Abstract
Transcriptome studies are revealing that the eukaryotic genome actively transcribes a diverse repertoire of large noncoding RNAs (ncRNAs), many of which are unannotated and distinct from the small RNAs that have garnered much attention in recent years. Why are they so pervasive, and do they have a function? X-chromosome inactivation (XCI) is a classic epigenetic phenomenon associated with many large ncRNAs. Here, I provide a perspective on how XCI is achieved in mice and suggest how this knowledge can be applied to the rest of the genome. Emerging data indicate that long ncRNAs can function as guides and tethers, and may be the molecules of choice for epigenetic regulation: First, unlike proteins and small RNAs, large ncRNAs remain tethered to the site of transcription, and can therefore uniquely direct allelic regulation. Second, ncRNAs command a much larger sequence space than proteins, and can therefore achieve very precise spatiotemporal control of development. These properties imply that long noncoding transcripts may ultimately rival small RNAs and proteins in their versatility as epigenetic regulators, particularly for locus- and allele-specific control.
Keywords: X-chromosome inactivation, chromatin modifier, dosage compensation, epigenetic regulation, noncoding RNA
Over the past several years, large-scale analyses of the mouse and human genomes have revealed that, while only 1.5% of the genome carries protein-coding information, 60%–80% of its nucleotides are transcribed. According to Unigene (Wheeler et al. 2008), there are 84,000 transcripts in the human genome, of which only 20,000–25,000 are associated with protein-coding genes. According to FANTOM3 (Carninci et al. 2005), 62% of the mouse genome is transcribed to make 181,000 transcripts. These large noncoding RNAs (ncRNAs) are often developmentally regulated, sometimes conserved, and frequently next to genes known to be subject to tight transcriptional control. A very small minority of the RNAs has previously described functions as catalysts and structural RNAs, distinctly different from the small effectors of the RNAi pathway (Cech 2009; Ghildiyal and Zamore 2009; Sharp 2009). A few also have been characterized as transcriptional scaffolds, a platform on which protein factors can be recruited to chromatin (Cam et al. 2009; Moazed 2009). However, the vast majority of these transcripts, which can range in size from 100 nucleotides (nt) to >100 kb and are apparently pervasive, has no obvious function. Some are antisense to known protein-coding genes (Katayama et al. 2005; He et al. 2008), whereas others originate in promoter regions and intergenic space (Claverie 2005; Kapranov et al. 2007a,b; Guttman et al. 2009; Mercer et al. 2009). In the post-genomic era, why so much cellular energy should be spent on RNA production has captured the imagination. Are they merely spurious transcripts—wasteful biproducts of genomic activity? Or do they encode useful information and serve crucial functions in the epigenome?
Although it is now very fashionable to study “macroRNAs,” knowledge of RNA behemoths dates back decades and has been pursued intensively by those studying the once-unusual epigenetic phenomena of genomic imprinting (Sleutels and Barlow 2002; Edwards and Ferguson-Smith 2007; Wan and Bartolomei 2008) and X-chromosome inactivation (XCI) (Lyon 1961; Wutz 2003; Lucchesi et al. 2005; Masui and Heard 2006; Payer and Lee 2008). Discovery of the 17-kb Xist RNA in 1991 marked the beginning of a long fascination with regulatory ncRNAs in this field (Borsani et al. 1991; Brown et al. 1991a, 1992; Brockdorff et al. 1992). Discovery of a second transcript—the 40-kb Tsix antisense RNA (Lee and Lu 1999; Lee et al. 1999a)—brought early recognition that untranslated RNAs may come to dominate regulation of XCI. Today, the Xist and Tsix RNA pair serves as a paradigm for understanding sense–antisense relationships in eukaryotes and for long-range chromatin control. Around the “X-inactivation center” (Xic) (Brown et al. 1991b; Lee et al. 1996; Simmler et al. 1996; Willard 1996; Chureau et al. 2002), more than seven distinct ncRNA loci have now been identified, and it is clear that Xist and Tsix are not the only ones with regulatory properties. For years, the unique aspects of Xist and Tsix were thought to be evolutionary deviations—possibly “X-centric” and relevant only for unusual epigenetic phenomena such as sex-chromosome inactivation and imprinting.
Below, I argue that lessons learned from the X can be applied elsewhere. In short vignettes, I illustrate models for ncRNA control of X and propose the idea of RNA as the molecule of choice for locus-specific and allelic control. I alert the reader to the fact that, for every aspect of XCI, there are many worthy contemporary models and that the goal of the following dissertation is not to review all of them, but rather to elaborate on current thinking about the role of ncRNA. I kindly refer interested parties to published works elsewhere, as cited, for in-depth discussion of alternative viewpoints.
X-inactivation, checkpoints, and ncRNA
XCI is the mechanism of dosage compensation in mammals by which one X chromosome is transcriptionally silenced in the female sex to ensure that XX and XY individuals have equivalent X-linked gene dosage (Lyon 1961; Wutz 2003; Lucchesi et al. 2005; Masui and Heard 2006; Payer and Lee 2008). Two forms of XCI occur in eutherian mammals. During preimplantation development, dosage compensation is imprinted to occur exclusively on the paternal X chromosome (XP) (Takagi 1974; Huynh and Lee 2003; Okamoto et al. 2004). Around the time of uterine implantation (mouse embryonic day 4.6–6.5 [E4.5–E6.5]), “imprinted XCI” is retained in the extraembryonic tissues (placenta) of the developing embryo. However, in the epiblast lineage (embryo proper), the imprint is erased, XP undergoes reactivation (Mak et al. 2004), and a second form of XCI takes hold. In “random XCI,” the two Xs in each female cell have an equal chance of being inactivated (Lyon 1961).
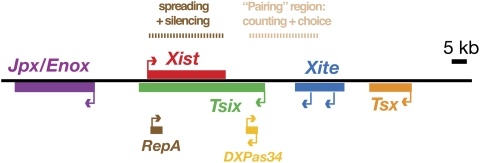
Random XCI is controlled by a specialized X-linked region that is replete with ncRNA (Fig. 1; Brown et al. 1991b; Lee et al. 1996; Simmler et al. 1996; Willard 1996; Chureau et al. 2002). Known as the Xic, the region has been genetically delineated to a <500-kb region, with a 100- to 200-kb subregion being sufficient to recapitulate many if not all steps of the inactivation process (Lee et al. 1996, 1999b; Heard et al. 1999; Migeon et al. 1999). At least seven distinct genes that make large ncRNAs have been identified within the Xic and surrounding regions, and several have been ascribed specific function during XCI. The 17-kb Xist RNA is produced exclusively from the Xi (inactive X) and is thought to initiate chromosome-wide silencing as it accumulates and blankets the X in cis (Fig. 2; Brockdorff et al. 1992; Brown et al. 1992; Clemson et al. 1996; Penny et al. 1996; Marahrens et al. 1997). Knockouts of Xist result in an X chromosome that is no longer capable of undergoing silencing. Xist is regulated by Tsix, Xist's 40-kb antisense counterpart that antagonizes Xist in cis (Lee and Lu 1999; Lee et al. 1999a; Lee 2000; Sado et al. 2001). Knocking out Tsix results in the inability to designate Xa (active X) in female cells. In turn, Tsix is regulated by Xite, an upstream activator that is associated with production of ncRNA within a 10- to 15-kb region immediately upstream of Tsix's major promoter (Ogawa and Lee 2003; Stavropoulos et al. 2005). More recently, a number of smaller transcripts have also emerged within the Xic. At the 5′ end of Tsix, short bidirectional transcripts (Cohen et al. 2007) originate within a microsatellite repeat element known as DXPas34 (Courtier et al. 1995; Debrand et al. 1999) and have been proposed to have both positive and negative regulatory effects on Tsix. At the 5′ end of Xist, a short RNA of 1.6 kb has been discovered within another repeat element, known as Repeat A (Zhao et al. 2008). Furthermore, very small RNAs on the order of 21–42 nt have also been reported to occur in the complementary region of Xist and Tsix (Ogawa et al. 2008).
Figure 1.
Noncoding genes of the Xic. The large noncoding elements Xist, Tsix, and Xite are now well established as regulators of XCI. More recently, shorter internal transcription units have been identified from the larger loci. These include the “sense” locus RepA and the bidirectionally transcribed locus DXPas34. The regions responsible for X-chromosome pairing, counting/choice, and spreading/silencing are shown above the map.
Figure 2.
Xist RNA is expressed only from the Xi, coats that chromosome in cis, and is thought to recruit the first silencing factors to the X. The photograph shows an RNA FISH experiment performed on a metaphase chromosome spread taken from a female fibroblast cell line. Xist RNA is labeled by a rhodamine dye (red). Chromosomes are counterstained with DAPI (blue, left panel; white, right panel). Arrow indicates Xi.
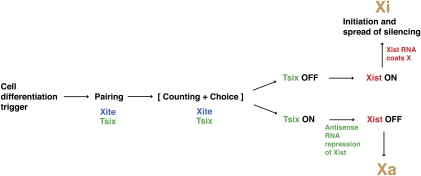
These ncRNAs collaborate to ensure orderly progression through the many crucial “checkpoints” of XCI (Fig. 3). These checkpoints have been investigated intensively in a mouse embryonic stem (ES) cell model that mimics the epiblast lineage from which it is derived and faithfully recapitulates random XCI when induced to differentiate in vitro. A cell differentiation signal based on changes in binding of pluripotency factors triggers XCI (Monk and Harper 1979; Navarro et al. 2008; Donohoe et al. 2009). During counting, the X-to-autosome ratio (X:A) is measured and XCI proceeds only if the X:A ratio is 1 or greater (Kay et al. 1994; Clerc and Avner 1998; Lyon 1999; Avner and Heard 2001; Boumil and Lee 2001; Morey et al. 2004; Lee 2005). Thus, XCI is initiated in female cells where X:A = 1, while XCI is blocked in male cells where X:A = 0.5. The genetic identities of the numerators (X) and denominators (A), how they interact to measure the X:A ratio, and what they target on the X chromosome have remained elusive, but evidence points to two ncRNA loci—Xite and Tsix—as X-linked dosage sensors (Morey et al. 2004; Lee 2005).
Figure 3.
Checkpoints of XCI regulated by noncoding genes of the Xic. See the text for detailed discussion.
Following the “counting” checkpoint, a choosing mechanism randomly selects one X chromosome as Xa and the other as Xi (Avner and Heard 2001; Boumil and Lee 2001). I favor the idea that “choice” occurs in a mutually exclusive manner to ensure that every event results in a nucleus with exactly one Xa and one Xi (Lee 2002, 2005). An alternative viewpoint posits that chromosome choice is not determined purposefully but is the outcome of a “stochastic” process—one that leads to the survival of cells that, by chance, selected one Xa and one Xi, and one that leads to the death of others that made the inappropriate selection of two Xa or two Xi (Monkhorst et al. 2008,2009). Still another model suggests that choice may be predetermined long before XCI actually initiates (Mlynarczyk-Evans et al. 2006). I refer readers to the cited works for further elaboration of these alternative models. Hereafter, discussion will focus on the preferred view that allelic choice occurs through a tightly controlled mechanism during XCI to achieve mutual exclusion of Xa and Xi without incurring cell death.
In my opinion, the precision with which choice is determined implies the existence of a second checkpoint—in the form of cross-talking or a feedback loop—to guarantee distinct fates of the two Xs (Lee 2005). Because the two female Xs are thought to be equivalent in the pre-XCI state, how their epigenetic symmetry is broken at the onset of cell differentiation, especially when the Xs are bathed in the same nucleoplasm, remains one of the big questions in the field. Below, I argue that somatic “pairing” facilitates symmetry break, and that the same ncRNAs, Xite and Tsix, play crucial roles during this process.
Once chosen, the Xi-elect must be distinctly marked from Xa-elect and initiate the process of chromosome-wide silencing in a strictly cis-limited fashion. The strict cis requirement presents several conceptual difficulties for the female cell. First, how is the initial silencer recruited only to one Xic when Xi and Xa-elect reside in the same nuclear milieu following the pairing and choice checkpoints? Second, how can silencing spread in an allele-specific fashion in cis along 150 Mb of sequence? As for all other checkpoints, the answers may lie in ncRNA: in this case, in the two overlapping transcripts, RepA (Zhao et al. 2008) and Xist RNA (Brown et al. 1992).
X-chromosome pairing controlled by transcription of ncRNA
Diverse as they are, ncRNAs of the Xic have been proposed to act in cis; i.e., on the chromosome that synthesizes them. Although cis-acting genes dominate the Xic, one viewpoint suggests that Xic function must also extend in trans (Marahrens 1999; Lee 2005). The requirement to select Xa and Xi in a mutually exclusive manner implies a means of interchromosomal communication or feedback to ensure that no cell befalls the lethal outcome of creating two Xa or two Xi. The idea of cross-talk was first supported by a homozygous knockout of Tsix, which led to a state in which choice became chaotic, with some female cells displaying two Xi, one Xi, or no Xi at all (Lee 2005). This result suggested that the noncoding Tsix gene, already known to be a negative regulator of Xist and a requirement for designating Xa (Lee and Lu 1999), must also play a role in the mutually exclusive choice of Xa and Xi.
The subsequent discovery of the transient pairing of Xs in ES cells led to the realization that physical association provides a unique opportunity for the Xs to communicate directly (Anguera et al. 2006; Bacher et al. 2006; Carrel 2006; Xu et al. 2006). By fluorescence in situ hybridization (FISH), it was observed that, at the onset of cell differentiation and just prior to the initiation of chromosome-wide silencing, the Xs briefly make contact at the Xic. Before cell differentiation, the Xs are epigenetically equivalent and both express Xist RNA at very low levels. The pairing event then brings the two Xs together and leads to the establishment of asymmetry, as Xist RNA grows to engulf Xi while simultaneously disappearing from Xa. Genetic or pharmacological interference with pairing results in aberrant XCI patterns and loss of cell viability (Xu et al. 2006, 2007).
The discovery of X–X interactions in ES cells has offered a convenient genetic model to understand homologous chromosome pairing, a phenomenon previously thought to be excluded in the mammalian soma. Pairing elements reside within a 15-kb region encompassing Xite and the 5′ end of Tsix (Fig. 1; Xu et al. 2006). Deleting a 3.7-kb region of Tsix that includes the promoter and DXPas34 abolishes pairing; likewise, deleting Xite—even just one allele of it—significantly affects pairing potential in female cells. Tsix and Xite are not only required but are also sufficient to induce pairing, as placing them ectopically on an autosome directs that autosome to pair with an X (Bacher et al. 2006; Xu et al. 2006). These results demonstrate that pairing elements reside within two ncRNA genes of the Xic. Transgene deletional analysis has also shown that pairing can be induced by very small fragments (Xu et al. 2007). In fact, even a 1.6-kb element corresponding to the DXPas34 repeat—an element made up of 29 tandem repeats of a 34mer and therefore of very low sequence complexity—could recapitulate pairing, at least when multimerized at the ectopic site. Closer examination revealed two features that are shared by most if not all fragments. First, they contain binding sites for Ctcf (Xu et al. 2007), a versatile transcription factor and chromatin insulator (Lobanenkov et al. 1990; Bell et al. 1999; Ohlsson et al. 2001) for which binding sites are found in abundance at the 5′ ends of both Xite and Tsix (Chao et al. 2002; Xu et al. 2007). When Ctcf protein was knocked down in ES cells using siRNAs, pairing was significantly abrogated.
Another common thread among the pairing-competent transgene fragments is the occurrence of cryptic and minor RNA polymerase II (Pol II) promoters with the potential to drive transcription independently of the major promoters (Sado et al. 2001; Ogawa and Lee 2003; Stavropoulos et al. 2005; Cohen et al. 2007; Xu et al. 2007). This observation raised the possibility that RNA may direct pairing. Although there is currently no direct evidence for the involvement of RNA per se, inhibiting Pol II activity using actinomycin D exerted discrete effects on the pairing process (Xu et al. 2007). Treating cells for 2.0–4.0 h abrogated X–X pairing, suggesting that pairing requires new transcription. Interestingly, time-course analysis indicated that treating with actinomycin D for as little as 0.5 h was sufficient to disrupt new pair formation but did not affect cohesion of previously formed pairs. Approximately 50% of preformed pairs still remained together at 1.0 h post-treatment, implying a pairing half-life of no more than 0.5–1.0 h. Together, these data argue that homologous X-chromosome interaction requires transcription of Tsix and Xite and, moreover, insinuate that the ncRNAs themselves may be involved in pairing.
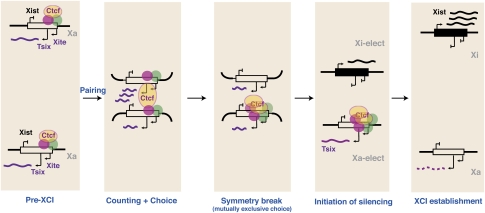
It therefore has been proposed that protein and RNA may collaborate to bring the Xs together and facilitate the breaking of chromosomal symmetry (Fig. 4; Xu et al. 2006, 2007; Nicodemi and Prisco 2007b). Latest analyses indicate that Oct4 also binds Tsix/Xite and controls initiation of XCI by determining the timing of X-chromosome pairing (Donohoe et al. 2009). In pre-XCI cells, it is suggested that Oct4 and Ctcf proteins bind multiple sites at the 5′ ends of Tsix and Xite to transactivate expression of the ncRNAs. The onset of cell differentiation made possible by falling Oct4 levels would then trigger trans-association of the two Xic's, possibly mediated by the RNA–protein “bridge” formed at Tsix and Xite. To ensure mutually exclusive choice, the trans-interactions must create physical differences between the two Xs and signal for one to become Xa and the other to become Xi. It has been postulated that proximity of the Xs enables direct cross-talk and results in the irreversible shift of protein factors, such as Oct4 and Ctcf, from one Tsix/Xite allele (future Xi) to the other (on future Xa) (Xu et al. 2006, 2007; Donohoe et al. 2009). Although direct experimental evidence is lacking so far, computational models suggest that such a scenario is possible and that the shift of factors to one chromosome would be thermodynamically favored and therefore stable under some conditions (Nicodemi and Prisco 2007a,b; Nicodemi et al. 2008). This shift of factors would then lead to the allele-specific expression of Tsix, which in turn would establish the asymmetric pattern of Xist expression on Xa and Xi.
Figure 4.
Symmetry break mediated by homologous chromosome pairing. Adapted from Anguera et al. (2006). The two X chromosomes are epigenetically identical and euchromatic in the pre-XCI stage. The two Xs are brought together by Ctcf, Tsix, and Xite (pairing) during cell differentiation to enable cross-talk and mutually exclusive choice of Xa and Xi. Because it is thermodynamically favorable to do so, transcription factors such as Oct4 and others (red, green circles) that were previously randomly distributed between the two Tsix/Xite alleles stochastically shift to one X, which would then become future Xa. This shift results in monoallelic Tsix expression and differential chromatin modifications within the Xist region that lead to repression of Xist on Xa and up-regulation of Xist on Xi.
Binary state of X specified by antisense RNA
By this model, the binary state of X is directly regulated by allele-specific expression patterns of Tsix (Lee and Lu 1999). How does Tsix act as the molecular switch and how does it specify Xa and Xi fates? Genetic analyses have shown that loss of Tsix expression on the Xi-elect enables transcriptional activation of Xist. Forced expression of the antisense RNA on an X chromosome renders that X incapable of expressing Xist (Luikenhuis et al. 2001; Stavropoulos et al. 2001), and deletion or truncation of the antisense RNA results in constitutively elevated Xist expression (Lee and Lu 1999; Sado et al. 2001; Morey et al. 2004; Shibata and Lee 2004; Ohhata et al. 2008). Conversely, continued expression of Tsix is required to maintain Xa activity during female cell differentiation.
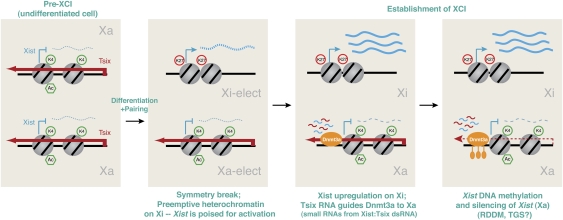
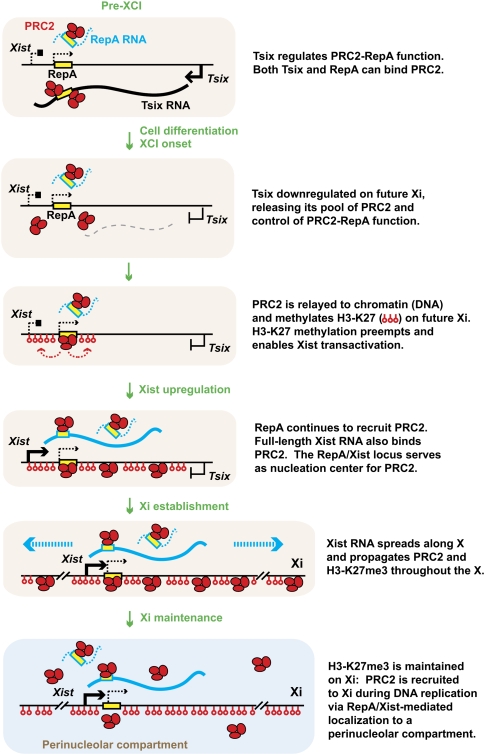
Chromatin immunoprecipitation (ChIP) analyses have revealed that changes in chromatin structure at or around the Xic are highly associated with events of XCI (Heard et al. 2001; Navarro et al. 2005, 2006; Sado et al. 2005; Sun et al. 2006). While differences in experimental systems have led to slightly different conclusions in some of these studies, most agreed that Tsix's effect on Xa and Xi involves changing the chromatin state of the Xist locus (Fig. 5; Navarro et al. 2005, 2006; Sado et al. 2005; Sun et al. 2006). I again refer readers to the cited literature for details of each system. In one study that tracked allelic differences in female ES cells before, during, or after the onset of XCI, it was observed that, in the pre-XCI state, Tsix RNA is expressed from both Xs and establishes the 40-kb region spanning the overlapping Tsix and Xist loci in an open chromatin state; at the onset of cell differentiation, the Xi-elect loses Tsix expression with the resulting effect of losing euchromatic marks along the 40-kb domain shared by Tsix and Xist (Sun et al. 2006). The 5′ end of Xist of Xi-elect becomes enriched for repressive chromatin marks such as histone H3 Lys-27 trimethylation (H3-K27me3). Intriguingly, this mark precedes Xist transactivation and is paradoxically followed by a 100-fold induction of Xist RNA. It therefore has been postulated that Xist requires repressive chromatin marks for its transactivation. The concept that Xist expression thrives in a heterochromatic environment seems to make sense from an evolutionary perspective: Not only must Xist initiate silencing, but it also must remain active in the otherwise heterochromatic environment of Xi. More recent analysis supports the idea that H3-K27me3 stimulates Xist induction (see below; Zhao et al. 2008).
Figure 5.
Tsix RNA as molecular switch for Xa and Xi. Adapted from Sun et al. (2006) with permission from Elsevier. Before XCI, biallelic expression of Tsix RNA enables the Tsix/Xist alleles on both Xs to remain euchromatic, as indicated by biallelic H3-K4 dimethylation and H4 acetylation. Paradoxically, the euchromatic state precludes Xist transcription. At the onset of XCI, silencing of Tsix on one X (future Xi) results in pre-emptive heterochromatin formation across the 40-kb Tsix/Xist locus, as indicated by loss of H3-K4me2 and H4 acetylation, and gain of H3-K27me3. Xist is poised and transactivated 100-fold upon cell differentiation. On the future Xa, persistent Tsix RNA expression maintains the 40-kb Tsix/Xist locus in cis in a euchromatic configuration. During the establishment phase, Tsix RNA also recruits the activity of Dnmt3a to the Xist promoter to methylate (lollipops) and lock in the silent state of Xist. Dnmt3a methylation is hypothetically mediated by small RNAs created from long Xist:Tsix dsRNA via the RNAi pathway in a manner similar to RDDM and TGS in yeast and plants. This idea remains to be tested.
Opposite events occur on Xa-elect. During cell differentiation, Xa-elect continues to express Tsix, which in turn maintains the Tsix/Xist domain in the euchromatic state enriched for H3-K4 methylation and H4 acetylation. This euchromatic state paradoxically correlates with Xist repression. By RNA ChIP, it was shown that Tsix RNA occurs in a specific complex containing the de novo methyltransferase Dnmt3a at the Xist promoter (Sun et al. 2006). Allele-specific analysis indicated that DNA methylation occurs predominantly on the Xist allele of Xa-elect (Sado et al. 2005; Sun et al. 2006), presumably induced in cis by the bound Tsix RNA. Thus, Tsix RNA elicits two specific effects on Xa-elect: First, the RNA (or the act of transcribing it) directs euchromatic modifications to the Xist locus and, in doing so, apparently prevents the activation of the Xist allele in cis even as cell differentiation signals trigger XCI on Xi-elect. Second, it associates with Dnmt3a at the Xist promoter and facilitates de novo CpG methylation and stable silencing of Xa's Xist allele.
These findings have led to the idea of RNA-directed DNA methylation (RDDM) and transcriptional gene silencing (TGS) on the X in mammals (Sun et al. 2006). Although both phenomena are well-established in yeast and plants (Volpe et al. 2002; Cam et al. 2009; Martienssen et al. 2008) and much less so in mammals (Morris et al. 2004; Kim et al. 2006; Moazed 2009), recent evidence indicated that Xist and Tsix RNAs can duplex in vivo, and that the long duplexes are processed to small RNAs during XCI (Ogawa et al. 2008). The small RNAs were 21–42 nt in size and occurred in the complementary regions of Tsix and Xist in ES cells, as well as within the promoter and 5′ end of Xist. In Dicer (Dcr)-deficient ES cells, production of the small RNAs was dramatically reduced during cell differentiation, suggesting that Dcr may be involved, directly or indirectly, in cleaving long duplexes to small RNAs. The possibility of endogenous siRNAs originating from overlapping long duplexes has also been proposed recently for mammalian oocytes (Tam et al. 2008). Intriguingly, when Dcr is deficient, female ES cells displayed reduced DNA methylation at the Xist promoter and, at the same time, elevated steady-state levels of Xist RNA in undifferentiated ES cells (Nesterova et al. 2008; Ogawa et al. 2008; Kanellopoulou et al. 2009). Because Dcr has pleiotropic effects on many aspects of cell physiology, indirect effects on Dnmt3a must be considered (Nesterova et al. 2008) alongside indirect effects of poor cell differentiation on XCI (Ogawa et al. 2008; Kanellopoulou et al. 2009). Nonetheless, the findings provide tantalizing, albeit preliminary evidence for RNAi's involvement in regulating DNA methylation and expression of Xist. For example, small RNAs produced from Xist and Tsix RNA may direct DNA methylation of the Xist promoter and silence it via an RNAi-mediated pathway.
Silencing factors targeted to the X by ncRNA
Xi-elect undergoes a cascade of changes that culminate in the outward spread of the ncRNA away from the Xic to blanket the rest of the X chromosome. Recent analysis has provided clues to the initial series of events. When Tsix RNA is down-regulated from Xi-elect, one of the first changes to appear is a concentration of H3-K27me3 at the 5′ end of Xist, concurrently with enrichment for Polycomb-repressive complex 2 (PRC2) (Sun et al. 2006; Zhao et al. 2008; BK Sun and JT Lee, unpubl.), the enzyme complex responsible for trimethylating H3-K27 (Cao et al. 2002; Czermin et al. 2002; Kuzmichev et al. 2002; Muller et al. 2002). PRC2 belongs to the Polycomb group (PcG) family of proteins that were originally discovered in fruit flies and are now known to have genome-wide repressive functions that are conserved from flies to mammals (Ringrose and Paro 2004; Schwartz and Pirrotta 2008). Intriguingly, the changes at the 5′ end of Xist occur prior to Xist up-regulation, but are followed rapidly by Xist transcriptional induction upon the initiation of cell differentiation (Sun et al. 2006).
The concentration of PRC2 at the 5′ end of Xist is intriguing because recruitment of PRC2 and H3-K27me3 are two of the earliest changes to occur on the X following Xist up-regulation (Plath et al. 2003; Silva et al. 2003; Kohlmaier et al. 2004). The question was raised whether the 5′ end of Xist could function as a PRC2 “nucleation site” akin to Polycomb response elements (PRE) in Drosophila melanogaster that have remained elusive in mammals (Ringrose and Paro 2004; Schwartz and Pirrotta 2008). If so, what might be recruiting PRC2 to this region? Given the abundance of ncRNA within the Xic, could an as yet unidentified ncRNA be the missing link?
Answers to these questions were facilitated by discovery of a novel 1.6-kb ncRNA located within the 5′ end of Xist (Zhao et al. 2008). Called “RepA,” this independent transcription unit encompasses Xist's Repeat A, a conserved motif composed of 7.5 tandem repeats of two stem–loop structures (Wutz et al. 2002). Genetic analyses have revealed that this repeat element is required for the initiation of X-chromosome silencing, as an X chromosome deleted for the repeat cannot initiate XCI (Wutz et al. 2002; Hoki et al. 2009). Prior to XCI, RepA RNA is produced from both Xs and occurs at a steady level of approximately six to seven copies per female cell—approximately twice the level of full-length Xist RNA in the pre-XCI state (Zhao et al. 2008). During XCI, RepA continues to be produced only from Xi. Unlike Xist RNA, however, RepA RNA is not up-regulated 100-fold during cell differentiation, and RNA FISH of autosomally produced RepA suggests that it localizes to the Xic without spreading along the X in cis.
Most importantly, RepA RNA was found to interact directly with PRC2 (Zhao et al. 2008). RNA immunoprecipitations (RIPs) using antibodies against two of PRC2's subunits, Ezh2 and Suz12, demonstrated that RepA RNA is complexed with PRC2 in vivo. Gel shift analysis using RepA oligos showed that a 28-nt conserved stem–loop structure interacts directly with the catalytic subunit Ezh2 in vitro. When a tet-inducible RepA transgene is placed at an ectopic autosomal location, its expression leads to increased recruitment of PRC2. Thus, RepA alone, without full-length Xist, is sufficient to recruit PRC2. Furthermore, the experiment suggests that transcription of RepA or the ncRNA itself is important, as the effect of recruitment is only observed when cells were treated with doxycyline (which results in expression of the RepA transgene). Additional analysis using shRNAs to destroy the RNA without compromising transcription demonstrated that the act of recruiting can be attributed to the ncRNA itself.
On the basis of these findings, it has been posited that RepA is the RNA cofactor that initially recruits PRC2 to the X chromosome. When RepA RNA is knocked down by shRNA, Xist RNA clusters and H3-K27me3 foci could not form on Xi-elect. By contrast, knockdown of a nonoverlapping region of Xist exon 1 displayed a lesser effect, suggesting that the effect of RepA shRNA resulted from loss of RepA RNA rather than Xist RNA. Knocking down Eed or Ezh2 protein also compromised Xist induction and H3-K27me3 of Xi-elect. These data argued that the PRC2–RepA complex's initial function may be to trimethylate H3-K27 locally at the 5′ end of Xist and to create the heterochromatic patch that has been proposed to be essential for Xist transactivation (Sun et al. 2006).
Previously, PRC2 seemed an unlikely binding partner for the Repeat A motif, as some studies reported no effects on PRC2 recruitment when Repeat A was deleted (Plath et al. 2003; Kohlmaier et al. 2004). However, one of the studies actually found that PRC2 recruitment dropped precipitously in the Repeat A mutants (Kohlmaier et al. 2004), supporting the idea that RepA is required to recruit PRC2. Other studies have also suggested that mutations in Eed do not affect the initiation of XCI (Kalantry and Magnuson 2006; Schoeftner et al. 2006). It is noteworthy that endogenous regulation of Xist activation was bypassed by fusion to a tet-inducible promoter in some studies (Kohlmaier et al. 2004; Schoeftner et al. 2006) (so that the effect on H3-K27me3 on the Xist promoter would have been masked). The lack of effect seen with both Eed mutants may indicate the functional redundancy between Polycomb proteins, as proposed by Schoeftner et al. (2006).
Tsix regulation of PRC2–RepA function
By pretreating RIP products with various nucleases, it was learned that the RNA bound by PRC2 possess both single-stranded (ss) and double-stranded (ds) character (Zhao et al. 2008). The RNA was destroyed in the presence of RNase I and RNase V1, which digest ssRNA and dsRNA respectively, whereas pretreatment with RNase H and DNase I had no effect on the RNA's ability to be reverse-transcribed and amplified by PCR. By inference, the bound transcript may be one RNA strand containing a secondary structure formed by intramolecular base-pairing, such as that computationally predicted to occur within the tandem elements of the Repeat A motif (Wutz et al. 2002). Alternatively, the bound RNA may be a duplex formed by intermolecular base-pairing between two complementary single strands, such as would occur should RepA and its antisense counterpart, Tsix, anneal to each other in vivo. Indeed, in vivo RNase protection assays have shown that dsRNA are detected in pre-XCI ES cells (Ogawa et al. 2008). Moreover, when RIP analysis was performed in a manner that could discern strandedness, both forward and reverse strands were detected (Zhao et al. 2008). Gel shift analysis confirmed that PRC2 could directly bind either sense or antisense RNA alone. Thus, PRC2 unexpectedly incorporates both RepA and Tsix RNA, although it is not known if each PRC2 complex binds both strands or if binding of sense and antisense RNAs is mutually exclusive.
These findings provoked ideas about how the PRC2–RepA complex might be regulated. Because Tsix is an established antagonist of Xist, Tsix RNA may directly control the function of PRC2–RepA in several ways. First, given that Tsix RNA could bind both RepA RNA and PRC2, Tsix RNA could prevent formation of the PRC2–RepA complex by titrating RepA away from PRC2, especially if the binding of Tsix or RepA RNA to PRC2 were mutually exclusive. Because Tsix RNA is present in a 10-fold to 100-fold molar excess over RepA/Xist RNA during the pre-XCI stage in vivo (Shibata and Lee 2003), binding to Tsix RNA would be thermodynamically favored.
Instead of blocking PRC2 binding to RepA, Tsix RNA could also interfere with the loading of precomplexed PRC2–RepA to chromatin or, alternatively, with the complex's catalytic activity on chromatin. Notably, some PRC2 already can be found to bind RepA in the pre-XCI state, although the extent to which this occurs is not known. RIP and ChIP studies showed that the RNA–protein interaction and the complex's contact with chromatin are biochemically separable (Zhao et al. 2008). While bound to RNA in the pre-XCI state, PRC2 was not enriched at the 5′ end of Xist and H3-K27me3 levels were accordingly low. At this time, Tsix RNA levels were high. Only upon cell differentiation and the initiation of XCI—when Tsix RNA was down-regulated from Xi-elect—did PRC2 bind Xist chromatin and methylate H3-K27.
In my model (Fig. 6), when Tsix is allele-specifically down-regulated on Xi-elect, RepA RNA productively engages PRC2 and directs H3-K27me3 of the Xist promoter region in cis (Zhao et al. 2008). This would account for the previously observed concentration of PRC2 and H3-K27me3 at the 5′ end of Xist, creating the local heterochromatic state proposed to be required for 100-fold induction of full-length Xist (Sun et al. 2006). Importantly, RIP analysis showed that full-length Xist RNA also binds PRC2, most probably also through the Repeat A motif shared with RepA RNA. Because Xist RNA also carries chromosome localization motifs, the spread of Xist RNA through Xi-elect would provide a plausible mechanism by which PRC2 and H3-K27me3 could be distributed throughout the X. My model therefore suggests that ncRNA controls Polycomb proteins in several ways: (1) RepA RNA first directs PRC2 to the Xic; (2) Xist RNA then spreads PRC2 along the future Xi; and (3) Tsix RNA blocks these pivotal activities on the future Xa by interfering with RepA–PRC2 function.
Figure 6.
The initiation of XCI controlled by interaction of Tsix, RepA, and Xist RNAs with PRC2. See the text in the figure for detailed discussion. Adapted from Zhao et al. (2008).
One of the most puzzling questions in the Polycomb field has been how the complexes are recruited to their target destinations. The idea of an RNA cofactor for PcG proteins has been suspected for quite some time, beginning with the curious observation that the stability of PcG complexes may be affected by RNase treatment (R Paro, pers. comm.). In Drosophila, PcG complexes are known to contain sequence-specific DNA-binding proteins such as Zeste, Pipsqueak (PSQ), and Pho, which could target PcG complexes to the genome (Ringrose and Paro 2004; Schwartz and Pirrotta 2008). Because mammalian complexes do not necessarily associate with such proteins, how the mammalian proteins can be targeted in a sequence-specific manner has been a point of major interest. It has been hypothesized that specific ncRNA may be the missing link that directs the chromatin modifier to its genomic target.
Why long ncRNAs make excellent guides and tethers for cis regulation
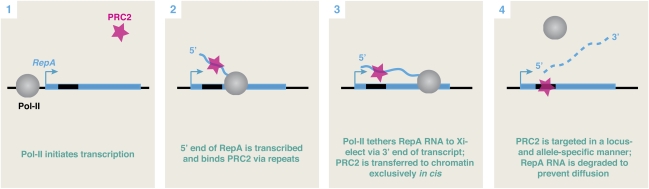
By virtue of the fact that long ncRNA remains tethered to its parent locus during the act of transcription, large RNAs may be the molecule of choice for regulatory systems challenged by the need to act in cis. No member of the proteome can subsume this function, as memory of allelic origin is always lost once mRNA exits the nucleus and is translated to protein. I argue that such ncRNAs are necessarily lengthy, with a 5′ business end that binds protein partners as soon as it is synthesized and a transcriptionally lagging 3′ end that therefore tethers the RNA to chromatin through Pol II during the act of transcription (Fig. 7). In the case of RepA and Xist RNA, perhaps it is no coincidence that the Repeat A motif is at the very 5′ end of both molecules. This arrangement would enable the RNAs to bind PRC2 cotranscriptionally and hold the RNA–protein complex in place for its exclusive cis action. If the RNA is degraded rapidly once transcribed—for example, via destabilization motifs at the 3′ end that would be revealed once Pol II reached the terminus—then the RNA could be prevented from diffusing away from the site of synthesis.
Figure 7.
Locus- and allele-specific recruitment of protein factors by long ncRNA. RepA recruitment of PRC2 is shown as an example of how sequence-specific long ncRNAs act as tethers to chromatin to restrict recruitment exclusively in cis. See ithe text in the figure for discussion.
Because RNA is also inherently sequence-specific and is transcribed in a developmentally specific manner, RNA would also be an ideal regulator of spatial and temporal specificity during development. Transcription factors and DNA-binding proteins usually interact with an entire network of genes (e.g., Oct4 binds an 8mer consensus that appears many times over in the genome), and therefore rarely specify a single location in the genome. In contrast, long ncRNA can identify a unique address. For instance, Tsix and RepA RNAs occur only once in the genome and are therefore uniquely positioned to attract specific chromatin modifiers to that location. With a virtually infinite number of unique addresses that can be specified by a combination of RNA length and nucleotide permutations, sequence space for the long ncRNA transcriptome must ultimately far exceed that of the proteome.
Here, I describe the potential for long transcripts to function as tethers and guides that recruit chromatin modifiers—and even entire chromosomes (Bacher et al. 2006; Xu et al. 2006; Zhang et al. 2007)—to a specific location in the genome. RNA conventionally has been regarded as information carriers, translating genetic code to protein sequence. The idea of RNA guidance takes this nucleic acid beyond its traditional role of messenger in the transition from genotype to phenotype and moves it into a more dynamic arena of reshaping the epigenome. The principles of RNA guidance need not be confined to the X. Indeed, there is very recent evidence of PRC2 being associated either directly or indirectly with other ncRNAs, such as HOTAIR (Rinn et al. 2007) or Kcnqt1ot1 (Pandey et al. 2008). There is also no reason to think that RNA guides would be restricted to Polycomb complexes. ChIP using antibodies against G9a (Nagano et al. 2008) and Dnmt3a (Sun et al. 2006) have also hinted at RNA in their complexes, although it remains to be seen whether there is direct RNA–protein interaction or whether, instead, the RNAs interact with the proteins indirectly via the underlying chromatin. A general RNA-based mechanism would rationally explain how a limited set of chromatin modifiers, which often lack subunits with sequence-specific DNA-binding activity but curiously possess putative RNA-binding domains (Denisenko et al. 1998; Bernstein and Allis 2005; Bernstein et al. 2006), could be directed to the mammalian genome in a spatially and temporally unique manner.
These unique properties of long ncRNA may explain why evolution, at the expense of protein-coding genes, has placed so many ncRNA genes at the Xic. It seems likely that strategies employed by the X will become recurrent themes throughout the epigenome, as indeed interchromosomal interactions are not confined to the X (LaSalle and Lalande 1996; Spilianakis et al. 2005; Lomvardas et al. 2006), many autosomal genes are now known to possess antisense partners (Katayama et al. 2005; He et al. 2008), and a large number of long ncRNAs are being discovered across the genome (Claverie 2005; Kapranov et al. 2007a,b; Guttman et al. 2009; Mercer et al. 2009). Even within the Xic, ncRNA's full capabilities have yet to be revealed, as most of the seven known ncRNA loci still remain to be studied. It seems probable that long ncRNAs ultimately will rival small RNAs and proteins in versatility. With its unique ability to function in cis and its inherent command of a large sequence space, long ncRNA may prove to be the molecule of choice for many challenges presented by epigenetic regulation.
Acknowledgments
I am grateful to all laboratory members for many valuable discussions, and especially thank Brian del Rosario, Daniel Kim, Stefan Pinter, Sha Sun, and Jing Zhao for thoughtful comments on the manuscript. I am an investigator of the HHMI.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1811209.
References
- Anguera MC, Sun BK, Xu N, Lee JT. X-chromosome kiss and tell: How the Xs go their separate ways. Cold Spring Harb Symp Quant Biol. 2006;71:429–437. doi: 10.1101/sqb.2006.71.012. [DOI] [PubMed] [Google Scholar]
- Avner P, Heard E. X-chromosome inactivation: Counting, choice and initiation. Nat Rev Genet. 2001;2:59–67. doi: 10.1038/35047580. [DOI] [PubMed] [Google Scholar]
- Bacher CP, Guggiari M, Brors B, Augui S, Clerc P, Avner P, Eils R, Heard E. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol. 2006;8:293–299. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Allis CD. RNA meets chromatin. Genes & Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani G, Tonlorenzi R, Simmler MC, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzny D, Lawrence C, et al. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- Boumil RM, Lee JT. Forty years of decoding the silence in X-chromosome inactivation. Hum Mol Genet. 2001;10:2225–2232. doi: 10.1093/hmg/10.20.2225. [DOI] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991a;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Lafreniere RG, Powers VE, Sebastio G, Ballabio A, Pettigrew AL, Ledbetter DH, Levy E, Craig IW, Willard HF. Localization of the X inactivation centre on the human X chromosome in Xq13. Nature. 1991b;349:82–84. doi: 10.1038/349082a0. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Cam HP, Chen ES, Grewal SI. Transcriptional scaffolds for heterochromatin assembly. Cell. 2009;136:610–614. doi: 10.1016/j.cell.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Carrel L. Molecular biology. ‘X’-rated chromosomal rendezvous. Science. 2006;311:1107–1109. doi: 10.1126/science.1124662. [DOI] [PubMed] [Google Scholar]
- Cech TR. Crawling out of the RNA world. Cell. 2009;136:599–602. doi: 10.1016/j.cell.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Chao W, Huynh KD, Spencer RJ, Davidow LS, Lee JT. CTCF, a candidate trans-acting factor for X-inactivation choice. Science. 2002;295:345–347. doi: 10.1126/science.1065982. [DOI] [PubMed] [Google Scholar]
- Chureau C, Prissette M, Bourdet A, Barbe V, Cattolico L, Jones L, Eggen A, Avner P, Duret L. Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine. Genome Res. 2002;12:894–908. doi: 10.1101/gr.152902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie JM. Fewer genes, more noncoding RNA. Science. 2005;309:1529–1530. doi: 10.1126/science.1116800. [DOI] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc P, Avner P. Role of the region 3′ to Xist exon 6 in the counting process of X-chromosome inactivation. Nat Genet. 1998;19:249–253. doi: 10.1038/924. [DOI] [PubMed] [Google Scholar]
- Cohen DE, Davidow LS, Erwin JA, Xu N, Warshawsky D, Lee JT. The DXPas34 repeat regulates random and imprinted X inactivation. Dev Cell. 2007;12:57–71. doi: 10.1016/j.devcel.2006.11.014. [DOI] [PubMed] [Google Scholar]
- Courtier B, Heard E, Avner P. Xce haplotypes show modified methylation in a region of the active X chromosome lying 3′ to Xist. Proc Natl Acad Sci. 1995;92:3531–3535. doi: 10.1073/pnas.92.8.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Debrand E, Chureau C, Arnaud D, Avner P, Heard E. Functional analysis of the DXPas34 locus, a 3′ regulator of Xist expression. Mol Cell Biol. 1999;19:8513–8525. doi: 10.1128/mcb.19.12.8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisenko O, Shnyreva M, Suzuki H, Bomsztyk K. Point mutations in the WD40 domain of Eed block its interaction with Ezh2. Mol Cell Biol. 1998;18:5634–5642. doi: 10.1128/mcb.18.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe ME, Silva SS, Pinter SF, Xu N, Lee JT. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature. 2009;460:128–132. doi: 10.1038/nature08098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19:281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: An expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Mongelard F, Arnaud D, Avner P. Xist yeast artificial chromosome transgenes function as X-inactivation centers only in multicopy arrays and not as single copies. Mol Cell Biol. 1999;19:3156–3166. doi: 10.1128/mcb.19.4.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- Hoki Y, Kimura N, Kanbayashi M, Amakawa Y, Ohhata T, Sasaki H, Sado T. A proximal conserved repeat in the Xist gene is essential as a genomic element for X-inactivation in mouse. Development. 2009;136:139–146. doi: 10.1242/dev.026427. [DOI] [PubMed] [Google Scholar]
- Huynh KD, Lee JT. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature. 2003;426:857–862. doi: 10.1038/nature02222. [DOI] [PubMed] [Google Scholar]
- Kalantry S, Magnuson T. The Polycomb group protein EED is dispensable for the initiation of random X-chromosome inactivation. PLoS Genet. 2006;2:e66. doi: 10.1371/journal.pgen.0020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Dimitrov SD, Chen X, Colin C, Plath K, Livingston DM. X chromosome inactivation in the absence of Dicer. Proc Natl Acad Sci. 2009;106:1122–1127. doi: 10.1073/pnas.0812210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007a;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007b;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kay GF, Barton SC, Surani MA, Rastan S. Imprinting and X chromosome counting mechanisms determine Xist expression in early mouse development. Cell. 1994;77:639–650. doi: 10.1016/0092-8674(94)90049-3. [DOI] [PubMed] [Google Scholar]
- Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2004;2:E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes & Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSalle JM, Lalande M. Homologous association of oppositely imprinted chromosomal domains. Science. 1996;272:725–728. doi: 10.1126/science.272.5262.725. [DOI] [PubMed] [Google Scholar]
- Lee JT. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell. 2000;103:17–27. doi: 10.1016/s0092-8674(00)00101-x. [DOI] [PubMed] [Google Scholar]
- Lee JT. Homozygous Tsix mutant mice reveal a sex-ratio distortion and revert to random X-inactivation. Nat Genet. 2002;32:195–200. doi: 10.1038/ng939. [DOI] [PubMed] [Google Scholar]
- Lee JT. Regulation of X-chromosome counting by Tsix and Xite sequences. Science. 2005;309:768–771. doi: 10.1126/science.1113673. [DOI] [PubMed] [Google Scholar]
- Lee JT, Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell. 1999;99:47–57. doi: 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- Lee JT, Strauss WM, Dausman JA, Jaenisch R. A 450 kb transgene displays properties of the mammalian X-inactivation center. Cell. 1996;86:83–94. doi: 10.1016/s0092-8674(00)80079-3. [DOI] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999a;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- Lee JT, Lu N, Han Y. Genetic analysis of the mouse X inactivation center defines an 80-kb multifunction domain. Proc Natl Acad Sci. 1999b;96:3836–3841. doi: 10.1073/pnas.96.7.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′ flaking sequence of the chicken c-myc gene. Oncogene. 1990;5:1743–1753. [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annu Rev Genet. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- Luikenhuis S, Wutz A, Jaenisch R. Antisense transcription through the Xist locus mediates Tsix function in embryonic stem cells. Mol Cell Biol. 2001;21:8512–8520. doi: 10.1128/MCB.21.24.8512-8520.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Imprinting and X chromosome inactivation. In: Ohlsson R, editor. Results and problems in cell differentiation. Springer-Verlag; Heidelberg: 1999. pp. 73–90. [DOI] [PubMed] [Google Scholar]
- Mak W, Nesterova TB, de Napoles M, Appanah R, Yamanaka S, Otte AP, Brockdorff N. Reactivation of the paternal X chromosome in early mouse embryos. Science. 2004;303:666–669. doi: 10.1126/science.1092674. [DOI] [PubMed] [Google Scholar]
- Marahrens Y. X-inactivation by chromosomal pairing events. Genes & Dev. 1999;13:2624–2632. doi: 10.1101/gad.13.20.2624. [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes & Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- Martienssen RA, Kloc A, Slotkin RK, Tanurdzic M. Epigenetic inheritance and reprogramming in plants and fission yeast. Cold Spring Harb Symp Quant Biol. 2008;73:265–271. doi: 10.1101/sqb.2008.73.062. [DOI] [PubMed] [Google Scholar]
- Masui O, Heard E. RNA and protein actors in X-chromosome inactivation. Cold Spring Harb Symp Quant Biol. 2006;71:419–428. doi: 10.1101/sqb.2006.71.058. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Migeon BR, Kazi E, Haisley-Royster C, Hu J, Reeves R, Call L, Lawler A, Moore CS, Morrison H, Jeppesen P. Human X inactivation center induces random X chromosome inactivation in male transgenic mice. Genomics. 1999;59:113–121. doi: 10.1006/geno.1999.5861. [DOI] [PubMed] [Google Scholar]
- Mlynarczyk-Evans S, Royce-Tolland M, Alexander MK, Andersen AA, Kalantry S, Gribnau J, Panning B. X chromosomes alternate between two states prior to random X-inactivation. PLoS Biol. 2006;4:e159. doi: 10.1371/journal.pbio.0040159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk M, Harper MI. Sequential X chromosome inactivation coupled with cellular differentiation in early mouse embryos. Nature. 1979;281:311–313. doi: 10.1038/281311a0. [DOI] [PubMed] [Google Scholar]
- Monkhorst K, Jonkers I, Rentmeester E, Grosveld F, Gribnau J. X inactivation counting and choice is a stochastic process: Evidence for involvement of an X-linked activator. Cell. 2008;132:410–421. doi: 10.1016/j.cell.2007.12.036. [DOI] [PubMed] [Google Scholar]
- Monkhorst K, de Hoon B, Jonkers I, Mulugeta Achame E, Monkhorst W, Hoogerbrugge J, Rentmeester E, Westerhoff HV, Grosveld F, Grootegoed JA, et al. The probability to initiate X chromosome inactivation is determined by the X to autosomal ratio and X chromosome specific allelic properties. PLoS One. 2009;4:e5616. doi: 10.1371/journal.pone.0005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey C, Navarro P, Debrand E, Avner P, Rougeulle C, Clerc P. The region 3′ to Xist mediates X chromosome counting and H3 Lys-4 dimethylation within the Xist gene. EMBO J. 2004;23:594–604. doi: 10.1038/sj.emboj.7600071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- Navarro P, Pichard S, Ciaudo C, Avner P, Rougeulle C. Tsix transcription across the Xist gene alters chromatin conformation without affecting Xist transcription: Implications for X-chromosome inactivation. Genes & Dev. 2005;19:1474–1484. doi: 10.1101/gad.341105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro P, Page DR, Avner P, Rougeulle C. Tsix-mediated epigenetic switch of a CTCF-flanked region of the Xist promoter determines the Xist transcription program. Genes & Dev. 2006;20:2787–2792. doi: 10.1101/gad.389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro P, Chambers I, Karwacki-Neisius V, Chureau C, Morey C, Rougeulle C, Avner P. Molecular coupling of Xist regulation and pluripotency. Science. 2008;321:1693–1695. doi: 10.1126/science.1160952. [DOI] [PubMed] [Google Scholar]
- Nesterova TB, Popova BC, Cobb BS, Norton S, Senner CE, Tang YA, Spruce T, Rodriguez TA, Sado T, Merkenschlager M, et al. Dicer regulates Xist promoter methylation in ES cells indirectly through transcriptional control of Dnmt3a. Epigenetics Chromatin. 2008;1:2. doi: 10.1186/1756-8935-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodemi M, Prisco A. Self-assembly and DNA binding of the blocking factor in x chromosome inactivation. PLoS Comput Biol. 2007a;3:e210. doi: 10.1371/journal.pcbi.0030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodemi M, Prisco A. Symmetry-breaking model for X-chromosome inactivation. Phys Rev Lett. 2007b;98:108104–108107. doi: 10.1103/PhysRevLett.98.108104. [DOI] [PubMed] [Google Scholar]
- Nicodemi M, Panning B, Prisco A. A thermodynamic switch for chromosome colocalization. Genetics. 2008;179:717–721. doi: 10.1534/genetics.107.083154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Lee JT. Xite, X-inactivation intergenic transcription elements that regulate the probability of choice. Mol Cell. 2003;11:731–743. doi: 10.1016/s1097-2765(03)00063-7. [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Sun BK, Lee JT. Intersection of the RNA interference and X-inactivation pathways. Science. 2008;320:1336–1341. doi: 10.1126/science.1157676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohhata T, Hoki Y, Sasaki H, Sado T. Crucial role of antisense transcription across the Xist promoter in Tsix-mediated Xist chromatin modification. Development. 2008;135:227–235. doi: 10.1242/dev.008490. [DOI] [PubMed] [Google Scholar]
- Ohlsson R, Renkawitz R, Lobanenkov VV. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;7:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Payer B, Lee JT. X chromosome dosage compensation: How mammals keep the balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sado T, Wang Z, Sasaki H, Li E. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development. 2001;128:1275–1286. doi: 10.1242/dev.128.8.1275. [DOI] [PubMed] [Google Scholar]
- Sado T, Hoki Y, Sasaki H. Tsix silences Xist through modification of chromatin structure. Dev Cell. 2005;9:159–165. doi: 10.1016/j.devcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr Opin Cell Biol. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Shibata S, Lee JT. Characterization and quantitation of differential Tsix transcripts: Implications for Tsix function. Hum Mol Genet. 2003;12:125–136. doi: 10.1093/hmg/ddg010. [DOI] [PubMed] [Google Scholar]
- Shibata S, Lee JT. Tsix transcription- versus RNA-based mechanisms in Xist repression and epigenetic choice. Curr Biol. 2004;14:1747–1754. doi: 10.1016/j.cub.2004.09.053. [DOI] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed–Enx1 polycomb group complexes. Dev Cell. 2003;4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Simmler MC, Cunningham DB, Clerc P, Vermat T, Caudron B, Cruaud C, Pawlak A, Szpirer C, Weissenbach J, Claverie JM, et al. A 94 kb genomic sequence 3′ to the murine Xist gene reveals an AT rich region containing a new testis specific gene Tsx. Hum Mol Genet. 1996;5:1713–1726. doi: 10.1093/hmg/5.11.1713. [DOI] [PubMed] [Google Scholar]
- Sleutels F, Barlow DP. The origins of genomic imprinting in mammals. In: Dunlap JC, Wu C-T, editors. Homology effects. Academic Press; San Diego: 2002. pp. 119–154. [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Stavropoulos N, Lu N, Lee JT. A functional role for Tsix transcription in blocking Xist RNA accumulation but not in X-chromosome choice. Proc Natl Acad Sci. 2001;98:10232–10237. doi: 10.1073/pnas.171243598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavropoulos N, Rowntree RK, Lee JT. Identification of developmentally specific enhancers for Tsix in the regulation of X chromosome inactivation. Mol Cell Biol. 2005;25:2757–2769. doi: 10.1128/MCB.25.7.2757-2769.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell. 2006;21:617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- Takagi N. Differentiation of X chromosomes in early female mouse embryos. Exp Cell Res. 1974;86:127–135. doi: 10.1016/0014-4827(74)90657-0. [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wan LB, Bartolomei MS. Regulation of imprinting in clusters: Noncoding RNAs versus insulators. Adv Genet. 2008;61:207–223. doi: 10.1016/S0065-2660(07)00007-7. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard HF. X chromosome inactivation, XIST, and pursuit of the X-inactivation center. Cell. 1996;86:5–7. doi: 10.1016/s0092-8674(00)80071-9. [DOI] [PubMed] [Google Scholar]
- Wutz A. RNAs templating chromatin structure for dosage compensation in animals. Bioessays. 2003;25:434–442. doi: 10.1002/bies.10274. [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- Xu N, Tsai CL, Lee JT. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311:1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- Xu N, Donohoe ME, Silva SS, Lee JT. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat Genet. 2007;39:1390–1396. doi: 10.1038/ng.2007.5. [DOI] [PubMed] [Google Scholar]
- Zhang LF, Huynh KD, Lee JT. Perinucleolar targeting of the inactive X during S phase: Evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]