Abstract
Oxidized bases are common types of DNA modifications. Their accumulation in the genome is linked to aging and degenerative diseases. These modifications are commonly repaired by the base excision repair (BER) pathway. Oxoguanine DNA glycosylase (OGG1) initiates BER of oxidized purine bases. A small number of protein interactions have been identified for OGG1, while very few appear to have functional consequences. We report here that OGG1 interacts with the recombination protein RAD52 in vitro and in vivo. This interaction has reciprocal functional consequences as OGG1 inhibits RAD52 catalytic activities and RAD52 stimulates OGG1 incision activity, likely increasing its turnover rate. RAD52 colocalizes with OGG1 after oxidative stress to cultured cells, but not after the direct induction of double-strand breaks by ionizing radiation. Human cells depleted of RAD52 via small interfering RNA knockdown, and mouse cells lacking the protein via gene knockout showed increased sensitivity to oxidative stress. Moreover, cells depleted of RAD52 show higher accumulation of oxidized bases in their genome than cells with normal levels of RAD52. Our results indicate that RAD52 cooperates with OGG1 to repair oxidative DNA damage and enhances the cellular resistance to oxidative stress. Our observations suggest a coordinated action between these proteins that may be relevant when oxidative lesions positioned close to strand breaks impose a hindrance to RAD52 catalytic activities.
Oxidative DNA damage is generated at high levels in mammalian cells, even in cells not exposed to exogenous sources of reactive oxygen species. Several kinds of DNA modifications are formed upon oxidative stress (8). The most prevalent modifications, quantitatively, are single-strand breaks and oxidized bases. Clustered DNA damage, when two or more modifications are closely positioned in opposite strands, is detectable after gamma irradiation and has recently been shown to be generated by normal oxidative metabolism (3, 35). One unique aspect of such clustered lesions is that they can be converted into double-strand breaks (DSB) if a DNA glycosylase removes the two opposite bases and an apurinic/apyrimidinic (AP)-endonuclease cleaves the resulting abasic sites. Thus, although quantitatively minor, DSB are possible outcomes of oxidative DNA damage.
Oxidized DNA bases are repaired primarily by the base excision repair pathway (BER) (22, 39). BER is initiated by a lesion-specific DNA N-glycosylase that recognizes and excises the damaged base. Eight-hydroxyguanine (8-oxoG) is one of the most abundant oxidized bases detected in cellular DNA. This adduct is easily bypassed by replicative polymerases; however, it can direct the misincorporation of adenine opposite 8-oxoG, thus leading to G·C-to-T·A transversion mutations (31). 8-oxoG accumulation has been causally associated with carcinogenesis and aging in several experimental models (1, 12). In eukaryotes, oxoguanine DNA glycosylase (OGG1) is the major 8-oxoG DNA glycosylase. OGG1 possesses an associated AP-lyase activity, such that it removes 8-oxoG and cleaves the DNA backbone. Human cells express two distinct OGG1 isoforms, α and β, which share the first 316 amino acids but differ significantly in their C termini (25). While OGG1-α is a bone fide DNA glycosylase (5) and localizes both to nuclei and mitochondria, OGG1-β localizes exclusively to mitochondria. We recently showed that the recombinant OGG1-β protein has no DNA glycosylase activity (13). The high degree of conservation of repair pathways for 8-oxoG, from bacteria to humans, along with epidemiological data correlating OGG1 polymorphisms and activity with predisposition to some cancers (11, 27, 33) attest to the biological importance of the repair of 8-oxoGs and other oxidative DNA lesions.
Until recently, distinct classes of DNA lesions were believed to be metabolized by different and independent repair pathways. However, experimental evidence indicates that these pathways can interact and that there is a considerable degree of overlap in their substrate specificity and in the proteins that participate in each pathway. Experiments using yeast strains lacking one or more distinct DNA repair genes suggest that DSB repair pathways may play a role in repair of oxidative DNA damage. Swanson et al. showed that while yeast cells lacking ntg1 and ntg2 (homologues of Escherichia coli endonuclease III, a DNA glycosylase specific for pyrimidine lesions formed by oxidation) and apn1 (the major yeast abasic site endonuclease) are not overtly sensitive to oxidative stress, the additional disruption of the rad52 gene significantly increases sensitivity to H2O2 and menadione (36). Similarly, yeast cells expressing decreased levels of frataxin, which leads to elevated oxidative stress, show accumulation of oxidative damage in nuclear DNA only in a rad52 mutant background (18). RAD52 is a member of the RAD51 epistatic group. These proteins are believed to be involved in the early steps of homologous recombination, contributing to homology search and strand invasion; disruption of the corresponding genes renders cells deficient in DSB repair and hyper-recombinogenic (19).
These results suggested a possible role for RAD52 in the repair of oxidative DNA damage. Moreover, an in vitro screening of protein partners that interact physically with OGG1-β performed in our lab (unpublished data) showed that human RAD52 strongly interacted with this glycosylase, again suggesting a possible function for RAD52 in the oxidative DNA damage response. Thus, we investigated whether RAD52 plays a role in the repair of oxidative DNA damage in human cells. We show here that human RAD52 physically interacts with both OGG1-α and -β, in vitro and in cell extracts. We also show that OGG1-α and -β inhibit RAD52 enzymatic activities. Conversely, RAD52 stimulates OGG1-α 8-oxoG incision activity. RAD52 colocalizes with OGG1-α in cells, and this colocalization increases after oxidative stress. Moreover, lower RAD52 expression, via gene knockdown (KD) or disruption of the RAD52 gene, render cells sensitive to oxidative stress. Based on our results, we discuss a model in which OGG1 and RAD52 cooperate to repair 8-oxoG lesions.
MATERIALS AND METHODS
Proteins and DNA substrates.
Purification of full-length, His6-tagged human RAD52 has been described previously (2). Purification of recombinant His-tagged OGG1-α from Sf9 insect cells and His- and glutathione S-transferase-tagged OGG1-β from E. coli has been described (13). Oligonucleotides were obtained from Midland Certified Reagent Co., and their sequences are shown in Table 1. 5′-End labeling was carried out using T4 polynucleotides kinase (New England Biolabs) and [γ-32P]ATP. Unincorporated [32P]ATP was removed with a Sephadex G25 spin column (GE Healthcare).
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′-3′)a |
|---|---|
| G50 | AATTCAATTCATATACCGCGGCCGGCCGATCAAGCTTATTAGCCGAGCCG |
| TTAAGTTAAGTATATGGCGCCGGCCGGCTAGTTCGAATAATCGGCTCGGC | |
| OG50 | AATTCAATTCATATACCGCGogCCGGCCGATCAAGCTTATTAGCCGAGCCG |
| TTAAGTTAAGTATATGGCGCCGGCCGGCTAGTTCGAATAATCGGCTCGGC | |
| OG30 | ATATACCGCGogCCGGCCGATCAAGCTTATT |
| TATATGGCGCCGGCCGGCATGTTCGAATAA | |
| C80 | GCTGATCAACCCTACATGTGTAGGTAACCCTAACCCTAACCCTAAGGACAACCCTAGTGAAGCTTGTAACCCTAGGAGCT |
| G80 | AGCTCCTAGGGTTACAAGCTTCACTAGGGTTGTCCTTAGGGTTAGGGTTAGGGTTACCTACACATGTAGGGTTGATCAGC |
| G80fork26 | CTTCACAGTCAGAGTCACAGTGTGCCGGGTTGTCCTTAGGGTTAGGGTTAGGGTTACCTACACATGTAGGGTTGATCAGC |
The 8-oxoG (OG) modifications are indicated by boldfacing and underlining. In the G80fork26 oligonucleotide, the unpaired region is underlined.
Cell culture.
HEK293 cells (from the American Type Culture Collection) harboring OGG1-α-FLAG or the empty vector were described earlier (16). The cells were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (Gibco-BRL) and 800 μg of Geneticin (Invitrogen)/ml at 37°C under an atmosphere of 5% CO2. HeLa cells were grown in DMEM supplemented with 10% fetal bovine serum, 50 μg of streptomycin/ml, and 50 U of penicillin/ml. Primary mouse skin fibroblasts (MSF) were prepared from mouse tail snips (2 to 5 cm) by collagenase (Sigma) digestion according to the protocol described by Maynard and Miller (21).
ELISA.
A 96-well enzyme-linked immunosorbent assay (ELISA) plate (50 μl/well) was coated with recombinant RAD52, at 1 ng/μl in carbonate buffer (0.016 M NaCO3, 0.034 M NaHCO3 [pH 9.6]) overnight at 4°C; control wells were coated with bovine serum albumin (BSA; Sigma). Wells were aspirated and blocked with 100 μl of blocking buffer (3% BSA in phosphate-buffered saline [PBS]) for 1 h at room temperature. Serial dilutions of OGG1-α or OGG1-β, diluted in blocking buffer, were added to appropriate wells and incubated for 2 h at 37°C. After five washes with 100 μl of washing buffer (PBS plus 0.1% Tween 20), primary monoclonal anti-OGG1-α (Assay Designs, Inc.) diluted 1:100 in blocking buffer or primary polyclonal anti-OGG1-β antibody (Novus Biologicals) diluted 1:1,000 in blocking buffer were added to appropriate wells, followed by incubation for 2 h at 37°C. The wells were washed and incubated with secondary antibodies (1:1,000 in blocking buffer) for 1 h at 37°C. The immunocomplexes were detected by using o-phenylenediamine dihydrochloride (OPD) substrate (Sigma) in developing solution (50 mM phosphate citrate buffer [pH 5.0] containing 0.02% hydrogen peroxide). The reactions were stopped after 3 min with 3 M H2SO4 (25 μl/well), and absorbance readings were taken at 490 nm. Dissociation constants were calculated by using Hill plot analyses of the ELISA data as described elsewhere (6).
DNA strand annealing assay.
DNA strand annealing activity was determined by using two complementary synthetic 50-mer oligonucleotides (G50 in Table 1) in which one strand was radioactively labeled at the 5′ end. The assay (50 μl) was carried out in buffer containing 40 mM Tris-HCl (pH 8.0), 50 mM NaCl, 1 mM dithiothreitol (DTT), and 0.1 mg of BSA/ml. Oligonucleotides (5.6 pmol of each) were equilibrated at room temperature before use. The purified RAD52 or the mixture of RAD52 and OGG1, preincubated in ice for 5 min, were combined with the unlabeled DNA strand, and the reactions were initiated by adding the labeled DNA strand, followed by incubation at room temperature. Aliquots (9 μl) were removed at 0, 1, 2, 4, and 8 min to tubes containing 7 μl of stop solution (2.5% sodium dodecyl sulfate [SDS], 2.5 mg of proteinase K/ml, and 20 pmol of unlabeled oligonucleotide 1), followed by incubation for 15 min at room temperature. Control reactions, initiated with enzyme dilution buffer (20 mM Tris-HCl [pH 7.4], 30 mM NaCl, and 0.1 mg of BSA/ml) were carried out to determine the spontaneous rate of annealing. Loading dye (16 μl, 25% glycerol, 0.04% bromophenol blue, and 0.04% xylene cyanol) was added to each sample, and the products were resolved on native 12% polyacrylamide gels, visualized by using a PhosphorImager, and analyzed by using ImageQuant software (GE Healthcare).
DNA strand exchange assay.
The strand exchange assay was performed as described previously (24), with some modifications. Briefly, RAD52 (32 nM) was preincubated with unlabeled G80 oligonucleotide (1 nM) for 5 min at 37°C, followed by the addition of the labeled fork substrate C80/G80fork26 (1 nM) incubated for 25 min at 37°C with or without the addition of increasing concentrations of OGG1-α or OGG1-β (32 and 128 nM). Reactions were terminated by the addition of 10 μl of stop buffer (35 mM EDTA, 0.6% SDS, 25% glycerol, 0.04% bromophenol blue, and 0.04% xylene cyanol). Products were separated by polyacrylamide gel electrophoresis (PAGE) on 10% native polyacrylamide gels, visualized by using a PhosphorImager, and quantified by using ImageQuant software.
DNA incision activity assay.
OGG1 incision activity was assayed as described previously (13), with a duplex DNA substrate containing an 8-oxoG/C lesion at the eleventh nucleotide position (OG30). The incision reaction (20 μl) contained 50 mM Tris-HCl (pH 7.4), 50 mM NaCl, 2 mM EDTA, 0.1 mg of BSA/ml, and the amounts of RAD52 and OGG1-α indicated in the figures. The reactions were initiated by adding 89 fmol of 8-oxoG/C containing oligonucleotide, followed by incubation at 37°C for 20 min. To promote complete strand cleavage at the abasic sites, the reactions were stopped with an equal volume (20 μl) of formamide loading dye supplemented with 100 mM NaOH and heated at 95°C for 5 min. The substrates and products were resolved in a 20% acrylamide-7 M urea gel, visualized by using a PhosphorImager, and analyzed with ImageQuant software. To detect DNA glycosylase-oligonucleotide complexes, 100 mM NaBH4 was added to the reaction buffer as for the DNA incision assay. After 15 min of incubation at 37°C, reactions were boiled in SDS-PAGE sample buffer, and the products were separated by SDS-PAGE and visualized by using a PhosphorImager.
Electrophoretic mobility shift assay.
32P-labeled oligonucleotide substrates (10 nM) were incubated with the indicated amounts of purified proteins in 10 μl of reaction buffer (50 mM Tris-HCl [pH 7.4], 50 mM NaCl, 2 mM EDTA, 100 μg of BSA/ml) on ice for 30 min. Samples were directly loaded onto a 5% polyacrylamide gel containing 5% glycerol in 1× Tris-borate-EDTA buffer, and the gel was run at 200 V constant for 2 h at 4°C. The radioactivity was analyzed as described above.
Immunoprecipitation assay.
HEK293 cells expressing FLAG-OGG1-α or empty vector (one 150-mm2 plate per condition) were grown to near confluence, treated as described in the figure legend, washed twice in PBS, and lysed at 4°C in lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and complete protease inhibitor cocktail [Roche]; 1.5 ml/plate). The lysates were centrifuged at 20,000 × g for 20 min at 4°C to remove cellular debris, and 1.5-ml portions of supernatant were collected. For the results presented in Fig. 6C and D, anti-FLAG M2 resin (Sigma) was washed with Tris-buffered saline (TBS), 0.1 M glycine, and TBS three more times to give a 50% slurry. Then, 50 μl of the anti-FLAG M2 resin slurry was added to 1.2 ml of cell lysate, followed by incubation overnight with rotation at 4°C. The resin was spun down at 800 × g for 3 min and washed three times with TBS. FLAG-OGG1-α and its binding proteins were eluted by incubating the resin for 15 min with 20 μl of FLAG peptide competitor (Sigma). The supernatant was mixed with 20 μl of 2× SDS sample buffer (Invitrogen) containing 50 mM DTT, and the samples were boiled 5 min. For the results of Fig. 6E, protein A-agarose (Santa Cruz) was washed three times with wash buffer (0.05 M Tris-HCl [pH 7.5], 0.15 M NaCl) to create a 50% bead slurry. The lysates were precleared with 20 μl of protein A-agarose bead slurry and 1 μg of normal mouse immunoglobulin G (IgG; Santa Cruz) for 2 h. Protein A-agarose slurry (30 μl) and monoclonal anti-RAD52 antibody (6 μg) (GeneTex) were added to 1.5 ml of lysate and incubated overnight with rotation at 4°C. Normal mouse IgG (6 μg) was used as a negative control. The immunoprecipitate was washed three times with wash buffer, and the final bead pellet (∼20 μl) was mixed with 20 μl of 2× SDS sample buffer (Invitrogen) containing 50 mM DTT and boiled for 5 min. In all experiments, the immunoprecipitates were resolved on Tris-glycine gels (Invitrogen), followed by electro blotting to polyvinylidene difluoride membrane, as directed by the manufacturer. The immunoblots were analyzed with anti-FLAG antibody (Sigma), anti-human OGG1 antibody (Novus Biologicals), and polyclonal RAD52 antibody (Santa Cruz) and visualized by using enhanced chemiluminescence (GE Healthcare) or SuperSignal West Femto maximum sensitivity substrate (Pierce) kits.
FIG. 6.
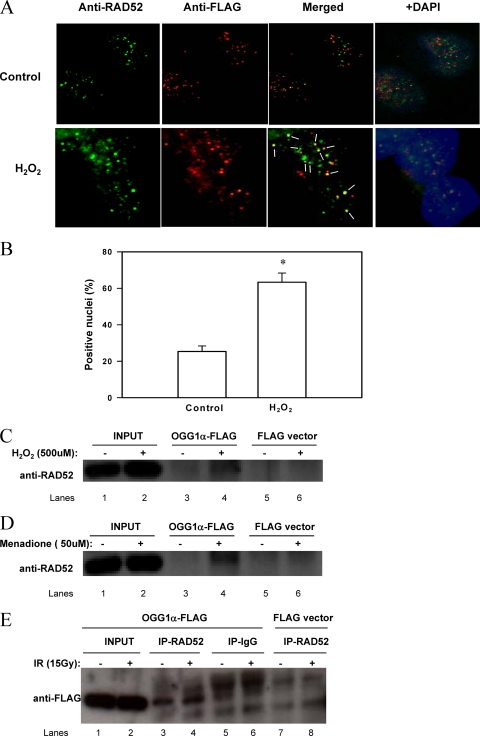
OGG1 and RAD52 colocalize in vivo after DNA damage. (A) HEK293 cells expressing FLAG-OGG1-α were exposed to either buffer alone (Control) or 500 μM H2O2 in medium for 0.5 h and fixed as described. Primary antibodies used were anti-RAD52 (Genetex) at 3.5 μg/ml and anti-FLAG (Sigma) at 2 μg/ml; the secondary antibodies were Alexa Fluor 488-conjugated anti-mouse IgG (green) and Alexa Fluor 568-conjugated anti-rabbit IgG (red) at 1:1,000 dilutions. Nuclei were stained with DAPI. Images were acquired by using a Zeiss fluorescence inverted microscope with a ×40 objective magnification for all images. (B) A total of 50 nuclei per experiment were scored for colocalization of RAD52 and FLAG-OGG1-α. Nuclei were counted as positive if 50% of OGG1-α foci colocalized with RAD52 foci. The data points show the averages ± the standard deviations of three independent experiments. The asterisk denotes significantly higher colocalization at P < 0.01. (C, D, and E) RAD52/OGG1-α complexes were immunoprecipitated from HEK293 cells expressing FLAG-OGG1-α, treated or not with 500 μM hydrogen peroxide for 0.5 h (C) or 50 μM menadione for 1 h (D). From cells irradiated (IR) with 15 Gy (E), protein complexes were precipitated using mouse anti-RAD52 antibodies (or normal IgG, as the negative control). Extracts from HEK293 transfected with empty vector were used as a negative control in all experiments.
Immunofluorescence.
For OGG1/RAD52 colocalization experiments, HEK293 cells expressing FLAG-OGG1-α were seeded in covered slides (Lab-Tek II chamber slide; Fisher) and exposed to 500 μM H2O2 or PBS alone for 30 min at 37°C. The cells were washed twice with fresh PBS and fixed with 3.7% formaldehyde (Polyscience) for 10 min at room temperature. The cells were permeabilized with 0.2% Triton X-100 in 1× PBS for 5 min and blocked overnight at 4°C with 10% normal goat serum in PBS. Anti-FLAG (2 μg/ml) (Sigma) or anti-RAD52 (3.5 μg/ml) (GeneTex) antibodies (in 10% normal goat serum/PBS) were added, and the slides were incubated for 1 h at 37°C. After five washes with PBS, Alexa Fluor 488 anti-mouse (Invitrogen) or Alexa Fluor 568 anti-rabbit (Invitrogen) antibodies were added at a 1:1,000 dilution, followed by incubation for 1 h at 37°C. The slides were mounted with DAPI (4′,6′-diamidino-2-phenylindole)-containing Vectashield (Vector Laboratories) and analyzed by using a Zeiss Axiovert 200M microscope, with a ×40 objective lens magnification for all images. For quantification of colocalization, nuclei were scored as positive if 50% or more of the OGG1-α-FLAG foci colocalized with RAD52 foci. The experiment was done in triplicate (three slides). For each slide, 50 nuclei were scored. For 8-oxoG immunocytochemistry, HeLa cells transfected with scrambled or RAD52 siRNAs were seeded into covered slides and fixed with ice-cold 1:1 methanol-acetone for 20 min. Slides were washed with PBS twice and then treated with RNase (100 μg/ml) in Tris buffer (pH 7.5) at 37°C for 1 h. After being washed with PBS, the slides were treated with proteinase K (10 μg/ml) for 7 min at room temperature. The slides were then rinsed in PBS, the DNA was denatured by treatment with 4 N HCl for 7 min at room temperature, and the pH was adjusted with 50 mM Trizma base for 5 min at room temperature. After being washed with PBS, the slides were blocked overnight at 4°C with 10% normal goat serum-PBS and then incubated in primary anti-8-oxoG antibody (Trevigen) at a 1:250 dilution in 10% goat serum-PBS. After five washes with PBS, the slides were treated with Alexa Fluor 488 anti-mouse (Invitrogen) at a 1:1,000 dilution for 1 h at 37°C. The slides were mounted with DAPI-containing Vectashield (Vector Laboratories) and analyzed by using a Zeiss Axiovert 200M microscope with a ×40 objective lens. Negative controls consisted of secondary antibody-only and normal mouse or rabbit IgG in place of the primary antibody. This procedure was also done on cells treated with 500 μM H2O2 for 0.5 h as a positive control.
siRNA transfections.
RAD52 and OGG1 expression in HeLa cells were transiently knocked down with duplex small interfering RNAs (siRNAs) obtained from Dharmacon (ON-TARGETplus SMARTpool Rad52 [L-011760-00] and OGG1 [M-005147-01]; nontargeting negative control 1 [D-001210-01]). The cells were seeded in six-well dishes at 500,000 per well; after 24 h, 50 nM concentrations of each siRNA were transfected into the cells by using DharmaFECT 1 (Dharmacon) according to the manufacturer's directions. The cells were harvested for further use 48 h after transfection.
Cell viability and growth assays.
For the cytotoxicity assays, 30,000 cells were plated in triplicate onto 96-well microtiter plates in complete growth medium. After 24 h of incubation the cells were washed in PBS and exposed to H2O2 and menadione (dissolved in DMEM), over the range of doses indicated in the figure legends, for 6 and 4 h, respectively. At the end of the treatments, the cells were washed with PBS and incubated in DMEM plus 2% BSA for 18 h to allow time for cell death to occur. The medium was removed, and then 100 μl of fresh medium was added, followed by 10 μl of WST-1 (Roche). After 3 h of incubation in WST-1, the absorbance at 450 nm was determined. For cell growth assays, 2,000 cells were plated in triplicate into 96-well microtiter plates. Cell numbers were measured each day by replacing growth medium with medium containing WST-1 as described above.
DNA analysis by GC-MS.
Genomic DNA was isolated by salting out, as described previously (15). The identification and quantification of FapyGua in DNA was performed by gas chromatography-mass spectrometry (GC-MS) after hydrolysis of the DNA samples by E. coli Fpg. Aliquots (50 μg) of DNA samples were supplemented with 7 pmol of FapyGua-13C,15N2 as internal standards and then hydrolyzed with 2 μg of Fpg. After ethanol precipitation, DNA pellets and supernatant fractions were separated by centrifugation. Supernatant fractions were lyophilized, trimethylsilylated, and analyzed by GC-MS (29). For identification and quantification, selected-ion monitoring was used to monitor the characteristic ions of trimethylsilylated FapyGua and its stable isotope-labeled analogues (9).
RESULTS
RAD52 interacts with OGG1-α and -β in vitro and in vivo.
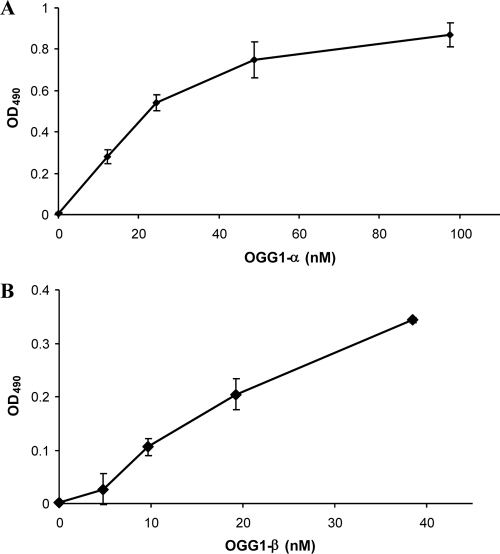
The physical interaction between RAD52 and OGG1 was detected by ELISA using recombinant purified proteins. Figure 1 shows that RAD52 interacts with high affinity with both isoforms of OGG1-α (Fig. 1A) and -β (Fig. 1B). The interaction is independent of DNA binding, since similar binding was observed in the presence of DNase I (data not shown). The apparent dissociation constants were calculated from the ELISA data using a Hill plot; Kd values of 18.4 and 39.7 nM were obtained for OGG1-α and -β, respectively. Negative control wells, coated with BSA only, showed no OGG1 binding (not shown). RAD52 also interacts directly with OGG1-α in vivo, as demonstrated by coimmunoprecipitation from whole-cell extracts from HEK293 cells expressing a FLAG-tagged OGG1-α, using an anti-RAD52 antibody (Fig. 6C and D).
FIG. 1.
Direct interaction between OGG1 and RAD52. Purified recombinant RAD52 was coated onto ELISA plates (20 nM). After blocking with 3% BSA in PBS buffer, the wells were incubated with increasing concentrations of purified OGG1-α (0 to 100 nM) or OGG1-β (0 to 40 nM) for 2 h at 37°C. The OGG1-RAD52 complexes were detected by ELISA using monoclonal antibody against OGG1-α (A) and polyclonal antibody against OGG1-β (B). Absorbance readings at each point were corrected by subtracting the background A490 reading generated by BSA-coated wells. The values presented are averages ± the standard errors of the mean (SEM) of triplicate determinations from two independent experiments.
OGG1 modulates RAD52 enzymatic activity.
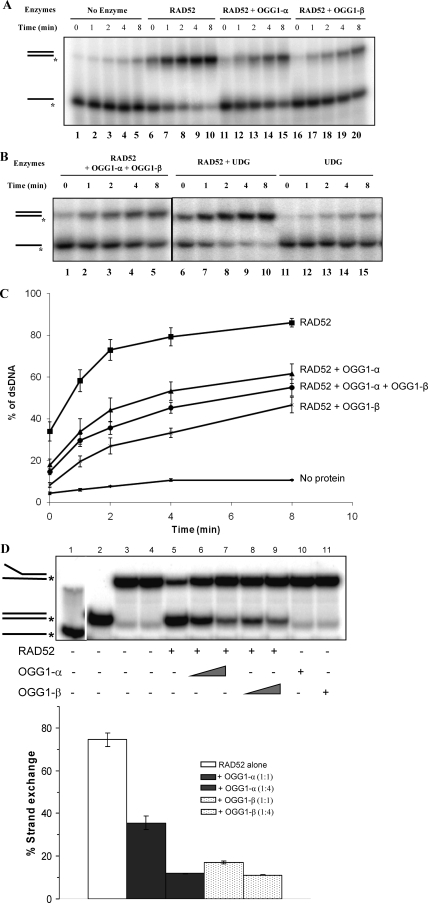
We next investigated whether the physical interaction between RAD52 and OGG1 translated into a functional interaction. RAD52 has two enzymatic activities: single-strand DNA annealing (23, 28) and DNA strand exchange (4, 20). Both activities are relevant to its role in homologous recombination and DSB repair. Recombinant RAD52 catalyzed rapid annealing of complementary single-stranded DNA (Fig. 2A, compare lanes 1 to 5 and lanes 6 to 10). The addition of OGG1-α (lanes 11 to 15) or -β (lanes 16 to 20) significantly inhibited RAD52-catalyzed strand annealing. Quantification of these data is presented in Fig. 2C. At a RAD52/OGG1 molar ratio of 1:2, OGG1-α inhibited annealing by 45% and OGG1-β by 70%. Both OGG1 isoforms inhibited RAD52 strand annealing activity by a similar mechanism, since the addition of equimolar amounts of each to the reaction showed only additive (50% inhibition for OGG1-α plus -β), but not a synergistic effect (Fig. 2B, lanes 1 to 5; Fig. 2C). This observation suggests that both the physical interaction and the RAD52 inhibition are mediated by a common domain to the two OGG1 isoforms, likely localized in the conserved 316 amino acids of the two proteins. This inhibitory effect was specific to the RAD52-OGG1 interaction because uracil DNA glycosylase (UDG), another DNA glycosylase, does not inhibit RAD52 activity (Fig. 2B, lanes 6 to 10). A similar inhibitory effect of both OGG1-α and -β was observed for RAD52-catalyzed strand exchange activity (Fig. 2D). The addition of OGG-α or -β significantly decreased strand exchange activity at a 1:1 molar ratio and almost completely abrogated the activity at a 1:4 molar ratio.
FIG. 2.
RAD52 enzymatic activities are attenuated by OGG1. (A) Strand annealing reactions were performed at room temperature for the indicated times using labeled G50 oligonucleotides and unlabeled complementary strand (112 nM each) without any addition (lanes 1 to 5) or with RAD52 (lanes 6 to 10), RAD52 plus OGG1-α (lanes 11 to 15), or OGG1-β (lanes 16 to 20). RAD52 was added at 56 nM, and OGG1 was added at 112 nM. (B) Reactions included RAD52 plus OGG1-α and OGG1-β (56 nM each) (lanes 1 to 5), RAD52 plus UDG (112 nM) (lanes 6 to 10), or UDG alone (lanes 11 to 15). (C) Quantification of percentage of double-stranded DNA generated by the indicated enzymes in panels A and B. The data points are the averages ± the SEM of three independent experiments. (D) Strand exchange reactions were carried out with 32 nM RAD52 alone or with increasing concentrations (32 and 128 nM) of OGG1-α or OGG1-β. Labeled heat-denatured G80/C80 (lane 1), G80/C80 blunt-end duplex (lane 2), and C80/G80fork26 (lanes 3 and 4) were used as markers. The graph shows the averages ± the SEM of three independent experiments.
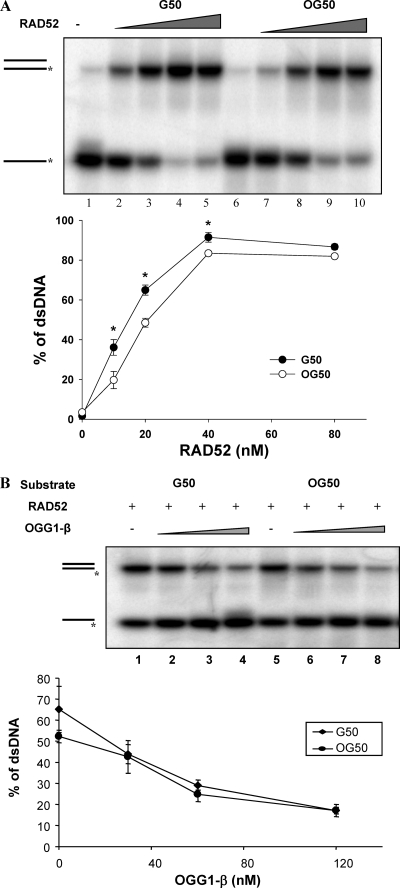
This inhibitory effect of OGG1 on RAD52 activity raised the question of whether oxidative DNA damage could impose a block to RAD52-catalyzed activities. We tested this hypothesis with a pair of oligonucleotides containing (OG50) or not (G50) a single 8-oxodG at nucleotide position 21 (Table 1). The presence of the 8-oxodG lesion partially, but significantly, inhibited RAD52-mediated single-strand annealing (Fig. 3A).
FIG. 3.
8-oxoG lesions inhibit strand annealing activity of RAD52. (A) Strand annealing activity of RAD52 was determined using a radiolabeled 50-mer control oligonucleotide (G50, lanes 1 to 5) or the same oligonucleotide with a single 8-oxoG lesion at the 21st position (OG50, lanes 6 to 10) (the oligonucleotide sequences are presented at Table 1). The reactions were carried out at room temperature for 8 min as described in the text, with 5 nM concentrations of oligonucleotides and increasing concentrations (10 to 80 nM) of RAD52. Lanes 1 and 6 show reactions without RAD52 protein. The graph shows the averages ± the SEM of three independent experiments. RAD52 annealing activity denoted by asterisks is significantly higher on G50 substrate than OG50 at P < 0.05. (B) Strand annealing reactions contained 5 nM G50 (lanes 1 to 4) or OG50 (lanes 5 to 8) substrates, 30 nM RAD52 alone (lanes 1 and 5), or increasing concentrations (30 to 120 nM) of OGG1-β (lanes 2 to 4 and lanes 6 to 8). The data points presented in the graph are the averages ± the SEM of three independent experiments.
Because OGG1 binds strongly to DNA containing 8-oxoG lesions, we next investigated whether the inhibitory effect of OGG1 on RAD52-catalyzed strand annealing was due to OGG1 binding and/or cleavage of the DNA, thus inhibiting RAD52 loading on to the substrate. We found that OGG1-β inhibited RAD52 strand-annealing activity in assays using either the G50 or the OG50 substrates (Fig. 3B). Since recombinant OGG1-β has neither incision nor DNA-binding activity (13), these results dissociate the inhibition of RAD52 from possible interactions of OGG1 with the DNA substrate and from OGG1's incision activity.
RAD52 modulates OGG1 incision activity.
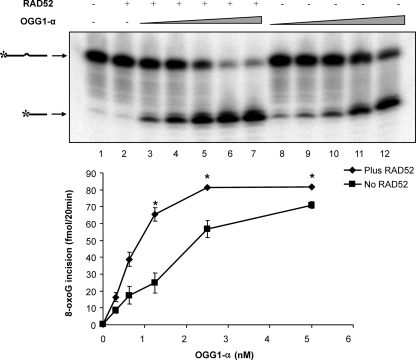
Since 8-oxoG lesions in the DNA substrate partially inhibited RAD52 strand annealing activity (Fig. 3), we hypothesized that RAD52 and OGG1 may cooperate to remove 8-oxoG lesions that constrain strand annealing. We thus tested the effect of RAD52 on OGG1-α incision activity, using a 30-mer oligonucleotide substrate containing a single 8-oxodG lesion (Table 1). Recombinant OGG1 cleaved the substrate at the 8-oxoG lesion in a concentration-dependent manner (Fig. 4). The addition of RAD52 significantly stimulated OGG1 incision activity, with a threefold increase in incision at the lower OGG1 concentrations. Together with our earlier observation that 8-oxoG inhibits RAD52 strand annealing activity, this result strongly indicates that RAD52 may cooperate with OGG1 to promote repair of oxidative lesions.
FIG. 4.
RAD52 stimulates OGG1-α 8-oxoG incision activity. OGG1 incision reactions (20 μl) were performed with serial dilutions of OGG1-α (0.3 to 5 nM) with (lanes 3 to 7) or without (lanes 8 to 12) 25 nM RAD52 at 37°C for 20 min as described in Materials and Methods. Lane 1 shows a reaction without enzymes, and lane 2 shows the reaction with RAD52 alone. The data points are the averages ± the SEM of three independent experiments. Asterisks denote incision activity significantly higher in the presence of RAD52 at P < 0.05.
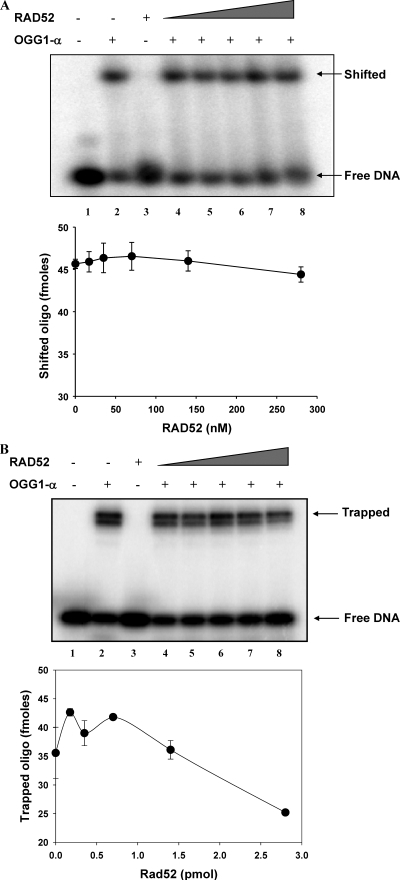
Under the same reaction conditions used for the incision assays, RAD52 did not increase OGG1 binding to its substrate (Fig. 5A, compare lanes 4 to 8 with lane 2), indicating that RAD52 does not participate in loading OGG1 onto its substrate. It is noteworthy that RAD52 alone did not bind to the double stranded oligonucleotide substrate (Fig. 5A, lane 3), which provides additional evidence that the stimulatory effect of RAD52 on OGG1's incision activity is associated with the direct protein-protein interaction and not to DNA-protein interactions.
FIG. 5.
RAD52 increases OGG1-α turnover rate. (A) Reactions (10 μl) containing 35 nM OGG1-α and increasing concentrations of RAD52 (17.5 to 280 nM) were incubated in ice for 10 min and directly loaded onto the EMSA gels. The percentage of the OG30 oligonucleotide (Table 1) bound was quantified, and the graph represents the averages ± the SEM of two independent experiments. (B) Reactions under conditions similar to those in panel A were incubated in the presence of 100 mM NaBH4 and resolved by SDS-PAGE. The percentage of oligonucleotide covalently bound was calculated, and the data points represent the averages ± the SEM of three independent experiments.
RAD52 significantly decreased the amount of OGG1-α enzyme-DNA complex trapped by sodium borohydride (Fig. 5B). This assay detects a covalent enzyme-DNA adduct generated by the reduction of the Schiff base intermediate formed during OGG1 nucleophilic attack of the abasic site (34). Since OGG1 AP-lyase activity is uncoupled from its faster glycosylase activity (41), this result suggests that RAD52 increases the OGG1 dissociation rate from its product and thus provides a molecular mechanism by which RAD52 stimulates OGG1 incision activity.
RAD52 and OGG1-α interaction increases after oxidative DNA damage.
In order to ascertain whether the interaction between OGG1-α and RAD52 is relevant in vivo, we exposed HEK293 cells stably transfected with a FLAG-tagged OGG1-α (16) to an oxidizing agent and examined the cellular localization of RAD52 and FLAG-OGG1. Figure 6A shows the immunofluorescence detection of RAD52 and FLAG-OGG1 in cells exposed or not to 500 μM H2O2. We observed a punctate localization pattern for both OGG1-α and RAD52, with the signals mostly restricted to the nuclei. Other groups have also observed a punctate signal for OGG1, even when using antibodies to the endogenous protein (7), suggesting that this pattern is not due to the FLAG epitope. In control cells, even though OGG1 and RAD52 foci were closely positioned and a few colocalized, the degree of colocalization was small, with only 25% of the scored nuclei containing more than 50% of the OGG1-α foci colocalized with RAD52 (Fig. 6B). In contrast, after H2O2 treatment, the number of nuclei with more than 50% of foci colocalizing increased more than twofold, to over 60% of the nuclei scored (Fig. 6B), indicating that the association of OGG1 and RAD52 is an active response to DNA damage.
We confirmed that the increased colocalization of OGG1 and RAD52 after oxidative stress corresponded to an increased physical association between the two proteins by immunoprecipitation. HEK293-FLAG-OGG1-α cells were exposed to 500 μM H2O2 for 30 min or 50 μM menadione for 1 h and immediately collected for cell extract preparation. Anti-FLAG M2 resin (Fig. 6C and D) was used to precipitate FLAG-OGG1-α. The anti-FLAG resin precipitated RAD52 along with the FLAG-OGG1-α in treated but not in untreated cells (compare the “-” and “+” lanes in Fig. 6C and D), indicating a new or increased physical interaction between the two proteins after oxidative stress. No RAD52 was detected in immunoprecipitates from empty vector cells (Fig. 6C and D, FLAG vector lanes) in both treated and untreated cells, indicating that RAD52 does not directly associate with the FLAG domain itself. Similar levels of FLAG-OGG1-α were detected in the precipitates in both treated and untreated cells (data not shown). In contrast to the situation for H2O2 and menadione, we did not observe increased physical interaction between OGG1 and RAD52 after 15 Gy of gamma irradiation (Fig. 6E, compare lanes 3 and 4) or 8-methoxy-psoralen (not shown), as shown by similar band intensities in control and treated lanes. Because these cells express high levels of FLAG-OGG1-α, we often saw a small immunoreactive signal from the preimmune IgG control (Fig. 6E, lanes 5 and 6); however, these signals were always much weaker than what we observed in the immunoprecipitates with anti-RAD52. The presence of RAD52 in the precipitates (Fig. 6E) was also confirmed by WB (data not shown).
RAD52 participates in the accumulation of oxidative lesions and confers cellular resistance to oxidative stress.
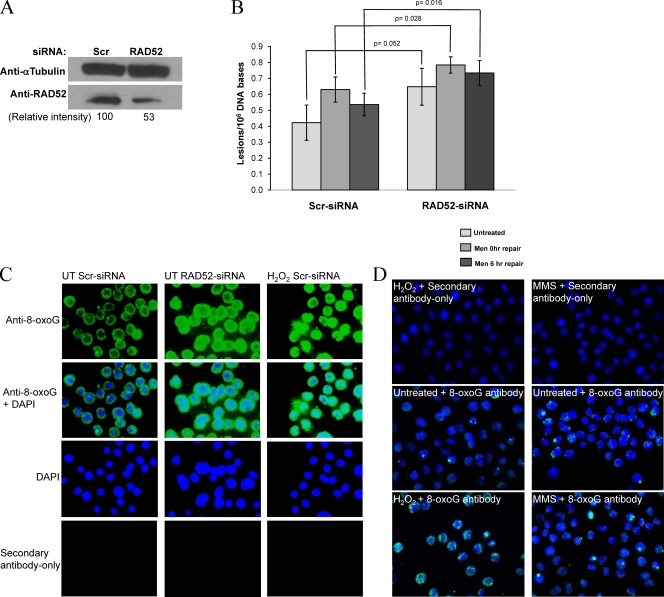
We next investigated whether RAD52 participated in the accumulation of oxidative lesions and cellular responses to oxidative stress in vivo. RAD52 levels were decreased by siRNA KD, to ca. 50% of that in cells transfected with a nontargeting scrambled siRNA (Fig. 7A). The cells were then exposed to menadione and the levels of an oxidized base that is removed by OGG1, FapyGua, were measured by GC-MS (Fig. 7B). RAD52-KD cells showed higher levels of FapyGua even in untreated cells, compared to the scrambled siRNA-transfected cells (Fig. 7B, UT). Immediately after the menadione treatment (0 h), cells lacking RAD52 accumulated significantly higher levels of FapyGua lesions than the scrambled siRNA (scr-siRNA) cells, and the levels were still significantly higher 6 h after the treatment. Although repair efficiency was relatively low under these conditions, with the scrambled-siRNA transfected cells removing ca. 15% of the initial damage introduced, the RAD52-KD cells showed no significant removal of FapyGua in the 6 h repair period, with a decrease of only 6% of the lesions introduced by the menadione treatment after 6 h. Interestingly, we had shown earlier (15) that OGG1 is the major repair enzyme for FapyGua in the nuclei and mitochondria in murine cells.
FIG. 7.
Cells with diminished RAD52 accumulate oxidative lesions. HeLa cells grown in large culture dishes were transfected with scrambled (Scr) or RAD52 siRNA. (A) After 48 h, the cells were harvested for Western blot analyses; 50 μg of protein was resolved by PAGE, and the proteins were detected by using anti-RAD52 (GeneTex) and antitubulin (Santa Cruz) antibodies. (B) For FapyGua determination, transfected cells were exposed to 50 μM menadione in PBS for 30 min. After the drug was removed and the cells washed, cultures were either harvested immediately (0 h) or replaced with fresh medium and allowed to repair (6 h). Cells from untreated (UT), 0 h and 6 h repair were harvested; the DNA isolated and analyzed by using GC/MS for FapyGu. The P values (Student t test) are given in the figure. For 8-oxoG determination, the transfected cells were exposed to 500 μM H2O2 in PBS for 30 min (C) or 100 μM H2O2 or 1 mM MMS in PBS for 60 min (D), washed, and passaged into covered slides. 8-oxoG levels were detected by using an anti-8-oxoG antibody (green fluorescence) as described in Materials and Methods. Nuclei were counterstained with DAPI (blue fluorescence). Slides incubated with secondary antibody alone are also shown. Representative slides from two independent experiments are presented.
We next evaluated the role of RAD52 in 8-oxoG accumulation in HeLa cells. 8-oxoG levels were detected using an anti-8-oxoG antibody in HeLa cells transfected either with scrambled or RAD52 siRNAs (Fig. 7C). The green fluorescence, representing the 8-oxoG signal, detected in the nucleus of RAD52-KD cells was much more intense than that detected in the scr-siRNA controls, indicating higher background levels of this modified base in the nucleus of the RAD52-KD cells, even in the absence of exogenous oxidative stress. For comparison, the fluorescence of scr-siRNA cells exposed to 500 μM H2O2 for 30 min, a situation that induces high levels of 8-oxoG, is presented. Other laboratories using the same methodology have also demonstrated a punctuate pattern, with the signal more intense on the periphery of the nucleus, probably at heterochromatic DNA (7, 32, 40; see also the Trevigen web site). For the control, the results for the DAPI signal and the secondary antibody-only control are also presented. To validate the specificity of the 8-oxoG antibody for 8-oxoG lesions, we tested the antibody on HeLa cells treated with both H2O2 and the alkylating agent methyl methanesulfonate (MMS) in parallel (Fig. 7D). H2O2 is known to cause extensive 8-oxoG lesions, whereas MMS does not (its main activity is methylation of DNA bases). Both of these lesion types are repaired by BER. Although the exposure time (50 ms) and H2O2 concentration (100 μM for 1 h) used were less than those used in the experiments shown in Fig. 7C (400 ms and 500 μM for 0.5 h, respectively), H2O2 caused a significant increase in the 8-oxoG signal (green signal), whereas MMS did not. The concentration of MMS used (1 mM for 1 h) was optimized beforehand as the maximum concentration that does not cause any cell death or any other morphological changes. Together, the data from Fig. 7 suggest that in RAD52-KD cells FapyGua and 8-oxoG lesions are either more slowly removed or accumulate faster.
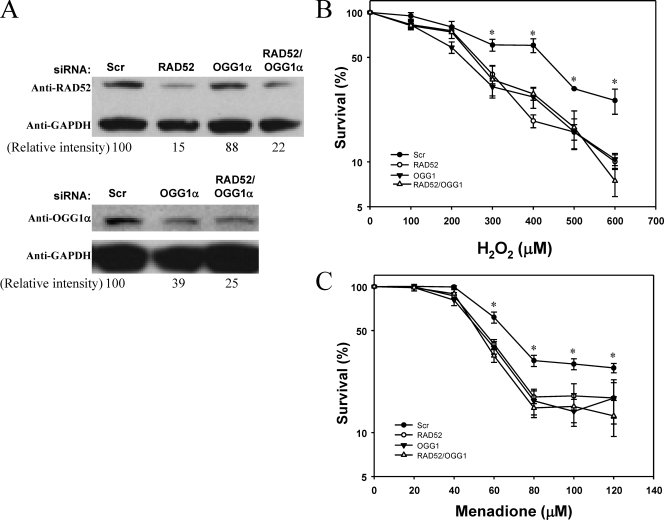
We then tested whether RAD52 levels, again diminished by siRNA KD (Fig. 8A), affected cellular sensitivity to oxidative stress induced by H2O2 or menadione, using the vital dye WST-1 to measure cell viability (Fig. 8B and C). In this experiment, we also sought to determine whether the concomitant depletion of RAD52 and OGG1-α would further sensitize the cells to oxidant injury. RAD52 levels were decreased by more than 70% in cells transfected with the RAD52 siRNA alone or together with the OGG1-α siRNA (Fig. 8A), after correction for the GAPDH levels. Likewise, OGG1-α-KD was more than 60% in the single-KD and double-KD cells. Cells depleted of either RAD52 or OGG1-α alone were significantly more sensitive to H2O2 and menadione than cells transfected with the scrambled (scr) siRNA (Fig. 8B and C, respectively). Interestingly, the double-KD cells, depleted of both RAD52 and OGG1-α, showed similar sensitivity to that of the single-KD cells. Initial cell viability experiments (data not shown), done in the same way but without the double KDs, also showed that the RAD52-depleted cells were more sensitive to H2O2 and menadione over the same range of concentrations.
FIG. 8.
RAD52 and OGG1-α-KD sensitize cells to oxidative stress. (A) HeLa cells were transfected with scrambled (Scr), RAD52, OGG1-α, or both RAD52 and OGG1-α siRNAs and harvested 48 h after transfection for Western blot analyses. A total of 50 μg of protein was resolved by PAGE, and the proteins were detected by using anti-RAD52 (GeneTex), anti-OGG1-α (Assay Designs), and anti-GAPDH (Santa Cruz) antibodies. HeLa cells, transfected as described above, were plated onto 96-well dishes and exposed to increasing concentrations of H2O2 (B) or menadione (C) as described in Materials and Methods. Survival, measured as WST-1 reduction, was calculated relative to the survival of control cells, exposed to buffer alone. The data points show the averages ± the standard deviations of triplicate determinations from two independent experiments. The asterisks denote significantly lower survival for all three KDs than control cells at P < 0.05.
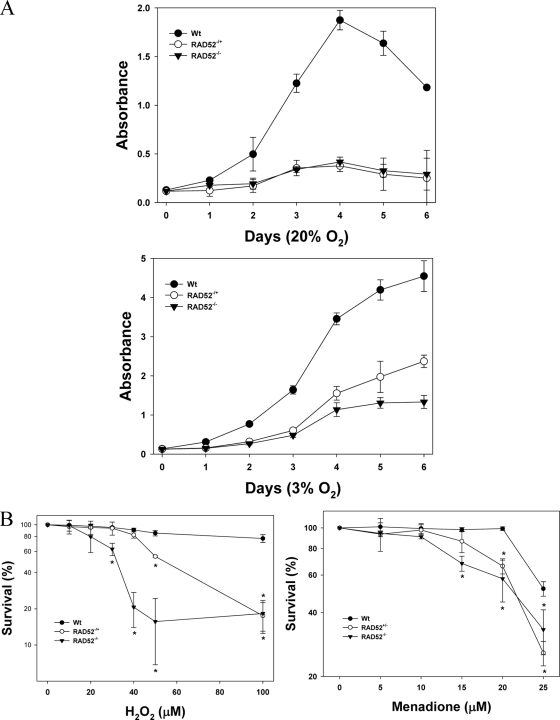
Because the experiments above were conducted with cells that still expressed residual amounts of the RAD52 protein, we tested cells obtained from RAD52 knockout mice, in which no form of the protein is expressed (30). We prepared adult MSFs from tails of wild-type (WT), Rad52−/+ (Het), and Rad52−/− (KO) mice. After establishing the cultures we initially attempted to maintain the cells under normal culture conditions, at atmospheric oxygen concentration (20%). We found that cells from both Het and KO mice grew extremely poorly (Fig. 9A, top panel). The cultures were then maintained on a 3% oxygen atmosphere, where we observed better growth of all cells, but particularly for the Het and KO fibroblasts (Fig. 9A, lower panel), as measured by WST-1 staining. Similar results were observed when cell growth was measured by directly counting the cell numbers (not shown). Because atmospheric oxygen levels are known to cause endogenous oxidative damage in cells in culture, these results indicate that RAD52 is important in cellular proliferation and response to oxidative stress.
FIG. 9.
MSFs lacking RAD52 are sensitive to oxidative stress. (A) MSFs from WT, RAD52 heterozygote (RAD52−/+), and RAD52 KO (RAD52−/−) animals were cultured in 20 or 3% oxygen atmosphere, and cell numbers were monitored for 6 days, measured as the WST-1 reduction. The data points show the averages ± the standard deviations of two independent experiments, performed in triplicate. (B) MSFs, at passages 2 and 3, were plated onto 96-well dishes and exposed to increasing concentrations of H2O2 or menadione. Survival, measured as the WST-1 reduction, was calculated relative to the survival of control cells exposed to buffer alone. The data points show the average ± the standard deviations of triplicate determinations from two independent experiments. The asterisks denote significantly lower survival than WT cells at P < 0.01.
MSF cultures maintained at 3% oxygen atmosphere were then tested for sensitivity to H2O2 or menadione as described above (Fig. 9B). We observed significant decreases in viability of the Het and KO cells for both agents tested, although a more striking difference was observed for H2O2 (left panel). Interestingly, cells from Het animals showed an intermediate sensitivity compared to the WT and KO cells, suggesting that the relative levels of RAD52 protein are important in the cellular response to oxidative stress.
DISCUSSION
Oxidative DNA base lesions are constantly generated at very high levels in normal cells. Such accumulation of DNA damage may cause severe biological consequences, ranging from mutagenesis to cell death. Thus far, only indirect evidence has implicated the homologous recombination protein RAD52 in oxidative stress resistance. For instance, in yeast mutants lacking BER enzymes, deletion of rad52 greatly increases sensitivity to H2O2 (36). Furthermore, sensitivity to stannous chloride, a metal salt believed to cause oxidative stress, is higher in rad52 mutants as in superoxide dismutase 1 and 2 mutants, which are directly involved in reactive oxygen species detoxification (37). In transforming growth factor alpha/c-myc double-transgenic mice, a model for oxidative stress-induced accelerated hepatocarcinogenesis, BER genes (OGG1 and NTH1) and RAD52 are constitutively upregulated, indicating a role for RAD52 in oxidative stress response in mammals as well. However, the mechanistic basis for RAD52's involvement remains unclear. It is unlikely that its role in DSB repair alone plays a major role in this, since H2O2 does not induce significant amounts of conventional DSB (38).
In the present study, we identified a new role for RAD52 in the repair of oxidative DNA damage through a direct stimulation of the DNA glycosylase OGG1. We show that RAD52 interacts with and modulates the activity of OGG1 (Fig. 4). The functional interaction between these two proteins is reciprocal, since OGG1 also modulates the two known RAD52 enzymatic activities (Fig. 2). The physical interaction of the recombinant proteins is strong in vitro; with low apparent Kd values. We detected the RAD52/OGG1-α interaction in vivo as well (Fig. 6), a finding indicative of a biologically significant protein interaction. More importantly, we directly show that cells lacking RAD52 accumulate more FapyGua lesions in their genomes than cells with normal levels of RAD52, even in the absence of any oxidative treatment (Fig. 7B). 8-oxoG levels, detected by immunocytochemistry, were also elevated in cells depleted of RAD52, even in the absence of exogenous oxidative stress (Fig. 7C).
This reciprocal functional interaction indicates that OGG1 and RAD52 may cooperate to repair oxidative damage. Because of the specific DNA substrate affinities of these two proteins it is possible that this interaction may have a more significant impact at sites of free DNA ends containing oxidized purine samples, such as Fapys and 8-oxoG. RAD52 binds preferentially to resected and tailed DNA structures, a finding consistent with an early role in DSB repair. We found that the presence of 8-oxoG lesions in DNA partially, but significantly, inhibited RAD52 strand annealing activity (Fig. 3A). Because 8-oxoGs positioned close to DNA ends are removed less efficiently by OGG1 (26), this scenario would result in reduced recombination resolution in the presence of high levels of oxidative DNA damage at DNA ends. Since homologous recombination is the preferred error-free pathway to repair DSB, high levels of oxidative DNA damage in the vicinity of free DNA ends could impose a significant hindrance to the resolution of these lesions. The observation that RAD52 stimulates OGG1 8-oxoG incision activity supports a role for their interaction in relieving 8-oxoG inhibition of RAD52 activity.
Another scenario in which this interaction may play an important biological role is at sites of clustered oxidative DNA lesions. Such clusters, of more than one oxidative lesion in close proximity, often in opposite strands, can generate DSB upon partial resolution by the BER pathway. Clustered oxidative DNA lesions can be easily detected not only in cells exposed to ionizing radiation but also in unexposed cells, likely as a result of oxidative stress generated as by-product of normal metabolism. The removal of the oxidized base and subsequent abasic site processing can lead directly to a DSB. The interaction of RAD52 with OGG1 could function to direct RAD52 to these break sites.
We found that OGG1's catalytic activity stimulation by RAD52 is the result of the direct interaction between the two proteins. RAD52 did not increase loading of OGG1 onto the substrate, since no increase in OGG1-DNA complexes was observed in the presence of RAD52 (Fig. 5A). On the other hand, we observed a protein concentration-dependent decrease in sodium borohydride-trapped OGG1-substrate complexes (Fig. 5B). Because OGG1's AP-lyase activity is uncoupled from its DNA glycosylase activity and sodium borohydride traps the intermediates generated during the nucleophilic attack of the abasic site, this result suggests that RAD52 may increase OGG1 turnover rate, and it is reminiscent of the molecular mechanism by which APE1 stimulates OGG1(14). The affinity of OGG1 for its intermediate product abasic site:C is significantly higher than for its substrate 8-oxoG:C pair (14). Thus, the enzyme tends to stay bound to the intermediate, which is detected as higher sodium borohydride trapping. RAD52 binding to OGG1 stimulates its AP-lyase activity, and consequently enzyme turnover, and thus decreases the number of sodium borohydride-trapped complexes.
Immunofluorescence microscopy of control cells showed limited colocalization of RAD52 and OGG1 (Fig. 6A) and little or no coimmunoprecipitation (Fig. 6C and D, “-” lanes). More importantly, however, we showed that the degree of colocalization and physical association between OGG1 and RAD52 increases significantly after oxidative stress, as detected both by the immunocytochemistry and by coimmunoprecipitation of RAD52 with FLAG-tagged OGG1-α (Fig. 6B, C, and D, treated lanes). These results indicate that this association is biologically important in the response to oxidative DNA damage and implicates RAD52 in this process. On the other hand, the RAD52-OGG1 association did not increase after treatment with agents that cause conventional DNA DSB, such as gamma irradiation (Fig. 6E) and 8-methoxy-psoralen (data not shown). Thus, our results suggest that RAD52 is part of the oxidative DNA damage response pathway but that OGG1 does not seem to participate directly in DSB repair. The observation that RAD52-KD results in enhanced accumulation of FapyGua and 8-oxoG lesions in genomic DNA (Fig. 7) also provides evidence for this hypothesis.
It is important to note that we have validated the specificity of the 8-oxoG antibody (Fig. 7C) that was used (Trevigen) in two ways. First, treatment of HeLa cells with the alkylating agent MMS (which does not generate 8-oxoG) resulted in no increase in green fluorescein isothiocyanate signal (Fig. 7D). Second, as part of our current unpublished work on oxidative damage monitoring, we have now demonstrated that addition of Fpg (an E. coli DNA glycosylase/AP-lyase that removes 8-oxoG from the damaged DNA) after fixation (and before addition of the antibody) results in more than 50% ablation of the green fluorescein isothiocyanate fluorescent signal produced by the 8-oxoG antibody, on H2O2-treated cells (based on the average green fluorescent intensities of 50 cells per condition, using Axiovision software [data not shown]). This is direct evidence of the specificity of the Trevigen 8-oxoG antibody for 8-oxoG lesions in vivo.
Our study also provides direct evidence connecting RAD52 to the molecular pathways protecting human and mouse cells from oxidative stress-induced cell death. Human cells partially depleted of RAD52 via siRNA KD (Fig. 8) or mouse cells expressing half or no RAD52 due to gene knockout (Fig. 9) were significantly more susceptible to H2O2 and menadione than their counterparts expressing normal levels of RAD52. Menadione is a redox compound that can generate large quantities of superoxide anions upon oxidation of the partially reduced molecule. It is important to note that, as opposed to H2O2, which has been shown to generate extensive single-strand DNA breakage in addition to base damage, menadione generates almost exclusively base modifications. The implication of the increased sensitivity of cells lacking RAD52 to menadione is that RAD52 may participate directly in the removal of such modifications. A direct role of RAD52 in removal of oxidized base is also suggested by the observation that untreated cells partially depleted of RAD52 accumulate more FapyGua than cells with normal levels of RAD52, and accumulate more 8-oxoG, even in the absence of exogenous oxidative stress (Fig. 7). Since OGG1-initiated BER is believed to be the major repair pathway for these oxidized bases (10), our in vitro observation of the functional interaction between RAD52 and OGG1 thus provide a mechanistic explanation for the biological effect seen in cells lacking RAD52.
We did not find, however, an additive effect of concomitantly lowering the expression levels of RAD52 and OGG1, since double-KD cells showed sensitivity to menadione and H2O2 similar to that of the single KDs. These results suggest that both proteins function in the same pathway of cellular response to oxidative stress. Moreover, because each individual KD still expressed some levels of the proteins (15% for RAD52 and 39% for OGG1), it may indicate that even small amounts of RAD52 are sufficient to stimulate OGG1 incision activity. This is supported by the very low apparent Kd for the physical association of the two proteins (Fig. 1) and by the stimulation of OGG1 incision activity in relatively low molar ratios, from 1:20 to 1:5 (Fig. 4).
The present study is the first to directly connect RAD52 to oxidative DNA damage response in mammalian cells. Moreover, these data demonstrate the interplay between what had been considered distinct DNA repair pathways. In yeast cells, it has recently become evident that a biological network exists for preventing genomic instability caused by oxidative stress, which involves other repair processes in addition to BER (17). It is now important to identify additional participants in this network and to further dissect the molecular mechanisms mediating these interactions.
Acknowledgments
This study was supported by the Intramural Research Program of the National Institutes of Health, National Institute on Aging.
We thank Marie Rossi and Yie Liu for the critical reading of the manuscript and Xiufen Sui and Regina Knight for technical help. RAD52 transgenic mouse tails were kindly provided by Maria Jasin, Memorial Sloan-Kettering Cancer Center, New York, NY. We also thank Miral Dizdaroglu, National Institute of Standards and Technology, Gaithersburg, MD, for the use of the GC-MS equipment.
Footnotes
Published ahead of print on 8 June 2009.
REFERENCES
- 1.Barja, G. 2004. Free radicals and aging. Trends Neurosci. 27595-600. [DOI] [PubMed] [Google Scholar]
- 2.Baynton, K., M. Otterlei, M. Bjoras, C. von Kobbe, V. A. Bohr, and E. Seeberg. 2003. WRN interacts physically and functionally with the recombination mediator protein RAD52. J. Biol. Chem. 27836476-36486. [DOI] [PubMed] [Google Scholar]
- 3.Bennett, P. V., N. S. Cintron, L. Gros, J. Laval, and B. M. Sutherland. 2004. Are endogenous clustered DNA damages induced in human cells? Free Radic. Biol. Med. 37488-499. [DOI] [PubMed] [Google Scholar]
- 4.Bi, B., N. Rybalchenko, E. I. Golub, and C. M. Radding. 2004. Human and yeast Rad52 proteins promote DNA strand exchange. Proc. Natl. Acad. Sci. USA 1019568-9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjoras, M., L. Luna, B. Johnsen, E. Hoff, T. Haug, T. Rognes, and E. Seeberg. 1997. Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. EMBO J. 166314-6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosh, R. M., Jr., D. K. Orren, J. O. Nehlin, P. H. Ravn, M. K. Kenny, A. Machwe, and V. A. Bohr. 1999. Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem. 27418341-18350. [DOI] [PubMed] [Google Scholar]
- 7.Conlon, K. A., D. O. Zharkov, and M. Berrios. 2003. Immunofluorescent localization of the murine 8-oxoguanine DNA glycosylase (mOGG1) in cells growing under normal and nutrient deprivation conditions. DNA Repair 21337-1352. [DOI] [PubMed] [Google Scholar]
- 8.Cooke, M. S., M. D. Evans, M. Dizdaroglu, and J. Lunec. 2003. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 171195-1214. [DOI] [PubMed] [Google Scholar]
- 9.Dizdaroglu, M. 1993. Quantitative determination of oxidative base damage in DNA by stable isotope-dilution mass spectrometry. FEBS Lett. 3151-6. [DOI] [PubMed] [Google Scholar]
- 10.Fortini, P., B. Pascucci, E. Parlanti, M. D'Errico, V. Simonelli, and E. Dogliotti. 2003. 8-Oxoguanine DNA damage: at the crossroad of alternative repair pathways. Mutat. Res. 531127-139. [DOI] [PubMed] [Google Scholar]
- 11.Gackowski, D., E. Speina, M. Zielinska, J. Kowalewski, R. Rozalski, A. Siomek, T. Paciorek, B. Tudek, and R. Olinski. 2003. Products of oxidative DNA damage and repair as possible biomarkers of susceptibility to lung cancer. Cancer Res. 634899-4902. [PubMed] [Google Scholar]
- 12.Hamilton, M. L., H. Van Remmen, J. A. Drake, H. Yang, Z. M. Guo, K. Kewitt, C. A. Walter, and A. Richardson. 2001. Does oxidative damage to DNA increase with age? Proc. Natl. Acad. Sci. USA 9810469-10474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashiguchi, K., J. A. Stuart, N. C. Souza-Pinto, and V. A. Bohr. 2004. The C-terminal αO helix of human Ogg1 is essential for 8-oxoguanine DNA glycosylase activity: the mitochondrial β-Ogg1 lacks this domain and does not have glycosylase activity. Nucleic Acids Res. 325596-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill, J. W., T. K. Hazra, T. Izumi, and S. Mitra. 2001. Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res. 29430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu, J., N. C. de Souza-Pinto, K. Haraguchi, B. A. Hogue, P. Jaruga, M. M. Greenberg, M. Dizdaroglu, and V. A. Bohr. 2005. Repair of formamidopyrimidines in DNA involves different glycosylases: role of the OGG1, NTH1, and NEIL1 enzymes. J. Biol. Chem. 28040544-40551. [DOI] [PubMed] [Google Scholar]
- 16.Hu, J., S. Z. Imam, K. Hashiguchi, N. C. Souza-Pinto, and V. A. Bohr. 2005. Phosphorylation of human oxoguanine DNA glycosylase (α-OGG1) modulates its function. Nucleic Acids Res. 333271-3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, M. E., and R. D. Kolodner. 2005. A biological network in Saccharomyces cerevisiae prevents the deleterious effects of endogenous oxidative DNA damage. Mol. Cell 17709-720. [DOI] [PubMed] [Google Scholar]
- 18.Karthikeyan, G., J. H. Santos, M. A. Graziewicz, W. C. Copeland, G. Isaya, B. Van Houten, and M. A. Resnick. 2003. Reduction in frataxin causes progressive accumulation of mitochondrial damage. Hum. Mol. Genet. 123331-3342. [DOI] [PubMed] [Google Scholar]
- 19.Krejci, L., L. Chen, S. Van Komen, P. Sung, and A. Tomkinson. 2003. Mending the break: two DNA double-strand break repair machines in eukaryotes. Prog. Nucleic Acids Res. Mol. Biol. 74159-201. [DOI] [PubMed] [Google Scholar]
- 20.Kumar, J. K., and R. C. Gupta. 2004. Strand exchange activity of human recombination protein Rad52. Proc. Natl. Acad. Sci. USA 1019562-9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maynard, S. P., and R. A. Miller. 2006. Fibroblasts from long-lived Snell dwarf mice are resistant to oxygen-induced in vitro growth arrest. Aging Cell 589-96. [DOI] [PubMed] [Google Scholar]
- 22.Mitra, S., I. Boldogh, T. Izumi, and T. K. Hazra. 2001. Complexities of the DNA base excision repair pathway for repair of oxidative DNA damage. Environ. Mol. Mutagen. 38180-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mortensen, U. H., C. Bendixen, I. Sunjevaric, and R. Rothstein. 1996. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl. Acad. Sci. USA 9310729-10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muftuoglu, M., S. Sharma, T. Thorslund, T. Stevnsner, M. M. Soerensen, R. M. Brosh, Jr., and V. A. Bohr. 2006. Cockayne syndrome group B protein has novel strand annealing and exchange activities. Nucleic Acids Res. 34295-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishioka, K., T. Ohtsubo, H. Oda, T. Fujiwara, D. Kang, K. Sugimachi, and Y. Nakabeppu. 1999. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol. Biol. Cell 101637-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parsons, J. L., D. O. Zharkov, and G. L. Dianov. 2005. NEIL1 excises 3′ end proximal oxidative DNA lesions resistant to cleavage by NTH1 and OGG1. Nucleic Acids Res. 334849-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paz-Elizur, T., M. Krupsky, D. Elinger, E. Schechtman, and Z. Livneh. 2005. Repair of the oxidative DNA damage 8-oxoguanine as a biomarker for lung cancer risk. Cancer Biomarkers 1201-205. [DOI] [PubMed] [Google Scholar]
- 28.Reddy, G., E. I. Golub, and C. M. Radding. 1997. Human Rad52 protein promotes single-strand DNA annealing followed by branch migration. Mutat. Res. 37753-59. [DOI] [PubMed] [Google Scholar]
- 29.Reddy, P., P. Jaruga, T. O'Connor, H. Rodriguez, and M. Dizdaroglu. 2004. Overexpression and rapid purification of Escherichia coli formamidopyrimidine-DNA glycosylase. Protein Expr. Purif. 34126-133. [DOI] [PubMed] [Google Scholar]
- 30.Rijkers, T., O. J. Van Den, B. Morolli, A. G. Rolink, W. M. Baarends, P. P. Van Sloun, P. H. Lohman, and A. Pastink. 1998. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol. 186423-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shibutani, S., M. Takeshita, and A. P. Grollman. 1991. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 349431-434. [DOI] [PubMed] [Google Scholar]
- 32.Soultanakis, R. P., R. J. Melamede, I. A. Bespalov, S. S. Wallace, K. B. Beckman, B. N. Ames, D. J. Taatjes, and Y. M. Janssen-Heininger. 2000. Fluorescence detection of 8-oxoguanine in nuclear and mitochondrial DNA of cultured cells using a recombinant Fab and confocal scanning laser microscopy. Free Radic. Biol. Med. 28987-998. [DOI] [PubMed] [Google Scholar]
- 33.Sugimura, H., T. Kohno, K. Wakai, K. Nagura, K. Genka, H. Igarashi, B. J. Morris, S. Baba, Y. Ohno, C. Gao, Z. Li, J. Wang, T. Takezaki, K. Tajima, T. Varga, T. Sawaguchi, J. K. Lum, J. J. Martinson, S. Tsugane, T. Iwamasa, K. Shinmura, and J. Yokota. 1999. hOGG1 Ser326Cys polymorphism and lung cancer susceptibility. Cancer Epidemiol. Biomarkers Prev. 8669-674. [PubMed] [Google Scholar]
- 34.Sun, B., K. A. Latham, M. L. Dodson, and R. S. Lloyd. 1995. Studies on the catalytic mechanism of five DNA glycosylases. Probing for enzyme-DNA imino intermediates. J. Biol. Chem. 27019501-19508. [DOI] [PubMed] [Google Scholar]
- 35.Sutherland, B. M., P. V. Bennett, N. S. Cintron, P. Guida, and J. Laval. 2003. Low levels of endogenous oxidative damage cluster levels in unirradiated viral and human DNAs. Free Radic. Biol. Med. 35495-503. [DOI] [PubMed] [Google Scholar]
- 36.Swanson, R. L., N. J. Morey, P. W. Doetsch, and S. Jinks-Robertson. 1999. Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 192929-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viau, C., C. Pungartnik, M. C. Schmitt, T. S. Basso, J. A. Henriques, and M. Brendel. 2006. Sensitivity to Sn2+ of the yeast Saccharomyces cerevisiae depends on general energy metabolism, metal transport, anti-oxidative defences, and DNA repair. Biometals 19705-714. [DOI] [PubMed]
- 38.Ward, J. F., J. W. Evans, C. L. Limoli, and P. M. Calabro-Jones. 1987. Radiation and hydrogen peroxide induced free radical damage to DNA. Br. J. Cancer 1987(Suppl. 8)105-112. [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson, D. M., III, T. M. Sofinowski, and D. R. McNeill. 2003. Repair mechanisms for oxidative DNA damage. Front. Biosci. 8d963-d981. [DOI] [PubMed] [Google Scholar]
- 40.Yarborough, A., Y. J. Zhang, T. M. Hsu, and R. M. Santella. 1996. Immunoperoxidase detection of 8-hydroxydeoxyguanosine in aflatoxin B1-treated rat liver and human oral mucosal cells. Cancer Res. 56683-688. [PubMed] [Google Scholar]
- 41.Zharkov, D. O., T. A. Rosenquist, S. E. Gerchman, and A. P. Grollman. 2000. Substrate specificity and reaction mechanism of murine 8-oxoguanine-DNA glycosylase. J. Biol. Chem. 27528607-28617. [DOI] [PubMed] [Google Scholar]