it peaked at 16 h compared with 12 h for cyclin E1 mRNA.
FIG. 1.
Cyclins E1 and E2 are differentially regulated by estrogen. MCF-7 cells were arrested for 48 h with 10 nM ICI 182780 and then treated with 100 nM 17β-estradiol (estrogen) or vehicle (EtOH), and lysates were collected over a time course of 24 h. Data are representative of at least two experiments, and graphs represent pooled results of analyses from at least two experiments. Error bars represent range or standard errors of the means (SEM), as appropriate. (A) Estrogen- or vehicle (EtOH)-treated cells were harvested for analysis of cell cycle phase by flow cytometry. Representative histograms are shown. (B) Cyclins E1 and E2 and a panel of cell cycle proteins were detected by Western blotting. GAPDH was used as a loading control. (C) Antiestrogen-arrested cells were treated for 6 h and 16 h with 100 nM, 50 nM, 10 nM, 1 nM, and 0.1 nM 17β-estradiol (estrogen) and cell lysates immunoprobed for cyclin D1, cyclin E1, cyclin E2, c-Myc, and β-actin. (D) Protein from replicate experiments (including that shown in panel B) was quantitated by densitometry and corrected for loading using GAPDH. (E) cyclin E1 and cyclin E2 total mRNA was measured by qPCR using RPLPO or β-2-microglobulin as an endogenous control.
Since cyclins E1 and E2 are induced to different levels by estrogen, we examined whether the differences in induction led to distinct activation of cyclin E-Cdk2 complexes. Both cyclins E1 and E2 formed active kinase complexes with Cdk2, with similar levels of induction of kinase activity and similar time courses of induction. Cyclin E1-Cdk2 activity peaked at 20 h and then declined, whereas cyclin E2-Cdk2 activity was still increasing 24 h after estrogen stimulation (Fig. 2A and B). Thus, estrogen induces similar kinase activities associated with cyclin E1 and cyclin E2, despite distinct inductions of cyclin E1 and E2 mRNA and protein.
Both cyclins E1 and E2 form active kinase complexes with Cdk2 that can be inhibited through binding of the CDK inhibitors p21Waf1/Cip1 and p27Kip1. We performed coimmunoprecipitation experiments to confirm that these complexes form after estrogen stimulation. Cyclin E1 coimmunoprecipitated Cdk2 and p21Waf1/Cip1/p27Kip1 in antiestrogen-arrested cells, and while Cdk2 coimmunoprecipitation was maintained after estrogen stimulation, p21Waf1/Cip1/p27Kip1 coimmunoprecipitation was substantially decreased (Fig. 2C). Cyclin E2 was only significantly precipitated following estrogen induction of cyclin E2 protein. Cdk2 effectively coprecipitated with cyclin E2 under these conditions, while small quantities of p21Waf1/Cip1/p27Kip1 were coimmunoprecipitated with cyclin E2. Thus, cyclin E2 upregulation following estrogen induction leads to the formation of a substantial quantity of cyclin E2-Cdk2 complexes, a small proportion of which also contain p21Waf1/Cip1/p27Kip1.
Immunoprecipitation of p21Waf1/Cip1 and p27Kip1 provided similar results; in estrogen-stimulated cells, only a small quantity of cyclins E1 and E2 coimmunoprecipitated with either CDK inhibitor (Fig. 2C). However, a substantial quantity of cyclin D1 coimmunoprecipitated with the CDK inhibitors (compared to the lysate control), and the proportion of cyclin D1 binding to p21Waf1/Cip1 increased following estrogen induction, despite a decrease in absolute p21Waf1/Cip1 levels. This indicates that cyclin D1 binds a significant proportion of p21Waf1/Cip1 following estrogen induction, in agreement with previous data showing that cyclin D1 acts as a sink for p21Waf1/Cip1, with a resultant decrease in the inhibition of cyclin E-Cdk2 complexes (52, 54-56).
Ablation of either cyclin E1 or cyclin E2 attenuates estrogen-induced proliferation.
The overexpression of either cyclin E1 or cyclin E2 is able to shorten G1 phase and increase overall proliferation (28), and the overexpression of cyclin E1 reduces acute sensitivity to antiestrogen arrest in MCF-7 cells (16, 33). This suggests that the absence of either E cyclin could potentially attenuate estrogen-induced proliferation.
Antiestrogen-arrested cells were treated with siRNAs to cyclin E1, cyclin E2, or pooled nontargeting control siRNAs or mock transfected, and the cells were treated with estrogen 48 h later. Western blotting confirmed specific knockdown of cyclins E1 and E2 by their respective siRNAs in exponentially proliferating cells (Fig. 3A) and the severely attenuated estrogen-mediated induction of cyclin E2 protein (Fig. 3B). Cell counts showed that control siRNA- or mock-transfected cells resumed exponential proliferation following estrogen treatment of antiestrogen-arrested cells (Fig. 3C). By comparison, cells treated with siRNAs to either cyclin E1 or cyclin E2 showed only slight increases (less than twofold) in cell number (Fig. 3C). Consequently, each E cyclin is required for the full proliferative effects of estrogen in this model.
Induction of c-Myc does not account for estrogen regulation of cyclin E2.
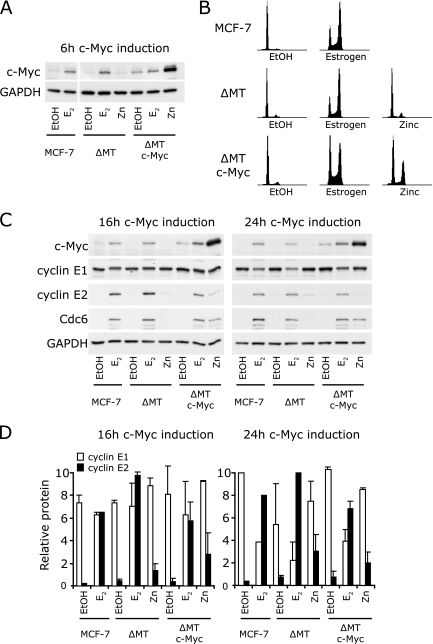
Estrogen acts on the cell cycle through two main regulatory nodes, c-Myc and cyclin D1 (55). Since cyclins E1 and E2 had distinct expression patterns after estrogen treatment, we investigated whether cyclins E1 and E2 were differentially targeted by these estrogen target genes. Mouse mammary tumor virus Myc-driven mammary tumors frequently overexpress cyclins E1 and E2, suggesting that the genes for them may be c-Myc responsive (27), and therefore we first examined whether the overexpression of c-Myc was able to induce the expression of cyclin E1 or E2 in antiestrogen-arrested cells. MCF-7 cells expressing c-Myc under the control of a zinc-inducible promoter, and the corresponding vector controls, were pretreated with antiestrogen and then treated with either vehicle, zinc, or 17β-estradiol for 16 and 24 h, time points when cyclin E2 expression was markedly increased (Fig. 1). Under these conditions, c-Myc protein is strongly induced within 6 h of either estrogen or zinc treatment (Fig. 4A) (48, 55), leading to rescue of antiestrogen arrest (Fig. 4B) (48, 55). Although after zinc treatment, near-maximal c-Myc levels are sustained until at least 24 h, after estrogen treatment, c-Myc levels decline from their maxima at 3 to 6 h, although they remain higher than in antiestrogen-arrested cells (48, 55, 56).
FIG. 4.
Overexpression of c-Myc does not mimic the estrogen-induced expression of cyclin E2. Parental MCF-7 cells and MCF-7 cells inducibly expressing c-Myc (ΔMT c-Myc) or the vector control (ΔMT) were arrested for 48 h with 10 nM ICI 182780 and then treated with 100 nM 17β-estradiol (estrogen [E2]), 65 μM zinc (Zn), or vehicle (EtOH). (A) Protein lysates were collected at 6 h and subjected to Western blotting for c-Myc and GAPDH. (B) Cells additionally treated with nocodazole to prevent cell division of stimulated cells were harvested for analysis of the cell cycle phase by flow cytometry. Representative histograms 32 h after estrogen, zinc, or EtOH treatment are shown. (C) Protein lysates were collected at 16 and 24 h and subjected to Western blotting for c-Myc, cyclin E1, cyclin E2, Cdc6, and GAPDH. (D) Protein was quantitated by densitometry, and quantities were corrected for loading using GAPDH. Data are pooled from two independent experiments; error bars represent ranges.
Cyclin E1 protein levels were not significantly regulated by either c-Myc induction or estrogen treatment at 16 h and were reduced by estrogen but not by zinc at 24 h (Fig. 4C and D). Consistently with data presented in Fig. 1, cyclin E2 levels were greatly increased (6- to 10-fold) by estrogen treatment at 16 h. However, cyclin E2 protein was increased only approximately threefold by c-Myc induction, despite much higher c-Myc induction by zinc than by estrogen, including at early time points (Fig. 4C and D). Furthermore, the vector control line also showed slight induction of cyclin E2 after the addition of zinc (∼1.5-fold), suggesting that the effect of c-Myc induction on cyclin E2 expression may be partially due to the nonspecific effects of zinc. By 24 h, c-Myc induction had no effect on the expression of cyclin E2 over the effects of zinc treatment in empty vector cells, whereas the increase in cyclin E2 following estrogen treatment was sustained (Fig. 4C and D). Thus, while induction of sustained high levels of c-Myc causes antiestrogen-arrested cells to reenter the cell cycle (Fig. 4B) (48, 55), it is able to induce only a transitory increase in cyclin E2 levels (Fig. 4C and D) rather than the high and sustained levels of cyclin E2 expression seen after treatment with estrogen. In contrast, a known target of c-Myc, Cdc6 (39), did exhibit sustained induction by c-Myc at 24 h, and Cdc6 was induced by c-Myc to levels only slightly lower than those following induction by estrogen (Fig. 1 and 4C) (48).
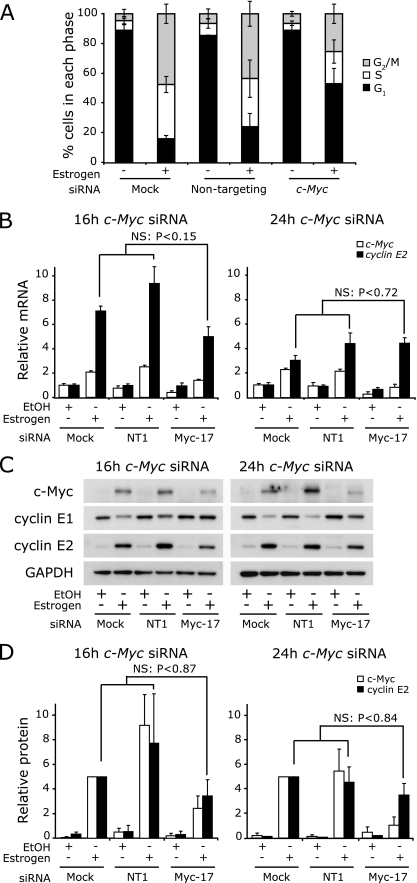
In complementary experiments, we treated MCF-7 cells with c-Myc siRNA during arrest with ICI 182780 and then rescued proliferation with estrogen. These experiments used the c-Myc siRNA Myc-17, which we have previously shown to reduce the peak of c-Myc induction in this model system by 50 to 70% (8, 48) and which also reduces estrogen induction of cell cycle progression by 50% (Fig. 5A) (8, 48). The effects of the c-Myc siRNA were sustained, so that the levels of c-Myc protein and mRNA remained reduced at both 16 and 24 h (Fig. 5B and D). However, 16 h and 24 h after estrogen treatment, the levels of expression of cyclin E2 mRNA and protein did not significantly differ between control conditions and following treatment with c-Myc siRNA (Fig. 5B and D). Collectively, the data in Fig. 4 and 5 argue that the estrogen induction of cyclin E2 is unlikely to be secondary to the induction of c-Myc.
FIG. 5.
Ablation of c-Myc does not significantly alter the estrogen-induced expression of cyclin E2. Growth-arrested MCF-7 cells were transfected with 100 nM Myc-17 siRNA or nontargeting control 1 (NT1) or mock transfected (mock). Cells were then stimulated with estrogen and RNA and protein lysates collected 16 and 24 h poststimulation. Data represent or are pooled from results of two to five independent experiments. Statistical analyses were performed using t tests. (A) Cells transfected with siRNA were cotreated with nocodazole at the time of estrogen/vehicle addition. Cells were harvested after 36 h for analysis of cell cycle phase by flow cytometry. Error bars represent standard deviations or ranges. (B) cyclin E1, cyclin E2, and c-Myc total mRNAs were measured by quantitative PCR using RPLPO or β-2-microglobulin as endogenous controls. Error bars represent SEM. (C) c-Myc, cyclin E1 and cyclin E2 were detected by Western blotting, with GAPDH as a loading control. (D) Protein was quantitated by densitometry, and quantities were corrected for loading using GAPDH. Error bars represent ranges. NS, not significant.
Cyclin E2 is upregulated by estrogen through cyclin D1.
Since a subset of estrogen effects are regulated by cyclin D1 in a manner distinct from that of c-Myc (48), we next examined whether cyclin E2 was a downstream target of cyclin D1. First, we examined the effect of cyclin D1 induction on the expression of cyclins E1 and E2 and compared the effect to the response after estrogen treatment. MCF-7 cells expressing cyclin D1 under the control of a zinc-inducible promoter and the corresponding vector controls were treated with either vehicle, zinc, or estrogen. The levels of cyclin D1 were similar following estrogen or zinc treatment of the cyclin D1-inducible cell line and estrogen treatment of the vector control cell line (Fig. 6A). Cyclin E1 levels were not significantly altered at 24 h by either cyclin D1 induction or estrogen treatment (Fig. 6A). In contrast, cyclin E2 was increased to similar levels by both cyclin D1 induction and estrogen treatment (Fig. 6A and B). Similar observations were made following 9 h of treatment with vehicle, zinc, or estrogen (data not shown). A second, independently derived MCF-7 clone also displayed increased cyclin E2 expression following zinc induction of cyclin D1, and this was not apparent following zinc treatment of an empty vector line derived in parallel (see Fig. 8E and F). The clonal MCF-7 cell line used for cyclin D1 induction in Fig. 4 displayed higher levels of both cyclin D1 and cyclin E2 in the absence of zinc treatment than the corresponding empty vector cells (Fig. 5A), confirming the previously observed leakiness of the inducible vector system (55) but also supporting the conclusion that cyclin E2 expression is responsive to cyclin D1 levels.
FIG. 6.
Cyclin D1 overexpression mimics estrogen effects on cyclin E2. MCF-7 or T-47D cells inducibly expressing cyclin D1 (ΔMT cyclin D1) or the vector control (ΔMT) were arrested for 48 h with 10 nM ICI 182780 and then treated with 100 nM 17β-estradiol (estrogen [E2]), 70 μM zinc (Zn), or vehicle (EtOH), and lysates were collected at 24 h. Data are representative of two experiments. (A to C) Data relating to MCF-7 cells; (D to F) data relating to T-47D cells. (A and D) Cyclins D1, E1, E2, and E2F1 and p21Waf1/Cip1 were detected by Western blotting. GAPDH was used as a loading control. (B and E) Protein was quantitated by densitometry, and quantities were corrected for loading using GAPDH. Data are pooled from two independent experiments; error bars represent ranges. (C and F) Cells were cotreated with nocodazole to prevent cell division of stimulated cells and were harvested for analysis of cell cycle phase by flow cytometry. Representative histograms 41 h after estrogen, zinc, or EtOH treatment are shown.
FIG. 8.
E2F transcription factors have altered association with the cyclin E2 promoter after estrogen stimulation and also after cyclin D1 induction. (A and B) Antiestrogen-arrested MCF-7 cells were stimulated with 100 nM 17β-estradiol and lysates collected over a time course of 24 h. Binding of E2F1 and E2F4 to the cyclin E1 and cyclin E2 gene promoters was determined by ChIP. Results are pooled analyses of the results of three independent experiments, and error bars represent SEM. (C and D) MCF-7 cells inducibly expressing cyclin D1 (ΔMT cyclin D1) or a vector control (ΔMT) were arrested for 48 h with 10 nM ICI 182780 and then treated with 100 μM zinc, and lysates were collected at 0 and 16 h. Binding of E2F1 and E2F4 to the cyclin E1 and cyclin E2 gene promoters was determined by ChIP. Results are pooled from two independent experiments, and error bars represent SEM from triplicate analyses. (E) MCF-7 cells inducibly expressing cyclin D1 (ΔMT cyclin D1), catalytically inactive cyclin D1 (ΔMT cyclin D1 K112E), or a vector control (ΔMT) were arrested for 48 h with 10 nM ICI 182780 and then treated with 100 nM 17β-estradiol (estrogen [E2]), 100 μM zinc (Zn), or vehicle (EtOH), and lysates were collected at 24 h. Data are representative of two experiments. (F) Protein was quantitated by densitometry, and quantities were corrected for loading using GAPDH. Error bars represent ranges.
To provide additional evidence for this conclusion, we repeated the experiments using another estrogen-responsive cell line, T-47D. Parental T-47D cells, a clonal derivative expressing cyclin D1 under the control of the same zinc-responsive promoter, and a corresponding clone of empty-vector-transfected cells (47) all displayed induction of cyclin E2 following estrogen treatment, although to a lesser extent than was observed in MCF-7 cells (Fig. 6D and data not shown). Despite the lower sensitivity of this model system, we also observed increased expression of cyclin E2 following zinc induction of cyclin D1 in T-47D cells (Fig. 6D and E), confirming that cyclin D1 expression is sufficient for upregulation of cyclin E2 to a degree that is comparable with the level of estrogen induction of cyclin E2.
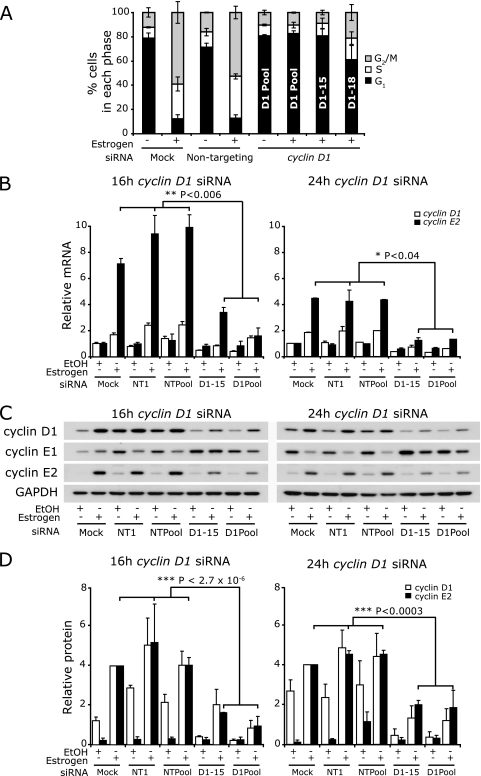
To provide further evidence of a role for cyclin D1 in estrogen induction of cyclin E2, we examined whether the treatment of cells with cyclin D1 siRNA would prevent the estrogen-induced upregulation of cyclin E2. MCF-7 cells were treated with siRNAs and simultaneously arrested with ICI 182780. Expression of cyclins D1, E1, and E2 was then examined after 16 or 24 h of estrogen stimulation. A pool of four siRNAs directed against cyclin D1 reduced estrogen-induced expression of cyclin D1 mRNA and protein by at least 50% at 16 to 24 h, and two of the component siRNAs (cyclin D1-15 and cyclin D1-18) also effectively reduced cyclin D1 expression (Fig. 7 and data not shown). The cyclin D1 siRNA treatments also effectively attenuated estrogen-induced proliferation by at least 35% (Fig. 7A).
FIG. 7.
Cyclin D1 siRNA attenuates estrogen-induced expression of cyclin E2. Growth-arrested MCF-7 cells were transfected with 20 nM pooled cyclin D1 siRNA (D1 Pool), cyclin D1-15 (D1-15), cyclin D1-18 (D1-18), nontargeting pooled siRNA (NTPool), or nontargeting control 1 (NT1) or mock transfected (mock). Cells were then stimulated with estrogen, and RNA and protein lysates were collected 16 and 24 h poststimulation. Some experiments were performed in parallel with the experiments whose results are shown in Fig. 5B and D and so have the same control samples. Data are representative of four experiments, and error bars represent SEM. Statistical analyses were performed using t tests. (A) Cells transfected with siRNA were cotreated with nocodazole at the time of estrogen/vehicle addition. Cells were harvested after 40 h for analysis of cell cycle phase by flow cytometry. Error bars represent standard deviations or ranges. (B) Total cyclin E1, cyclin E2, and cyclin D1 mRNAs were measured by quantitative PCR using RPLPO or β-2-microglobulin as endogenous controls. Error bars represent SEM. (C) Cyclin D1, cyclin E1, and cyclin E2 were detected by Western blotting, with GAPDH as a loading control. (D) Protein was quantitated by densitometry, and quantities were corrected for loading using GAPDH.
Treatment of cells with estrogen in the presence of cyclin D1 siRNA (pool or D1-15) led to the attenuation of induction of cyclin E2 mRNA and protein. This was apparent 9 h after estrogen treatment (data not shown) and was sustained until at least 16 to 24 h, when levels of cyclin E2 mRNA and protein were reduced by 55 to 70% compared with levels in the mock-infected and nontargeting controls (Fig. 7B and D). An additional cyclin D1 siRNA (D1-18) gave similar results (data not shown). Thus, the reduction in cyclin D1 induction led to a proportional inhibition of cyclin E2 induction.
The experiments whose results are shown in Fig. 6 and 7 show that cyclin D1 is necessary for the estrogen-induced expression of cyclin E2 and that the induction of cyclin D1 alone is sufficient for sustained cyclin E2 expression. This contrasts with c-Myc effects, where c-Myc siRNA does not significantly affect the induction of cyclin E2, and c-Myc induction, even at levels higher than those following estrogen treatment, does not lead to sustained induction of cyclin E2 (Fig. 4 and 5). Consequently, cyclin E2 expression appears to be targeted mainly for induction by estrogen via cyclin D1 and not via c-Myc.
Role of E2F in the estrogen regulation of cyclin E2 via cyclin D1.
Cyclins E1 and E2 are well-known E2F targets (34, 74), and we therefore investigated the role of E2F in the estrogen regulation of cyclin E2 downstream of cyclin D1. Previous studies of E2F regulation by estrogen have found strong transcriptional regulation of E2F1 but little or no effect on the expression of E2F2-5 (66, 68, 72). However, antiestrogen treatment equivalent to the pretreatment used in experiments discussed in this paper leads to decreased E2F1 expression, increased E2F4 expression, and a marked increase in E2F4-p130 complexes (12). We therefore focused on E2F1 and E2F4 as the most relevant representative “activating” and “repressive” E2F family members. Consistent with data obtained using other experimental designs, the addition of estrogen to antiestrogen-pretreated cells led to the induction of E2F1 over a time course that paralleled the induction of cyclin E2 (Fig. 1B).
To determine the time course of E2F association with E2F-responsive elements in the cyclin E1 and cyclin E2 promoters following estrogen treatment, we performed ChIP using the E2F1 and E2F4 antibodies. In control antiestrogen-pretreated cells (i.e., at time zero for estrogen treatment), both the well-characterized E2F binding site in the cyclin E1 promoter and the consensus E2F sites 550 to 600 bp upstream of the cyclin E2 transcription start site were associated with E2F4 and smaller, but still readily detectable, amounts of E2F1 (Fig. 8A and B). E2F association remained at similar levels between 0 and 9 h. However, at 16 h—a time point when both E2F1 and cyclin E2 levels are maximal—E2F1 binding to both promoters increased approximately threefold and E2F4 binding to both promoters decreased by ∼50% (Fig. 8A and B).
To test the hypothesis that estrogen might cause these effects via cyclin D1 regulation of E2F expression or activity, we first examined E2F1 expression after cyclin D1 induction. Zinc induction of cyclin D1 led to increased E2F1 expression in both MCF-7 and T-47D cells, to levels comparable with those following estrogen treatment (Fig. 6A and D and 8E). ChIP after 16 h of zinc treatment of MCF-7 cells expressing cyclin D1 revealed increased E2F1 binding at both the cyclin E1 and E2 promoters, and this was not apparent in vector-transfected cells treated in parallel (Fig. 8C and D). In contrast with the decreased E2F4 binding following estrogen treatment, there was no decrease in E2F4 binding after zinc induction of cyclin D1 (Fig. 8C and D).
Since cyclin D1-Cdk4 phosphorylation of pRb allows relief of pRb-mediated repression and activation of E2F target genes, including the E2F1 gene (41), we compared the effects of induction of wild-type cyclin D1 with those of a well-characterized “kinase-dead” point mutant (with a K112E mutation). Cyclin D1 K112E binds but does not activate Cdk4 and Cdk6 (32) and is consequently unable to phosphorylate Rb (25). In a clonal MCF-7 cell line transfected with cyclin D1 (K112E) under the control of a zinc-inducible promoter, zinc treatment did not lead to induction of E2F1 or cyclin E2, although both were induced when cells expressing wild-type cyclin D1 were treated with zinc in parallel (Fig. 8E and F). Thus, the effects of cyclin D1 on cyclin E2 expression require Cdk4-Cdk6 activation.
CHD8 is required for cyclin E2 upregulation.
Although E2F1 is recruited to the promoters of both cyclin E1 and cyclin E2 after estrogen stimulation, only cyclin E2 is significantly upregulated. The chromatin remodelling factor CHD8 modulates transcriptional elongation through interactions with RNA polymerase II (58, 64) and has recently been identified to specifically target a subset of E2F1 target genes that include cyclin E2 but not cyclin E1 (58). Consequently, we investigated whether estrogen induction of cyclin E2 requires CHD8.
Antiestrogen-arrested cells were treated with siRNA to CHD8 or cyclin D1 or controls and rescued with estrogen for 16 h. CHD8 gene expression was not changed with estrogen stimulation in control samples but was decreased by ∼80% following CHD8 siRNA treatment (Fig. 9A). CHD8 siRNA treatment significantly attenuated both cyclin E2 gene and protein induction (Fig. 9B and D), indicating that cyclin E2 gene transcription is affected by CHD8 in this model system. In contrast, the low-level induction of cyclin E1 by estrogen was maintained in the presence of CHD8 siRNA, as was the induction of cyclin D1 and E2F1 (Fig. 9B and D). Thus, CHD8 is necessary for cyclin E2 induction, but this is independent of effects on cyclin D1 or E2F1 regulation.
FIG. 9.
CHD8 is necessary for cyclin E2 induction following estrogen treatment. Growth-arrested MCF-7 cells were transfected with 20 nM concentrations of the CHD8 siRNA pool, cyclin D1 siRNA pool, Dharmacon nontargeting pool, or Ambion Silencer Select pool or mock transfected (mock). Cells were then stimulated with estrogen, and RNA and protein lysates were collected 16 h poststimulation. Results are pooled from analyses of two independent experiments in which nontargeting siRNA data were pooled (NT). (A) CHD8 and cyclin D1 total mRNAs were measured by quantitative PCR using RPLPO as an endogenous control. Error bars represent SEM. (B) Cyclin D1, cyclin E1, cyclin E2, and cyclin E2F1 were detected by Western blotting, with β-actin as a loading control. NT samples are of the Dharmacon Nontargeting pool. (C) cyclin E1 and cyclin E2 mRNAs were measured by quantitative PCR using RPLPO as an endogenous control. Error bars represent SEM. (D) Protein was quantitated by densitometry, and quantities were corrected for loading using β-actin. Error bars represent ranges.
DISCUSSION
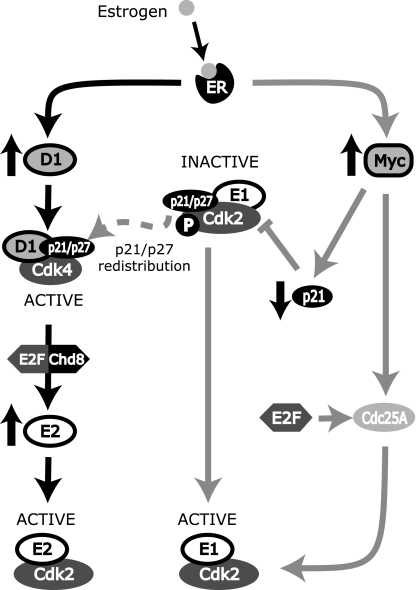
Estrogen stimulates proliferation through the modulation of a panel of cell cycle proteins, resulting in rapid entry into the cell cycle from quiescence. In breast cancer cells, these effects converge on the activation of cyclin E-Cdk2, which is essential for estrogen-mediated cell cycle progression (55, 70) and occurs via decreased p21Waf1/Cip1 and p27Kip1 availability through sequestration to cyclin D1-Cdk4 complexes (52), repression of p21Waf1/Cip1 expression via c-Myc (43, 54, 55), and induction of Cdc25A, leading to Cdk2 activation (22, 23). Here, we identify an additional conduit for the estrogen activation of cyclin E-Cdk2 and the G1/S-phase transition through CHD8-dependent regulation of cyclin E2 downstream of cyclin D1 (Fig. 10). This model is supported by evidence that an siRNA-mediated reduction in cyclin D1 expression during estrogen stimulation leads to a proportional decrease in induction of cyclin E2 and that induction of cyclin D1 is sufficient for increased cyclin E2 expression and E2F1 recruitment to the cyclin E2 gene promoter. Recent evidence that cyclin E2 expression is increased when cyclin D1, D2, or D3 is transiently expressed in hepatocytes provides further support (44).
FIG. 10.
Cyclin E1-Cdk2 and cyclin E2-Cdk2 are activated by distinct pathways during estrogen-mediated proliferation. Estrogen, via the estrogen receptor, upregulates c-Myc and cyclin D1. Furthermore, cyclin D1 upregulates E2F transcription factors, leading to E2F/CHD8-mediated transcription of cyclin E2 mRNA and consequent formation of active cyclin E2-Cdk2 complexes. Upregulation of cyclin D1 also results in the sequestration of p21Waf1/Cip1/p27Kip1 into cyclin D1-Cdk4 complexes, contributing to the activation of cyclin E1-Cdk2 complexes. c-Myc represses the expression of p21Waf1/Cip1 and induces Cdc25A, which together allow cyclin E-Cdk2 complex activation. Cdc25A is also positively regulated by E2F transcription factors.
The >10-fold increase in cyclin E2 expression following estrogen treatment appeared to have effects on the overall pool of active cyclin E-Cdk2 complexes in addition to simply increasing the total number of cyclin E-Cdk2 complexes. Cyclin E2-Cdk2 complexes bind p21Waf1/Cip1 and p27Kip1 (37) (Fig. 2C) and therefore contribute indirectly to the activation of cyclin E1-Cdk2 and, although to a lesser extent, cyclin D1-Cdk4. We noted a delay in the peak activity of cyclin E2-Cdk2 compared to that of cyclin E1-Cdk2, consistent with previous observations of the timing of the activations of these kinases (37), suggesting that cyclin E2-Cdk2 may provide an additional window of cyclin E-Cdk2 activity late in the G1/S-phase transition. Certainly, cyclin E2 appears to play a role that is equivalent to, but independent of, the role of cyclin E1 in estrogen-induced proliferation, as the ablation of either E cyclin leads to similar attenuations of estrogen-induced proliferation (Fig. 3).
Cyclins E1 and E2 have generally been assumed to be coregulated (51), except in a few rare circumstances (76). Both E cyclins are known E2F targets (34, 50, 74), providing a likely mechanism for coregulation. In our experiments, after estrogen stimulation, E2F1 binding to both the cyclin E1 and cyclin E2 gene promoters was increased and E2F4 binding was decreased. E2F4 forms a repressor complex with p130 that prevents transcription of gene subsets during quiescence (12) and is present following antiestrogen pretreatment (12). Although cyclin D1-Cdk4 phosphorylates E2F4 and p130, thereby promoting disassociation of the E2F4/p130 complex and release of E2F4 from DNA binding (9, 20, 59), cyclin D1 induction did not lead to the release of E2F4 from the cyclin E1 and cyclin E2 gene promoters. However, this did not impair E2F1 recruitment to these promoters or cyclin E2 induction, suggesting that the major E2F effect on cyclin E2 transcription in this model is E2F1-mediated activation rather than relief of E2F4-mediated repression.
E2F1 expression is responsive to a number of transcription factors, including E2F (53) and the ER (72). In the absence of estrogen stimulation, cyclin D1 induction led to substantial upregulation of E2F1 (Fig. 6). This is likely via phosphorylation of Rb and consequent activation of E2F-mediated transcription, since induction of a cyclin D1 mutant unable to activate Cdk4/6 did not affect E2F1 expression. After estrogen stimulation, E2F1 expression did not increase until 9 to 12 h, coincident with the increase in cyclin E2 expression (Fig. 1) but following the peak of Cdk4 activation at 6 h in this model system (56). Thus, despite direct ER-mediated activation of E2F1 transcription following estrogen stimulation (72), the major induction of E2F1 expression and activation of E2F-mediated transcription appear to occur as a consequence of cyclin D1-Cdk4 activation. This conclusion is supported by our observation that estrogen stimulation of E2F1 expression is attenuated in the presence of cyclin D1 siRNA (Fig. 9B).
Estrogen treatment led to an upregulation of cyclin E2 that was significantly greater than its effects on cyclin E1, despite increased E2F1 binding to the promoters of both cyclin E1 and cyclin E2. Several microarray studies document a greater increase in cyclin E2 transcription than in cyclin E1 transcription following expression of the activating E2Fs (E2F1 to -3) (3, 65, 74), suggesting that cyclin E2 gene transcription may be inherently more sensitive to E2F activation than cyclin E1 transcription. CHD8 is a chromatin remodelling factor that facilitates efficient RNA polymerase II transcript elongation of a subset of genes including E2F1 targets such as cyclin E2, but not cyclin E1 (58), and it is required for the upregulation of cyclin E2 as cells pass through the G1/S-phase transition. Similarly, in our model of estrogen rescue of antiestrogen-arrested cells, CHD8 is necessary for the upregulation of the cyclin E2 transcript. Rodriguez-Paredes et al. (58) propose that E2F and CHD8 may interact at the promoter of E2F-responsive genes. The dependence of estrogen induction of cyclin E2 on both cyclin D1 and CHD8 is consistent with such a model (Fig. 10).
Although both cyclin D1 and cyclin E2 are c-Myc-responsive in rodent fibroblasts (5, 29), in antiestrogen-treated cells, induction of c-Myc is sufficient for reinitiation of cell cycle progression (48, 55), but this occurs in the absence of a significant increase in cyclin D1 expression or cyclin D1-Cdk4 activity, and only a small (less-than-threefold), transient increase in cyclin E2 expression. c-Myc can activate E2F in rodent fibroblasts, but this is independent of cyclin E-Cdk2 activation (2) and does not depend on E2F1 but rather on E2F2 and E2F3 (38), which are not strongly associated with estrogen action. In preliminary experiments, we did not see increased expression of E2F1 following c-Myc expression in antiestrogen-arrested cells. Although activation of cyclin E1-Cdk2 downstream of c-Myc leads to pRb phosphorylation (48, 55) and is therefore likely to activate E2F-mediated transcription, this is apparently not sufficient for the sustained induction of cyclin E2. Consequently, the principal mechanisms for c-Myc regulation of the G1-to-S-phase transition during estrogen stimulation of proliferation appear to be the repression of p21Waf1/Cip1 and the induction of Cdc25A, which together allow effective activation of cyclin E-Cdk2 complexes (22, 24, 43, 54) (Fig. 10). Cdc25A may also be targeted for upregulation by E2F in other model systems (71), but its upregulation appears to occur independently of cyclin D1 after estrogen stimulation (22). The failure of c-Myc to substantially induce cyclin E2 implies that the peak in cyclin E2 expression coincident with S-phase transit is not simply the result of cell cycle position but rather represents a specific response to cyclin D1 induction, emphasizing the role of cyclin D1 and its downstream targets in the regulation of cyclin E2.
The model of estrogen regulation of cyclin E-Cdk2 activity presented in Fig. 10 identifies several differences between the mechanisms by which cyclin E1-Cdk2 and cyclin E2-Cdk2 are activated. Although transcriptional regulation of cyclin E1 does occur, decreased p21Waf1/Cip1 synthesis appears to play a more major role in initiating cyclin E1-Cdk2 activation than does increased synthesis of cyclin E1 (54). In contrast, the temporal correspondence between increased cyclin E2 expression and increased cyclin E2-Cdk2 activity suggests that the major mechanism for cyclin E2-Cdk2 activation is the transcriptional regulation of the cyclin E2 gene. Furthermore, although cyclin E1-Cdk2 is activated downstream of both c-Myc and cyclin D1 by effects on p21Waf1/Cip1 expression or availability (55), cyclin E2 activation following estrogen treatment is dependent on cyclin D1 but not c-Myc. This contrasts with the observations of Geng et al., who identified high levels of cyclin E1 and cyclin E2 mRNAs in c-Myc-driven mouse mammary tumors but low levels of both transcripts in Ras-driven tumors (27), which commonly display increased levels of cyclin D1 and are dependent on cyclin D1 (75). However, the expression of the cyclin E1 and cyclin E2 genes is responsive to factors in addition to estrogen, most prominently E2F, that could affect their expression in vivo.
Studies of mice have identified that both cyclin E1 and cyclin E2 are not required for normal cell cycles in mouse embryonic fibroblasts and that mouse embryonic fibroblasts with the combined cyclin E1−/− and E2−/− knockout show only slightly impaired proliferation (26). However, the presence of at least one E cyclin is required for the reentry of cells into the cell cycle after quiescence (25, 26). Antiestrogen treatment of MCF-7 cells results in a quiescent-like state, which can be reversed by the addition of estrogen (12). Complete cell cycle reentry from quiescence depends on the activation of both cyclin D1 and c-Myc and, as shown here, the presence of their downstream targets, cyclin E1 and cyclin E2. Overexpression of either cyclin D1 or c-Myc can recapitulate estrogen effects on proliferation in breast cancer cells. However, these two oncogenes have distinct associations with ER-positive breast cancer (7), presumably through the activation of distinct subsets of genes and pathways as well as preferential deregulation in different subtypes of breast cancer. Similarly, cyclins E1 and E2 are independently associated with breast cancer survival (60), perhaps reflecting their distinct regulation by estrogen. Moreover, since cyclin E2 frequently appears in gene signatures of therapeutic responsiveness and patient outcome in breast cancer that do not include the cyclin E1 gene (62, 69, 73), cyclin E2 may be targeted independently of cyclin E1 in other contexts relevant to breast cancer. High cyclin E2 expression is slightly more common in ER-negative than ER-positive patients, consistent with the observation that ER-negative breast cancers commonly display increased expression of many estrogen-responsive genes (1). The cyclin E2 gene has been associated with poor outcome in ER-positive but not ER-negative cancers (60), reminiscent of the poorer outcome associated with cyclin D1 overexpression in ER-positive but not ER-negative breast cancers (36). The differences that we have observed in the regulation of the activity of the two E-type cyclins raise questions about previous conclusions made about cyclin “E.” Previously, cyclin E1 expression in breast cancer has been compared to cyclin D1 expression, with the conclusion that patients with high levels of cyclin E and high levels of cyclin D1 segregate into distinct groups, often with distinct patient outcomes (30, 40). Based on our study, it would be appropriate to reexamine these associations and to determine whether there is an association between cyclin D1 and cyclin E2 and any relationship with disease progression and response to therapy in breast cancer.
Acknowledgments
This research was supported by the National Health and Medical Research Council of Australia, the Cancer Institute NSW, the Australian Cancer Research Foundation (ACRF Unit for the Molecular Genetics of Cancer), KWF Kankerbestrijding, the Petre Foundation, and the R. T. Hall Trust. J.S.C. acknowledges support from the University of Cambridge, Cancer Research UK, and Hutchison Whampoa Limited. E.A.M. is a Cancer Institute NSW Fellow.
Footnotes
Published ahead of print on 29 June 2009.
REFERENCES
- 1.Alles, C. M., M. Gardiner-Garden, D. J. Nott, Y. Wang, J. A. Foekens, R. L. Sutherland, E. A. Musgrove, and C. J. Ormandy. 2009. Meta-analysis and gene set enrichment relative to ER status reveal elevated activity of MYC and E2F in the “basal” breast cancer subgroup. PLoS ONE 4e4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beier, R., A. Burgin, A. Kiermaier, M. Fero, H. Karsunky, R. Saffrich, T. Moroy, W. Ansorge, J. Roberts, and M. Eilers. 2000. Induction of cyclin E-cdk2 kinase activity, E2F-dependent transcription and cell growth by Myc are genetically separable events. EMBO J. 195813-5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieda, M., X. Xu, M. A. Singer, R. Green, and P. J. Farnham. 2006. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 16595-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bortner, D. M., and M. P. Rosenberg. 1997. Induction of mammary gland hyperplasia and carcinomas in transgenic mice expressing human cyclin E. Mol. Cell. Biol. 17453-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouchard, C., J. Marquardt, A. Bras, R. H. Medema, and M. Eilers. 2004. Myc-induced proliferation and transformation require Akt-mediated phosphorylation of FoxO proteins. EMBO J. 232830-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butt, A. J., C. E. Caldon, C. M. McNeil, A. Swarbrick, E. A. Musgrove, and R. L. Sutherland. 2008. Cell cycle machinery: links with genesis and treatment of breast cancer Adv. Exp. Med. Biol. 630189-205. [DOI] [PubMed] [Google Scholar]
- 7.Butt, A. J., C. M. McNeil, E. A. Musgrove, and R. L. Sutherland. 2005. Downstream targets of growth factor and oestrogen signalling and endocrine resistance: the potential roles of c-Myc, cyclin D1 and cyclin E. Endocr. Relat. Cancer 12S47-S59. [DOI] [PubMed] [Google Scholar]
- 8.Butt, A. J., C. M. Sergio, C. K. Inman, L. R. Anderson, C. M. McNeil, A. J. Russell, M. Nousch, T. Preiss, A. V. Biankin, R. L. Sutherland, and E. A. Musgrove. 2008. The estrogen and c-Myc target gene HSPC111 is over-expressed in breast cancer and associated with poor patient outcome. Breast Cancer Res. 10R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calbo, J., M. Parreno, E. Sotillo, T. Yong, A. Mazo, J. Garriga, and X. Grana. 2002. G1 cyclin/cyclin-dependent kinase-coordinated phosphorylation of endogenous pocket proteins differentially regulates their interactions with E2F4 and E2F1 and gene expression. J. Biol. Chem. 27750263-50274. [DOI] [PubMed] [Google Scholar]
- 10.Carroll, J. S., X. S. Liu, A. S. Brodsky, W. Li, C. A. Meyer, A. J. Szary, J. Eeckhoute, W. Shao, E. V. Hestermann, T. R. Geistlinger, E. A. Fox, P. A. Silver, and M. Brown. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 12233-43. [DOI] [PubMed] [Google Scholar]
- 11.Carroll, J. S., C. A. Meyer, J. Song, W. Li, T. R. Geistlinger, J. Eeckhoute, A. S. Brodsky, E. K. Keeton, K. C. Fertuck, G. F. Hall, Q. Wang, S. Bekiranov, V. Sementchenko, E. A. Fox, P. A. Silver, T. R. Gingeras, X. S. Liu, and M. Brown. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 381289-1297. [DOI] [PubMed] [Google Scholar]
- 12.Carroll, J. S., O. W. J. Prall, E. A. Musgrove, and R. L. Sutherland. 2000. A pure estrogen antagonist inhibits cyclin E-cdk2 activity in MCF-7 breast cancer cells and induces accumulation of p130-E2F4 complexes characteristic of quiescence. J. Biol. Chem. 27538221-38229. [DOI] [PubMed] [Google Scholar]
- 13.Cicatiello, L., R. Addeo, A. Sasso, L. Altucci, V. B. Petrizzi, R. Borgo, M. Cancemi, S. Caporali, S. Caristi, C. Scafoglio, D. Teti, F. Bresciani, B. Perillo, and A. Weisz. 2004. Estrogens and progesterone promote persistent CCND1 gene activation during G1 by inducing transcriptional derepression via c-Jun/c-Fos/estrogen receptor (progesterone receptor) complex assembly to a distal regulatory element and recruitment of cyclin D1 to its own gene promoter. Mol. Cell. Biol. 247260-7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coser, K. R., J. Chesnes, J. Hur, S. Ray, K. J. Isselbacher, and T. Shioda. 2003. Global analysis of ligand sensitivity of estrogen inducible and suppressible genes in MCF7/BUS breast cancer cells by DNA microarray. Proc. Natl. Acad. Sci. USA 10013994-13999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desmedt, C., F. E. Ouriaghli, V. Durbecq, A. Soree, M. A. Colozza, E. Azambuja, M. Paesmans, D. Larsimont, M. Buyse, A. Harris, M. Piccart, P. Martiat, and C. Sotiriou. 2006. Impact of cyclins E, neutrophil elastase and proteinase 3 expression levels on clinical outcome in primary breast cancer patients. Int. J. Cancer 1192539-2545. [DOI] [PubMed] [Google Scholar]
- 16.Dhillon, N. K., and M. Mudryj. 2002. Ectopic expression of cyclin E in estrogen responsive cells abrogates antiestrogen mediated growth arrest. Oncogene 214626-4634. [DOI] [PubMed] [Google Scholar]
- 17.Dubik, D., and R. P. C. Shiu. 1992. Mechanism of estrogen activation of c-myc oncogene expression. Oncogene 71587-1594. [PubMed] [Google Scholar]
- 18.Eeckhoute, J., J. S. Carroll, T. R. Geistlinger, M. I. Torres-Arzayus, and M. Brown. 2006. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 202513-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ekholm-Reed, S., J. Mendez, D. Tedesco, A. Zetterberg, B. Stillman, and S. I. Reed. 2004. Deregulation of cyclin E in human cells interferes with prereplication complex assembly. J. Cell Biol. 165789-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farkas, T., K. Hansen, K. Holm, J. Lukas, and J. Bartek. 2002. Distinct phosphorylation events regulate p130- and p107-mediated repression of E2F-4. J. Biol. Chem. 27726741-26752. [DOI] [PubMed] [Google Scholar]
- 21.Foster, J. S., D. C. Henley, S. Ahamed, and J. Wimalasena. 2001. Estrogens and cell-cycle regulation in breast cancer. Trends Endocrinol. Metab. 12320-327. [DOI] [PubMed] [Google Scholar]
- 22.Foster, J. S., D. C. Henley, A. Bukovsky, P. Seth, and J. Wimalasena. 2001. Multifaceted regulation of cell cycle progression by estrogen: regulation of Cdk inhibitors and Cdc25A independent of cyclin D1-Cdk4 function. Mol. Cell. Biol. 21794-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galaktionov, K., X. Chen, and D. Beach. 1996. Cdc25 cell-cycle phosphatase as a target of c-myc. Nature 382511-517. [DOI] [PubMed] [Google Scholar]
- 24.Gartel, A. L., X. Ye, E. Goufman, P. Shianov, N. Hay, F. Najmabadi, and A. L. Tyner. 2001. Myc represses the p21(WAF1/CIP1) promoter and interacts with Sp1/Sp3. Proc. Natl. Acad. Sci. USA 984510-4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geng, Y., Y.-M. Lee, M. Welcker, J. Swanger, A. Zagozdzon, J. D. Winer, J. M. Roberts, P. Kaldis, B. E. Clurman, and P. Sicinski. 2007. Kinase-independent function of cyclin E. Mol. Cell 25127-139. [DOI] [PubMed] [Google Scholar]
- 26.Geng, Y., Q. Yu, E. Sicinska, M. Das, J. E. Schneider, S. Bhattacharya, W. M. Rideout, R. T. Bronson, H. Gardner, and P. Sicinski. 2003. Cyclin E ablation in the mouse. Cell 114431-443. [DOI] [PubMed] [Google Scholar]
- 27.Geng, Y., Q. Yu, W. Whoriskey, F. Dick, K. Y. Tsai, H. L. Ford, D. K. Biswas, A. B. Pardee, B. Amati, T. Jacks, A. Richardson, N. Dyson, and P. Sicinski. 2001. Expression of cyclins E1 and E2 during mouse development and in neoplasia. Proc. Natl. Acad. Sci. USA 9813138-13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gudas, J. M., M. Payton, S. Thukral, E. Chen, M. Bass, M. O. Robinson, and S. Coats. 1999. Cyclin E2, a novel G1 cyclin that binds Cdk2 and is aberrantly expressed in human cancers. Mol. Cell. Biol. 19612-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo, Q. M., R. L. Malek, S. Kim, C. Chiao, M. He, M. Ruffy, K. Sanka, N. H. Lee, C. V. Dang, and E. T. Liu. 2000. Identification of c-Myc responsive genes using rat cDNA microarray. Cancer Res. 605922-5928. [PubMed] [Google Scholar]
- 30.Han, S., K. Park, B.-N. Bae, K. H. Kim, H.-J. Kim, Y.-D. Kim, and H.-Y. Kim. 2003. Prognostic implication of cyclin E expression and its relationship with cyclin D1 and p27Kip1 expression on tissue microarrays of node negative breast cancer. J. Surg. Oncol. 83241-247. [DOI] [PubMed] [Google Scholar]
- 31.Henderson, B. E., R. Ross, and L. Bernstein. 1988. Estrogens as a cause of human cancer: the Richard and Hinda Rosenthal Foundation Award lecture. Cancer Res. 48246-253. [PubMed] [Google Scholar]
- 32.Hinds, P. W., S. F. Dowdy, E. N. Eaton, A. Arnold, and R. A. Weinberg. 1994. Function of a human cyclin gene as an oncogene. Proc. Natl. Acad. Sci. USA 91709-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hui, R., G. L. Finney, J. S. Carroll, C. S. L. Lee, E. A. Musgrove, and R. L. Sutherland. 2002. Constitutive overexpression of cyclin D1 but not cyclin E confers acute resistance to antiestrogens in T-47D breast cancer cells. Cancer Res. 626916-6923. [PubMed] [Google Scholar]
- 34.Hwang, H. C., and B. E. Clurman. 2005. Cyclin E in normal and neoplastic cell cycles. Oncogene 242776-2786. [DOI] [PubMed] [Google Scholar]
- 35.Kawamura, K., H. Izumi, Z. Ma, R. Ikeda, M. Moriyama, T. Tanaka, T. Nojima, L. S. Levin, K. Fujikawa-Yamamoto, K. Suzuki, and K. Fukasawa. 2004. Induction of centrosome amplification and chromosome instability in human bladder cancer cells by p53 mutation and cyclin E overexpression. Cancer Res. 644800-4809. [DOI] [PubMed] [Google Scholar]
- 36.Kenny, F. S., R. Hui, E. A. Musgrove, J. M. W. Gee, R. W. Blamey, R. I. Nicholson, R. L. Sutherland, and J. F. R. Robertson. 1999. Overexpression of cyclin D1 messenger RNA predicts for poor prognosis in estrogen receptor-positive breast cancer. Clin. Cancer Res. 52069-2076. [PubMed] [Google Scholar]
- 37.Lauper, N., A. R. Beck, S. Cariou, L. Richman, K. Hofmann, W. Reith, J. M. Slingerland, and B. Amati. 1998. Cyclin E2: a novel CDK2 partner in the late G1 and S phases of the mammalian cell cycle. Oncogene 172637-2643. [DOI] [PubMed] [Google Scholar]
- 38.Leone, G., R. Sears, E. Huang, R. Rempel, F. Nuckolls, C.-H. Park, P. Giangrande, L. Wu, H. I. Saavedra, S. J. Field, M. A. Thompson, H. Yang, Y. Fujiwara, M. E. Greenberg, S. Orkin, C. Smith, and J. R. Nevins. 2001. Myc requires distinct E2F activities to induce S phase and apoptosis. Mol. Cell 8105-113. [DOI] [PubMed] [Google Scholar]
- 39.Li, Z., S. Van Calcar, C. Qu, W. K. Cavenee, M. Q. Zhang, and B. Ren. 2003. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc. Natl. Acad. Sci. USA 1008164-8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loden, M., M. Stighall, N. H. Nielsen, G. Roos, S. O. Emdin, H. Ostlund, and G. Landberg. 2002. The cyclin D1 high and cyclin E high subgroups of breast cancer: separate pathways in tumorogenesis based on pattern of genetic aberrations and inactivation of the pRb node. Oncogene 214680-4690. [DOI] [PubMed] [Google Scholar]
- 41.Lundberg, A. S., and R. A. Weinberg. 1998. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol. Cell. Biol. 18753-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsumoto, Y., and J. L. Maller. 2004. A centrosomal localization signal in cyclin E required for Cdk2-independent S phase entry. Science 306885-888. [DOI] [PubMed] [Google Scholar]
- 43.Mukherjee, S., and S. E. Conrad. 2005. c-Myc suppresses p21WAF1/CIP1 expression during estrogen signaling and antiestrogen resistance in human breast cancer cells. J. Biol. Chem. 28017617-17625. [DOI] [PubMed] [Google Scholar]
- 44.Mullany, L. K., P. White, E. A. Hanse, C. J. Nelsen, M. M. Goggin, J. E. Mullany, C. K. Anttila, L. E. Greenbaum, K. H. Kaestner, and J. H. Albrecht. 2008. Distinct proliferative and transcriptional effects of the D-type cyclins in vivo. Cell Cycle 72215-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mumberg, D., K. Haas, T. Moroy, R. Niedenthal, J. H. Hegemann, M. Funk, and R. Muller. 1996. Uncoupling of DNA replication and cell cycle progression by human cyclin E. Oncogene 132493-2497. [PubMed] [Google Scholar]
- 46.Musgrove, E. A., L.-J. K. Hunter, C. S. L. Lee, A. Swarbrick, R. Hui, and R. L. Sutherland. 2001. Cyclin D1 overexpression induces progestin resistance in T-47D breast cancer cells despite p27Kip1 association with cyclin E-Cdk2. J. Biol. Chem. 27647675-47683. [DOI] [PubMed] [Google Scholar]
- 47.Musgrove, E. A., C. S. L. Lee, M. F. Buckley, and R. L. Sutherland. 1994. Cyclin D1 induction in breast cancer cells shortens G1 and is sufficient for cells arrested in G1 to complete the cell cycle. Proc. Natl. Acad. Sci. USA 918022-8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Musgrove, E. A., C. M. Sergio, S. Loi, C. K. Inman, L. R. Anderson, M. C. Alles, M. Pinese, C. E. Caldon, J. Schutte, M. Gardiner-Garden, C. J. Ormandy, G. McArthur, A. J. Butt, and R. L. Sutherland. 2008. Identification of functional networks of estrogen- and c-Myc-responsive genes and their relationship to response to tamoxifen therapy in breast cancer. PLoS One 3e2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mussman, J. G., H. F. Horn, P. E. Carroll, M. Okuda, P. Tarapore, L. A. Donehower, and K. Fukasawa. 2000. Synergistic induction of centrosome hyperamplification by loss of p53 and cyclin E overexpression. Oncogene 191635-1646. [DOI] [PubMed] [Google Scholar]
- 50.Ohtani, K., J. DeGregori, and J. R. Nevins. 1995. Regulation of the cyclin E gene by transcription factor E2F1. Proc. Natl. Acad. Sci. USA 9212146-12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Payton, M., and S. Coats. 2002. Cyclin E2, the cycle continues. Int. J. Biochem. Cell Biol. 34315-320. [DOI] [PubMed] [Google Scholar]
- 52.Planas-Silva, M. D., and R. A. Weinberg. 1997. Estrogen-dependent cyclin E-cdk2 activation through p21 redistribution. Mol. Cell. Biol. 174059-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Polager, S., and D. Ginsberg. 2008. E2F—at the crossroads of life and death. Trends Cell Biol. 18528-535. [DOI] [PubMed] [Google Scholar]
- 54.Prall, O. W. J., J. S. Carroll, and R. L. Sutherland. 2001. A low abundance pool of nascent p21WAF1/Cip1 is targeted by estrogen to activate cyclin E-Cdk2. J. Biol. Chem. 27645433-45442. [DOI] [PubMed] [Google Scholar]
- 55.Prall, O. W. J., E. M. Rogan, E. A. Musgrove, C. K. W. Watts, and R. L. Sutherland. 1998. c-Myc or cyclin D1 mimics estrogen effects on cyclin E-Cdk2 activation and cell cycle reentry. Mol. Cell. Biol. 184499-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prall, O. W. J., B. Sarcevic, E. A. Musgrove, C. K. W. Watts, and R. L. Sutherland. 1997. Estrogen-induced activation of Cdk4 and Cdk2 during G1-S phase progression is accompanied by increased cyclin D1 expression and decreased cyclin-dependent kinase inhibitor association with cyclin E-Cdk2. J. Biol. Chem. 27210882-10894. [DOI] [PubMed] [Google Scholar]
- 57.Rasband, W. S. 1997-2007. ImageJ. U.S. National Institutes of Health, Bethesda, MD. http://rsb.info.nih.gov/ij/.
- 58.Rodriguez-Paredes, M., M. Ceballos-Chavez, M. Esteller, M. Garcia-Dominguez, and J. C. Reyes. 2009. The chromatin remodeling factor CHD8 interacts with elongating RNA polymerase II and controls expression of the cyclin E2 gene. Nucleic Acids Res. doi: 10.1093/nar/gkp101. [DOI] [PMC free article] [PubMed]
- 59.Scime, A., L. Li, G. Ciavarra, and P. Whyte. 2008. Cyclin D1/cdk4 can interact with E2F4/DP1 and disrupts its DNA-binding capacity. J. Cell. Physiol. 214568-581. [DOI] [PubMed] [Google Scholar]
- 60.Sieuwerts, A. M., M. P. Look, M. E. Meijer-van Gelder, M. Timmermans, A. M. Trapman, R. R. Garcia, M. Arnold, A. J. Goedheer, V. de Weerd, H. Portengen, J. G. Klijn, and J. A. Foekens. 2006. Which cyclin E prevails as prognostic marker for breast cancer? Results from a retrospective study involving 635 lymph node-negative breast cancer patients. Clin. Cancer Res. 123319-3328. [DOI] [PubMed] [Google Scholar]
- 61.Smith, A. P., M. Henze, J. A. Lee, K. G. Osborn, J. M. Keck, D. Tedesco, D. M. Bortner, M. P. Rosenberg, and S. I. Reed. 2006. Deregulated cyclin E promotes p53 loss of heterozygosity and tumorigenesis in the mouse mammary gland. Oncogene 257245-7259. [DOI] [PubMed] [Google Scholar]
- 62.Sotiriou, C., P. Wirapati, S. Loi, A. Harris, S. Fox, J. Smeds, H. Nordgren, P. Farmer, V. Praz, B. Haibe-Kains, C. Desmedt, D. Larsimont, F. Cardoso, H. Peterse, D. Nuyten, M. Buyse, M. J. Van de Vijver, J. Bergh, M. Piccart, and M. Delorenzi. 2006. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst. 98262-272. [DOI] [PubMed] [Google Scholar]
- 63.Spruck, C. H., K. A. Won, and S. I. Reed. 1999. Deregulated cyclin E induces chromosome instability. Nature 401297-300. [DOI] [PubMed] [Google Scholar]
- 64.Srinivasan, S., J. A. Armstrong, R. Deuring, I. K. Dahlsveen, H. McNeill, and J. W. Tamkun. 2005. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA polymerase II. Development 1321623-1635. [DOI] [PubMed] [Google Scholar]
- 65.Stanelle, J., T. Stiewe, C. C. Theseling, M. Peter, and B. M. Putzer. 2002. Gene expression changes in response to E2F1 activation. Nucleic Acids Res. 301859-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stender, J. D., J. Frasor, B. Komm, K. C. Chang, W. L. Kraus, and B. S. Katzenellenbogen. 2007. Estrogen-regulated gene networks in human breast cancer cells: involvement of E2F1 in the regulation of cell proliferation. Mol. Endocrinol. 212112-2123. [DOI] [PubMed] [Google Scholar]
- 67.Swarbrick, A., C. S. L. Lee, R. L. Sutherland, and E. A. Musgrove. 2000. Cooperation of p27Kip1 and p18INK4c in progestin-mediated cell cycle arrest in T-47D breast cancer cells. Mol. Cell. Biol. 202581-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tu, Z., S. Prajapati, K. J. Park, N. J. Kelly, Y. Yamamoto, and R. B. Gaynor. 2006. IKK alpha regulates estrogen-induced cell cycle progression by modulating E2F1 expression. J. Biol. Chem. 2816699-6706. [DOI] [PubMed] [Google Scholar]
- 69.van 't Veer, L. J., H. Dai, M. J. van de Vijver, Y. D. He, A. A. Hart, M. Mao, H. L. Peterse, K. van der Kooy, M. J. Marton, A. T. Witteveen, G. J. Schreiber, R. M. Kerkhoven, C. Roberts, P. S. Linsley, R. Bernards, and S. H. Friend. 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415530-536. [DOI] [PubMed] [Google Scholar]
- 70.Varma, H., A. J. Skildum, and S. E. Conrad. 2007. Functional ablation of pRb activates Cdk2 and causes antiestrogen resistance in human breast cancer cells. PLoS One 2e1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vigo, E., H. Muller, E. Prosperini, G. Hateboer, P. Cartwright, M. C. Moroni, and K. Helin. 1999. CDC25A phosphatase is a target of E2F and is required for efficient E2F-induced S phase. Mol. Cell. Biol. 196379-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang, W., L. Dong, B. Saville, and S. Safe. 1999. Transcriptional activation of E2F1 gene expression by 17beta-estradiol in MCF-7 cells is regulated by NF-Y-Sp1/estrogen receptor interactions. Mol. Endocrinol. 131373-1387. [DOI] [PubMed] [Google Scholar]
- 73.Wang, Y., J. G. Klijn, Y. Zhang, A. M. Sieuwerts, M. P. Look, F. Yang, D. Talantov, M. Timmermans, M. E. Meijer-van Gelder, J. Yu, T. Jatkoe, E. M. Berns, D. Atkins, and J. A. Foekens. 2005. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365671-679. [DOI] [PubMed] [Google Scholar]
- 74.Young, A. P., R. Nagarajan, and G. D. Longmore. 2003. Mechanisms of transcriptional regulation by Rb-E2F segregate by biological pathway. Oncogene 227209-7217. [DOI] [PubMed] [Google Scholar]
- 75.Yu, Q., Y. Geng, and P. Sicinski. 2001. Specific protection against breast cancers by cyclin D1 ablation. Nature 4111017-1021. [DOI] [PubMed] [Google Scholar]
- 76.Zariwala, M., J. Liu, and Y. Xiong. 1998. Cyclin E2, a novel human G1 cyclin and activating partner of CDK2 and CDK3, is induced by viral oncoproteins. Oncogene 172787-2798. [DOI] [PubMed] [Google Scholar]